Summary
Human immunodeficiency viral (HIV-1) fusion is mediated by the viral envelope gp120/gp41 complex (ENVelope glycoprotein). After gp120 shedding, gp41 is exposed and elicits membrane fusion via a cascade of conformational changes. In contrast to pre-fusion and post-fusion conformation, little is known about any intermediate conformation. We report on a solution NMR investigation of homotrimeric HIV-1 gp4127–194, comprising the transmembrane region and reconstituted in dodecyl phosphocholine (DPC) micelles. The protein is mainly α-helical but experiences internal dynamics on the nanosecond and micro-to millisecond time scale and transient α-helical behavior for certain residues in the N-terminal heptad repeat (NHR). Strong lipid interactions are observed, in particular for C-terminal residues of the NHR and imunodominant loop region connecting NHR and C-terminal heptad repeat (CHR). Our data indicate an extended conformation with features anticipated for a pre-fusion intermediate, presumably in exchange with a lowly populated post-fusion six-helical bundle conformation.
Introduction
Fusion of the Human immunodeficiency virus (HIV-1) and the human T-cell is elicited by the envelope glycoprotein (Env) trimer on the viral surface. The Env trimer is composed of gp120/gp41 heterodimers. Upon binding to the CD4 receptor and a co-receptor on the surface of the human T cell, gp120 sheds from the viral spike, exposing gp41 which can then bind to the host cell membrane via its fusion peptide (FP), while the C-terminal transmembrane (TM) region remains anchored in the viral membrane (Blumenthal et al., 2012; Harrison, 2005). This elongated conformation of gp41, bridging the gap between HIV viral and host cell membrane, has been dubbed pre-hairpin intermediate conformation (Furuta et al., 1998; Harrison, 2005). HIV infection is characterized by a relatively long lag-phase, where prior to fusion and cell entry, gp41 is in an exposed pre-hairpin conformation (Dimitrov and Blumenthal, 1994; Munoz-Barroso et al., 1998) which is accessible to the humoral immune response. Hence, gp41 has been the target of a new class of antiviral drugs, called fusion inhibitors, with T-20 (Enfuvirtide, Fuzeon, Roche) being the first FDA-approved and most prominent one (Caffrey, 2011; Douek et al., 2006; Wild et al., 1994).
In the current model, fusion is initiated by a conformational change of the central gp41 ectodomain region from the pre-hairpin intermediate conformation to an antiparallel six-helical (6HB) bundle formed by a trimer of hairpins. In this way, viral and host-cell membrane are closely juxtaposed and hemifusion can be initiated (Blumenthal et al., 2012; Chan and Kim, 1998; Harrison, 2005, 2008). The current model is mainly based on X-ray crystallographic studies of the soluble ectodomain composed of an N- and Cterminal heptad repeat (NHR and CHR) region (Buzon et al., 2010; Chan et al., 1997; Weissenhorn et al., 1997). Besides that, high-resolution structural data for gp41 have remained limited to isolated domains only, such as fusion peptide (FP) and membrane proximal external region (MPER) (Jaroniec et al., 2005; Kim et al., 2011; Lai et al., 2012; Li and Tamm, 2007; Qiang et al., 2009; Sun et al., 2008). Cryo-electron microscopy studies have provided low-resolution models of the non-activated native viral spike (Zanetti et al., 2006; Zhu et al., 2006) and, at higher resolution, for reconstituted constructs of the ENV trimer, both in soluble form (Bartesaghi et al., 2013; Lyumkis et al., 2013) and reconstituted in detergent micelles (Mao et al., 2013b; Mao et al., 2012). The latter study suggested the gp41 ectodomain comprised smaller helical entities – in contrast to the two single α-helices for the NHR and CHR domain observed for the post-fusion X-ray structure of the isolated soluble NHR and CHR peptides. This discrepancy has been claimed artefactual due to technical issues and has been a matter of intense debate (Henderson, 2013; Mao et al., 2013a; Subramaniam, 2013; van Heel, 2013).
Recently, an X-ray structure of a of a soluble cleaved HIV-1 Env trimer (gp140, lacking the gp41 transmembrane (TM) and MPER region) in complex with a CD4 binding site broadly neutralizing antibody was obtained at 4.7 Å resolution (Julien et al., 2013). The structure was consistent with a high-resolution cryo-EM reconstruction and structural model of a similar construct (Lyumkis et al., 2013). In this model, the ectodomain arrangement differs significantly from the post-fusion 6HB arrangement: A more extended structure is observed where the NHR regions form a three helical bundle with the neighboring protomers in the trimer core, analogous to the post-fusion 6HB and the proposed open activated state determined by single particle EM (Tran et al., 2012). However, the CHR does not fold back along the NHR trimer in an anti-parallel manner but extends away from the bottom of the trimer.
Here we present an NMR study in solution of the structure and dynamics of gp4127–194, comprising residues 27–194 of gp41, corresponding to 538–705 in the HXB2 Env precursor, reconstituted in dodecyl phosphocholine (DPC) micelles. Compared to full-length gp41, this construct lacks the cytoplasmic tail (CT) and fusion peptide (FP) as well as fusion peptide proximal region (FPPR), but comprises the NHR and CHR region, connecting immunodominant loop (IL) region as well as membrane proximal external region (MPER) and transmembrane region (TM). Recently, we described studies on an analogous construct (gp411–194) which included in addition the FP and FPPR domains (Lakomek et al., 2013). Whilst this allowed the important assessment of secondary structure and dynamics of these domains, their addition compromised both the expression (lowered) and physical properties (less soluble, requiring higher detergent concentrations) of the protein. The gp4127–194 was physicochemically superior and this has resulted in more detailed NMR analyses and also allowed additional interpretation of the more complex gp411–194 spectra. We have used the detergent DPC as it has been found to be a suitable mimic for the membrane environment of gp41 (Lai et al., 2012; Li and Tamm, 2007; Sun et al., 2008), and despite extensive study we have not found a better replacement.
We show that gp4127–194, although α-helical for the NHR and visible part of the CHR region, contains several transient α-helical notches in the NHR region. High internal dynamics are observed with properties expected for an extended pre-fusion intermediate. Strong lipid interactions for the IL region and for several residues in the NHR region are observed, suggesting that they may play a more complex role in driving the fusion process. A monomer-trimer equilibrium for gp41 is observed that strongly depends on lipid interactions and this may also have a mechanistic impact on the fusion process.
Results
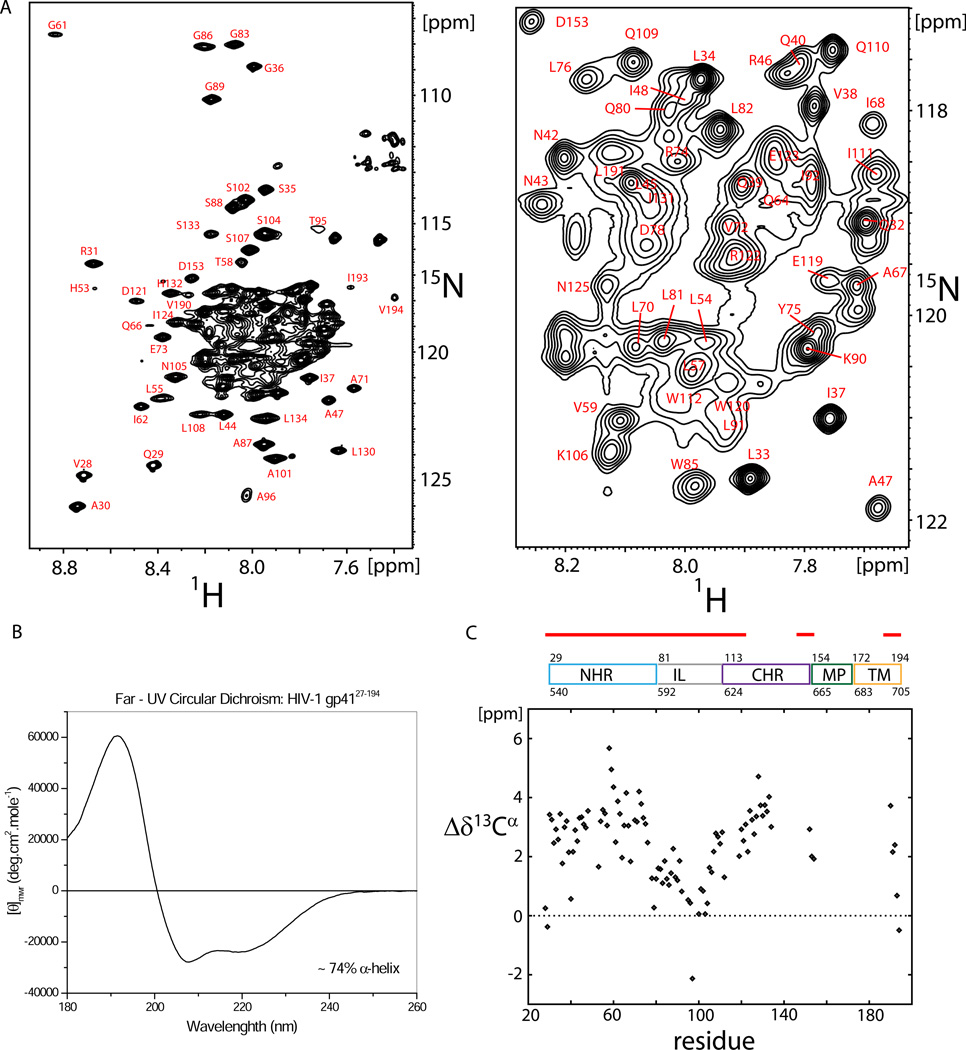
Secondary structure analysis
A representative 15N-1H TROSY-HSQC spectrum of uniformly 15N13C2H -enriched gp4127–194 (1mM in 50 mM NaAc pH 4, 25mM KCl, 138 mM DPC) is shown in Figure 1A. About 115 out of 165 expected resonances were detected in a 3D HNCO spectrum, of which approximately 85% could be assigned unambiguously, including an almost complete assignment of the NHR and IL regions and a major part of the CHR region as well as the C-terminus of the TM region (compare Table S3). Resonances for the C-terminal CHR, MPER and N-terminal TM region appear line broadened below the detection threshold (as discussed further below).
Figure 1. Secondary structure propensity of gp4127–194.
a) 800 MHz 15N-1H TROSY-HSQC spectrum of 1.0 mM 2H15N13C gp4127–194 in DPC micelles, 50mM sodium acetate, pH 4.0, 25 mM KCl, recorded at 40 °C (left) and where the most crowded region has been magnified for clarity (right). b) Far-UV spectrum of gp4127–194. The mean residue ellipticity θmwr [degrees cm2mol–1] is plotted versus the wave length [nm]. c) 13Cα secondary chemical shifts of gp4127–194 in DPC micelles.
The α-helical content of gp4127–194 was determined by far-UV CD spectroscopy and estimated to be about 74% (see Figure 1B). Both NHR and CHR region are α-helical, as indicated by large positive NMR 13Cα secondary chemical shifts (Figure 1C, Table S1). In detail, observed 13Cα secondary chemical shifts (Δδ13Cα) show a strong α-helical signature for residues A30 to D78 of the NHR region with an average value of < Δδ13Cα > ≈ 3.1 ± 0.9 ppm which is characteristic for stable α-helices. The central CHR region (starting from residue W120) also shows a strong α-helical signature (<Δδ13Cα > ≈ 3.3 ± 0.6 ppm), while residues D153-K154 at the transition line between CHR and MPER show reduced α-helical propensity. For the C-terminus, the α-helical propensity gradually decreases from V190 to V194, as indicated by decreasing Δδ13Cα chemical shifts.
It should be noted, however, that the strong α-helical propensity in the NHR helix is interrupted by several outliers such as G36, Q40, H53, G61 and Q64 that show decreased Δδ13Cα chemical shifts, whereas the region T58-W60 shows the strongest α-helical propensity. Residue V97 in the IL region shows a negative secondary chemical shift as expected due to nearest neighbor effects of P98 (Wang and Jardetzky, 2002).
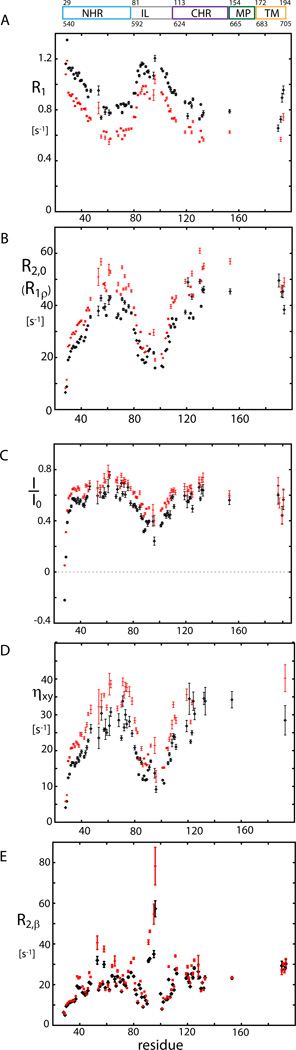
Internal dynamics
To assess the internal dynamics of gp41, 15N R1, R2, 15N-{1H} NOE and transverse 15N CSA-dipolar cross-correlated correlation rates, ηxy, were recorded (Figure 2A–2D, Table S4). Data were analyzed by the extended model-free approach (Clore et al., 1990), for details refer to Experimental Procedures and the Supplemental Information. Internal dynamics are characterized by a generalized order parameter, S2, and a correlation time τ, both for the fast, picosecond (Sf2, τf) and slower, low nanosecond time scale (Ss2, τs). As summarized in Table 1 (compare also Table S2), all domains of gp4127–194 show dynamics on a 2–5 ns time scale: moderate amplitude for the NHR and CHR regions as characterized by an order parameter of Ss2=0.76, high amplitude for the IL region connecting both domains (Ss2=0.48) and the TM region appears most rigid on the same time scale (Ss2=0.79). On a molecular level, this means that NHR and CHR are (stochastically) sampling different orientations relative to each other, the extensions of these motions are, however, (on the low ns time scale) restricted to an average semi-cone opening angle of 24±2° (in the model of motion in a cone).
Figure 2. 15N relaxation data of gp4127–194 in DPC micelles.
a) 15N R1 relaxation data recorded at 600 MHz (black) and 800 MHz (red) are highly consistent for both fields. b) R2,0 relaxation data (derived from R1ρ with a 2 kHz RF field; R1 contribution corrected) at 600 MHz (black) and 800 MHz (red). c) 15N- {1H} NOE values. d) Transverse CSA–dipolar cross-correlated relaxation rates ηxy. e) Hahn echo transverse relaxation rates, R2,β, at 600 MHz (black) and 800 MHz (red) for the slowly relaxing component of the 15N-{1H} doublets. Unlike in the R1ρ experiment, all conformational-exchange effects, Rex, contribute to R2,β and are not refocused.
Table 1.
Results of the extended model-free analysis of 15N relaxation data.
| Cluster | τc [ns][a] | S2f[b] | τf [ps][c] | τs [ns][d] | S2s[e] |
|---|---|---|---|---|---|
| 55–78 | 41±2 | 0.90±0.02 | 50 | 4.2±0.9 | 0.76±0.03 |
| 83–102 | 41±2 | 0.67±0.03 | 50 | 3.1±0.7 | 0.48±0.04 |
| 119–133 | 41±2 | 0.88±0.02 | 50 | 3.7±1.1 | 0.76±0.03 |
| 190–194 | 41±2 | 0.90±0.04 | 50 | 2.4±0.7 | 0.79±0.04 |
Rotational correlation time.
Order parameter for the fast time scale, order parameters can range between 0 and 1 with 0 being fully mobile and 1 entirely rigid.
The correlation time of the fast time scale motion has been fixed to 50 ps during the fitting.
Correlation time of the slow time-scale internal motion (<< τc).
Order parameter for the slow time scale.
Apart from the IL region, all domains, NHR, CHR and TM, show little dynamics on the fast time scale as characterized by order parameters equal or close to Sf2=0.90. Dynamics on the fast picosecond to nanosecond time-scale can be considered as internal flexibility. The finding of (on average) low flexibility of the NHR, CHR and TM region confirms the presence of well-ordered helical structure as already indicated by Δδ13Cα secondary chemical shifts (with exception of the two N- and two C-terminal residues displaying random-coil like Δδ13Cα).
Observing dynamics on the µs-ms time scale by the field dependent Hahn-echo experiment (Lakomek et al., 2013), we find strong exchange contributions, Rex, for H53, T58 and in particular for residues in the IL region near A96 (Figure 2E). In addition, there are moderate exchange contributions for residues around Q40, Q64, D78 and residues K106 to T128. Exchange effects, Rex, are due to conformational changes on the µs to ms time scale. As Rex contributions largely disappear when a spin-lock of νRF = 2kHz RF field strength is applied, most of the conformational changes must occur on a time scale much slower than about 80 µs. Several of those residues that are subject to Rex effects show reduced Cα secondary chemical shifts in an otherwise strongly α-helical surrounding, such as Q40, H53 and Q64. This finding points to transient α-helical character on the µs-ms time scale. Hence, the otherwise strongly α-helical NHR seems to be interrupted by transient α-helical notches allowing conformational rearrangements on the µs-ms time scale.
The IL region connecting NHR and CHR is characterized by high internal flexibility (fast ps-ns motions), high mobility on a 2–5 ns time scale and conformational dynamics on the µs-ms time scale (see above). Transitions between conformational states which are separated by a higher energy barrier will occur less frequently, meaning at lower rate and on a slower time scale, as described by transition state theory. It is of interest that the IL shows dynamics on three different time scales. Such hierarchy of time scales has been observed previously for the enzyme adenylate kinase (Henzler-Wildman et al., 2007). The low ns dynamics will not be connected to large conformational re-arrangements but rather to a stochastic sampling of different loop conformations within a certain energy well on the protein’s energy landscape. In contrast, the µs-ms dynamics observed for the region around A96 will be linked with larger conformational transitions such as, for example, between pre- and post-fusions intermediates or between different pre-fusion conformations.
Lipid interactions and solvent-accessibility
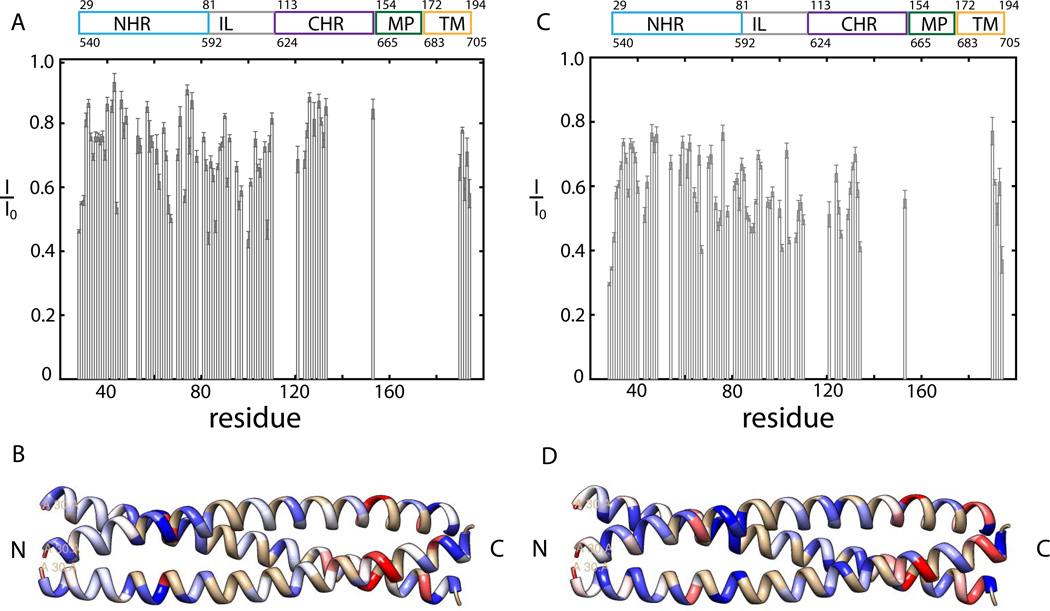
Lipid interactions of gp4127–194 have been investigated by adding 5-DOXYL-stearic acid (5-DSA), a lipophilic paramagnetic spin-label to the DPC detergent buffer. Paramagnetic relaxation enhancement (PRE) introduced by the 5-DSA label will lead to intensity reduction of the gp41 signals as compared to the (un-treated) diamagnetic reference sample (Hilty et al., 2004), for details refer to the Experimental Procedures.
Those residues that show the strongest decrease in normalized intensities (I/I0) experience the strongest lipid interactions, while residues not associated with lipid will show normalized intensities (I/I0) close to 1. The C-terminal TM region is the helical region with the strongest lipid interaction (compare Figure 3A, Table S5), as expected for a membrane anchor.
Figure 3. Normalized intensities (I/I0) for the lipid PRE and solvent PRE sample.
Intensities are normalized by comparison to the intensity of the reference sample I0, the smaller the ratio, the stronger the PRE effect is. I/I0 of 0.1 mM gp4127–194, 50 mM Na Ac pH4, 25mM KCl, 35 mM DPC plus a) 0.5 mM 5-DSA (final concentration) and c) 5 mM Gadodiamide (Omniscan) (final concentration). Normalized intensities are visualized for the NHR domain of 1ENV. Residues are color coded according to their lipid interactions and solvent exposure: b) blue for little lipid interactions (I/I0> 0.93) to white for medium lipid interactions (I/I0=0.71) to red for strong lipid interactions (I/I0< 0.5) and d) blue for little solvent exposure (I/I0> 0.77) to white for medium solvent exposure (I/I0=0.61) to red for strong solvent exposure (I/I0< 0.44).
Several residues in the NHR and IL regions exhibit strong lipid interaction (except S88-K90 and I92 which are all weak). In particular, strong interactions are observed in the C-terminal portion of NHR (I62, Q66, A67 and E73), whereas the N-terminal portion shows less interaction with only L44 showing strong binding. The lipid interaction sites are highlighted (Figure 3C) using the gp41 ectodomain X ray structure (pdb 1ENV. The actual structure of the NHR in gp4127–194 is likely to differ slightly). In the CHR region, residues N125-S133 show little lipid interaction, whereas, residue L134, at the interface to the invisible portion of the CHR region shows strong lipid interactions (discussed further below).
To complement the lipid PRE measurements, solvent PREs were measured using a soluble Gadoliniumbased spin label (compare Figure 3B, Table S5, see also Experimental Procedures). Solvent PREs are expected to behave complementary to lipid PREs, as only solvent accessible sites, not embedded in detergent, experience increased relaxation. The IL region (L81-Q110) shows the strongest intensity reduction compared to a reference sample with <I/I0>= 0.55±0.08. Solvent exposure of the loop region is expected. Also the CHR region shows significant solvent exposure with <I/I0>= 0.56±0.09. The strong solvent exposure of the CHR, together with the increased internal dynamics observed, indicates that the CHR (in the main conformation) will be detached from the NHR rather than forming a stable 6HB.
In contrast to the CHR, the more N-terminal part of the NHR region (L33-Q39, R46-I62) seems on average more protected from the solvent with <I/I0>= 0.70±0.05, while the C-terminal section of NHR seems more solvent accessible (compare Figure 3D, Table S5). However, within the NHR region several residues were identified that show increased solvent interactions as indicated by decreased I/I0, namely G36 and Q40-L44 and Q64. These residues show strongly decreased 13Cα secondary chemical shifts as well, indicating transient α-helical behavior in an otherwise strongly α-helical region. Another residue H53, which shows transient α-helical behavior, is broadened below the detection threshold in the solvent PRE spectra. In the NHR, residues Q40, H53 and Q64 all show Rex effects which indicate conformational dynamics on the intermediate µs-ms time scale.
Ideally, membrane bound and soluble paramagnetic agents produce mirror images of each other as solvent and lipid interactions are mutually exclusive. For gp4127–194, however, we find several residues, in particular in the IL region and C-terminal NHR, which show both solvent exposure and lipid interactions. This suggests transient behavior and sampling of at least two different conformations, one being solvent-exposed and the other one interacting with lipid.
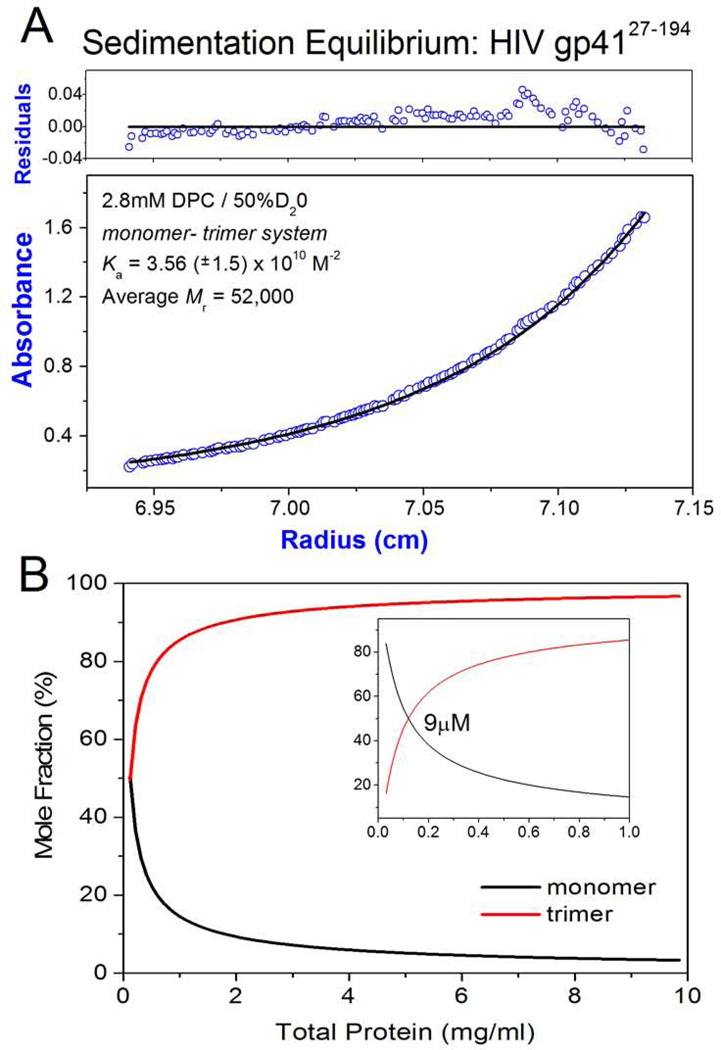
Monomer-Trimer Equilibrium
The molecular weight of gp4127–194 and associative properties were directly determined using sedimentation equilibrium centrifugation. To negate the density effect of bound detergent, centrifugation was performed in D2O (Noy et al., 2003). The average molecular weight was determined as 52 kDa and transformation of the equilibrium profile indicated a molecular weight increase as a function of concentration, indicating an associative process. Based on molecular weights approaching a trimer, we treated the equilibrium data as a monomer – trimer system and obtained an association constant (Ka) of 3.6 × 1010 M−2 (Figure 4 A). Thus, at NMR concentrations of 1 mM gp41 (~ 20mg/ml) the sample will be populated with close to 100% trimer (Figure 4B). This can be compared to gp4127–154 (also known as the HIV ectodomain) which, in the absence of the C-terminal MPER and TM regions as well as detergent, is a tighter trimer (Figure S1, A; Wingfield et al., 1997). However, when DPC is added at concentrations > CMC (1.5 mM) there is reduction in the Ka, which is now similar to that of gp4127–194 (Figure S1, B). This suggests that the detergent weakening of the trimeric association in gp4127–194 may be due to binding to shared sites with the longer construct, namely, as detailed above, the NHR and IL regions. To further delineate the effect of domain contributions, we studied a gp4127–154 construct (gp41 LD) where the IL loop was replaced with a six residue flexible linker. In the absence of detergent, this protein was also a monomer - trimer system with a Ka, similar to the parent gp4127–154 (Figure S1, C). However, in detergent micelles, removal of the IL loop significant destabilized the trimer potential of gp41 LD with a two order of magnitude reduction in Ka compared to the parent molecule (Figure S1, D). A similar ectodomain construct with an IL loop deletion was described recently (Roche et al., 2014) which also has a low trimer potential.
Figure 4. Analytical Ultracentrifugation.
a) Sedimentation equilibrium ultracentrifugation of HIV-1 gp4127–194: Panels are absorbance (bottom panel) and residuals (upper panel). Opened circles show 280nm absorbance gradients in the centrifuge cell. The solid line indicates the calculated fit for the monomer-trimer association. Residuals show the difference in the fitted and experimental values as a function of radial position. The data shown refers to a 15N13C2H labelled protein with a calculated monomeric mass of 22,000 (sample used directly for NMR analyses). The concentration range of the gradient (open circles) at equilibrium is ~ 1.0 µM – 20 µM. b) The monomer – trimer potential of gp4127–194 is indicated as mole fraction of monomer and trimer plotted as function of total protein concentration. Profiles were constructed using the Ka value indicated in (A). The insert is at a tenfold lower protein concentration and the indicated protein molarity corresponds to ~ 50% monomer and 50% trimer composition. The molecular weight of the non-labelled gp4127–194 construct is 19.65 kDa (1mg/ml = 50.9µM) and the NMR measurements described in the study were performed at ~ 20mg/ml (1mM).
From these results, it is clear that while detergent weakens the trimeric association of gp4127–194, it does so in a manner consistent with the behavior of the ectodomain core (gp4127–154) which in the presence of detergent is also a weaker trimer. The reversible dissociation of gp4127–194 would transiently populate monomeric domains, as for example the CHR regions, while maintaining a trimeric subunit structure at high protein concentrations. Furthermore, the IL region which we have shown can interact with detergent / lipid (Figure 3) has a stabilizing impact on the trimeric association of the ectodomain.
Under the conditions of our study, it should be emphasized that although detergent weakens the trimer potential of the various gp41 constructs they remain on the average mostly trimeric as can be appreciated by examining the profiles which show the mole fractions of monomer and trimer as a function of total protein concentration (Figures 4B, S2). How these concentrations relate to in-vivo values is hard to say, but in general terms, although average cellular values could be low, in membrane associated systems local concentrations can be very high.
Comparison of gp4127–194 and gp411–194 including the fusion peptide
The NMR backbone assignment and investigation of internal dynamics for gp4127–194 are more complete than achieved previously for the longer gp411–194 construct which included the FP and fusion peptide proximal region (FPPR) (Lakomek et al., 2013). Moreover, the 1H, 15N and 13Cα chemical shift correlations of gp4127–194 are almost identical to those of gp411–194 (compare Figure S3). Using the backbone assignment of gp4127–194, several additional resonances in gp411–194 were identified and additional relaxation data was extracted (Figure S4). The 15N relaxation data for gp411–194 are very similar compared to gp4127–194, as can be seen in Figure S5. Major differences occur only for residues V28-R45 which are N-terminal in gp4127–194 and this is probably due to the truncation. Relaxation data indicate overall very similar internal dynamics for both constructs, especially the TM region.
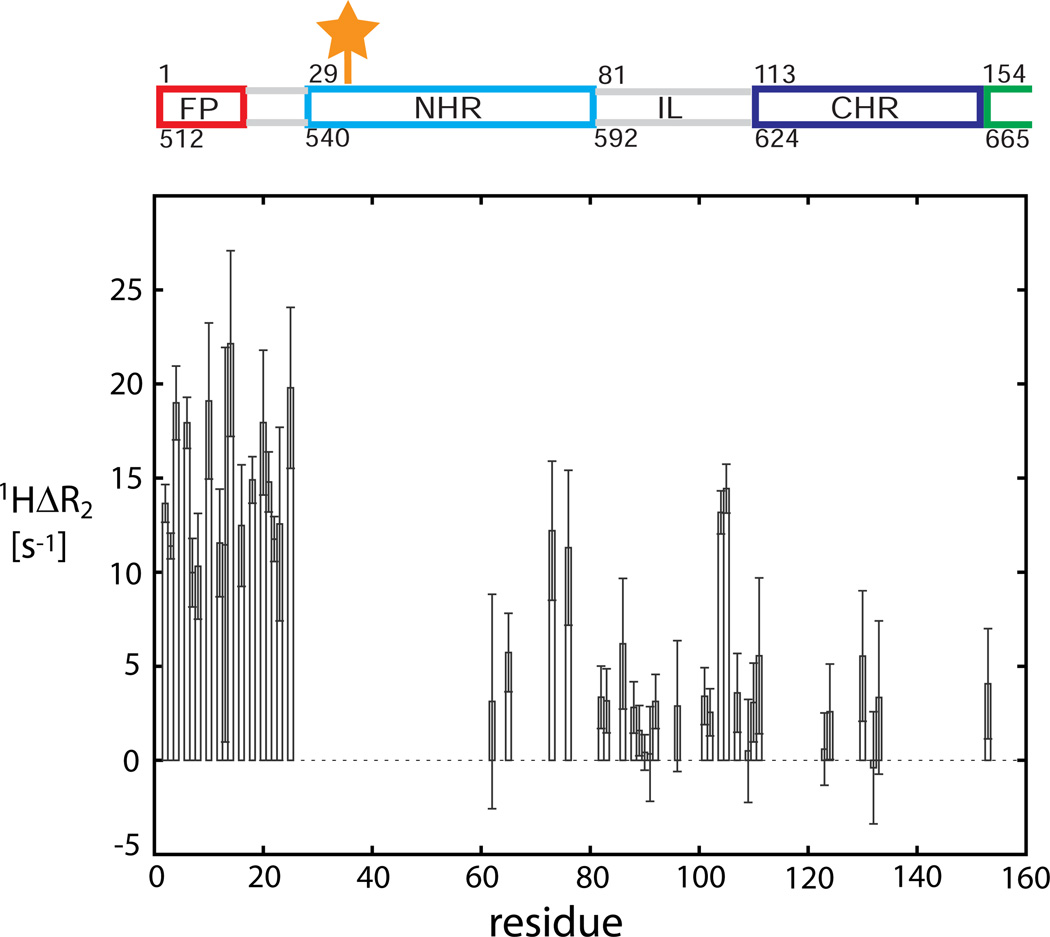
In gp411–194, we found that the α-helical FP (A1-F12) is connected to the NHR by a highly flexible and transient α-helical FPPR linker region (residues G13-L26). High amplitude movements of the FP relative to the NHR helical axis are observed (Lakomek et al., 2013). As the 15N relaxation data for gp411–194 and gp4127–194 are very similar, we conclude that the FP shows independent large amplitude motions and does not interact with the TM region, at least on the fast ns time-scale. Indeed, the TM region shows dynamic properties that are very different from the FP, making it highly unlikely that FP and TM region are located in the same micelle (or membrane). As the TM and FP regions seem topographically separated, it appears unlikely that gp411–194 is in a post-fusion conformation (6HB) where FP and TM would be in close proximity. Rather, both gp4127–194 and gp411–194 constructs are significantly populated with less restricted conformations with high domain mobility, as predicted for a pre-fusion intermediate (Blumenthal et al., 2012; Harrison, 2005; Julien et al., 2013). When positioning a paramagnetic nitroxide (MTSL) spin label at residue 35 using a S35C mutant of gp411–194, an increase in 1H R2 relaxation rates for the FP and FPPR region is observed (compare Figure 5), which indicates that, at least transiently, the highly dynamic FP must be visiting the proximity of the N-terminal NHR region. This also is incompatible with the presence of a highly populated post-fusion conformation, because FP interaction with NHR in the 6HB would be blocked due to shielding by CHR. These findings relate to recent structural studies from the Ward and Wilson laboratories which observed additional low intensity electron density proximal to the NHR region and attributed it to the FP and FPPR region (Julien et al., 2013; Lyumkis et al., 2013). This additional electron density we suggest is weak and diffuse because of the dynamic and transient α-helical character of the FP and FPPR regions as described herein.
Figure 5. PRE measurements for the paramagnetic nitroxide spin-labeled S35C mutant of gp411–194.
The difference of 1H R2 rates is taken with respect to the reference sample, reduced by ascorbic acid. An increased difference rate 1H ΔR2 indicates (transient) proximity. The position of the paramagnetic spinlabel is indicated by the orange star.
Line broadening of C-terminus
Resonances for the C-terminal CHR, MPER and N-terminal TM region appear line broadened below the detection threshold. Increased line widths are due to elevated 1H and/or 15N R2 relaxation rates, and can result from either slow tumbling in the absence of large internal motions, or from conformational exchange processes in the micro- to millisecond time scale that add an exchange contribution, Rex, to R2. Although the visible part of the CHR region indicate little exchange effects, a strong Rex effect can be observed in the IL loop region around A96 (which also shows lipid interaction), suggesting a transition between different conformations, such as, for example, an extended pre-fusion intermediate conformation and a more compact post-fusion conformation (with 6HB conformation of the ectodomain). Alternatively, the transition could be between two different pre-hairpin conformations.
Discussion
Structural properties and internal dynamics
We have shown that HIV-1 gp4127–194 embedded in lipid-mimicking dodecyl phosphocholine (DPC) detergent environment shows well-defined α-helical structure for both NHR and the (visible) N-terminal CHR region and that the CHR region appears detached from the NHR.
NMR relaxation data indicate high internal dynamics of the individual gp41 domains on different time scales. The FP and TM domains experience very different dynamics. While the FP is the most mobile structured region of the protein (on the 2–5 ns time scale), the TM region is the most rigid. This is incompatible with a compact 6HB arrangement of the ectodomain which would place FP and TM in close proximity where they would be expected to experience similar dynamics. Instead it indicates a more extended conformation of gp41 and one more comparable to that postulated for a pre-fusion intermediate state bridging viral and host-cell membranes. The FP, connected to the NHR by a highly flexible and transient α-helical FPPR region, samples a conformational space proximal to the N-terminal NHR region, as shown by paramagnetic relaxation enhancement data. The strong Rex effects for the region around A96 in the IL indicate conformational dynamics on the µs-ms time scale. This could reflect an exchange between two different pre-hairpin conformations, and together with the observed line-broadening for the C-terminal CHR, MPER and N-terminal TM regions and PRE data on gp411–194, indicates that the TM region transiently visits the proximity of the FP (Lakomek et al., 2013). We interpret these conformational dynamics as reflecting an exchange process between a highly populated pre-fusion intermediate and a lowly populated post-fusion conformation (for which FP and TM region will presumably be located in close proximity).
Several residues in the NHR display transient α-helical behavior as inferred from reduced Δδ13Cα secondary chemical shift, such as G36, Q40, H53, G61 and Q64. Apart from G61, these residues show increased solvent accessibility in an otherwise solvent protected region as indicated from solvent PRE measurements. Also, residues Q40, H53 and Q64, show Rex effects which indicate conformational dynamics on the µs-ms time scale. Thus, the NHR domain will transiently form a non-contiguous α-helix with notches at (G36), Q40, H53, (G61) and Q64. These residues are spaced by multiples of n ∙(3 – 4) corresponding to the helical turns across the NHR region (α- helix has ~ 3.6 residues per turn). Roche et al. (2014), studying a truncated ectodomain core, also observed a kink in the NHR helix at position Q51. This residue is close to H53, which in gp4127–194 shows Rex effects, and which is adjacent to the invisible stretch I48-Q52 of the NHR region, suggesting a conformational exchange process on the intermediate time scale for the stretch I48-H53 of the NHR. A cryo-EM study of detergent embedded ENV by Sodroski and co-workers, indicated that the NHR and CHR helices in the un-liganded, non-activated pre-fusion state are broken up into smaller helical entities (Mao et al., 2013b; Mao et al., 2012). This is in contrast to the two single α-helices for the NHR and CHR domain observed for the post-fusion structure of the gp41 ectodomain and for the native viral spike and soluble constructs of ENV (Chan et al., 1997; Tran et al., 2012; Weissenhorn et al., 1997). Although these discrepancies have been the matter of intense debate (Henderson, 2013; Mao et al., 2013a; Subramaniam, 2013; van Heel, 2013), we cannot exclude the presence of breaking points in the NHR helix at position Q40, H53 and Q64 that would enable a transition between shorter helices and a single long NHR helix at an intermediate µs- ms time scale. Further, it is noteworthy that H53 and Q64 are adjacent to residues forming the rim of a highly conserved hydrophobic groove of the NHR which accommodates three hydrophobic residues of the CHR region (I124, W120 and W117) (Chan et al., 1997). While the former residues show transient α-helical character allowing increased plasticity, T58, which is part of the floor of the hydrophobic groove and shows the highest α- helical propensity of all observed residues in the protein, is also involved in a conformational exchange process as inferred from a strong Rex effect.
Detergent / lipid binding interactions
We have shown that the IL region as well as several residues predominantly in the C-terminal portion of the NHR, have lipid binding potential, implying that these regions are more intimately involved in the fusion process than originally thought. Lipid interactions involving the NHR have also been observed by other groups (Kliger et al., 2000; Sackett and Shai, 2002; Roche et al. 2014). Our results with gp4127–194 extend these studies as our construct includes the IL, MPER and TM regions. The strong solvent exposure of the CHR, together with the high internal dynamics observed, indicate that the CHR (in the main conformation) is detached from the NHR rather than forming the stable 6HB (associated with post-fusion). A structure with an extended CHR with MPER would be expected for a pre-hairpin intermediate bridging viral and host cell membranes. This is supportive of structural studies by Julien et al. (2013) and Lyumkis et al. (2013) on a soluble pre-fusion conformation of the ENV construct (lacking the MPER and TM region) who observed an extended arrangement of the CHR. Furthermore, our results suggest that the extended arrangement of the CHR cannot be explained as a result of truncating the lipid anchor, namely the TM region.
Monomer – Trimer Associations
In the various gp41 constructs we have described, detergent binding (analogous to potential in-vivo lipid interactions) weakens the trimeric interactions as indicated by decreases in the association constant Ka (Figures S2 and S3). This is not unexpected, as lipid interactions compete with hydrophobic inter-residue interactions stabilizing the trimer. However, over the concentration range of our study, gp4127–194 remains mostly trimeric in the presence of lipid-mimicking dodecyl phosphocholine (Figure 4B). Comparison with the shorter ectodomain construct gp4127–154, which gives similar Ka values, indicates that the N-terminal NHR region is the main trimerization domain (see Figures S1 and S2). The IL region, which interacts with lipid, does stabilize the trimeric association of gp4127–154. This stabilization could arise by the NHR-IL linkage conformationally restricting the interactions of NHR at the detergent/solvent interface. In the presence of IL, the lipid interactions of the NHR do not exclude trimer formation, as long as the lipid binding residues of NHR remain surface exposed and not buried in the trimer. In Figure 3C, residues of the NHR trimer of (pdb code 1ENV) have been color-coded according to their degree of lipid interactions. Residues with strong lipid interactions and side-chain pointing to the interior of the NHR (as for example L44) would disturb the trimeric arrangement. This discrepancy could be resolved by a slight reorientation of the NHR helices in the pre-fusion intermediate ectodomain structure (which is unknown) or by transient kinks and notches within the NHR.
The observed monomer-trimer equilibrium and lipid interaction sides in the NHR and IL region suggest they play a fundamental role during the fusion process, presumably being regulated by local changes in lipid composition and concentration gradients upon viral membrane and host-cell encounter.
The Fusion Pathway
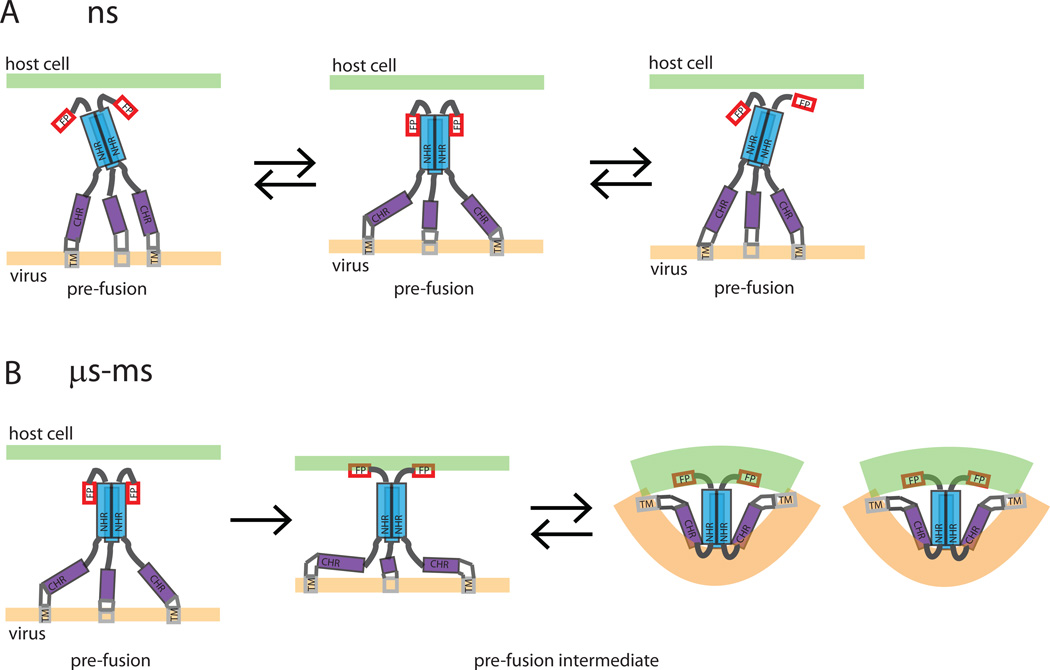
In the transition pathway from a non-activated pre-fusion to pre-fusion intermediate to post-fusion conformation the behavior of the gp41 C-terminal region is of key importance. The fusion event should relate the gp41 structural transitions to the energy landscape which drives the fusion process. Breaking up of a trimeric arrangement would incur a high energetic penalty during membrane fusion. The observed monomer-trimer equilibrium, as well as strong interaction of gp41 with the lipid environment, may play a key role in fusion by guaranteeing an energetically downhill (favorable) reaction pathway. A schematic model of the initial stages of gp41 mediated membrane fusion compatible with our data is shown in Figure 6: On the 2–5 ns time scale NHR and CHR regions are stochastically sampling different orientations relative to each other while the FP exhibits larger amplitude motion relative to the orientation of the NHR axis. On the µs-ms time scale larger conformational changes between different intermediate pre-hairpin conformations and a lowly populated post-fusion hairpin (6HB) conformation occur, eventually driving membrane fusion.
Figure 6. A schematic model of the initial stages of gp41 mediated membrane fusion.
A schematic model of the initial stages of gp41 mediated membrane fusion compatible with our data. a) The gp41 domains exhibit stochastic dynamics of smaller (NHR and CHR) and higher amplitude on a 2–5 ns timescale. b) Larger conformational changes that require transition of a higher energy barrier occur on a µs-ms time scale. (The TM and MPER region adjacent to the CHR region are drawn for completeness, although our data do not provide any direct information on the arrangement of those two domains. Also, the transient α-helical notches in the NHR region are not included in the scheme).
Inherent conformational flexibility of gp41 has been postulated previously and its associated meta-stability appears linked to function by energetically driving membrane fusion when transitioning to the post-fusion 6HB (Julien et al., 2013; Lu et al., 1995). The conformational dynamics of gp41 during the fusion process further complicate the search for broadly neutralizing antibodies targeted to gp41, as binding epitopes may only be transiently exposed. Thus, the presence of conformational flexibility must be considered in the search of gp41 targeted fusion inhibitors. This study provides a first insight on the complexity of gp41 conformational space and we hope it increases awareness of complications which might arise during the design of fusion inhibitors. On the other hand, a clearer understanding of the conformational dynamics of gp41 could significantly increase chances of gripping HIV at its weak point and help in the design of highly potent fusion inhibitors. Eventually, we may also be able to identify specific mutations which would fix (or stabilize) the pre-fusion intermediate conformation which is the actual drug target, thus simplifying inhibitor discovery. In conclusion, NMR spectroscopy, both in solution and in the solid-state, can provide very useful information for understanding conformational transitions in membrane proteins.
Experimental Procedures
Protein expression and purification
Protein expression and purification were performed essentially as described previously for the HIV-1 gp411–194 construct (Lakomek et al., 2013). In brief, the expression plasmid HIV-1 gp4127–194 corresponding to HXB2 Env residues 538 – 706 was amplified by polymerase chain reaction and cloned into the T7 expression vector pet11a. N-terminal methionine was added and Cys 87 and 93 were exchanged to Ala by PCR mutagenesis. Uniformly 2H, 15N and 13C enriched HIV-1 gp4127–194 was expressed in BL21-DE3 cells using a fermenter, as described previously (Yamazaki et al., 1996).
Expressed protein was extracted using SDS and purified by a combination of gel filtration and hydroxyapatite chromatographies. The protein was reconstituted by exchanging SDS for dodecyl phosphocholine (DPC, Fos-Choline-12, Affymetrix, USA) and further purified by size exclusion chromatography. Protein dissolved in 50 mM sodium acetate, pH 4.0, 25 mM KCl, 0.1% (w/v) DPC (starting concentration) was concentrated to ~ 1mM (20 mg/ml) with an Amicon-Ultra 15ml, 30-kD concentrator (EMD Millipore).
Circular Dichroism
The overall conformation of the protein was measured by far-UV CD (compare Figure 1A). Spectra of ~12 µM gp4127–194 (~ 0.25mg/ml) in 50 mM sodium acetate, pH 4.0, containing 3mM n-dodecylphosphocholine (DPC) were recorded at 20°C on a Jasco J-715 spectropolarimeter using a 0.02-cm pathlength cell. Secondary structures were estimated using the CONTIN program (Provencher and Glockner, 1981) and Self Consistent methods (http://dichroweb.cryst.bbk.ac.uk) (Sreerama and Woody, 1993; Whitmore and Wallace, 2008). Calculation of the secondary structure from the spectrum indicated about 74% α-helix which is close to that predicted (~ 80%) assuming that the 33 residues of the loop region are non-helical and remaining 134 residues helical.
Thermal stability of the protein was measured by far-UV CD at 222 nm: no thermal melting, corresponding to a helix-coil transition, was observed. This is in contrast to many gp41 constructs which are assembled from separate helical N- and C-terminal fragments, see for example Buzon et al.(Buzon et al., 2010). The inferred high stability of the gp4127–194 construct was confirmed by limited tryptic digestion which specifically clipped within the loop region. The digested protein was still intact as judged by gel filtration but now exhibited a thermal transition with a melting temperature of 83°C (data not shown).
Analytical Ultracentrifugation
A Beckman Optima XL-I analytical ultracentrifuge, absorption optics, an An-60 Ti rotor and standard double-sector centerpiece cells were used. Equilibrium measurements were taken at 20°C and the concentration profile recorded after 20 hours at 18,500 rpm. The baseline was established by over-speeding at 45,000 rpm for 3 hours. Uniformly 15N 13C 2H enriched gp4127–194 was dissolved in 50 mM sodium acetate, pH 4.0 containing ~ 5mM DPC. The mass contribution of the detergent was eliminated with D20 as described by (Noy et al., 2003). The monomeric molecular mass of uniformly 15N 13C 2H isotopically enriched gp4127–194 monomer was calculated to be 22 kDa.
The molecular weight of the complex was determined by sedimentation equilibrium, to negate the density effect of bound detergent the centrifugation was performed in 50% D2O (Noy et al., 2003). An average molecular weight of 52 kDa was determined by fitting the equilibrium profile using an ideal single species model (not shown) which made no assumption about associative behavior. As the molecular weight increased as a function of concentration, an associative process was indicated. Based on a molecular weight approaching that calculated for a 15N13C2H labeled trimer (66 kDa) we treated the equilibrium data as a monomer – trimer system and obtained a Ka of 3.6×1010M-2 (Figure 4A).
NMR spectroscopy
Backbone assignment and 15N relaxation measurements
NMR backbone assignment and 15N relaxation measurements were carried out on a uniformly 2H15N13C-enriched sample of 1.0 mM homotrimeric gp4127–194 in 50 mM sodium acetate buffer, pH 4.0, 25 mM KCl and 138 mM DPC. Spectra were recorded at 40°C on a 600 MHz Bruker Avance III, 800 MHz Avance III, and 900 MHz Bruker Avance II system, all equipped with cryo probe head technology.
Backbone assignment experiments were performed as described in (Lakomek et al., 2013). Secondary chemical shifts were corrected for 2H isotope effects using the program TALOS+ (Shen et al., 2009) and random coil values of (Maltsev et al., 2012). Internal backbone dynamics of gp4127–194 trimer were studied by 15N relaxation. 15N R1 and R1ρ relaxation rates as well as 15N-{1H} NOE and transverse 15N CSA-dipolar cross-correlated correlation rates, ηxy, were measured at both 600 and 800 MHz, using TROSY-based methods optimized for perdeuterated proteins (Lakomek et al., 2012). Limited sensitivity did not permit the use of 3D measurement methods and only data for amides that could be resolved in the 2D 15N-1H TROSY-HSQC spectrum were analyzed. Conformational changes on the micro- to millisecond time scale give rise to exchange contributions to R2. To identify all conformational changes that occur on this time scale and give rise to chemical shift changes, Rex was measured on the slowly relaxing 15N-{1H} doublet component, using a Hahn-echo experiment at two different field strengths, 600 and 800 MHz. All 15N relaxation experiments were performed as described in (Lakomek et al., 2013).
Paramagnetic relaxation enhancement (PRE) measurements
To investigate lipid interactions, lipid PREs of gp4127–194 were measured by adding 0.5 mM (final concentration) 5-DOXYL-stearic acid (5-DSA, Sigma-Aldrich, Germany, final concentration) to a defined volume (500 µl) of gp41 in DPC detergent buffer to a ratio of 5-DSA:DPC =1:70. 5-DSA contains a paramagnetic label but is otherwise highly lipophilic (Hilty et al., 2004). Thus, 5-DSA will integrate itself in a lipid (or lipid mimicking environment like DPC) and distribute evenly. Paramagnetic relaxation enhancement introduced by the 5-DSA label will lead to intensity reduction of the gp41 signals when the corresponding gp41 region is proximal to the lipid spin label. Intensities were measured for 1H 15N TROSY-HSQC spectra (recorded at 900 MHz, 40°C, 9h) and normalized by taking the ratio to intensities measured for a (untreated, diamagnetic) reference sample of same gp41 and detergent concentration. Those residues that show the strongest decrease in normalized intensities (I/I0) experience the strongest lipid interactions, while residues in far distance from the lipid will show normalized intensities (I/I0) close to 1.
Solvent PREs were measured by adding 5 mM Gadodiamide (Omniscan, Selleckchem, USA) to a sample of 0.1 mM 2H15N13C-enriched gp4127–194, 50 mM Na Ac pH4, 25mM KCl, 35 mM DPC. Solvent PREs are expected to behave complementary to lipid PREs as only solvent accessible sites, not embedded in detergent, experience increased relaxation.
Paramagnetic relaxation enhancement (PRE) measurements for transient proximity to the N-terminal NHR region were performed by covalently attaching a paramagnetic nitroxide spin-label (S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate, MTSL, Toronto Research Chemicals Inc., USA) at residue 35 for a S35C mutant of gp411–194. 1H R2 relaxation measurements for S35C gp411–194 were carried out as described in (Lakomek et al., 2013). Reference data were obtained after reducing the paramagnetic label with a 10-fold excess of ascorbic acid.
NMR data analysis
Spectra were processed and analyzed using the NMRPipe software package (Delaglio et al. 1995). Spectral intensities and relaxation rates were extracted with NMRpipe as described in (Lakomek et al., 2013; Lakomek et al., 2012). 15N relaxation data, including the transverse cross-correlated relaxation rate, ηxy, were analyzed by the extended model-free approach (Clore et al., 1990). Clusters of similar dynamics were identified (L55-D78, G83-S102, E119-S133, V190-V194), for which error-weighted average values were calculated (Table S1). Fitting of the data was only possible when an internal motion on the nanosecond time scale was invoked (Korzhnev et al., 1997). All input data could be reproduced within the experimental error (Table S1). The extended model-free analysis has been implemented using a homewritten MATLAB (MathWorks, Inc., USA) script.
Structural Analysis
Structural analysis and visualization of lipid interactions and solvent exposure has been performed using UCSF Chimera (Pettersen et al., 2004).
Supplementary Material
Highlights.
HIV-1 gp41 shows high internal dynamics on different timescales (low ns and µs-ms).
Extended (dynamic) helical conformation as expected for a pre-fusion intermediate
Strong lipid interactions of immunodominant loop region and C-terminal NHR
Transient α-helical notches in NHR allow breaking points for conformational changes
Acknowledgements
This research was funded by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (NIH) and the Intramural AIDS-Targeted Antiviral Program of the Office of the Director, NIH as well as the DFG/NIH Research Career Transition Award Program (to N.L., LA 2724/1-1). N.L. thanks Dr. Ad Bax and Prof. Dr. Christian Griesinger for allowing him to use their NMR facilities at NIH, Bethesda, MD, USA and the Max-Planck Institute for Biophysical Chemistry, Göttingen, Germany. N.L. is very grateful to Dr. Ad Bax for his continuous support, for receiving an excellent training in NMR spectroscopy and for allowing him to continue independent research on this research project.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
None of the authors have any competing financial interests related to this work.
References
- Bartesaghi A, Merk A, Borgnia MJ, Milne JL, Subramaniam S. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nature structural & molecular biology. 2013;20:1352–1357. doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal R, Durell S, Viard M. HIV Entry and Envelope Glycoprotein-mediated Fusion. Journal of Biological Chemistry. 2012;287:40841–40849. doi: 10.1074/jbc.R112.406272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzon V, Natrajan G, Schibli D, Campelo F, Kozlov MM, Weissenhorn W. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS pathogens. 2010;6:e1000880. doi: 10.1371/journal.ppat.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey M. HIV envelope: challenges and opportunities for development of entry inhibitors. Trends in microbiology. 2011;19:191–197. doi: 10.1016/j.tim.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- Chan DC, Kim PS. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- Clore GM, Szabo A, Bax A, Kay LE, Driscoll PC, Gronenborn AM. Deviations from the Simple 2-Parameter Model-Free Approach to the Interpretation of N-15 Nuclear Magnetic-Relaxation of Proteins. Journal of the American Chemical Society. 1990;112:4989–4991. [Google Scholar]
- Dimitrov DS, Blumenthal R. Photoinactivation and kinetics of membrane fusion mediated by the human immunodeficiency virus type 1 envelope glycoprotein. Journal of virology. 1994;68:1956–1961. doi: 10.1128/jvi.68.3.1956-1961.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douek DC, Kwong PD, Nabel GJ. The rational design of an AIDS vaccine. Cell. 2006;124:677–681. doi: 10.1016/j.cell.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Furuta RA, Wild CT, Weng Y, Weiss CD. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- Harrison SC. Mechanism of membrane fusion by viral envelope proteins. Advances in virus research. 2005;64:231–261. doi: 10.1016/S0065-3527(05)64007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC. Viral membrane fusion. Nature structural & molecular biology. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R. Avoiding the pitfalls of single particle cryo-electron microscopy: Einstein from noise. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:18037–18041. doi: 10.1073/pnas.1314449110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M, Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450:913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- Hilty C, Wider G, Fernandez C, Wuthrich K. Membrane protein-lipid interactions in mixed micelles studied by NMR spectroscopy with the use of paramagnetic reagents. Chembiochem. 2004;5:467–473. doi: 10.1002/cbic.200300815. [DOI] [PubMed] [Google Scholar]
- Jaroniec CP, Kaufman JD, Stahl SJ, Viard M, Blumenthal R, Wingfield PT, Bax A. Structure and dynamics of micelle-associated human immunodeficiency virus gp41 fusion domain. Biochemistry. 2005;44:16167–16180. doi: 10.1021/bi051672a. [DOI] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. Crystal Structure of a Soluble Cleaved HIV-1 Envelope Trimer. Science. 2013 doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Sun ZY, Rand KD, Shi X, Song L, Cheng Y, Fahmy AF, Majumdar S, Ofek G, Yang Y, et al. Antibody mechanics on a membrane-bound HIV segment essential for GP41-targeted viral neutralization. Nature structural & molecular biology. 2011;18:1235–1243. doi: 10.1038/nsmb.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliger Y, Peisajovich SG, Blumenthal R, Shai Y. Membrane-induced conformational change during the activation of HIV-1 gp41. Journal of molecular biology. 2000;301:905–914. doi: 10.1006/jmbi.2000.4004. [DOI] [PubMed] [Google Scholar]
- Korzhnev DM, Orekhov VY, Arseniev AS. Model-free approach beyond the borders of its applicability. J Magn Reson. 1997;127:184–191. doi: 10.1006/jmre.1997.1190. [DOI] [PubMed] [Google Scholar]
- Lai AL, Moorthy AE, Li Y, Tamm LK. Fusion activity of HIV gp41 fusion domain is related to its secondary structure and depth of membrane insertion in a cholesterol-dependent fashion. Journal of molecular biology. 2012;418:3–15. doi: 10.1016/j.jmb.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakomek NA, Kaufman JD, Stahl SJ, Louis JM, Grishaev A, Wingfield PT, Bax A. Internal Dynamics of the Homotrimeric HIV-1 Viral Coat Protein gp41 on Multiple Time Scales. Angew Chem Int Edit. 2013;52:3911–3915. doi: 10.1002/anie.201207266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakomek NA, Ying JF, Bax A. Measurement of N-15 relaxation rates in perdeuterated proteins by TROSY-based methods. Journal of biomolecular NMR. 2012;53:209–221. doi: 10.1007/s10858-012-9626-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tamm LK. Structure and plasticity of the human immunodeficiency virus gp41 fusion domain in lipid micelles and bilayers. Biophysical journal. 2007;93:876–885. doi: 10.1529/biophysj.106.102335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Blacklow SC, Kim PS. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, et al. Cryo-EM Structure of a Fully Glycosylated Soluble Cleaved HIV-1 Envelope Trimer. Science. 2013 doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltsev AS, Ying J, Bax A. Deuterium isotope shifts for backbone (1)H, (1)(5)N and (1)(3)C nuclei in intrinsically disordered protein alpha-synuclein. Journal of biomolecular NMR. 2012;54:181–191. doi: 10.1007/s10858-012-9666-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Castillo-Menendez LR, Sodroski JG. Reply to Subramaniam, van Heel, and Henderson: Validity of the cryo-electron microscopy structures of the HIV-1 envelope glycoprotein complex. Proceedings of the National Academy of Sciences of the United States of America. 2013a;110:E4178–E4182. doi: 10.1073/pnas.1316666110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Wang L, Gu C, Herschhorn A, Desormeaux A, Finzi A, Xiang SH, Sodroski JG. Molecular architecture of the uncleaved HIV-1 envelope glycoprotein trimer. Proceedings of the National Academy of Sciences of the United States of America. 2013b;110:12438–12443. doi: 10.1073/pnas.1307382110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Wang L, Gu C, Herschhorn A, Xiang SH, Haim H, Yang X, Sodroski J. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nature structural & molecular biology. 2012;19:893–899. doi: 10.1038/nsmb.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Barroso I, Durell S, Sakaguchi K, Appella E, Blumenthal R. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. The Journal of cell biology. 1998;140:315–323. doi: 10.1083/jcb.140.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy D, Calhoun JR, Lear JD. Direct analysis of protein sedimentation equilibrium in detergent solutions without density matching. Anal Biochem. 2003;320:185–192. doi: 10.1016/s0003-2697(03)00347-6. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Provencher SW, Glockner J. Estimation of Globular Protein Secondary Structure from Circular-Dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- Qiang W, Sun Y, Weliky DP. A strong correlation between fusogenicity and membrane insertion depth of the HIV fusion peptide. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15314–15319. doi: 10.1073/pnas.0907360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche J, Louis JM, Grishaev A, Ying J, Bax A. Dissociation of the trimeric gp41 ectodomain at the lipid-water interface suggests an active role in HIV-1 Env-mediated membrane fusion. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:3425–3430. doi: 10.1073/pnas.1401397111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackett K, Shai Y. The HIV-1 gp41 N-terminal heptad repeat plays an essential role in membrane fusion. Biochemistry. 2002;41:4678–4685. doi: 10.1021/bi0255322. [DOI] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. Journal of biomolecular NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreerama N, Woody RW. A Self-Consistent Method for the Analysis of Protein Secondary Structure from Circular-Dichroism. Anal Biochem. 1993;209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- Subramaniam S. Structure of trimeric HIV-1 envelope glycoproteins. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E4172–E4174. doi: 10.1073/pnas.1313802110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZY, Oh KJ, Kim M, Yu J, Brusic V, Song L, Qiao Z, Wang JH, Wagner G, Reinherz EL. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity. 2008;28:52–63. doi: 10.1016/j.immuni.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Tran EE, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JL, Subramaniam S. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS pathogens. 2012;8:e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M. Finding trimeric HIV-1 envelope glycoproteins in random noise. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E4175–E4177. doi: 10.1073/pnas.1314353110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jardetzky O. Investigation of the neighboring residue effects on protein chemical shifts. Journal of the American Chemical Society. 2002;124:14075–14084. doi: 10.1021/ja026811f. [DOI] [PubMed] [Google Scholar]
- Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- Whitmore L, Wallace BA. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield PT, Stahl SJ, Kaufman J, Zlotnick A, Hyde CC, Gronenborn AM, Clore GM. The extracellular domain of immunodeficiency virus gp41 protein: Expression in Escherichia coli, purification, and crystallization. Protein Science. 1997;6:1653–1660. doi: 10.1002/pro.5560060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Hinck AP, Wang YX, Nicholson LK, Torchia DA, Wingfield P, Stahl SJ, Kaufman JD, Chang CH, Domaille PJ, et al. Three-dimensional solution structure of the HIV-1 protease complexed with DMP323, a novel cyclic urea-type inhibitor, determined by nuclear magnetic resonance spectroscopy. Protein Sci. 1996;5:495–506. doi: 10.1002/pro.5560050311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti G, Briggs JA, Grunewald K, Sattentau QJ, Fuller SD. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS pathogens. 2006;2:e83. doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Liu J, Bess J, Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.