Abstract
T-cell intracellular antigen-1 (TIA-1) is a key DNA/RNA binding protein that regulates translation by sequestering target mRNAs in stress granules (SG) in response to stress conditions. TIA-1 possesses three RNA recognition motifs (RRM) along with a glutamine-rich domain, with the central domains (RRM2 and RRM3) acting as RNA binding platforms. While the RRM2 domain, which displays high affinity for U-rich RNA sequences, is primarily responsible for interaction with RNA, the contribution of RRM3 to bind RNA as well as the target RNA sequences that it binds preferentially are still unknown. Here we combined nuclear magnetic resonance (NMR) and surface plasmon resonance (SPR) techniques to elucidate the sequence specificity of TIA-1 RRM3. With a novel approach using saturation transfer difference NMR (STD-NMR) to quantify protein–nucleic acids interactions, we demonstrate that isolated RRM3 binds to both C- and U-rich stretches with micromolar affinity. In combination with RRM2 and in the context of full-length TIA-1, RRM3 significantly enhanced the binding to RNA, particularly to cytosine-rich RNA oligos, as assessed by biotinylated RNA pull-down analysis. Our findings provide new insight into the role of RRM3 in regulating TIA-1 binding to C-rich stretches, that are abundant at the 5′ TOPs (5′ terminal oligopyrimidine tracts) of mRNAs whose translation is repressed under stress situations.
Keywords: DNA/RNA binding protein (D/RBP), RNA recognition motifs (RRM), scaffold independent analysis (SIA), surface plasmon resonance (SPR), saturation transfer difference-NMR (STD-NMR), TIA-1
Introduction
Stress granules (SG) formation is a protective mechanism that eukaryotic cells implement in response to environmental stresses that require repression of mRNA translation.1,2 Stress conditions such as heat shock, UV irradiation, oxidative stress, or starvation trigger SG assembly in the cytoplasm, where mRNAs are sequestered by their associated proteins and protected from degradation for the duration of the stress.
T-cell intracellular antigen-1 (TIA-1) and TIA-1 related (TIAR) are DNA/RNA binding proteins (D/RBPs) that shuttle between the nucleus and the cytoplasm so as to regulate transcriptional and post-transcriptional processes.3-6 Under stress conditions, these proteins accumulate in the cytoplasm and regulate the translation and the turnover of an ample set of mRNAs.7-10 In fact, TIA-1/TIAR are key proteins in SG formation,3,11 as they first bind mRNAs, and then self-associate to initiate SG assembly.12 Some of these mRNAs are defined by the presence of A/U-rich elements (AREs)13,14 and others by their C-rich 5′ terminal oligopyrimidine tracts (5′ TOPs)10,15,16 at the 3′- and 5′-untranslated regions (UTRs), respectively. In addition, the binding specificity of TIA-1/TIAR proteins to the UTRs-motifs has been detected and analyzed in several in vitro studies.17-21
The D/RBP TIA-1 shows high-sequence homology with TIAR.22 It is comprised of three RNA recognition motifs (RRM) followed by a glutamine-rich prion-related domain (PRD) at the C-terminal region. Notably, each single TIA-1 domain is characterized by different DNA/RNA specificities and affinities. RRM1 binds to T-rich DNA,20,23 and not to RNA, but enhances TIA-1 binding to U-rich sequences.17,21 The PRD domain seems to be essential for SG assembly as it mediates TIA-1/TIAR aggregation12 and is involved in splicing together with RRM1.24 Among all the TIA-1 constituent-domains, RRM2 is necessary, and actually the major contributor, to the U-rich/T-rich RNA/DNA binding sequences.17,21 The C-terminal RRM domain, RRM3, was described to bind RNA sequences that differ from U-rich motifs,17 but little is known about its sequence specificity and contribution to TIA-1/RNA interactions. Recent studies show that RRM3 enhances RRM2 binding to U-rich elements, suggesting a significant role in RNA binding.21
TIA-1 RRM domains 1 and 2 exhibit the typical βαββαβ RRM fold,25,26 whereas RRM3 shows a non-canonical N-terminal α-helix using the TIAR RRM3 domain (PDB ID 1X4G) as a template.27 This extra element in the third RRM domain has also been identified in the homologous Sacharomyces cerevisiae Pub1p protein.28 In addition, these proteins share some other unusual features such as a Trp-Gly-[Arg/Lys] motif and a distorted β-sheet. The presence of these features suggests that a new type of RRM family called TIA-1 C-terminal domain like RRM could be considered (TRRM).27,28
Given its singular structural features along with the evidence of contribution to the overall binding to RNA, the present work investigates the RNA specificity and affinity of RRM3 in the context of TIA-1 function. Here, an NMR approach based on a Scaffold-independent analysis (SIA)29-31 allowed us to decipher the RNA sequence specificity of the TIA-1 RRM3 domain. As an outcome, a remarkable affinity for C-rich RNA was identified, which was further corroborated by saturation transfer difference NMR spectroscopy (STD-NMR). This technique is a powerful tool to analyze protein–ligand interactions that provides quantitative information about the binding affinity by monitoring the ligand signals.32-34 In addition, STD-NMR can be used to measure KD for protein–ligand interactions over a wide range of affinities, from nM to high mM values.32 STD-NMR has been used before to detect protein–DNA/RNA interactions,35,36 although binding affinities were not determined. In fact, this is a novel approach to study protein–RNA interactions as the use of STD-NMR allows not only to detect the interactions and confirm SIA data, but also to obtain KD values and discriminate among the tested sequences. Finally, the influence of RRM3 on TIA-1 binding to RNA was evaluated in the context of RRM2 by SPR, as well as in the full-length TIA-1 protein in pull-down assays using proteins expressed in human cervical carcinoma (HeLa) cells. The presence of RRM3 significantly enhanced the RNA binding of TIA-1 to C-rich motifs. The RNA sequence specificity elucidated in this work along with the influence on RRM2 binding suggest that RRM3 confers additional RNA sequence specificity to TIA-1/RNA binding, with a potential role in the regulation of mRNAs containing 5′ TOPs regions defined by C-rich elements.
Results
TIA-1 RRM3 significantly binds to C-rich RNA sequences
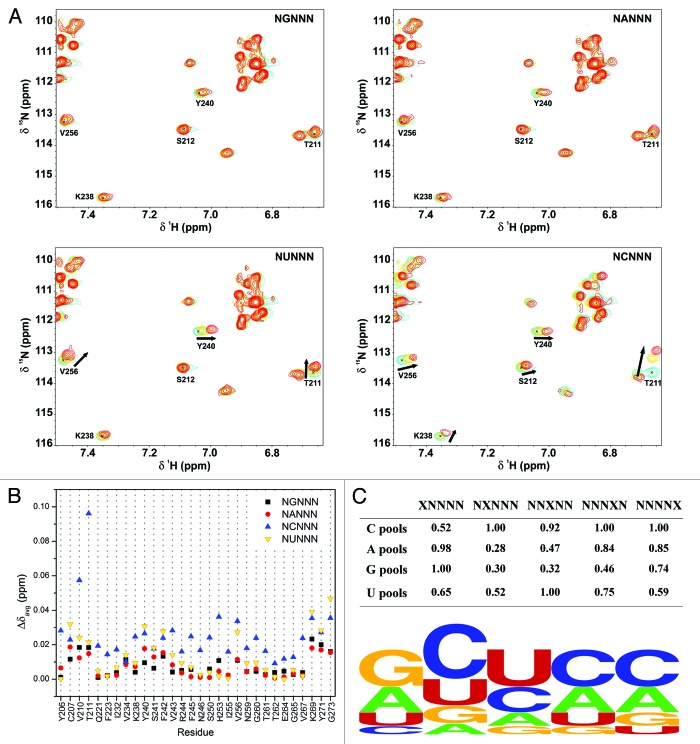
The SIA method was applied to TIA-1 RRM3 in order to determine its specificity for particular nucleotide sequences (Fig. 1). This method compares interactions between an array of short quasi-randomized RNA sequences and a RRM domain detected by NMR providing the preferred RNA binding sequence of the protein domain. Oligonucleotides comprising five bases were used to maximize the interaction of an RRM domain and minimize the possibility of multiple binding. The prevalence of a nucleotide in a specific position of the RNA sequence is indicated by a score ranging between 0–1 and the comparison of these scores for the four possible nucleotides in each position are used to evaluate the binding preference of a RRM for a target RNA.29-31
Figure 1. SIA assays for TIA-1 RRM3. (A) Overlays of a region from 15N-HSQC spectra recorded along the titration of the TIA-1 RRM3 domain with the NGNNN, NANNN, NUNNN, and NCNNN pools. Each panel shows the superimposition of the three spectra corresponding to the free protein (blue), 1:1 (yellow), and 1:4 (red) protein:RNA ratios. (B) Averaged chemical-shift changes (Δδavg) of selected RRM3 residues in the four protein:RNA titrations with the non-randomized pools in the second position (NXNNN pools). Δδavg for the 1:4 protein:RNA ratio is plotted vs. the residue number. (C) Top, comparison of the final score for the four nucleotides in a specific position on a 0–1 scale. This provides an evaluation of the binding sequence preference(s) of the protein. Bottom, graphic representation of the TIA-1 RRM3 RNA sequence preference. The picture was obtained by plotting SIA data with the Weblogo server (http://weblogo.berkeley.edu/logo.cgi).
15N RRM3 NMR titrations (Fig. 1A) were performed according to the protocol described in Materials and Methods with 20 RNA pools to follow protein–RNA interactions. To analyze nucleotide preference of RRM3 in a particular position of its RNA target, the scores for the fixed position pools were obtained and classified (Materials and Methods, Fig. 1B and C). In the case of the second position, as an example, NCNNN yielded the highest score (1.00) in comparison to the other three NA/G/UNNN pools, thus the preferred nucleotide in the second position of RNA sequence is a C. Interestingly, SIA showed that the RRM3 domain preferentially bound Cs at the second, the fourth, and the fifth positions of RNA targets, whereas the first and the third positions were occupied by G/A and U/C, respectively.
To validate the SIA method and assess binding using NMR (Fig. 2), several 5-mer RNA oligonucleotides were designed, according to the RNA preference results (Fig. 1C). GCUCC and ACUCC were chosen among the “best” RRM3 targets, while CAGGU was used as the “worst” target sequence. In addition, several “intermediate-affinity” targets were designed for testing possible discrimination between nucleotides in the first and the third RNA positions like ACCCC and GCCCC. RNA binding was monitored by acquiring 15N HSQC spectra (Materials and Methods, Fig. 2) of TIA-1 RRM3 titrated with the described oligonucleotides. Most observed chemical-shifts perturbations were small for the affected residues in all sets of assays with the exception of CAGGU titration that showed no signal changes even at a 1:4 RRM3:RNA ratio (Fig. 2). Notably, GCUCC and ACUCC showed larger chemical-shifts perturbations than ACCCC or GCCCC, suggesting that RRM3 preferentially binds a U at the third position over a C, in agreement with SIA results. Nevertheless, RRM3 binding affinities between ACUCC/GCUCC or ACCCC/GCCCC pairs might be similar based on the magnitude of chemical-shift perturbations, suggesting that G or A at the first position were equally probable. The RNA interaction platform was mainly located at the central RRM3 β strands (β1 and β3), along with the distorted β4 and the C-terminal stretch from Gly273 to Tyr276 (Fig. S1) as it has been recently described for TIA-1 and the homologous Pub1p.21,28,37
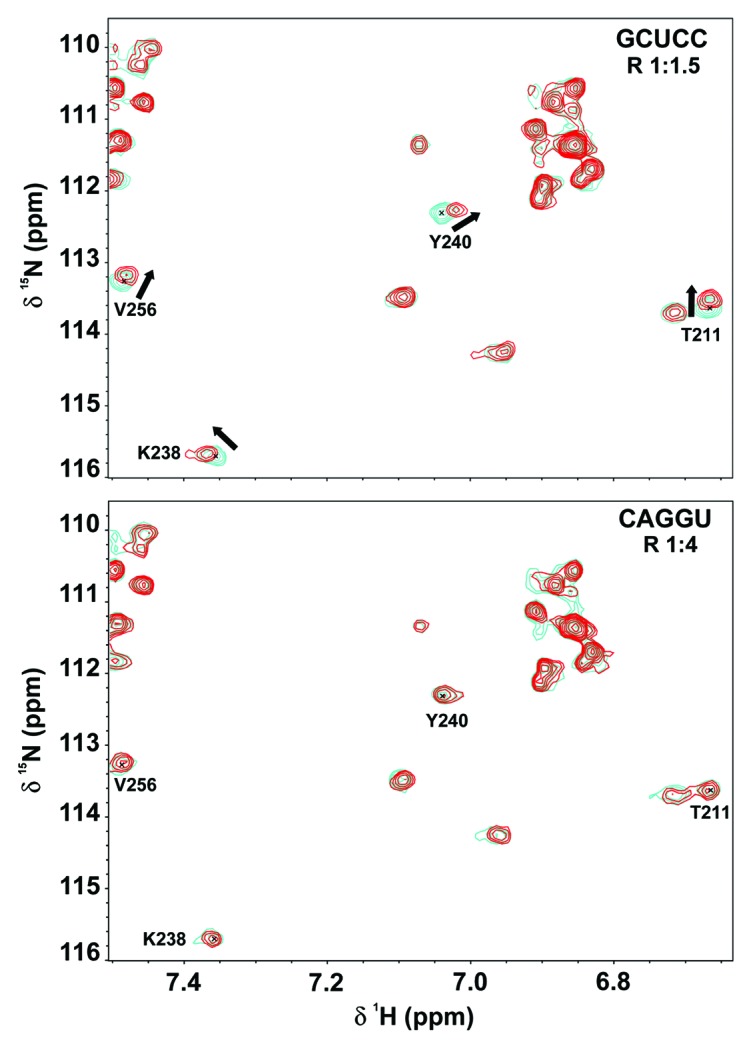
Figure 2. Binding of TIA-1 RRM3 to one of the “best” (GCUCC) and the “worst” (CAGGU) RNA targets resulting from SIA scores. Overlays of a region from 15N-HSQC spectra recorded along the titration of RRM3 domain with GCUCC (upper panel) and CAGGU (lower panel) RNA oligos. Each panel shows the superimposition of the spectra corresponding to free protein (blue) and 1:1.5 (red) protein:RNA ratio for the GCUCC oligo and 1:4 for the CAGGU.
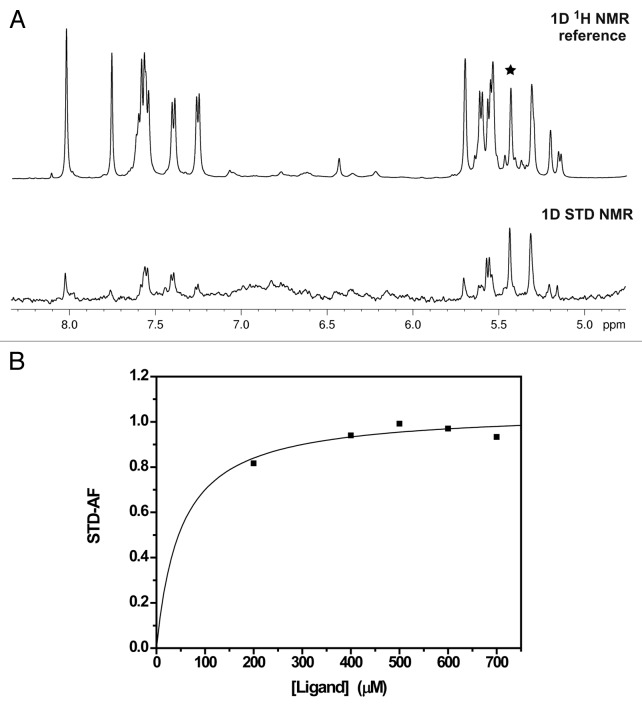
As the magnitudes of the chemical-shifts perturbations were too small to be used for quantitative assessment of binding affinities (Fig. 2; Fig. S1), STD-NMR was applied to measure the TIA-1 RRM3-RNA target equilibrium dissociation constants (Fig. 3; Fig. S2; Table 1). Several RNA oligos were tested as potential binders of TIA-1 RRM3 protein as described in Materials and Methods. The STD results shown in Table 1 were consistent with SIA data confirming not only that GCUCC/ACUCC and U-rich are good RRM3 targets but also that CAGGU does not bind to RRM3 at all. According to the KD values determined for the tested targets, RRM3 binds C’s in several RNA sequence positions. RRM3 binds ACUCC with a KD ≈50 μM (Fig. 3; Fig. S2; Table 1), ~20-fold stronger than ACCCC in agreement with the observed trends in the NMR titrations. Similar results were observed for the GCUCC and GCCCC KD values. Thus, STD-NMR was able to discriminate preferential binding of a U at the third position over a C. With respect to the first position at the RNA oligo, KD values of A/GCUCC pair indicated that A was favored over G, although both purines arise with similar SIA scores (Fig. 1C).
Figure 3. STD-NMR experiments for the determination of RRM3-RNA target binding affinity. (A) Expansion of the aromatic region of the 1D 1H NMR reference spectrum (top) and STD NMR spectrum (bottom) of RRM3 with ACUCC RNA at a protein: RNA ratio of 1:13 recorded at 1 s of saturation time and 5 °C. Due to the ligand excess, the signals in the reference spectrum correspond to aromatic protons of the RNA oligonucleotide. A black star stands for the signal monitored in the isotherm. (B) Binding isotherm from STD-AF values during the titration of RRM3 with the ACUCC oligo.
Table 1. Dissociation constants for RRM3-RNA targets complexes by STD NMR.
| Construct | RNA | KD (μM) |
|---|---|---|
| RRM3 | ACUCC | 50 ± 20 |
| ACCCC | 1100 ± 480 | |
| GCUCC | 155 ± 41 | |
| GCCCC | ~600 | |
| UUUUU | 140 ± 60 | |
| CAGGU | No binding |
C- and U-rich motifs are both RRM3 targets
To evaluate the significance of RRM3 binding to C-rich sequences compared with the canonical U-rich targets,17,18,21 STD-NMR titration with the UUUUU oligo was performed. RRM3 bound to the 5-mer U-rich with ~3-fold weaker affinity than to the best target ACUCC (KD ≈140 μM and 50 μM, respectively), but with a similar KD as GCUCC oligo (KD ≈155 μM; Table 1). The differences in KD values between the sequences ACUCC/GCUCC suggest that the presence of one adenine at the first position of these RNA motifs is clearly favored. Moreover, the central U flanked is essential for RRM3 binding, as inferred from STD-NMR measurements comparing ACUCC/ACCCC and GCUCC/GCCCC pairs. Thus, although C- and U-rich RNA stretches are targets of TIA-1 RRM3 domain, the protein shows slight preference by ACUCC sequences.
RRM3 enhances the overall binding of TIA-1 to C-rich RNA targets
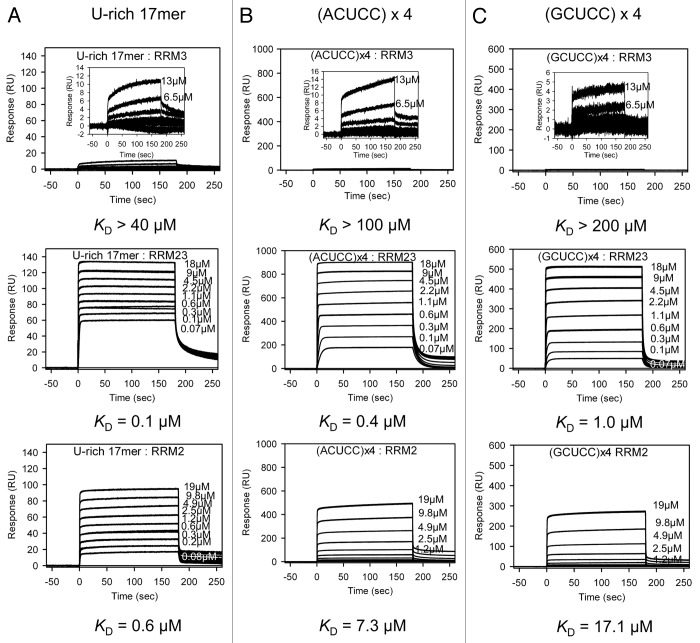
In order to use another method to validate the comparative binding affinities of RRM3 to the C-rich sequences, we also utilized SPR. This approach involved tethering tetrameric repeats of the target RNA sequences to the surface of the chip to ensure that the binding site would be available without steric hindrance from the 5′-tether and to effectively enhance the affinity for quantitative comparison. Figure 4 shows a series of RRM3 sensorgrams at a range of concentrations binding to the ACUCC, GCUCC, and a U-rich sequence for comparison.
Figure 4. SPR Analysis of the interactions of different TIA-1 constructs with RNA. The binding of TIA-1 RRM3 (top), TIA-1 RRM23 (middle), and TIA-1 RRM2 (bottom) to three difference RNA sequences is shown. (A) U-rich RNA, (B) (ACUCC)x4 RNA sequence, and (C) (GCUCC)x4 RNA, biotinylated at the 5′-end, were captured on SA coated sensor chips in parallel. Each protein was injected across the four flow cells (blank cell and a cell for each RNA sequence) at a range of concentrations and in triplicate. Injections were performed for 180 s (association phase), followed by a 360 s flow of running buffer to assess dissociation. The data were used to construct binding curves for KD determination or approximation (where steady-state binding was not achieved). The data shown in the figure are presented with comparative scales. The top row sensorgrams show inset graphs with a reduced scale to reflect the critical point of TIA-1 RRM3 binding.
Binding affinities of RRM3 were at the lower limit of detection for this method (in the mid-micromolar range; Fig. 4, Table 2), but were consistent with the affinities estimated by STD-NMR. Importantly, RRM3 binding to ACUCC gave rise to higher responses than those to GCUCC. Binding to the poly-U sequence seemed to be higher affinity than any of the C-rich sequences determined by the SIA method. However, this is likely to reflect the enhancement of the association-rate that occurs when there are many possible binding registers at the target, rather than reflecting a binding preference for U-rich RNA, as shown using NMR above.
Table 2. Dissociation constants for TIA-1 RRM domains-RNA complexes by SPR.
| Construct | RNA | KD (μM) |
|---|---|---|
| RRM2 | U-rich 17mer | 0.646 ± 0.005 |
| (ACUCC)x4 | 7.34 ± 0.07 | |
| (GCUCC)x4 | 17.1 ± 0.1 | |
| C-rich TIAR* | > 30 | |
| RRM3 | U-rich 17mer | > 40 |
| (ACUCC)x4 | > 100 | |
| (GCUCC)x4 | > 200 | |
| RRM23 | U-rich 17mer | 0.1042 ± 0.0007 |
| (ACUCC)x4 | 0.418 ± 0.001 | |
| (GCUCC)x4 | 0.958 ± 0.002 | |
| C-rich TIAR* | 3.50 ± 0.01 |
The C-rich TIAR RNA oligo refers to the UUGCCACCUCCUGCUCCUGCCCAGA biological sequence of TIAR reported in reference 18.
In order to also consider the contribution to binding that RRM3 makes in the context of an adjacent RRM2, the equivalent SPR measurements were made with TIA-1 RRM23 and RRM2 alone. Binding of TIA-1 RRM23 to all three sequences was of sub- micromolar affinity (Fig. 4, Table 2). Binding to RRM2, in each case, was of slightly lower affinity. By comparing these measurements, the contribution made by RRM3 within the RRM23 construct could thus be ascertained. In the case of poly-U binding, RRM3 enhanced binding by 6-fold. In the case of ACUCC and GCUCC binding, RRM3 enhanced binding by 18- and 17-fold, respectively.
Similar results were obtained from the assays performed with the C-rich motif (UUGCCACCUC CUGCUCCUGC CCAGA), which is present in several putative target mRNAs of TIAR protein18 (Table 2; Fig. S3). In these experiments, the RRM23 domain affinity by such oligo (KD = 3.50 ± 0.01 μM) was slightly lower than those from the other C-rich RNA targets, although RRM3 enhanced binding over 9-fold too. Thus, SPR confirms the ability of RRM3 to contribute to binding specifically to these C-rich sequences.
To further evaluate the RRM3 domain effect over the TIA-1 protein binding to RNA and their biological significance, biotin pull-down assays were performed (Fig. 5). TIA-1 full-length (TIA-1 FL) and TIA-1 devoid of RRM3 domain (TIA-1-ΔRRM3) were first expressed in HeLa cells transfected with the corresponding vectors, and the lysates were used to study protein binding to biotinylated RNA. TIA-1 FL preferentially bound to poly-U and certain C-rich motifs in contrast to the others, as those enriched with G. Among the C-rich oligos tested, (ACUCC)x5 sequence was the more effective in binding to TIA-1 FL protein, and the affinity significantly decreased when the TIA-1-ΔRRM3 mutant was tested. The specificity of TIA-1 FL by the (ACUCC)x5 and the consensus target-sequence of TIAR18 RNA sequences is clear in comparison to the affinity observed by the (ACCCC)x5 motif. In fact, C-rich motifs negligibly bound to the mutant while the affinity for the U-rich oligo was nearly the same as that seen with the wild-type protein, highlighting the “enhancer effect” of RRM3 domain over the TIA-1 binding to C-rich sequences. Importantly, these observations fully corroborate the SPR results with RRM2 and RRM23 domains, as well as the remarkable binding of the TIA-1 RRM3 domain by ACUCC RNA motifs elucidated by SIA and STD-NMR.
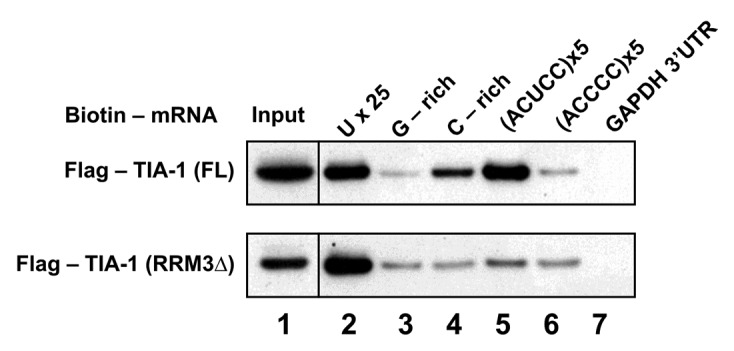
Figure 5. Immunoprecipitation of TIA-1 FL and TIA-1-ΔRRM3 proteins with RNA oligos. Western blot detection of Flag-tagged TIA-1 FL and TIA-1-ΔRRM3 present in the pull-down assays cell extracts. Lane 1 corresponds to the input fraction of TIA-1 FL and TIA-1-ΔRRM3 expressed in HeLa cells while lanes 2–7 are purified fractions of such TIA-1 proteins bound to the RNA sequences tested. Sequence GGGGGCGGGG GCGGGGCGGG GG was used in lane 3, UUGCCACCUC CUGCUCCUGC CCAGA18 was used in lane 4, and negative control GAPDH 3`UTR was used in lane 7.
Discussion
The contribution of the third RRM domain of TIA-1 and its homologs to the RNA recognition process has been hinted in recent studies, which suggest a key role for RRM3 in RNA binding and mRNA transcript selection.21,28 In the current study, the specific binding to certain RNA sequences by this domain was determined by the SIA method and evaluated by a combination of STD-NMR and SPR techniques. Further, the RRM3 influence in the context of RRM2 domain and the full-length TIA-1—using a tandem construct of RRM23 for SPR and the TIA-1 FL and TIA-1-ΔRRM3 mutant for pull-down assays with mammalian cells—was analyzed to better understand the TIA-1/RNA interaction.
Regarding the NMR-based approach, we first determined the RNA sequence specificity of TIA-1 RRM3 domain, and later confirmed that it binds to C-rich and U-rich motifs in the sub-micromolar range (Fig. 1 and Table 1). The RNA interaction platform is located at the canonical β1, β3, and the distorted β4 strands along with the C-terminal stretch from Gly273 to Tyr276 (Fig. S1). Since the magnitude of the chemical-shift perturbations were too small for accurate quantitative analysis, STD-NMR was used to evaluate the RRM3–RNA interactions detected by the SIA method. In the STD-NMR experiments, no isotopic labeling of the molecules under study was required. Rather, the 1H resonances of the RNA nitrogen-bases, either in their protein-bound or free states, were followed. Protein-nucleic acids complexes have been previously studied by Cross Saturation measurements.38 However, this NMR method monitored only the protein resonances instead of the nucleic acid ones. Previous works used the STD-NMR method to detect protein–DNA/RNA interactions, although binding affinities were not determined.35,36 To our knowledge, this is the first instance in which such an approach has been used to quantify the affinity of protein–RNA complexes (Fig. 3, Table 1). In this system, the TIA-1 RRM3 domain was a particularly demanding protein for STD NMR analysis due to its small size (~14 kDa) that limits the sensitivity of the experiment because it could not be effectively saturated by spin diffusion, hindering the intra- and intermolecular saturation transfer. To overcome this problem, low temperature (5 °C) was used in the titrations to optimize the relaxation properties of the protein and favor the saturation process. In addition, the RNA concentration employed in the titrations was adjusted to optimize the signal-to-noise ratios of STD-NMR spectra and quantify the binding affinities of TIA-1 RRM3 domain. The accuracy of this approach is further corroborated by the use of SPR and biotin pull-down assays. This analysis revealed that ACUCC was the preferred RNA target by RRM3 (with KD in the micromolar range), with the most specific positions of the RNA located at the first and the middle nitrogen-bases. SPR measurements performed on C-rich motifs were in agreement with STD data, although the affinity constants were not as accurate because RRM3 bound more weakly. These data are consistent with Dember and co-workers who concluded that TIA-1 RRM2 binds to U-rich RNA sequences, and suggested that RRM3-RNA binding specificity could be distinct from U-rich motifs.17 Our results also correlate with more recent works that describe TIAR protein binding not only to U-rich RNA motifs, but also AU-rich and C-rich stretches with lower but significant binding affinities.18,19 Interestingly, enhanced binding to an AU-rich sequence of TIAR123 (three-domain construct) over TIAR12 (two-domain construct) was also reported,19 whereas this effect was negligible for U-rich elements. The high homology between TIAR and TIA-1,22 along with TIAR in vitro studies, allows one to speculate that RRM3 may influence the binding affinity and sequence preference of TIA-1 by RNA oligos. In fact, the SIA and STD-NMR data highlight the fact that the TIA-1 RRM3 domain exhibits a sequence preference, which goes clearly beyond the canonical U-rich sequences, namely C-rich motifs.
The contribution of the C-terminal RRM3 domain in the binding of TIA-1 protein to C-rich (A/GCUCC) or U-rich RNA motifs was first analyzed by SPR assays (Fig. 4, Table 2) using either combined or isolated RRM domains, and then in the context of mammalian cells transfected with the gen coding for the TIA-1 full-length protein. As was expected, the SPR assays showed that the RRM2 domain was mainly responsible for TIA-1 binding to RNA in terms of affinity, with the U-rich motifs being preferred (KD of 0.6 μM) over the C-rich motifs. The addition of RRM3 fused to RRM2 revealed enhanced binding to all RNA sequences. However, this effect was dramatically stronger (17- or 18-fold) for C-rich sequences in comparison to U-rich sequences (6-fold). Pull-down assays, performed with TIA-1 FL protein and the TIA-1-ΔRRM3 mutant expressed in HeLa cells and incubated with the RNA sequences elucidated by the former NMR experiments, confirmed these findings in a biological context. These experiments indicated that the presence of RRM3 domain was essential for the TIA-1 binding to C-rich motifs and further supported the preference for ACUCC sequences.
The role of RRM3 as an RNA binding-enhancer for TIA-1 was previously suggested only for U-rich stretches.21,39 Nevertheless, our study reveals that such a role strongly depends on the RNA sequence. Although the RNA binding of TIA-1 is mainly dictated by RRM2, we conclude that RRM3 significantly enhances the affinity, especially for certain sequences such as C-rich motifs.
Under stress conditions, TIA-1/TIAR proteins are translational repressors that target specific mRNAs sequestering them into SG.1,8 TIA-1 interacts with these mRNAs by recognizing certain motifs at the 3′ or 5′ UTRs, such as AREs and 5′ TOPs, respectively.7,10 The mRNAs containing 5′ TOPs, which are C-rich stretches, encode ribosomal proteins and translation factors15,40,41 that are selectively repressed in response to nutrient deprivation or amino acid starvation.42,43 The results reported herein confirm the RNA binding specificity of TIA-1 for C-rich RNA sequences. Actually, RRM3 shows a preference for ACUCC, which substantially increases the TIA-1 specificity for this kind of sequences over the U-rich ones. Overall, this work sheds light on the role of the RRM3 domain of TIA-1 in regulating and enhancing the protein binding to C-rich RNA stretches. The different contribution of each TIA-1 domain to the RNA/DNA recognition seems to be related by the specific RNA sequence, with RRM3 possibly enhancing TIA-1 binding to C-rich elements at 5′ TOP mRNAs10 for repressing translation.
Materials and Methods
Protein and RNA oligonucleotides preparation
Single- and two-domain constructs of TIA-1, named RRM2 (residues 95–184), RRM3 (residues 190–288), and RRM23 (residues 80–288), were obtained from the plasmid containing full-length TIA-1 provided by Prof P Anderson (Harvard Medical School) as previously described.27 15N-labeled and unlabeled proteins were expressed in Escherichia coli BL21 (DE3) as His-tagged proteins and purified using nickel affinity chromatography according to well-established protocols.27 The His-tag was not removed from the constructs for NMR experiments since no differences between the secondary structure of TIA-1 domains with or without His-tag were observed.27 The His-tag was cleaved from the proteins using TEV protease and removed with a second nickel affinity step only for SPR experiments. Protein concentration was spectrophotometrically determined using predicted extinction coefficients and protein purity was assessed by SDS-PAGE.
All RNA oligonucleotides were chemically synthesized (IDT, Integrated DNA Technologies; 1 μmol synthesis). The 20 randomized RNA pools (designed as described in the next section) were solubilized in 20 mM potassium phosphate buffer pH 7.0 and 50 mM KCl to a final concentration in the range 3–3.5 mM. To test the reliability of the RNA randomized-based method, the 5-mer RNA oligos GCUCC, ACUCC, ACCCC, GCCCC, UUUUU, and CAGGU were redissolved in the same conditions as the pools. Those oligos used in STD-NMR experiments were prepared with 99.9% D2O buffer containing 20 mM potassium phosphate pH 7.0 and 50 mM KCl in the range of 8–11 mM.
Scaffold-independent analysis (SIA)
NMR titrations of 50 μM RRM3 samples with 20 randomized 5-mer RNA pools were performed to analyze the RNA binding sequence preference of this TIA-1 domain. Each of the 20 pools contained 256 different RNA sequences that shared a single nucleotide at a single position. Four of the five oligo positions were occupied by a randomized mixture of four bases (N) and the fifth position—which could be either position one, two, three, four, or five—were occupied by A, G, U, or C (i.e., NANNN, NGNNN, NUNNN, NCNNN).29 15N-HSQC spectra were recorded at protein–RNA ratios of 1:0, 1:1, and 1:4 and chemical-shift perturbations were evaluated for the peaks affected upon binding (Fig. 1A). The assignments of TIA-1 RRM3 backbone resonances were previously performed with standard 3D NMR experiments (HNCACB, HNCA, and HNCO) and were already deposited at the BMRB with the accession number 18829. Thirty protein peaks affected by RNA binding that could be followed in all titrations were selected and the largest chemical-shift changes were observed for NCNNN titration. The averaged chemical-shift changes upon addition of 20 pools to the RRM3 domain were plotted in five histograms (Fig. 1B). To evaluate the differences between the RNA pools and the RNA binding preference, the four shifts for a given nucleotide position displayed in each histogram per residue were normalized to the highest of the four. Then, normalized values were averaged over all 30 residues to obtain a set of scores as an outcome. Comparison and evaluation of the final scores are shown in Figure 1C.
NMR titrations
Solutions of 50 μM 15N-labeled samples of RRM3 in 90% H2O/10% D2O buffer containing 20 mM potassium phosphate pH 7.0 and 50 mM KCl were titrated with the 5-mer RNA oligonucleotides inferred from SIA analysis at 25 °C in the protein:RNA ratio from 1:0 to 1:4. It was not possible to reach higher ratios than 1:1.5 with the GCUCC and ACUCC sequences, ratios 1:3.5 with the GCCCC and ACCCC oligos, and ratio 1:4 with CAGGU in samples prepared in 90% H2O/10% D2O due to protein precipitation. 15N-HSQC spectra were recorded at each point of the titrations on a 800 MHz Varian INOVA spectrometer and were processed using the NMRPipe package.44 Further, the spectra were analyzed and superimposed with Sparky.45 The pH value of the samples was verified after each titration step. Averaged chemical-shift differences were measured and calculated as follows: Δδavg = (([ΔδH]2 + [ΔδN/5]2) / 2)1/2, being ΔδH and ΔδN chemical-shift increments in 1H and 15N, respectively. The Origin 8.5 program (OriginLab) was used for data manipulation.
The assigned chemical shifts of RRM3 backbone deposited at the BMRB with accession number 18829 were used to generate a three-dimensional protein structure at the CS23D2.0 web server.46 This structure was used for data discussion and to build Figure S1.
Saturation transfer difference NMR (STD-NMR) spectroscopy
Solutions of 60 μM unlabeled TIA-1 RRM3 protein in 99.9% D2O buffer containing 20 mM potassium phosphate pH 7.0 and 50 mM KCl were titrated with the RNA oligos derived from SIA analysis (GCUCC, ACUCC, ACCCC, GCCCC, UUUUU, and CAGGU; protein-RNA ratios from 1:0 to 1:13) at 5 °C recording 1D 1H STD NMR spectra on a 500 MHz Bruker Avance DRX spectrometer.
Very briefly, STD NMR experiments consist of two 1D 1H NMR spectra, in which we apply a selective low power radiofrequency before the observation 90 pulse in two different spectral regions, respectively. In the “off-resonance” experiment it is applied at a frequency where no NMR resonances, either from the protein or from the ligand, are present (typically 40 ppm), so no effect on any intensity is expected, and a normal 1D spectrum is obtained. In the “on-resonance” experiment, the radiofrequency is applied at a frequency characteristic of protein signals, but avoiding any frequency of the ligand. This produces a selective saturation of the protein signals, which in the end is spread all over the macromolecule due to the efficient process of spin diffusion, leading to a more or less homogeneous distribution of saturation over the whole protein. The binding of the smaller molecule to the protein-binding site brings some ligand protons to a very close spatial position related to the protein protons, and a process of intermolecular NOE transfer then part of the saturation from the protein protons to the ligand ones. The exchange between the bound and free states of the ligand allows the accumulation of that saturation in the bulk solution, and the transferred saturation is observed on the ligand resonances. The difference between the “off” and “on” experiments give rise to a 1D spectrum containing only the signals that received saturation from the protein in the “on” experiment.
For selective saturation of the protein signals, a train of 20 low power Gaussian-shaped pulses of 49 ms, separated each by a 1 ms delay, was applied at 0.7 ppm (on-resonance frequency), whereas for the reference spectrum, the frequency was shifted to 40 ppm (off-resonance). Spectra were processed and analyzed with TopSpin 3.0. The pH value of the samples was verified after each titration step.
In STD NMR titration experiments, the STD intensity (ηSTD) reflects the concentration of ligand–protein complex present in solution33,47 as it depends on the fraction of bound ligand. Multiplying the observed STD by the molar excess of ligand over protein (ε) the STD intensity is converted into a factor, the STD amplification factor, defined by STD-AF = ε (Io-Isat)/Io = ε ηSTD. This factor is proportional to the fraction of bound protein.48 The evolution of the STD-AF, along a TIA-1 RRM3 titration experiment, gave an association isotherm that could be fitted to a Langmuir equation: STD-AF([L]) = (STD-AFmax·[L])/(KD +[L]), where STD-AFmax is the maximum STD-AF value during the titration (plateau), [L] is the unbound ligand concentration, and KD is the dissociation constant. To determine the KD values of the TIA-1 RRM3-RNA complexes from titration experiments, the STD-AF values were plotted vs. the concentration of ligand, using a selected ligand STD signal (well-isolated in the spectrum). To avoid potential effects of fast ligand rebinding on the measured KD, the lowest saturation time that led to observable ligand signals was used (tsat = 1 s).49 The Origin 8.5 program (OriginLab) was used for data manipulation.
Protein–RNA binding affinity determined by surface plasmon resonance (SPR)
SPR was performed using a BIAcore T100 (GE Healthcare). RNA oligomers 5′–UUUUUUUUUU UUUUUUU–3′, 5′–ACUCCACUCC ACUCCACUCC–3′, 5′–GCUCCGCUCC GCUCCGCUCC–3′, 5′–UUGCCACCUC CUGCUCCUGC CCAGA–3′, synthesized with 5′ Biotin tags (Dharmacon Research), were diluted to 2 nM in 10 mM HEPES, 150 mM NaCl, 1 mM DTT, 0.025% P20, pH 7.4, and were immobilized on flow cells of a BIAcore SA series S sensor chip (GE Life Science) to levels 48 RU, 109 RU, 89 RU, and 114 RU, respectively. Flow cell 1 was left blank as a reference. Triplicate samples in 10 mM HEPES, 150 mM NaCl, 1 mM DTT, 0.025% P20, pH 7.4 of TIA-1 RRM2 (0–19 µM), TIA-1 RRM3 (0–13 µM), and TIA-1 RRM23 (0–18 µM) were injected for 180 s at 50 µL per min, with a 6 min dissociation, using 10 mM HEPES, 150 mM NaCl, 1 mM DTT, 0.025% P20, pH 7.4 as running buffer. Experiments were run at 10 °C. Results were analyzed using Scrubber2 (BioLogic Software). The data was double referenced. Equilibrium binding responses were calculated using averaging over a 91 s window starting 57 s after injection and used to determine equilibrium affinities.
Biotin pull-down assays
HeLa cells were transfected with plasmids pCI-neo-3XFlag-TIA-1 (FL) and pCI-neo-3XFlag-TIA-1(ΔRRM3) (kind gifts from RN Singh and NN Singh).50 Twenty-four h later, whole-cell lysates were prepared in a buffer containing 20 mM TRIS-HCl at pH 7.5, 100 mM KCl, 5 mM MgCl2, and 0.5% NP-40 by incubation on ice for 10 min and centrifugation at 10 000 × g for 15 min at 4 °C. RNAs pre-labeled with 5′-end biotin [5′–UUUUUUUUUU UUUUUUUUUU UUUUU–3′, 5′–GGGGGCGGGG GCGGGGGCGG GGG–3′, 5′–UUGCCACCUC CUGCUCCUGC CCAGA–3′,18 5′–ACUCCACUCC ACUCCACUCC ACUCC–3′, and 5′–ACCCCACCCC ACCCCACCCC ACCCC–3′] were purchased from IDT. Biotinylated GAPDH 3′UTR was synthesized by PCR amplification of cDNA with primers (CCAAGCTTCT AATACGACTC ACTATAGGGA GACCTCAACG ACCACTTTGT CA and GGTTGAGCAC AGGGTACTTT ATT) in the presence of biotinylated CTP and T7 RNA polymerase, as described previously.51,52
Lysates (150 μg per sample) were incubated with biotinylated RNA (1 μg per sample) for 30 min at room temperature, and complexes were isolated with streptavidin-coupled Dynabeads (Invitrogen). Proteins present in the pull-down material were studied by western blot analysis as described previously.52,53
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the Andalusian Government (P07-CVI-02896, P11-CVI-7216, and BIO198) and by the Australian Research Council (DP110102056 awarded to Wilce JA). Cruz-Gallardo I was supported by European Social Fund (ERDF 2007–2013) and Andalusian Government (fellowship associated to P08-CVI-3876). Aroca Á was awarded with a FEBS Short-Term fellowship in 2011. The financial support in the form of Access to the Bio-NMR Research Infrastructure is co-funded under the 7th Framework Programme of the EC (FP7/2007–2013) grant agreement 261863. Gorospe M and Yoon J-H were supported by the NIA-IRP, NIH.
The authors would like to thank Dr P.Anderson (Harvard Medical School) and RN Singh and NN Singh;50 for kindly provide the plasmid containing the TIA-1 full-length protein and RRM3 mutant. The authors thank the technical assistance provided by the NMR services at the Centro de Investigación Tecnología e Innovación de la Universidad de Sevilla (CITIUS). We are also grateful to Prof Miguel A De la Rosa and Dr Antonio Díaz-Quintana for critical reading of the manuscript.
Glossary
Abbreviations:
- 5′ TOPs
5′ terminal oligopyrimidine tracts
- AREs
A/U-rich elements
- D/RBPs
DNA/RNA binding proteins
- KD
equilibrium dissociation constant
- HSQC
heteronuclear single-quantum correlation
- NMR
nuclear magnetic resonance
- PRD
prion-related domain
- RRM
RNA recognition motifs
- STD-NMR
saturation transfer difference NMR
- SIA
scaffold-independent analysis
- SG
stress granules
- SPR
surface plasmon resonance
- TIA-1
T-cell intracellular antigen-1
- TIA-1 FL
TIA-1 full-length
- TIAR
TIA-1 related
- UTRs
untranslated regions
References
- 1.Anderson P, Kedersha N. Stressful initiations. J Cell Sci. 2002;115:3227–34. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- 2.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–50. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–68. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Förch P, Puig O, Kedersha N, Martínez C, Granneman S, Séraphin B, Anderson P, Valcárcel J. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol Cell. 2000;6:1089–98. doi: 10.1016/S1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 5.Zhang T, Delestienne N, Huez G, Kruys V, Gueydan C. Identification of the sequence determinants mediating the nucleo-cytoplasmic shuttling of TIAR and TIA-1 RNA-binding proteins. J Cell Sci. 2005;118:5453–63. doi: 10.1242/jcs.02669. [DOI] [PubMed] [Google Scholar]
- 6.Reyes R, Alcalde J, Izquierdo JM. Depletion of T-cell intracellular antigen proteins promotes cell proliferation. Genome Biol. 2009;10:R87. doi: 10.1186/gb-2009-10-8-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.López de Silanes I, Galbán S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol. 2005;25:9520–31. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mazan-Mamczarz K, Lal A, Martindale JL, Kawai T, Gorospe M. Translational repression by RNA-binding protein TIAR. Mol Cell Biol. 2006;26:2716–27. doi: 10.1128/MCB.26.7.2716-2727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamasaki S, Stoecklin G, Kedersha N, Simarro M, Anderson P. T-cell intracellular antigen-1 (TIA-1)-induced translational silencing promotes the decay of selected mRNAs. J Biol Chem. 2007;282:30070–7. doi: 10.1074/jbc.M706273200. [DOI] [PubMed] [Google Scholar]
- 10.Damgaard CK, Lykke-Andersen J. Translational coregulation of 5’TOP mRNAs by TIA-1 and TIAR. Genes Dev. 2011;25:2057–68. doi: 10.1101/gad.17355911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 α to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–42. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–98. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen CYA, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–70. doi: 10.1016/S0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 14.von Roretz C, Di Marco S, Mazroui R, Gallouzi IE. Turnover of AU-rich-containing mRNAs during stress: a matter of survival. Wiley Interdiscip Rev RNA. 2011;2:336–47. doi: 10.1002/wrna.55. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton TL, Stoneley M, Spriggs KA, Bushell M. TOPs and their regulation. Biochem Soc Trans. 2006;34:12–6. doi: 10.1042/BST0340012. [DOI] [PubMed] [Google Scholar]
- 16.Ivanov P, Kedersha N, Anderson P. Stress puts TIA on TOP. Genes Dev. 2011;25:2119–24. doi: 10.1101/gad.17838411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dember LM, Kim ND, Liu KQ, Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J Biol Chem. 1996;271:2783–8. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- 18.Kim HS, Kuwano Y, Zhan M, Pullmann R, Jr., Mazan-Mamczarz K, Li H, Kedersha N, Anderson P, Wilce MCJ, Gorospe M, et al. Elucidation of a C-rich signature motif in target mRNAs of RNA-binding protein TIAR. Mol Cell Biol. 2007;27:6806–17. doi: 10.1128/MCB.01036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim HS, Wilce MCJ, Yoga YMK, Pendini NR, Gunzburg MJ, Cowieson NP, Wilson GM, Williams BRG, Gorospe M, Wilce JA. Different modes of interaction by TIAR and HuR with target RNA and DNA. Nucleic Acids Res. 2011;39:1117–30. doi: 10.1093/nar/gkq837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HS, Headey SJ, Yoga YMK, Scanlon MJ, Gorospe M, Wilce MCJ, Wilce JA. Distinct binding properties of TIAR RRMs and linker region. RNA Biol. 2013;10:579–89. doi: 10.4161/rna.24341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bauer WJ, Heath J, Jenkins JL, Kielkopf CL. Three RNA recognition motifs participate in RNA recognition and structural organization by the pro-apoptotic factor TIA-1. J Mol Biol. 2012;415:727–40. doi: 10.1016/j.jmb.2011.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawakami A, Tian Q, Duan X, Streuli M, Schlossman SF, Anderson P. Identification and functional characterization of a TIA-1-related nucleolysin. Proc Natl Acad Sci U S A. 1992;89:8681–5. doi: 10.1073/pnas.89.18.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suswam EA, Li YY, Mahtani H, King PH. Novel DNA-binding properties of the RNA-binding protein TIAR. Nucleic Acids Res. 2005;33:4507–18. doi: 10.1093/nar/gki763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Förch P, Puig O, Martínez C, Séraphin B, Valcárcel J. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 2002;21:6882–92. doi: 10.1093/emboj/cdf668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar AO, Swenson MC, Benning MM, Kielkopf CL. Structure of the central RNA recognition motif of human TIA-1 at 1.95A resolution. Biochem Biophys Res Commun. 2008;367:813–9. doi: 10.1016/j.bbrc.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuwasako K, Takahashi M, Tochio N, Abe C, Tsuda K, Inoue M, Terada T, Shirouzu M, Kobayashi N, Kigawa T, et al. Solution structure of the second RNA recognition motif (RRM) domain of murine T cell intracellular antigen-1 (TIA-1) and its RNA recognition mode. Biochemistry. 2008;47:6437–50. doi: 10.1021/bi7024723. [DOI] [PubMed] [Google Scholar]
- 27.Aroca A, Díaz-Quintana A, Díaz-Moreno I. A structural insight into the C-terminal RNA recognition motifs of T-cell intracellular antigen-1 protein. FEBS Lett. 2011;585:2958–64. doi: 10.1016/j.febslet.2011.07.037. [DOI] [PubMed] [Google Scholar]
- 28.Santiveri CM, Mirassou Y, Rico-Lastres P, Martínez-Lumbreras S, Pérez-Cañadillas JM. Pub1p C-terminal RRM domain interacts with Tif4631p through a conserved region neighbouring the Pab1p binding site. PLoS One. 2011;6:e24481. doi: 10.1371/journal.pone.0024481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beuth B, García-Mayoral MF, Taylor IA, Ramos A. Scaffold-independent analysis of RNA-protein interactions: the Nova-1 KH3-RNA complex. J Am Chem Soc. 2007;129:10205–10. doi: 10.1021/ja072365q. [DOI] [PubMed] [Google Scholar]
- 30.García-Mayoral MF, Díaz-Moreno I, Hollingworth D, Ramos A. The sequence selectivity of KSRP explains its flexibility in the recognition of the RNA targets. Nucleic Acids Res. 2008;36:5290–6. doi: 10.1093/nar/gkn509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cukier CD, Hollingworth D, Martin SR, Kelly G, Díaz-Moreno I, Ramos A. Molecular basis of FIR-mediated c-myc transcriptional control. Nat Struct Mol Biol. 2010;17:1058–64. doi: 10.1038/nsmb.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer B, Peters T. NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew Chem Int Ed Engl. 2003;42:864–90. doi: 10.1002/anie.200390233. [DOI] [PubMed] [Google Scholar]
- 33.Angulo J, Nieto PM. STD-NMR: application to transient interactions between biomolecules-a quantitative approach. Eur Biophys J. 2011;40:1357–69. doi: 10.1007/s00249-011-0749-5. [DOI] [PubMed] [Google Scholar]
- 34.Bhunia A, Bhattacharjya S, Chatterjee S. Applications of saturation transfer difference NMR in biological systems. Drug Discov Today. 2012;17:505–13. doi: 10.1016/j.drudis.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 35.Xia Y, Zhu Q, Jun K-Y, Wang J, Gao X. Clean STD-NMR spectrum for improved detection of ligand-protein interactions at low concentration of protein. Magn Reson Chem. 2010;48:918–24. doi: 10.1002/mrc.2687. [DOI] [PubMed] [Google Scholar]
- 36.Harris KA, Shekhtman A, Agris PF. Specific RNA-protein interactions detected with saturation transfer difference NMR. RNA Biol. 2013;10:1307–11. doi: 10.4161/rna.25948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cruz-Gallardo I, Aroca Á, Persson C, Karlsson BG, Díaz-Moreno I. RNA binding of T-cell intracellular antigen-1 (TIA-1) C-terminal RNA recognition motif is modified by pH conditions. J Biol Chem. 2013;288:25986–94. doi: 10.1074/jbc.M113.489070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramos A, Kelly G, Hollingworth D, Pastore A, Frenkiel T. Mapping the interfaces of protein-nucleic acid complexes using cross-saturation. J Am Chem Soc. 2000;122:11311–4. doi: 10.1021/ja002233w. [DOI] [Google Scholar]
- 39.McLaughlin KJ, Jenkins JL, Kielkopf CL. Large favorable enthalpy changes drive specific RNA recognition by RNA recognition motif proteins. Biochemistry. 2011;50:1429–31. doi: 10.1021/bi102057m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem. 2000;267:6321–30. doi: 10.1046/j.1432-1327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 41.Iadevaia V, Caldarola S, Tino E, Amaldi F, Loreni F. All translation elongation factors and the e, f, and h subunits of translation initiation factor 3 are encoded by 5′-terminal oligopyrimidine (TOP) mRNAs. RNA. 2008;14:1730–6. doi: 10.1261/rna.1037108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Avni D, Shama S, Loreni F, Meyuhas O. Vertebrate mRNAs with a 5′-terminal pyrimidine tract are candidates for translational repression in quiescent cells: characterization of the translational cis-regulatory element. Mol Cell Biol. 1994;14:3822–33. doi: 10.1128/mcb.14.6.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Avni D, Biberman Y, Meyuhas O. The 5′ terminal oligopyrimidine tract confers translational control on TOP mRNAs in a cell type- and sequence context-dependent manner. Nucleic Acids Res. 1997;25:995–1001. doi: 10.1093/nar/25.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 45.Goddard TD, Kneller DG. SPARKY 3. 2006. University of California, San Francisco. [Google Scholar]
- 46.Wishart DS, Arndt D, Berjanskii M, Tang P, Zhou J, Lin G. CS23D: a web server for rapid protein structure generation using NMR chemical shifts and sequence data. Nucleic Acids Res. 2008;36:W496–502. doi: 10.1093/nar/gkn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mayer M, Meyer B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew Chem Int Ed. 1999;38:1784–8. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 48.Mayer M, Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J Am Chem Soc. 2001;123:6108–17. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 49.Angulo J, Enríquez-Navas PM, Nieto PM. Ligand-receptor binding affinities from saturation transfer difference (STD) NMR spectroscopy: the binding isotherm of STD initial growth rates. Chemistry. 2010;16:7803–12. doi: 10.1002/chem.200903528. [DOI] [PubMed] [Google Scholar]
- 50.Singh NN, Seo J, Ottesen EW, Shishimorova M, Bhattacharya D, Singh RN. TIA1 prevents skipping of a critical exon associated with spinal muscular atrophy. Mol Cell Biol. 2011;31:935–54. doi: 10.1128/MCB.00945-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tominaga K, Srikantan S, Lee EK, Subaran SS, Martindale JL, Abdelmohsen K, Gorospe M. Competitive regulation of nucleolin expression by HuR and miR-494. Mol Cell Biol. 2011;31:4219–31. doi: 10.1128/MCB.05955-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoon J-H, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–55. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.