Abstract
Anophelines harbour a diverse microbial consortium that may represent an extended gene pool for the host. The proposed effects of the insect microbiota span physiological, metabolic and immune processes. Here we synthesise how current metagenomic tools combined with classical culture-dependent techniques provide new insights in the elucidation of the role of the Anopheles-associated microbiota. Many proposed malaria control strategies have been based upon the immunomodulating effects that the bacterial components of the microbiota appear to exert and their ability to express anti-Plasmodium peptides. The number of identified bacterial taxa has increased in the current “omics” era and the available data are mostly scattered or in “tables” that are difficult to exploit. Published microbiota reports for multiple anopheline species were compiled in an Excel® spreadsheet. We then filtered the microbiota data using a continent-oriented criterion and generated a visual correlation showing the exclusive and shared bacterial genera among four continents. The data suggested the existence of a core group of bacteria associated in a stable manner with their anopheline hosts. However, the lack of data from Neotropical vectors may reduce the possibility of defining the core microbiota and understanding the mosquito-bacteria interactive consortium.
Keywords: Anopheles, core microbiota, paratransgenesis, Plasmodium, metagenomics, databank
Insects are by far the most diverse and abundant clade of metazoans (Basset et al. 2012). Their success is partly attributable to the beneficial members of their associated microbiome. The term microbiota refers to the microbial communities that stably or transiently colonise the insect epithelium and their intracellular compartments and target organs. These communities vary from bacteria to viruses, yeasts and protists. The bacterial component of this ecosystem is the most studied and characterised (Ng et al. 2011b, Gendrin & Christophides 2013, Minard et al. 2013). These symbiotic microbiotas or consortia are beneficial to their insect hosts in many ways (Dillon & Dillon 2004, Azambuja et al. 2005, Engel & Moran 2013), including dietary supplementation, enhancement of digestive mechanisms, tolerance of environmental perturbations, protection from parasites (Degnan & Moran 2008, Weiss & Aksoy 2011) and pathogens (Nartey et al. 2013) and maintenance and/or enhancement of host immune system homeostasis. Furthermore, the absence or elimination of the microbial fauna and even the modification of its composition can reduce the fitness of the host insect (Toft & Andersson 2010). This influence of the microbiota on its host has been referred as the “extended phenotype” (Koch & Schmid-Hempel 2011), which can range from mutualism to parasitism. Notably, bacterial communities appear to be associated with their invertebrate hosts in a highly specific manner that resembles the host phylogeny (Bauman 2005). How these associations are determined remains unclear, but species-specific antimicrobial peptides likely shape these co-evolving species-specific bacterial communities. The species that are vital to the host are stably associated with it even under different environmental conditions (Franzenburg et al. 2013).
Studies of microorganisms living in insect guts have recently increased considerably. In the last decade, multiple relevant studies have been published, ranging from diversity screening using metagenomic surveys (Baumann 2005, Lindh et al. 2005, Carpi et al. 2011, Djadid et al. 2011, Lindh & Lehane 2011, Ng et al. 2011a, b, Chavshin et al. 2012) to molecular studies of the interaction of gut bacteria with the host immune system and the response to infection (Azambuja et al. 2005, Chouaia et al. 2010, Cirimotich et al. 2011, Boissière et al. 2012). In addition to the native gut microbiota, symbiotic bacteria occupy other key organs, tissues or structures (e.g., salivary glands, ovaries, haemolymph). Primary endosymbionts, which are transmitted vertically from mother to offspring, are found in bacteriocytes, specialised cells that provide nutrients to bacteria (Wernegreen 2012). Secondary endosymbionts are transmitted horizontally, vertically or via the environment (Eleftherianos et al. 2013) and can be located in the haemocoel (Wernegreen 2012). Both surface-colonising consortia and intracellular symbiotic bacteria live under the pressure of an active immune system and therefore must devise strategies that enable them to withstand the adverse effects of the host immune response (Douglas 2011, Weiss & Aksoy 2011).
An exhaustive review of metagenomic tools or the architecture and dynamics of this microecosystem within Culicidae vectors is outside the scope of this analysis. Recent reviews have covered both of these topics substantially and creatively (Dillon & Dillon 2004, Abdul-Ghani et al. 2012, Suenaga 2012, Engel & Moran 2013, Gendrin & Christophides 2013, Minard et al. 2013, Segata et al. 2013). Rather, we summarise basic topics of this computational field and how its interaction with vector biology has expanded our knowledge of vector mosquitoes and their associated microbiota. From our perspective, computational sciences offer a means to coordinate, analyse and visualise the growing body of anopheline microbiota-related data in an organised manner that is readily available and understandable to the community. For instance, we propose to continue the effort of Minard et al. (2013), who generated a databank encompassing the reported microbiota data pertaining to several mosquito vectors. We could use this databank as a foundation for Anopheles-oriented expansion and, depending on the filtering criteria applied, the identification of underlying patterns.
First, we will briefly explore the concept of metagenomics and summarise recent advancements in malaria vector control based on the exploitation of the anopheline microbiota and its impact on vector competence traits. The use of metataxonomic computational tools has enabled the identification of insect-associated microbial species. Strategies have been developed based on these species to block malarial transmission via the expression of anti-Plasmodium peptides, immunomodulation or via induced dysbiosis (Coutinho-Abreu et al. 2010, Ricci et al. 2012, Wang & Jacobs-Lorena 2013).
Due to the vast amount of taxonomic profiling data provided by next-generation sequencing (NGS), the categorisation and filtering parameters selected for databank searching are critical for a better understanding of the mosquito as a holobiont (Brucker & Bordenstein 2013).
On metagenomics and insect vectors - Metagenomics emerged as a derivative of classic microbial genomics; the key difference between these disciplines is that metagenomics bypasses the requirement for obtaining pure cultures for sequencing (Olsen et al. 1986, Glass et al. 2010, Kim et al. 2013). Genomic information can be obtained directly from microbial communities in their natural habitats, enabling these communities to be studied in a concerted manner to describe their species composition and even predict their potential genomic functions and metabolic capabilities (Wooley et al. 2010, Huttenhower et al. 2012, Williamson & Yooseph 2012).
Metagenomics refers to the sequencing of whole-community DNA or a range of techniques; tools and approaches that attempt to identify microorganisms, genes and variants, metabolic pathways or functions within a community (Segata et al. 2013). The sequencing of whole-community DNA can be complemented by metatranscriptomics (cDNA sequencing), metaproteomics and metabolomics (Wilmes & Bond 2006, Gilbert & Hughes 2011, McCarthy et al. 2011). Both metagenomics and metatranscriptomics aim to elucidate the genomic composition, diversity and potential within and across microenvironments by applying culture-independent techniques that include targeting rRNA gene sequences (16S for bacteria, 18S for eukaryotes and internal transcribed spacers for fungi) and whole-metagenomic shotgun sequencing (Huson et al. 2007, Segata et al. 2013).
Determining the identity and proportions of the members of a microbial community is the typical primary objective of amplicon sequencing investigations such as 16S rRNA gene surveys (Suenaga 2012). Such surveys are currently the predominant approach of insect vector metagenomics. According to Segata et al. (2013), when performing metagenomic profiling, shotgun-sequencing approaches can provide an information profile comparable to that obtained from 16S rRNA-based surveys, often with better resolution. Taxonomic profiling can be achieved by either the de novo binning of microbial sequences (based upon intrinsic sequence properties) or relying on information from sequenced microbial genome databases (extrinsic information) (Thomas et al. 2012). Taking into consideration sequencing coverage, read quality, read length and experimental design, a high-throughput shotgun-driven survey may enable the user to assess the diversity of genes and biochemical pathways that are represented in the consortium (Abubucker et al. 2012).
As NGS platforms develop, our potential to generate genomic data has increased exponentially (Ansorge 2009); benchmarking has thus gained relevance in providing guidance to experimental biologists confronted with a myriad of bioinformatic tools, which include GAAS (Angly et al. 2009), Glimmer (Delcher et al. 2007), CARMA3 (Gerlach & Stoye 2011), MG-RAST (Glass et al. 2010), MEGAN (Huson et al. 2007) and TAMER (Jiang et al. 2012). When designing experiments that encompass metagenomic data generation, it is imperative to consider factors such as sampling techniques, DNA/RNA extraction protocols, sequencing platforms, assembly, taxonomic binning and gene annotation tools, statistical analysis and data/meta-data sharing formats (Wommack et al. 2008, Tanenbaum et al. 2010, Wooley et al. 2010, Thomas et al. 2012). The availability of standardised procedures (Field et al. 2008, Tanenbaum et al. 2010) and platforms for data storage and sharing are increasingly important to ensure that the output of individual projects can be assessed, shared and compared (Thomas et al. 2012).
In this meta-omic era, NGS screening assays are used to determine the diversity of microorganisms and viruses residing in arthropod vectors of medical importance. This information enables human health agencies and research groups to monitor endemic infections, perform real-time surveillance of newly emerging zoonotic pathogens, identify etiological agents and determine how these agents associate with and within their hosts (Bishop-Lilly et al. 2010, Carpi et al. 2011, Ng et al. 2011a, b, Mokili et al. 2012, Hall-Mendelin et al. 2013).
Modulating effects of bacteria on the Plasmodium-anopheline interaction - Due to their importance as malarial vectors, anopheline mosquitoes have been the targets of multiple microbiota profiling efforts (Gendrin & Christophides 2013). Underlying these efforts is the knowledge that bacteria living in the midgut can modulate the response of mosquitoes toward Plasmodium infection (Dong et al. 2009, Eappen et al. 2013). Thus, these bacteria have the potential to block infections and be used as genetic transformation vehicles (Pumpuni et al. 1996, Weiss & Aksoy 2011, Ricci et al. 2012).
Midgut-colonising microbial organisms have been identified in both laboratory-cultivated and field-collected Anopheles mosquitoes. For example, field populations of Anopheles gambiae and Anopheles funestus were found to contain 16 bacterial species spanning 14 genera (Lindh et al. 2005). Laboratory populations of An. gambiae and Anopheles stephensi have been found to harbour a wide variety of bacteria, particularly those belonging to the genera Asaia, Enterobacter, Mycobacterium, Sphingomonas, Serratia and Chryseobacterium (Favia et al. 2007, Dong et al. 2009, Rani et al. 2009).
Bacterial components of the anopheline midgut microenvironment modulate vectorial competence initially by inhibiting the sporogonic development of malaria parasites when infection is established (Pumpuni et al. 1993, 1996, Gonzalez-Ceron et al. 2003, Dong et al. 2009, Cirimotich et al. 2011). In addition, Gram-negative, but not Gram-positive bacteria inhibit oocyst formation in whole or in part (Pumpuni et al. 1993, 1996). Evidence regarding the influence of midgut microbiota in the life cycle of parasites has also been found in other insects such as sandflies and tsetse flies (Schlein et al. 1985, Welburn & Maldlin 1999).
Recent studies suggest that Enterobacter species isolated from Anopheles arabiensis from Zambia act directly on Plasmodium falciparum, hindering the development of the parasite in an An. gambiae model. This effect is due to the bacterial production of reactive oxygen species that interfere with the development of the parasite and cause parasite death before invasion of the intestinal epithelium (Cirimotich et al. 2011).
Bacteria colonising the midgut lumen modify the intestinal environment and inhibit the development of parasites by modulating the immune system and inducing the expression rate of immunity genes encoding antimicrobial peptides (Ratcliffe & Whitten 2004, Michel & Kafatos 2005, Meister et al. 2009). These peptides are likely to play key roles in suppressing pathogenic bacteria and modulating the abundance of symbiotic bacteria and may also play a role in the response towards parasite infections (Beard et al. 2001, Boulanger et al. 2004).
The inducible synthesis of nitric oxide metabolites is an important component of the immune response of anopheline mosquitoes to limit the development of Plasmodium parasites (Peterson et al. 2007). Using a comparative genomics approach, Wang et al. (2012) proposed that certain bacterial taxa may induce a reductive environment within the mosquito midgut, thus aiding in the detoxification of reactive oxygen and nitrogen species. This predicted bacterial redox capacity would enable a more aggressive mosquito immune response mediated by nitrogen oxides upon Plasmodium infection.
These observations have motivated interest in exploiting the genomic power of the bacterial passengers within the mosquito or modifying the inner environment of the mosquito midgut to develop novel transmission blocking tools. Briefly, we summarise the results of five examples of such control strategies.
Delivering anti-plasmodium effector molecules into the anopheline midgut - Paratransgenesis can be defined as the set of strategies for the use of engineered symbiotic bacteria from the mosquito midgut to deliver effector genes that encode molecules that inhibit parasite development (Durvasula et al. 1997). A recent literature review by Wang and Jacobs-Lorena (2013) (and references therein) lists known anti-Plasmodium molecules and classifies them as parasite killers, parasite-interacting, mosquito midgut or salivary gland epithelia-interacting and immunomodulators.
The potential success of such strategies relies on the fact that most of the mosquito microbiome resides in the same compartment in which the most vulnerable stages of malaria parasite development occur (Whitten et al. 2006); at that key intersection of space and time, bacterial loads increase due to blood meal ingestion. Thus, when expressed, the yield of anti-Plasmodium molecules should increase proportionally with this increase in bacterial load. Several research groups have explored paratransgenesis to interfere with the Plasmodium cycle within the anopheline host. The proposed paratransgenic vectors are discussed below.
Asaia - Bacteria from the genus Asaia have been evaluated as paratransgenic vectors to control malaria transmission (Favia et al. 2007, Damiani et al. 2010). Bacteria from this genus are considered viable candidates due to their stable association with anopheline mosquitoes throughout the body and different life stages; they are also easily cultivated and transformed. Engineered Asaia are capable of colonising laboratory-reared An. stephensi mosquitoes via vertical and horizontal transmission routes. Based on a series of experiments, Capone et al. (2013) reported three key conclusions. First, the physiological changes occurring in the mosquito midgut after an infected or uninfected blood meal had no negative effect and in fact appeared to benefit the Asaia population. Boissière et al. (2012) also observed a strong positive correlation between the presence of Enterobacteriaceae and an infected blood meal. Second, Asaia can act as an immunomodulator within the mosquito by inducing the production of antimicrobial peptides that may interfere with the course of infection, particularly during the invasion of epithelial tissues and translocation to the salivary glands. Asaia may also act as up-regulators of immunity genes. Third and more relevant to genetic manipulation strategies, Capone et al. (2013) demonstrated that modified strains of Asaia were widely spread in the midgut and salivary glands of infected mosquitoes and that they co-localised with Plasmodium parasites. Exogenous recombinant bacteria of this genus replicated and spread within the mosquito and colonised key organs in a sustained manner. When engineered to release anti-Plasmodium peptides, these strains could interfere with the parasite’s life cycle, thus reducing its vectorial competence.
Furthermore, the presence of this genus in laboratory-reared and field-collected An. gambiae from Burkina Faso (Damiani et al. 2010), Anopheles maculipennis from Italy (Favia et al. 2007) and in laboratory-reared Aedes aegypti (Crotti et al. 2009) suggests that Asaia cross-colonises species and genera. The bacterial association with widely distributed vectors could signify that, if used as a biological control strategy, Asaia would bypass genetic barriers among reproductively isolated mosquito populations (Damiani et al. 2010).
These are promising findings that support the efforts to identify a bacterial symbiont that negatively impacts vectorial competence without detrimental effects on the mosquito holobiont. However, Djadid et al. (2011) and Rani et al. (2009) did not identify bacteria from the genus Asaia in field-collected or laboratory-reared Asian malaria vectors (An. stephensi and An. maculipennis from Iran and India) using culture-dependent or culture-independent methods. Although it is possible that the predominant bacterial taxa in these anopheline species may have changed during the transition from their natural habitat to a laboratory colony (Gendrin & Christophides 2013), their apparent absence in field-collected Asian vectors could hinder the use of this model. The colony from which the main observations of these studies have been generated was established approximately two decades ago at the University of Camerino in Italy (1985-1988) (Favia et al. 2007, Damiani et al. 2008). What are the implications of a non-existent natural and steady relationship between this Acetobacteraceae and field-collected Asian anophelines? Both Djadid et al. (2011) and Rani et al. (2009) discuss the possibility of this absence being a “rounded zero” observation due to “low coverage” of the NGS data. Furthermore, the size of the continent and the ecological niches in which these two species are distributed across Asia and Eurasia in the case of the maculipennis subgroup (Di Luca et al. 2009, Sinka et al. 2010, 2012) could partly explain the absence of this genus in the sampled mosquitoes. It was not until 2013 that Manguin et al. reported Asaia in Southeast Asia anophelines (non-An. stephensi mosquitoes from Thailand and Vietnam). The intention of this narrative is to emphasise the need for a data repository to record such events and ease their interpretation upon publication as well as what we consider a fundamental step in vector metagenomics - the need for coordinated efforts to produce technically homogeneous and comparable data across continents, species or tested conditions regardless of the metataxonomic profiling technique. If Asaia is to become a successful component of the bacterial malaria control arsenal, further testing to determine if other relevant anophelines harbour it naturally (or in a laboratory model) would be beneficial. Neotropical anophelines are of particular interest and have been largely neglected when developing such strategies. There are no records of this genus in American anophelines.
Serratia - Strategies for controlling malaria through the genetic manipulation of midgut bacterial symbionts primarily involve mechanisms that act by killing Plasmodium ookinete/oocysts in the mosquito midgut, the “location” in which the cycle within the mosquito undergoes a severe bottleneck (Wang & Jacobs-Lorena 2013). However, resistance of the Plasmodium parasite to effector molecules may arise during paratransgenesis within the mosquito because the principal mode of action is to attack and kill the pathogen itself (Bando et al. 2013).
Serratia marcescens HB3, which was isolated from laboratory-reared An. stephensi mosquitoes, inhibits Plasmodium development within the mosquito midgut by interrupting ookinete invasion through the midgut epithelial cells. Phenotypic variation at the cellular and structural levels was observed and directly correlated with the ability to induce resistance against Plasmodium invasion (Bando et al. 2013).
To evaluate whether the intraspecific diversity of S. marcescens is associated with vectorial competency of human malaria vectors in endemic areas, Bando et al. (2013) profiled the microbiota in the midgut of wild An. gambiae mosquitoes collected in Burkina Faso. Six strains of S. marcescens were identified based on their 16S rRNA sequences. To determine if the isolated strains were able to influence Plasmodium development within the mosquito midgut, they were tested in the An. stephensi-Plasmodium berghei model. A significant inverse correlation between cell length and oocyst number was recorded for five different Serratia strains.
A blocking strategy denying parasite entry into the epithelial cells could be effective for Plasmodium development inhibition. This would broaden the arsenal of biological control strategies and enable these strategies to be used in synergy. Finally, the authors of this study noted that the S. marcescens HB3 strain must be continuously introduced to the vector to exert this action. This would indicate that there may be active stress factors, such as innate immunity, that modulate the dynamics of Serratia populations within the enclosed environment of the midgut.
Pantoea - The natural mosquito symbiont Pantoea can cross-colonise several mosquito species and is readily transformed and cultured; thus, Pantoea has been proposed for paratransgenic applications (Bisi & Lampe 2011, Djadid et al. 2011).
A new strategy to deliver anti-Plasmodium effector molecules into the mosquito midgut was recently described. Wang et al. (2012) engineered a Pantoea agglomerans strain secreting five different antimalaria proteins in the mosquito midgut. They observed suppression of the development of P. falciparum and P. berghei in their respective mosquito models (An. gambiae and An. stephensi) of up to 98% and 83%, respectively. The authors stressed that the data collected on prevalence are more important than the inhibition of oocyst formation. The authors observed a dramatic reduction of P. falciparum-infected mosquitoes when scorpine or (EPIP)4 were the expressed anti-Plasmodium effectors (from 90% in controls to 14% and 18%, respectively). This reduction represents an 80-84% transmission blocking potential. The authors aim to reduce the probability of resistant parasite selection by combining multiple effector proteins. In addition, the anti-Plasmodium effects of the tested molecules were equally effective when tested in An. gambiae (African vector) and An. stephensi (Asian vector). Thus, paratransgenesis is promising as a universal control strategy because the effectiveness of the molecules tested in this study was not influenced by the mosquito species. Introducing engineered Plasmodium agglomerans strains into multiple species in the wild and their establishment as dominant members of the midgut ecosystem is a future logistical challenge and an interesting field within micro-ecology studies.
The potential role of viruses - There is little information regarding the association of viruses with anophelines and the available information primarily pertains to the use of viruses as tools to control malaria. An early claim on the use of viruses to control Plasmodium infections was made by Bird et al. (1972), who reported the presence of cytoplasmic polyhedrosis virus in adult An. stephensi during the sporogonic phase of infection with P. bergei yoeli. They associated the presence of the virus with an observed reduction in the transmission of the parasite in this model and speculated on whether the virus had an impact on both the mosquito and developing parasite. According to their microscopic findings, the majority of the sporozoites exhibited deformed structure and/or vacuolation.
An entomovirus with high specificity towards anophelines would provide the research community with a new set of tools to develop and implement more integral vector eradication policies. Such viruses had not been reported abundantly and remained unexplored until 2008, when Ren et al. discovered and described the An. gambiae densonucleosis virus (AgDNV). This entomovirus has a narrow host range and is readily transmitted between generations in natural populations. AgDNV is transmitted vertically and horizontally from adult mosquitoes to larvae. AgDNV apparently exhibits replication tropism towards adult tissues over larvae and pupae tissue, making it a potential selective biopesticide of adult anophelines. In addition, an exogenous green fluorescent protein (EGFP) gene was expressed by infection of An. gambiae cells and adult mosquitoes with recombinant AgDNV (Ren & Rasgon 2010). The recombinant virions readily infected mosquitoes, expressed EGFP in key tissues involved in the infective cycle of Plasmodium parasites and were transmitted to offspring in a manner similar to that of the wild-type virus. Recombinant AgDNV could be used as a paratransgenic tool to express anti-Plasmodium peptides or insect-specific toxins (Ren & Rasgon 2010).
Wolbachia - Wolbachia is a genus of intracellular α-proteobacteria that has been estimated to infect 40-75 % of all arthropod species and several nematodes (Hilgenboecker et al. 2008, Zug & Hammerstein 2012, Rainey et al. 2014). These bacteria propagate through insects partly through the modulation of host reproduction (Moreira et al. 2009). Wolbachia can inhibit the replication and spread of arbovirus in vector mosquitoes, particularly dengue, Chikungunya, West Nile and yellow fever viruses (Moreira et al. 2009, Bian et al. 2010, Walker et al. 2011, Lu et al. 2012, van den Hurk et al. 2012, Hussain et al. 2013). The mechanism underlying these antiviral effects is unknown (Rainey et al. 2014). Recently, Bourtzis et al. (2014) published a concise review of mosquito-Wolbachia symbiosis, with an emphasis on the Aedes albopictus model. The mechanism underlying Wolbachia-mediated pathogen interference is likely strain-specific and unique to each parasitic challenge. The authors suggest that the involved modes of action could be the priming of the mosquito innate immune system and competition for access to key metabolites (limiting the cellular resources that are required for pathogen and symbiont replication).
An inhibitory effect of Wolbachia towards Plasmodium infection has been reported in anopheline laboratory models. While exploring its use as a dengue virus-blocking agent, Moreira et al. (2009) tested the effect of this Wolbachia endosymbiont in an avian malaria transmission system. The wMelPop-CLA Wolbachia strain reduced Plasmodium gallinaceum infection in Ae. aegypti. Because Anopheles mosquitoes are not naturally infected with Wolbachia, transinfection and ex vivo models have been tested to determine if Wolbachia can establish an infection and replicate within human malaria vectors (Jin et al. 2009, Kambris et al. 2010, Hughes et al. 2011b, 2012). The virulent Wolbachia strain wMelPop can survive and replicate when injected into female An. gambiae adults to establish a somatic avirulent infection (Jin et al. 2009). When Wolbachia (wMelPop and wAlbB strains)-infected An. gambiae mosquitoes were challenged with P. falciparum, a significant reduction in P. falciparum oocyst levels was detected. This somatic bacterial infection disseminated widely throughout the mosquito, reaching the fat body, head, sensory organs and other tissues, with the notable exception of the gut and ovaries (Hughes et al. 2011b).
These results have increased interest in viable techniques for establishing stable transinfections in major human malaria vectors. Although not fully understood, this Wolbachia-infected Anopheles line possesses a reduced ability to maintain Plasmodium transmission due to multiple strain-dependent mechanisms (Hughes et al. 2011a). A Wolbachia-based approach would represent a new vector control option to target dominant malaria vectors, even though human malaria is transmitted globally by multiple, highly specialised and sometimes sympatric species.
Recently, Bian et al. (2013) described the stable establishment of Wolbachia infection in laboratory-reared An. stephensi. The Wolbachia wAlbB strain displayed maternal transmission and high levels of cytoplasmic incompatibility. Furthermore, the seeding of infected female An. stephensi into a population of uninfected mosquitoes resulted in Wolbachia invasion into the uninfected population. In agreement with previous observations of refractoriness towards malaria transmission, the stable infection of wAlb Wolbachia conferred An. stephensi resistance to P. falciparum infection. The immense potential of this development could aid in the gradual reduction of vectorial capacity amongst major anopheline vectors. Nevertheless, it remains unknown if Plasmodium parasites will develop resistance towards the inhibitory mechanisms triggered by the Wolbachia-mosquito association.
The findings of this study and the Wolbachia-mediated strategy were recently analysed by four experts in the field (Killeen et al. 2013), all of whom noted that the observed inhibitory effect is most likely too modest to block malaria transmission in the field. However, they consider this approach a promising biological weapon against malaria transmission if it can be established in other anophelines to induce similar infection outcomes.
Following this line of thought, all applicable possibilities should be explored and assessed. The capacity of Wolbachia endosymbionts to substantially affect Plasmodium infection was also reported by Zelé et al. (2012), who observed that the Wolbachia pipientis strains wPip(SI) and wPip(Mc) naturally found in Culex pipiens protected the mosquitoes from infection by Plasmodium relictum parasites. Interestingly, this research group later attempted to validate this observation in a natural infection model (Zelé et al. 2014). Their aim was to determine if natural Wolbachia infections have the same parasite-inhibiting properties observed in laboratory models. Natural partnerships between Wolbachia and mosquito species with a long evolutionary history are key to understanding the effects of several generations of co-evolution on Wolbachia-transfected mosquitoes. For this purpose, they used an entirely natural mosquito-Wolbachia-Plasmodium combination. In contrast to most previous studies centred on the quantification of the midgut stages of Plasmodium, they measured a parasitaemia value that related directly to transmission potential by following infections to the salivary gland stages. They found that the presence of Wolbachia (of any density inside the mosquito) increased the susceptibility of Cx. pipiens mosquitoes to P. relictum, significantly increasing the prevalence of salivary gland-stage infections. Strikingly, these results suggest that mosquitoes that have been naturally infected with Wolbachia may be better malaria vectors than Wolbachia-free mosquitoes.
The anopheline (dynamic) core microbiota - Our knowledge of the anopheline internal ecosystem indicates that there is no natural obligate endosymbiont in the Anopheles genus, in contrast to what has been observed in other haematophagous insects such as the tsetse fly (Wigglesworthia spp) (Aksoy 1995), head louse (Riesia pediculicola) (Kirkness et al. 2010), Ae. albopictus-Wolbachia (walbA and wAlbB) (Armbruster et al. 2003) and Aedes polynesiensis-Wolbachia (wPolA) (Andrews et al. 2012). Secondary symbionts may have more recently associated with their insect host and are thus not obligate, although they may fulfill a role in host biology (Minard et al. 2013).
Studies on the microcosmos of anophelines have generated information that can appear to be contradictory, such as the presence (Cirimotich et al. 2011) or absence (Boissière et al. 2012) of Enterobacter sp. in field-collected African anophelines or the unexpected absence of Asaia in Indo/middle eastern field-collected or laboratory-reared anophelines (Rani et al. 2009, Djadid et al. 2011, Chavshin et al. 2012). Notwithstanding, predominant taxa (at the family and sometimes genus level) have consistently been observed in both laboratory-reared and natural mosquito populations. These observations suggest (Wang et al. 2012, Gendrin & Christophides 2013) that laboratory colonies may retain their association with the predominant bacterial taxons established prior to laboratory colonisation.
The proposed existence of a “core group” of anopheline gut-associated bacteria mirrors the concept of the bacterial human enterotype (Turnbaugh et al. 2007, Evans et al. 2013); this core group may be critical to the host mosquito both as a metabolic exchanger/enhancer (Russell et al. 2013) and as a key element of its immune response (Nyholm & Graf 2012). The identity of this core microbiota has been referred to in several recent publications (Boissière et al. 2012, Gendrin & Christophides 2013). Wang et al. (2012) determined that at a family taxonomic level, Enterobacteriaceae, Flavobacteriaceae and Pseudomonadaceae encompass 80.9% and 94.3% of the An. gambiae microbiota in laboratory-reared and field-collected Kenyan samples after a blood meal, respectively. However, many other bacterial genera or families may be involved; although multiple detection techniques have been utilised, they primarily focused on a single anopheline species and organ. Although An. gambiae may be the most important, efficient and anthropophilic malaria vector, it cannot be assumed that the true composition of this “core” consortium can be elucidated using a single species. Furthermore, the patterns of fluctuation in the bacterial genera abundance within a “core family”, and among different anopheline species and life stages are unknown and should be explored in more depth to strengthen the hypothesis that the bacterial population is similar/stable among laboratory-reared and wild adult Anopheles mosquitoes.
It is enticing to suggest that anopheline mosquitoes harbour their microbiota in a selective manner. If microbiota equilibrium could be reached, bacteria from a single genus could be engineered to serve as a universal paratransgenic vehicle in multiple anopheline species. The results obtained and expertise involved in the Eliminate Dengue project [based upon the interaction of Ae. aegypti mosquitoes and a bacterial endosymbiont (Wolbachia)] suggests a feasible partnership. Nevertheless, recent observations oppose the existence of a constant inter-species bacterial consortium associated with the insect gut environment (Wong et al. 2013).
If the anopheline holobiont is to be understood and other potential dysbiosis techniques are to be developed in the arms race against the malaria parasite, multiple avenues of research may be required to address current questions. The data generated by Boissière et al. (2012) and Wang et al. (2012) demonstrate that this microbiome is dynamic and that some of its members correlate to particular characteristics of the An. gambiae model, such as laboratory rearing and the abundance of the Flavobacteriaceae family and the association of the genus Enterobacter with Plasmodium infection. Simple questions could be answered by compiling the data from the reported microbiome surveys and mining the relevant text. How many bacterial families or genera have been reported for anophelines to date? Is there an anopheline species with clear data enrichment? Can filtering and building groups reveal an underlying presence/absence pattern? Minard et al. (2013) generated an index with microbiota data that encompassed multiple mosquito vectors. We undertook the challenge of reviewing this index and complementing it with additional recent and prior reports. The data collection is an ongoing process and may lack genera from other recently published sources (Supplementary data 1).
Filtering and visualising the anopheline microbiota data - The reports included in the databank were selected based on a simple criterion: that they accounted exclusively for bacterial genera in anopheline mosquitoes that were field collected or reared on the continent in which they act as a malaria vector. The listed taxa are accompanied by their respective citation. Key sources were employed as guides to track each of the included genera reports: Abdul-Ghani et al. (2012), Apte-Deshpande et al. (2012), Gendrin and Christophides (2013), Manguin et al. (2013) and Minard et al. (2013). For our purpose, the inclusion criterion excluded important observations of identified taxons in laboratory models such as those by Pumpuni et al. (1996).
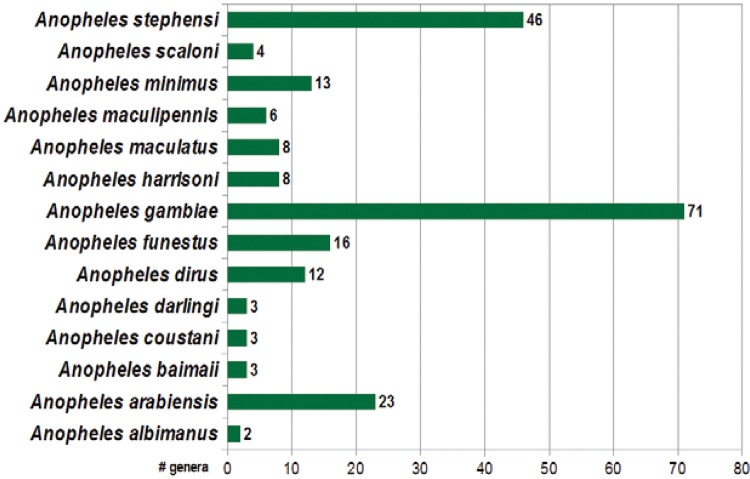
By filtering the compiled data regarding bacterial genera in each anopheline species, we identified 109 unique genera among the 218 reports in the literature (Fig. 1).
Fig. 1. : number of bacterial genera reported per anopheline human malaria vectors. Current count on bacterial genera reported for multiple anopheline species according to our inclusion criterion. A total of 109 unique genera were identified amongst 218 published reports.
The anopheline species with the most reported associated bacterial genera was An. gambiae (71 genera), followed by the Asian human malaria vector An. stephensi (46 genera). These findings are based on multiple detection techniques that range from plate culture to 16S rRNA surveys with multiple platforms. Although malaria transmission is a current burden for developing regions or countries in Latin America, there are only five reported bacterial genera representing two primary Neotropical vectors (Anopheles darlingi and Anopheles albimanus).
The recent publication of the An. darlingi genome (Marinotti et al. 2013) mentions the presence (as contamination or associated) of bacterial reads and genes. The assembly of the Aeromonas hydrophila genome amidst the anopheline NGS data should be explored further. Genome sequencing projects present opportunities for anopheline microbiome research that should be carefully exploited while fully considering the lack of information available for American malaria vectors.
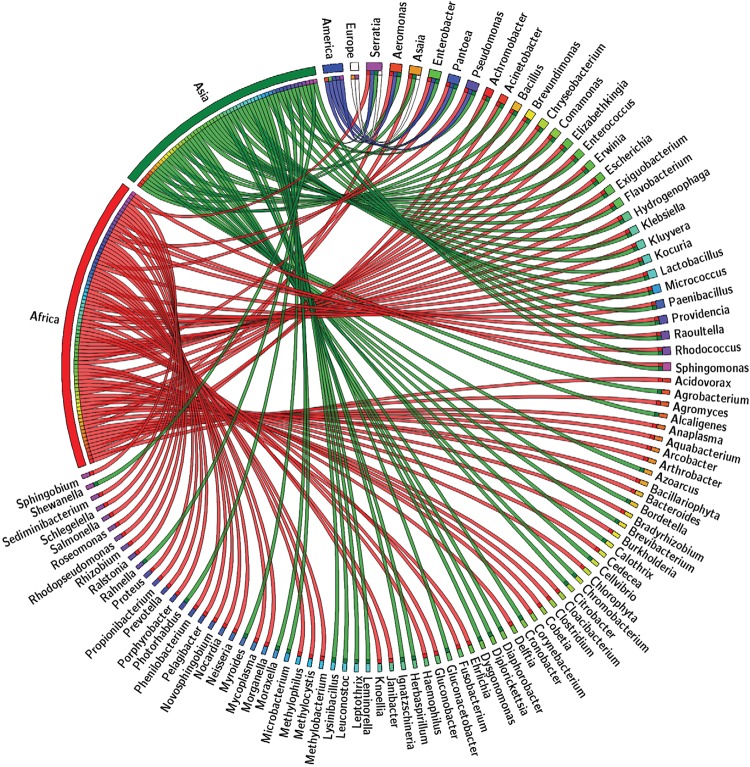
The species-enriched composition of the compiled data motivated us to develop another strategy to “interrogate” it from a broader scope to identify patterns and a core bacterial assembly across the screened Anopheles species. A classification according to continent criterion was applied to the compiled data and the groups obtained represent the bacterial genera identified in the continents of Africa, Asia, Europe and America. The correlation between each continent and bacterial genera was plotted using the online tool Circos: mkweb.bcgsc.ca/tableviewer/visualise/ (Krzywinski et al. 2009). Due to constraints on the table size allowed by the tool, only a subset of the filtered data is shown in Fig. 2. The matrix built after filtering the data is shown in Supplementary data 2. This file is provided only to exemplify the format that Circos requires.
Fig. 2. : Anopheles-associated microbiota distributed by continent. Circular web plotted from a subset of the anopheline microbiota databank filtered in order to show the correspondence between bacterial genus and its anopheline host according to the continent in which the mosquito exerts vectorial activity. The upper right quadrant contains the bacterial genera shared by at least two continents. The bacterial genera exclusive to each continent are not fully shown due to table size constrain on the online tool Circos (mkweb.bcgsc.ca/tableviewer/visualize/). By prospecting the metabolic potential within the genome of the “continent-specific” bacterial genera, we could explore if it is linked to specific functional relations built throughout mosquito-bacteria co-evolution in ecologically differentiated niches. To further depict and quantify the shared and exclusive genera relations obtained with the “continent” filtering criteria, a Venn diagram was generated (Fig. 3) with the online tool Venny: bioinfogp.cnb.csic.es/tools/venny/ (Oliveros 2007). The distribution of the 109 unique genera reported in the literature is the following: Africa (82), Asia (58), America (5), Europe (3), being 46.78% exclusive to Africa, 0% exclusive to America, 24.77% exclusive to Asia, 22.01% shared by Africa and Asia, 3.66% shared by America, Africa and Asia, 1.83% shared by Africa, Asia and Europe and 0.91% shared by America, Africa, Asia and Europe.
Tables and lists are often difficult to follow and interpret. Visual inspection of the available data permits the elucidation of relationships in a more efficient manner. The circular web plotted (Fig. 2) by “continent filtering” the compiled data not only synthesises the information, but also displays it in a useful biological and geographical sense. In examining circular web, we can ask: is there an interpretable presence/absence pattern? Could we use it to further deepen our knowledge of anopheline bionomics and identify other candidate bacterial genera for paratransgenesis?
As shown in Fig. 2, the paratransgenesis candidates Serratia and Pantoea are present in malaria-endemic countries in Africa, Asia and America. However, data regarding America remain limited due to the absence of the Asaia genus in American mosquito-related literature. Significant efforts and progress have been made in exploring the potential of this genus as a paratransgenesis vehicle and attention should now focus on identifying Asaia in Neotropical vectors.
By prospecting the metabolic potential within the genome of the “continent-specific” bacterial genera, we could explore if this potential is linked to specific functional relationships developed throughout mosquito-bacteria co-evolution in ecologically differentiated niches.
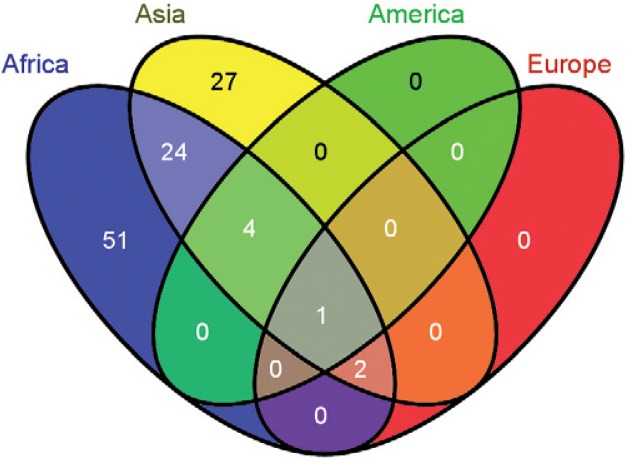
To further visualise and quantify the shared and exclusive genera relationships obtained with the “continent” filtering criteria (refer to Supplementary data 1, spreadsheet 2), a Venn diagram was generated (Fig. 3) using the online tool Venny: bioinfogp.cnb.csic.es/tools/venny/ (Oliveros 2007). The distribution of the 109 unique genera reported in the literature was as follows: Africa (82), Asia (58), America (5) and Europe (3) (46.78% exclusive to Africa, 0% exclusive to America, 24.77% exclusive to Asia, 22.01% shared by Africa and Asia, 3.66% shared by America, Africa and Asia, 1.83% shared by Africa, Asia and Europe and 0.91% shared by America, Africa, Asia and Europe).
Fig. 3. : shared and exclusive bacterial genera identified in anophelines by continent. Using a continent-oriented filtering criterion, the 109 unique bacterial genera reported to date in multiple anophelines distribute as follows: Africa (82), Asia (58), America (5), Europe (3), being 46.78% exclusive to Africa, 0% exclusive to America, 24.77% exclusive to Asia, 22.01% shared by Africa and Asia, 3.66% shared by America, Africa and Asia, 1.83% shared by Africa, Asia and Europe and 0.91% shared by America, Africa, Asia and Europe. Venn diagram generated with the online tool Venny (bioinfogp.cnb.csic.es/tools/venny/).
Five genera are shared among Africa, Asia and America: Enterobacter, Aeromonas, Pantoea, Pseudomonas and Serratia. At the family taxonomic level, these core genera represent the Pseudomonadaceae, Aeromonadaceae and Enterobacteriaceae families. Wang et al. (2012) suggested a stable association between laboratory and field-collected An. gambiae and Enterobacteriaceae, Flavobacteriaceae and Pseudomonadaceae. Thus, these families may be present despite geographical barriers. However, the lack of data for Neotropical vectors could reduce this list of core bacterial families and, concomitantly, our understanding of this consortium and its impact on vector ecology.
Due to the skewed composition of the current data available regarding anopheline microbiota, we considered this broad filtering criterion to be suitable. Although it may deviate from the true nature of the anopheline-microbiome correlation, this exercise was primarily intended to draw the research community’s attention to the construction of an online public repository of such information. This task is both necessary and attainable. Understanding the microbial ecology within insect vectors is feasible as long as reliable and formatted data are made available to the research community.
As multiple anopheline metagenomic profiles become available, network association approaches to evaluate taxa co-occurrence and co-exclusion in microbial consortia across anopheline species, tissues or considering biogeographical parameters could be performed, as has been performed for the human microbiome (Faust et al. 2012, Friedman & Alm 2012).
Substantial advancements in vector (micro) biology have been made in recent years, many of which have been attained using “synergic” approaches in which computational science is a key element. The advent of high-throughput, meta-omic techniques ranging from metagenomics to metabolomics has exposed the large number of genes, transcripts, proteins and metabolites that compose the intricate web formed between the host and its inner ecosystem. Many interesting theories are now being discussed and explored regarding the hologenomic basis of speciation (Rosenberg & Zilber-Rosenberg 2011, Brucker & Bordenstein 2013) and how bacteria and viruses may have shaped and continue to impact the genome architecture and phenotypes of harbouring organisms (Gorski et al. 2003, Crochu et al. 2004, Nikoh et al. 2008, Klasson et al. 2009, Rosario & Breitbart 2011, Ni et al. 2012).
The Brazilian Malaria Network is currently focusing its efforts on establishing a viable malaria infection laboratory model using Anopheles aquasalis as the initial target species. Based on its bionomical characteristics and the fact that there are one million bacteria and 10 million viral particles per millilitre of surface seawater (Suttle 2005, Rosario & Breitbart 2011), maximising the NGS sequencing data generated in a genome or transcriptome project becomes an opportunity to explore many of these new avenues. The An. aquasalis genome project initiated a series of studies, one of which aims to elucidate and describe the composition of the potentially associated microcosmos. The diversity of this consortium and its impact on the genome during the co-evolution of the host and microbial members are unknown. In particular, we are interested in the extent to which larval development in brackish coastal waters may differentiate An. aquasalis from traditionally studied anopheline vectors.
ACKNOWLEDGEMENTS
To Dr Guilherme Oliveira and the Genomics and Computational Biology Group at CPqRR, for valuable assistance and computational infrastructure support.
Supplementary data 1
Combined databank generated by reviewing sources cited by Minard et al. (2013) and other authors cited within the DB as well.
| Africa (82) | Asia (58) | America (5) | Europa (3) |
|---|---|---|---|
| Achromobacter | Achromobacter | Aeromonas | Asaia |
| Acidovorax | Acinetobacter | Enterobacter | Staphylococcus |
| Acinetobacter | Aeromonas | Pantoea | Serratia |
| Aeromonas | Agrobacterium | Pseudomonas | |
| Agromyces | Alcaligenes | Serratia | |
| Anaplasma | Asaia | ||
| Aquabacterium | Azoarcus | ||
| Arcobacter | Bacillus | ||
| Arthrobacter | Bordetella | ||
| Asaia | Brevibacterium | ||
| Bacillariophyta | Brevundimonas | ||
| Bacillus | Calothrix | ||
| Bacteroides | Cellvibrio | ||
| Bradyrhizobium | Chromobacterium | ||
| Brevundimonas | Chryseobacterium | ||
| Burkholderia | Citrobacter | ||
| Cedecea | Comamonas | ||
| Chlorophyta | Cronobacter | ||
| Chryseobacterium | Diaphorobacter | ||
| Cloacibacterium | Diplorickettsia | ||
| Clostridium | Dysgonomonas | ||
| Cobetia | Elizabethkingia | ||
| Comamonas | Enterobacter | ||
| Corynebacterium | Enterococcus | ||
| Delftia | Erwinia | ||
| Ehrlichia | Escherichia | ||
| Elizabethkingia | Exiguobacterium | ||
| Enterobacter | Flavobacterium | ||
| Enterococcus | Gluconacetobacter | ||
| Erwinia | Herbaspirillum | ||
| Escherichia | Hydrogenophaga | ||
| Exiguobacterium | Ignatzschineria | ||
| Flavobacterium | Klebsiella | ||
| Fusobacterium | Kluyvera | ||
| Gluconobacter | Kocuria | ||
| Haemophilus | Lactobacillus | ||
| Hydrogenophaga | Leminorella | ||
| Janibacter | Leptothrix | ||
| Klebsiella | Leuconostoc | ||
| Kluyvera | Lysinibacillus | ||
| Knoellia | Microbacterium | ||
| Kocuria | Micrococcus | ||
| Lactobacillus | Moraxella | ||
| Methylobacterium | Myroides | ||
| Methylocystis | Paenibacillus | ||
| Methylophilus | Pantoea | ||
| Micrococcus | Photorhabdus | ||
| Morganella | Providencia | ||
| Mycoplasma | Pseudomonas | ||
| Neisseria | Rahnella | ||
| Nocardia | Raoultella | ||
| Novosphingobium | Rhodococcus | ||
| Paenibacillus | Serratia | ||
| Pantoea | Shewanella | ||
| Pelagibacter | Sphingomonas | ||
| Phenilobacterium | Staphylococcus | ||
| Porphyrobacter | Stenotrophomonas | ||
| Prevotella | Xenorhabdus | ||
| Propionibacterium | |||
| Proteus | |||
| Providencia | |||
| Pseudomonas | |||
| Ralstonia | |||
| Raoultella | |||
| Rhizobium | |||
| Rhodococcus | |||
| Rhodopseudomonas | |||
| Roseomonas | |||
| Salmonella | |||
| Schlegelella | |||
| Sediminibacterium | |||
| Serratia | |||
| Sphingobium | |||
| Sphingomonas | |||
| Spiroplasma | |||
| Staphylococcus | |||
| Stenotrophomonas | |||
| Streptococcus | |||
| Sulfurospirillum | |||
| Thorsellia | |||
| Vibrio | |||
| Zymobacter |
Supplementary data 2
Bacterial genera per continent.
| Genera | 255,0,0 | 0,128,0 | 0,0,255 | 255,250,250 |
|---|---|---|---|---|
|
| ||||
| Genera | Africa | Asia | America | Europe |
| Achromobacter | 1 | 1 | 0 | 0 |
| Acidovorax | 1 | 0 | 0 | 0 |
| Acinetobacter | 1 | 1 | 0 | 0 |
| Aeromonas | 1 | 1 | 1 | 0 |
| Agrobacterium | 0 | 1 | 0 | 0 |
| Agromyces | 1 | 0 | 0 | 0 |
| Alcaligenes | 0 | 1 | 0 | 0 |
| Anaplasma | 1 | 0 | 0 | 0 |
| Aquabacterium | 1 | 0 | 0 | 0 |
| Arcobacter | 1 | 0 | 0 | 0 |
| Arthrobacter | 1 | 0 | 0 | 0 |
| Asaia | 1 | 1 | 0 | 1 |
| Azoarcus | 0 | 1 | 0 | 0 |
| Bacillariophyta | 1 | 0 | 0 | 0 |
| Bacillus | 1 | 1 | 0 | 0 |
| Bacteroides | 1 | 0 | 0 | 0 |
| Bordetella | 0 | 1 | 0 | 0 |
| Bradyrhizobium | 1 | 0 | 0 | 0 |
| Brevibacterium | 0 | 1 | 0 | 0 |
| Brevundimonas | 1 | 1 | 0 | 0 |
| Burkholderia | 1 | 0 | 0 | 0 |
| Calothrix | 0 | 1 | 0 | 0 |
| Cedecea | 1 | 0 | 0 | 0 |
| Cellvibrio | 0 | 1 | 0 | 0 |
| Chlorophyta | 1 | 0 | 0 | 0 |
| Chromobacterium | 0 | 1 | 0 | 0 |
| Chryseobacterium | 1 | 1 | 0 | 0 |
| Citrobacter | 0 | 1 | 0 | 0 |
| Cloacibacterium | 1 | 0 | 0 | 0 |
| Clostridium | 1 | 0 | 0 | 0 |
| Cobetia | 1 | 0 | 0 | 0 |
| Comamonas | 1 | 1 | 0 | 0 |
| Corynebacterium | 1 | 0 | 0 | 0 |
| Cronobacter | 0 | 1 | 0 | 0 |
| Delftia | 1 | 0 | 0 | 0 |
| Diaphorobacter | 0 | 1 | 0 | 0 |
| Diplorickettsia | 0 | 1 | 0 | 0 |
| Dysgonomonas | 0 | 1 | 0 | 0 |
| Ehrlichia | 1 | 0 | 0 | 0 |
| Elizabethkingia | 1 | 1 | 0 | 0 |
| Enterobacter | 1 | 1 | 1 | 0 |
| Enterococcus | 1 | 1 | 0 | 0 |
| Erwinia | 1 | 1 | 0 | 0 |
| Escherichia | 1 | 1 | 0 | 0 |
| Exiguobacterium | 1 | 1 | 0 | 0 |
| Flavobacterium | 1 | 1 | 0 | 0 |
| Fusobacterium | 1 | 0 | 0 | 0 |
| Gluconacetobacter | 0 | 1 | 0 | 0 |
| Gluconobacter | 1 | 0 | 0 | 0 |
| Haemophilus | 1 | 0 | 0 | 0 |
| Herbaspirillum | 0 | 1 | 0 | 0 |
| Hydrogenophaga | 1 | 1 | 0 | 0 |
| Ignatzschineria | 0 | 1 | 0 | 0 |
| Janibacter | 1 | 0 | 0 | 0 |
| Klebsiella | 1 | 1 | 0 | 0 |
| Kluyvera | 1 | 1 | 0 | 0 |
| Knoellia | 1 | 0 | 0 | 0 |
| Kocuria | 1 | 1 | 0 | 0 |
| Lactobacillus | 1 | 1 | 0 | 0 |
| Leminorella | 0 | 1 | 0 | 0 |
| Leptothrix | 0 | 1 | 0 | 0 |
| Leuconostoc | 0 | 1 | 0 | 0 |
| Lysinibacillus | 0 | 1 | 0 | 0 |
| Methylobacterium | 1 | 0 | 0 | 0 |
| Methylocystis | 1 | 0 | 0 | 0 |
| Methylophilus | 1 | 0 | 0 | 0 |
| Microbacterium | 0 | 1 | 0 | 0 |
| Micrococcus | 1 | 1 | 0 | 0 |
| Moraxella | 0 | 1 | 0 | 0 |
| Morganella | 1 | 0 | 0 | 0 |
| Mycoplasma | 1 | 0 | 0 | 0 |
| Myroides | 0 | 1 | 0 | 0 |
| Neisseria | 1 | 0 | 0 | 0 |
| Nocardia | 1 | 0 | 0 | 0 |
| Novosphingobium | 1 | 0 | 0 | 0 |
| Paenibacillus | 1 | 1 | 0 | 0 |
| Pantoea | 1 | 1 | 1 | 0 |
| Pelagibacter | 1 | 0 | 0 | 0 |
| Phenilobacterium | 1 | 0 | 0 | 0 |
| Photorhabdus | 0 | 1 | 0 | 0 |
| Porphyrobacter | 1 | 0 | 0 | 0 |
| Prevotella | 1 | 0 | 0 | 0 |
| Propionibacterium | 1 | 0 | 0 | 0 |
| Proteus | 1 | 0 | 0 | 0 |
| Providencia | 1 | 1 | 0 | 0 |
| Pseudomonas | 1 | 1 | 1 | 0 |
| Rahnella | 0 | 1 | 0 | 0 |
| Ralstonia | 1 | 0 | 0 | 0 |
| Raoultella | 1 | 1 | 0 | 0 |
| Rhizobium | 1 | 0 | 0 | 0 |
| Rhodococcus | 1 | 1 | 0 | 0 |
| Rhodopseudomonas | 1 | 0 | 0 | 0 |
| Roseomonas | 1 | 0 | 0 | 0 |
| Salmonella | 1 | 0 | 0 | 0 |
| Schlegelella | 1 | 0 | 0 | 0 |
| Sediminibacterium | 1 | 0 | 0 | 0 |
| Serratia | 1 | 1 | 1 | 1 |
| Shewanella | 0 | 1 | 0 | 0 |
| Sphingobium | 1 | 0 | 0 | 0 |
| Sphingomonas | 1 | 1 | 0 | 0 |
| Spiroplasma | 1 | 0 | 0 | 0 |
| Staphylococcus | 1 | 1 | 0 | 1 |
| Stenotrophomonas | 1 | 1 | 0 | 0 |
| Streptococcus | 1 | 0 | 0 | 0 |
| Sulfurospirillum | 1 | 0 | 0 | 0 |
| Thorsellia | 1 | 0 | 0 | 0 |
| Vibrio | 1 | 0 | 0 | 0 |
| Xenorhabdus | 0 | 1 | 0 | 0 |
| Zymobacter | 1 | 0 | 0 | 0 |
Footnotes
Financial support: Bill & Melinda Gates Foundation (TransEpi Study), FIOCRUZ, Strategic Programme for Supporting Health Research (PAPES V), CNPq, FAPEMIG, FAPEAM
REFERENCES
- Abdul-Ghani R, Al-Mekhlafi AM, Alabsi MS. Microbial control of malaria: biological warfare against the parasite and its vector. Acta Trop. 2012;121:71–84. doi: 10.1016/j.actatropica.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, Rodriguez-Mueller B, Zucker J, Thiagarajan M, Henrissat B, White O, Kelley ST, Methé B, Schloss PD, Gevers D, Mitreva M, Huttenhower C. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8: doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksoy S. Wigglesworthia gen. nov . and Wigglesworthia glossini- dia sp. nov., taxa consisting of the mycetocyte-associated, primary endosymbionts of tsetse flies. . Int J Syst Bacteriol. 1995;45:848–851. doi: 10.1099/00207713-45-4-848. [DOI] [PubMed] [Google Scholar]
- Andrews ES, Crain PR, Fu Y, Howe DK, Dobson SL. Reactive oxygen species production and Brugia pahangi survivorship in Aedes polynesiensis with artificial Wolbachia infection types. PLoS Pathog. 2012;8: doi: 10.1371/journal.ppat.1003075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angly FE, Willner D, Prieto-Davó A, Edwards RA, Schmieder R, Vega-Thurber R, Antonopoulos DA, Barott K, Cottrell MT, Desnues C, Dinsdale EA, Furlan M, Haynes M, Henn MR, Hu Y, Kirchman DL, McDole T, McPherson JD, Meyer F, Miller RM, Mundt E, Naviaux RK, Rodriguez-Mueller B, Stevens R, Wegley L, Zhang L, Zhu B, Rohwer F. The GAAS metagenomic tool and its estimations of viral and microbial average genome size in sour major biomes. PLoS Comput Biol. 2009;5: doi: 10.1371/journal.pcbi.1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansorge WJ. Next-generation DNA sequencing techniques. N Biotechnol. 2009;25:195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Apte-Deshpande A, Paingankar M, Gokhale MD, Deobagkar DN. Serratia odorifera a midgut inhabitant of Aedes aegypti mosquito enhances its susceptibility to dengue-2 virus. PLoS ONE. 2012;7: doi: 10.1371/journal.pone.0040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster P, Damsky WE, Jr, Giordano R, Birungi J, Munstermann LE, Conn JE. Infection of New and Old-World Aedes albopictus (Diptera: Culicidae) by the intracellular parasite Wolbachia: implications for host mitochondrial DNA evolution. J Med Entomol. 2003;40:356–360. doi: 10.1603/0022-2585-40.3.356. [DOI] [PubMed] [Google Scholar]
- Azambuja P, Garcia ES, Ratcliffe NA. Gut microbiota and parasite transmission by insect vectors. Trends Parasitol. 2005;21:568–572. doi: 10.1016/j.pt.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Bando H, Okado K, Guelbeogo WM, Badolo A, Aonuma H, Nelson B, Fukumoto S, Xuan X, Sagnon N, Kanuka H. Intra-specific diversity of Serratia marcescens in Anopheles mosquito midgut defines Plasmodium transmission capacity. . 1641Sci Rep. 2013;3 doi: 10.1038/srep01641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basset Y, Cizek L, Cuénoud P, Didham RK, Guilhaumon F, Missa O, Novotny V, Ødegaard F, Roslin T, Schmidl J, Tishechkin AK, Winchester NN, Roubik DW, Aberlenc HP, Bail J, Barrios H, Bridle JR, Castaño-Meneses G, Corbara B, Curletti G, Rocha WD, Bakker D, Delabie JH, Dejean A, Fagan LL, Floren A, Kitching RL, Medianero E, Miller SE, Oliveira EG, Orivel J, Pollet M, Rapp M, Ribeiro SP, Roisin Y, Schmidt JB, Sørensen L, Leponce M. Arthropod diversity in a tropical forest. Science. 2012;338:1481–1484. doi: 10.1126/science.1226727. [DOI] [PubMed] [Google Scholar]
- Baumann P. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol. 2005;59:155–189. doi: 10.1146/annurev.micro.59.030804.121041. [DOI] [PubMed] [Google Scholar]
- Beard CB, Dotson EM, Pennington PM, Eichler S, Cordon-Rosales C, Durvasula RV. Bacterial symbiosis and paratransgenic control of vector-borne Chagas disease. Int J Parasitol. 2001;31:621–627. doi: 10.1016/s0020-7519(01)00165-5. [DOI] [PubMed] [Google Scholar]
- Bian G, Joshi D, Dong Y, Lu P, Zhou G, Pan X, Xu Y, Dimopoulos G, Xi Z. Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science. 2013;340:748–751. doi: 10.1126/science.1236192. [DOI] [PubMed] [Google Scholar]
- Bian G, Xu Y, Lu P, Xie Y, Xi Z. The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog. 2010;6: doi: 10.1371/journal.ppat.1000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird RG, Draper CC, Ellis DS. A cytoplasmic polyhedrosis virus in midgut cells of Anopheles stephensi and in the sporogonic stages of Plasmodium berghei yoelii . . Bull World Health Organ. 1972;46:337–343. [PMC free article] [PubMed] [Google Scholar]
- Bishop-Lilly KA, Turell MJ, Willner KM, Butani A, Nolan NME, Lentz SM, Akmal A, Mateczun A, Brahmbhatt TN, Sozhamannan S, Whitehouse CA, Read TD. Arbovirus detection in insect vectors by rapid, high-throughput pyrosequencing. PLoS Negl Trop Dis. 2010;4: doi: 10.1371/journal.pntd.0000878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisi DC, Lampe DJ. Secretion of anti-Plasmodium effector proteins from a natural Pantoea agglomerans isolate by using PelB and HlyA secretion signals. Appl Environ Microbiol. 2011;77:4669–4675. doi: 10.1128/AEM.00514-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissière A, Tchioffo MT, Bachar D, Abate L, Marie A, Nsango SE, Shahbazkia HR, Awono-Ambene PH, Levashina EA, Christen R, Morlais I. Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog. 2012;8: doi: 10.1371/journal.ppat.1002742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger N, Lowenberger C, Volf P, Ursic R, Sigutova L, Sabatier L, Svobodova M, Beverley SM, Späth G, Brun R, Pesson B, Bulet P. Characterization of a defensin from the sand fly Phlebotomus duboscqi induced by challenge with bacteria or the protozoan parasite Leishmania major. Infect Immun. 2004;72:7140–7146. doi: 10.1128/IAI.72.12.7140-7146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtzis K, Dobson SL, Xi Z, Rasgon JL, Calvitti M, Moreira LA, Bossin HC, Moretti R, Baton LA, Hughes GL, Mavingui P, Gilles JR. Harnessing mosquito- Wolbachia symbiosis for vector and disease control. . Acta Trop. 2014;132(Suppl.):S150–S163. doi: 10.1016/j.actatropica.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Brucker RM, Bordenstein SR. The capacious hologenome. Zoology (Jena) 2013;116:260–261. doi: 10.1016/j.zool.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Capone A, Ricci I, Damiani C, Mosca M, Rossi P, Scuppa P, Crotti E, Epis S, Angeletti M, Valzano M, Sacchi L, Bandi C, Daffonchio D, Mandrioli M, Favia G. Interactions between Asaia, Plasmodium and Anopheles: new insights into mosquito symbiosis and implications in malaria symbiotic control. 182Parasit Vectors. 2013;6 doi: 10.1186/1756-3305-6-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpi G, Cagnacci F, Wittekindt NE, Zhao F, Qi J, Tomsho LP, Drautz DI, Rizzoli A, Schuster SC. Metagenomic profile of the bacterial communities associated with Ixodes ricinus ticks. PLoS ONE. 2011;6: doi: 10.1371/journal.pone.0025604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavshin AR, Oshaghi MA, Vatandoost H, Pourmand MR, Raeisi A, Enayati AA, Mardani N, Ghoorchian S. Identification of bacterial microflora in the midgut of the larvae and adult of wild caught Anopheles stephensi : a step toward finding suitable paratransgenesis candidates. . Acta Trop. 2012;121:129–134. doi: 10.1016/j.actatropica.2011.10.015. [DOI] [PubMed] [Google Scholar]
- Chouaia B, Rossi P, Montagna M, Ricci I, Crotti E, Damiani C, Epis S, Faye I, Sagnon N, Alma A, Favia G, Daffonchio D, Bandi C. Molecular evidence for multiple infections as revealed by typing of Asaia bacterial symbionts of four mosquito species. Appl Environ Microbiol. 2010;76:7444–7450. doi: 10.1128/AEM.01747-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirimotich CM, Dong Y, Clayton AM, Sandiford SL, Souza JA, Neto, Mulenga M, Dimopoulos G. Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science. 2011;332:855–858. doi: 10.1126/science.1201618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho-Abreu IV, Zhu KY, Ramalho-Ortigão M. Transgenesis and paratransgenesis to control insect-borne diseases: current status and future challenges. Parasitol Int. 2010;59:1–8. doi: 10.1016/j.parint.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crochu S, Cook S, Attoui H, Charrel RN, Chesse R, Belhouchet M, Lemasson JJ, Micco P, Lamballerie X. Sequences of flavivirus-related RNA viruses persist in DNA form integrated in the genome of Aedes spp mosquitoes. J Gen Virol. 2004;85:1971–1980. doi: 10.1099/vir.0.79850-0. [DOI] [PubMed] [Google Scholar]
- Crotti E, Damiani C, Pajoro M, Gonella E, Rizzi A, Ricci I, Negri I, Scuppa P, Rossi P, Ballarini P, Raddadi N, Marzorati M, Sacchi L, Clementi E, Genchi M, Mandrioli M, Bandi C, Favia G, Alma A, Daffonchio D. Asaia , a versatile acetic acid bacterial symbiont, capable of cross-colonizing insects of phylogenetically distant genera and orders. . Environ Microbiol. 2009;11:3252–3264. doi: 10.1111/j.1462-2920.2009.02048.x. [DOI] [PubMed] [Google Scholar]
- Damiani C, Ricci I, Crotti E, Rossi P, Rizzi A, Scuppa P, Capone A, Sagnon NF, Faye I, Kang A, Whitehorn C, Moussa GW, Esposito F, Sacchi L, Bandi C, Daffonchio D, Favia G. Mosquito-bacteria symbiosis: the case of Anopheles gambiae and Asaia. Microb Ecol. 2010;60:644–654. doi: 10.1007/s00248-010-9704-8. [DOI] [PubMed] [Google Scholar]
- Damiani C, Ricci I, Crotti E, Rossi P, Rizzi A, Scuppa P, Esposito F, Bandi C, Daffonchio D, Favia G. Paternal transmission of symbiotic bacteria in malaria vectors. Curr Biol. 2008;18:R1087–R1088. doi: 10.1016/j.cub.2008.10.040. [DOI] [PubMed] [Google Scholar]
- Degnan P, Moran N. Diverse phage-encoded toxins in a protective insect endosymbiont. Appl Environ Microbiol. 2008;74:6782–6791. doi: 10.1128/AEM.01285-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Luca M, Boccolini D, Severini F, Toma L, Barbieri FM, Massa A, Romi R. A 2-year entomological study of potential malaria vectors in central Italy. Vector Borne Zoonotic Dis. 2009;9:703–711. doi: 10.1089/vbz.2008.0129. [DOI] [PubMed] [Google Scholar]
- Dillon RJ, Dillon VM. The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol. 2004;49:71–92. doi: 10.1146/annurev.ento.49.061802.123416. [DOI] [PubMed] [Google Scholar]
- Djadid ND, Jazayeri H, Raz A, Favia G, Ricci I, Zakeri S. Identification of the midgut microbiota of An. stephensi and An. maculipennis for their application as a paratransgenic tool against malaria. PLoS ONE. 2011;6: doi: 10.1371/journal.pone.0028484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5: doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AE. Lessons from studying insect symbioses. Cell Host Microbe. 2011;10:359–367. doi: 10.1016/j.chom.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durvasula RV, Gumbs A, Panackal A, Kruglov O, Aksoy S, Merrifield RB, Richards FF, Beard CB. Prevention of insect-borne disease: an approach using transgenic symbiotic bacteria. Proc Natl Acad Sci USA. 1997;94:3274–3278. doi: 10.1073/pnas.94.7.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eappen AG, Smith RC, Jacobs-Lorena M. Enterobacter-activated mosquito immune responses to Plasmodium involve activation of SRPN6 in Anopheles stephensi. PLoS ONE. 2013;8: doi: 10.1371/journal.pone.0062937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleftherianos I, Atri J, Accetta J, Castillo JC. Endosymbiotic bacteria in insects: guardians of the immune system? 46Front Physiol. 2013;4 doi: 10.3389/fphys.2013.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel P, Moran NA. The gut microbiota of insects - diversity in structure and function. FEMS Microbiol Rev. 2013;37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- Evans JM, Morris LS, Marchesi JR. The gut microbiome: the role of a virtual organ in the endocrinology of the host. J Endocrinol. 2013;218:R37–R47. doi: 10.1530/JOE-13-0131. [DOI] [PubMed] [Google Scholar]
- Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol. 2012;8: doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favia G, Ricci I, Damiani C, Raddadi N, Crotti E, Marzorati M, Rizzi A, Urso R, Brusetti L, Borin S, Mora D, Scuppa P, Pasqualini L, Clementi E, Genchi M, Corona S, Negri I, Grandi G, Alma A, Kramer L, Esposito F, Bandi C, Sacchi L, Daffonchio D. Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. Proc Natl Acad Sci USA. 2007;104:9047–9051. doi: 10.1073/pnas.0610451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D, Garrity GM, Sansone SA, Sterk P, Gray T, Kyrpides N, Hirschman L, Glöckner FO, Kottmann R, Angiuoli S, White O, Dawyndt P, Thomson N, Gil IS, Morrison N, Tatusova T, Mizrachi I, Vaughan R, Cochrane G, Kagan L, Murphy S, Schriml L, Consortium Genomic Standards. Meeting report: the fifth Genomic Standards Consortium (GSC) workshop. OMICS. 2008;12:109–113. doi: 10.1089/omi.2008.A3B3. [DOI] [PubMed] [Google Scholar]
- Franzenburg S, Walter J, Künzel S, Wang J, Baines JF, Bosch TCG, Fraune S. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. PNAS. 2013;110:E3730–E3738. doi: 10.1073/pnas.1304960110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8: doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrin M, Christophides GK. The Anopheles mosquito microbiota and their impact on pathogen transmission, Anopheles mosquitoes - new insights into malaria vectors. 2013 intechopen.com/books/anopheles-mosquitoes-new-insights-into-malaria-vectors/the-anopheles-mosquito-microbiota-and-their-impact-on-pathogen-transmission
- Gerlach W, Stoye J. Taxonomic classification of metagenomic shotgun sequences with CARMA3. Nucleic Acids Res. 2011;39: doi: 10.1093/nar/gkr225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Hughes M. Gene expression profiling: metatranscriptomics. Methods Mol Biol. 2011;733:195–205. doi: 10.1007/978-1-61779-089-8_14. [DOI] [PubMed] [Google Scholar]
- Glass EM, Wilkening J, Wilke A, Antonopoulos D, Meyer F. Using the metagenomics RAST server (MG-RAST) for analyzing shotgun metagenomes. Cold Spring Harb Protoc. 20102010 doi: 10.1101/pdb.prot5368. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Ceron L, Santillan F, Rodriguez MH, Mendez D, Hernandez-Avila JE. Bacteria in midguts of field-collected Anopheles albimanus block Plasmodium vivax sporogonic development. J Med Entomol. 2003;3:371–374. doi: 10.1603/0022-2585-40.3.371. [DOI] [PubMed] [Google Scholar]
- Gorski A, Dabrowska K, Switala-Jeleń K, Nowaczyk M, Weber-Dabrowska B, Boratynski J, Wietrzyk J, Opolski A. New insights into the possible role of bacteriophages in host defense and disease. 2Med Immunol. 2003;2 doi: 10.1186/1476-9433-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Mendelin S, Allcock R, Kresoje N, van den Hurk AF, Warrilow D. Detection of arboviruses and other micro-organisms in experimentally infected mosquitoes using massively parallel sequencing. PLoS ONE. 2013;8: doi: 10.1371/journal.pone.0058026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. How many species are infected with Wolbachia ? - A statistical analysis of current data. . FEMS Microbiol Lett. 2008;281:215–220. doi: 10.1111/j.1574-6968.2008.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes GL, Koga R, Xue P, Fukatsu T, Rasgon JL. Wolbachia infections are virulent and inhibit the human malaria parasite Plasmodium falciparum in Anopheles gambiae. PLoS Pathog. 2011a;7: doi: 10.1371/journal.ppat.1002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes GL, Pike AD, Xue P, Rasgon JL. Invasion of Wolbachia into Anopheles and other insect germlines in an ex vivo organ culture system. PLoS ONE. 2012;7: doi: 10.1371/journal.pone.0036277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes GL, Ren X, Ramirez JL, Sakamoto JM, Bailey JA, Jedlicka AE, Rasgon JL. Wolbachia infections in Anopheles gambiae cells: transcriptomic characterization of a novel host-symbiont interaction. PLoS Pathog. 2011b;7: doi: 10.1371/journal.ppat.1001296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M, Lu G, Torres S, Edmonds JH, Kay BH, Khromykh AA, Asgari S. Effect of Wolbachia on replication of West Nile virus in a mosquito cell line and adult mosquitoes. . J Virol. 2013;87:851–858. doi: 10.1128/JVI.01837-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, FitzGerald MG, Fulton RS, Giglio MG, Hallsworth-Pepin K, Lobos EA, Madupu R, Magrini V, Martin JC, Mitreva M, Muzny DM, Sodergren EJ, Versalovic J, Wollam AM, Worley KC, Wortman JR, Young SK, Zeng Q, Aagaard KM, Abolude OO, Allen-Vercoe E, Alm EJ, Alvarado L, Andersen GL, Anderson S, Appelbaum E, Arachchi HM, Armitage G, Arze CA, Ayvaz T, Baker CC, Begg L, Belachew T, Bhonagiri V, Bihan M, Blaser MJ, Bloom T, Bonazzi V, Brooks J, Buck GA, Buhay CJ, Busam DA, Campbell JL, Canon SR, Cantarel BL, Chain PS, Chen IM, Chen L, Chhibba S, Chu K, Ciulla DM, Clemente JC, Clifton SW, Conlan S, Crabtree J, Cutting MA, Davidovics NJ, Davis CC, Santis TZ, Deal C, Delehaunty KD, Dewhirst FE, Deych E, Ding Y, Dooling DJ, Dugan SP, Dunne WM, Durkin A, Edgar RC, Erlich RL, Farmer CN, Farrell RM, Faust K, Feldgarden M, Felix VM, Fisher S, Fodor AA, Forney LJ, Foster L, Di Francesco V, Friedman J, Friedrich DC, Fronick CC, Fulton LL, Gao H, Garcia N, Giannoukos G, Giblin C, Giovanni MY, Goldberg JM, Goll J, Gonzalez A, Griggs A, Gujja S, Haake SK, Haas BJ, Hamilton HA, Harris EL, Hepburn TA, Herter B, Hoffmann DE, Holder ME, Howarth C, Huang KH, Huse SM, Izard J, Jansson JK, Jiang H, Jordan C, Joshi V, Katancik JA, Keitel WA, Kelley ST, Kells C, King NB, Knights D, Kong HH, Koren O, Koren S, Kota KC, Kovar CL, Kyrpides NC, La Rosa PS, Lee SL, Lemon KP, Lennon N, Lewis CM, Lewis L, Ley RE, Li K, Liolios K, Liu B, Liu Y, Lo CC, Lozupone CA, Lunsford R, Madden T, Mahurkar AA, Mannon PJ, Mardis ER, Markowitz VM, Mavromatis K, McCorrison JM, McDonald D, McEwen J, McGuire AL, McInnes P, Mehta T, Mihindukulasuriya KA, Miller JR, Minx PJ, Newsham I, Nusbaum C, O’Laughlin M, Orvis J, Pagani I, Palaniappan K, Patel SM, Pearson M, Peterson J, Podar M, Pohl C, Pollard KS, Pop M, Priest ME, Proctor LM, Qin X, Raes J, Ravel J, Reid JG, Rho M, Rhodes R, Riehle KP, Rivera MC, Rodriguez-Mueller B, Rogers YH, Ross MC, Russ C, Sanka RK, Sankar P, Sathirapongsasuti J, Schloss JA, Schloss PD, Schmidt TM, Scholz M, Schriml L, Schubert AM, Segata N, Segre JA, Shannon WD, Sharp RR, Sharpton TJ, Shenoy N, Sheth NU, Simone GA, Singh I, Smillie CS, Sobel JD, Sommer DD, Spicer P, Sutton GG, Sykes SM, Tabbaa DG, Thiagarajan M, Tomlinson CM, Torralba M, Treangen TJ, Truty RM, Vishnivetskaya TA, Walker J, Wang L, Wang Z, Ward DV, Warren W, Watson MA, Wellington C, Wetterstrand KA, White JR, Wilczek-Boney K, Wu Y, Wylie KM, Wylie T, Yandava C, Ye L, Ye Y, Yooseph S, Youmans BP, Zhang L, Zhou Y, Zhu Y, Zoloth L, Zucker JD, Birren BW, Gibbs RA, Highlander SK, Methé BA, Nelson KE, Petrosino JF, Weinstock GM, Wilson RK, White O. Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, An L, Lin SM, Feng G, Qiu Y. A statistical framework for accurate taxonomic assignment of metagenomic sequencing reads. PLoS ONE. 2012;7: doi: 10.1371/journal.pone.0046450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Ren X, Rasgon JL. The virulent Wolbachia strain w MelPop efficiently establishes somatic infections in the malaria vector Anopheles gambiae . . Appl Environ Microbiol. 2009;75:3373–3376. doi: 10.1128/AEM.00207-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambris Z, Blagborough AM, Pinto SB, Blagrove MS, Godfray HC, Sinden RE, Sinkins SP. Wolbachia stimulates immune gene expression and inhibits Plasmodium development in Anopheles gambiae. PLoS Pathog. 2010;6: doi: 10.1371/journal.ppat.1001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killeen GF, Barillas-Mury C, Thomas MB, Greenwood B. Modulating malaria with Wolbachia. Nat Med. 2013;19:974–975. doi: 10.1038/nm.3298. [DOI] [PubMed] [Google Scholar]
- Kim M, Lee KH, Yoon SW, Kim BS, Chun J, Yi H. Analytical tools and databases for metagenomics in the next-generation sequencing era. Genomics Inform. 2013;11:102–113. doi: 10.5808/GI.2013.11.3.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkness EF, Haas BJ, Sun W, Braig HR, Perotti MA, Clark JM, Lee SH, Robertson HM, Kennedy RC, Elhaik E, Gerlach D, Kriventseva EV, Elsik CG, Graur D, Hill CA, Veenstra JA, Walenz B, Tubío JM, Ribeiro JM, Rozas J, Johnston JS, Reese JT, Popadic A, Tojo M, Raoult D, Reed DL, Tomoyasu Y, Kraus E, Mittapalli O, Margam VM, Li HM, Meyer JM, Johnson RM, Romero-Severson J, Vanzee JP, Alvarez-Ponce D, Vieira FG, Aguadé M, Guirao-Rico S, Anzola JM, Yoon KS, Strycharz JP, Unger MF, Christley S, Lobo NF, Seufferheld MJ, Wang N, Dasch GA, Struchiner CJ, Madey G, Hannick LI, Bidwell S, Joardar V, Caler E, Shao R, Barker SC, Cameron S, Bruggner RV, Regier A, Johnson J, Viswanathan L, Utterback TR, Sutton GG, Lawson D, Waterhouse RM, Venter JC, Strausberg RL, Berenbaum MR, Collins FH, Zdobnov EM, Pittendrigh BR. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc Natl Acad Sci USA. 2010;107:12168–12173. doi: 10.1073/pnas.1003379107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L, Kambris Z, Cook PE, Walker T, Sinkins SP. Horizontal gene transfer between Wolbachia and the mosquito Aedes aegypti. 33BMC Genomics. 2009;10 doi: 10.1186/1471-2164-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch H, Schmid-Hempel P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc Natl Acad Sci USA. 2011;108:19288–19292. doi: 10.1073/pnas.1110474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindh JM, Lehane MJ. The tsetse fly Glossina fuscipes fuscipes (Diptera: Glossina) harbours a surprising diversity of bacteria other than symbionts. Antonie Van Leeuwenhoek. 2011;99:711–720. doi: 10.1007/s10482-010-9546-x. [DOI] [PubMed] [Google Scholar]
- Lindh JM, Terenius O, Faye I. 16S rRNA gene-based identification of midgut bacteria from field-caught Anopheles gambiae sensu lato and A. funestus mosquitoes reveals new species related to known insect symbionts. Appl Environ Microbiol. 2005;71:7217–7223. doi: 10.1128/AEM.71.11.7217-7223.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Bian G, Pan X, Xi Z. Wolbachia induces density-dependent inhibition to dengue virus in mosquito cells. PLoS Negl Trop Dis. 2012;6: doi: 10.1371/journal.pntd.0001754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manguin S, Ngo CT, Tainchum K, Juntarajumnong W, Chareonviriyaphap T, Michon A-L, Jumas-Bilak E. Bacterial biodiversity in midguts of Anopheles mosquitoes, malaria vectors in Southeast Asia. Anopheles mosquitoes - new insights into malaria vectors. 2013 intechopen.com/books/anopheles-mosquitoes-new-insights-into-malaria-vectors/bacterial-biodiversity-in-midguts-of-anopheles-mosquitoes-malaria-vectors-in-southeast-asia
- Marinotti O, Cerqueira GC, Almeida LG, Ferro MI, Loreto EL, Zaha A, Teixeira SM, Wespiser AR, Silva AA, Schlindwein AD, Pacheco AC, Silva AL, Graveley BR, Walenz BP, Lima BA, Ribeiro CA, Nunes-Silva CG, Carvalho CR, Soares CM, Menezes CB, Matiolli C, Caffrey D, Araújo DA, Oliveira DM, Golenbock D, Grisard EC, Fantinatti-Garboggini F, Carvalho FM, Barcellos FG, Prosdocimi F, May G, Azevedo GM, Jr, Guimarães GM, Goldman GH, Padilha IQ, Batista JS, Ferro JA, Ribeiro JM, Fietto JL, Dabbas KM, Cerdeira L, Agnez-Lima LF, Brocchi M, Carvalho MO, Teixeira MM, Maia MMD, Goldman MH, Schneider MPC, Felipe MS, Hungria M, Nicolás MF, Pereira M, Montes MA, Cantão ME, Vincentz M, Rafael MS, Silverman N, Stoco PH, Souza RC, Vicentini R, Gazzinelli RT, Neves RO, Silva R, Astolfi S, Filho, Maciel TE, Urményi TP, Tadei WP, Camargo EP, Vasconcelos AT. The genome of Anopheles darlingi , the main Neotropical malaria vector. . Nucleic Acids Res. 2013;41:7387–7400. doi: 10.1093/nar/gkt484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy CB, Diambra LA, Pomar RVR. Metagenomic analysis of taxa associated with Lutzomyia longipalpis , vector of visceral leishmaniasis, using an unbiased high-throughput approach. . PLoS Negl Trop Dis. 2011;5: doi: 10.1371/journal.pntd.0001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister S, Agianian B, Turlure F, Relógio A, Morlais I, Kafatos FC, Christophides GK. Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PLoS Pathog. 2009;5: doi: 10.1371/journal.ppat.1000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel K, Kafatos FC. Mosquito immunity against Plasmodium. Insect Biochem Mol Biol. 2005;35:677–689. doi: 10.1016/j.ibmb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Minard G, Mavingui P, Moro CV. Diversity and function of bacterial microbiota in the mosquito holobiont. 146Parasit Vectors. 2013;6 doi: 10.1186/1756-3305-6-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokili JL, Rohwer F, Dutilh BE. Metagenomics and future perspectives in virus discovery. Curr Opin Virol. 2012;2:63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, Hedges LM, Rocha BC, Hall-Mendelin S, Day A, Riegler M, Hugo LE, Johnson KN, Kay BH, McGraw EA, van den Hurk AF, Ryan PA, O’Neill SL. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya and Plasmodium. Cell. 2009;139:1268–1278. doi: 10.1016/j.cell.2009.11.042. [DOI] [PubMed] [Google Scholar]
- Nartey R, Owusu-Dabo E, Kruppa T, Baffour-Awuah S, Annan A, Oppong S, Becker N, Obiri-Danso K. Use of Bacillus thuringiensis var israelensis as a viable option in an Integrated Malaria Vector Control Programme in the Kumasi Metropolis, Ghana. 116Parasit Vectors. 2013;6 doi: 10.1186/1756-3305-6-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TFF, Duffy S, Polston JE, Bixby E, Vallad GE, Breitbart M. Exploring the diversity of plant DNA viruses and their satellites using vector-enabled metagenomics on whiteflies. PLoS ONE. 2011a;6: doi: 10.1371/journal.pone.0019050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TFF, Willner DL, Lim YW, Schmieder R, Chau B, Nilsson C, Anthony S, Ruan Y, Rohwer F, Breitbart M. Broad surveys of DNA viral diversity obtained through viral metagenomics of mosquitoes. PLoS ONE. 2011b;6: doi: 10.1371/journal.pone.0020579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni T, Yue J, Sun G, Zou Y, Wen J, Huang J. Ancient gene transfer from algae to animals: mechanisms and evolutionary significance. 83BMC Evol Biol. 2012;12 doi: 10.1186/1471-2148-12-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh N, Tanaka K, Shibata F, Kondo N, Hizume M, Shimada M, Fukatsu T. Wolbachia genome integrated in an insect chromosome: evolution and fate of laterally transferred endosymbiont genes. Genome Res. 2008;18:272–280. doi: 10.1101/gr.7144908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Graf J. Knowing your friends: invertebrate innate immunity fosters beneficial bacterial symbioses. Nat Rev Microbiol. 2012;10:815–827. doi: 10.1038/nrmicro2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveros JC. VENNY. An interactive tool for comparing lists with Venn diagrams. 2007 bioinfogp.cnb.csic.es/tools/venny/index.html
- Olsen GJ, Lane DJ, Giovannoni SJ, Pace NR, Stahl DA. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- Peterson TM, Gow AJ, Luckhart S. Nitric oxide metabolites induced in Anopheles stephensi control malaria parasite infection. . Free Radic Biol. 2007;42:132–142. doi: 10.1016/j.freeradbiomed.2006.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumpuni CB, Beier MS, Nataro JP, Guers LD, Davis JR. Plasmodium falciparum: inhibition of sporogonic development in Anopheles stephensi by Gram-negative bacteria. Exp Parasitol. 1993;77:195–199. doi: 10.1006/expr.1993.1076. [DOI] [PubMed] [Google Scholar]
- Pumpuni CB, Demaio J, Kent M, Davis JR, Beier JC. Bacterial population dynamics in three anopheline species: the impact on Plasmodium sporogonic development. Am J Trop Med Hyg. 1996;54:214–218. doi: 10.4269/ajtmh.1996.54.214. [DOI] [PubMed] [Google Scholar]
- Rainey SM, Shah P, Kohl A, Dietrich I. Understanding the Wolbachia-mediated inhibition of arboviruses in mosquitoes: progress and challenges. J Gen Virol. 2014;95:517–530. doi: 10.1099/vir.0.057422-0. [DOI] [PubMed] [Google Scholar]
- Rani A, Sharma A, Rajagopal R, Adak T, Bhatnagar RK. Bacterial diversity analysis of larvae and adult midgut microflora using culture-dependent and culture-independent methods in lab-reared and field-collected Anopheles stephensi - an Asian malarial vector. 96BMC Microbiol. 2009;9 doi: 10.1186/1471-2180-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe NA, Whitten MMA. Gillespie SH, Smith GL, Osbourn A. Microbe-vector interactions in vector-borne diseases. Cambridge University Press; Cambridge: 2004. Vector immunity. pp. 199–262. [Google Scholar]
- Ren X, Hoiczyk E, Rasgon JL. Viral paratransgenesis in the malaria vector Anopheles gambiae. PLoS Pathog. 2008;4: doi: 10.1371/journal.ppat.1000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Rasgon JL. Potential for the Anopheles gambiae densonucleosis virus to act as an “evolution-proof” biopesticide. . J Virol. 2010;84:7726–7729. doi: 10.1128/JVI.00631-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci I, Damiani C, Capone A, Defreece C, Rossi P, Favia G. Mosquito/microbiota interactions: from complex relationships to biotechnological perspectives. Curr Opin Microbiol. 2012;15:278–284. doi: 10.1016/j.mib.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Rosario K, Breitbart M. Exploring the viral world through metagenomics. Curr Opin Virol. 2011;1:289–297. doi: 10.1016/j.coviro.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Rosenberg E, Zilber-Rosenberg I. Symbiosis and development: the hologenome concept. Birth Defects Res C Embryo Today. 2011;93:56–66. doi: 10.1002/bdrc.20196. [DOI] [PubMed] [Google Scholar]
- Russell CW, Bouvaine S, Newell PD, Douglas AE. Shared metabolic pathways in a coevolved insect-bacterial symbiosis. Appl Environ Microbiol. 2013;79:6117–6123. doi: 10.1128/AEM.01543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlein Y, Polacheck I, Yuval B. Mycoses, bacterial infections and antibacterial activity in sandflies (Psychodidae) and their possible role in the transmission of leishmaniasis. Parasitology. 1985;90:57–66. doi: 10.1017/s0031182000049015. [DOI] [PubMed] [Google Scholar]
- Segata N, Boernigen D, Tickle TL, Morgan XC, Garrett WS, Huttenhower C. Computational meta’omics for microbial community studies. 666Mol Syst Biol. 2013;9 doi: 10.1038/msb.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinka ME, Bangs MJ, Manguin S, Coetzee M, Mbogo CM, Hemingway J, Patil AP, Temperley WH, Gething PW, Kabaria CW, Okara RM, Van Boeckel T, Godfray HC, Harbach RE, Hay SI. The dominant Anopheles vectors of human malaria in Africa, Europe and the Middle East: occurrence data, distribution maps and bionomic précis. . 117Parasit Vectors. 2010;3 doi: 10.1186/1756-3305-3-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinka ME, Bangs MJ, Manguin S, Rubio-Palis Y, Chareonviriyaphap T, Coetzee M, Mbogo CM, Hemingway J, Patil AP, Temperley WH, Gething PW, Kabaria CW, Burkot TR, Harbach RE, Hay SI. A global map of dominant malaria vectors. 69Parasit Vectors. 2012;5 doi: 10.1186/1756-3305-5-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suenaga H. Targeted metagenomics: a high-resolution metagenomics approach for specific gene clusters in complex microbial communities. Environ Microbiol. 2012;14:13–22. doi: 10.1111/j.1462-2920.2011.02438.x. [DOI] [PubMed] [Google Scholar]
- Suttle CA. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- Tanenbaum DM, Goll J, Murphy S, Kumar P, Zafar N, Thiagarajan M, Madupu R, Davidsen T, Kagan L, Kravitz S, Rusch DB, Yooseph S. The JCVI standard operating procedure for annotating prokaryotic metagenomic shotgun sequencing data. Stand Genomic Sci. 2010;2:229–237. doi: 10.4056/sigs.651139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Gilbert J, Meyer F. Metagenomics - a guide from sampling to data analysis. 3Microb Inform Exp. 2012;2 doi: 10.1186/2042-5783-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toft C, Andersson SG. Evolutionary microbial genomics: insights into bacterial host adaptation. Nat Rev Genet. 2010;11:465–475. doi: 10.1038/nrg2798. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hurk AF, Hall-Mendelin S, Pyke AT, Frentiu FD, McElroy K, Day A, Higgs S, O’Neill SL. Impact of Wolbachia on infection with chikungunya and yellow fever viruses in the mosquito vector Aedes aegypti . . PLoS Negl Trop Dis. 2012;6: doi: 10.1371/journal.pntd.0001892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, McMeniman CJ, Leong YS, Dong Y, Axford J, Kriesner P, Lloyd AL, Ritchie SA, O’Neill SL, Hoffmann AA. The w Mel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. . Nature. 2011;476:450–453. doi: 10.1038/nature10355. [DOI] [PubMed] [Google Scholar]
- Wang S, Ghosh AK, Bongio N, Stebbings KA, Lampe DJ, Jacobs-Lorena M. Fighting malaria with engineered symbiotic bacteria from vector mosquitoes. Proc Natl Acad Sci USA. 2012;109:12734–12739. doi: 10.1073/pnas.1204158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Jacobs-Lorena M. Genetic approaches to interfere with malaria transmission by vector mosquitoes. Trends Biotechnol. 2013;31:185–193. doi: 10.1016/j.tibtech.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B, Aksoy S. Microbiome influences on insect host vector competence. Trends Parasitol. 2011;27:514–522. doi: 10.1016/j.pt.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn SC, Maudlin I. Tsetse-trypanosome interactions: rites of passage. Parasitol Today. 1999;15:399–403. doi: 10.1016/s0169-4758(99)01512-4. [DOI] [PubMed] [Google Scholar]
- Wernegreen JJ. Strategies of genomic integration within insect-bacterial mutualisms. Biol Bull. 2012;223:112–122. doi: 10.1086/BBLv223n1p112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitten MM, Shiao SH, Levashina EA. Mosquito midguts and malaria: cell biology, compartmentalization and immunology. Parasite Immunol. 2006;28:121–130. doi: 10.1111/j.1365-3024.2006.00804.x. [DOI] [PubMed] [Google Scholar]
- Williamson SJ, Yooseph S. From bacterial to microbial ecosystems (metagenomics). Methods Mol Biol. 2012;804:35–55. doi: 10.1007/978-1-61779-361-5_3. [DOI] [PubMed] [Google Scholar]
- Wilmes P, Bond PL. Metaproteomics: studying functional gene expression in microbial ecosystems. Trends Microbiol. 2006;14:92–97. doi: 10.1016/j.tim.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Wommack KE, Bhavsar J, Ravel J. Metagenomics: read length matters. Appl Environ Microbiol. 2008;74:1453–1463. doi: 10.1128/AEM.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AC, Chaston JM, Douglas AE. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. . ISME J. 2013;7:1922–1932. doi: 10.1038/ismej.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooley JC, Godzik A, Friedberg I. A primer on metagenomics. PLoS Comput Biol. 2010;6: doi: 10.1371/journal.pcbi.1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zélé F, Nicot A, Berthomieu A, Weill M, Duron O, Rivero A. Wolbachia increases susceptibility to Plasmodium infection in a natural system. Proc Biol Sci. 2014;281 doi: 10.1098/rspb.2013.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zélé F, Nicot A, Duron O, Rivero A. nfection with Wolbachia protects mosquitoes against Plasmodium-induced mortality in a natural system. J Evol Biol. 2012;25:1243–1252. doi: 10.1111/j.1420-9101.2012.02519.x. [DOI] [PubMed] [Google Scholar]
- Zug R, Hammerstein P. Still a host of hosts for Wolbachia : analysis of recent data suggests that 40% of terrestrial arthropod species are infected. . PLoS ONE. 2012;7: doi: 10.1371/journal.pone.0038544. [DOI] [PMC free article] [PubMed] [Google Scholar]