Abstract
Plasmodium vivax infects human erythrocytes through a major pathway that requires interaction between an apical parasite protein, the Duffy binding protein (PvDBP) and its receptor on reticulocytes, the Duffy antigen/receptor for chemokines (DARC). The importance of the interaction between PvDBP (region II, DBPII) and DARC to P. vivax infection has motivated our malaria research group at Oswaldo Cruz Foundation (state of Minas Gerais, Brazil) to conduct a number of immunoepidemiological studies to characterise the naturally acquired immunity to PvDBP in populations living in the Amazon rainforest. In this review, we provide an update on the immunology and molecular epidemiology of PvDBP in the Brazilian Amazon - an area of markedly unstable malaria transmission - and compare it with data from other parts of Latin America, as well as Asia and Oceania.
Keywords: malaria, Plasmodium vivax, Duffy binding protein, naturally acquired antibodies, genetic variability
Plasmodium vivax malaria in the Brazilian Amazon - Malaria is still a major public health problem in Brazil, with 244,000 cases registered in 2012 (WHO 2013), with 99.9% of them distributed through the Amazon Basin, including the states of Pará, Rondônia, Amazonas (AM), Mato Grosso, Amapá, Acre and Roraima (saude.gov.br/sivep_malaria). In these areas, the spatial distribution of malaria is not homogenous and changes over time at individual, community, state and national scales (Tauil & Daniel-Ribeiro 1998, de Castro et al. 2006, 2007, Barbieri & Sawyer 2007, da Silva-Nunes et al. 2008, Oliveira-Ferreira et al. 2010). In the Brazilian Amazon, the instability of transmission is the dominant feature of malaria (Camargo et al. 1994), with exposed populations consisting mostly of migrants from malaria-free areas. In these individuals, the infection is generally accompanied by clinical symptoms of variable degrees of intensity. Nevertheless, during the past few years, epidemiologic studies carried out among individuals with long-term exposure to malaria in Brazil clearly show the existence of symptomless malaria infections (Camargo et al. 1999a, Alves et al. 2002, da Silva-Nunes et al. 2008, Ladeia-Andrade et al. 2009).
Of the five species of malaria parasites known to infect humans, three species occur in Brazil: P. vivax, Plasmodium falciparum and Plasmodium malariae. Until 1990, the prevalence of P. falciparum and P. vivax infections was similar with roughly 50% of each species (Marques et al. 1986, Tauil & Daniel-Ribeiro 1998, Loiola et al. 2002), while the prevalence of P. malariae was very low. After that time, the incidence of both P. falciparum and P. vivax has decreased, probably due to the intensification of malaria control measures, which included early diagnosis and treatment (Loiola et al. 2002). However, certain features of the biology of P. vivax give this species greater resilience than P. falciparum. Whereas P. falciparum parasites invade blood cells at various stages of development, P. vivax infects reticulocytes and the latter parasite species seems to be more transmissible at low parasite densities (Boyd & Kitchen 1937). Furthermore, P. vivax parasites are associated with the early appearance of infective sexual stages (gametocytes) in the blood and can remain in the liver as dormant hypnozoites responsible for relapses. These unique characteristics of the biology of P. vivax make its management and elimination particularly challenging. In fact, as control measures become more effective, the residual malaria burden is increasingly shifting towards P. vivax malaria (Mendis et al. 2001). Consequently, the number of P. vivax cases has increased over the years and this malaria parasite species is now responsible for roughly 80% of all malaria cases in the Brazilian Amazon Region (Camargo et al. 1999b, Ladeia-Andrade et al. 2009, WHO 2012). Although P. vivax malaria is often regarded as benign due to its low mortality, its morbidity is high, reducing the prosperity of affected populations. Of note, in the last few years, complicated P. vivax clinical malaria has been reported around the world (Tjitra et al. 2008, Anstey et al. 2012), including the Brazilian Amazon area (Alexandre et al. 2010, Lacerda et al. 2012).
Finally, in the Amazon, local P. vivax populations are extremely genetically diverse and also show substantial genetic differentiation among populations (Ferreira et al. 2007, Rezende et al. 2009, 2010, Orjuela-Sanchez et al. 2010, Sousa et al. 2010). Beyond the complexity of parasite population, it has been proposed that asymptomatic parasite carriage and massive environmental changes - that affect vector abundance and behaviour - are major contributors to malaria transmission in the epidemiologically diverse areas across the Amazon Basin (da Silva-Nunes et al. 2012). It may explain why malaria has proved so difficult to control in the Amazon Basin, where transmission rates remain far below those recorded in tropical Africa.
The rationale behind P. vivax Duffy binding protein (PvDBP) as a blood-stage vaccine candidate - The invasion of red blood cells (RBCs) by Plasmodium merozoites - an essential event in the life cycle of all malaria parasites - is a highly complex, multistep process that is dependent on a cascade of specific molecular interactions (Gaur et al. 2004). Despite this complexity, time-lapse microscopy of live parasites demonstrates that parasite entry into RBCs is a rapid process that is completed, on average, within 30 s after primary contact of the merozoite (Gilson & Crabb 2009). This multistep invasion process requires coordinated activities of host cell attachment, reorientation placing the apical end of the parasite adjacent to the erythrocyte membrane and active penetration of the host cell. Central to this process is the establishment of a structure called a tight or moving junction, which forms a tight connection between the invading parasite and host cell membranes (Aikawa et al. 1978). For P. vivax, the formation of this irreversible junction is mediated by the PvDBP, a protein of approximately 140 kDa localised in merozoite apical organelles called micronemes (Wertheimer & Barnwell 1989, Adams et al. 1990, 1992, Fang et al. 1991). During invasion, PvDBP is secreted from the micronemes and binds to its cognate host receptor on the reticulocyte surface, the Duffy antigen/receptor for chemokines (DARC) (Wertheimer & Barnwell 1989, Adams et al. 1990). Although P. vivax can infect and cause disease in DARC-negative individuals (Ryan et al. 2006, Cavasini et al. 2007, Menard et al. 2013), this situation seems to be rare and/or occur only in specific areas; so far, no other alternative ligand for P. vivax binding to reticulocytes has been identified, which makes the PvDBP one of the most promising P. vivax vaccine targets.
PvDBP belongs to the Duffy binding-like erythrocyte-binding protein (DBL-EBP) family, which encompasses other micronemal proteins, such as the DBP of the simian malaria parasite Plasmodium knowlesi and the P. falciparum ligands EBA-175, EBA-181/JESEBL and EBA-140/BAEBL (Sim et al. 1990, Adams et al. 1992, 2001, Mayer et al. 2001, Gilberger et al. 2003). The members of the DBL-EBP family share a similar gene structure and this homology was used to define six extracellular regions (I-VI) followed by a type I transmembrane domain and a short cytoplasmic tail (Adams et al. 1992, 2001). Common to all EBPs are the two cysteine-rich domains (regions II and VI) in the ectodomain, with the erythrocyte ligand-binding domain lying within region II (DBPII) (Fig. 1). In P. falciparum, the EBPs and the reticulocyte-binding-like protein homologues play an important role in phenotypic variation, allowing different parasite isolates to utilise alternative erythrocyte invasion pathways (Orlandi et al. 1992, Rayner et al. 2000, Triglia et al. 2001, Duraisingh et al. 2003). Until recently, the gene that encodes PvDBP was described as single copy gene (Carlton et al. 2008). However, new whole genome sequences from field isolates provides evidence for a duplication of the dbp gene in P. vivax (Menard et al. 2013). Interestingly, the frequency of the dbp duplication was highest in geographical regions where the highest frequencies of P. vivax-infected Duffy-negative people were reported. These data suggest that PvDBP is rapidly evolving, possibly in response to constraints imposed by erythrocyte DARC-negativity in some human populations.
Fig. 1. : schematic drawing of the Plasmodium vivax Duffy binding protein gene structure. Exons are represented as blocks and drawn to scale. Exon 1 encodes a peptide signal sequence, exon 3 encodes a transmembrane domain and exons 4 and 5 encode a cytoplasmic domain. Exon 2 encodes a large protein domain that contains six regions (Roman numerals) as defined by amino acid (aa) sequence identity to Duffy binding-like erythrocyte-binding protein of other Plasmodium species (Adams et al. 1992). The erythrocyte-binding domain lies in the 5’ cysteine-rich region (region II) and the critical binding residues have been mapped to a 170-aa stretch between cysteines 4-7 (Ranjan & Chitnis 1999).

The PvDBP ligand domain (DBPII) is a 330 amino acid (aa) region characterised by 12 conserved cysteine residues (Chitnis & Miller 1994). The critical binding residues have been mapped to a central 170-aa stretch that includes cysteines 4-7 (Ranjan & Chitnis 1999, Singh et al. 2003, VanBuskirk et al. 2004b, Hans et al. 2005, Batchelor et al. 2011, Bolton & Garry 2011, Sampath et al. 2013). This is the minimal domain responsible for binding to DARC-positive human reticulocytes. In 2006, the structure of the P. knowlesi DBL domain was determined by X-ray crystallography and characterised as an all-helical, monomeric module containing 12 helices spread over three distinct subdomains (SD1-SD3) that are stabilised by intra-SD disulfide bridges (Singh et al. 2006). Essential and invariant residues required for recognition of DARC on human erythrocytes were identified within a region on SD2 (Singh et al. 2003, 2006, VanBuskirk et al. 2004b, Hans et al. 2005). Recently, the crystal structure of DBPII was elucidated and a model was proposed of receptor recognition through PvDBP dimerisation upon receptor binding, leading to the formation of a complex composed of two PvDBP and two DARC molecules (Batchelor et al. 2011). Despite the conserved nature of regions spanning the DARC-binding groove and dimer interface, many residues in DBPII are variable and these polymorphisms map to multiple non-functional regions of the protein (Tsuboi et al. 1994, Ampudia et al. 1996, Xainli et al. 2000, Kho et al. 2001, VanBuskirk et al. 2004b, Sousa et al. 2006, Gosi et al. 2008, Babaeekho et al. 2009, Batchelor et al. 2011, Premaratne et al. 2011, Chenet et al. 2012, Ju et al. 2012, 2013). Some of these naturally-occurring polymorphisms flank critical residues and it is suggested that protective antibodies that target the functional regions in DBPII lead to disruption of dimerisation and/or prevention of receptor binding (Batchelor et al. 2011). This pattern of diversity is consistent with strong immune selection pressure on DBPII and suggests that allelic variation is an important mechanism of immune evasion (Tsuboi et al. 1994, Cole-Tobian & King 2003, Martinez et al. 2004, Sousa et al. 2010). Although PvDBP is a vital ligand for blood-stage infection, its use for vaccine development poses significant challenges. These include: (i) brief exposure of PvDBP to the host immune system, due to its micronemal location (Adams et al. 1990) and the rapid kinetics of parasite invasion (Dvorak et al. 1975, Gilson & Crabb 2009) and (ii) polymorphisms in PvDBP, which seem to be critical for the evasion of host immune response (VanBuskirk et al. 2004a), as further discussed below.
Naturally-acquired antibodies against PvDBP - The goal in developing PvDBP as a vaccine directed against asexual blood-stage of P. vivax is to elicit an antibody response that inhibits parasite adhesion to DARC-positive human reticulocytes and thereby abrogate merozoite invasion. Unfortunately, the available data on the functional properties of anti-PvDBP antibodies in human populations are still limited (Michon et al. 2000, Ceravolo et al. 2008, King et al. 2008, Souza-Silva et al. 2010, Chootong et al. 2012), partly due to constraints on performing in vitro functional assays in the absence of a continuous culture method for P. vivax blood-stages (Ntumngia et al. 2012). Consequently, many field studies of immunity to PvDBP have focused on measuring antibodies to recombinant antigens, but paid less attention to approaches that evaluate functionally important immune mechanisms.
Anti-PvDBP IgG antibodies measured by conventional serology - Given the limitations of performing functional assays, ELISAs using different recombinant PvDBP proteins have been useful in evaluating the level of PvDBP IgG antibodies in different endemic populations. Using conventional serology, we demonstrated that PvDBP is naturally immunogenic in the Amazon area and that the proportions of PvDBP IgG-positive subjects increased with exposure to malaria, reaching a peak in those subjects with long-term exposure in the Amazon (Table) (Ceravolo et al. 2005). Of importance, this study provided an additional insight by demonstrating for the first time that cumulative exposure is a determinant that acts independently of host age in the generation of anti-PvDBP IgG response. In fact, we demonstrated that each additional year of exposure to malaria increased the probability of having anti-PvDBP IgG antibodies by 2% (Souza-Silva et al. 2010). While these previous studies included subjects who were migrants from malaria-free areas of Brazil - whose ages did not correlate with exposure to malaria - further studies were carried-out with native populations of the Amazon area (Kano et al. 2012). In this area, a well-established frontier settlement located in AM, a significant proportion of the community (50%) had acquired anti-PvDBP antibodies, with the subject’s age being the only strong predictor of seropositivity to PvDBP. Together, these data reinforce the variety of malaria transmission patterns in communities from the Amazon area.
TABLE. Summary of previous studies reporting naturally-acquired IgG antibody responses to Plasmodium vivax Duffy binding protein (PvDBP)a .
| Region [city/state (n)] | Time of malaria exposure [months (m) or years (y)] | Frequency of responders - ELISA (%) | Recombinant PvDBPb | Subjects with inhibitory antibodiesc (%) | References |
|---|---|---|---|---|---|
| Latin America (low transmission) | |||||
| Brazilian Amazon | |||||
| Belém/Pará (36) | < 1 m | 14 | II-IV | 0 | Ceravolo et al. (2005, 2008) |
| Terra Nova do Norte/Mato Grosso (47) | 10 y | 38 | - | 12/87 (14) | |
| Apiacás/Mato Grosso (37) | 17 y | 65 | - | ||
| Buritis/Rondônia (30) | 7 y | 43 | II | NA | Tran et al. (2005) |
| Colina/Rondônia (87) | 21 y | 70 | - | NA | |
| Ribeirinha/Rondônia (177) | 28 y | 70 | - | NA | |
| Several areas/Pará (220) | - | 44.5 | II | NA | Barbedo et al. (2007) |
| Granada/Acre (≈360) | 14 y | 9-18.6 | II-IV | 14-16/50 (28-32) | Souza-Silva et al. (2010) |
| Rio Pardo/Amazonas (432) | 19 y | 49.5 | II-IV | NA | Kano et al. (2012) |
| Colombia | |||||
| Pacific Coast/Valle (92) | - | 40.2 | II-IV | NA | Michon et al. (1998) |
| Pacific Coast/Chocó and Valle (214) | - | ≈8-12 | II | NA | Herrera et al. (2005) |
| Caribbean Coast/Antioquia (185) | > 5 y | 9 | II | NA | Maestre et al. (2010) |
| Western Pacific | |||||
| Papua New Guinea (high transmission) | |||||
| Madang (100) | - | 60 | II-IV | NA | Fraser et al. (1997) |
| Liksul/Madang (551) | - | 80 | II-IV and II | NA | Xainli et al. (2003) |
| Mugil and Megiar/Madang (206) | - | 72 | II | 58/206 (28) | King et al. (2008), Cole-Tobian et al. (2009) |
a: previous studies not included here: Ceravolo et al. (2009), performed with samples from a small outbreak outside endemic area of Brazil, and Chootong et al. (2012), which only analysed acutely infected patients; b: region of the recombinant PvDBP used in the studies [the PvDBP Sal-1 variant was used in all studies; other variants were used by Ceravolo et al. (2008) and King et al. (2008)]; c: > 50% inhibitory activity; NA: not available.
So far, few studies have investigated anti-PvDBP antibody response in Latin America and data are still restricted to endemic areas of Brazil and Colombia (Table). In general, the pattern of antibody response described in these studies corroborated our data that anti-PvDBP antibodies increase with exposure to P. vivax. In addition, PvDBP antibodies seem to be biased toward the cytophilic subclasses IgG1 and IgG3 (Tran et al. 2005, Maestre et al. 2010).
From our experience in the Amazon area, it has become evident that PvDBP has relatively low immunogenicity, similar to what has been described in other epidemiological contexts. For example, in the Colombian Pacific coastal region, an area of unstable malaria transmission and mainly composed of Afro-Colombian individuals, as well in the Caribbean Coast, less than 40% of the total number of patients sampled responded to PvDBP (Michon et al. 1998, Herrera et al. 2005, Maestre et al. 2010). This antibody response profile is quite different from those described in highly endemic areas for malaria, such as Papua New Guinea (PNG), where antibody responses to PvDBP seem to be much more common (60-80%) (Table) and reach a plateau at ages of 15 years and older (Xainli et al. 2003). In the latter endemic regions, the proportion of individuals developing T cell responses to PvDBP increased rapidly within the first four years of life such that by five-nine years of age 80% of children responded (Xainli et al. 2002). The cellular response against PvDBP has not yet been evaluated in Latin America.
DBPII binding inhibitory antibodies (BIAbs) - Currently, few reports have examined functional antibodies in malaria-exposed populations and most of them were carried-out in the highly endemic areas of PNG (Michon et al. 2000, King et al. 2008), which might not be representative of many P. vivax endemic regions. Consequently, our goal was to characterise the DBPII BIAbs response in individuals from an area of markedly unstable malaria transmission, as found in the Brazilian Amazon (Table). Our results indicate that long-term exposure to malaria in the Brazilian Amazon elicits DBP-specific antibodies that inhibit the binding of different DBPII variants to erythrocytes (Ceravolo et al. 2008, Souza-Silva et al. 2010). However, this inhibitory activity was detected only in one third of malaria-exposed subjects, with a moderate correlation between DBPII BIAbs and ELISA anti-PvDBP antibodies.
Despite significant epidemiological and host/parasite genetic differences between the Amazon Basin and PNG, the relatively low frequency of DBPII BIAbs described among long-term residents of the Amazon area (~30%) was also found in PNG (Table). In fact, in the latter highly endemic area, less than 10% of immune children had acquired a high level of DBPII BIAbs (King et al. 2008). It is very intriguing that in the Amazon and PNG, the DBPII BIAbs response was remarkably stable over time (King et al. 2008, Souza-Silva et al. 2010). These findings imply that although the majority of people naturally-exposed to P. vivax do not develop antibodies that inhibit the DBPII-DARC interaction, once they are acquired, these inhibitory antibodies seem to be stable under continuous exposure to malaria transmission.
A plausible explanation for the low immunogenicity of PvDBP is the fact that this protein is localised in the micronemes until the beginning of the process of erythrocyte invasion by merozoites (Adams et al. 1990). As a consequence of its brief exposure, the host immune system seems to have little opportunity to produces an efficient antibody response. However, the “just-in-time” hypothesis of PvDBP exposure (Singh et al. 2006) does not completely explain the large proportion of individuals who remain unresponsive to PvDBP after prolonged exposure to malaria. The reasons for this are not clear, but may relate to the complexity of the immune response, in terms of genetic diversity of both the parasite and human populations. With regard to the genetic diversity of the parasite, several studies now indicate the existence of strain-specificity in the natural immune response against PvDBP (Ceravolo et al. 2009, Cole-Tobian et al. 2009, Chootong et al. 2012). In a study conducted during a malaria outbreak outside of the Brazilian endemic area, we demonstrated that the majority of responders had developed inhibitory antibodies against the homologous DBPII sequence identified in the outbreak isolate (Ceravolo et al. 2009). These findings provided the first clear evidence that naturally-acquired inhibitory antibodies to DBPII are biased towards a specific allele in individuals with no previous exposure to malaria infection (Ceravolo et al. 2009). In Thailand, an area of low unstable transmission of P. vivax, the inhibitory antibody responses against DBPII also correlated with homologous protection (Chootong et al. 2012). Similarly, a profile of strain-specific inhibitory activity was frequently observed among asymptomatic children from PNG (Cole-Tobian et al. 2009).
Even though the current data clearly demonstrated strain-specific immunity, we and others have also described strain-transcendent inhibitory responses to DBPII (King et al. 2008, Souza-Silva et al. 2010). In the Brazilian Amazon area, only individuals with long-term exposure to malaria (roughly 20 years) acquired DBPII BIAbs against different DBPII variants (Ceravolo et al. 2008). Similarly, only 9% of asymptomatic children residing in a P. vivax hyperendemic area had acquired a significant anti-PvDBP inhibitory antibody response that transcended strain-specificity (King et al. 2008). These findings highlight the complexity of the immune responses to DBPII, which includes both a strain-specific and strain-transcending component (Cole-Tobian et al. 2009).
Besides PvDBP allelic variation, recent evidence suggests that host genetic polymorphisms might also affect humoral immunity against PvDBP (Maestre et al. 2010, King et al. 2011). The most relevant finding was that DARC allelic polymorphisms are thought to affect the ability of anti-PvDBP antibodies to block parasite invasion (King et al. 2011). DARC is encoded by two codominant allele groups FY*A and FY*B, which differ by a single point mutation. Of particular interest is that we recently demonstrated that DBPII inhibitory antibody responses were approximately 50% lower in FY*A/FY*A and FY*B/FY*B homozygous individuals when compared with individuals heterozygous for FY*A or FY*B alleles, suggesting a gene-dosage effect (Souza-Silva et al. 2014). Due to the relevance of these findings for vaccines now in development, it would be pertinent to investigate whether such an association exists in other P. vivax malaria endemic countries. In this context, it would be relevant to determine if PvDBP non-responsiveness could be major histocompatibility complex-linked. So far, only a single study investigated the association between human leukocyte antigen (HLA) class II and PvDBP antibodies (Storti-Melo et al. 2012). Although that study was unable to demonstrate an association between HLA type and PvDBP antibodies, the low number of individuals studied (PvDBP IgG sera, n = 48) precludes any solid conclusions about the highly polymorphic HLA class II and PvDBP antibodies. In this context, follow-up studies are currently in progress in the Amazon area (LH Carvalho, unpublished observations).
Genetic diversity of PvDBP in the Amazon area - Analysis of the genetic variability of P. vivax isolates from the field revealed that the PvDBP binding domain (region II, DBPII) is highly polymorphic, similar to most blood-stage malaria vaccine candidates, which may facilitate parasite escape from host immune detection. Based on field-studies, it seems clear that this extensive diversity might hamper vaccine development, since variable residues could alter immune recognition of the protein (Ceravolo et al. 2009, Cole-Tobian et al. 2009). Thus, for development of PvDBP-based vaccine it is important to assess the levels of DBPII genetic diversity among P. vivax field isolates. The first data on the variability of PvDBP were obtained for isolates of the parasite from PNG and Colombia (Tsuboi et al. 1994, Ampudia et al. 1996). Further analyses showed that most of the variable residues lie within a 170-aa region of DBPII and, at that time, there was no evidence that variable residues could interfere with erythrocyte receptor recognition (Xainli et al. 2000). Later, the same authors confirmed the influence of DBPII polymorphism as a mechanism of immune evasion (VanBuskirk et al. 2004a). Aiming to contribute to efforts towards vaccine development, we initially investigated in the Brazilian Amazon those eight single nucleotide polymorphisms previously suggested to be involved in the immune recognition of PvDBP (VanBuskirk et al. 2004a). Brazilian P. vivax isolates showed variability in almost all residues and a strong association of residues at positions 417, 424 and 437, suggesting a synergistic functional effect of these aas, possibly constituting a discontinuous epitope in DBPII (Sousa et al. 2006) (Fig. 2). The synergistic effect of the duo 417 and 424, including residue 419 were reinforced by the findings that they are part of a block of high linkage disequilibrium and show a clear signature of positive natural selection among Brazilian isolates (Sousa et al. 2010). In accordance, these three residues were found to be part of an in silico DBPII predicted epitope that is recognised by DBPII BIAbs present in sera from PNG (Chootong et al. 2010, Sousa et al. 2010).
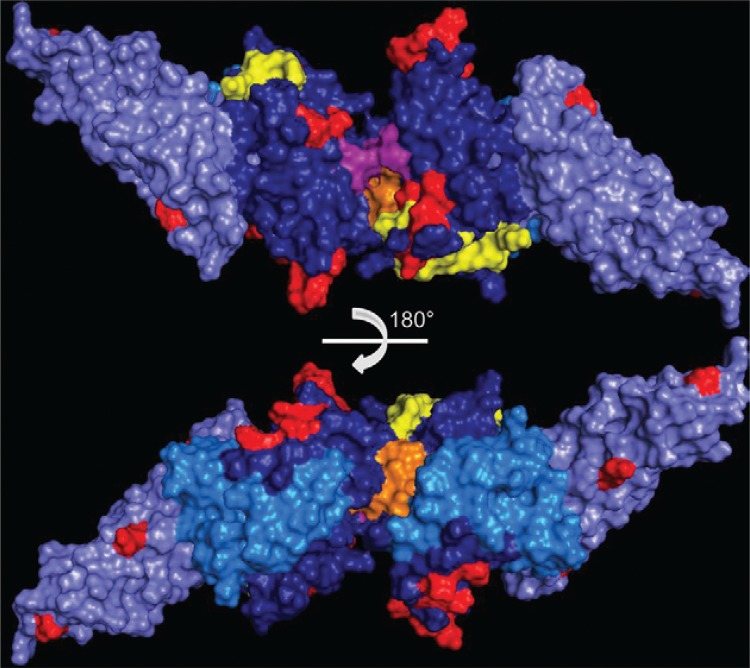
Fig. 2. : Duffy binding protein (DBPII) dimeric structure. Subdomain (SD1) (sky blue), SD2 (dark blue) and SD3 (light blue). Critical binding residues are coloured yellow (Asn291, Tyr295, Asn296, Lys297, Phe299 and Val365, Lys366, Lys367, Arg368, Leu369, Phe373, Ile374, Ile376) (VanBuskirk et al. 2004b, , Hans et al. 2005, , Bolton & Garry 2011, , Sampath et al. 2013). Residues that form the putative sulfotyrosine-binding pocket at the dimer interface are coloured purple (Lys273, Arg274 and Gln356). Residues that make contact creating the dimeric architecture are coloured orange (Phe267, Leu270, Ile277, Tyr278, Val282, Tyr363 and Arg274, Glu249) (Batchelor et al. 2011). Polymorphic residues in Brazilian isolates are coloured red (N305N, R308S, L333F, K371E, N375D, R378R, G384D, E385K, K386N, H390R, S398T, T404R, N417K, I419M, L424I, W437R, I464I, Q486E, I503K) (Sousa et al. 2006, 2010, 2010). Images modelled in PyMol on the DBPII dimer structure (Batchelor et al. 2011).
Subsequently, we compared the DBPII diversity among P. vivax isolates from different countries worldwide, including samples from the Brazilian Amazon. In general, high levels of haplotype diversity were observed among isolates independent of the malaria endemicity (Nóbrega de Sousa et al. 2011). These data on the nucleotide diversity of DBPII also provided evidence that recombination plays an important role in determining the haplotype structure of DBPII (Martinez et al. 2004, Sousa et al. 2010). In addition, it was possible to demonstrate that natural selection acts differentially across the DBPII sequence, with neutrally evolving codons as well as codons evolving under diversifying selection (Cole-Tobian & King 2003, Martinez et al. 2004, Sousa et al. 2010). Of importance, positive natural selection preferentially acts on epitopes in DBPII, which also have greater nucleotide diversity (Cole-Tobian & King 2003, Sousa et al. 2010, Ju et al. 2012, 2013). This is in agreement with the hypothesis that immune selection is the major evolutionary force that drives the generation of new PvDBP variants. In accordance with these findings, field-studies carried-out in different malaria endemic areas showed that naturally-acquired inhibitory antibodies to DBPII are biased towards a specific allele (Ceravolo et al. 2009, Cole-Tobian et al. 2009). This is relevant because current vaccine development is based only on the DBPII haplotype of the P. vivax laboratory-adapted strain Sal-1 (Yazdani et al. 2004, Arevalo-Herrera et al. 2005, Moreno et al. 2008). However, this haplotype has been found at only a low frequency in most Amazon regions and, of note, it seems to be largely restricted to some geographical areas of the world (Nóbrega de Sousa et al. 2011).
Recently, crystallographic and functional studies allowed the identification of the structural regions of PvDBP targeted by inhibitory antibodies (Chootong et al. 2010, Batchelor et al. 2011, Sampath et al. 2013). The findings suggest that some epitopes recognised by BIAbs lie close to the predicted DARC interaction site, suggesting that cause disruption of PvDBP dimerisation and/or prevention of PvDBP receptor binding (Batchelor et al. 2011, Sampath et al. 2013). Despite cumulative knowledge about DBPII structure, there is limited understanding about the molecular basis for protection and immune evasion. Research to date has underlined the importance of the SD2 of DBPII for DARC binding and immune selection (Singh et al. 2006, Batchelor et al. 2011, Sampath et al. 2013). Nevertheless, Siddiqui et al. (2012) showed that the SD3 is also important for binding to DARC. Furthermore, inhibitory murine monoclonal antibodies mapped to SD3 recognise epitopes that are strain transcendent (Siddiqui et al. 2012). Since this SD is relatively conserved it could form the basis of a strain-transcending vaccine against P. vivax.
Aiming towards universal, strain-transcending anti-PvDBP immunity, the current strategies of vaccine development have been focusing on immune responses against more conserved DBPII epitopes. An interesting approach was based on the hypothesis that the polymorphic residues, which are not functionally important for erythrocyte-binding, but flank the receptor binding motif of DBPII, comprise variant epitopes that tend to divert the immune response away from more conserved epitopes (Ntumngia & Adams 2012). In this respect, these authors demonstrated that immunisation with a synthetic antigen lacking the immunodominant DBPII variant epitopes enabled the development of immune responses towards the more conserved neutralising epitopes, which are the potential targets of a strain-transcending immunity. Another promising vaccine approach should be immunisation with a multiple component vaccine representing the major DBPII haplotypes (Ntumngia et al. 2013). In this context, a preliminary analysis of the worldwide DBPII sequences allowed us to determine that seven haplotypes should be the minimum number of haplotypes to be included in a DBP-based vaccine of broad coverage (Nóbrega de Sousa et al. 2011).
Concluding remarks - PvDBP is likely to be exposed on the merozoite surface during invasion, enabling it to bind to its receptor and, thus, making it accessible to serum antibodies. While measuring antibodies to recombinant PvDBP by ELISA is a simple and robust procedure widely used in human population studies, it provides little functional information when used alone. As a consequence, field-studies on PvDBP immune response should include assays to evaluate the functional proprieties of anti-PvDBP antibodies. Cross-sectional and cohort studies carried-out in the Amazon area provide evidence that the majority of people naturally-exposed to P. vivax do not develop antibodies that inhibit the DBPII-DARC interaction. Nevertheless, once they are acquired, these inhibitory antibodies seem to be stable under continuous exposure to malaria transmission. These results are intriguing and seem to be a common phenomenon in other endemic areas, including those highly endemic areas of PNG, where most individuals have developed clinical immunity to malaria (King et al. 2008). The results presented here also provide strong evidence that DARC interaction site and epitopes on PvDBP have sufficient overlap for antibodies to disrupt dimerisation and/or inhibit binding (Batchelor et al. 2011, Sampath et al. 2013) and provide support for the role of allelic diversity in anti-PvDBP immune responses. Of note, while our findings point to allelic variation eliciting a strain-specific immunity, individuals with long-term exposure in the Amazon area acquired DBPII antibodies that inhibit in vitro binding of different DBPII variants to erythrocytes. Future challenges include understanding why only few malaria exposed-individuals develop an immune response able to inhibit DBPII-DARC interaction and to establish whether PvDBP inhibitory immune response predicts partial protection from infection and/or disease, as suggested by others (King et al. 2008). We hope that these findings from unstable malaria transmission areas contribute to current efforts towards vaccine development and may facilitate future clinical trials in areas of unstable malaria transmission.
ACKNOWLEDGEMENTS
To Luke Baton, for reviewing the paper.
Footnotes
Financial support: FAPEMIG, CNPq, FIOCRUZ (FIOCRUZ/CNPq/PAPES), PRONEX-Malária/CNPq/DECIT/MS
REFERENCES
- Adams J, Hudson D, Torii M, Ward G, Wellems T, Aikawa M, Miller L. The Duffy receptor family of Plasmodium knowlesi is located within the micronemes of invasive malaria merozoites. Cell. 1990;63:141–153. doi: 10.1016/0092-8674(90)90295-p. [DOI] [PubMed] [Google Scholar]
- Adams JH, Blair PL, Kaneko O, Peterson DS. An expanding ebl family of Plasmodium falciparum. Trends Parasitol. 2001;17:297–299. doi: 10.1016/s1471-4922(01)01948-1. [DOI] [PubMed] [Google Scholar]
- Adams JH, Sim BKL, Dolan SA, Fang XD, Kaslow DC, Miller LH. A family of erythrocyte binding-proteins of malaria parasites. Proc Natl Acad Sci USA. 1992;89:7085–7089. doi: 10.1073/pnas.89.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aikawa M, Miller LH, Johnson J, Rabbege J. Erythrocyte entry by malarial parasites. A moving junction between erythrocyte and parasite. J Cell Biol. 1978;77:72–82. doi: 10.1083/jcb.77.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre MA, Ferreira CO, Siqueira AM, Magalhães BL, Mourão MP, Lacerda MV, Alecrim MG. Severe Plasmodium vivax malaria, Brazilian Amazon. Emerg Infect Dis. 2010;16:1611–1614. doi: 10.3201/eid1610.100685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves FP, Durlacher RR, Menezes MJ, Krieger H, Silva LH, Camargo EP. Plasmodium vivax and Plasmodium falciparum infections in native Amazonian populations. Am J Trop Med Hyg. 2002;66:641–648. doi: 10.4269/ajtmh.2002.66.641. [DOI] [PubMed] [Google Scholar]
- Ampudia E, Patarroyo MA, Patarroyo ME, Murillo LA. Genetic polymorphism of the Duffy receptor binding domain of Plasmodium vivax in Colombian wild isolates. Mol Biochem Parasitol. 1996;78:269–272. doi: 10.1016/s0166-6851(96)02611-4. [DOI] [PubMed] [Google Scholar]
- Anstey NM, Douglas NM, Poespoprodjo JR, Price RN. Plasmodium vivax: clinical spectrum, risk factors and pathogenesis. Adv Parasitol. 2012;80:151–201. doi: 10.1016/B978-0-12-397900-1.00003-7. [DOI] [PubMed] [Google Scholar]
- Arevalo-Herrera M, Castellanos A, Yazdani SS, Shakri AR, Chitnis CE, Dominik R, Herrera S. Immunogenicity and protective efficacy of recombinant vaccine based on the receptor-binding domain of the Plasmodium vivax Duffy binding protein in Aotus monkeys. Am J Trop Med Hyg. 2005;73:25–31. doi: 10.4269/ajtmh.2005.73.5_suppl.0730025. [DOI] [PubMed] [Google Scholar]
- Babaeekho L, Zakeri S, Djadid ND. Genetic mapping of the Duffy binding protein (DBP) ligand domain of Plasmodium vivax from unstable malaria region in the Middle East. Am J Trop Med Hyg. 2009;80:112–118. [PubMed] [Google Scholar]
- Barbedo MB, Ricci R, Jimenez MCS, Cunha MG, Yazdani SS, Chitnis CE, Rodrigues MM, Soares IS. Comparative recognition by human IgG antibodies of recombinant proteins representing three asexual erythrocytic stage vaccine candidates of Plasmodium vivax. Mem Inst Oswaldo Cruz. 2007;102:335–339. doi: 10.1590/s0074-02762007005000040. [DOI] [PubMed] [Google Scholar]
- Barbieri AF, Sawyer DO. Heterogeneity of malaria prevalence in alluvial gold mining areas in northern Mato Grosso state, Brazil. Cad Saude Publica. 2007;23:2878–2886. doi: 10.1590/s0102-311x2007001200009. [DOI] [PubMed] [Google Scholar]
- Batchelor JD, Zahm JA, Tolia NH. Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nat Struct Mol Biol. 2011;18:908–914. doi: 10.1038/nsmb.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton MJ, Garry RF. Sequence similarity between the erythrocyte binding domain 1 of the Plasmodium vivax Duffy binding protein and the V3 loop of HIV-1 strain MN reveals binding residues for the Duffy antigen receptor for chemokines. 45Virol J. 2011;8 doi: 10.1186/1743-422X-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd MF, Kitchen SF. Simultaneous inoculation with Plasmodium vivax and Plasmodium falciparum. Am J Trop Med Hyg. 1937;17:855–861. [Google Scholar]
- Camargo EP, Alves F, Silva LH. Symptomless Plasmodium vivax infections in native Amazonians. Lancet. 1999a;353:1415–1416. doi: 10.1016/s0140-6736(99)00941-1. [DOI] [PubMed] [Google Scholar]
- Camargo LM, Ferreira MU, Krieger H, Camargo EP, Silva LP. Unstable hypoendemic malaria in Rondônia (western Amazon Region, Brazil): epidemic outbreaks and work-associated incidence in an agro-industrial rural settlement. Am J Trop Med Hyg. 1994;51:16–25.. doi: 10.4269/ajtmh.1994.51.16. [DOI] [PubMed] [Google Scholar]
- Camargo LM, Noronha E, Salcedo JM, Dutra AP, Krieger H, Silva LH, Camargo EP. The epidemiology of malaria in Rondônia (western Amazon Region, Brazil): study of a riverine population. Acta Trop. 1999b;72:1–11. doi: 10.1016/s0001-706x(98)00082-5. [DOI] [PubMed] [Google Scholar]
- Carlton J, Adams J, Silva J, Bidwell S, Lorenzi H, Caler E, Crabtree J, Angiuoli S, Merino E, Amedeo P, Cheng Q, Coulson R, Crabb B, Del Portillo H, Essien K, Feldblyum T, Fernandez-Becerra C, Gilson P, Gueye A, Guo X, Kang’a S, Kooij T, Korsinczky M, Meyer E, Nene V, Paulsen I, White O, Ralph S, Ren Q, Sargeant T, Salzberg S, Stoeckert C, Sullivan S, Yamamoto M, Hoffman S, Wortman J, Gardner M, Galinski M, Barnwell J, Fraser-Liggett C. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavasini CE, Mattos LC, Couto AAD, Bonini-Omingos CR, Valencia SH, Neiras WCD, Alves RT, Rossit ARB, Castilho L, Machado RLD. Plasmodium vivax infection among Duffy antigen-negative individuals from the Brazilian Amazon Region: an exception? T Roy Soc Trop Med H. 2007;101:1042–1044. doi: 10.1016/j.trstmh.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Ceravolo IP, Bruna-Romero O, Braga EM, Fontes CJF, Brito CFA, Souza JM, Krettli AU, Adams JH, Carvalho LH. Anti-Plasmodium vivax Duffy binding protein antibodies measure exposure to malaria in the Brazilian Amazon. Am J Trop Med Hyg. 2005;72:675–681. [PubMed] [Google Scholar]
- Ceravolo IP, Sanchez BA, Sousa TN, Guerra BM, Soares IS, Braga EM, McHenry AM, Adams JH, Brito CF, Carvalho LH. Naturally acquired inhibitory antibodies to Plasmodium vivax Duffy binding protein are short-lived and allele-specific following a single malaria infection. Clin Exp Immunol. 2009;156:502–510. doi: 10.1111/j.1365-2249.2009.03931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceravolo IP, Souza-Silva FA, Fontes CJF, Braga EM, Madureira AP, Krettli AU, Souza JM, Brito CFA, Adams JH, Carvalho LH. Inhibitory properties of the antibody response to Plasmodium vivax Duffy binding protein in an area with unstable malaria transmission. Scand J Immunol. 2008;67:270–278. doi: 10.1111/j.1365-3083.2007.02059.x. [DOI] [PubMed] [Google Scholar]
- Chenet SM, Tapia LL, Escalante AA, Durand S, Lucas C, Bacon DJ. Genetic diversity and population structure of genes encoding vaccine candidate antigens of Plasmodium vivax. 68Malar J. 2012;11 doi: 10.1186/1475-2875-11-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis CE, Miller LH. Identification of the erythrocyte binding domains of Plasmodium vivax and Plasmodium knowlesi proteins involved in erythrocyte invasion. J Exp Med. 1994;180:497–506. doi: 10.1084/jem.180.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chootong P, Ntumngia F, Vanbuskirk K, Xainli J, Cole-Tobian J, Campbell C, Fraser T, King C, Adams J. Mapping epitopes of the Plasmodium vivax Duffy binding protein with naturally acquired inhibitory antibodies. Infect Immun. 2010;78:1089–1095. doi: 10.1128/IAI.01036-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chootong P, Panichakul T, Permmongkol C, Barnes SJ, Udomsangpetch R, Adams JH. Characterization of inhibitory anti-Duffy binding protein II immunity: approach to Plasmodium vivax vaccine development in Thailand. PLoS ONE. 2012;7: doi: 10.1371/journal.pone.0035769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole-Tobian J, King CL. Diversity and natural selection in Plasmodium vivax Duffy binding protein gene. Mol Biochem Parasitol. 2003;127:121–132. doi: 10.1016/s0166-6851(02)00327-4. [DOI] [PubMed] [Google Scholar]
- Cole-Tobian JL, Michon P, Biasor M, Richards JS, Beeson JG, Mueller I, King CL. Strain-specific Duffy binding protein antibodies correlate with protection against infection with homologous compared to heterologous Plasmodium vivax strains in Papua New Guinean children. Infect Immun. 2009;77:4009–4017. doi: 10.1128/IAI.00158-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Nunes M, Codeço C, Malafronte R, Silva N, Juncansen C, Muniz P, Ferreira M. Malaria on the Amazonian frontier: transmission dynamics, risk factors, spatial distribution and prospects for control. Am J Trop Med Hyg. 2008;79:624–635. [PubMed] [Google Scholar]
- Silva-Nunes M, Moreno M, Conn JE, Gamboa D, Abeles S, Vinetz JM, Ferreira MU. Amazonian malaria: asymptomatic human reservoirs, diagnostic challenges, environmentally driven changes in mosquito vector populations and the mandate for sustainable control strategies. Acta Trop. 2012;121:281–291. doi: 10.1016/j.actatropica.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro MC, Monte-Mor RL, Sawyer DO, Singer BH. Malaria risk on the Amazon frontier. Proc Natl Acad Sci USA. 2006:2452–2457. doi: 10.1073/pnas.0510576103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro MC, Sawyer DO, Singer BH. Spatial patterns of malaria in the Amazon: implications for surveillance and targeted interventions. Health Place. 2007;13:368–380. doi: 10.1016/j.healthplace.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Duraisingh M, Triglia T, Ralph S, Rayner J, Barnwell J, McFadden G, Cowman A. Phenotypic variation of Plasmodium falciparum merozoite proteins directs receptor targeting for invasion of human erythrocytes. EMBO J. 2003;22:1047–1057. doi: 10.1093/emboj/cdg096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak JA, Miller LH, Whitehouse WC, Shiroishi T. Invasion of erythrocytes by malaria merozoites. Science. 1975;187:748–750. doi: 10.1126/science.803712. [DOI] [PubMed] [Google Scholar]
- Fang X, Kaslow D, Adams J, Miller L. Cloning of the Plasmodium vivax Duffy receptor. Mol Biochem Parasitol. 1991;44:125–132. doi: 10.1016/0166-6851(91)90228-x. [DOI] [PubMed] [Google Scholar]
- Ferreira MU, Karunaweera ND, Silva-Nunes M, Silva NS, Wirth DF, Hartl DL. Population structure and transmission dynamics of Plasmodium vivax in rural Amazonia. J Infect Dis. 2007;195:1218–1226. doi: 10.1086/512685. [DOI] [PubMed] [Google Scholar]
- Fraser T, Michon P, Barnwell JW, Noe AR, AlYaman F, Kaslow DC, Adams JH. Expression and serologic activity of a soluble recombinant Plasmodium vivax Duffy binding protein. Infect Immun. 1997;65:2772–2777. doi: 10.1128/iai.65.7.2772-2777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur D, Mayer D, Miller L. Parasite ligand-host receptor interactions during invasion of erythrocytes by Plasmodium merozoites. Int J Parasitol. 2004;34:1413–1429. doi: 10.1016/j.ijpara.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Gilberger TW, Thompson JK, Triglia T, Good RT, Duraisingh MT, Cowman AF. A novel erythrocyte binding antigen-175 paralogue from Plasmodium falciparum defines a new trypsin-resistant receptor on human erythrocytes. J Biol Chem. 2003;278:14480–14486. doi: 10.1074/jbc.M211446200. [DOI] [PubMed] [Google Scholar]
- Gilson PR, Crabb BS. Morphology and kinetics of the three distinct phases of red blood cell invasion by Plasmodium falciparum merozoites. Int J Parasitol. 2009;39:91–96. doi: 10.1016/j.ijpara.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Gosi P, Khusmith S, Khalambaheti T, Lanar D, Schaecher K, Fukuda M, Miller S. Polymorphism patterns in Duffy binding protein among Thai Plasmodium vivax isolates. 112Malar J. 2008;7 doi: 10.1186/1475-2875-7-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans D, Pattnaik P, Bhattacharyya A, Shakri AR, Yazdani SS, Sharma M, Choe H, Farzan M, Chitnis CE. Mapping binding residues in the Plasmodium vivax domain that binds Duffy antigen during red cell invasion. Mol Microbiol. 2005;55:1423–1434. doi: 10.1111/j.1365-2958.2005.04484.x. [DOI] [PubMed] [Google Scholar]
- Herrera S, Gomez A, Vera O, Vergara J, Valderrama-Aguirre A, Maestre A, Mendez F, Wang RB, Chitnis CE, Yazdani SS, Arevalo-Herrera M. Antibody response to Plasmodium vivax antigens in Fy-negative individuals from the Colombian Pacific Coast. Am J Trop Med Hyg. 2005;73:44–49. doi: 10.4269/ajtmh.2005.73.44. [DOI] [PubMed] [Google Scholar]
- Ju HL, Kang JM, Moon SU, Bahk YY, Cho PY, Sohn WM, Park YK, Park JW, Kim TS, Na BK. Genetic diversity and natural selection of Duffy binding protein of Plasmodium vivax Korean isolates. Acta Trop. 2013;125:67–74. doi: 10.1016/j.actatropica.2012.09.016. [DOI] [PubMed] [Google Scholar]
- Ju HL, Kang JM, Moon SU, Kim JY, Lee HW, Lin K, Sohn WM, Lee JS, Kim TS, Na BK. Genetic polymorphism and natural selection of Duffy binding protein of Plasmodium vivax Myanmar isolates. 60Malar J. 2012;11 doi: 10.1186/1475-2875-11-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano FS, Sanchez BA, Sousa TN, Tang ML, Saliba J, Oliveira FM, Nogueira PA, Gonçalves AQ, Fontes CJ, Soares IS, Brito CF, Rocha RS, Carvalho LH. Plasmodium vivax Duffy binding protein: baseline antibody responses and parasite polymorphisms in a well-consolidated settlement of the Amazon Region. Trop Med Int Health. 2012;17:989–1000. doi: 10.1111/j.1365-3156.2012.03016.x. [DOI] [PubMed] [Google Scholar]
- Kho W, Chung J, Sim E, Kim D, Chung W. Analysis of polymorphic regions of Plasmodium vivax Duffy binding protein of Korean isolates. Korean J Parasitol. 2001;39:143–150.. doi: 10.3347/kjp.2001.39.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C, Michon P, Shakri A, Marcotty A, Stanisic D, Zimmerman P, Cole-Tobian J, Mueller I, Chitnis C. Naturally acquired Duffy binding protein-specific binding inhibitory antibodies confer protection from blood-stage Plasmodium vivax infection. Proc Natl Acad Sci USA. 2008;105:8363–8368. doi: 10.1073/pnas.0800371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CL, Adams JH, Xianli J, Grimberg BT, McHenry AM, Greenberg LJ, Siddiqui A, Howes RE, Silva-Nunes M, Ferreira MU, Zimmerman PA. Fy(a)/Fy(b) antigen polymorphism in human erythrocyte Duffy antigen affects susceptibility to Plasmodium vivax malaria. Proc Natl Acad Sci USA. 2011;108:20113–20118. doi: 10.1073/pnas.1109621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerda MV, Mourao MP, Alexandre MA, Siqueira AM, Magalhaes BM, Martinez-Espinosa FE, Filho FS, Brasil P, Ventura AM, Tada MS, Couto VS, Silva AR, Silva RS, Alecrim MG. Understanding the clinical spectrum of complicated Plasmodium vivax malaria: a systematic review on the contributions of the Brazilian literature. 12Malar J. 2012;11 doi: 10.1186/1475-2875-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladeia-Andrade S, Ferreira MU, Carvalho ME, Curado I, Coura JR. Age-dependent acquisition of protective immunity to malaria in riverine populations of the Amazon Basin of Brazil. Am J Trop Med Hyg. 2009;80:452–459. [PubMed] [Google Scholar]
- Loiola CC, Silva CJ, Tauil PL. Malaria control in Brazil: 1965 to 2001. Rev Panam Salud Publica. 2002;11:235–244. doi: 10.1590/s1020-49892002000400005. [DOI] [PubMed] [Google Scholar]
- Maestre A, Muskus C, Duque V, Agudelo O, Liu P, Takagi A, Ntumngia FB, Adams JH, Sim KL, Hoffman SL, Corradin G, Velez ID, Wang R. Acquired antibody responses against Plasmodium vivax infection vary with host genotype for Duffy antigen receptor for chemokines (DARC). PLoS ONE. 2010;5: doi: 10.1371/journal.pone.0011437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques A, Pinheiro E, Souza A. Dispersion of cases of malaria in Brazil. Rev Bras Malariol Doencas Trop. 1986;38:51–75. [PubMed] [Google Scholar]
- Martinez O, Suarez CF, Cardenas PP, Patarroyo MA. Plasmodium vivax Duffy binding protein: a modular evolutionary proposal. Parasitology. 2004;128:353–366. doi: 10.1017/s0031182003004773. [DOI] [PubMed] [Google Scholar]
- Mayer DC, Kaneko O, Hudson-Taylor DE, Reid ME, Miller LH. Characterization of a Plasmodium falciparum erythrocyte-binding protein paralogous to EBA-175. Proc Natl Acad Sci USA. 2001;98:5222–5227. doi: 10.1073/pnas.081075398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard D, Chan ER, Benedet C, Ratsimbasoa A, Kim S, Chim P, Do C, Witkowski B, Durand R, Thellier M, Severini C, Legrand E, Musset L, Nour BY, Mercereau-Puijalon O, Serre D, Zimmerman PA. Whole genome sequencing of field isolates reveals a common duplication of the Duffy binding protein gene in Malagasy Plasmodium vivax strains. PLoS Negl Trop Dis. 2013;7: doi: 10.1371/journal.pntd.0002489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendis K, Sina BJ, Marchesini P, Carter R. The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg. 2001;64:97–106. doi: 10.4269/ajtmh.2001.64.97. [DOI] [PubMed] [Google Scholar]
- Michon P, Fraser T, Adams JH. Naturally acquired and vaccine-elicited antibodies block erythrocyte cytoadherence of the Plasmodium vivax Duffy binding protein. Infect Immun. 2000;68:3164–3171. doi: 10.1128/iai.68.6.3164-3171.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michon PA, Arevalo-Herrera M, Fraser T, Herrera S, Adams JH. Serologic responses to recombinant Plasmodium vivax Duffy binding protein in a Colombian village. Am J Trop Med Hyg. 1998;59:597–599. doi: 10.4269/ajtmh.1998.59.597. [DOI] [PubMed] [Google Scholar]
- Moreno A, Caro-Aguilar I, Yazdani S, Shakri A, Lapp S, Strobert E, McClure H, Chitnis C, Galinski M. Preclinical assessment of the receptor-binding domain of Plasmodium vivax Duffy binding protein as a vaccine candidate in rhesus macaques. Vaccine. 2008;26:4338–4344. doi: 10.1016/j.vaccine.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nóbrega de Sousa T, Carvalho LH, Brito CF. Worldwide genetic variability of the Duffy binding protein: insights into Plasmodium vivax vaccine development. PLoS ONE. 2011;6: doi: 10.1371/journal.pone.0022944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntumngia FB, Adams JH. Design and immunogenicity of a novel synthetic antigen based on the ligand domain of the Plasmodium vivax Duffy binding protein. Clin Vaccine Immunol. 2012;19:30–36. doi: 10.1128/CVI.05466-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntumngia FB, King CL, Adams JH. Finding the sweet spots of inhibition: understanding the targets of a functional antibody against Plasmodium vivax Duffy binding protein. Int J Parasitol. 2012;42:1055–1062. doi: 10.1016/j.ijpara.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntumngia FB, Schloegel J, McHenry AM, Barnes SJ, George MT, Kennedy S, Adams JH. Immunogenicity of single versus mixed allele vaccines of Plasmodium vivax Duffy binding protein region II. Vaccine. 2013;31:4382–4388. doi: 10.1016/j.vaccine.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira-Ferreira J, Lacerda MV, Brasil P, Ladislau JL, Tauil PL, Daniel-Ribeiro CT. Malaria in Brazil: an overview. 115Malar J. 2010;9 doi: 10.1186/1475-2875-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjuela-Sanchez P, Karunaweera ND, Silva-Nunes M, Silva NS, Scopel KK, Goncalves RM, Amaratunga C, Sa JM, Socheat D, Fairhust RM, Gunawardena S, Thavakodirasah T, Galapaththy GL, Abeysinghe R, Kawamoto F, Wirth DF, Ferreira MU. Single-nucleotide polymorphism, linkage disequilibrium and geographic structure in the malaria parasite Plasmodium vivax: prospects for genome-wide association studies. 65BMC Genet. 2010;11 doi: 10.1186/1471-2156-11-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi P, Klotz F, Haynes J. A malaria invasion receptor, the 175-kilodalton erythrocyte binding antigen of Plasmodium falciparum recognizes the terminal Neu5Ac(alpha 2-3)Gal- sequences of glycophorin A. J Cell Biol. 1992;116:901–909. doi: 10.1083/jcb.116.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premaratne PH, Aravinda BR, Escalante AA, Udagama-Randeniya PV. Genetic diversity of Plasmodium vivax Duffy binding protein II (PvDBPII) under unstable transmission and low intensity malaria in Sri Lanka. Infect Genet Evol. 2011;11:1327–1339. doi: 10.1016/j.meegid.2011.04.023. [DOI] [PubMed] [Google Scholar]
- Ranjan A, Chitnis C. Mapping regions containing binding residues within functional domains of Plasmodium vivax and Plasmodium knowlesi erythrocyte-binding proteins. Proc Natl Acad Sci USA. 1999;96:14067–14072. doi: 10.1073/pnas.96.24.14067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JR, Stoute JA, Amon J, Dunton RF, Mtalib R, Koros J, Owour B, Luckhart S, Wirtz RA, Barnwell JW, Rosenberg R. Evidence for transmission of Plasmodium vivax among a Duffy antigen negative population in western Kenya. Am J Trop Med Hyg. 2006;75:575–581. [PubMed] [Google Scholar]
- Rayner JC, Galinski MR, Ingravallo P, Barnwell JW. Two Plasmodium falciparum genes express merozoite proteins that are related to Plasmodium vivax and Plasmodium yoelii adhesive proteins involved in host cell selection and invasion. Proc Natl Acad Sci USA. 2000;97:9648–9653. doi: 10.1073/pnas.160469097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezende A, Tarazona-Santos E, Fontes C, Souza J, Couto A, Carvalho L, Brito C. Microsatellite loci: determining the genetic variability of Plasmodium vivax. Trop Med Int Health. 2010;15:718–726. doi: 10.1111/j.1365-3156.2010.02535.x. [DOI] [PubMed] [Google Scholar]
- Rezende AM, Tarazona-Santos E, Couto AD, Fontes CJ, Souza JM, Carvalho LH, Brito CF. Analysis of genetic variability of Plasmodium vivax isolates from different Brazilian Amazon areas using tandem repeats. Am J Trop Med Hyg. 2009;80:729–733. [PubMed] [Google Scholar]
- Sampath S, Carrico C, Janes J, Gurumoorthy S, Gibson C, Melcher M, Chitnis CE, Wang R, Schief WR, Smith JD. Glycan masking of Plasmodium vivax Duffy binding protein for probing protein binding function and vaccine development. PLoS Pathog. 2013;9: doi: 10.1371/journal.ppat.1003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui AA, Xainli J, Schloegel J, Carias L, Ntumngia F, Shoham M, Casey JL, Foley M, Adams JH, King CL. Fine specificity of Plasmodium vivax Duffy binding protein binding engagement of the Duffy antigen on human erythrocytes. Infect Immun. 2012;80:2920–2928. doi: 10.1128/IAI.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sim BK, Orlandi PA, Haynes JD, Klotz FW, Carter JM, Camus D, Zegans ME, Chulay JD. Primary structure of the 175K Plasmodium falciparum erythrocyte binding antigen and identification of a peptide which elicits antibodies that inhibit malaria merozoite invasion. J Cell Biol. 1990;111:1877–1884. doi: 10.1083/jcb.111.5.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Hora R, Belrhali H, Chitnis C, Sharma A. Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain. Nature. 2006;439:741–744. doi: 10.1038/nature04443. [DOI] [PubMed] [Google Scholar]
- Singh SK, Singh AP, Pandey S, Yazdani SS, Chitnis CE, Sharma A. Definition of structural elements in Plasmodium vivax and P. knowlesi Duffy binding domains necessary for erythrocyte invasion. Biochem J. 2003;374:193–198. doi: 10.1042/BJ20030622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa TN, Ceravolo IP, Fontes CJF, Couto A, Carvalho LH, Brito CFA. The pattern of major polymorphisms in the Duffy binding protein ligand domain among Plasmodium vivax isolates from the Brazilian Amazon area. Mol Biochem Parasitol. 2006;146:251–254. doi: 10.1016/j.molbiopara.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Sousa TN, Tarazona-Santos EM, Wilson DJ, Madureira AP, Falcao PR, Fontes CJ, Gil LH, Ferreira MU, Carvalho LH, Brito CF. Genetic variability and natural selection at the ligand domain of the Duffy binding protein in Brazilian Plasmodium vivax populations. 334Malar J. 2010;9 doi: 10.1186/1475-2875-9-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza-Silva FA, Silva-Nunes M, Sanchez BA, Ceravolo IP, Malafronte RS, Brito CF, Ferreira MU, Carvalho LH. Naturally acquired antibodies to Plasmodium vivax Duffy binding protein (DBP) in Brazilian Amazon. Am J Trop Med Hyg. 2010;82:185–193. doi: 10.4269/ajtmh.2010.08-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza-Silva FA, Torres LM, Santos-Alves JR, Tang ML, Sanchez BA, Sousa TN, Fontes CJ, Nogueira PA, Rocha RS, Brito CF, Adams JH, Kano FS, Carvalho LH. Duffy antigen receptor for chemokine (DARC) polymorphisms and its involvement in acquisition of inhibitory anti-Duffy binding protein II (DBPII) immunity. PLoS ONE. 2014;9: doi: 10.1371/journal.pone.0093782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storti-Melo LM, Costa DR, Souza-Neiras WC, Cassiano GC, Couto VS, Povoa MM, Soares Ida S, Carvalho LH, Arevalo-Herrera M, Herrera S, Rossit AR, Cordeiro JA, Mattos LC, Machado RL. Influence of HLA-DRB-1 alleles on the production of antibody against CSP, MSP-1, AMA-1 and DBP in Brazilian individuals naturally infected with Plasmodium vivax. Acta Trop. 2012;121:152–155. doi: 10.1016/j.actatropica.2011.10.009. [DOI] [PubMed] [Google Scholar]
- Tauil P, Daniel-Ribeiro C. Some aspects of epidemiology of malaria in Brazil. Rev Iber Parasitol. 1998;58:163–167. [Google Scholar]
- Tjitra E, Anstey N, Sugiarto P, Warikar N, Kenangalem E, Karyana M, Lampah D, Price R. Multidrug-resistant Plasmodium vivax associated with severe and fatal malaria: a prospective study in Papua, Indonesia. PLoS Med. 2008;5: doi: 10.1371/journal.pmed.0050128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TM, Oliveira-Ferreira J, Moreno A, Santos F, Yazdani SS, Chitnis CE, Altman JD, Meyer EVS, Barnwell JW, Galinski MR. Comparison of IgG reactivities to Plasmodium vivax merozoite invasion antigens in a Brazilian Amazon population. Am J Trop Med Hyg. 2005;73:244–255. [PubMed] [Google Scholar]
- Triglia T, Thompson J, Caruana S, Delorenzi M, Speed T, Cowman A. Identification of proteins from Plasmodium falciparum that are homologous to reticulocyte binding proteins in Plasmodium vivax. Infect Immun. 2001;69:1084–1092. doi: 10.1128/IAI.69.2.1084-1092.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi T, Kappe SHI, Alyaman F, Prickett MD, Alpers M, Adams JH. Natural variation within the principal adhesion domain of the Plasmodium vivax Duffy binding protein. Infect Immun. 1994;62:5581–5586. doi: 10.1128/iai.62.12.5581-5586.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuskirk KM, Cole-Tobian JL, Baisor M, Sevova ES, Bockarie M, King CL, Adams JH. Antigenic drift in the ligand domain of Plasmodium vivax Duffy binding protein confers resistance to inhibitory antibodies. J Infect Dis. 2004a;190:1556–1562. doi: 10.1086/424852. [DOI] [PubMed] [Google Scholar]
- VanBuskirk KM, Sevova E, Adams JH. Conserved residues in the Plasmodium vivax Duffy binding protein ligand domain are critical for erythrocyte receptor recognition. Proc Natl Acad Sci USA. 2004b;101:15754–15759. doi: 10.1073/pnas.0405421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheimer S, Barnwell J. Plasmodium vivax interaction with the human Duffy blood group glycoprotein: identification of a parasite receptor-like protein. Exp Parasitol. 1989;69:340–350. doi: 10.1016/0014-4894(89)90083-0. [DOI] [PubMed] [Google Scholar]
- WHO - World Health Organization World Malaria Report 2012. 2012 who.int/malaria/publications/world_malaria_report_2012/en/
- WHO - World Health Organization World Malaria Report 2013. 2013 who.int/malaria/publications/world_malaria_report_2013/en/
- Xainli J, Adams J, King C. The erythrocyte binding motif of Plasmodium vivax Duffy binding protein is highly polymorphic and functionally conserved in isolates from Papua New Guinea. Mol Biochem Parasitol. 2000;111:253–260. doi: 10.1016/s0166-6851(00)00315-7. [DOI] [PubMed] [Google Scholar]
- Xainli J, Baisor M, Kastens W, Bockarie M, Adams J, King C. Age-dependent cellular immune responses to Plasmodium vivax Duffy binding protein in humans. J Immunol. 2002;169:3200–3207. doi: 10.4049/jimmunol.169.6.3200. [DOI] [PubMed] [Google Scholar]
- Xainli J, Cole-Tobian JL, Baisor M, Kastens W, Bockarie M, Yazdani SS, Chitnis CE, Adams JH, King CL. Epitope-specific humoral immunity to Plasmodium vivax Duffy binding protein. Infect Immun. 2003;71:2508–2515. doi: 10.1128/IAI.71.5.2508-2515.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdani SS, Shakri AR, Mukherjee P, Baniwal SK, Chitnis CE. Evaluation of immune responses elicited in mice against a recombinant malaria vaccine based on Plasmodium vivax Duffy binding protein. Vaccine. 2004;22:3727–3737. doi: 10.1016/j.vaccine.2004.03.030. [DOI] [PubMed] [Google Scholar]