Abstract
Neuronal exocytosis is mediated by a Ca2+-triggered membrane fusion event that joins synaptic vesicles and presynaptic membrane. In this event, synaptotagmin I plays a key role as a Ca2+ sensor protein that binds to and bends the presynaptic membrane with its C2B domain, and thereby initiates membrane fusion. We report free energy calculations according to which C2B-induced membrane bending is preceded by a Ca2+- and membrane-dependent conformational transition. In this transition C2B attaches to the membrane, moves its C-terminal helix from the orientation seen in the available (but membrane-free) crystal/NMR structures as pointing away from the membrane (helix-up), to an orientation pointing toward the membrane (helix-down). In the C2B helix-down state, lipid tails in the proximal membrane bilayer leaflet interact with the moved helix and become disordered, whereas tails in the distal leaflet, to keep in contact with the proximal leaflet, become stretched and ordered. The difference in lipid tail packing between the two leaflets results in an imbalance of pressure across the membrane, and thereby causes membrane bending. The lipid-disordering monitored in the simulations is well suited to facilitate Ca2+-triggered membrane fusion.
Introduction
Protein-induced membrane bending and remodeling govern many cellular processes, including cell division, growth, movement, and cell-cell communication (1, 2, 3, 4, 5, 6). The event of neurotransmitter release is a typical example for a process involving a significant change in membrane shape. When a signaling membrane potential propagates to a neuron axon terminal, Ca2+ channels open and the local intracellular Ca2+ concentration rises, triggering a membrane fusion event that joins awaiting synaptic vesicles with the presynaptic membrane. Thus, neurotransmitters, stored within the vesicles, are released into the synaptic cleft through a fusion pore. Synaptotagmin I (syt1) has been claimed as a Ca2+ sensor protein (7, 8, 9, 10, 11) that can cause, with the help of soluble n-ethylmaleimide-sensitive-factor attachment protein receptors (SNAREs) (12, 13), complexins (14, 15), and other key proteins (16), membrane fusion in response to Ca2+ concentration increase (17, 18).
Despite intense efforts, the molecular mechanism of vesicle fusion, and how synaptotagmin I may regulate the fusion step, is far from clear. Although syt1 is the Ca2+ sensor in the Ca2+-regulated fusion process, SNARE complex can by itself trigger low-efficiency membrane fusion in vitro without syt1 and Ca2+ (19). It has been proposed that syt1 exerts its function by sensing (20) the vesicle membrane curvature, binding to both the vesicle and presynaptic membrane and bringing them close enough together (21) to catalyze SNARE complex formation that by itself completes fusion (22).
However, highly efficient membrane fusion and large fusion pore formation can only be observed in the presence of synaptotagmin I (23, 24), indicating syt1’s critical function in the final step toward fusion pore formation. Studies (18, 25, 26, 27) showed that, with Ca2+ bound, syt1 induces presynaptic membrane bending, which can be essential to fusion pore formation. If syt1’s membrane-bending activity is abolished, it does not trigger membrane fusion (26). The N-BAR domain of endophilin, which is known to cause membrane bending, can rescue the membrane fusion activity of a membrane-bending-defective syt1 (26). Syt1 was observed by cryo-electron microscopy to cause presynaptic membrane protrusion, which can be further enhanced in the presence of the SNARE complex (28). Therefore, it is of interest to study the molecular mechanism underlying syt1-induced membrane bending with the help of atomistic simulations, and to elucidate how the two membrane remodeling processes, bending and fusion, are linked.
Mechanistic insight into syt1’s membrane bending can be obtained from comparing the two Ca2+ binding domains in syt1, namely, C2A and C2B. Despite the domains’ similarity in structure, sequence, and membrane binding affinity with bound Ca2+, isolated C2B can bend membranes effectively, but isolated C2A cannot (26). The two C2 domains have a very similar β-sandwich structure (the backbone root-mean-square displacement (RMSD) measures only ∼1.6 Å), except that C2A binds three Ca2+ ions whereas C2B binds two Ca2+ ions (29, 30), and that C2B has a C-terminal helix, which is missing in C2A. The binding domains C2A and C2B have 40% sequence identity and 61% sequence similarity when the C2B C-terminal helix is ignored. Both isolated C2A and isolated C2B make contact with anionic lipid bilayers upon Ca2+ binding (27, 31, 32, 33, 34, 35). The two domains’ Ca2+-loops insert into the membrane surface at a similar depth (31, 36, 37, 38, 39), indicating that the wedge-insertion (40) of the domains’ Ca2+-loop is unlikely to cause the difference in membrane-bending capability. Both domains have a poly-basic region at the side of the β-sandwich. C2A has four lysines and C2B has five lysines and one arginine.
The most obvious difference between the C2A and C2B domains is that the C-terminal helix arises only in C2B. Because this helix (409VEEEVDAMLAVKK421) has many hydrophobic and positive lysine residues that can interact with the negatively charged membrane, as well as negative residues that are close to the Ca2+-loop, the C-terminal helix might generate membrane curvature upon Ca2+ binding. However, according to the C2B crystal (PDB:1TJX) (41) and NMR (PDB:1K5W) (42) structures, both showing the protein without membrane presence, the C-terminal helix is pointing parallel to the C2B β-sandwich structure, in a direction that we define as “up”. Given the upright orientation of the β-sandwich structure on the membrane surface as shown in electron paramagnetic resonance (EPR) studies (39), the C-terminal helix in its orientation shown in the crystallographic and NMR structures points away from the membrane and does not interact with membrane lipids. If the C-terminal helix is going to play a role in membrane bending, a conformational transition is required.
Actually, protein conformational transitions upon membrane binding are rather common. The membrane surface, in comparison to bulk solvent, can affect protein secondary (43) and tertiary structures (44), as well as domain-domain arrangements (45), due to protein-lipid interactions and due to the heterogeneity of the dielectric environment near the membrane surface. Although many studies have focused on the C2B domain bound to a membrane (for instance, on the domain orientation relative to the membrane (38, 39, 46), on Ca2+ loop insertion depth (31, 36, 37, 38, 39), and on the domain’s membrane binding affinity in regard to ionic strength (37, 47, 48)), all experimental data were interpreted based on the known crystal/NMR structure without membrane. A possible C2B conformational transition upon membrane binding was not considered.
We suggest that a conformational transition can give a satisfactory explanation for C2B-induced membrane bending and explain why C2A and C2B exhibit distinct membrane-bending activity (26). According to this suggestion, a conformational transition in C2B’s C-terminal helix occurs when C2B with Ca2+ bound attaches to the membrane. The helix, instead of pointing up as in the membrane-free crystal/NMR structures, according to our suggestion turns into the opposite direction, defined here as “down”, to interact with both membrane and the Ca2+ ion bound to the domain. Indeed, our potential of mean force calculations, described in detail below, show that, on an anionic membrane, the helix-down C2B conformation is energetically more stable than the helix-up conformation. Because of the suggested conformational transition, the C-terminal helix comes to lie on top of the membrane surface; lipid tails in the proximal membrane leaflet reposition themselves to interact with the helix and become disordered, whereas lipid tails in the distal leaflet become stretched and ordered to remain in close contact with the proximal leaflet. As a result, a pressure imbalance between the proximal and distal membrane leaflets arises and the membrane becomes bent.
According to the simulations reported below and in agreement with experimental observation, neither the C2A domain without a C-terminal helix, nor the C2B domain with the C-terminal helix positioned up, nor a C2B domain with the C-terminal helix truncated, cause membrane bending, but the C2B with the C-terminal helix in the down-orientation does. The rearrangement of proximal membrane leaflet lipid tails induced by the downward-pointing C2B C-terminal helix should not only lead to membrane bending, but should also facilitate lipid stalk formation between two membranes, a process that leads to hemifusion, a key intermediate step in membrane fusion and neurotransmitter release.
Methods
System composition
The systems simulated were typically composed of a membrane patch and a synaptotagmin I domain (isolated C2A or C2B domain). All domains in the simulations had Ca2+ bound. The membrane was composed of 50% POPC and 50% DOPS, the composition being adopted from the corresponding experimental tubulation studies (26). NaCl ions were added at a molarity of 0.15 M and extra Na+ ions were added to neutralize the system. Systems simulated and their composition are listed in Table 1.
Table 1.
Simulations carried out
| Property studied | Lipid | Number of lipids | Membrane | Simulation length (ns) |
|---|---|---|---|---|
| Domain binding | HMMM | 72 | Infinite membrane bilayer | 400 |
| Equilibration | HMMM | 360 | Infinite membrane bilayer | 400 |
| Potential of mean force | HMMM | 72 | Infinite membrane bilayer | 50/window (34 windows) |
| Lipid distribution | HMMM | 2560 | Infinite membrane bilayer | 250 |
| Membrane bending | Full lipid | 360 | Membrane bilayer 250 Å-wide in the x direction and infinite in the y direction | 1000 |
| Pressure profile | Full lipid | 72 | Infinite membrane bilayer | 200 |
HMMM, highly mobile membrane mimetic (56) model.
All simulations were carried out with the software NAMD, Ver. 2.9 (http://www.ks.uiuc.edu/Research/namd/2.9/ug/) (49). The particle-mesh Ewald algorithm (50) was used for long-range electrostatic interactions. The r-RESPA multiple time-step integrator (51) was applied with time steps of 2 and 4 fs for short- and long-range interactions. The SETTLE algorithm (52) maintained water rigid geometry whereas RATTLE (53) constrained the length of covalent bonds for the rest of the system. Temperature was set at 300 K for all systems by Langevin thermostat and pressure was kept constant at 1 atm by the Langevin piston method (54). For systems including full lipid molecules, four independent simulations were performed for sampling purposes.
Steering C2B into the conformation with C-terminal helix down
The conformational transition from helix-up to helix-down in C2B was induced in a series of steered molecular-dynamics (MD) simulations (55). In the first step, the helix was slowly pulled by increasing the angle between helix and C2B’s β-sandwich structure through an elastic constraining force with a force constant of k = 2 kcal/(mol⋅degree2) and a pulling velocity of 4° per nanosecond. After the C-terminal helix reached the down-conformation, a harmonic steering force was applied between Ca2+ (the Ca2+ ion that is away from C2B’s loop-2) and each of the four negatively charged helix residues, i.e., E410, E411, E412, and D414, with a force constant of k = 3 kcal/(mol⋅Å2) and a pulling velocity of 0.2 Å/ns. For the sake of sampling, four independent steered MD simulations (55) were carried out for each negative helix residue (i.e., 16 steered simulations in total). During the steered MD simulations, the C-terminal helix backbone dihedral angles were constrained to maintain helix secondary structure. Subsequently, 200-ns simulations without constraint were carried out for each of the 16 resulting structures to test the stability of the final structures. The most stable Ca2+ binding sites for the helix were found to be E411 and D414, and the corresponding structures were employed in later simulations.
Enhancing lipid diffusion through the HMMM model
Due to lipid tail-tail entanglement, lipid redistribution is too slow to be described computationally. The recently developed highly mobile membrane mimetic (HMMM) model (56), replacing long-tail lipids with short-tail ones and adding into the resulting empty space an organic solvent between the membrane leaflets, greatly accelerates lipid redistribution while maintaining detailed lipid headgroup interactions. Compared to the original long-tail lipids, the model enables faster protein-membrane binding and lipid mixture redistribution around proteins. The HMMM model was applied at the first stage of our simulations, to study syt1 domains’ membrane binding and lipid PE/PS distribution around the domains. At the second stage, the HMMM lipids were converted to long-tail lipids to study membrane bending induced by the syt1 domain.
We first prepared an HMMM membrane by randomly placing short-tail DVPC and DVPS lipids on the proximal and distal membrane leaflets. With an empty region in-between the short-tail lipids, the distance between the two leaflets’ phosphorous atoms was set to 40 Å, i.e., to the thickness of a POPC membrane. For each lipid type we placed the same amount of lipid on either membrane leaflet in the HMMM membrane, as arises in the POPC/DOPS membrane. Then a layer of organic solvent, DCLE (1,1-dichloroethane), was added between the short-tail lipids to fill the empty region. Thereby, we constructed a membrane mimetic model composed of short-tail lipids and DCLE to mimic a long-tail POPC and DOPS lipid bilayer.
An isolated domain (C2A or C2B) was placed 5 Å above the HMMM membrane and MD simulations were carried out to give the domains an opportunity to attach to the membrane spontaneously. The CHARMM C27 force field (57) was employed for the protein and the CHARMM C36 force field for the short-tail lipids (58). The HMMM simulation protocol was the same as reported in the original HMMM article (56). After the domain attached to the membrane, ∼400 ns of further simulation was performed to equilibrate the HMMM membrane, allowing the short-tail PC and PS lipids to redistribute in response to domain binding.
The potential of mean force for the transition between the helix-up and helix-down C2B structures was calculated via umbrella sampling. An umbrella potential with force constant k = 15 kcal/(mol⋅Å2) was applied between the backbone center of mass (COM) of residues E411–A418 in the C-terminal helix and the backbone COM of residues Q356–V359 and K375–V378 in the C2B β-sandwich structure. Starting from the helix-down conformation, in which the helix contacts the membrane, 34 simulations with umbrella potential minima separated by 0.5 Å from each other were carried out. Backbone dihedral angles in the C-terminal helix were constrained so that the helix maintained its secondary structure. As noted above, the HMMM membrane model was employed to enhance lipid diffusion; however, use of the HMMM model prevents membrane bending during the calculation.
After the domain attached itself to the membrane and lipid redistributed in the context of the HMMM model, the short-tail lipids were elongated and converted to POPC and DOPS long-tail lipids. For this purpose, the HMMM’s DCLE solvent was removed and straight hydrocarbon tails were attached to short lipid tails toward the membrane center, converting DVPC/DVPS to POPC/DOPS. Next, both protein and lipid headgroups were restrained and a high temperature (800 K) MD simulation was carried out for 40 ns to ensure lipid tails to assume liquid phase ordering. Finally, all restraints were removed and the whole system was relaxed for another 40 ns in an NPxy PzT thermodynamic ensemble at 300 K and at Pxy = Pz = 1 atm to achieve full equilibration, where lipid tail-order parameters recovered to reported values (58). Membrane mechanical properties were seen to be recovered with the full-tail lipids, permitting now a study of protein-induced membrane bending.
Simulations of membrane with long-tail lipids
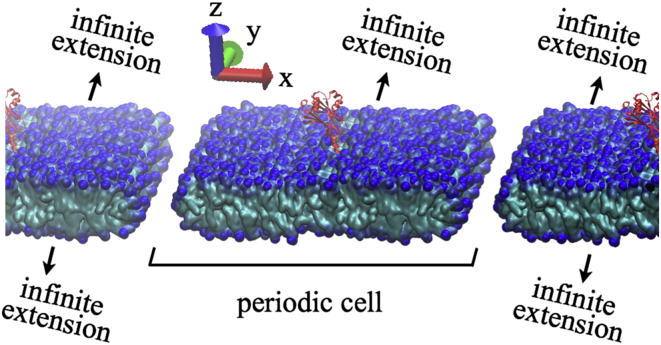
MD simulations of membranes with long-tail lipids were carried out with a single C2A or C2B domain bound to the membrane, to study the domain’s capability for membrane bending. A membrane stripe that extends to infinity along the y direction, but is finite in the x direction, can bend along the latter direction. For this reason, such a membrane stripe (or ribbon) was used, rather than a membrane patch infinite in two dimensions as typically used in membrane simulations. Actually, due to the use of periodic boundary conditions, the simulations were dealing only with finite systems. Fig. 1 shows the actual system simulated.
Figure 1.
View onto a flat membrane stripe with a C2B domain attached on top. Shown is the system simulated to study C2B-induced membrane curvature. The membrane is continuous in the ±y directions, but forms a 250 Å-wide stripe in the x direction. Periodic boundary conditions are employed in all directions but water molecules are added in the x direction to permit bending along the x direction. Shown is one periodic cell in the center and half of a periodic cell to the left and right. (Red) C2B domain. (Blue) Lipid headgroups. (Green) Lipid tails. For the sake of clarity, water and ions are not shown. To see this figure in color, go online.
A flat two-dimensional extended membrane patch served first to determine the local lipid tail-order parameter and membrane lateral pressure. The order parameter is defined as
where θ is the angle between a lipid tail carbon-carbon vector and the membrane normal. The S value for the nth carbon is defined through the vector between carbon n + 1 and carbon n − 1. The flat membrane patch, before any curvature generation, defined the membrane normal. The method adopted for calculating the membrane lateral pressure (59) also requires a flat reference membrane to define z-coordinate positions for all membrane components, namely, headgroups and hydrophobic tails.
Results
C2B conformational transition is required to trigger membrane bending
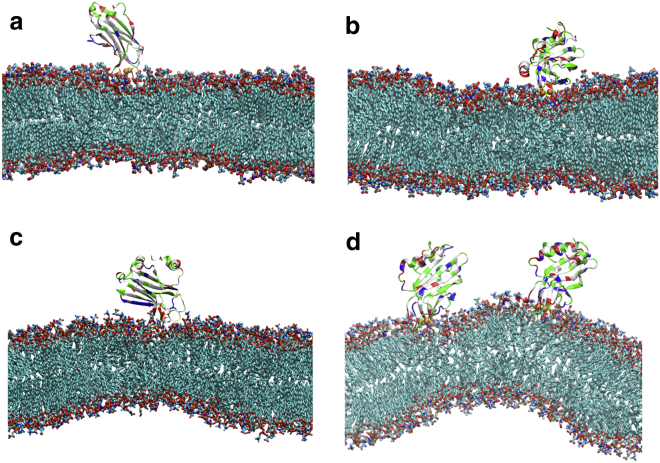
MD simulations were carried out with isolated C2A and C2B domains starting from C2A’s NMR structure (PDB:1BYN) and C2B’s crystal structure (PDB:1TJX, C-terminal helix pointing up), respectively. Membrane bending was not observed for either domain as can be seen in Fig. 2, a and b. The C2A and C2B domains maintain their initial structure as reflected by backbone RMSDs of 0.8 and 0.7 Å, respectively. The RMSDs in the Ca2+ loop regions (loop-1 and loop-3) are 0.9 and 0.7 Å for C2A and C2B, respectively. Both domains’ Ca2+-loops show a membrane insertion depth close to that seen in the EPR experiments (38, 39): for both C2A and C2B, the loop-1 regions stay at the same location as the lipid phosphate groups, whereas their loop-3 regions are ∼3 Å below and their loop-2 regions are ∼5 Å (7 Å in case of C2B) above the lipid phosphate groups.
Figure 2.
Membrane-bending activities of synaptotagmin I C2A and C2B domains. (a) No membrane bending with an isolated C2A domain. (b) No membrane bending with an isolated C2B domain, with the C-terminal helix in the helix-up conformation as in the crystal structure. (c) Membrane bending with an isolated C2B domain, with the C-terminal helix in the helix-down conformation. (d) Strong membrane bending with two isolated C2B domains, each with its C-terminal helix in the helix-down conformation. See Movie S1 for panel c and Movie S2 for panel d, illustrating the dynamics of C2B-induced membrane bending as seen in the simulations. All snapshots are taken from the fully equilibrated simulations at 1000 ns. For the C2B domain: (red) negative residues; (blue) positive residues; (green) polar residues; (white) hydrophobic residues; and (orange) Ca2+ ions. For lipid headgroups: (blue) positive charge; (red) negative charge; (green) tails. For the sake of clarity, water, hydrogen, and ions are not shown. To see this figure in color, go online.
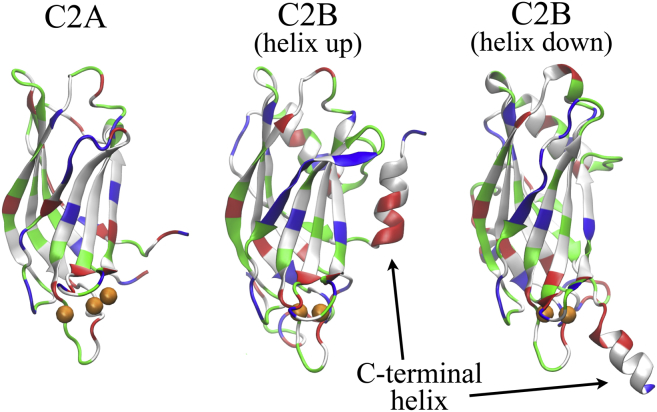
The most straightforward explanation for the difference between simulation and observation in regard to C2B’s membrane-bending activity is that C2B’s C-terminal helix, absent in the case of C2A as shown in Fig. 3, changes its position when C2B is placed on a suitable membrane, namely a membrane with a key fraction of negatively charged lipids. Interestingly, the C-terminal helix (409VEEEVDAMLAVKK421) is rich in both hydrophobic and lysine residues, which exhibit a strong affinity to the membrane bilayer. Accordingly, the helix might alter its orientation from pointing away from the membrane to an orientation pointing toward the membrane, thereby engaging its relevant side chains with lipid headgroups and tails. Such reorientation would also be favored by the negative residues in the helix, 3 Glu and 1 Asp, which have the potential to interact with Ca2+ bound to C2B’s Ca2+ loop. Indeed, NMR chemical shift changes were observed, upon Ca2+ binding, in the helix’ negative residues (42), indicating strong interaction between the Ca2+ ion and the C-terminal helix. The EPR spectrum of the helix residue A415 is different from the typical helix spectrum, indicating large motional average occurs in the helix spectrum (39). We suggest, therefore, that the C2B C-terminal helix undergoes a conformational transition from a helix-up conformation to a helix-down conformation, as shown in Fig. 3.
Figure 3.
Simulated structures of individual C2A and C2B domains with Ca2+ bound. The C-terminal helix is present only in C2B, not in C2A. See Movie S3 for an illustration of the C2B helix-up to helix-down conformational transition. The domains are colored as in Fig. 2, except with (orange spheres) Ca2+ ions. To see this figure in color, go online.
We carried out an MD simulation that placed the C2B domain onto a membrane with its C-terminal helix in a down-orientation. The simulation showed that C2B in this state is very stable and keeps interacting with the membrane for an entire 1 μs. The C2B domain β-sandwich structure remained the same as in the C2B helix-up state, with an RMSD value of the β-sandwich between up- and down-states of <1.6 Å.
Helix-down conformation energetically favorable on anionic membrane
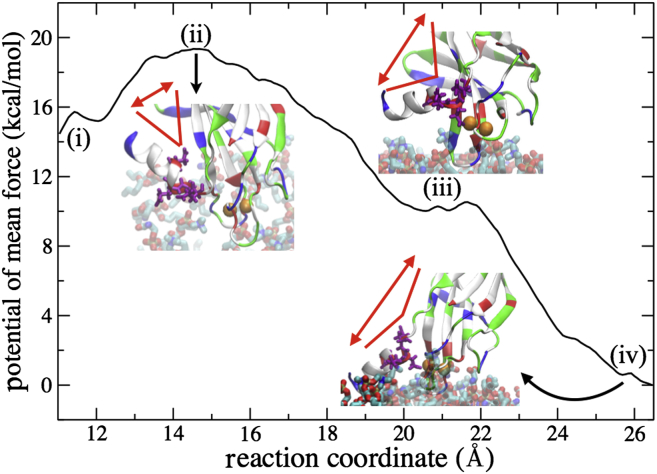
To demonstrate the feasibility of C2B’s helix-down conformation, we determined the potential of mean force (PMF) for the helix-up to helix-down transition. For the sake of computational convenience, the reaction coordinate enforced was the distance between backbone COM of residues E411–A418 in the C-terminal helix and backbone COM of residues Q356–V359 and K375–V378 in the domain β-sandwich structure. When the distance is small (11 Å), the helix points up as in the crystal structure; when the distance is large (26.5 Å), the helix points down and makes contact with the membrane surface. A path along the reaction coordinate was first enforced by gradually reducing the distance between helix and β-sandwich structure, starting from the helix-down conformation. Then umbrella potentials were added evenly along the path, yielding 34 calculation windows; the distance distribution along the reaction coordinate for each window was collected to determine the associated PMF profile employing the weighted histogram analysis method (60).
Fig. 4 shows the PMF. In the helix up-orientation, the PMF exhibits a shallow minimum labeled (i) in Fig. 4. One can recognize from the overall PMF that C2B’s helix-down conformation is more stable energetically than its helix-up conformation. To reach the helix-down conformation, a low energy barrier (labeled (ii) in Fig. 4) of ∼5 kcal/mol needs to be overcome to separate the C-terminal helix from the side of C2B’s β-sandwich structure. The existence of this small energy barrier is consistent with the degeneracy of many C-terminal helix positions shown in solution NMR structures (42).
Figure 4.
Potential of mean force connecting C2B’s C-terminal helix up- and down-conformations. The reaction coordinate enforced for the calculation is the distance between the backbone COM of helix residues E411–A418 and the backbone COM of residues Q356–V359 and K375–V378 in C2B’s β-sandwich structure. For labels (i –iv), see text. The C-terminal helix separates from the β-sandwich of the C2B domain at a reaction coordinate value of 15 Å. The polyacidic residues (highlighted in purple) start interacting with Ca2+ at a reaction coordinate value of 21 Å (see also Fig. 5). The C-terminal helix interacts with both Ca2+ and membrane at a reaction coordinate value of 26.5 Å. Colors as in Fig. 2. To see this figure in color, go online.
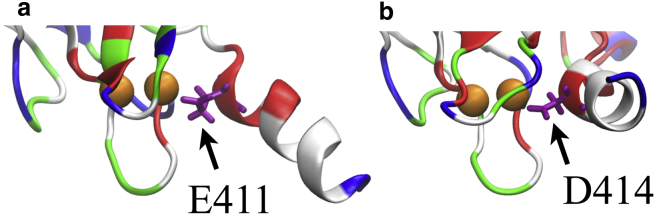
Energy barrier (ii) leads to an energy minimum (iii) corresponding to the point when the negative residues of the C-terminal helix Glu411 and Asp414 begin to interact with the Ca2+-loop of the C2B domain as shown in Fig. 5. Both residues, residue Glu411 and Asp414, have high affinity for binding to Ca2+; in the case that one of these residues is mutated to alanine, the C-terminal helix rotates slightly and the other negative residue interacts with the Ca2+ ion. The two alternative Ca2+ binding sites in the helix explain why a single point mutation on the helix polyacidic region does not alter synaptotagmin-I membrane fusion activity (61).
Figure 5.
Closeup view showing negative residues of the C-terminal helix, Glu411 and Asp414 (highlighted in purple), interacting with the Ca2+ loop of the C2B domain. Both interactions arise when C2B attaches to the membrane. Colors as in Fig. 2. To see this figure in color, go online.
At point (iv) of the PMF, the helix becomes attached to the membrane surface, its hydrophobic residues interact with lipid tails, and its lysine residues interact with anionic lipid headgroups, distributing the latter around the helix. Both the hydrophobic and electrostatic interactions stabilize the helix-down conformation. In strengthening the interaction with the hydrophobic residues of the helix, the lipid tails reorient. A closeup view showing how the C-terminal helix positions itself on the membrane is included in Movie S4 in the Supporting Material. Our simulations also showed that the helix binds to a 50% PS and 50% PC membrane, but not to a 100% PC membrane. The results indicate the importance of membrane charge density, and agree with EPR studies that demonstrated different C2B docking on membranes with PI(4,5)P2 and low PS content (39, 62).
Even though the helix-down conformation is thermodynamically more favorable than the helix-up conformation, the conformational transition from up to down does not take place spontaneously in simulations due to slow kinetics. The relatively long timescales for the C-terminal helix negative residues to locate Ca2+ and for the lipid headgroups and tails to reorganize in response to binding to the helix are inaccessible to simulation. The corresponding conformational transitions can only be described through the addition of steering forces, a method that has been successfully applied in many other cases (63). Slow kinetics in the helix conformational transition arises here on a the timescale of approximately microseconds. However, compared to the dynamics of kiss-and-run exocytosis, which typically occurs on an approximately millisecond timescale or a bit shorter (64), the conformational transition discussed here is actually a fast process.
C2B-induced membrane bending
With the C-terminal helix in the helix-down conformation, C2B-induced membrane bending occurred spontaneously in our simulation, as shown in Fig. 2 c. The bent membrane adopted curvature within ∼100 ns, characterized through a ∼102-nm diameter that was maintained throughout the rest of the simulation (1000 ns). The 100-ns timescale of curvature generation is comparable to timescales characterizing the function of other membrane-bending proteins, such as N-BAR domain (65, 66) and F-BAR domain (67). The membrane-bending process is illustrated in Movie S1. When two isolated C2B domains, both in a C-terminal helix-down conformation, were placed on a membrane, a curvature with ∼75-nm diameter formed (Fig. 2 d), indicating that multiple C2B domains can cooperatively enhance curvature and regulate membrane fusion (24, 26, 68). The membrane also becomes thinner between the two domains, indicating a potential tendency of membrane breaking when the two domains are getting too close or are chemically linked (26). The enhanced membrane-bending process is illustrated in Movie S2.
Lipid-tail arrangement around the C-terminal helix
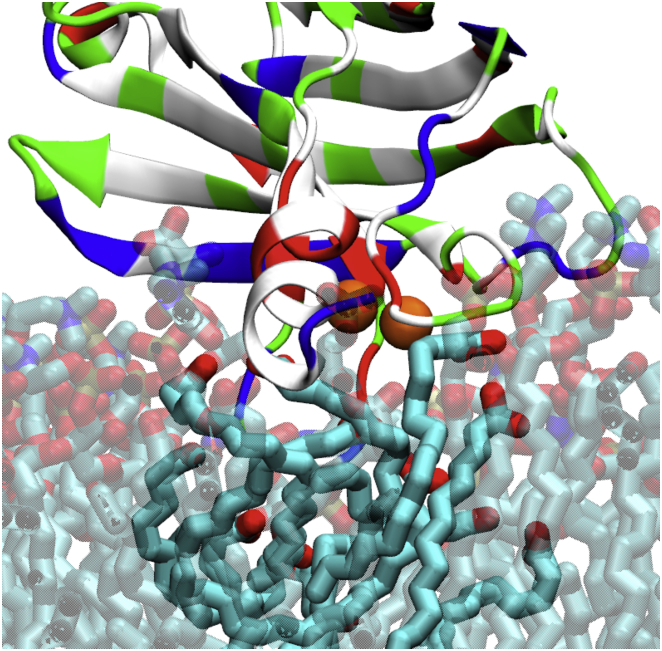
The C-terminal helix in C2B, in its helix-down conformation, is essential for triggering membrane bending. Because of the charged residues at both ends, the helix (409VEEEVDAMLAVKK421) does not insert deeply into the membrane, but acts from the membrane interface. Lipid tails in the proximal membrane leaflet approach the helix hydrophobic residues as can be seen in Fig. 6, the tails coming to lie eventually parallel to the membrane surface. The lipid tail behavior induced by C2B’s C-terminal helix is closely related to a similar lipid tail behavior found around many fusion peptides (69, 70, 71), indicating a shared functionality of the C2B C-terminal helix and fusion peptides.
Figure 6.
Lipid tails in membrane upper leaflet, having moved toward the C2B C-terminal helix. Colors as in Fig. 2. A closeup view of the lipid tails below the helix is shown in Movie S5. To see this figure in color, go online.
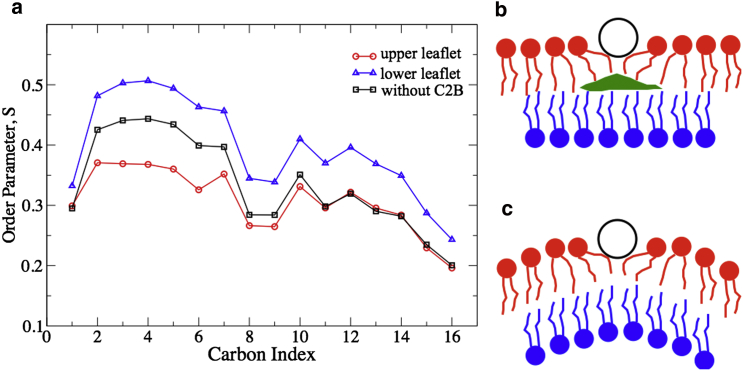
It is important to note that the helix-membrane interaction causes different arrangements for lipid tails in the proximal and in the distal membrane leaflet. Whereas lipid tails in the proximal membrane leaflet move to interact with the C-terminal helix and become more disordered, the ones in the distal membrane leaflet become more stretched and ordered, compared to lipid tails without C2B attached. The difference in local lipid-tail ordering, a process depending of course not only on lipid-helix but also on lipid-lipid interaction, is illustrated by the local lipid-tail order parameter, S. The unsaturated lipid tails within a cylinder of 13 Å radius around the α-carbon of Met416, namely under the C-terminal helix, were measured on the flat membrane before membrane curvature developed. For small S values, lipid tails are random and nearly parallel to the membrane surface; for large S values, tails tend to be ordered and oriented perpendicular to the membrane surface. Shown in Fig. 7 a, lipid tails in the proximal membrane leaflet exhibit for their carbon-carbon vectors relatively small S values compared to a membrane without C2B domain; on the other hand, lipid tails in the distal membrane leaflet assume relatively large S values. Fig. 7, b and c, provides a schematic representation of the latter lipid-tail behavior: tails in the distal leaflet have to stretch up to fill the otherwise empty space between the two membrane leaflets, thus becoming more ordered compared to tails in the other regions of the membrane.
Figure 7.
(a) Calculated order parameter S for lipid tail carbon-carbon vectors. (b) Schematic representation for helix (open circle)-induced lipid-tail arrangement consistent with the proximal leaflet S values. Without interaction between lipids in the two membrane leaflets, an open region (green) would develop. (c) Schematic representation for the actual lipid-tail arrangement induced by the C-terminal helix. Lipid tails of the distal membrane leaflet stretch out to fill the open region. The behavior of the lipids depends not only on lipid-helix interaction, but also on lipid-lipid interaction. To see this figure in color, go online.
Membrane bending is induced by an imbalance of pressure across the membrane
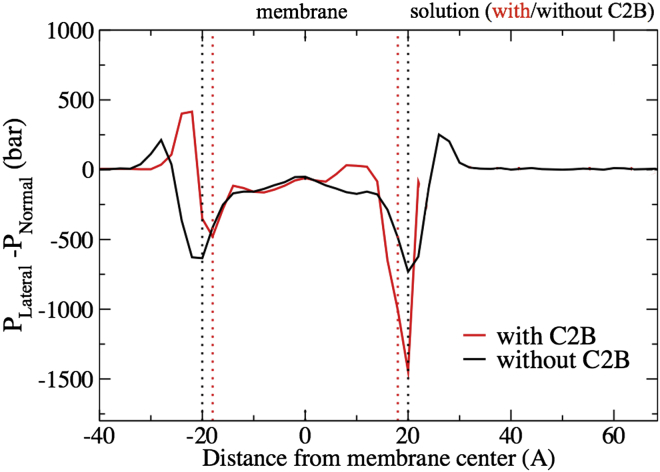
The difference in lipid-tail ordering between proximal and distal membrane leaflets generates a pressure imbalance across the bilayers. This imbalance becomes the driving force for membrane bending. Compared to a membrane without a C2B domain bound in its helix-down-state, lipid tails in the distal membrane leaflet are more ordered and, therefore, pack together more easily, resulting in less lateral pressure. In contrast, lipids in the proximal membrane leaflet are more disordered and, therefore, pack together less easily, resulting in stronger lateral pressure. The imbalance of pressure across the membrane bilayer generated by the C2B C-terminal helix (shown in Fig. 8), drives the membrane to bend.
Figure 8.
Pressure profile across the membrane. C2B binds to the membrane at a distance >20 Å from the membrane center. (Vertical dotted lines) Position of membrane phosphorous atoms, defining membrane location. For the calculation of the membrane lateral pressure, the method of Lindahl and Edholm (59) was employed. To see this figure in color, go online.
Discussion
C2B’s conformational transition is essential for membrane bending
In this study, we conclude that C2B-induced membrane bending is preceded by a Ca2+- and membrane-dependent conformational transition. In particular, C2B’s C-terminal helix reorients itself from pointing up, as seen in the membrane-free NMR/crystal structures, to a down-position where it interacts with both membrane and Ca2+. The proposed conformational transition agrees with the PMF calculation, which shows on an anionic membrane the conformation with the C-terminal helix orienting down toward the membrane to be more favorable than the one with the helix pointing up. The suggested conformational transition can also explain why C2B’s membrane-binding affinity depends on the ionic strength in solution. It has been reported that C2B’s membrane binding becomes substantially weakened when the ionic strength is increased (37), whereas C2A’s membrane binding is less sensitive to ionic strength (kd ∼ 10 nM) (47, 48). With C2B’s helix-down structure, the interaction between Ca2+ and the helix negative residues should decrease when the ionic strength increases such that the helix-down structure becomes less stable; this reduces the helix contribution to the total membrane binding energy.
With the resulting C2B helix-down conformation, the helix-membrane hydrophobic interaction is found to be the origin of C2B’s membrane-bending activity. The C-terminal helix changes lipid-tail packing in the lipid membrane bilayer: Lipid tails in the proximal membrane leaflet interact with the helix hydrophobic residues and become disordered, whereas tails in the distal leaflet become stretched and ordered, to remain in close contact with the proximal leaflet. An imbalance of pressure across the membrane is resulted, and the membrane bends.
To further demonstrate the importance of the C-terminal helix in membrane bending, simulations were performed with C2B domains with their C-terminal helix truncated. Starting from a membrane bent by two intact C2B domains (in the helix-down conformation), the simulation is extended, but for the C2B mutant with the C-terminal helix eliminated (from 409V to 421K). Several ions were removed to maintain system charge neutrality. Truncated C2B domains did not maintain the membrane curvature initially present, demonstrating the essential role of helix-membrane interactions in membrane bending. Movie S6 shows the membrane unbending.
The simulation with the C-terminal helix truncated also examined whether the distribution of negatively charged lipids around the C2B domain plays a role in membrane bending. Our simulations with the HMMM model revealed that the negatively charged PS lipids tend to colocalize in regions close to the C2B Ca2+-loop and helix lysine residues. With the helix truncated from the bent membrane, the system will have the same lipid PC and PS distribution, C2B domain binding position, and membrane curvature as before. Therefore, the membrane unbending indicates that the distribution of negatively charged lipids is not the major factor for membrane bending, because the lipid PC and PS distribution will not change dramatically over a 100-ns simulation involving long-tail lipids (56).
C2B bends membrane through a hydrophobic insertion mechanism
Campelo et al. (40) have proposed a shallow hydrophobic insertion mechanism for protein-induced membrane bending. With an elastic membrane model, a helix induces largest membrane curvature when it inserts slightly into a membrane. In the reported calculation (40), the helix radius was assumed to be r = 0.5 nm, a value that also characterizes the C-terminal α-helix in this study. One of the membrane monolayer thicknesses tested in the previous work was assumed to be h = 2.0 nm, the same thickness as seen in the simulated membrane of this study. The interface between lipid polar headgroups and tails was defined at z0 = 2/3h = 1.3 nm. From the reported calculation, the coupled membrane bilayer reaches maximum curvature when the helix center is located at zinc = 1.7 nm from the bilayer center. Taking the helix radius into account, the lower surface of the helix lies at zlow = 1.2 nm from the membrane center. Thus, the helix penetrates through the entire lipid polar headgroup region, and embeds slightly (z0 – zlow = 0.1 nm) into the membrane hydrophobic core.
The helix position from the elastic model calculation (40) agrees with the C2B C-terminal helix insertion in our MD simulations. Shown in both Fig. 6 and Movie S4, the C-terminal helix penetrates through the region of lipid headgroups, and has its membrane-inserted hydrophobic side chains interact with lipid hydrophobic tails. However, the atomistic simulations in the present study provide molecular details regarding the helix-induced lipid tail behavior. Such detail cannot be derived from an elastic model description of the shallow insertion mechanism. As the helix shallowly inserts into the membrane, the induced tensions in membrane proximal and distal leaflets not only comes from volume exclusion, but also from helix-lipid tail interaction that modifies lipid tail ordering. If the helix insertion only has an effect on volume exclusion, the pressure imbalance across the membrane may be balanced out by lipid diffusion from the proximal membrane leaflet to the distal leaflet, and no membrane curvature may occur. Therefore, membrane bending according to our simulations results mainly from the helix-induced change in lipid tail ordering, which causes pressure imbalance across membrane and generates local membrane curvature.
C-terminal helix-membrane interaction facilitates membrane fusion
It has been reported that C2B-induced membrane bending is essential to syt1’s membrane fusion activity. When C2B’s membrane-bending activity is abolished, syt1 does not trigger membrane fusion (26). It is of interest then to consider how membrane bending is linked to membrane fusion. In particular, it is believed that there is a high energy penalty if two membranes approach each other: Lipid headgroups will be dehydrated at the point where two membranes are in close contact. Fortunately, according to the molecular mechanism for C2B-induced membrane bending, membrane lipids are reorganized by the C-terminal helix in such a way that lipid headgroup dehydration will not occur when the respective membranes approach each other. Lipids in the proximal membrane leaflet redistribute in response to the membrane binding of the C2B domain and its C-terminal helix. Because of interactions between helix hydrophobic residues and lipid tails of the proximal membrane leaflet, a region free of lipid headgroups develops around the helix. As shown in Fig. 9, an ∼40 × 15 Å2 region with low headgroup density is observed in the HMMM simulations. Lipid tails within the region are more solvent-exposed than they are in other membrane regions. Movie S4 and Movie S5 illustrate the solvent-exposed lipid tails around the C-terminal helix.
Figure 9.
Two-dimensional lipid headgroup density of the proximal leaflet around the C2B domain (top view) resulting from simulations employing the HMMM model. (Shown to scale on the left) C2B domain with its protruding C-terminal helix. Colors of the C2B domain are the same as in Fig. 2. To see this figure in color, go online.
The results presented in Fig. 9 agree with fluorescence resonance energy transfer measurements, which show a larger distance from the helix (residue A415 in the middle of the helix) to lipid headgroups than from other membrane-attached residues in the Ca2+-loop (V304, N333, and I367) (46). The radial distribution function (not shown) gives the closest distribution between amino-acid α-carbon and lipid phosphate at 5 Å for V304, then ∼10 Å for N333 and I367, but 15 Å for A415. However, rather than concluding from this measurement that the helix points away from the membrane surface as suggested in Hui et al. (46), we conclude, based on our simulations, that the difference in distance results from unevenly distributed lipid headgroups.
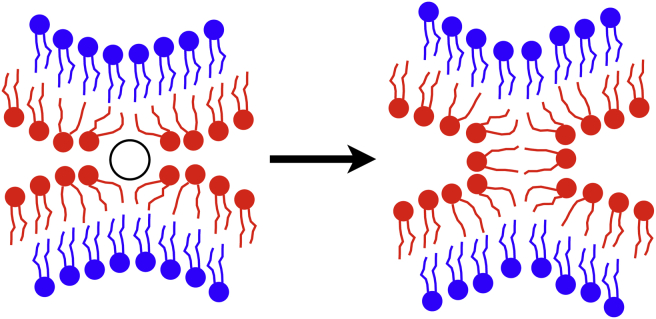
The hypothesis had been voiced earlier that solvent-exposed lipid tails facilitate membrane fusion (71, 72, 73). Indeed, several fusion peptides, such as melittin (69), influenza hemagglutinin (70, 71), and their mutants, interact with lipid tails of the proximal membrane leaflet and drive them to the membrane surface. Similar solvent-exposed lipid tails are also found in the fusion process induced by the transmembrane helix of the SNARE complex on small vesicles (with highly bent vesicle membranes) as revealed in a coarse-grained MD description (73). One may conclude, therefore, that solvent-exposed lipid tails can fuse two membranes without lipid headgroup dehydration by forming first a lipid stalk between membranes (72, 73), as illustrated schematically in Fig. 10. Accordingly, the solvent-exposed lipid tails induced by C2B’s C-terminal helix can drive both membrane bending and membrane fusion, which explains the experimentally observed correlation between the two membrane remodeling processes (26).
Figure 10.
Schematic representation of solvent-exposed lipid tails on the membrane surface induced by C2B’s C-terminal helix (open circle). Such tails can facilitate stalk formation, namely hemifusion, between two membranes. To see this figure in color, go online.
Synaptotagmin C2B is different from other C2 domains
Although >200 C2 domains have been identified to date, only some of them can induce positive membrane curvature. Even though synaptotagmins C2A domain and C2B domain are similar in both sequence and structure, only the C2B domain can bend a membrane. The distinct membrane-bending activity indicates that one cannot take for granted that the C2 domains are capable of membrane bending, and one should not expect a similar membrane-bending mechanism for all C2 domains. Different C2 domains could have their unique motifs and mechanism to induce membrane curvature, although they might not have the C-terminal helix as in syt1 C2B. For example, cytosolic phospholipase A2’s Ca2+-loop structure could induce membrane curvature (74), and its membrane-bending activity could have originated from the helical structure in the Ca2+-loop region. For Doc2b, it is the C2A domain that bends the membrane (75), and the calcium-loop sequence is different from syt1 C2B.
According to sequence alignment, the C-terminal helix of C2B is conserved in syt1 among different species. However, the helix appears mostly only in syt1, and not in other C2 domains. Although many proteins have C2 domains, only some of them are Ca2+-sensitive and only syt1 senses Ca2+ at the right concentration and the right timescale to regulate neurotransmitter release. We believe syt1’s uniqueness comes from its unusual sequence and structure.
Conclusion
Synaptotagmin I is a key player in Ca2+-ion-triggered fusion of neurotransmitter vesicles with the presynaptic membrane (17, 18). The mechanism by which it acts is still unknown, though a membrane-free crystal structure (41) and a related NMR structure (42) permit MD simulations that may reveal the mechanism. Based on such simulations, we suggest now that a key detail of the available structures, namely the orientation of the C-terminal helix in one of the two synaptotagmin domains, C2B, becomes altered before inducing membrane bending as a primary step to fusion. Membrane bending arises only after the helix alters its orientation from pointing away (up) from the membrane surface to an orientation pointing toward the membrane (down). In the down-orientation, the helix makes contact with lipid tails and reorders them. Our suggestion is based specifically on the observation, both in experiment (26) and simulation, that, of the two domains of synaptotagmin I, the C2A domain has no ability to bend a membrane, but the C2B domain has that ability with anionic lipids. However, simulations show that the latter domain bound with Ca2+ induces membrane bending only when the C-terminal helix points down; removal of the C-terminal helix abolishes membrane bending.
Simulations determining the free energy barrier for the needed conformational transition of the C-terminal helix of the C2B domain find a rather low (5 kcal/mol) barrier and reveal that for a membrane-bound C2B domain, the helix-down state has lower free energy than the helix-up state, arguing strongly for the feasibility of the suggested membrane-bending mechanism. Although the insertion of the C2B Ca2+ loops has been explored in previous experimental studies (31, 36, 37, 39), the potential conformational change in C2B’s C-terminal helix has not been tested. We anticipate a decrease of membrane-bending activity in C2B will be observed in mutation experiments where the C2B’s C-terminal helix is truncated. With the help of double electron-electron resonance (76, 77), distance distribution from the C-terminal helix to C2B’s β-sandwich could be compared between the C2B structure in solution and its structure after binding to an anionic membrane.
An analysis of the effect of the C-terminal helix on the membrane shows a strong reordering of lipid tails that differs significantly between the membrane bilayer leaflet proximal to the C2B domain and the one distal. Lipids in the proximal leaflet move their tails toward the membrane surface, exposing them to the C-terminal helix and solvent; lipid tails in the distal leaflet instead straighten up to fill the void left in the proximal leaflet, and become more ordered. The different ordering results in an increase in lateral pressure in the proximal leaflet and a decrease in the distal leaflet, and results in membrane bending.
The reordering of lipid tails can not only be an initial step for membrane bending, but also for so-called stalk formation and hemifusion, which eventually leads to membrane fusion. Indeed, the lipid tail reordering is consistent with the behavior of lipids around oligopeptides that facilitate membrane fusion (72, 73). The simulation results explain naturally the close correlation seen experimentally between synaptotagmin’s ability to bend and to fuse a membrane.
Acknowledgments
Authors are grateful for discussions with Qiang Cui, Arun Yethiraj, Emad Tajkhorshid, Edwin R. Chapman, and David S. Cafiso.
The research has been supported by the National Science Foundation (grant No. PHY-0822613) and by the National Institutes of Health (grants No. 9P41GM104601 and No. 1R01GM067887). The authors also acknowledge supercomputer time on STAMPEDE provided by the Texas Advanced Computing Center at the University of Texas at Austin through Extreme Science and Engineering Discovery Environment (XSEDE) grant No. MCA93S028, as well as supercomputer time on Blue Waters as part of the Petascale Computational Resource (PRAC) grant “The Computational Microscope”, which is supported by the National Science Foundation (OCI-0832673).
Editor: David Cafiso.
Footnotes
Seven movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(14)00786-3.
Supporting Material
References
- 1.Marsh M., McMahon H.T. The structural era of endocytosis. Science. 1999;285:215–220. doi: 10.1126/science.285.5425.215. [DOI] [PubMed] [Google Scholar]
- 2.Kirchhausen T. Clathrin. Annu. Rev. Biochem. 2000;69:699–727. doi: 10.1146/annurev.biochem.69.1.699. [DOI] [PubMed] [Google Scholar]
- 3.Cho W., Stahelin R.V. Membrane-protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys. Biomol. Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 4.McMahon H.T., Gallop J.L. Membrane curvature and mechanisms of dynamic cell membrane remodeling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 5.Lecuit T., Pilot F. Developmental control of cell morphogenesis: a focus on membrane growth. Nat. Cell Biol. 2003;5:103–108. doi: 10.1038/ncb0203-103. [DOI] [PubMed] [Google Scholar]
- 6.McMahon H.T., Mills I.G. COP and clathrin-coated vesicle budding: different pathways, common approaches. Curr. Opin. Cell Biol. 2004;16:379–391. doi: 10.1016/j.ceb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Südhof T.C. A molecular machine for neurotransmitter release: synaptotagmin and beyond. Nat. Med. 2013;19:1227–1231. doi: 10.1038/nm.3338. [DOI] [PubMed] [Google Scholar]
- 8.Südhof T.C. Calcium control of neurotransmitter release. Cold Spring Harb. Perspect. Biol. 2012;4:a011353. doi: 10.1101/cshperspect.a011353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernández-Chacón R., Königstorfer A., Südhof T.C. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–49. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]
- 10.Xu J., Pang Z.P., Südhof T.C. Synaptotagmin-1 functions as a Ca2+ sensor for spontaneous release. Nat. Neurosci. 2009;12:759–766. doi: 10.1038/nn.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee H.-K., Yang Y., Yoon T.-Y. Dynamic Ca2+-dependent stimulation of vesicle fusion by membrane-anchored synaptotagmin 1. Science. 2010;328:760–763. doi: 10.1126/science.1187722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohrmann R., de Wit H., Sørensen J.B. Synaptotagmin interaction with SNAP-25 governs vesicle docking, priming, and fusion triggering. J. Neurosci. 2013;33:14417–14430. doi: 10.1523/JNEUROSCI.1236-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vrljic M., Strop P., Brunger A.T. Molecular mechanism of the synaptotagmin-SNARE interaction in Ca2+-triggered vesicle fusion. Nat. Struct. Mol. Biol. 2010;17:325–331. doi: 10.1038/nsmb.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaub J.R., Lu X., McNew J.A. Hemifusion arrest by complexin is relieved by Ca2+-synaptotagmin I. Nat. Struct. Mol. Biol. 2006;13:748–750. doi: 10.1038/nsmb1124. [DOI] [PubMed] [Google Scholar]
- 15.Tang J., Maximov A., Südhof T.C. A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis. Cell. 2006;126:1175–1187. doi: 10.1016/j.cell.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 16.Ma C., Su L., Rizo J. Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science. 2013;339:421–425. doi: 10.1126/science.1230473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jahn R., Fasshauer D. Molecular machines governing exocytosis of synaptic vesicles. Nature. 2012;490:201–207. doi: 10.1038/nature11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chapman E.R. How does synaptotagmin trigger neurotransmitter release? Annu. Rev. Biochem. 2008;77:615–641. doi: 10.1146/annurev.biochem.77.062005.101135. [DOI] [PubMed] [Google Scholar]
- 19.Maximov A., Südhof T.C. Autonomous function of synaptotagmin 1 in triggering synchronous release independent of asynchronous release. Neuron. 2005;48:547–554. doi: 10.1016/j.neuron.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Saludes J.P., Morton L.A., Yin H. Detection of highly curved membrane surfaces using a cyclic peptide derived from synaptotagmin-I. ACS Chem. Biol. 2012;7:1629–1635. doi: 10.1021/cb3002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Severin P.M., Zou X., Gaub H.E. Effects of cytosine hydroxymethylation on DNA strand separation. Biophys. J. 2013;104:208–215. doi: 10.1016/j.bpj.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honigmann A., van den Bogaart G., Jahn R. Phosphatidylinositol 4,5-bisphosphate clusters act as molecular beacons for vesicle recruitment. Nat. Struct. Mol. Biol. 2013;20:679–686. doi: 10.1038/nsmb.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geppert M., Goda Y., Südhof T.C. Synaptotagmin I: a major Ca2+ sensor for transmitter release at a central synapse. Cell. 1994;79:717–727. doi: 10.1016/0092-8674(94)90556-8. [DOI] [PubMed] [Google Scholar]
- 24.Lai Y., Diao J., Shin Y.-K. Fusion pore formation and expansion induced by Ca2+ and synaptotagmin 1. Proc. Natl. Acad. Sci. USA. 2013;110:1333–1338. doi: 10.1073/pnas.1218818110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martens S., Kozlov M.M., McMahon H.T. How synaptotagmin promotes membrane fusion. Science. 2007;316:1205–1208. doi: 10.1126/science.1142614. [DOI] [PubMed] [Google Scholar]
- 26.Hui E., Johnson C.P., Chapman E.R. Synaptotagmin-mediated bending of the target membrane is a critical step in Ca2+-regulated fusion. Cell. 2009;138:709–721. doi: 10.1016/j.cell.2009.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai J., Tucker W.C., Chapman E.R. PIP2 increases the speed of response of synaptotagmin and steers its membrane-penetration activity toward the plasma membrane. Nat. Struct. Mol. Biol. 2003;11:36–44. doi: 10.1038/nsmb709. [DOI] [PubMed] [Google Scholar]
- 28.Bharat T.A.M., Malsam J., Briggs J.A.G. SNARE and regulatory proteins induce local membrane protrusions to prime docked vesicles for fast calcium-triggered fusion. EMBO Rep. 2014 doi: 10.1002/embr.201337807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nalefski E.A., Falke J.J. The C2 domain calcium-binding motif: structural and functional diversity. Protein Sci. 1996;5:2375–2390. doi: 10.1002/pro.5560051201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sutton R.B., Davletov B.A., Sprang S.R. Structure of the first C2 domain of synaptotagmin I: a novel Ca2+/phospholipid-binding fold. Cell. 1995;80:929–938. doi: 10.1016/0092-8674(95)90296-1. [DOI] [PubMed] [Google Scholar]
- 31.Chapman E.R., Davis A.F. Direct interaction of a Ca2+-binding loop of synaptotagmin with lipid bilayers. J. Biol. Chem. 1998;273:13995–14001. doi: 10.1074/jbc.273.22.13995. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z., Hui E., Jackson M.B. Phosphatidylserine regulation of Ca2+-triggered exocytosis and fusion pores in PC12 cells. Mol. Biol. Cell. 2009;20:5086–5095. doi: 10.1091/mbc.E09-08-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reference deleted in proof.
- 34.Seven A.B., Brewer K.D., Rizo J. Prevalent mechanism of membrane bridging by synaptotagmin-1. Proc. Natl. Acad. Sci. USA. 2013;110:E3243–E3252. doi: 10.1073/pnas.1310327110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vennekate W., Schröder S., Walla P.J. Cis- and trans-membrane interactions of synaptotagmin-1. Proc. Natl. Acad. Sci. USA. 2012;109:11037–11042. doi: 10.1073/pnas.1116326109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bai J., Earles C.A., Chapman E.R. Membrane-embedded synaptotagmin penetrates cis or trans target membranes and clusters via a novel mechanism. J. Biol. Chem. 2000;275:25427–25435. doi: 10.1074/jbc.M906729199. [DOI] [PubMed] [Google Scholar]
- 37.Hui E., Bai J., Chapman E.R. Ca2+-triggered simultaneous membrane penetration of the tandem C2-domains of synaptotagmin I. Biophys. J. 2006;91:1767–1777. doi: 10.1529/biophysj.105.080325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frazier A.A., Roller C.R., Cafiso D.S. Membrane-bound orientation and position of the synaptotagmin I C2A domain by site-directed spin labeling. Biochemistry. 2003;42:96–105. doi: 10.1021/bi0268145. [DOI] [PubMed] [Google Scholar]
- 39.Rufener E., Frazier A.A., Cafiso D.S. Membrane-bound orientation and position of the synaptotagmin C2B domain determined by site-directed spin labeling. Biochemistry. 2005;44:18–28. doi: 10.1021/bi048370d. [DOI] [PubMed] [Google Scholar]
- 40.Campelo F., McMahon H.T., Kozlov M.M. The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophys. J. 2008;95:2325–2339. doi: 10.1529/biophysj.108.133173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng Y., Sequeira S.M., Patel D.J. Crystallographic identification of Ca2+ and Sr2+ coordination sites in synaptotagmin I C2B domain. Protein Sci. 2004;13:2665–2672. doi: 10.1110/ps.04832604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernandez I., Araç D., Rizo J. Three-dimensional structure of the synaptotagmin 1 C2B-domain: synaptotagmin 1 as a phospholipid binding machine. Neuron. 2001;32:1057–1069. doi: 10.1016/s0896-6273(01)00548-7. [DOI] [PubMed] [Google Scholar]
- 43.Im W., Brooks C.L., 3rd Interfacial folding and membrane insertion of designed peptides studied by molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 2005;102:6771–6776. doi: 10.1073/pnas.0408135102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grobler J.A., Essen L.-O., Hurley J.H. C2 domain conformational changes in phospholipase C-δ1. Nat. Struct. Biol. 1996;3:788–795. doi: 10.1038/nsb0996-788. [DOI] [PubMed] [Google Scholar]
- 45.Arkhipov A., Shan Y., Shaw D.E. Architecture and membrane interactions of the EGF receptor. Cell. 2013;152:557–569. doi: 10.1016/j.cell.2012.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hui E., Gaffaney J.D., Chapman E.R. Mechanism and function of synaptotagmin-mediated membrane apposition. Nat. Struct. Mol. Biol. 2011;18:813–821. doi: 10.1038/nsmb.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davletov B.A., Südhof T.C. A single C2 domain from synaptotagmin I is sufficient for high affinity Ca2+/phospholipid binding. J. Biol. Chem. 1993;268:26386–26390. [PubMed] [Google Scholar]
- 48.Wang P., Wang C.-T., Chapman E.R. Mutations in the effector binding loops in the C2A and C2B domains of synaptotagmin I disrupt exocytosis in a nonadditive manner. J. Biol. Chem. 2003;278:47030–47037. doi: 10.1074/jbc.M306728200. [DOI] [PubMed] [Google Scholar]
- 49.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Darden T., York D., Pedersen L.G. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 51.Tuckerman M., Berne B.J., Martyna G.J. Reversible multiple time scale molecular dynamics. J. Chem. Phys. 1992;97:1990–2001. [Google Scholar]
- 52.Miyamoto S., Kollman P.A. SETTLE: an analytical version of the SHAKE and RATTLE algorithm for rigid water molecules. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 53.Andersen H.C. RATTLE: a “velocity” version of the SHAKE algorithm for molecular dynamics calculations. J. Chem. Phys. 1983;52:24–34. [Google Scholar]
- 54.Feller S.E., Zhang Y., Brooks B.R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 55.Izrailev S., Stepaniants S., Schulten K. Steered molecular dynamics. In: Deuflhard P., Hermans J., Leimkuhler B., Mark A.E., Reich S., Skeel R.D., editors. Computational Molecular Dynamics: Challenges, Methods, Ideas, Vol. 4, Lecture Notes in Computational Science and Engineering. Springer-Verlag; Berlin, Germany: 1998. pp. 39–65. [Google Scholar]
- 56.Ohkubo Y.Z., Pogorelov T.V., Tajkhorshid E. Accelerating membrane insertion of peripheral proteins with a novel membrane mimetic model. Biophys. J. 2012;102:2130–2139. doi: 10.1016/j.bpj.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MacKerell A.D., Jr., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 58.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lindahl E., Edholm O. Spatial and energetic-entropic decomposition of surface tension in lipid bilayers from molecular dynamics simulations. J. Chem. Phys. 2000;113:3882–3893. [Google Scholar]
- 60.Kumar S., Rosenberg J.M., Kollman P.A. The weighted histogram analysis method for free-energy calculations on biomolecules. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- 61.Gaffaney J.D., Dunning F.M., Chapman E.R. Synaptotagmin C2B domain regulates Ca2+-triggered fusion in vitro: critical residues revealed by scanning alanine mutagenesis. J. Biol. Chem. 2008;283:31763–31775. doi: 10.1074/jbc.M803355200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuo W., Herrick D.Z., Cafiso D.S. Phosphatidylinositol 4,5-bisphosphate alters synaptotagmin 1 membrane docking and drives opposing bilayers closer together. Biochemistry. 2011;50:2633–2641. doi: 10.1021/bi200049c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Isralewitz B., Gao M., Schulten K. Steered molecular dynamics and mechanical functions of proteins. Curr. Opin. Struct. Biol. 2001;11:224–230. doi: 10.1016/s0959-440x(00)00194-9. [DOI] [PubMed] [Google Scholar]
- 64.Valtorta F., Meldolesi J., Fesce R. Synaptic vesicles: is kissing a matter of competence? Trends Cell Biol. 2001;11:324–328. doi: 10.1016/s0962-8924(01)02058-x. [DOI] [PubMed] [Google Scholar]
- 65.Arkhipov A., Yin Y., Schulten K. Four-scale description of membrane sculpting by BAR domains. Biophys. J. 2008;95:2806–2821. doi: 10.1529/biophysj.108.132563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cui H., Ayton G.S., Voth G.A. Membrane binding by the endophilin N-BAR domain. Biophys. J. 2009;97:2746–2753. doi: 10.1016/j.bpj.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu H., Schulten K. Membrane sculpting by F-BAR domains studied by molecular dynamics simulations. PLOS Comput. Biol. 2013;9:e1002892. doi: 10.1371/journal.pcbi.1002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee J., Guan Z., Littleton J.T. Genetic analysis of synaptotagmin C2 domain specificity in regulating spontaneous and evoked neurotransmitter release. J. Neurosci. 2013;33:187–200. doi: 10.1523/JNEUROSCI.3214-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bernèche S., Nina M., Roux B. Molecular dynamics simulation of melittin in a dimyristoylphosphatidylcholine bilayer membrane. Biophys. J. 1998;75:1603–1618. doi: 10.1016/S0006-3495(98)77604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lagüe P., Roux B., Pastor R.W. Molecular dynamics simulations of the influenza hemagglutinin fusion peptide in micelles and bilayers: conformational analysis of peptide and lipids. J. Mol. Biol. 2005;354:1129–1141. doi: 10.1016/j.jmb.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 71.Larsson P., Kasson P.M. Lipid tail protrusion in simulations predicts fusogenic activity of influenza fusion peptide mutants and conformational models. PLOS Comput. Biol. 2013;9:e1002950. doi: 10.1371/journal.pcbi.1002950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smirnova Y.G., Marrink S.-J., Knecht V. Solvent-exposed tails as prestalk transition states for membrane fusion at low hydration. J. Am. Chem. Soc. 2010;132:6710–6718. doi: 10.1021/ja910050x. [DOI] [PubMed] [Google Scholar]
- 73.Risselada H.J., Kutzner C., Grubmüller H. Caught in the act: visualization of SNARE-mediated fusion events in molecular detail. ChemBioChem. 2011;12:1049–1055. doi: 10.1002/cbic.201100020. [DOI] [PubMed] [Google Scholar]
- 74.Ward K.E., Ropa J.P., Stahelin R.V. C2 domain membrane penetration by group IVA cytosolic phospholipase A2 induces membrane curvature changes. J. Lipid Res. 2012;53:2656–2666. doi: 10.1194/jlr.M030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu H., Rathore S.S., Shen J. Doc2b promotes GLUT4 exocytosis by activating the SNARE-mediated fusion reaction in a calcium- and membrane bending-dependent manner. Mol. Biol. Cell. 2013;24:1176–1184. doi: 10.1091/mbc.E12-11-0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schiemann O., Prisner T.F. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Q. Rev. Biophys. 2007;40:1–53. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- 77.Fanucci G.E., Cafiso D.S. Recent advances and applications of site-directed spin labeling. Curr. Opin. Struct. Biol. 2006;16:644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.