Background: Selenocysteine incorporation into selenoprotein P (SEPP1), which contains 10 Sec residues, is unique.
Results: Processive Sec incorporation can be reconstituted in vitro, independently of the conserved non-SECIS portions of the 3′-UTR.
Conclusion: Processivity is intrinsic, but efficiency is governed by selenium levels.
Significance: SEPP1 synthesis is essential for male fertility and proper neurologic function.
Keywords: Selenium, Selenocysteine, Selenocysteine Insertion Sequence (SECIS), Selenoprotein, Translation, SEPP1, Channeling
Abstract
Selenoproteins are unique as they contain selenium in their active site in the form of the 21st amino acid selenocysteine (Sec), which is encoded by an in-frame UGA stop codon. Sec incorporation requires both cis- and trans-acting factors, which are known to be sufficient for Sec incorporation in vitro, albeit with low efficiency. However, the abundance of the naturally occurring selenoprotein that contains 10 Sec residues (SEPP1) suggests that processive and efficient Sec incorporation occurs in vivo. Here, we set out to study native SEPP1 synthesis in vitro to identify factors that regulate processivity and efficiency. Deletion analysis of the long and conserved 3′-UTR has revealed that the incorporation of multiple Sec residues is inherently processive requiring only the SECIS elements but surprisingly responsive to the selenium concentration. We provide evidence that processive Sec incorporation is linked to selenium utilization and that reconstitution of known Sec incorporation factors in a wheat germ lysate does not permit multiple Sec incorporation events, thus suggesting a role for yet unidentified mammalian-specific processes or factors. The relationship between our findings and the channeling theory of translational efficiency is discussed.
Introduction
Selenoproteins are known for their role in catalyzing redox reactions and defending cells from oxidative stress (reviewed in Ref. 1). Selenoproteins incorporate selenium into their catalytic site in the form of the 21st amino acid selenocysteine (Sec), which is encoded by UGA, a naturally occurring stop codon. Targeted deletion of the mouse selenocysteine tRNA gene causes embryonic lethality indicating an essential role for selenoproteins (2). Specific incorporation of Sec into the nascent polypeptide of selenoproteins requires the presence of a cis-element known as Sec incorporation sequence (SECIS),2 which in eukaryotes is located in the 3′-UTR of all selenoprotein mRNAs (3). SECIS elements share a basic stem loop structure wherein the internal loop that is comprised of an AUGA positioned opposite to a GA dinucleotide is referred to as the SECIS core. In addition to the SECIS, three other factors are required: SECIS binding protein 2 (SBP2), a dedicated eukaryotic elongation factor (eEFSec), and the selenocysteyl tRNASec (Sec-tRNASec) (4–7).
In bacteria, the maximum Sec incorporation efficiency has been reported to be 7–10% (8). The efficiency of Sec incorporation in the rabbit reticulocyte lysate in vitro translation system was also reported to be ∼10% (9), but the frequency of early termination at the Sec codon for endogenous selenoproteins in mammals remains unknown. In animals, several endogenous selenoproteins (particularly GPX4 in testes, GPX1 in liver and kidney, and SEPP1 in liver) are expressed in abundance, implying that selenoprotein synthesis in vivo is likely to be efficient (10–12). To date, several factors such as tissue type, selenium levels, mRNA stability, SECIS-binding proteins (nucleolin, L30, and eIF4A3), and UGA codon position and context, have been shown to influence selenoprotein expression (13–18). In the case of selenoprotein N, a cis-element known as the Sec codon redefinition element, which is located in the coding region, stimulates Sec incorporation efficiency (19, 20). Very recently, the 3′-UTR of selenoprotein S was also shown to contain sequences that regulated Sec incorporation efficiency both upstream and downstream of the SECIS in the 3′-UTR (21). Thus, in addition to the essential components required for Sec incorporation, cis-acting efficiency elements are crucial determinants of selenoprotein expression. Identification of these elements is key to understanding the mechanistic details of Sec incorporation.
Among the 25 known selenoproteins, selenoprotein P (SEPP1), primarily synthesized in the liver is the only human selenoprotein that contains multiple Sec residues (10) and two SECIS elements in its 3′-UTR (22, 23). SEPP1 ablation in mice has demonstrated that full-length SEPP1 serves to transport selenium into extrahepatic tissues (24, 25). Besides the full-length SEPP1, three other SEPP1 isoforms resulting from termination at the 2nd, 3rd, and 7th UGA have been purified from rat plasma (22). The isoform resulting from termination at the second UGA has been shown to have enzymatic activity and may have functional significance (26). The co-production of multiple SEPP1 isoforms from a single open reading frame indicates that SEPP1 synthesis in vivo is tightly regulated. However, despite the tight regulation and multiple Sec residues, this protein is secreted at a high rate into plasma (27). It is the most abundant plasma selenoprotein at a concentration of 26–30 μg/ml in selenium adequate rats (28). In animals, the plasma half-life of SEPP1 has been shown to be 3–4 h (as opposed to 12 h observed for plasma glutathione peroxidase 3), thereby suggesting that its synthesis and turnover in vivo is rapid and efficient (29). Several studies have shown that administration of selenium into selenium deficient animals initiates SEPP1 recovery within 1–3 h, prior to other selenoproteins (30).
As a whole, therefore, the in vivo data suggests that SEPP1 synthesis is efficient and processive. However, the initial in vitro translation study that modeled SEPP1 synthesis using a Dual-Luciferase construct with multiple UGA codons and the SEPP1 3′-UTR resulted in very inefficient synthesis of full-length product (32). It also led to the idea that Sec incorporation in SEPP1 synthesis may be non-processive. More recently, Sec incorporation into a similar Dual-Luciferase construct was shown to be up to ∼70% efficient once the inefficient Sec incorporation event at the first UGA codon was completed (31). Similarly, when the zebrafish version of SEPP1 was transfected into human HEK293 cells, it was found that the first UGA codon was a “bottleneck,” preventing processive and efficient incorporation downstream (50). Thus, multiple Sec incorporation within SEPP1, unlike single Sec incorporation, may involve several levels of regulation. To further interrogate the mechanism of SEPP1 synthesis, we have developed here an in vitro translation system that makes use of radiolabeled selenium incorporation into the native rat SEPP1 protein. We show that the conserved non-SECIS sequences in the SEPP1 3′-UTR are not required for processive Sec incorporation but that selenium supplementation stimulates multiple but not single Sec incorporation events by >4-fold. We have also extended our analysis to the wheat germ lysate system, which lacks the Sec incorporation machinery, and found it to be unable to support processive Sec incorporation into SEPP1.
EXPERIMENTAL PROCEDURES
Plasmid Construction and mRNA Synthesis
The firefly luciferase construct containing a Sec codon at position 258 followed by the SEPP1 3′-UTR was created as described previously (Fig. 1A) (9, 18). The native rat SEPP1 cDNA containing both the 5′-UTR and the full-length 3′-UTR was PCR amplified from total RNA isolated from McArdle 7777 cells and subcloned by TOPO-TA cloning into pcDNA3.1 (Invitrogen). Site-directed mutagenesis was used to insert FLAG tags and generate serially mutated SEPP1 mutants annotated as U1C to U1-10C in which each consecutive Sec (U) was replaced with Cys (C) (the U1–10C version is termed CysSEPP1) (Fig. 2A). A natural Pac1 site located at 83 nt downstream of the SEPP1 termination codon and the vector NotI site were used to ligate the GPX4 SECIS containing Pac1 and Not1 linkers into the native SEPP1 and U1C construct. Mutants were verified using automated DNA sequencing. Native rat SEPP1, luciferase, and all mutant plasmids were linearized with NotI and then used as templates for in vitro transcription with T7 RNA polymerase in the presence of m7G(5′)ppp(5′)G (mMessage mMachine Ambion).
FIGURE 1.
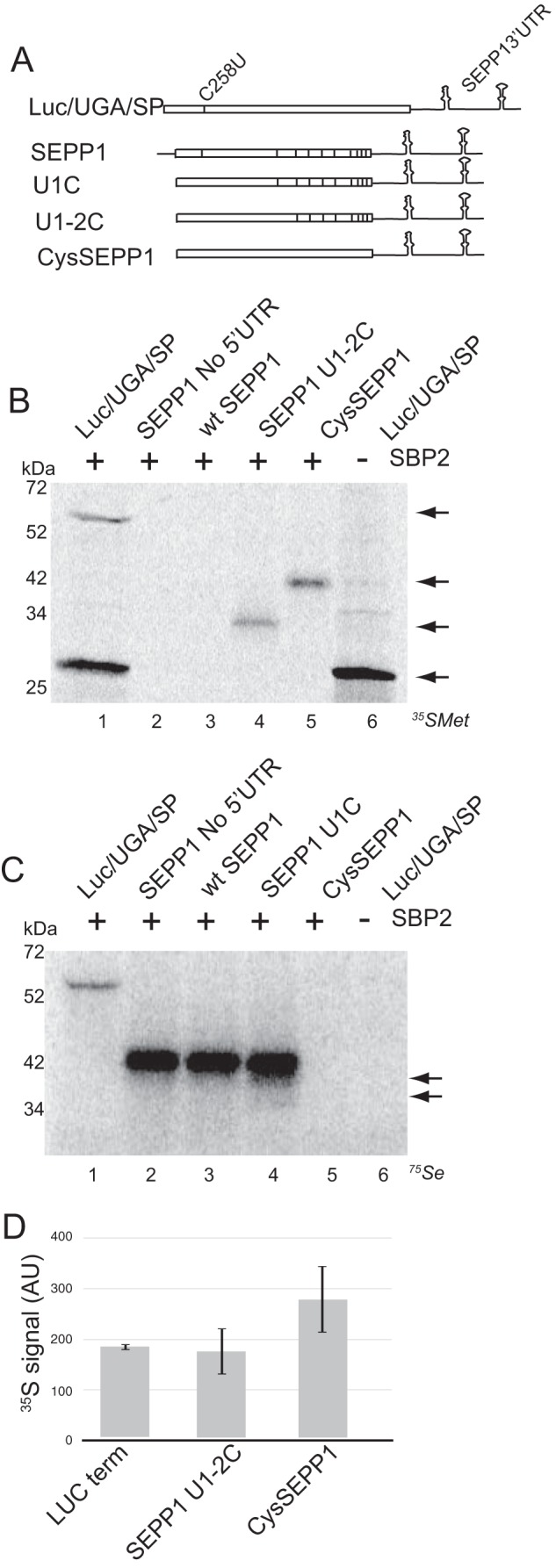
In vitro SEPP1 synthesis is processive but inefficient. A, schematic representation of luciferase and SEPP1 mRNAs. The Sec codon in the luciferase mRNA (Luc/UGA/SP) is represented by a vertical line within the coding region. Black vertical lines within the coding region of SEPP1 mRNAs represent the 10 Sec codons. B, Luc/UGA/SP, SEPP1, U1–2C, and CysSEPP1 mRNAs were translated in RRL supplemented with or without SBP2 and analyzed using [35S]Met labeling. Arrows (from top to bottom) indicate full-length Luc/UGA/SP, CysSEPP1, termination product of U1–2C, and termination product for Luc/UGA/SP. Radiolabeled proteins were resolved by SDS-PAGE and detected by PhosphorImager analysis. C, Luc/UGA/SP, SEPP1, and U1C mRNAs were translated in RRL supplemented with and without SBP2 and [75Se]selenite. The two arrows on the right represent truncated products of U1C translation. D, quantitative analysis of the data in lanes 1, 4, and 5 from A. The [35S]Met signal for each protein in A was calculated and normalized for the number of Met residues. Quantitative data were obtained using ImageQuant. Data are plotted as the average arbitrary density units (AU) ± S.D. for for three independent experiments. All RRL reactions for all figures were supplemented with the C-terminal fragment of SBP2 bearing an N-terminal Xpress/His tag (XH-CTSBP2).
FIGURE 2.

SEPP1 synthesis is processive. A, schematic representation of SEPP1 mRNAs with each Sec residue progressively mutated to cysteine. Black vertical lines within the coding region represent the Sec residues of SEPP1. B, all progressively mutated SEPP1 Sec to Cys mRNAs were translated in RRL and analyzed using [35S]Met labeling. Lane 10 represents translation of full-length CysSEPP1. C, same as that in A, except the RRL was supplemented with 400 nm 75Se. Lane 10 represents translation of full-length native SEPP1. D, graphical representation of data from B (in dark gray) and C (in light gray). Data from B and C was adjusted for the maximum number of Met or Sec residues in each mRNA and then normalized to CysSEPP1 or SEPP1, respectively. Radiolabeled protein analysis was performed as described for Fig. 1.
In Vitro Rabbit Reticulocyte Lysate (RRL) Translation
Translation reaction volumes were 12.5 μl containing 48% RRL (Promega) supplemented with capped mRNA, 4 pmol of SBP2 (where indicated), RNAsin (Promega), and an amino acid mixture lacking either Met or Cys depending on the source of radioactive amino acid as indicated. Translation reactions were incubated for 1 h at 30 °C. 4 μl of labeled translation products were resolved by SDS-PAGE on a 12% gel and quantitated by PhosphorImager analysis (GE Healthcare).
RRL translation reactions with 75Se labeling were performed exactly as above with the exception that complete amino acid mixture was used and also the reaction was supplemented with 400 nm 75Se. ImageQuant (GE Healthcare) was used to quantitate relative protein amounts, and the values were normalized either for the number of methionine, cysteine, or selenocysteine residues.
Efficiency of Sec Incorporation
Efficiency in RRL assays using either [35S]Met or [35S]Cys labeling was calculated as described previously (9). In brief, the amount of full-length protein was divided by the total amount of translation (termination product plus full-length) and normalized for the number of either methionines or cysteines, respectively.
In Vitro Wheat Germ Lysate (WGL) Translation
All wheat germ lysate translation reactions were performed using the optimized protocol described previously (33), except that in place of total testes tRNA, they were supplemented with 1.25 μg of total tRNA purified from HEPG2 cells that were grown in medium supplemented with 100 nm 75Se for 48 h before being lysed. In addition, our prior batch of WGL required the addition of mammalian ribosomes to permit Sec incorporation (33), but for the new lot that was used in this study, ribosome supplementation was not necessary for the Sec incorporation assays shown here. The basis for this discrepancy is currently being investigated.
Purification of CTSBP2
The recombinant C-terminal half of Xpress/His-tagged (XH-CTSBP2) was prepared as described previously (9).
Preparation of [75Se]Sec tRNASec
75Se-labeled Sec-tRNASec was prepared using HepG2 cells that were grown in Eagle's minimal essential medium containing Earle's balanced salt solution and l-glutamine and supplemented with 10% FBS. At ∼70% confluence, the medium was changed to serum-free medium containing 100 nm 75Se. Cells were grown for 72 h and then collected. Total tRNA was extracted from these cells using acidic phenol and ethanol precipitation (35). The aminoacylation of Sec-tRNASec was confirmed by resolving the purified total tRNA on an acid-urea denaturing gel and exposing to a PhosphorImager screen. The tRNA was quantitated using absorbance at 260 nm.
RESULTS
Mammalian SEPP1 Synthesis in Rabbit Reticulocytes Occurs Processively but with Low Efficiency
To evaluate Sec incorporation into SEPP1 in its native context, we translated 5′-capped full-length rat SEPP1 mRNA in RRL in the presence of [35S]Met. However, the rat SEPP1 protein contains only two Met residues, not including the initiator Met, which is naturally removed during post-translational processing. One is located before the 1st UGA and the other between the 2nd and 3rd UGA. Thus, the early termination at the first UGA in wild type SEPP1 results in a 6-kDa product that contains a single Met residue, which is difficult to detect and separate from free [35S]Met. To obtain a robust signal for the first termination product, thus allowing calculation of efficiency (as described under “Experimental Procedures”), the first and second SEPP1 UGA were mutated to UGC, encoding Cys (U1-2C; Fig. 1A). Termination at the 3rd Sec codon position would therefore yield a 32-kDa termination product containing both Met residues. For comparison, we also utilized a luciferase reporter mRNA that possesses an in-frame UGA codon at position 258 followed by the SEPP1 3′-UTR (Luc/UGA/SP) (Fig. 1A). The translation of Luc/UGA/SP in the presence of [35S]Met results in a ∼28-kDa termination product and a ∼63-kDa band corresponding to the Sec-containing full-length protein (Fig. 1B, lane 1). Sec incorporation into this reporter has been confirmed previously by 75Se labeling and by virtue of its dependence on a SECIS element and added SBP2, which is extremely limiting in rabbit reticulocyte lysate (9). Consistent with our prior results, translation of Luc/UGA/SP mRNA showed 10% Sec incorporation efficiency (9, 18). Surprisingly, no full-length product was detectable for native SEPP1 (Fig. 1B, lanes 2–4) except in the case where all Sec residues were changed to Cys (CysSEPP1; Fig. 1B, lane 5). The U1-2C mutant yielded only a termination product at 32 kDa.
To circumvent the limitations associated with standard [35S]Met labeling for SEPP1, we optimized the RRL assay for 75Se labeling. As shown in Fig. 1C, the translation of Luc/UGA/SP mRNA in the presence of 400 nm [75Se]sodium selenite yielded a distinct SBP2-dependent ∼63-kDa band corresponding to full-length luciferase. A broad but intense 42-kDa band corresponding to the molecular weight of full-length SEPP1 is visible for the wild type (both with and without the 5′-UTR) and U1C mutant SEPP1 (Fig. 1C, lanes 2–4). Interestingly, the lack of any early termination products for native rat SEPP1 indicates highly processive Sec incorporation. However, the U1C mutant does show faint termination products that correspond to early termination at the fourth and sixth Sec codon (Fig. 1C, lane 4, arrows), indicating that the first UGA position may influence processive Sec incorporation. The total canonical (non Sec-related) translation was plotted in Fig. 1D, showing that each of these mRNAs was translated with equal efficiency prior to the ribosomes encountering a UGA codon.
Together, these results indicate that the incorporation of multiple Sec residues into the native SEPP1 protein is highly processive even though inefficient. Moreover, the position or context of the first Sec codon within SEPP1 may be a determinant for processivity.
SEPP1 Synthesis in Vitro Yields a Mixture of Termination and Full-length Products
To determine whether the 42-kDa 75Se-labeled band in Fig. 1C corresponds to full-length SEPP1, we generated nine mutant versions in which we progressively changed each of the 10 Sec (U) codons to encode Cys (C) annotated as U1C to U1-9C (Fig. 2A). To observe the predominant termination products that are the result of the inherently inefficient Sec incorporation in RRL, we first translated each of these mutant mRNAs in the presence of [35S]Met (Fig. 2B, lanes 1–10). This allowed detection of truncation products corresponding to termination at Sec codons 2 to 10 (Fig. 2B, lanes 1–9). This result clearly shows that we cannot easily resolve termination events that occur between codons 7–10 (Fig. 2B, compare lanes 6–9). Interestingly, when 75Se labeling was performed for the same mRNAs (Fig. 2C), only a higher molecular mass 42-kDa band is observed, albeit migrating at a slightly lower average molecular weight for U1C to U1-5C (lanes 1–5) than for U1-6C to U1-9C (lanes 6–9). The overall comparison of the progressively mutated versions of SEPP1 (Fig. 2C, lanes 1–9) to wild-type SEPP1 (Fig. 2C, lane 10) shows that most of the product likely results from termination between Sec codons 7 and 10, but very little termination is detectable that would correspond to Sec codons upstream of position 7. To further examine what fraction of the SEPP1 translation product corresponds to bona fide full-length protein, we used a C-terminally FLAG-tagged SEPP1 mRNA and purified its translation product from RRL using anti-FLAG magnetic beads. The yield of full-length SEPP1 was calculated to be 13% (data not shown). Thus, the band at ∼42 kDa in Fig. 1C corresponds to a mixed species of SEPP1 translation products with ∼13% being full length.
Interestingly, although the serially mutated SEPP1 mRNAs possess decreasing numbers of Sec codons, increased 75Se labeling is observed. For example, the U1C product shown in Fig. 2C has nine Sec residues but the band intensity is significantly lower than for U1-8C, which has only two (Fig. 2C, compare lanes 1 and 8). As shown graphically in Fig. 2D, this increase in Sec incorporation does not correlate with an overall increase in translation because the levels of truncation products observed in Fig. 2B do not significantly increase after U1-4C, whereas the amount of Sec incorporation occurring per codon increases by ∼11-fold. Overall, these data indicate that processive Sec incorporation occurs for at least the first six Sec codons in SEPP1 but that most of the product of in vitro translation is the result of termination prior to the natural stop codon. These data may suggest that the early incorporation events are fundamentally different than those that occur in the latter portion of the protein.
Selenium Supplementation Stimulates Efficiency of SEPP1 Synthesis
To robustly evaluate Sec incorporation efficiency into SEPP1, we repeated the in vitro translation shown in Fig. 1A using 35S-labeled Cys instead of [35S]Met to label the 18 Cys residues in SEPP1. Fig. 3A shows that in addition to the full-length luciferase protein (lane 1), we are also able to detect a [35S]Cys labeled band that corresponds to the molecular weight of full-length SEPP1 (lanes 2 and 3). Using the U1C mRNA (lane 4), we were able to calculate an efficiency of Sec incorporation into SEPP1 of ∼1% compared with 10% for the Luc/UGA/SP. Interestingly, we also observe several smaller bands that we conclude are early termination products because they appear only in the presence of SBP2 (compare lanes 3 to lane 8). The presence of these early termination products for SEPP1 was surprising as from Fig. 1C, it is evident that when 75Se is used, these products are not detected. To determine whether this discrepancy is due to the addition of selenium, we performed [35S]Cys labeling with increasing concentrations of non-radioactive selenium (sodium selenite) for Luc/UGA/SP, SEPP1, and SEPP1 U1C mRNAs. Interestingly, the yield of full-length wild type SEPP1 protein increased by >4-fold with added selenium, but the efficiency of Sec incorporation for Luc/UGA/SP did not increase (Fig. 3B). Although selenium addition increased the yield of the 42-kDa translation product for both native SEPP1 and U1C mutant, there was no major reduction in the early termination product at 29 kDa for U1C (Fig. 3B, third panel). Fig. 3C shows a quantitation of the effect of selenium supplementation on SEPP1 versus Luc/UGA/SP synthesis plotted as the fold increase in Sec incorporation efficiency.
FIGURE 3.
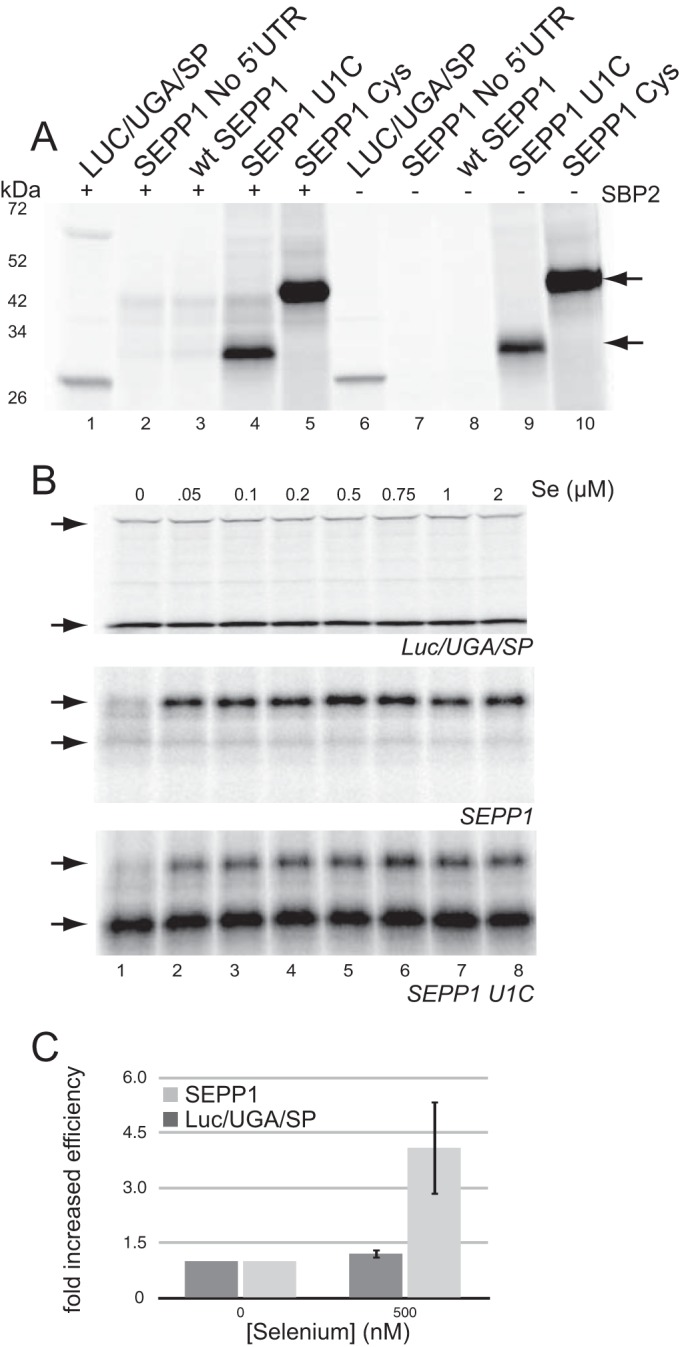
SEPP1 synthesis is stimulated by selenium supplementation. A, Luc/UGA/SP, SEPP1, U1C, and CysSEPP1 mRNAs were translated in RRL analyzed using [35S]Cys labeling. The lower arrow represents SEPP1 product from termination at the second UGA, and the upper arrow marks full-length SEPP1. B, Luc/UGA/SP, SEPP1, and SEPP1 U1C mRNAs were translated in RRL with increasing concentrations of selenium and analyzed using [35S]Cys labeling (top, middle, and bottom panel, respectively). Upper and lower arrows mark full-length and termination products, respectively. C, graphical representation of data obtained in B for the 500 nm selenite reaction was plotted as described in Fig. 1.
These results indicate that the efficiency of Sec incorporation into SEPP1 but not luciferase can be dramatically increased by selenium supplementation. The specificity for SEPP1 and prior work showing that the addition of purified Sec-tRNASec to reticulocyte lysate does not enhance Sec incorporation (36) indicates that the stimulatory effect of selenium is not simply due to an increase in available Sec-tRNASec. Rather, this may indicate a coupling of Sec-tRNASec synthesis to the efficiency of translation elongation.
SECIS-1 Alone Is Essential for Processive Sec Incorporation
Given the long and conserved nature of the SEPP1 3′-UTR, we wanted to determine whether non-SECIS cis-elements located in the 3′-UTR possess any regulatory function. For this purpose, we created nine deletion mutants of the SEPP1 3′-UTR and analyzed the effects on [75Se]selenite labeling in RRL. Simultaneously, we also created the same deletion mutants of SEPP1 3′-UTR in the U1C mutant construct, thus allowing us to analyze the effect of the deletions on both Sec incorporation by 75Se labeling and translation by [35S]Met labeling. When these SEPP1 mRNAs harboring 3′-UTR deletions were translated in RRL, only the SECIS-1 deletion mutant resulted in a significant reduction of Sec incorporation (Fig. 4A, lane 4). Similar to the results obtained for native rat SEPP1, deletion of SECIS-1 in U1C had the most deleterious effect on expression and showed early termination products (Fig. 4B, lane 4), which were entirely dependent on the presence of a functional SECIS-2 (data not shown). To control for changes in general translation efficiency, we also labeled the U1C mRNAs with [35S]Met, but we detected no significant differences (Fig. 4C). Together, these results indicate that neither SECIS-2 nor the conserved non-SECIS portions of the SEPP1 3′-UTR are essential for processive Sec incorporation in vitro.
FIGURE 4.
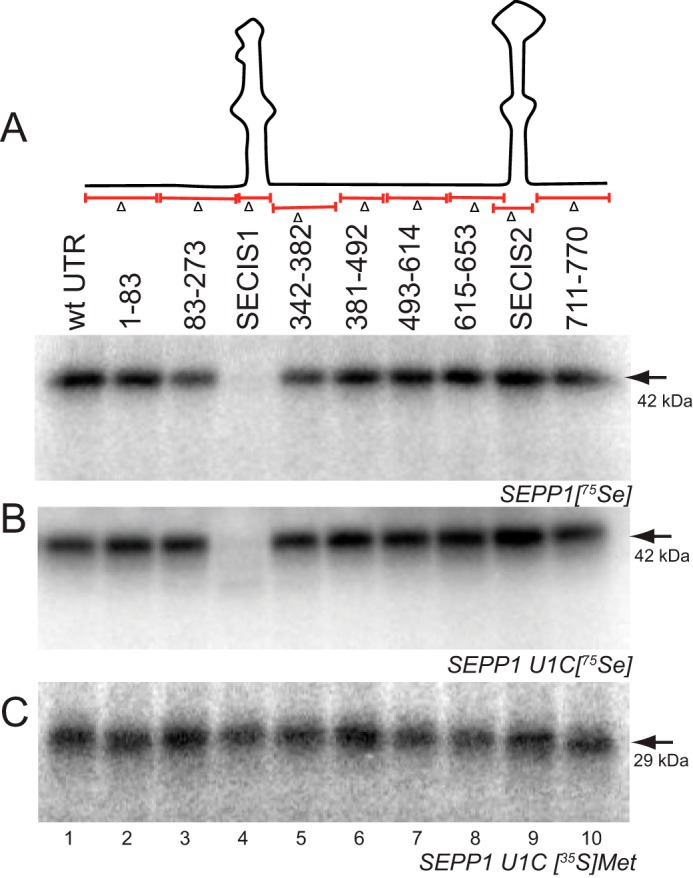
Most of the SEPP1 3′-UTR is not required for Sec incorporation. A, top panel, schematic diagram of the 3′-UTR deletion mutants. Bottom panel, SEPP1 mRNAs harboring the indicated deletion mutations in the 3′-UTR were translated in RRL and analyzed using 75Se labeling. The arrow indicates the 42-kDa product. B, U1C SEPP1 mRNAs harboring the indicated deletion mutations in the 3′-UTR were translated in RRL and analyzed using 75Se labeling. The arrow indicates the 42-kDa product. C, same as in B except with [35S]Met labeling. Radiolabeled proteins were analyzed as described in Fig. 1. The arrow indicates the 29-kDa product that corresponds to termination at the second UGA in U1C construct.
SECIS Elements Are Inherently Processive
As SECIS-1 was the only portion of the SEPP1 3′-UTR that was determined to be essential, we subsequently tested whether a single unrelated SECIS element could promote processive Sec incorporation in vitro. To examine this, we analyzed the ability of the GPX4 SECIS element to support processive Sec incorporation in RRL using 75Se labeling. We replaced the full-length SEPP1 3′-UTR in native rat SEPP1 mRNA with a 105-nt SECIS derived from rat GPX4 (SEPP1-GPX4). Because Sec incorporation activity has been shown to be inhibited by placing SECIS elements <52 nucleotides downstream from the UGA Sec codon (37), the GPX4 SECIS was also ligated further downstream of the stop codon in wild-type and U1C SEPP1 at a naturally occurring PacI site. This yielded a 3′-UTR that contained the 5′ 83 nt of the SEPP1 3′-UTR followed by 105 nt of the GPX4 SECIS element (SEPP1-GPX4+83). These chimeric mRNAs were translated in RRL supplemented with 75Se. As seen in Fig. 5A (lanes 3 and 5), native rat SEPP1 and U1C mRNAs both produced a distinct band migrating at size of full length SEPP1 (42 kDa). SEPP1-GPX4+83 and U1C-GPX4+83 mRNAs also yielded the large 42-kDa product, although with 38 and 45% lower efficiency (Fig. 5A, lanes 4 and 6, and Fig. 5B). The U1C-GPX4+83 mRNA also produced early termination products that were easily detectable (Fig. 5A, compare lanes 4 and 6). In addition, the mRNA that contained only the GPX4 SECIS element without the 83-nt spacer also yielded full-length SEPP1 protein (Fig. 5A, lane 1). This clearly suggests that SECIS elements may generally be intrinsically processive but may differ in their efficiency. Overall, these results indicate that the non-SECIS elements of SEPP1 3′-UTR are not essential for processivity and that processive Sec incorporation into native SEPP1 can be supported by a single unrelated SECIS element.
FIGURE 5.
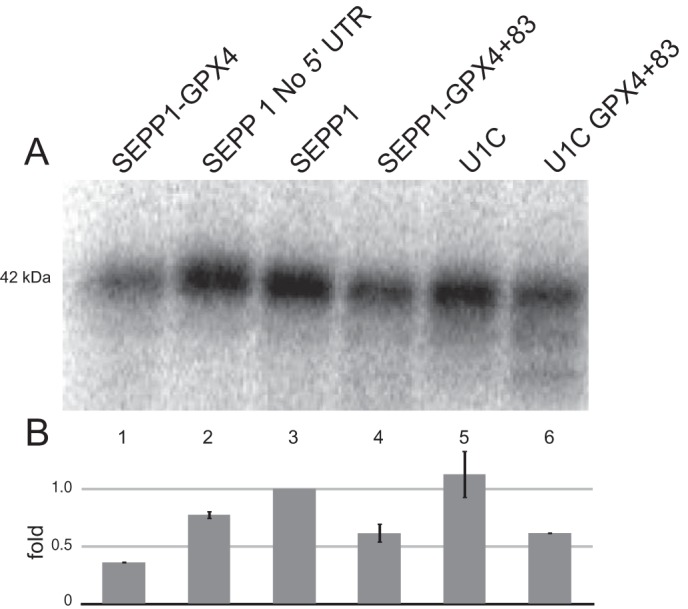
The GPX4 SECIS is sufficient for processive Sec incorporation. A, wild type and mutants of native rat SEPP1 containing just GPX4 SECIS (SEPP1-GPX4) or the GPX4 SECIS placed 83 nt downstream of the SEPP1 and U1C stop codon (SEPP1-GPX4 + 83 or U1C-GPX4 + 83) were translated in RRL in the presence of 75Se. Radiolabeled proteins were analyzed as described in Fig. 1. B, data from A was adjusted for the maximum number of Sec residues then normalized to native SEPP1 and plotted as described in Fig. 1.
Known Sec Incorporation Factors Are Insufficient for Processive SEPP1 Synthesis
Given that the GPX4 SECIS alone was sufficient for processive SEPP1 synthesis in RRL, we subsequently tested whether other factors may be required for processivity. We recently reconstituted Sec incorporation in WGL, which like all higher plants is devoid of any Sec incorporation factors, and we showed that the known Sec incorporation factors were sufficient for Sec incorporation into our luciferase reporter (33). To test whether this held true for processive Sec incorporation into native SEPP1, we used the same WGL assay and translated SEPP1 and Luc/UGA/SP mRNAs with added recombinant SBP2, eEFSec, and total HepG2 tRNA containing [75Se]Sec-tRNASec. The production of full-length luciferase was clearly visible as a 75Se-labeled band (Fig. 6, lane 1), but SEPP1 mRNA yielded a band that corresponded to termination at the second UGA with only barely detectable full-length SEPP1 product (Fig. 6, compare lane 2–4). To determine whether the dominance of SECIS-1 is also observed in a reconstituted system, we translated the SEPP1 mRNA that lacked SECIS-2. This also yielded an equally intense band that corresponded to termination at the first UGA (Fig. 6, lane 4), indicating that the dominance of SECIS-1 is intrinsic to the known factors. Overall, these results suggest that known Sec incorporation factors were sufficient for single Sec incorporation in both Luc/UGA/SP and SEPP1 (Fig. 6, compare lanes 1, 3, and 4). However, the efficient synthesis of full-length SEPP1 may require additional factors or processes such as active aminoacylation.
FIGURE 6.
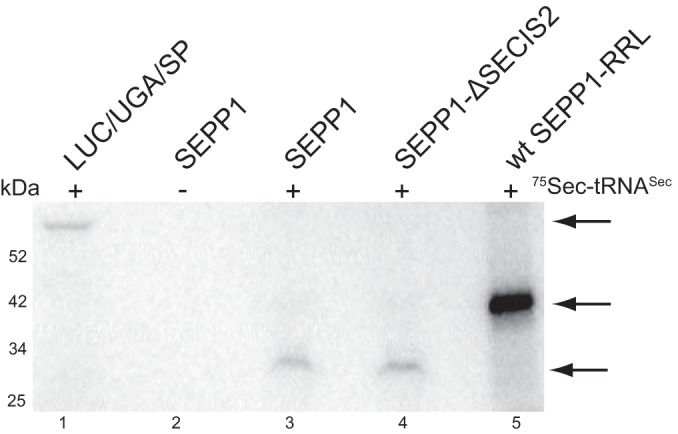
Known Sec incorporation factors are not be sufficient for full-length SEPP1 synthesis. Luc/UGA/SP, SEPP1, and SEPP1 with SECIS-2 deletion (Δ) mRNAs were translated in WGL assay supplemented with recombinant XH-CTSBP2, FLAG-tagged eEFSec, and 75Se-labeled total tRNA from HepG2 cells. 75Se-labeled SEPP1 translated in RRL was loaded in lane 5. The top arrow marks full-length Luc/UGA/SP, the middle arrow marks full-length SEPP1 from the RRL assay, and the bottom arrow represents termination product of SEPP1 at the second UGA.
DISCUSSION
In this study, we investigated the processivity and efficiency of Sec incorporation into SEPP1 and probed for potential regulatory elements within the 3′-UTR. Prior work with reporter constructs containing multiple Sec codons has shown that Sec incorporation is generally inefficient. However, one of these studies elegantly showed that Sec incorporation efficiency at all subsequent Sec codons after the first UGA, increased substantially by almost 10-fold, thereby suggesting that after the first UGA, Sec incorporation is very efficient. In addition, because the amount of termination product resulting from termination at UGA codons downstream from the first UGA was reduced after the first UGA, processivity was also increased (31). Despite these findings, it is clear from the prior work that early termination products are always observed when multiple Sec codons are present (18, 31, 32). Likewise, when we used [35S]Cys to label SEPP1 products, we also observed a significant fraction of termination products, but in the presence of added selenium, the relative fraction of termination products was significantly reduced. This may explain why in 75Se-labeling experiments we exclusively observe ∼42-kDa SEPP1 products that correspond to termination events at UGA codons 7–10. It is striking, however, that even in the presence of added selenium, we observe a substantial SBP2-dependent [35S]Cys-labeled product that corresponds to termination at the second UGA, which is not detectable by 75Se labeling (Fig. 3B). Although it is possible that this is simply due to the reduced number of selenium atoms per molecule of protein, it is also possible that the product is the result of SECIS-dependent Cys incorporation as has been described recently (38). This may be further evidence that Sec incorporation at the first UGA is a unique event. In addition, we have shown here that changing the first Sec codon to Cys results in an increase in early termination products (Fig. 1C). The importance of the first UGA has been illustrated earlier in a zebrafish SEPP1 model, wherein maximum termination was observed at the first UGA, thus indicating that it may serve as a bottleneck in SEPP1 synthesis. Further analysis using polysome profiling led to the model that the first UGA in SEPP1 may serve as a checkpoint to regulate the number of ribosomes approaching the Sec codons located on the 3′-end (50). Given the evidence on the relevance of the first UGA in SEPP1 and also its high level of conservation among all vertebrates wherein it always serves as the 59th amino acid, it can be speculated that its role may be maintained in mammals. Remarkably, the progressive mutation of each Sec codon has shown that Sec incorporation efficiency increases by >11-fold as the number of Sec codons is reduced. This is consistent with recent evidence that UGA position can have a substantial effect on the amount of Sec incorporation into thioredoxin reductase, but the mechanism by which this phenomenon is regulated is unknown (39). Although it is tempting to suppose that the sequence context surrounding the UGA is key, prior evidence indicates that the codon context surrounding the 10th UGA codon in SEPP1 is not sufficient to increase efficiency (18). Interestingly, in vitro translation of wild-type SEPP1 mRNA in RRL yielded a single band that we determined only contained 13% full-length protein. Prior mass spectrometric analysis of SEPP1 purified from rat plasma consisted of full-length protein plus three isoforms resulting from termination at the second, third, and seventh UGA codons (22, 40), but the natural ratio of full-length to truncation products was not determined. Although we cannot distinguish between termination at the seventh UGA and the native stop codon, we do not observe termination at codons 1–6 in vitro for native SEPP1. We did, however, observe early truncation (likely at the fourth and sixth Sec codons) either when SECIS-1 was deleted or when the first Sec codon was mutated to Cys. These results clearly suggest that the processivity of incorporation into the first ∼6 Sec codons is regulated by several potentially mechanistically distinct variables. Ideally these results would be interpreted in the context of the known ratio of full-length to truncated SEPP1 in vivo, but there is considerable difficulty in experimentally determining this. This is in part because most studies of SEPP1 production in vivo have involved a purification step that would likely bias the ratios of full-length to truncated proteins (11, 22, 28, 40–45). In addition, it is impossible to assess the ratio of termination events when the relative stabilities of the fragments versus full-length protein are not known. Other efforts have examined human tissue or serum by immunoblot where two equally intense bands at 55–60 kDa were observed (46, 47), but it is not possible to discern differential glycosylation from termination events. Taken together, however, most SEPP1 expression studies are consistent with the idea that the majority of the protein runs as a single band corresponding to roughly full-length along with usually small amounts of one major truncated product, likely derived from termination at the second UGA. This is also the pattern that has been observed in [75Se]selenite labeling of endogenous SEPP1 in HepG2 cells (48). We can therefore conclude that our results are concordant with the nature of SEPP1 production in vivo, with full-length or nearly full-length protein being predominant albeit with some level of termination occurring at the second UGA codon. Whether the bulk of termination events that occur in vivo are upstream of the native stop codon (e.g. at codon 7), as is the case in our experiments in vitro, will require further investigation.
In mice, the expression of the SEPP1b transcript variant is regulated by microRNAs that target a unique structure located on SEPP1 5′-UTR (49). We therefore used a 75Se-labeled RRL assay to examine whether there existed any regulatory elements within the rat SEPP1 5′-UTR. We established that the 75-nt 5′-UTR we obtained on all of our SEPP1 clones from rat hepatoma cells (McArdle RH-7777) was not necessary for SEPP1 synthesis in vitro, so we proceeded to examine 3′-UTR deletion mutants. With the exception of the SECIS-1 deletion, none of the other deletion mutants had any dramatic effect on SEPP1 synthesis. Consistent with prior work in transfected cells (50), the deletion of SECIS-2 had no effect on SEPP1 synthesis in vitro, but it was able to support inefficient Sec incorporation into the first few Sec codons. Thus, the role of SECIS-2 remains to be determined.
The absence of any effect on SEPP1 synthesis upon deletion of the non-SECIS conserved sequences within the SEPP1 3′-UTR, leaves open the question of what these sequences may be regulating. The fact that the GPX4 SECIS element can effectively replace the entire SEPP1 3′-UTR, albeit with slightly lower efficiency, indicates that basal SEPP1 synthesis does not require the complexity that is found in the native SEPP1 3′-UTR. We can only speculate, therefore, that these conserved sequences are likely to be involved in the regulation of SEPP1 synthesis in vivo, either through regulated mRNA stability, localization, or translation. It is also possible that the non-SECIS sequences are efficiency elements recruiting cellular factors that may be missing or highly variable in RRL. This could explain the discrepancy between this work and our earlier study that reported ∼40% total Sec incorporation (full-length and truncation products), with different batches of RRL likely being the key variable (9).
In our study, we found that selenium supplementation increased native SEPP1 translation by 6-fold. Selenium administration has been shown in animal studies to stimulate and initiate SEPP1 synthesis prior to other selenoprotein synthesis (30, 51, 52). The shift of the relative abundance of the SEPP1 isoforms toward full-length synthesis in humans upon selenium supplementation has also been reported (45). Although much of these effects are likely due to simply increasing the liver Sec-tRNASec concentration, this stimulation could also be in part due to the efficient channeling of Sec-tRNASec to the UGA codon. Our study provides direct evidence for the first time that exogenously added selenium specifically stimulates processive SEPP1 synthesis but not incorporation into a single Sec codon in our luciferase reporter. We speculate that this effect could be due to the postulated channeling mechanism. Studies on the mammalian translation system have led to the idea that its components in vivo are highly organized on the cytoskeleton so as to allow all the intermediates in the process to interact and channel from one to another, without dissociating (53–55). The initial channeling theory suggested a need for the cellular framework to increase translation efficiency, but using continuous flow cell-free systems, another study showed that tRNAs and translation initiation and elongation factors all remain bound to the protein synthesis machinery and do not exist as freely diffusible molecules (34). Interestingly, this process has been proposed to be the basis for the low efficiency of Sec incorporation in E. coli due to the potential disruption of channeling that must occur during the delivery of SelB by Sec-tRNASec (8). Here, we propose the opposite effect where increased aminoacylation of tRNASec that may result from the addition of exogenous selenium might increase the efficiency of SEPP1 synthesis. Indeed, the lack of active aminoacylation in wheat germ lysate, which does not contain any of the Sec incorporation machinery, may explain why we do not observe processive SEPP1 synthesis in that system. Another potential cause for absence of SEPP1 in wheat germ lysate could be the inefficient interaction between the supplemented Sec incorporation factors and the endogenous translation factors and/or ribosomes in the wheat germ lysate. Taken one step further, the channeling effect may be enhanced within an intact cell, perhaps explaining why Sec incorporation is inefficient in vitro.
CONCLUSIONS
Based on the results shown here, we conclude that the highly conserved non-SECIS sequences in the SEPP1 3′-UTR are surprisingly not involved in specifically promoting the processivity or efficiency of Sec incorporation in vitro. It is clear, therefore, that the 3′-UTR is imbued with more complex regulatory roles that may influence mRNA stability or localization. Interestingly, however, the finding that the efficiency of processive Sec incorporation can be significantly enhanced by the addition of exogenous selenium suggests that this process is linked to selenium utilization, likely through the factors required for Sec-tRNASec synthesis. This finding opens a new chapter in the search for all of the potential regulatory points for the essential process of Sec incorporation in mammals.
Acknowledgment
We thank Aditi Dubey for critical reading of the manuscript.
This work was supported by National Institutes of Health Grant GM077073 (to P. R. C.)
- SECIS
- Sec incorporation sequence
- Sec
- selenocysteine
- SEPP1
- selenoprotein P
- SECIS
- selenocysteine insertion sequence element
- nt
- nucleotide(s)
- Sec-tRNASec
- selenocysteine tRNA aminoacylated with selenocysteine
- SBP2
- SECIS binding protein 2
- RRL
- rabbit reticulocyte lysate
- WGL
- wheat germ lysate.
REFERENCES
- 1. Reeves M. A., Hoffmann P. R. (2009) The human selenoproteome: recent insights into functions and regulation. Cell Mol. Life Sci. 66, 2457–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bösl M. R., Takaku K., Oshima M., Nishimura S., Taketo M. M. (1997) Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp). Proc. Natl. Acad. Sci. U.S.A. 94, 5531–5534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berry M. J., Kieffer J. D., Harney J. W., Larsen P. R. (1991) Selenocysteine confers the biochemical properties characteristic of the type I iodothyronine deiodinase. J. Biol. Chem. 266, 14155–14158 [PubMed] [Google Scholar]
- 4. Copeland P. R., Fletcher J. E., Carlson B. A., Hatfield D. L., Driscoll D. M. (2000) A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 19, 306–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fagegaltier D., Hubert N., Yamada K., Mizutani T., Carbon P., Krol A. (2000) Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 19, 4796–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tujebajeva R. M., Copeland P. R., Xu X. M., Carlson B. A., Harney J. W., Driscoll D. M., Hatfield D. L., Berry M. J. (2000) Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 1, 158–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee B. J., Rajagopalan M., Kim Y. S., You K. H., Jacobson K. B., Hatfield D. (1990) Selenocysteine tRNA[Ser]Sec gene is ubiquitous within the animal kingdom. Mol. Cell. Biol. 10, 1940–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Suppmann S., Persson B. C., Böck A. (1999) Dynamics and efficiency in vivo of UGA-directed selenocysteine insertion at the ribosome. EMBO J. 18, 2284–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mehta A., Rebsch C. M., Kinzy S. A., Fletcher J. E., Copeland P. R. (2004) Efficiency of mammalian selenocysteine incorporation. J. Biol. Chem. 279, 37852–37859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ursini F., Heim S., Kiess M., Maiorino M., Roveri A., Wissing J., Flohé L. (1999) Dual function of the selenoprotein PHGPx during sperm maturation. Science 285, 1393–1396 [DOI] [PubMed] [Google Scholar]
- 11. Yang J. G., Morrison-Plummer J., Burk R. F. (1987) Purification and quantitation of a rat plasma selenoprotein distinct from glutathione peroxidase using monoclonal antibodies. J. Biol. Chem. 262, 13372–13375 [PubMed] [Google Scholar]
- 12. Cheng W. H., Ho Y. S., Ross D. A., Han Y., Combs G. F., Jr., Lei X. G. (1997) Overexpression of cellular glutathione peroxidase does not affect expression of plasma glutathione peroxidase or phospholipid hydroperoxide glutathione peroxidase in mice offered diets adequate or deficient in selenium. J. Nutr. 127, 675–680 [DOI] [PubMed] [Google Scholar]
- 13. Christensen M. J., Cammack P. M., Wray C. D. (1995) Tissue specificity of selenoprotein gene expression in rats. J. Nutr. Biochem. 6, 367–372 [DOI] [PubMed] [Google Scholar]
- 14. Sun Q. A., Kirnarsky L., Sherman S., Gladyshev V. N. (2001) Selenoprotein oxidoreductase with specificity for thioredoxin and glutathione systems. Proc. Natl. Acad. Sci. U.S.A. 98, 3673–3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Budiman M. E., Bubenik J. L., Miniard A. C., Middleton L. M., Gerber C. A., Cash A., Driscoll D. M. (2009) Eukaryotic initiation factor 4a3 is a selenium-regulated RNA-binding protein that selectively inhibits selenocysteine incorporation. Mol. Cell 35, 479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chavatte L., Brown B. A., Driscoll D. M. (2005) Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat. Struct. Mol. Biol. 12, 408–416 [DOI] [PubMed] [Google Scholar]
- 17. Miniard A. C., Middleton L. M., Budiman M. E., Gerber C. A., Driscoll D. M. (2010) Nucleolin binds to a subset of selenoprotein mRNAs and regulates their expression. Nucleic Acids Res. 38, 4807–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gupta M., Copeland P. R. (2007) Functional analysis of the interplay between translation termination, selenocysteine codon context, and selenocysteine insertion sequence-binding protein 2. J. Biol. Chem. 282, 36797–36807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maiti B., Arbogast S., Allamand V., Moyle M. W., Anderson C. B., Richard P., Guicheney P., Ferreiro A., Flanigan K. M., Howard M. T. (2009) A mutation in the SEPN1 selenocysteine redefinition element (SRE) reduces selenocysteine incorporation and leads to SEPN1-related myopathy. Hum. Mutat. 30, 411–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Howard M. T., Moyle M. W., Aggarwal G., Carlson B. A., Anderson C. B. (2007) A recoding element that stimulates decoding of UGA codons by Sec tRNA[Ser]Sec. RNA 13, 912–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bubenik J. L., Miniard A. C., Driscoll D. M. (2013) Alternative transcripts and 3′-UTR elements govern the incorporation of selenocysteine into selenoprotein S. PLoS One 8, e62102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma S., Hill K. E., Caprioli R. M., Burk R. F. (2002) Mass spectrometric characterization of full-length rat selenoprotein P and three isoforms shortened at the C terminus. Evidence that three UGA codons in the mRNA open reading frame have alternative functions of specifying selenocysteine insertion or translation termination. J. Biol. Chem. 277, 12749–12754 [DOI] [PubMed] [Google Scholar]
- 23. Hill K. E., Lloyd R. S., Yang J. G., Read R., Burk R. F. (1991) The cDNA for rat selenoprotein P contains 10 TGA codons in the open reading frame. J. Biol. Chem. 266, 10050–10053 [PubMed] [Google Scholar]
- 24. Hill K. E., Zhou J., McMahan W. J., Motley A. K., Atkins J. F., Gesteland R. F., Burk R. F. (2003) Deletion of selenoprotein P alters distribution of selenium in the mouse. J. Biol. Chem. 278, 13640–13646 [DOI] [PubMed] [Google Scholar]
- 25. Schomburg L., Schweizer U., Holtmann B., Flohé L., Sendtner M., Köhrle J. (2003) Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem. J. 370, 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kurokawa S., Eriksson S., Rose K. L., Wu S., Motley A. K., Hill S., Winfrey V. P., McDonald W. H., Capecchi M. R., Atkins J. F., Arnér E. S., Hill K. E., Burk R. F. (2014) Sepp1(UF) forms are N-terminal selenoprotein P truncations that have peroxidase activity when coupled with thioredoxin reductase-1. Free Radic Biol. Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Burk R. F., Hill K. E., Motley A. K. (2003) Selenoprotein metabolism and function: evidence for more than one function for selenoprotein P. J. Nutr. 133, 1517S-1520S [DOI] [PubMed] [Google Scholar]
- 28. Read R., Bellew T., Yang J. G., Hill K. E., Palmer I. S., Burk R. F. (1990) Selenium and amino acid composition of selenoprotein P, the major selenoprotein in rat serum. J. Biol. Chem. 265, 17899–17905 [PubMed] [Google Scholar]
- 29. Burk R. F. (1991) Molecular biology of selenium with implications for its metabolism. FASEB J. 5, 2274–2279 [DOI] [PubMed] [Google Scholar]
- 30. Motsenbocker M. A., Tappel A. L. (1984) Effect of dietary selenium on plasma selenoprotein P, selenoprotein P1 and glutathione peroxidase in the rat. J. Nutr 114, 279–285 [DOI] [PubMed] [Google Scholar]
- 31. Fixsen S. M., Howard M. T. (2010) Processive selenocysteine incorporation during synthesis of eukaryotic selenoproteins. J. Mol. Biol. 399, 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nasim M. T., Jaenecke S., Belduz A., Kollmus H., Flohé L., McCarthy J. E. (2000) Eukaryotic selenocysteine incorporation follows a nonprocessive mechanism that competes with translational termination. J. Biol. Chem. 275, 14846–14852 [DOI] [PubMed] [Google Scholar]
- 33. Gupta N., DeMong L. W., Banda S., Copeland P. R. (2013) Reconstitution of selenocysteine incorporation reveals intrinsic regulation by SECIS elements. J. Mol. Biol. 425, 2415–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baranov V. I., Spirin A. S. (1993) Gene expression in cell-free system on preparative scale. Methods Enzymol. 217, 123–142 [DOI] [PubMed] [Google Scholar]
- 35. Gonzalez-Flores J. N., Gupta N., DeMong L. W., Copeland P. R. (2012) The selenocysteine-specific elongation factor contains a novel and multi-functional domain. J. Biol. Chem. 287, 38936–38945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kinzy S. A., Caban K., Copeland P. R. (2005) Characterization of the SECIS binding protein 2 complex required for the co-translational insertion of selenocysteine in mammals. Nucleic Acids Res. 33, 5172–5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martin G. W., Harney J. W., Berry M. J. (1996) Selenocysteine incorporation in eukaryotes: insights into mechanism and efficiency from sequence, structure, and spacing proximity studies of the type 1 deiodinase SECIS element. RNA 2, 171–182 [PMC free article] [PubMed] [Google Scholar]
- 38. Turanov A. A., Xu X. M., Carlson B. A., Yoo M. H., Gladyshev V. N., Hatfield D. L. (2011) Biosynthesis of selenocysteine, the 21st amino Acid in the genetic code, and a novel pathway for cysteine biosynthesis. Adv. Nutr 2, 122–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Turanov A. A., Lobanov A. V., Hatfield D. L., Gladyshev V. N. (2013) UGA codon position-dependent incorporation of selenocysteine into mammalian selenoproteins. Nucleic Acids Res. 41, 6952–6959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chittum H. S., Himeno S., Hill K. E., Burk R. F. (1996) Multiple forms of selenoprotein P in rat plasma. Arch. Biochem. Biophys. 325, 124–128 [DOI] [PubMed] [Google Scholar]
- 41. Mostert V., Lombeck I., Abel J. (1998) A novel method for the purification of selenoprotein P from human plasma. Arch. Biochem. Biophys. 357, 326–330 [DOI] [PubMed] [Google Scholar]
- 42. Motchnik P. A., Tappel A. L. (1989) Rat plasma selenoprotein P properties and purification. Biochim. Biophys. Acta 993, 27–35 [DOI] [PubMed] [Google Scholar]
- 43. Motchnik P. A., Tappel A. L. (1990) Multiple selenocysteine content of selenoprotein P in rats. J. Inorg Biochem. 40, 265–269 [DOI] [PubMed] [Google Scholar]
- 44. Himeno S., Chittum H. S., Burk R. F. (1996) Isoforms of selenoprotein P in rat plasma. Evidence for a full-length form and another form that terminates at the second UGA in the open reading frame. J. Biol. Chem. 271, 15769–15775 [DOI] [PubMed] [Google Scholar]
- 45. Méplan C., Nicol F., Burtle B. T., Crosley L. K., Arthur J. R., Mathers J. C., Hesketh J. E. (2009) Relative abundance of selenoprotein P isoforms in human plasma depends on genotype, Se intake, and cancer status. Antioxid. Redox Signal. 11, 2631–2640 [DOI] [PubMed] [Google Scholar]
- 46. Bellinger F. P., He Q. P., Bellinger M. T., Lin Y., Raman A. V., White L. R., Berry M. J. (2008) Association of selenoprotein p with Alzheimer's pathology in human cortex. J. Alzheimers Dis. 15, 465–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Renko K., Werner M., Renner-Müller I., Cooper T. G., Yeung C. H., Hollenbach B., Scharpf M., Köhrle J., Schomburg L., Schweizer U. (2008) Hepatic selenoprotein P (SePP) expression restores selenium transport and prevents infertility and motor-incoordination in Sepp-knockout mice. Biochem. J. 409, 741–749 [DOI] [PubMed] [Google Scholar]
- 48. Tujebajeva R. M., Harney J. W., Berry M. J. (2000) Selenoprotein P expression, purification, and immunochemical characterization. J. Biol. Chem. 275, 6288–6294 [DOI] [PubMed] [Google Scholar]
- 49. Dewing A. S., Rueli R. H., Robles M. J., Nguyen-Wu E. D., Zeyda T., Berry M. J., Bellinger F. P. (2012) Expression and regulation of mouse selenoprotein P transcript variants differing in non-coding RNA. RNA Biol. 9, 1361–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stoytcheva Z., Tujebajeva R. M., Harney J. W., Berry M. J. (2006) Efficient incorporation of multiple selenocysteines involves an inefficient decoding step serving as a potential translational checkpoint and ribosome bottleneck. Mol. Cell. Biol. 26, 9177–9184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Deagen J. T., Butler J. A., Zachara B. A., Whanger P. D. (1993) Determination of the distribution of selenium between glutathione peroxidase, selenoprotein P, and albumin in plasma. Anal. Biochem. 208, 176–181 [DOI] [PubMed] [Google Scholar]
- 52. Yang J. G., Hill K. E., Burk R. F. (1989) Dietary selenium intake controls rat plasma selenoprotein P concentration. J. Nutr. 119, 1010–1012 [DOI] [PubMed] [Google Scholar]
- 53. Sivaram P., Deutscher M. P. (1990) Existence of two forms of rat liver arginyl-tRNA synthetase suggests channeling of aminoacyl-tRNA for protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 87, 3665–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Negrutskii B. S., Deutscher M. P. (1991) Channeling of aminoacyl-tRNA for protein synthesis in vivo. Proc. Natl. Acad. Sci. U.S.A. 88, 4991–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stapulionis R., Deutscher M. P. (1995) A channeled tRNA cycle during mammalian protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 92, 7158–7161 [DOI] [PMC free article] [PubMed] [Google Scholar]


