Background: The assembly of Photosystem I requires additional protein factors.
Results: Arabidopsis RNAi mutants of PPD1 were characterized for Photosystem I function and assembly.
Conclusion: The PPD1 protein serves as a factor to facilitate functional assembly of Photosystem I.
Significance: RNAi suppression of PPD1 provides new details on its function in Photosystem I assembly.
Keywords: Arabidopsis, Membrane Protein, Photosynthesis, Plant Biochemistry, Protein Assembly, Photosystem I
Abstract
Photosystem I (PS I) is a multisubunit membrane protein complex that functions as a light-driven plastocyanin-ferredoxin oxidoreductase. The PsbP domain protein 1 (PPD1; At4g15510) is located in the thylakoid lumen of plant chloroplasts and is essential for photoautotrophy, functioning as a PS I assembly factor. In this work, RNAi was used to suppress PPD1 expression, yielding mutants displaying a range of phenotypes with respect to PS I accumulation and function. These PPD1 RNAi mutants showed a loss of assembled PS I that was correlated with loss of the PPD1 protein. In the most severely affected PPD1 RNAi lines, the accumulated PS I complexes exhibited defects in electron transfer from plastocyanin to the oxidized reaction center P700+. The defects in PS I assembly in the PPD1 RNAi mutants also had secondary effects with respect to the association of light-harvesting antenna complexes to PS I. Because of the imbalance in photosystem function in the PPD1 RNAi mutants, light-harvesting complex II associated with and acted as an antenna for the PS I complexes. These results provide new evidence for the role of PPD1 in PS I biogenesis, particularly as a factor essential for proper assembly of the lumenal portion of the complex.
Introduction
The thylakoid membranes of oxygenic organisms contain the membrane protein complexes Photosystem II (PS II),2 cytochrome b6f complex, Photosystem I (PS I), ferredoxin-NAPD+ reductase, ATP synthase, NADPH dehydrogenase, and two separate light-harvesting complexes (LHC I and LHC II). These complexes function together to oxidize water, reduce NADP+, and synthesize ATP. All of these complexes contain numerous protein subunits and cofactors that must be coordinately assembled to form functional ensembles.
PS I catalyzes the light-driven transfer of electrons from the lumenal protein plastocyanin to the stromal protein ferredoxin (1, 2). Cyanobacterial PS I is composed of 12 protein subunits, whereas higher plant PS I contains at least 15 protein subunits (2, 3). Additionally, numerous cofactors are associated with the complex. Photochemical charge separation in PS I occurs at the reaction center P700, a special chlorophyll pair coordinated to the core PS I subunits PsaA and PsaB. The stromal side of the photosystem contains the extrinsic proteins PsaC, PsaD, and PsaE, which facilitate binding of oxidized ferredoxin and association with ferredoxin-NAPD+ reductase. On the lumenal side of PS I, the extrinsic domains of the PsaA, PsaB, and PsaF proteins along with the soluble subunit PsaN form the docking site for reduced plastocyanin, the soluble protein responsible for transferring electrons from the cytochrome b6f complex to PS I. In plants, the assembled PS I complex associates with its antenna complex LHC I, comprising chlorophyll proteins Lhca1–Lhca4, to form the PS I-LHC I supercomplex (4).
Although a great deal is known about PS II assembly and repair (5), much less is known about the steps required to form functional PS I (2). One PS I assembly intermediate from plants, containing the subunits PsaA-F, PsaH-J, and PsaL, has been identified (6). Labeling experiments suggest that PS I assembly occurs rapidly (2, 6), which complicates experimental efforts to identify additional PS I assembly intermediates. A number of assembly factors have been identified that transiently associate with PS I to facilitate the incorporation of various PS I subunits into the functional complex. The Ycf3 protein is involved in the assembly of the stromal side of PS I and is required for stable complex accumulation (7–9). The Y3IP1 protein was identified as an interacting partner of Ycf3. Mutations in this protein also result in the loss of PS I accumulation (10); however, the precise role of Y3IP1 remains unclear. The Ycf4 protein is essential for PS I assembly in Chlamydomonas reinhardtii but not in plants or the cyanobacterium Synechocystis (7, 11, 12). Pyg7 is a tetratricopeptide repeat protein that associates with PS I and is required for accumulation of PS I subunits and complexes (13). Proper integration of cofactors into PS I also affects assembly of the complex. The rubA mutant in Synechococcus sp. PCC 7002 does not properly assemble the Fx iron-sulfur cluster (14). As a result, there is a decrease in the amount of PS I complexes, and those that do assemble lack the PsaC, PsaD, and PsaE proteins on the stromal side of the complex (15).
PsbP domain protein 1 (PPD1), a member of the PsbP protein family found in the thylakoid lumen of plants, has also been reported to be a PS I assembly factor (16, 17). Arabidopsis ppd1 T-DNA mutants do not contain detectable levels of the PPD1 protein, are not photoautotrophic, and do not assemble detectable levels of PS I (16). To further elucidate the role of the PPD1 protein in PS I assembly, RNAi silencing was used to suppress PPD1 expression in Arabidopsis. This technique yields individuals exhibiting a range of phenotypes correlated to the suppression of the targeted gene and is particularly useful for gaining additional functional information when null alleles result in severe or lethal phenotypes (18–20). Here, we report that the suppression of the PPD1 protein in PPD1 RNAi plant lines appears highly correlated with the loss of assembled PS I. The accumulation of the PsaF, PsaG, and PsaN subunits and plastocyanin (PC) was the most affected in the PPD1 RNAi plants. This observation was consistent with observed defects in LHC I antenna coupling to PS I and electron transfer to P700+. In addition to these PS I defects, LHC II antenna proteins functionally associated with the PS I complexes which did accumulate in the PPD1 RNAi plants. The role of the PPD1 protein as a PS I assembly factor is discussed.
EXPERIMENTAL PROCEDURES
Plant Growth Conditions
Surface-sterilized seeds of wild-type A. thaliana var. Columbia (Col-0) were germinated on solid Murashige-Skoog medium (21) containing 2% sucrose and 0.7% agar and then incubated for 2 days at 4 °C in the dark. The seedlings were transferred to soil 14 days later and grown at 20 °C under 50–80 μmol of photons m−2 s−1 white light under 8-h light/16-h dark diurnal conditions.
RNAi Line Construction
The pHANNIBAL vector (22) was used to construct an intron-spliced hairpin RNA (RNAi construct). Because PPD1 is a member of an extended gene family, a region of the gene unique to PPD1 with little sequence similarity to other genes within the PsbP family was chosen as the RNAi target. The sequence (+628 to +790) of the PPD1 gene (At4g15510) was chosen for expression suppression (see Fig. 1A). This construct will be referred to as PPD1 RNAi. The primers were 5′-GCTCTAGAGAGCATGATGATGGGCTTGCTC-3′ and 5′-CCATCGATGCACCAGCTCCTCGGACTTGG-3′ for the sense fragment and 5′-CCGCTCGAGGAGCATGATGATGGGCTTGCTC-3′ and 5′-GGGGTACCGCACCAGCTCCTCGGACTTGG-3′ for the antisense fragment. After construction and verification by sequencing, the expression region was excised from the pHANNIBAL vector with NotI and then subcloned into the pART27 plasmid. The resultant PPD1 RNAi plasmid was introduced into Agrobacterium (strain GV3101) by the freeze-thaw method (23). Healthy, flowering WT plants were then transformed by the floral dip method as described previously (24). Surface-sterilized seeds were spread on solid Murashige-Skoog medium containing 2% sucrose, 0.7% agar, 50 mg/liter kanamycin, and 400 mg/liter carbenicillin and then incubated for 2 days at 4 °C in the dark. The transgenic kanamycin-resistant seedlings were transferred to soil after the first true leaves appeared and grown as described above. It should be noted that the more severely affected PPD1 RNAi strains grew extremely poorly and did not set seed. Consequently, all characterization was performed on primary transformant plants.
FIGURE 1.

PPD1 gene structure and PPD1 RNAi plant growth. A, PPD1 gene structure showing RNAi target location. B, 3-week-old wild-type and mutant PPD1 RNAi-E, -F, -H, and -J plants. Scale bar, 1 cm.
Chlorophyll Fluorescence Measurements
Chlorophyll fluorescence measurements were performed using a Joliot Type Spectrophotometer (JTS-10, Bio-Logic Science Instruments) in fluorescence mode with a 520 nm actinic light source. Detached leaves were incubated in the dark for 5 min and subsequently illuminated at the indicated light intensity for 4 min. Chlorophyll fluorescence was collected over the entire time course. The fluorescence parameters are defined as follows: F0, the chlorophyll fluorescence of the dark-adapted sample; Fm, the maximal chlorophyll fluorescence upon actinic light illumination; Fs, the steady-state chlorophyll fluorescence of light-adapted samples; and Fv/Fm, (Fm − F0)/Fm. Data were analyzed using Origin version 8.1 and proprietary software provided by Bio-Logic Science Instruments.
Steady-state P700 Measurements
Measurements of steady-state P700 oxidation and reduction were also performed with the JTS-10 in absorbance mode using the “pulse of dark” method. Detached leaves were dark-incubated for 5 min prior to each measurement. Samples were illuminated, and the absorbance changes at 705 nm were measured to assay the oxidation of the PS I P700 reaction center during illumination followed by reduction of P700+ in the subsequent dark period. To specifically excite PS I and the LHC I antenna, a 720 nm actinic light source with 20-s illumination was used. To excite LHC II as well as PS I-LHC I, a broadband orange light source with a peak of 630 nm with 5-s illumination was used. Data were analyzed using Origin version 8.1 and proprietary software provided by Bio-Logic Science Instruments.
Protein Gel Electrophoresis and Detection
For analysis of the thylakoid membrane protein complement, thylakoid membranes were isolated from wild-type and PPD1 RNAi mutant plants as described previously (19). Chlorophyll concentration was determined by the method of Arnon (25). Protein concentration was determined using the bicinchoninic acid (BCA) protein assay (Pierce) on 1% SDS-extracted thylakoid membranes after being diluted to 0.1% SDS. LiDS-polyacrylamide gel electrophoresis (PAGE) was performed on 12.5–20% polyacrylamide gradient gels with samples loaded based on equivalent total protein amounts (26). The resolved proteins were electroblotted onto PVDF membranes (Immobilon-P, Millipore Corp.). After blocking for 2 h with 5% nonfat dry milk in 150 mm NaCl, 10 mm Tris-HCl, pH 7.4 (TS), the blots were washed extensively with TS. The blots were then incubated with diluted primary antibody followed by incubation with an anti-rabbit IgG-peroxidase conjugate (Sigma). Immobilized antibodies were detected with a chemiluminescent substrate (SuperSignal West Pico chemiluminescent substrate, Pierce), and the blots were exposed to x-ray film. For semiquantitative analysis of protein amounts, a standard curve for the chemiluminescence signal for a series of wild-type thylakoid amounts was generated for each protein signal using NIH ImageJ software (27). The integrated chemiluminescence signals from a dilution series of wild-type membrane samples were used to determine the relative abundance of various proteins in the PPD1 RNAi mutant samples. The antibodies used were directed against the AtpB, PsaB, PsaF, PsaG, PsaN, PC, Lhca1, Lhcb1, cytochrome f, and CP47 proteins. The PPD1-specific antibody was raised in rabbit against a recombinantly expressed PPD1 protein containing a C-terminal histidine tag.
Blue native (BN) PAGE was performed as follows. For thylakoid solubilization, membranes were resuspended in buffer A (25 mm BisTris-HCl, pH 7.0, 20% (w/v) glycerol, and 0.25 mg of 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride/ml) to a final concentration of 1 mg of chlorophyll/ml, and an equal volume of 1.6% (w/v) dodecyl β-d-maltoside (Anatrace) freshly prepared in buffer A was added. The final concentrations were 0.8% (w/v) dodecyl β-d-maltoside and 0.5 mg of chlorophyll/ml. Thylakoids were then incubated on ice for 5 min and centrifuged at 14,100 × g at 4 °C for 15 min. For solubilization with digitonin, thylakoid membranes were resuspended in buffer A as described above, and an equal volume of 3% digitonin in buffer A was added for a final concentration of 1.5% digitonin and 0.5 mg of chlorophyll/ml. This mixture was incubated at 4 °C with gentle agitation for 1 h and then centrifuged at 14,100 × g at 4 °C for 15 min. The supernatant was supplemented with volume of loading buffer (100 mm BisTris-HCl, pH 7.0, 0.5 m ϵ-amino-n-caproic acid, 30% (w/v) sucrose, and 50 mg of Serva Blue G/ml) and loaded on a gel with a 5.5–15% gradient of acrylamide in the separation gel. Electrophoresis was performed at 4 °C at constant voltage for a total of 21 h beginning with 4 h at 70 V followed by 17 h at 120 V. Upon increasing the voltage, the cathode buffer was exchanged for buffer lacking the dye. For subsequent LiDS-PAGE, slices from BN-polyacrylamide gels were incubated with loading buffer (5% LiDS, 15% sucrose, 12.5% β-mercaptoethanol, and 62.5 mm Tris-HCl, pH 6.8) for 2 h, and proteins were electrophoresed and blotted as described above. Two-dimensional (BN-LiDS) polyacrylamide gels were silver-stained as described (28).
RESULTS
PPD1 RNAi Individuals Exhibit a Range of Phenotypes for Plant Growth and PS I Function
A previous report characterized a T-DNA insertion mutant of PPD1 in Arabidopsis, and these plants were devoid of PS I (16). Our work characterizes RNAi lines that suppress PPD1 expression and allows the recovery of autotrophic transformants representing a range of phenotypes with respect to plant growth and PS I function. Fig. 1B shows wild-type and PPD1 RNAi mutant plants. Individual PPD1 RNAi-E, -F, and -H plants showed significantly stunted growth relative to wild type, whereas PPD1 RNAi-J was closer in size to wild type. Note that because individual primary transformants were characterized in this study, only a subset of experiments could be performed on each individual plant due to the small plant size and limiting sample size for the more severely affected mutant plants.
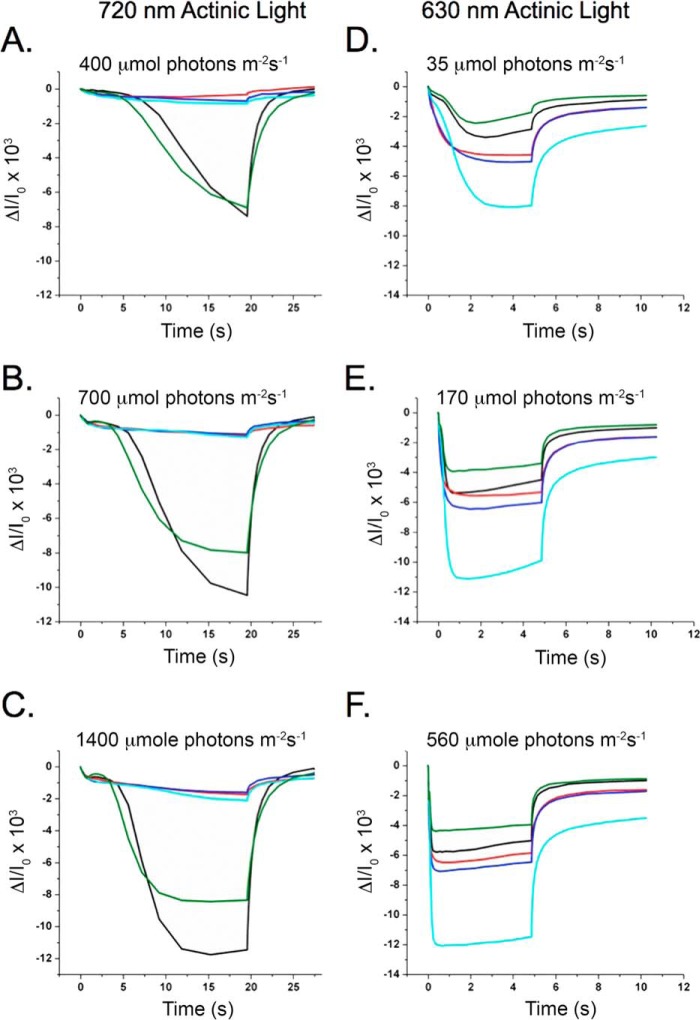
In correlation with the plant growth phenotype, P700 oxidation and reduction measurements using far red (720 nm) actinic illumination showed a significant decrease in the formation of P700+ (Fig. 2, A–C). The extent of the absorbance change at 705 nm for PPD1 RNAi-E, -F, and -H plants was only ∼15% that of wild-type plants for all actinic light intensities examined. The PPD1 RNAi-J individual had an absorbance change at 705 nm similar to wild type at the lowest light intensity, but for higher actinic light intensities, PPD1 RNAi-J exhibited an absorbance change of 70% of wild type. These results indicate that suppression of PPD1 expression by RNAi yielded individual transformants with a range of growth and P700+ accumulation defects.
FIGURE 2.
Steady-state P700 oxidation and reduction. Left, P700 oxidation and reduction measured using 720 nm actinic light at 400 (A), 700 (B), and 1400 μmol of photons m−2 s−1 (C). Right, P700 oxidation and reduction measured using 630 nm actinic illumination at 35 (D), 170 (E), and 560 μmol of photons m−2 s−1 (F). Colors correspond to plants as follows: wild type (black) and mutant PPD1 RNAi-E (red), -F (blue), -H (cyan), and -J (green).
Interestingly, although low amounts of P700+ were observed upon illumination at 720 nm (Fig. 2, A–C), equivalent or greater P700+ was observed for the PPD1 RNAi mutants upon illumination with 630 nm actinic light (Fig. 2, D–F). These differences suggest alterations in antenna coupling in the PPD1 RNAi mutants because these wavelengths differentially excite the two different antenna systems within the thylakoid. Although 720 nm illumination primarily excites LHC I, 630 nm illumination excites LHC II in addition to LHC I. Based on these P700 data, it appears that LHC I is poorly coupled to PS I in the PPD1 RNAi mutants. The 630 nm data suggest that LHC II is functioning as an antenna complex for PS I in the PPD1 RNAi mutants. The redox state of the plastoquinone pool balances electron transfer through the two photosystems by controlling function of the LHC II antenna. Under State I conditions (oxidized plastoquinone pool), LHC II transfers energy to PS II, whereas under State II conditions (reduced plastoquinone pool), LHC II also transfers energy to PS I. Defects in the accumulation of PS I would be expected to lead to oxidation of the plastoquinone pool, an increased accumulation of plastoquinol, and consequently State II-like conditions (29–31). Therefore, it is reasonable to expect that LHC II serves as an antenna for PS I in the PPD1 RNAi mutants.
Measurements of chlorophyll fluorescence also provide evidence of an imbalance in photosystem function leading to a reduced plastoquinone pool. Steady-state fluorescence values (Fs) in the light are indicative of an over-reduced plastoquinone pool and consistent with diminished PS I function (32). Fig. 3 shows the F0-normalized chlorophyll fluorescence inductions for wild type and the PPD1 RNAi-E, -F, -H, and -J individuals measured at three different light intensities. The extent of the Fs increase is inversely correlated to the PS I function measured with 720 nm actinic light in Fig. 2. Notably, measurements taken at different actinic light intensities show that the Fs increase over the course of the measurement is ameliorated with increasing light intensity. This result also points to a possible antenna defect in the PPD1 RNAi mutants. The accumulation of a reduced plastoquinone pool appears to be a consequence of diminished PS I function leading to an imbalance in photosystem function and alterations in the chlorophyll fluorescence induction curves.
FIGURE 3.
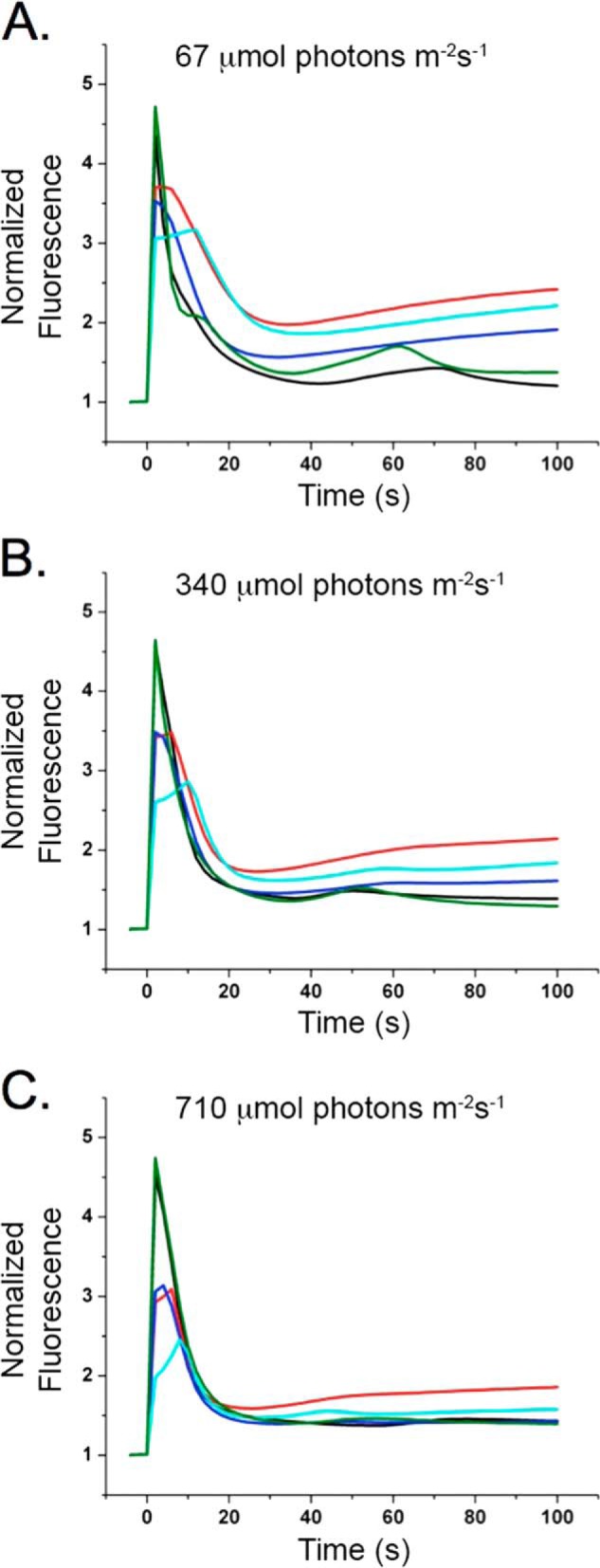
Chlorophyll fluorescence induction measurements. Wild-type and mutant PPD1 RNAi-E, -F, -H, and -J chlorophyll fluorescence induction at 67 (A), 340 (B), and 710 μmol of photons m−2 s−1 (C). Values were normalized to F0. Colors correspond to plants as follows: wild type (black) and mutant PPD1 RNAi-E (red), -F (blue), -H (cyan), and -J (green).
Loss of PPD1 Results in Altered Thylakoid Membrane Protein Supercomplexes
To obtain further details on the function of PPD1, native and denaturing gel electrophoreses were used to investigate the levels of accumulation of protein complexes as well as individual protein subunits within the thylakoid membranes in the PPD1 RNAi mutants. To have sufficient plant material for these biochemical experiments, PPD1 RNAi primary transformants were pooled according to their P700 signal under far red (720 nm) illumination (Fig. 4A). The P700 absorbance changes relative to wild type for the PPD1 RNAi mutants were grouped as follows: I, 80–100%; II, 50–60%; III, 30–45%; and IV, <25%. Note that the absorbance changes for measurements using orange (630 nm) illumination were equivalent for wild type and all PPD1 RNAi groups (Fig. 4B). Pictures of representative plants for wild type and PPD1 RNAi Groups I–IV are shown in Fig. 4C. It should be noted that the individual PPD1 RNAi-E, -F, and -H would correspond to Group IV, whereas the individual PPD1 RNAi-J signal would correspond to Group II based on their P700 absorbance change using far red illumination. Thylakoid membranes were isolated from these groups of individuals and used for protein analysis by BN- and LiDS-PAGE.
FIGURE 4.
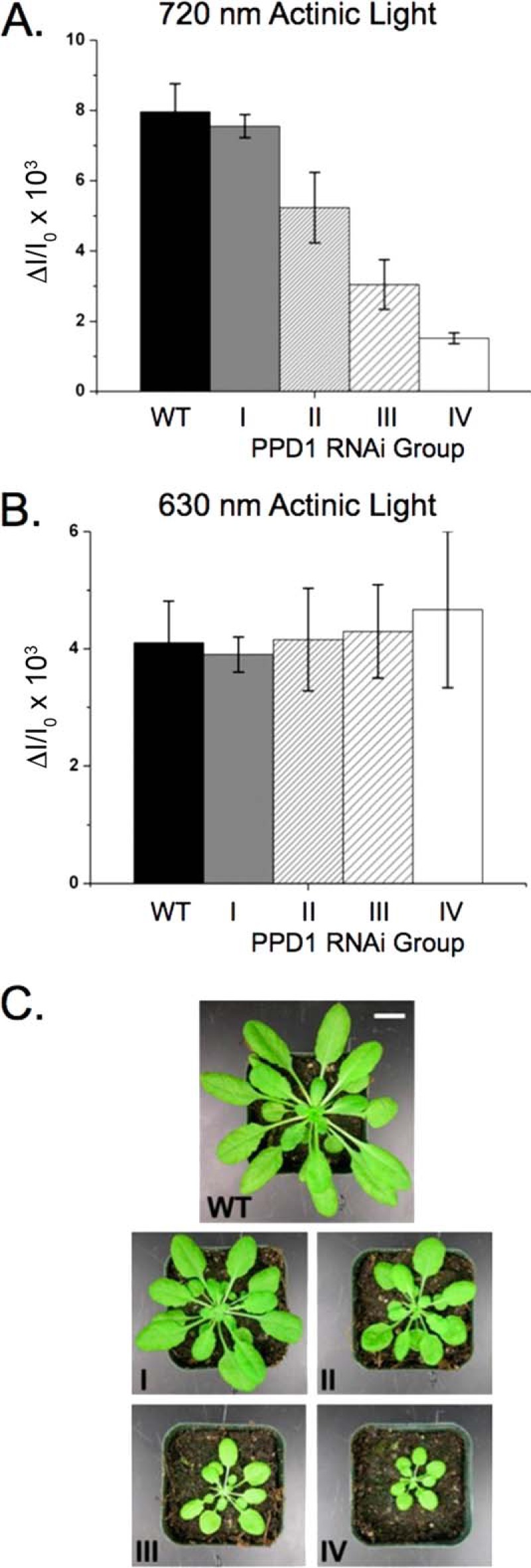
PPD1 RNAi mutant groups according to P700+ measurements. A, P700 oxidation using 720 nm actinic illumination. B, P700 oxidation using 630 nm actinic illumination. Individual PPD1 RNAi mutant plants were grouped according to their P700+ content using 720 nm actinic illumination as follows: I, 80–100% WT; II, 50–60% WT; III, 30–45% WT; and IV, <25% WT. C, pictures of 5-week-old representative plants of wild type and PPD1 RNAi Groups I–IV. Scale bar,1 cm. Error bars represent S.D. of 4–7 replicates.
Native gel electrophoresis was performed to assay the accumulation of protein complexes in the thylakoid membranes of wild-type and PPD1 RNAi plants. It should be noted that semiquantitative immunoblotting using LiDS-PAGE could not be performed for PPD1 detection because the PPD1 protein co-migrates with the abundant Lhcb proteins, which occlude its signal. However, on native gels, the PPD1 protein could be reliably detected for semiquantitative analysis. The PPD1 protein migrated as part of a complex with a molecular mass of ∼300 kDa near the bands for PS II monomer and the cytochrome b6f complex, separate from the PS I and PS I-LHC I bands (Fig. 5). BN-PAGE showed a direct relationship between the loss of PPD1 and decreased accumulation of PS I complexes (Fig. 6). In the most severely affected PPD1 RNAi lines (Group IV), the PPD1 accumulation was ∼38% that of wild-type plants (Fig. 6B). Similar decreases were observed for the PS I subunit PsaF and the LHC I subunit Lhca1 at the position of the PS I-LHC I complex molecular weight. Clearly, the loss of PPD1 is correlated with the loss of PS I and PS I-LHC I complex accumulation.
FIGURE 5.
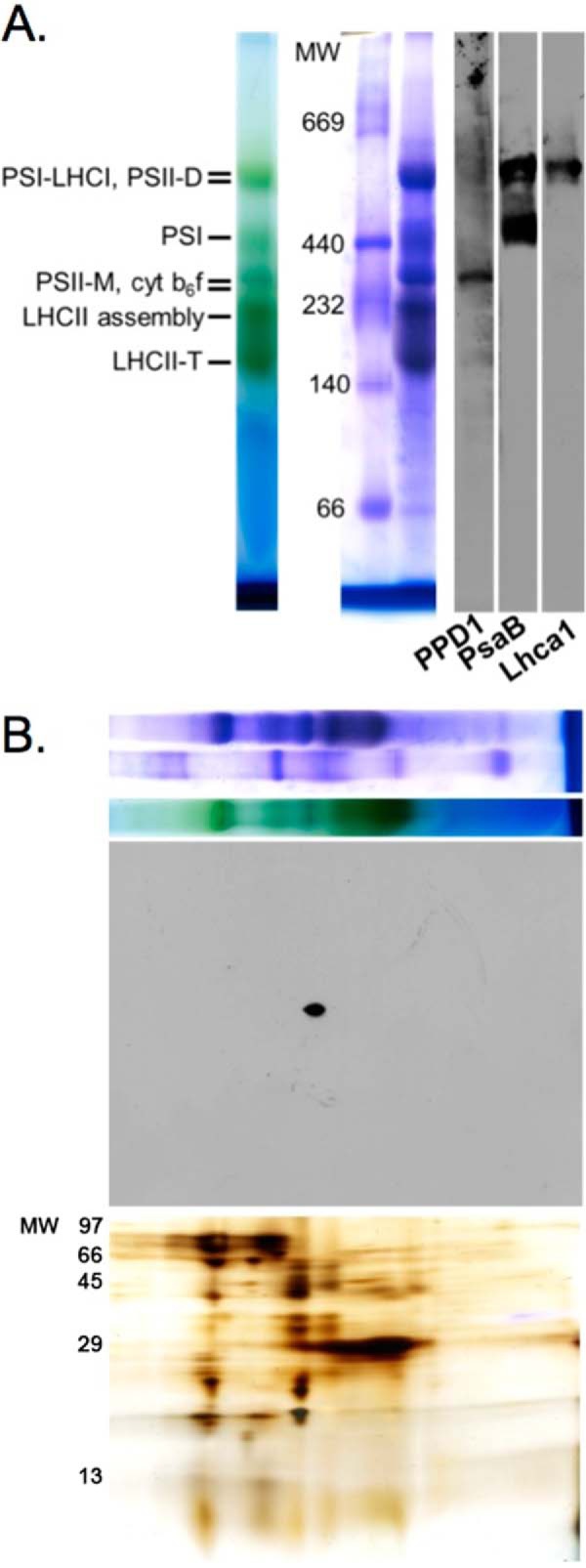
Detection of PPD1 in a protein complex. A, BN-PAGE separation of dodecyl β-d-maltoside-solubilized wild-type thylakoid membranes (unstained, Coomassie-stained, and immunodetection of PPD1, PsaB, and LhcaI proteins). B, two-dimensional (BN-LiDS) PAGE separation of wild-type thylakoids with the first dimension BN-PAGE panel (top) shown above the PPD1 immunodetection panel (middle) and silver-stained panel (bottom). cyt, cytochrome; M, monomer; D, dimer; T, trimer.
FIGURE 6.
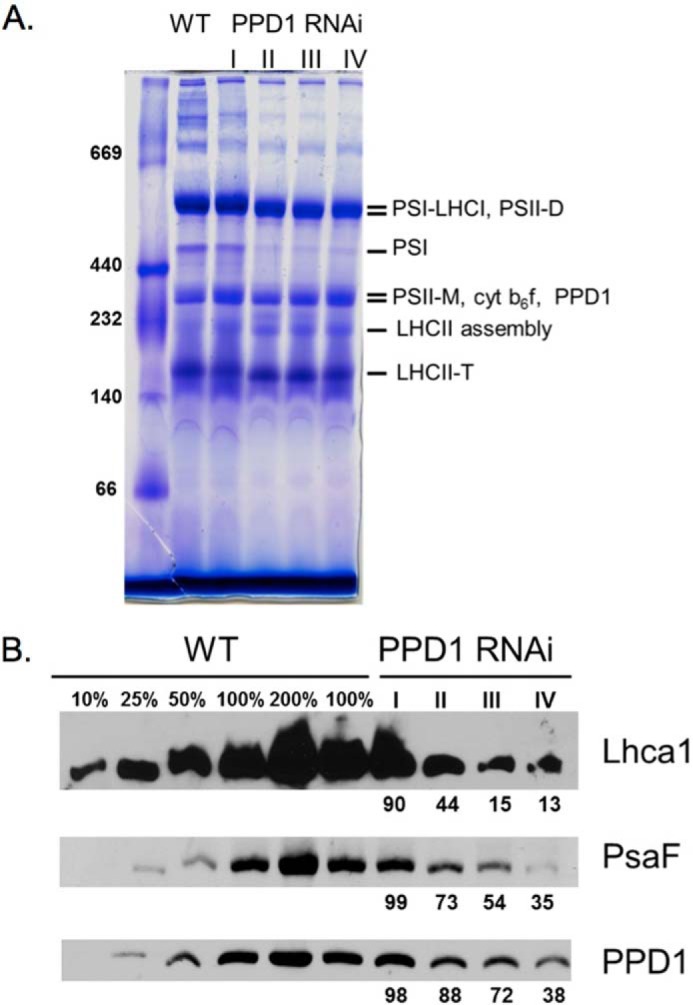
Accumulation of protein complexes. A, Coomassie-stained BN-polyacrylamide gel of wild-type and PPD1 RNAi mutant thylakoid membranes solubilized with dodecyl β-d-maltoside. B, immunodetection of Lhca1, PsaF, and PPD1 from BN-PAGE. PPD1 RNAi Groups I–IV have been defined previously. Values are given for complex accumulation in PPD1 RNAi Groups I–IV based on the signal from immunodetection. cyt, cytochrome; M, monomer; D, dimer; T, trimer.
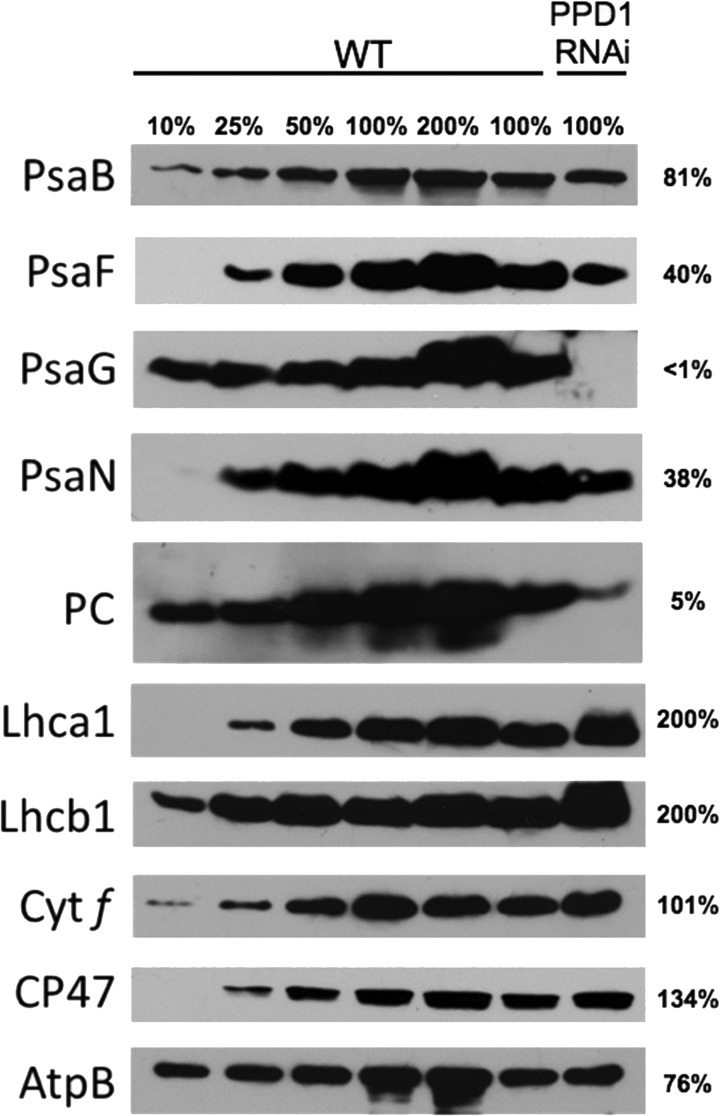
Semiquantitative immunoblotting of LiDS-PAGE gels was performed to determine the total accumulation of individual protein subunits in the thylakoid membranes from wild type and PPD1 RNAi mutants from the most severely affected plants (Group IV). Despite the significant reduction in PS I and PS I-LHC I complexes detected in native gel electrophoresis, the overall accumulation of thylakoid protein subunits was quite variable especially with respect to PS I subunits (Fig. 7). There was little loss of accumulation of the PsaB protein, but the amounts of the PsaF, PsaG, and PsaN proteins were significantly decreased in the PPD1 RNAi mutants. Thus, the PsaB and Lhca1 proteins accumulated in the thylakoid membranes but did not accumulate in the form of PS I-LHC I complexes as indicated by immunodetection of BN-polyacrylamide gels. The soluble electron carrier PC was also significantly decreased in abundance in the PPD1 RNAi mutants. The accumulation of cytochrome f (101%) was the same as observed in wild type, whereas more CP47 accumulated (134%) and less AtpB (76%) was present than in the wild-type plants. The amounts of the antenna proteins Lhca1 (LHC I) and Lhcb1 (LHC II) increased 2-fold in the PPD1 RNAi mutant plants. These antenna complex subunits accumulate independently of the photosystem proteins (33). Thus, the total amount of Lhca1 protein in the thylakoid membranes increased (Fig. 7), but the assembly of Lhca1 into PS I-LHC I complexes decreased in the PPD1 RNAi mutants. These results indicate that PPD1 is required specifically for the accumulation of some PS I subunits and PC.
FIGURE 7.
Semiquantitative immunoblots for thylakoid proteins separated by LiDS-PAGE. The proteins PsaB, PsaF, PsaG, PsaN, PC, Lhca1, Lhcb1, cytochrome (Cyt) f, CP47, and ATP synthase β subunit (AtpB) were detected for a series of wild-type and PPD1 RNAi mutant (Group IV) thylakoids. Values for protein amounts in the PPD1 RNAi mutants are given to the right of the blot panels.
BN-PAGE of thylakoid samples solubilized with the detergent digitonin was also performed to detect PS I-LHC I-LHC II “megacomplexes,” which contain the LHC II antenna in addition to LHC I (34). Fig. 8A shows the comparison of protein complexes present in wild-type and PPD1 RNAi digitonin-solubilized thylakoids. The differences in PS I complex distribution between wild-type and the PPD1 RNAi plants can also be seen in two-dimensional (BN-LiDS) PAGE (Fig. 8B). The PS I complexes in wild-type thylakoids were predominantly the PS I-LHC I and PS I monomer forms (corresponding to bands 2 and 4 in Fig. 8B). In contrast, although lower amounts of the PS I complexes are present in the PPD1 RNAi mutant thylakoids, the complexes that do form appeared to partition equally between the PS I-LHC I-LHC II and PS I-LHC I forms (corresponding to bands 1 and 2 in Fig. 8B). These results show that in wild type PS I exists predominantly as PS I-LHC I with very little PS I-LHC I-LHC II. The PPD1 RNAi mutants have an increased amount of PS I-LHC I-LHC II relative to wild type (Fig. 8B, compare the amounts of band 1 for the WT and PPD1 RNAi panels), consistent with the P700 data indicating that LHC II functions as a PS I antenna in these mutants. The accumulation of PS I-LHC I-LHC II complexes has been reported previously for other PS I subunit mutants, consistent with the hypothesis that defects in PS I function mimic State II-like conditions favoring LHC II association with PS I (34).
FIGURE 8.
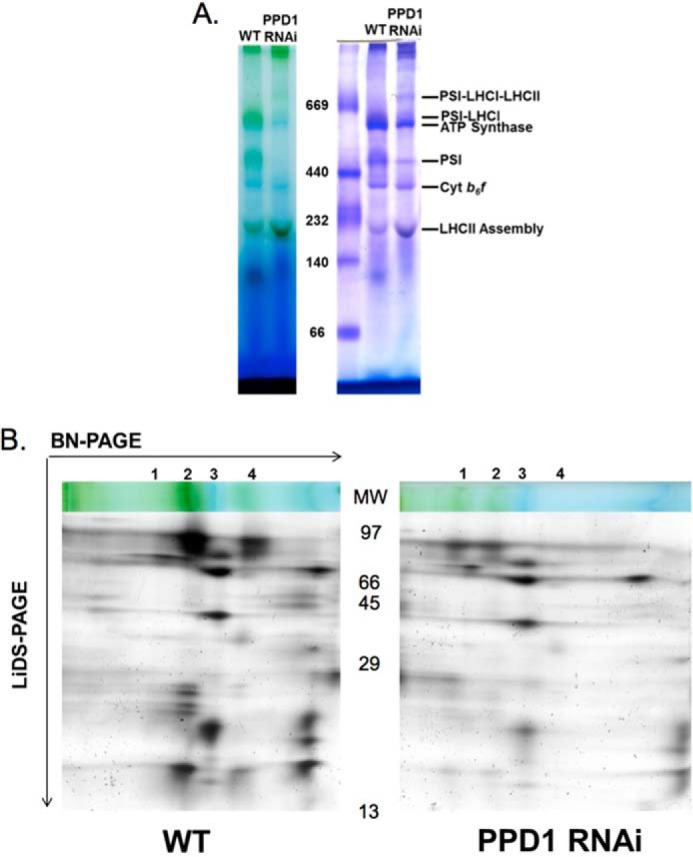
Comparison of PS I supercomplexes in wild-type and PPD1 RNAi mutant thylakoids. A, unstained and Coomassie-stained BN-polyacrylamide gels of digitonin-solubilized thylakoid membranes from wild type and PPD1 RNAi mutant (Group IV). B, silver-stained two-dimensional (BN-LiDS) PAGE of digitonin-solubilized thylakoid membranes from wild type and PPD1 RNAi mutant (Group IV). The truncated BN-PAGE dimension, corresponding to molecular mass ∼250 kDa and above, is shown for visual reference, and complexes are numbered as follows: 1, PS I-LHC I-LHC II; 2, PS I-LHC I; 3, ATP synthase; 4, PS I. Cyt, cytochrome.
Loss of PPD1 Leads to Specific Defects on the Lumenal Side of PS I
The PsaF and PsaN subunits contribute to the docking site of PC in PS I (2, 35). Because all of these components were significantly decreased in abundance in the PPD1 RNAi plants (Fig. 7), one would predict differences in the kinetics of electron transfer from PC to the P700 reaction center. Indeed, the steady-state P700 measurements of PPD1 RNAi plants identified kinetic differences in PS I electron transfer. The t½ values for P700 oxidation and P700+ reduction were determined from the P700 measurements performed using 630 nm light (Table 1). 630 nm actinic illumination was used for these measurements because the signal amplitude for all of the samples (wild type and the RNAi mutants) was similar. The PS I complexes in the severely affected PPD1 RNAi mutants had significant defects in electron transfer from plastocyanin to P700+. With increasing suppression of PPD1, the reduction of P700+ was slowed by a factor of 2 for the PPD1 RNAi Group IV plants, which exhibited the lowest PPD1 expression level relative to wild type. A similar trend was observed for the Group II and III plants, which accumulated intermediate levels of PPD1. The P700 oxidation rates were not significantly different between wild type and the various PPD1 RNAi groups.
TABLE 1.
P700 oxidation and reduction t½ values
t½ values are given in ms from n = 4–7 replicates of P700+ reduction measurements using 630 nm excitation at 560 μmol of photons m−2 s−1 (Fig. 4).
| P700 oxidation | P700+ reduction | |
|---|---|---|
| ms | ms | |
| WT | 89 ± 10 | 59 ± 6 |
| PPD1 RNAi Group I | 88 ± 7 | 50 ± 13 |
| PPD1 RNAi Group II | 88 ± 11 | 72 ± 9a |
| PPD1 RNAi Group III | 82 ± 21 | 87 ± 13a |
| PPD1 RNAi Group IV | 68 ± 22 | 118 ± 15a |
a Significantly different from wild type at p ≤ 0.05 (Student's t test).
These P700 measurements do not represent the intrinsic oxidation and reduction rates of P700 that occur on a picosecond time scale. These steady-state measurements monitor the balance between P700 oxidation and reduction that occurs under continuous illumination conditions. The steady-state accumulation of P700+ is a consequence of a more rapid oxidation of P700 and a relatively slower reduction of P700+.
It is important to note that the loss of PS I in the PPD1 RNAi lines should shift the redox equilibrium toward an over-reduced plastocyanin pool. Consequently, one would hypothesize that the P700+ reduction rates would be apparently faster in the PPD1 RNAi lines under these conditions if the only role of PPD1 were in PS I complex accumulation. Our data contradict this hypothesis because electron transfer in the PS I complexes was slower in the most severely affected mutants. It is unlikely that the differences in the megacomplex composition of the PPD1 RNAi mutants (i.e. stable State II-like conditions) affect P700+ reduction kinetics. Other PS I subunit mutants (psae1-3 and psad1-1) have been reported to have State II-like properties (over-reduced plastoquinone pool, phosphorylated LHC II, and PS I-LHC I-LHC II megacomplex formation) similar to those observed for the PPD1 RNAi mutants, but these exhibit no effects on P700+ reduction kinetics (36, 37). Altogether, these results are consistent with a role for PPD1 in the formation of a functional PS I lumenal side.
DISCUSSION
The results from the PPD1 RNAi plants provide additional insights on the role of PPD1 because a range of phenotypes with respect to PS I accumulation and function was observed. Although the original description of the ppd1 insertion mutant was the complete loss of PS I accumulation (16), the PPD1 RNAi lines showed a correlated loss of PS I assembly with loss of PPD1. Our results show that, in the presence of small amounts of PPD1, fewer PS I complexes accumulate, and these complexes have defects in coupling of LHC I to PS I as well as in electron transfer from plastocyanin to P700+. This suggests that decreased amounts of PPD1 result in defects in the assembly of the lumenal side of PS I.
P700+ reduction is dependent on both the availability of reduced PC and the efficient docking of PC to the lumenal side of the PS I complex. In the PPD1 RNAi plants, both of these factors appear to be affected. The amount of detectable PC is decreased as well as the PS I subunits that contribute to the PC docking site of the lumenal side of PS I. The N-terminal extension of the plant PsaF protein along with the lumenal PsaN protein and lumenal loops of the PsaA and PsaB proteins serves as the docking site for reduced PC (1, 38, 39). The PPD1 RNAi mutants that exhibit decreased accumulation of PsaF and PsaN are phenotypically similar to mutants deficient in these two components. Like previous reports for PsaF- and PsaN-deficient plants, the PPD1 RNAi plants have decreased PS I accumulation, loss of energetic coupling of LHC I to PS I, and slower electron transfer from plastocyanin (40, 41). It should also be noted that PPD1 RNAi plants had significantly decreased levels of the PsaG protein, which has also been reported to stabilize the association of LHC I with PS I complexes (42). Thus, the defects in LHC I coupling to PS I in the PPD1 RNAi plants is likely a combinatorial effect of the loss of PsaG and PsaF. No direct interaction between PPD1 and PsaF, PsaG, or PsaN has been reported, but our results are consistent with the observation that PPD1 may associate with the lumenal loop regions of the PsaA and PsaB proteins, which had been reported earlier (16). Our data provide the first direct evidence that the lumen-localized PPD1 protein contributes to PS I function as well as accumulation. We propose a role for PPD1 in the optimal assembly of the lumenal portions of PS I to ensure its proper assembly and function.
The PS I assembly defects in the PPD1 RNAi mutants induce secondary effects with respect to LHC II function. The loss of PPD1 leads to an apparent reduction of the plastoquinone pool because of the increased activity of PS II relative to PS I. In an apparent attempt to compensate for this imbalance, there is a change in antenna function to reduce the excitation pressure on PS II and provide more excitation energy to the small amount of PS I present. There are several lines of evidence consistent with this interpretation of photosynthetic activity in the PPD1 RNAi mutants. There were significant decreases in P700 oxidation for the PPD1 RNAi mutants relative to wild type upon illumination at 720 nm, which primarily excites LHC I. However, equivalent P700 oxidation was observed for wild type and mutants upon illumination at 630 nm, which excites both LHC II and LHC I. It has been shown that the reduced state of the plastoquinone pool activates the STN7 kinase, which phosphorylates LHC II and triggers its detachment from PS II and subsequent association with PS I (34, 43). Our data indicate that the PPD1 RNAi plants showed a relative increase in the amount of PS I-LHC I-LHC II megacomplexes. Consequently, the most severely affected PPD1 RNAi mutants contain a larger proportion of PS I-LHC I-LHC II relative to PS I-LHC I compared with wild-type plants.
The effects on PS I assembly described above indicate that the PPD1 protein does not merely have a catalytic role in PS I assembly where small protein amounts are required for biogenesis efficiency. Our data suggest that PPD1 is required for PS I accumulation as well as proper function with respect to electron transfer on the lumenal side of the complex. In this capacity, PPD1 may associate with PS I assembly intermediates to ensure proper integration and folding of the lumenal side of the complex, which contains domains of the PsaA, PsaB, PsaF, and PsaN proteins. Without sufficient amounts of the PPD1 protein, PS I complexes are not properly assembled and are likely subject to degradation (16). Also, defects in the integration of PsaF and PsaG contribute to the loss of energetic coupling of LHC I to PS I.
This putative role is consistent with the correlation between PPD1 accumulation and stable PS I accumulation seen in the PPD1 RNAi mutants. The electron transfer data presented in this work and previous data showing an interaction of PPD1 with the lumenal loops of PsaA and PsaB are also consistent with PPD1 functioning as an assembly factor for the lumenal portion of PS I (16).
Finally, in this work, PPD1 was shown to be predominately part of a complex with an apparent molecular mass of 300 kDa. It is unknown whether the PPD1-containing complex is the same as the PS I assembly intermediate identified by Ozawa et al. (6), but the sizes are similar. Future studies will focus on identifying the components of the PPD1-containing complex to determine when its assembly occurs during PS I biogenesis.
This work was supported by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the United States Department of Energy through Grant DE-FG02-98ER20310 (to T. M. B. and L. K. F.).
- PS II
- Photosystem II
- PPD1
- PsbP domain protein 1
- BN
- blue native
- LHC I
- light-harvesting complex I
- LHC II
- light-harvesting complex II
- LiDS
- lithium dodecyl sulfate
- PS I
- Photosystem I
- PC
- plastocyanin
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Busch A., Hippler M. (2011) The structure and function of eukaryotic Photosystem I. Biochim. Biophys. Acta 1807, 864–877 [DOI] [PubMed] [Google Scholar]
- 2. Schöttler M. A., Albus C. A., Bock R. (2011) Photosystem I: its biogenesis and function in higher plants. J. Plant Physiol. 168, 1452–1461 [DOI] [PubMed] [Google Scholar]
- 3. Grotjohann I., Fromme P. (2005) Structure of cyanobacterial Photosystem I. Photosynth. Res. 85, 51–72 [DOI] [PubMed] [Google Scholar]
- 4. Amunts A., Drory O., Nelson N. (2007) The structure of a plant Photosystem I supercomplex at 3.4 Å resolution. Nature 447, 58–63 [DOI] [PubMed] [Google Scholar]
- 5. Nickelsen J., Rengstl B. (2013) Photosystem II assembly: from cyanobacteria to plants. Annu. Rev. Plant Biol. 64, 609–635 [DOI] [PubMed] [Google Scholar]
- 6. Ozawa S., Onishi T., Takahashi Y. (2010) Identification and characterization of an assembly intermediate subcomplex of Photosystem I in the green alga Chlamydomonas reinhardtii. J. Biol. Chem. 285, 20072–20079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boudreau E., Takahashi Y., Lemieux C., Turmel M., Rochaix J. D. (1997) The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the Photosystem I complex. EMBO J. 16, 6095–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ruf S., Kössel H., Bock R. (1997) Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded Photosystem I-related gene. J. Cell Biol. 139, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Naver H., Boudreau E., Rochaix J. D. (2001) Functional studies of Ycf3: its role in assembly of Photosystem I and interactions with some of its subunits. Plant Cell 13, 2731–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Albus C. A., Ruf S., Schöttler M. A., Lein W., Kehr J., Bock R. (2010) Y3IP1, a nucleus-encoded thylakoid protein, cooperates with the plastid-encoded Ycf3 protein in Photosystem I assembly of tobacco and Arabidopsis. Plant Cell 22, 2838–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wilde A., Härtel H., Hübschmann T., Hoffmann P., Shestakov S. V., Börner T. (1995) Inactivation of a Synechocystis sp strain PCC 6803 gene with homology to conserved chloroplast open reading frame 184 increases the Photosystem II-to-Photosystem I ratio. Plant Cell 7, 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Krech K., Ruf S., Masduki F. F., Thiele W., Bednarczyk D., Albus C. A., Tiller N., Hasse C., Schöttler M. A., Bock R. (2012) The plastid genome-encoded Ycf4 protein functions as a nonessential assembly factor for Photosystem I in higher plants. Plant Physiol. 159, 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stöckel J., Bennewitz S., Hein P., Oelmüller R. (2006) The evolutionarily conserved tetratrico peptide repeat protein pale yellow green7 is required for Photosystem I accumulation in Arabidopsis and copurifies with the complex. Plant Physiol. 141, 870–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shen G., Antonkine M. L., van der Est A., Vassiliev I. R., Brettel K., Bittl R., Zech S. G., Zhao J., Stehlik D., Bryant D. A., Golbeck J. H. (2002) Assembly of Photosystem I. II. Rubredoxin is required for the in vivo assembly of FX in Synechococcus sp. PCC 7002 as shown by optical and EPR spectroscopy. J. Biol. Chem. 277, 20355–20366 [DOI] [PubMed] [Google Scholar]
- 15. Shen G., Zhao J., Reimer S. K., Antonkine M. L., Cai Q., Weiland S. M., Golbeck J. H., Bryant D. A. (2002) Assembly of photosystem I. I. Inactivation of the rubA gene encoding a membrane-associated rubredoxin in the cyanobacterium Synechococcus sp. PCC 7002 causes a loss of Photosystem I activity. J. Biol. Chem. 277, 20343–20354 [DOI] [PubMed] [Google Scholar]
- 16. Liu J., Yang H., Lu Q., Wen X., Chen F., Peng L., Zhang L., Lu C. (2012) PsbP-domain protein1, a nuclear-encoded thylakoid lumenal protein, is essential for Photosystem I assembly in Arabidopsis. Plant Cell 24, 4992–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bricker T. M., Roose J. L., Zhang P., Frankel L. K. (2013) The PsbP family of proteins. Photosynth. Res. 116, 235–250 [DOI] [PubMed] [Google Scholar]
- 18. Yi X., Hargett S. R., Liu H., Frankel L. K., Bricker T. M. (2007) The PsbP protein is required for Photosystem II complex assembly/stability and photoautotrophy in Arabidopsis thaliana. J. Biol. Chem. 282, 24833–24841 [DOI] [PubMed] [Google Scholar]
- 19. Yi X., Hargett S. R., Frankel L. K., Bricker T. M. (2008) The effects of simultaneous RNAi suppression of PsbO and PsbP protein expression in Photosystem II of Arabidopsis. Photosynth. Res. 98, 439–448 [DOI] [PubMed] [Google Scholar]
- 20. Yi X., Hargett S. R., Frankel L. K., Bricker T. M. (2009) The PsbP protein, but not the PsbQ protein, is required for normal thylakoid architecture in Arabidopsis thaliana. FEBS Lett. 583, 2142–2147 [DOI] [PubMed] [Google Scholar]
- 21. Murashige T., Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco cultures. Physiol. Plant. 15, 473–479 [Google Scholar]
- 22. Wesley S. V., Helliwell C. A., Smith N. A., Wang M. B., Rouse D. T., Liu Q., Gooding P. S., Singh S. P., Abbott D., Stoutjesdijk P. A., Robinson S. P., Gleave A. P., Green A. G., Waterhouse P. M. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27, 581–590 [DOI] [PubMed] [Google Scholar]
- 23. Holsters M., de Waele D., Depicker A., Messens E., van Montagu M., Schell J. (1978) Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. 163, 181–187 [DOI] [PubMed] [Google Scholar]
- 24. Clough S. J., Bent A. F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 25. Arnon D. I. (1949) Copper enzymes in isolated chloroplasts. Polyphenol oxidase in Beta vulgaris. Plant Physiol. 24, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Delepelaire P., Chua N. H. (1979) Lithium dodecyl sulfate/polyacrylamide gel electrophoresis of thylakoid membranes at 4°C: characterizations of two additional chlorophyll a-protein complexes. Proc. Natl. Acad. Sci. U.S.A. 76, 111–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schneider C. A., Rasband W. S., Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wray W., Boulikas T., Wray V. P., Hancock R. (1981) Silver staining of proteins in polyacrylamide gels. Anal. Biochem. 118, 197–203 [DOI] [PubMed] [Google Scholar]
- 29. Dietzel L., Bräutigam K., Pfannschmidt T. (2008) Photosynthetic acclimation: state transitions and adjustment of photosystem stoichiometry—functional relationships between short-term and long-term light quality acclimation in plants. FEBS J. 275, 1080–1088 [DOI] [PubMed] [Google Scholar]
- 30. Lemeille S., Rochaix J. D. (2010) State transitions at the crossroad of thylakoid signalling pathways. Photosynth. Res. 106, 33–46 [DOI] [PubMed] [Google Scholar]
- 31. Minagawa J. (2011) State transitions—the molecular remodeling of photosynthetic supercomplexes that controls energy flow in the chloroplast. Biochim. Biophys. Acta 1807, 897–905 [DOI] [PubMed] [Google Scholar]
- 32. Hohmann-Marriott M. F., Takizawa K., Eaton-Rye J. J., Mets L., Minagawa J. (2010) The redox state of the plastoquinone pool directly modulates minimum chlorophyll fluorescence yield in Chlamydomonas reinhardtii. FEBS Lett. 584, 1021–1026 [DOI] [PubMed] [Google Scholar]
- 33. Takahashi Y., Yasui T. A., Stauber E. J., Hippler M. (2004) Comparison of the subunit compositions of the PSI-LHCI supercomplex and the LHCI in the green alga Chlamydomonas reinhardtii. Biochemistry 43, 7816–7823 [DOI] [PubMed] [Google Scholar]
- 34. Pesaresi P., Hertle A., Pribil M., Kleine T., Wagner R., Strissel H., Ihnatowicz A., Bonardi V., Scharfenberg M., Schneider A., Pfannschmidt T., Leister D. (2009) Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. Plant Cell 21, 2402–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jensen P. E., Bassi R., Boekema E. J., Dekker J. P., Jansson S., Leister D., Robinson C., Scheller H. V. (2007) Structure, function and regulation of plant Photosystem I. Biochim. Biophys. Acta 1767, 335–352 [DOI] [PubMed] [Google Scholar]
- 36. Ihnatowicz A., Pesaresi P., Varotto C., Richly E., Schneider A., Jahns P., Salamini F., Leister D. (2004) Mutants for Photosystem I subunit D of Arabidopsis thaliana: effects on photosynthesis, Photosystem I stability and expression of nuclear genes for chloroplast functions. Plant J. 37, 839–852 [DOI] [PubMed] [Google Scholar]
- 37. Ihnatowicz A., Pesaresi P., Leister D. (2007) The E subunit of Photosystem I is not essential for linear electron flow and photoautotrophic growth in Arabidopsis thaliana. Planta 226, 889–895 [DOI] [PubMed] [Google Scholar]
- 38. Farah J., Rappaport F., Choquet Y., Joliot P., Rochaix J. D. (1995) Isolation of a psaF-deficient mutant of Chlamydomonas reinhardtii: efficient interaction of plastocyanin with the Photosystem I reaction center is mediated by the PsaF subunit. EMBO J. 14, 4976–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hippler M., Drepper F., Haehnel W., Rochaix J. D. (1998) The N-terminal domain of PsaF: precise recognition site for binding and fast electron transfer from cytochrome c6 and plastocyanin to Photosystem I of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 95, 7339–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Haldrup A., Naver H., Scheller H. V. (1999) The interaction between plastocyanin and Photosystem I is inefficient in transgenic Arabidopsis plants lacking the PSI-N subunit of Photosystem I. Plant J. 17, 689–698 [DOI] [PubMed] [Google Scholar]
- 41. Haldrup A., Simpson D. J., Scheller H. V. (2000) Down-regulation of the PSI-F subunit of Photosystem I (PSI) in Arabidopsis thaliana. The PSI-F subunit is essential for photoautotrophic growth and contributes to antenna function. J. Biol. Chem. 275, 31211–31218 [DOI] [PubMed] [Google Scholar]
- 42. Jensen P. E., Rosgaard L., Knoetzel J., Scheller H. V. (2002) Photosystem I activity is increased in the absence of the PSI-G subunit. J. Biol. Chem. 277, 2798–2803 [DOI] [PubMed] [Google Scholar]
- 43. Pesaresi P., Hertle A., Pribil M., Schneider A., Kleine T., Leister D. (2010) Optimizing photosynthesis under fluctuating light: the role of the Arabidopsis STN7 kinase. Plant Signal. Behav. 5, 21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]