Abstract
A new paradigm of gene expression regulation has emerged recently with the discovery of microRNAs (miRNAs). Most, if not all, miRNAs are thought to control gene expression, mostly by base pairing with miRNA-recognition elements (MREs) found in their messenger RNA (mRNA) targets. Although a large number of human miRNAs have been reported, many of their mRNA targets remain unknown. Here we used a combined bioinformatics and experimental approach to identify important rules governing miRNA-MRE recognition that allow prediction of human miRNA targets. We describe a computational program, “DIANA-microT”, that identifies mRNA targets for animal miRNAs and predicts mRNA targets, bearing single MREs, for human and mouse miRNAs.
Keywords: MicroRNA, microRNA targets, microRNP, Argonaute, bioinformatics, RNAi
MicroRNAs (miRNAs) are derived from endogenous genes that are initially transcribed as longer RNA transcripts (Lee et al. 1993; Wightman et al. 1993; Reinhart et al. 2000; Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001; Mourelatos et al. 2002; for review, see Nelson et al. 2003; Bartel 2004). In mammals, the primary miRNA transcripts (pri-miRNAs) are processed by the nuclease Drosha (Lee et al. 2003) into ∼70-nt precursor miRNAs (pre-miRNAs) that are exported by exportin-5 to the cytoplasm (Yi et al. 2003; Bohnsack et al. 2004; Lund et al. 2004). The Dicer nuclease excises the mature miRNAs from pre-miRNAs (Grishok et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001; Knight and Bass 2001). miRNAs are bound to proteins that belong to the Argonaute family and, in humans, may also assemble with other proteins, including the Gemin3 and Gemin4 proteins, to form micro-ribonucleoprotein complexes (miRNPs; Mourelatos et al. 2002; Nelson et al. 2004). Dicer also processes another class of ∼22-nt RNAs termed short interfering RNAs (siRNAs; Hamilton and Baulcombe 1999; Elbashir et al. 2001) from double-stranded RNAs (Bernstein et al. 2001). Analogous to miRNAs, siRNAs are bound to Argonaute proteins (Hammond et al. 2001; Martinez et al. 2002) and may also assemble with additional proteins to form RNA-induced silencing complexes (RISCs; Hammond et al. 2001). siRNAs and miRNAs (and RISCs and miRNPs) are functionally equivalent, and the main difference between the two classes of small RNAs are the fact that miRNAs are derived from endogenous genes (Ambros et al. 2003a).
Many miRNAs and siRNAs function by base pairing with miRNA-recognition elements (MREs) found in their mRNA targets and direct either target RNA endonucleolytic cleavage (Elbashir et al. 2001; Hutvagner and Zamore 2002) or translational repression (Olsen and Ambros 1999; Seggerson et al. 2002; Zeng et al. 2002; Doench et al. 2003). The manner by which a miRNA or siRNA base pairs with its mRNA target correlates with its function: if the complementarity between a miRNA and its target is extensive, the RNA target is cleaved (Hutvagner and Zamore 2002; Llave et al. 2002; Rhoades et al. 2002; Tang et al. 2003; Xie et al. 2003); if the complementarity is partial, the stability of the target mRNA in not affected but its translation is repressed (Olsen and Ambros 1999; Seggerson et al. 2002; Zeng et al. 2002; Doench et al. 2003). However, how general this correlation is and the factors and mechanisms that determine the function of any given miRNA are unknown.
In plants, the computational identification of miRNA targets was facilitated by the extensive complementarity between plant miRNAs and their mRNA targets (Llave et al. 2002; Rhoades et al. 2002). Plant miRNA targets have been verified experimentally (Llave et al. 2002; Aukerman and Sakai 2003; Kasschau et al. 2003; Palatnik et al. 2003; Xie et al. 2003; Chen 2004; for review, see Bartel and Bartel 2003). Two mouse miRNAs (miR-127 and miR-136) show perfect antisense complementarity with the coding region of a retrotransposon-like gene (Rtl1; Seitz et al. 2003). However, most animal miRNAs are thought to recognize their mRNA targets via partial antisense complementarity (Lee et al. 1993; Wightman et al. 1993; Moss et al. 1997; Olsen and Ambros 1999; Reinhart et al. 2000; Zeng et al. 2002; Doench et al. 2003). Because of this partial complementarity, simple homology-based searches have failed to uncover targets for miRNAs in organisms other than plants (Ambros et al. 2003b; Bartel and Bartel 2003). Animal miRNA targets were initially identified in genetic screens. In particular, genetic dissection of the heterochronic gene pathway in Caenorhabditis elegans identified the lin-14 and lin-28 mRNAs as targets for the lin-4 miRNA (Lee et al. 1993; Wightman et al. 1993; Moss et al. 1997), and the lin-41 mRNA as a target for the let-7 miRNA (Reinhart et al. 2000). In Drosophila, the bantam miRNA regulates the pro-apoptotic gene hid (Brennecke et al. 2003). Importantly, these and other studies demonstrated that MRE sequences are necessary and sufficient to confer miRNA-dependent gene expression regulation in MRE-bearing target mRNAs (Moss et al. 1997; Reinhart et al. 2000; Zeng et al. 2002; Doench et al. 2003; Vella et al. 2004). Putative targets for other miRNAs have been proposed (Lai 2002; Abrahante et al. 2003; Lin et al. 2003; Xu et al. 2003), but these are predominantly based on visual inspection of putative mRNA targets for partial complementarity with miRNAs and lack experimental verification of specific miRNA:MRE interactions.
Very recently, carefully designed bioinformatic approaches have been used to predict mRNA targets for Drosophila (Enright et al. 2003; Stark et al. 2003) and mammalian miRNAs (Lewis et al. 2003). In particular, Bartel, Burge and colleagues have presented a robust bioinformatics strategy that allows prediction of conserved, mammalian miRNA targets along with accurate estimates of false positive rates (at 31% for miRNA targets identified in human mouse and rat and 22% for targets identified in mammals and in pufferfish) and experimental validation of 11 (out of 15 tested) predicted targets (Lewis et al. 2003). Most of the targets identified by Lewis et al. contain multiple MREs for the same miRNA or are regulated by more that one miRNA. The targets reported for Drosophila miRNAs also contain, for the most part, multiple MREs (Enright et al. 2003; Stark et al. 2003). However, the rules guiding single miRNA: MRE (target mRNA) interactions have not been investigated, and as a result, predictions of miRNA targets containing single MREs are lacking.
Here we describe experimentally derived rules that guide single miRNA:MRE (target mRNA) recognition. Incorporation of these rules in computational algorithms allows prediction of human and mouse miRNA targets containing single MREs.
Results
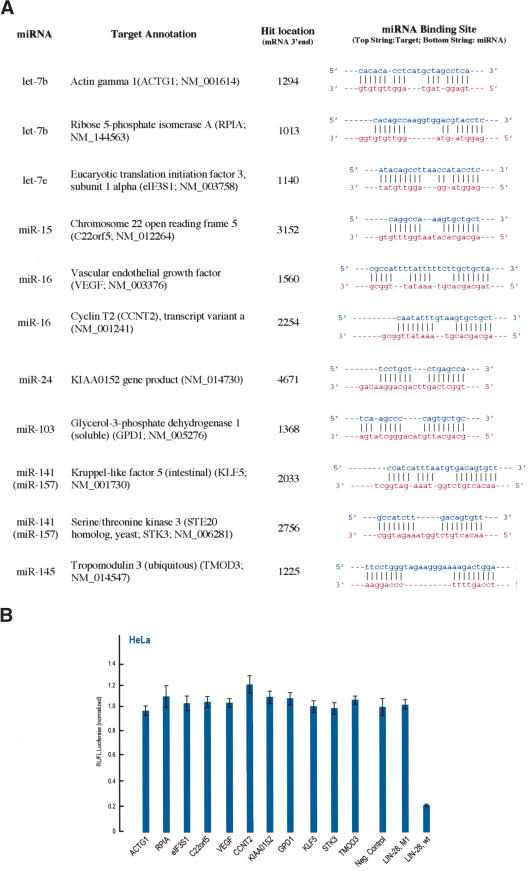
To search for human miRNA targets we initially employed a bioinformatics approach. We limited our searches to the 3′-UTRs of human mRNAs, extracted from the annotated Reference mRNA Sequences (Ref-Seq) database (Pruitt et al. 2003), comprising a total of 14,180,360 bases from 16,759 mRNAs. 3′-UTR sequences were used because the experimentally identified MREs for the C. elegans lin-4 and let-7 miRNAs are present in the 3′-UTR of their mRNA targets (Lee et al. 1993; Wightman et al. 1993; Moss et al. 1997; Reinhart et al. 2000). Repetitive elements, such as Alu transposable elements that are embedded in a random fashion in ∼5% of all human mRNAs, were filtered out before running the searches (leaving a total of 12,642,810 bases). In this initial search we used 10 miRNAs (let-7b, let-7e, miR-141, miR-24, miR-145, miR-23a, miR-15a, miR-16, miR-199b, and miR-103) which were arbitrarily chosen except for being conserved between humans and mice. We hypothesized that miRNA:MRE interactions might be guided by two factors. The first might be high-affinity interactions, based on binding energies, between a miRNA and its cognate MRE. To address this, we designed an algorithm that allowed us to identify putative miRNA:MRE interactions based on binding energies between two RNAs paired imperfectly. We implemented a modified dynamical programming algorithm that calculated free energies of both canonical (Watson-Crick) and G-U wobble dinucleotide base pairs (Tinoco et al. 1973) for two RNAs paired in trans. To identify putative MREs, we used a window of 38 nt that “slid” over the mRNA sequence and calculated the minimum binding energy between the miRNAs and sequences in the human 3′-UTR database. Mismatches were allowed, and binding energies were calculated for every three consecutive nucleotide pairs. We hypothesized that MREs might be evolutionary conserved, and we determined for each human miRNA all the hits that were conserved in the 3′-UTRs of the corresponding mouse ortholog mRNAs. Calculations were performed on a cluster of 128 dual-processor Linux machines.
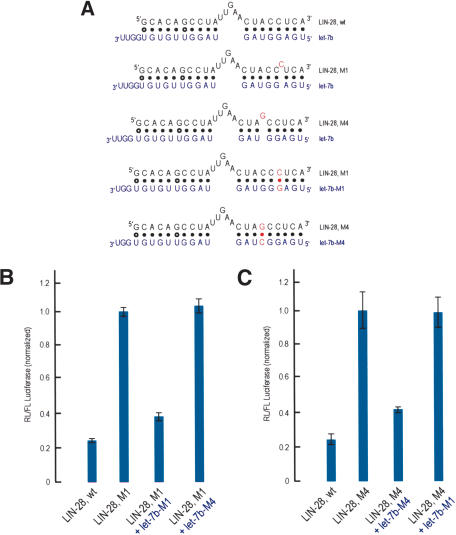
A second factor that may guide miRNA:MRE (target mRNA) bindings is miRNA-associated protein(s) that impose restraints on the position and sizes of loops and nucleotide bulges between miRNAs and their cognate MREs. miRNP proteins and in particular the Argonaute family of proteins represent excellent candidates for guiding such miRNA:target mRNA interaction (Nelson et al. 2004). In this case there may exist a general set of rules that are applicable to miRNA:MRE bindings and that may be deduced experimentally by testing various miRNA:MRE configurations. Based on this premise we tested a number of putative miRNA:MRE interactions that our initial algorithm had predicted. As a starting point for choosing putative miRNA:MRE interactions for further experiments, we considered miRNA:MRE pairs that had a central bulge or loop of the miRNA or its cognate mRNA. This was based on the experimentally verified C. elegans lin4:lin-14, lin-4:lin-28 and let-7:lin-41 miRNA:target mRNA interactions (Lee et al. 1993; Wightman et al. 1993; Moss et al. 1997; Reinhart et al. 2000). Our experimental strategy consisted of cloning the putative MREs (as single copies) into the 3′-UTR of a reporter construct. Because MREs are necessary and sufficient to confer mi/siRNA-dependent translational repression (Moss et al. 1997; Reinhart et al. 2000; Zeng et al. 2002; Doench et al. 2003), we reasoned that placement of predicted MREs for specific miRNAs in the 3′-UTR of a reporter construct, followed by transfections in cells expressing the miRNAs that recognize the MREs, should lead to a decrease of the reporter protein levels. By visual inspection we collected a number of hypothetical MREs (shown in Figs. 1, 3, 4; see below) from the list of conserved human/mouse hits for experimentation. One of the predicted MREs for let-7b was found in the 3′-UTR of both the human and mouse mRNAs that code for the human/mouse homolog of the C. elegans LIN-28 protein, a putative RNA-binding protein (see Fig. 1). This finding is particularly interesting because the lin-28 and let-7 genes function in the same developmental pathway in C. elegans (Moss et al. 1997). The Moss laboratory recently showed that the expression of LIN-28 protein is developmentally regulated in Drosophila, mouse, and Xenopus and in various human and mouse cell lines (Moss and Tang 2003). LIN-28 protein is present early in development and is absent from terminally differentiated cells, a pattern which is similar to the expression pattern of LIN-28 protein in C. elegans (Moss and Tang 2003). Interestingly, HeLa cells do not express LIN-28 protein, a result consistent with the likely repression of lin-28 mRNA translation by let-7b. The Moss laboratory also identified the same MRE for let-7b in the 3′-UTR of the human and mouse lin-28 mRNA (Moss and Tang 2003). We decided to investigate more thoroughly this putative interaction by extensive mutagenesis, as detailed below.
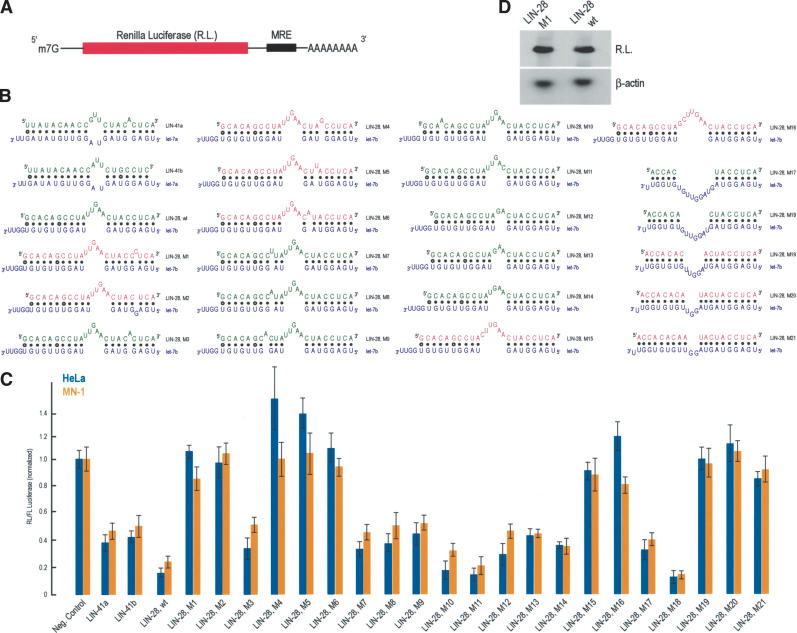
Figure 1.
Experimental verification of a predicted miRNA recognition element (MRE) and deduction of the miRNA binding rules. (A) Schematic representation of the reporter construct. (red) Coding region. (B) Potential base pairing between predicted MREs derived from the indicated mRNAs and their cognate miRNAs (blue). (wt) Wild-type sequence. MRE sequences that repressed the expression of luciferase are shown in green; the sequences that that did not are shown in red. (C) HeLa human cells (blue bars) or MN-1 mouse cells (orange bars) were cotransfected with Renilla luciferase (RL) constructs bearing the indicated MREs in the 3′-UTR, along with firefly luciferase (FL). Results shown are average values (with standard deviations) of normalized RL/FL activities obtained from six separate experiments. (D) HeLa cells were transfected with indicated constructs, total RNA was isolated, and RL and β-actin (as a normalization control) mRNAs were visualized with Northern blots.
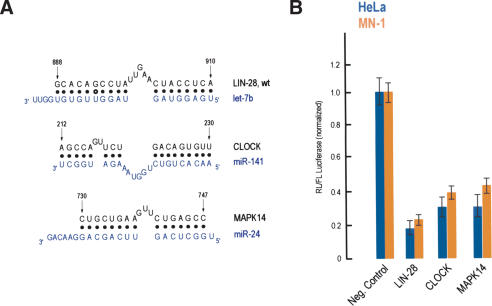
Figure 3.
Predicted miRNA targets. (A) Potential base pairing between predicted MREs derived from the indicated mRNAs (black) and their cognate miRNAs (blue). Numbers refer to nucleotide positions after the stop codon, based on the human mRNAs. (B) HeLa (blue bars) or MN-1 (orange bars) cells were cotransfected with Renilla luciferase (RL) constructs bearing the indicated MREs in the 3′-UTR, along with firefly luciferase (FL). Results shown are average values (with standard deviations) of normalized RL/FL activities obtained from six separate experiments.
Figure 4.
miRNA:MRE configurations that do not repress translation. (A) HeLa cells were cotransfected with Renilla luciferase (RL) constructs bearing the indicated MREs in the 3′-UTR, along with firefly luciferase (FL). (B) Results shown are average values (with standard deviations) of normalized RL/FL activities obtained from three separate experiments.
We cloned the predicted lin-28 MRE into the 3′-UTR of a Renilla luciferase (RL) reporter construct. As a positive control, we generated two RL constructs, each bearing in the 3′-UTR one of the two reported MREs for let-7, derived from the C. elegans lin-41 mRNA, an experimentally verified let-7 target (Reinhart et al. 2000). As a negative control, the sequence of the lin-28 MRE was scrambled and placed in the 3′-UTR of RL. We cotransfected the RL-MRE-bearing constructs along with a plasmid encoding firefly luciferase (FL) in two different cells lines: HeLa cells (a human epithelial cell line) and MN-1 cells (a mouse motor neuronal cell line). These cell lines normally express let-7 paralogs, which are conserved between humans and mice (Lagos-Quintana et al. 2001; Dostie et al. 2003; M. Kiriakidou and Z. Mourelatos, unpubl.). Eighteen hours after transfection we quantitated the levels of normalized RL/FL using standard luminometric assays. As shown in Figure 1, we consistently observed an approximately fivefold reduction in the protein levels of RL bearing the lin-28 MRE versus RL bearing the scrambled MRE (negative control), an effect which is stronger than that of the two positive control MREs derived from lin-41 (LIN-41a and LIN-41b, Fig. 1). Similar results were obtained with both cell lines when the luminometric assays were performed 16, 24, or 48 h after transfections (M. Kiriakidou, P.T. Nelson, and Z. Mourelatos, unpubl.). These results confirm the validity of the predicted lin-28 MRE. We have further demonstrated that in a human neuronal cell line, a Gemin3-Gemin4-Argonaute-let-7b-containing miRNP associates physically with endogenous lin-28 mRNA in polyribosomes only (Nelson et al. 2004), suggesting that there is an in vivo interaction between let-7b and lin-28 mRNA. Because other human let-7 paralogs show extensive homology to let-7b (Lagos-Quintana et al. 2001), they are also likely to recognize the lin-28 mRNA. Collectively, these findings strongly suggest that human lin-28 mRNA constitutes a target for let-7b and its paralogs.
We next wished to investigate further the rules governing miRNA:MRE interactions by generating a series of mutant lin-28 MREs with varying potentials to base pair with the human/mouse let-7b miRNA (Fig. 1B). These MREs were tested as described above, in HeLa and MN-1 cells. As shown in Figure 1C, with the exception of LIN-28-M3 MRE, single nucleotide bulges between let-7b and the lin-28 mutant MREs that map toward the 5′-end of let-7b abolish repression of RL expression (mutants LIN-28-M1, LIN-28-M2, LIN-28-M4, LIN-28-M5, and LIN-28-M6). The single nucleotide bulge of LIN28-M3 MRE is symmetrically placed between the beginning of the loop and the beginning of base pairing between the 5′-most let-7b nucleotide with LIN-28-M3 (i.e., this single nucleotide bulge is surrounded by an equal number of base-paired nucleotides). A similarly placed single nucleotide bulge is found between let-7a and LIN-41a (one of the two LIN-41 MREs present in the 3′-UTR of the C. elegans lin-41 mRNA; Fig. 1). The activities of these MREs are similar (Fig. 1C, cf. LIN-41a and LIN-28, M3). These results show that near perfect complementarity between the first ∼9 nt (from the 5′-end) of a miRNA and its cognate MRE is required for miRNA function, and that the 5′-most nucleotide of miRNAs is not required to base pair with MREs (see bindings between LIN-41a or LIN-41b MREs with let-7a in Fig. 1B). We refer to this region of the miRNA as the proximal region. Analysis of published work on si/miRNAs provides further support for this claim, based on the following 10 points: (1) In the experimentally verified MREs for lin-4 and let-7, there is perfect base pairing between the MREs and the first seven or eight (starting from the 5′ end of the miRNA) nucleotides of each miRNA with no or only a single symmetrically placed nucleotide bulge, and the 5′-most nucleotide of lin-4 and let-7 mayormay not base pair with MREs (Moss et al. 1997; Reinhart et al. 2000). (2) Two loss-of-function mutants of lin-4 and let-7 miRNAs, identified in genetic screens, are caused by single-point mutations mapping in the first six nucleotides in both miRNAs and are predicted to disrupt base pairing in the proximal region (Lee et al. 1993; Moss et al. 1997; Reinhart et al. 2000). (3) In the experimentally verified target for bantam miRNA, there is perfect complementarity between the MREs and the proximal region of each miRNA (Brennecke et al. 2003). (4) The 5′ end of siRNAs sets the “ruler” for target RNA cleavage, implying that recognition of the 5′ end of siRNAs is essential for their function (Elbashir et al. 2001). (5) A genetic, single-point mutation, present in the MRE of the Arabidopsis PHAVOLUTA (PHV) mRNA, disrupts base pairing with the fifth nucleotide of its cognate miR-165/166 miRNA and dramatically reduces the miR-165/166-mediated cleavage of the mutant phv mRNA (Tang et al. 2003). (6) Single point mutations mapping in the first seven nucleotides of an siRNA reduce siRNA activity, whereas point mutations mapping toward the 3′ end of the siRNA have no or much smaller effects (Amarzguioui et al. 2003). (7) A subset of Drosophila miRNAs show perfect complementarity between their proximal region and 3′-UTR elements that are known to mediate negative posttranscriptional regulation in flies (Lai 2002). (8) Computational prediction and experimental verification of six Drosophila miRNA targets, using reporter constructs, shows that perfect complementarity of the proximal miRNA region is required for repression of reporter expression (Stark et al. 2003). (9) Computational prediction of mammalian miRNA targets and experimental verification of 11 human miRNA targets shows that complementarity between nucleotides 2 and 8 (proximal region) of mammalian miRNAs and their targets is critical for target recognition by miRNAs (Lewis et al. 2003). (10) Translational repression of miRNA targets bearing multiple MREs is largely determined by perfect complementarity between the MREs and the proximal miRNA region (Doench and Sharp 2004).
In contrast to the strict requirements for base pairing at the proximal region, nucleotide bulges between lin-28 mutant MREs and the 3′ end of let-7b (a region that we refer to as the distal region) are tolerated and decrease by approximately twofold the activities of the mutant lin-28 MREs (Fig. 1; LIN-28-M7, LIN-28-M8, and LIN-28-M9). The activity of LIN-28-M10, which bears a single nucleotide mismatch away from the central bulge and close to the 3′-end of let-7b, is essentially the same as that of the wild-type lin-28 MRE. We next determined the requirements for the size and position of the central bulges between let-7b and mutant lin-28 MREs. The optimal length of the central bulge found in the wild-type lin-28 MRE is 5 nt. As shown in Figure 1, lin-28 mutant MREs with single, symmetrically placed central bulges varying in size from 2 nt to 4 nt were still active (LIN-28-M12 to LIN-28-M14), whereas a single nucleotide substitution of the lin-28 central bulge had the same activity as the wild-type lin-28 MRE (LIN-28-M11). However, lin-28 mutant MREs with central bulges longer than five nucleotides were unable to repress the Renilla luciferase activity (LIN-28-M15, LIN-28-M16). Finally, mutant lin-28 MREs were designed that allowed for a single let-7b central bulge of varying sizes. As shown in Figure 1, MREs with a 9-nt or 7-nt let-7b central bulge were active (LIN-28-M17, LIN-28-M18), but MREs with a let-7b central bulge of less than 5 nt were inactive (LIN-28-M19 to LIN-28-M21). In fact, the activity of LIN-28-M18 MRE is identical to the wild-type LIN-28 MRE, and resembles the binding characteristics between the C. elegans lin-4 miRNA and its lin-28 mRNA target (Moss et al. 1997; see also Fig. 6C, below). To verify that the reduction of the Renilla luciferase activity was due to let-7b-mediated translational repression, we performed Northern blots on total RNA isolated from HeLa cells that had been transfected with LIN-28-wt or LIN-28-M1 constructs. As shown in Figure 1D, the mRNA levels between these two constructs were unchanged, ruling out the possibility that the observed reduction of the Renilla luciferase activity in the construct bearing the wild-type LIN-28 MRE (LIN-28-wt) was secondary to destabilization of its mRNA.
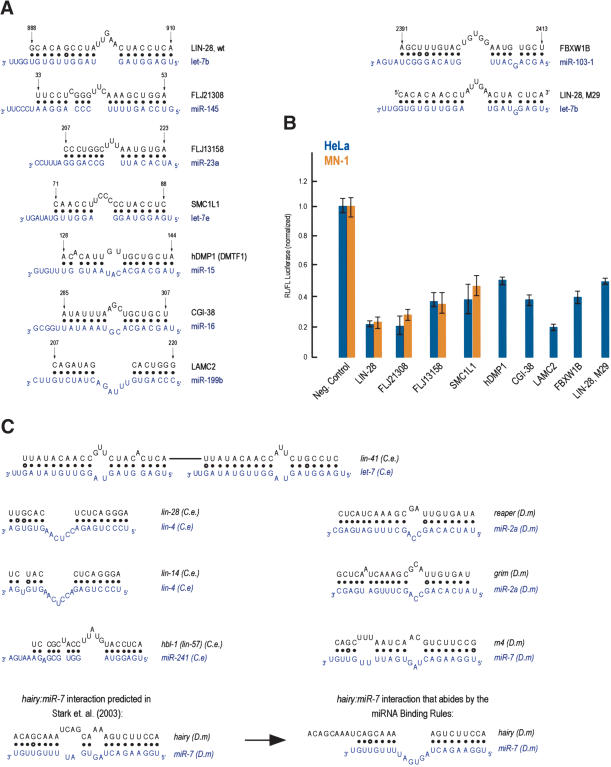
Figure 6.
(A,B) Additional, predicted miRNA targets. Details as in the legend for Figure 3. The LIN-28, M29:let-7b was designed to mimic the binding characteristics of FBXWIB:miR-103 (which allows for a symmetrically placed single nucleotide bulge of the miRNA at the proximal region of the miRNA:MRE binding). (C) miRNA:MRE (target mRNA) interactions from C. elegans (C.e) or Drosophila melanogaster (D.m) targets that abide by the miRNA binding rules.
These experiments demonstrate that there are discernible rules that govern miRNA:target mRNA interactions, which may be generally applicable. We note that the repressing properties of a miRNA may depend on the way it interacts with its mRNA target. A miRNA:MRE (target mRNA) interaction with a central bulge of optimal length (LIN-28, wt and LIN-28, M18; Fig. 1) is more potent than two small opposing loops (LIN-41a or LIN-41b; Fig. 1). This finding may explain the requirement, for optimal repression, of two MREs for let-7 in the 3′-UTR of the C. elegans lin-41 mRNA (Reinhart et al. 2000). On the other hand, a single MRE for lin-4 in the 3′-UTR of the C. elegans lin-28 mRNA suffices because it contains a single, 6-nt central bulge (Moss et al. 1997). The degree of miRNA-mediated translational repression may ultimately depend on additional factors such as the miRNA and target mRNA concentrations, the presence of multiple MREs on target mRNAs, the accessibility of MREs, and other cis elements. Indeed, in vivo repression of the C. elegans lin-41 mRNA expression by let-7 requires, in addition to the two MREs recognized by let-7, a stretch of 27 nucleotides between the two MREs. This finding suggests that the context of MREs is also important for miRNA-mediated regulation (Vella et al. 2004).
The finding of a let-7b MRE in the 3′-UTR of the human/mouse lin-28 mRNA and the fact that endogenous human lin-28 mRNA associates with a let-7b-containing miRNP in polyribosomes (Nelson et al. 2004) along with the regulation of LIN-28 protein expression as reported by the Moss lab (Moss and Tang 2003) strongly suggest that human let-7b and lin-28 are part of the same pathway, which may be functionally related to the C. elegans heterochronic gene pathway. The C. elegans lin-28 mRNA is predominantly regulated by lin-4 (Moss et al. 1997). Although a direct role for let-7 in the regulation of C. elegans lin-28 mRNA has not been shown, lin-28 is also regulated by a lin-4 independent pathway (Seggerson et al. 2002). There are four let-7 paralogs in C. elegans, and it is possible that one of them regulates lin-28.
To test further the validity of our findings, we made synthetic let-7b siRNAs (let-7b-M1 and let-7b-M4) carrying point mutations designed to compensate the point mutations found in two of the lin-28 MRE mutants (LIN-28-M1 and LIN-28-M4 respectively; see Fig. 2A). Sharp and colleagues have demonstrated the feasibility of transfecting siRNAs that function as miRNAs (Doench et al. 2003). LIN-28 constructs (Fig. 2) were transfected with or without these siRNAs, and the Renilla luciferase activity was measured 18 h after transfection. As shown in Figure 2B, cotransfection of the LIN-28-M1 construct with let-7b-M1 siRNA repressed the levels of luciferase, whereas cotransfection of LIN-28-M1 with let-7b-M4 siRNA had no effect. Similarly, cotransfection of the LIN-28-M4 construct with let-7b-M4 siRNA repressed the levels of luciferase, whereas cotransfection of LIN-28-M4 with let-7b-M1 siRNA had no effect (Fig. 2C). The suppression of the luciferase activity with siRNAs carrying compensatory mutations is significant, specific, and reproducible, but it is not as pronounced as the one seen with the endogenous let-7b miRNA targeting the wild-type lin-28 MRE. This may reflect inefficient incorporation of exogenous siRNAs in miRNPs/RISCs. These siRNA duplexes were designed prior to the reports by Khvorova et al. (2003) and Zamore and colleagues (Schwarz et al. 2003) describing the functional asymmetry of siRNA duplexes and siRNA-like duplexes (derived from Dicer processing of pre-miRNAs). Those studies showed that there is preferential incorporation in RISCs of siRNAs and miRNAs whose 5′ end is more loosely paired with its antisense (Khvorova et al. 2003; Schwarz et al. 2003). We note that, for both siRNA duplexes used in the Figure 2 experiment, the 5′ end of the antisense siRNA strand (which is predicted to base-pair with the MRE) starts with a uridine, and the 5′ end of the sense siRNA strand starts with a cytosine. In that regard, our siRNA duplexes conform to the design rules that maximize incorporation of siRNAs in RISCs. However, for the siRNA-like duplexes that are derived from pre-miRNAs (including pre-let-7b), an unpaired 5′ end of the miRNA is best suited for efficient incorporation in miRNPs/RISCs (Khvorova et al. 2003; Schwarz et al. 2003). Our siRNA duplexes might have been more efficient in repressing the expression of the reporter constructs had we used unpaired 5′ ends of the antisense strands. In summary, these experiments demonstrate the validity of the miRNA binding rules and the exquisite specificity of the miRNA:MRE interaction.
Figure 2.
Restoration of miRNA-mediated translational repression of LIN-28 mutant MREs by synthetic let-7b siRNAs that carry compensatory mutations. (A) Potential base pairing between LIN-28 MREs (black) and endogenous let-7b or synthetic let-7b siRNAs (blue). Mutated nucleotides are shown in red. (B,C) HeLa cells were cotransfected with Renilla luciferase (RL) constructs bearing the indicated MREs in the 3′-UTR, along with firefly luciferase (FL) and with or without the indicated synthetic siRNAs (30 nM). Results shown are average values (with standard deviations) of normalized RL/FL activities obtained from three separate experiments.
In parallel, we experimentally tested all hypothetical miRNA:MRE configurations shown in Figures 3 and 4. The miRNAs (let-7b, let-7e, miR-141, miR-24, miR-145, miR-23a, miR-15a, miR-16, miR-199b, and miR-103) for these putative targets are present in both HeLa and MN-1 cells (Lagos-Quintana et al. 2001, 2002; Mourelatos et al. 2002; Dostie et al. 2003; Z. Mourelatos, unpubl.). miR-141 was originally cloned from mouse (Lagos-Quintana et al. 2002). Human miR-141, containing two additional terminal nucleotides, has also been cloned from human colonic mucosa (Michael et al. 2003; originally deposited in the GenBank database under accession no. AJ535825). We have cloned miR-141 from HeLa and MN-1 cells and confirmed the presence of the two additional nucleotides, as shown in Figure 3A (M. Kiriakidou and Z. Mourelatos, unpubl.). Whether the longer or shorter miR-141 is more prevalent in cells is unknown. These miRNA:MRE configurations were chosen based on their general resemblance to the experimentally verified C. elegans miRNA:target mRNA interactions, as described above, and prior to the completion of the mutational analysis presented in Figure 1. All of these putative MREs were also conserved in the mouse. Only two (in addition to lin-28) of these putative MREs suppressed the expression of luciferase (Fig. 3), whereas 11 hypothetical MREs failed to do so (Fig. 4). These findings are entirely consistent with the results of our mutational analysis of the let-7b:lin-28 interaction, and further demonstrate the high specificity of the miRNA:MRE bindings. For example, the main difference in binding characteristics between miR-141 and its true MRE found in the Clock mRNA (which suppresses the levels of the reporter; Fig. 3B) versus mir-141 and STK3 (which does not suppress the levels of the reporter; Fig. 4) is in the number of nucleotides of the miRNA central bulge. In the second case (which is inactive), that bulge is five nucleotides. This result is expected based on the results of our mutational analysis, because a lin-28 mutant that has the same configuration (LIN-28, M19; Fig. 1) as that of miR-141:STK3 is unable to repress the expression of the reporter. Based on these findings and the results of the mutational analysis, a general set of miRNA binding rules may be formulated and is shown in Figure 5A.
Figure 5.
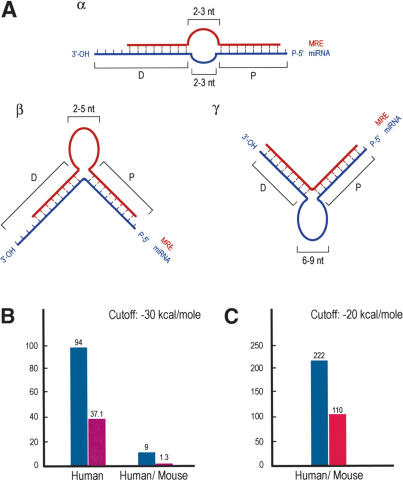
MicroRNA Binding Rules and Statistics. (A) Schematic representation of miRNA:MRE (target mRNA) bindings (miRNA binding rules). (Blue) miRNAs; (red) MRE; (P) proximal (relative to 5′-end of miRNA) region of miRNA:MRE binding; (D) distal region of binding. (Panel α) Loop, length (on each sequence) = 2–3 nt. (Panel β) Single MRE central bulge, length = 2–5 nt. (Panel γ) Single miRNA central bulge, length = 6–9 nt. Proximal binding characteristics are ≥7-nt base pairing between miRNA and MRE; the 5′-most nucleotide of the miRNA may or may not base pair with MRE; one symmetric single nucleotide bulge allowed (i.e., the single nucleotide bulge is surrounded by an equal number of base-paired nucleotides). Distal binding characteristics are ≥5-nt base pairing between miRNA and MRE; nucleotide bulges allowed. The last (toward the 3′ end) nucleotides of the miRNA may or may not base pair with the MRE. (B) Hits between 10 human miRNAs (blue bar) or shuffled RNAs (purple bar) and the 3′-UTR database of annotated human mRNAs or the conserved human/mouse 3′-UTR database (initial analysis). (C) Hits between 94 human miRNAs (blue bar) or shuffled controls (with the same compositional properties as the authentic miRNAs; red bar) and the conserved human/mouse 3′-UTR database extracted using EnsMart.
We subsequently combined our initial algorithm with an “MRE filter” to create the computational program “DIANA-microT” (see Supplemental Material), which predicted 94 human MREs for the 10 miRNAs (using a cutoff of –30 kcal/mole; Fig. 5B). Nine of the predicted human MREs are also conserved in the 3′-UTRs of the corresponding mouse ortholog mRNAs. To evaluate the statistical significance of our computational algorithm, we created a cohort of “negative control” sequences by randomly shuffling the sequence of each of the 10 real miRNAs, 10 times. All 10 of the randomized sequences for each miRNA (amounting to a total of 100 randomized sequences) were used for the computational searches. In addition, 371 “MREs” were predicted for these 100 randomized sequences, 13 of which were also conserved in the mouse. Normalizing the numbers for the 10 miRNAs, the total number of predicted “MREs” for the randomized RNA sequences was 37.1 (human) or 1.3 (conserved human/mouse; Fig. 5B). Thus, the average number of human MREs predicted for each real miRNA is 9.4 versus 3.7 for each shuffled. It is important to note that this significant difference between the number of predicted MREs for the real miRNAs versus the shuffled sequences is seen only when the miRNA binding rules are implemented in the computational algorithm. Using the –30kcal/mole energy cut-off as the only criterion for MRE prediction revealed an average number of 5094 predicted human MREs for each miRNA versus 4974 for each shuffled. No differences were detected when we determined the numbers of those MREs that were conserved in the mouse: 168 on average for each real miRNAs versus 158 for each shuffled. The predicted mRNA targets for the 10 human miRNAs are shown in Supplementary Table 1. We also determined which of the human MREs for the queried miRNAs were conserved in other species. The results of this analysis are shown in Supplementary Table 2 (conserved human-mouse MREs) and Supplementary Table 3 (conserved human-other mammals MREs).
During this initial analysis, we observed that many putative human MRE sequences were not detected in the 3′-UTR of homologous mRNAs from mouse because of the small mouse 3′-UTR database that we used. Conservation of predicted MREs provides a strong indication for the biological importance of these sites. During the next phase of our analysis, we used 84 additional miRNAs (for a total of 94 miRNAs) that are nonredundant and perfectly conserved between humans and mice (shown in Supplementary Table 5). These include the 79 miRNAs that Lewis et al. (2003) used in their study. We also created a program that generates shuffled miRNA controls that have the same compositional properties with the authentic miRNAs, and take into account the nucleotide composition biases of mammalian 3′-UTRs (Nussinov 1981; Lewis et al. 2003; described in Supplemental Material; shuffled sequences for all miRNAs are shown in Supplementary Table 5). We generated a new set of 3′-UTR databases by extracting the conserved human and mouse 3′-UTRs from orthologous genes using EnsMart (see Supplemental Material; Kasprzyk et al. 2004). This new approach significantly increased the number of orthologous genes with conserved 3′-UTRs (derived from 13,272 human transcripts, versus 4035 in our previous database). Finally, we used the DIANA-microT program to predict conserved human/mouse targets using the new datasets along with the new control (shuffled) sequences and using an energy threshold cutoff of –20 kcal/mole. We predicted 5031 human targets for the 94 miRNAs; 222 of these targets are also conserved in the mouse. The ratio of conserved:all hits for the 94 authentic miRNAs is 1:22.6. For the control sequences (376 shuffled controls; 4 for each miRNA), we obtained 15,620 human hits, 441 of which were conserved in the mouse. The ratio of conserved:all hits for the 376 shuffled control sequences is 1:35.4. Normalizing the numbers for the 94 miRNAs, the total number of conserved hits for the shuffled controls is 110 (Fig. 5C; numbers of hits per authentic or shuffled miRNA are also shown in Supplementary Table 5). The ratio of the conserved human/mouse targets for the authentic versus the shuffled miRNAs is thus 2:1, similar to the value obtained by Lewis et al. (2003) for the conserved human/mouse hits. We note that of the 94 miRNAs, conserved MREs were found for 73 miRNAs (Supplementary Table 4), suggesting that miRNA targets containing single MREs are less prevalent than targets containing multiple MREs and that some miRNAs may recognize only targets with multiple MREs. However, it is also very likely that additional rules exist for miRNAs that recognize single MREs. Thus, our current analysis may underestimate the number of miRNA targets bearing single MREs.
To further validate the results of the final computational analysis, we experimentally verified MREs that were predicted based on the miRNA binding rules. We chose to verify one predicted MRE for each of the miRNAs that we had previously tested with “MREs” that did not abide by the miRNA binding rules (and thus did not suppress the levels of the luciferase reporter as shown in Fig. 4). Seven MREs that abided by the miRNA binding rules were selected, two of which (FLJ13158 and SMCL1) are found in the 3′-UTR of human mRNAs only, whereas the remainder are also conserved in the 3′-UTR of their mouse orthologs. As shown in Figure 6A and B, all of these MREs suppressed the levels of the luciferase reporter. These findings provide further evidence for the validity of the miRNA binding rules. Among the mRNAs that may be regulated by miRNAs are the Clock transcription factor, which is critical for circadian rhythms, and the mitogen-activated protein kinase 14 (MAPK14, also known as p38α kinase). MAPK14 has pleiotropic cellular effects; it is a key regulator of stress-induced signaling, cell proliferation, and apoptosis and development (see OMIM entry 600289). Targets for miR-15a and miR-16 are particularly interesting because these miRNAs may play a role in the pathogenesis of B-cell chronic lymphocytic leukemia (Calin et al. 2002). miR-15a may regulate the known tumor suppressor gene cyclin D-binding Myb-like protein (DMP1/DMTF1), whereas miR-16 may regulate, among other genes, the mRNA coding for a ∼25-Kda protein (CGI-38) which is conserved in worms, flies, rodents, and humans. An MRE for miR-145 is found in the 3′-UTR of both human and mouse mRNAs coding for a hypothetical 501-amino acid protein termed FLJ21308 in humans and D13Ertd275e in mouse. FLJ21308 contains a putative poly (ADP-ribose) polymerase catalytic domain, suggesting that it may function in DNA damage control. For let-7e and miR-23 miRNAs, MREs were found in the 3′-UTR of human mRNAs coding for the structural maintenance of chromosome 1-like 1 protein (SMC1L1) and a 324-amino acid hypothetical protein termed FLJ13158, respectively. SMCL1 functions in sister chromatid cohesion during mitosis (see OMIM entries 606462 and 300040). The FLJ13158 protein contains a 120-amino acid domain of unknown function (termed the DUF738 domain), which is highly conserved in worm, fly, rodent, and human proteins. It is possible that some miRNAs may regulate mRNA targets in humans that are not regulated by the same miRNAs in mouse or other species. The length of 3′-UTRs increases with evolutionary age and organism complexity (with human mRNAs having the longest 3′-UTRs (Pesole et al. 2002; Mazumder et al. 2003). The longer 3′-UTRs may provide more regulatory elements that may contribute to a more complex posttranscriptional regulation of mRNAs in humans. However, predicted MREs that are not conserved in other species (Supplementary Table 1) should be approached cautiously. Ultimately, the contribution of a given miRNA in the regulation of its mRNA target may depend on multiple factors, including the presence of other cis regulatory elements in the 3′-UTR of the mRNA target.
We next wished to determine which of the previously identified (and experimentally verified) worm and fly miRNA targets (Lee et al. 1993; Wightman et al. 1993; Moss et al. 1997; Reinhart et al. 2000; Brennecke et al. 2003; Lin et al. 2003; Abrahante et al. 2003; Stark et al. 2003) contained MREs that abided by the miRNA binding rules. All three prototypical C. elegans miRNA targets (lin-14, lin-28, and lin-41 mRNAs; Lee et al. 1993; Wightman et al. 1993; Moss et al. 1997; Reinhart et al. 2000) contain miRNA:MRE interactions that abide by the miRNA binding rules (Fig. 6C). The C. elegans ortholog of the Drosophila hunchback gene (hbl-1, also known as lin-57) was recently identified as a heterochronic gene that may be regulated by miRNAs. Multiple potential sites (putative MREs) that may be recognized by various miRNAs have been proposed (Abrahante et al. 2003; Lin et al. 2003). We searched for putative hbl-1 MREs with our computational algorithm and found that an MRE for the C. elegans miR-241 is found in the 3′-UTR of hbl-1 (Fig. 6C). This MRE is also conserved in the 3′-UTR of the C. briggsae hbl-1 (data not shown). Interestingly, miR-241 is a paralog of let-7 and has a temporal expression pattern identical to that of let-7 (Lim et al. 2003). It is likely that miR-241, like let-7, controls heterochronic genes. Our algorithm predicts that hbl-1 is likely regulated by miR-241.
Putative miRNA targets for 75 Drosophila miRNAs were recently proposed (Stark et al. 2003). The computational algorithm used in that study was based on binding energies and required complementarity between the first eight nucleotides of the miRNA with its putative target (Stark et al. 2003). The database used for the searches consisted of the conserved D. melanogaster and D. pseudoobscura 3′-UTRs, and hits were scored based on the binding energy of the predicted miRNA:MRE interaction, the presence of multiple miRNA sites in the 3′-UTR of putative mRNA targets, and the conservation of the sites in a third genome (i.e., Anopheles gambiae; Stark et al. 2003). The presence of multiple MREs for the same miRNA in any given target correlated with high-scoring hits. However, this approach failed to accurately predict single conserved sites (MREs) for miRNAs, because there was no statistically significant difference between the real miRNAs versus randomized sequences (Stark et al. 2003). Stark et al. presented experimental verification for six of their predicted Drosophila miRNA targets. Their assay monitored the miRNA-dependent down-regulation of a reporter construct that contained the entire 3′-UTR from the putative miRNA target (Stark et al. 2003). Four of the tested targets (m4 and hairy mRNAs, targeted by miR-7; and reaper and grim mRNAs, targeted by miR-2) were predicted to harbor only a single site for their cognate miRNAs (Stark et al. 2003) and repressed the levels of the reporter. Do these sites abide by the miRNA binding rules as our study would have predicted? Figure 6C shows the putative base pairing between these four targets and their cognate miRNAs. Three of these target:miRNA interactions (reaper:miR-2a, grim:miR-2a, and m4:miR-7) abide by the miRNA binding rules (and in particular the configuration α shown in Fig. 5A). The hairy:miR-7 interaction (as depicted in Stark et al. 2003; see also the left side of Fig. 6C) does not, at first inspection, seem to abide by our miRNA binding rules. A closer analysis, however, shows that an alternative configuration between hairy and miR-7 may be adopted (see Fig. 6C, right side), which does abide by the miRNA binding rules (and in particular the configuration γ shown in Fig. 5A). An experimentally validated miRNA target that does not contain MREs that follow the miRNA binding rules, is the Drosophila hid mRNA that is regulated by the bantam miRNA (Brennecke et al. 2003). We note that hid mRNA contains multiple sites that show partial complementarity, especially with the proximal region of bantam. We expect that the miRNA binding rules are more “lax” if multiple MREs are present in the 3′-UTR of target genes. In such cases the cumulative effect of many “weaker” miRNA: MRE interactions may lead to robust repression of target gene expression. Indeed, cooperativity of multiple MREs has been demonstrated (Ha et al. 1996; Doench et al. 2003). Similarly, most of the predicted targets for Drosophila (Enright et al. 2003; Stark et al. 2003) and mammalian (Lewis et al. 2003) miRNAs contain multiple MREs; in that case the most critical aspect of miRNA: target RNA interactions is perfect base pairing between the proximal region of miRNAs and their targets (Lewis et al. 2003; Stark et al. 2003).
Discussion
Recently, Bartel, Burge, and colleagues (Lewis et al. 2003) have presented a computational algorithm that allows prediction of conserved, mammalian miRNA targets along with accurate estimates of false positive rates and experimental validation of 11 (out of 15 tested) predicted targets. The targets identified by Lewis et al. contain multiple MREs for the same miRNA or are regulated by more than one miRNA and are very different from the ones that we report. The targets reported for Drosophila miRNAs also contain, mostly, multiple MREs (Enright et al. 2003; Stark et al. 2003). In contrast, our study uncovers predominantly targets that contain single MREs. This is due to the different strategies employed and underscores the importance of using multiple independent approaches to uncover the full gamut of miRNA targets. Our approach is based on the experimental deduction of rules by evaluating the significance of individual miRNA bases on target RNA recognition. Our validation assay for putative MREs utilizes a single MRE in the 3′-UTR of the RL reporter, which is under the control of the relatively strong Herpes simplex thymidine kinase promoter. We deliberately chose to insert a single MRE to avoid extraneous effects of longer sequences that may have arisen if multiple MRE copies or if the entire 3′-UTR of the target genes were used. At the same time, our assay may not be sensitive enough to detect “weaker” miRNA:MRE interactions that may become apparent when multiple MREs are used. This may be true for miRNAs that are expressed at low levels or for “low-affinity” miRNA:MRE interactions. Many miRNAs are surprisingly abundant (Lim et al. 2003), and we expect that a single MRE should suffice to detect “high-affinity” miRNA:MRE interactions. As a result, our rules predict miRNA targets that contain, in the vast majority of cases, single MREs. In contrast, the computational approaches employed by Lewis et al. (2003), Stark et al. (2003), and Enright et al. (2003) were geared toward identifying targets containing multiple MREs.
An important point that emerges from this study and those of Lewis et al. (2003) and Stark et al. (2003) is the significance of the 5′ end of the miRNA for target RNA recognition. The manner by which the remainder of the miRNA base-pairs with its target becomes more important in cases where a single MRE is used. It is also becoming apparent that, in general, miRNAs recognize more targets containing multiple MREs (Enright et al. 2003; Lewis et al. 2003; Stark et al. 2003) rather than single MREs (as shown in this study). However, we note that additional miRNA binding rules (for miRNAs recognizing single MREs) are likely to exist and the putative targets that we propose may represent just a fraction of the total number of targets bearing single MREs.
We anticipate that, ultimately, numerous mRNAs may be regulated by miRNAs. We also expect that the degree of miRNA-mediated repression on the expression of target mRNAs may vary depending, among other factors, upon the binding characteristics between miRNAs and their targets. The presented miRNA targets remain predictions until they are verified in the context of the whole organism. Nevertheless, we anticipate that these predictions may prove valuable for guiding investigations of miRNA function.
Materials and methods
Computational analysis
Details of the computational analysis are found in the supplement.
Plasmids and siRNAs
MREs were cloned in the 3′-UTR of the pRL-TK vector (coding for Renilla luciferase; Promega), embedded in a common DNA backbone containing sites for the restriction enzymes XbaI, NdeI, XhoI, and NotI. Briefly, for each MRE, two complementary DNA oligos were synthesized (sense, 5′-CTA GAGACTAAATGACTCCATATGACA; sense MRE, ACGCTC GAGGC-3′; antisense, 5′-GGCCGCCTCGAGCGT; antisense MRE, TGTCATATGGAGTCATTTAGTCT-3′), annealed, and cloned in the XbaI-NotI sites of the pRL-TK vector. Sequences of siRNA duplexes were: let-7b-M1, antisense: 5′-UGAGGGU AGUAGGUUGUGUGGU-3′, sense: 5′-CACACAACCUACU ACCCUCAUU-3′; let-7b-M4, antisense: 5′-UGAGGCUAGUA GGUUGUGUGGU-3′, sense: 5′-CACACAACCUACUAGCC UCAUU-3′.
Transfections, dual luciferase assays, and Northern blots
pRL-TK plasmids bearing MREs in the 3′-UTR (0.4 μg) were cotransfected along with PGL-3 reporter plasmid (coding for firefly luciferase; Promega, 0.4 μg) into HeLa S3 cells (∼1 × 105) using Lipofectamine 2000 (Invitrogen). Luciferase activities were determined using a Dual Luciferase Reporter Assay System (Promega). For siRNA transfections, 30 nM of each siRNA duplex was transfected using Lipofectamine 2000.
For Northern blots, HeLa cells (∼1 × 106) were cotransfected with pRL-TK-LIN-28 or pRL-TK-M1 plasmids (2 μg) and pGL3 plasmid (2 μg) in 6-well plates. Then, 18 h after transfection, total cell lysates were assayed for luciferase activities and RNA was isolated. To control for transfection efficiency, samples with the same firefly luminescence values were used. Total RNA (10 μg) from each sample was fractionated on a 1% formaldehyde-agarose gel, transferred to a nylon membrane (Amersham), and probed with [α-32P]-dCTP-labeled DNA probe against Renilla luciferase. After autoradiography, the blot was stripped and reprobed with a radiolabeled probe against β-actin (Ambion). Probes were labeled with a random primed DNA labeling kit from Roche.
Acknowledgments
We thank A. Bernal, M. Megraw, and G. Grant for their help with statistical calculations during the early stages of this work; Malik Yousef for improving the parallel implementation of “DIANA-microT”; and N. Henke and D. Widyono, administrators of the Liniac cluster, Penn Genomics Institute, for their continuous support during the exhaustive runs of our calculations. We are grateful to Drs. H. Kazazian and J.-C. Oberholtzer for critical review of the manuscript. We thank two anonymous reviewers for bringing to our attention the dinucleotide composition biases in mammalian UTRs. This work was supported by grants from the NIH to M.K. (T32-AR07442), P.N. (T32-AG00255), A.K. (HG00046), and Z.M. (NS02199), the NSF to A.H. (DBI-0238295) and by a University of Pennsylvania Genomics Institute award to A.H. and Z.M.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1184704.
Corresponding authors.
References
- Abrahante J.E., Daul, A.L., Li, M., Volk, M.L., Tennessen, J.M., Miller, E.A., and Rougvie, A.E. 2003. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell 4: 625–637. [DOI] [PubMed] [Google Scholar]
- Amarzguioui M., Holen, T., Babaie, E., and Prydz, H. 2003. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 31: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V., Bartel, B., Bartel, D.P., Burge, C.B., Carrington, J.C., Chen, X., Dreyfuss, G., Eddy, S.R., Griffiths-Jones, S., Marshall, M., et al. 2003a. A uniform system for microRNA annotation. RNA 9: 277–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V., Lee, R.C., Lavanway, A., Williams, P.T., and Jewell, D. 2003b. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 13: 807–818. [DOI] [PubMed] [Google Scholar]
- Aukerman M.J. and Sakai, H. 2003. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bartel B. and Bartel, D.P. 2003. MicroRNAs: At the root of plant development? Plant Physiol. 132: 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- Bohnsack M.T., Czaplinski, K., and Gorlich, D. 2004. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10: 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36. [DOI] [PubMed] [Google Scholar]
- Calin G.A., Dumitru, C.D., Shimizu, M., Bichi, R., Zupo, S., Noch, E., Aldler, H., Rattan, S., Keating, M., Rai, K., et al. 2002. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. 99: 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. 2004. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G. and Sharp, P.A. 2004. Specificity of microRNA target selection in translational repression. Genes & Dev. 18: 504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Petersen, C.P., and Sharp, P.A. 2003. siRNAs can function as miRNAs. Genes & Dev. 17: 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J., Mourelatos, Z., Yang, M., Sharma, A., and Dreyfuss, G. 2003. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA 9: 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel, W., and Tuschl, T. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 15: 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A.J., John, B., Gaul, U., Tuschl, T., Sander, C., and Marks, D.S. 2003. MicroRNA targets in Drosophila. Genome Biol. 5: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34. [DOI] [PubMed] [Google Scholar]
- Ha I., Wightman, B., and Ruvkun, G. 1996. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes & Dev. 10: 3041–3050. [DOI] [PubMed] [Google Scholar]
- Hamilton A.J. and Baulcombe, D.C. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Boettcher, S., Caudy, A.A., Kobayashi, R., and Hannon, G.J. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150. [DOI] [PubMed] [Google Scholar]
- Hutvagner G. and Zamore, P.D. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060. [DOI] [PubMed] [Google Scholar]
- Hutvagner G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T., and Zamore, P.D. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838. [DOI] [PubMed] [Google Scholar]
- Kasprzyk A., Keefe, D., Smedley, D., London, D., Spooner, W., Melsopp, C., Hammond, M., Rocca-Serra, P., Cox, T., and Birney, E. 2004. EnsMart: A generic system for fast and flexible access to biological data. Genome Res. 14: 160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev. Cell 4: 205–217. [DOI] [PubMed] [Google Scholar]
- Ketting R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 15: 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A., Reynolds, A., and Jayasena, S.D. 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216. [DOI] [PubMed] [Google Scholar]
- Knight S.W. and Bass, B.L. 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293: 2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut, R., Lendeckel, W., and Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12: 735–739. [DOI] [PubMed] [Google Scholar]
- Lai E.C. 2002. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 30: 363–364. [DOI] [PubMed] [Google Scholar]
- Lau N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. [DOI] [PubMed] [Google Scholar]
- Lee R.C. and Ambros, V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. [DOI] [PubMed] [Google Scholar]
- Lee Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Shih, I.H., Jones-Rhoades, M.W., Bartel, D.P., and Burge, C.B. 2003. Prediction of mammalian microRNA targets. Cell 115: 787–798. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau, N.C., Weinstein, E.G., Abdelhakim, A., Yekta, S., Rhoades, M.W., Burge, C.B., and Bartel, D.P. 2003. The microRNAs of Caenorhabditis elegans. Genes & Dev. 17: 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.Y., Johnson, S.M., Abraham, M., Vella, M.C., Pasquinelli, A, Gamberi, C., Gottlieb, E., and Slack, F.J. 2003. The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev. Cell 4: 639–650. [DOI] [PubMed] [Google Scholar]
- Llave C., Xie, Z., Kasschau, K.D., and Carrington, J.C. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056. [DOI] [PubMed] [Google Scholar]
- Lund E., Guttinger, S., Calado, A., Dahlberg, J.E., and Kutay, U. 2004. Nuclear export of microRNA precursors. Science 303: 95–98. [DOI] [PubMed] [Google Scholar]
- Martinez J., Patkaniowska, A., Urlaub, H., Luhrmann, R., and Tuschl, T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574. [DOI] [PubMed] [Google Scholar]
- Mazumder B., Seshadri, V., and Fox, P.L. 2003. Translational control by the 3′-UTR: The ends specify the means. Trends Biochem. Sci. 28: 91–98. [DOI] [PubMed] [Google Scholar]
- Michael M.Z., O'Connor, S.M., van Holst Pellekaan, N.G., Young, G.P., and James, R.J. 2003. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 1: 882–891. [PubMed] [Google Scholar]
- Moss E.G. and Tang, L. 2003. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol. 258: 432–442. [DOI] [PubMed] [Google Scholar]
- Moss E.G., Lee, R.C., and Ambros, V. 1997. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88: 637–646. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M., and Dreyfuss, G. 2002. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 16: 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson P., Kiriakidou, M., Sharma, A., Maniataki, E., and Mourelatos, Z. 2003. The microRNA world: Small is mighty. Trends Biochem. Sci. 28: 534–540. [DOI] [PubMed] [Google Scholar]
- Nelson P.T., Hatzigeorgiou, A.G., and Mourelatos, Z. 2004. miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA 10: 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R. 1981. Nearest neighbor nucleotide patterns. Structural and biological implications. J. Biol. Chem. 256: 8458–8462. [PubMed] [Google Scholar]
- Olsen P.H. and Ambros, V. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 216: 671–680. [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. 2003. Control of leaf morphogenesis by microRNAs. Nature 425: 257–263. [DOI] [PubMed] [Google Scholar]
- Pesole G., Liuni, S., Grillo, G., Licciulli, F., Mignone, F., Gissi, C., and Saccone, C. 2002. UTRdb and UTRsite: Specialized databases of sequences and functional elements of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Update 2002. Nucleic Acids Res. 30: 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K.D., Tatusova, T., and Maglott, D.R. 2003. NCBI Reference Sequence project: Update and current status. Nucleic Acids Res. 31: 34–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. [DOI] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. 2002. Prediction of plant microRNA targets. Cell 110: 513–520. [DOI] [PubMed] [Google Scholar]
- Schwarz D.S., Hutvagner, G., Du, T., Xu, Z., Aronin, N., and Zamore, P.D. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208. [DOI] [PubMed] [Google Scholar]
- Seggerson K., Tang, L., and Moss, E.G. 2002. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol. 243: 215–225. [DOI] [PubMed] [Google Scholar]
- Seitz H., Youngson, N., Lin, S.P., Dalbert, S., Paulsen, M., Bachellerie, J.P., Ferguson-Smith, A.C., and Cavaille, J. 2003. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat. Genet. 34: 261–262. [DOI] [PubMed] [Google Scholar]
- Stark A., Brennecke, J., Russell, R.B., and Cohen, S.M. 2003. Identification of Drosophila MicroRNA Targets. Plos Biology 1: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. 2003. A biochemical framework for RNA silencing in plants. Genes & Dev. 17: 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Borer, P.N., Dengler, B., Levin, M.D., Uhlenbeck, O.C., Crothers, D.M., and Bralla, J. 1973. Improved estimation of secondary structure in ribonucleic acids. Nat. New Biol. 246: 40–41. [DOI] [PubMed] [Google Scholar]
- Vella M.C., Choi, E.Y., Lin, S.Y., Reinert, K., and Slack, F.J. 2004. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes & Dev. 18: 132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B., Ha, I., and Ruvkun, G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862. [DOI] [PubMed] [Google Scholar]
- Xie Z., Kasschau, K.D., and Carrington, J.C. 2003. Negative feedback regulation of dicer-like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13: 784–789. [DOI] [PubMed] [Google Scholar]
- Xu P., Vernooy, S.Y., Guo, M., and Hay, B.A. 2003. The Drosophila MicroRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 13: 790–795. [DOI] [PubMed] [Google Scholar]
- Yi R., Qin, Y., Macara, I.G., and Cullen, B.R. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Dev. 17: 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Wagner, E.J., and Cullen, B.R. 2002. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9: 1327–1333. [DOI] [PubMed] [Google Scholar]