Significance
Bacteria control biogeochemical cycles of elements and fluxes of energy in the ocean. Discovery of a membrane photoprotein widespread in marine bacteria—proteorhodopsin—expanded their potential importance for global energy budgets, providing a novel mechanism to harness light energy. Yet, how proteorhodopsin-derived energy is used for cell metabolism remains largely unexplored. We combined experiments in a model marine bacterium with gene expression analyses. Light-stimulated growth coincided with a shift in carbon acquisition pathways, with anaplerotic CO2 fixation providing up to one-third of the cell carbon. Exposure to light resulted in the up-regulation of several central metabolism genes, including the glyoxylate shunt. Thus, light provides proteorhodopsin-containing bacteria with wider means to adapt to environmental variability than previously recognized.
Keywords: microbial ecology, gene expression regulation, quantitative PCR
Abstract
Proteorhodopsin (PR) is present in half of surface ocean bacterioplankton, where its light-driven proton pumping provides energy to cells. Indeed, PR promotes growth or survival in different bacteria. However, the metabolic pathways mediating the light responses remain unknown. We analyzed growth of the PR-containing Dokdonia sp. MED134 (where light-stimulated growth had been found) in seawater with low concentrations of mixed [yeast extract and peptone (YEP)] or single (alanine, Ala) carbon compounds as models for rich and poor environments. We discovered changes in gene expression revealing a tightly regulated shift in central metabolic pathways between light and dark conditions. Bacteria showed relatively stronger light responses in Ala compared with YEP. Notably, carbon acquisition pathways shifted toward anaplerotic CO2 fixation in the light, contributing 31 ± 8% and 24 ± 6% of the carbon incorporated into biomass in Ala and YEP, respectively. Thus, MED134 was a facultative double mixotroph, i.e., photo- and chemotrophic for its energy source and using both bicarbonate and organic matter as carbon sources. Unexpectedly, relative expression of the glyoxylate shunt genes (isocitrate lyase and malate synthase) was >300-fold higher in the light—but only in Ala—contributing a more efficient use of carbon from organic compounds. We explored these findings in metagenomes and metatranscriptomes and observed similar prevalence of the glyoxylate shunt compared with PR genes and highest expression of the isocitrate lyase gene coinciding with highest solar irradiance. Thus, regulatory interactions between dissolved organic carbon quality and central metabolic pathways critically determine the fitness of surface ocean bacteria engaging in PR phototrophy.
Proteorhodopsins (PRs) are membrane-embedded light-driven proton pumps discovered more than a decade ago in marine bacteria (1, 2). The gene encoding PR is widely distributed among bacterial taxa, shows a large sequence divergence, and is abundant throughout the world oceans (1–7). PR is highly expressed in marine environments both at the RNA and the protein level (1, 2, 8–10) and is thus expected to have a significant impact on surface ocean energy budgets. PR gene expression can be induced by light, as observed both in model bacteria and in some natural marine bacterial communities (11–14). The functional characterization of PR has been done mainly using Escherichia coli or Shewanella oneidensis as heterologous experimentation systems (1, 15–19), but also in native marine bacterioplankton assemblages (2, 9). Although these studies provide a fairly complete picture of the biochemistry of PR in the form of proton pumping and spectral tuning, little is known about its energy contribution to marine bacteria and how this relates to cellular metabolism or carbon acquisition strategies.
Thus far, experiments with marine bacteria have demonstrated that PR phototrophy provides sufficient amounts of energy for significantly promoting growth in the Flavobacteriia Dokdonia sp. MED134 and Psychroflexus torquis and for improving survival during starvation in the Gammaproteobacteria Vibrio sp. AND4 and Vibrio campbellii BAA-1116 (11, 12, 20–22). If this were the case in natural seawater, PR light harvesting could indeed be quantitatively important. Further, work on MED134 showed that the net benefit of PR phototrophy was larger in seawater with lower concentrations of dissolved organic carbon (DOC), indicating that exposure to light confers a stronger selective advantage in oligotrophic environments (11). This finding was confirmed by Kimura et al. (12), who extended the analysis to include inhibitors of biosynthesis of the light-harvesting cofactor retinal, thereby providing direct evidence that PR light harvesting accounts for the light-induced growth response. In other marine bacteria, the PR-mediated light response is less clear. For example, first analyses of PR-containing bacteria belonging to the ubiquitous SAR11 and SAR92 clades showed no responses to light (9, 23). However, subsequent comprehensive analyses of the SAR11 clade representative Candidatus Pelagibacter ubique strain HTCC1062 showed that oxygen consumption decreased considerably in the light compared with darkness when DOC availability became limiting. Consequently, ATP generation from PR phototrophy potentially provides an alternative to respiration under starvation conditions (24). The continued study of PR phototrophy in bacteria with different genome contexts promises to reveal novel strategies for PR-bearing bacteria to adapt to life in oligotrophic environments.
Working on the PR-containing Polaribacter sp. MED152 (Flavobacteriia), González et al. (25) observed higher rates of anaplerotic CO2 fixation in the light even though no growth differences between light and darkness were found. They thus proposed that light-induced inorganic carbon fixation could be a means for PR-bearing bacteria to make more efficient use of dissolved organic matter. Further genomic analyses revealed that both Polaribacter sp. MED152 and Dokdonia sp. MED134 contain several enzymes involved in anaplerotic reactions that replenish intermediates in the tricarboxylic acid (TCA) cycle when they are used for biosynthesis (25, 26). Recently, Kimura et al. (12) used a genome-wide transcriptomics approach in an attempt to obtain more detailed information on the metabolic response to light in Dokdonia sp. MED134. They showed that 601 of a total of 2,944 protein coding genes were differentially expressed in light compared with darkness, including several genes involved in central metabolic pathways (e.g., glycolysis, TCA cycle). Collectively, these results suggest that critical insights into the quantitative importance and regulation of PR phototrophy could be gained from linking gene expression analyses with ecophysiological response experiments in marine bacteria.
Although the benefit of PR phototrophy is dependent on DOC concentrations (11), the potential role of DOC quality in determining the response of PR-containing bacteria to light remains unexplored. This lack of knowledge potentially hampers our understanding of the role of PR light energy harvesting in surface waters, considering that the concentrations of different DOC components are known to vary substantially in a spatiotemporal manner (27). In this context, quality of DOC with respect to bacteria includes considerations both of the actual chemical composition of DOC, i.e., which compounds are there or not, and the ease and/or benefit with which available compounds are used, e.g., compound lability, energetic value in chemical bonds or number of carbon atoms available for bacteria.
In this study, we investigated light-induced differences in growth performance and expression patterns of selected genes in the PR-containing Dokdonia sp. MED134 during organic carbon-limited growth in seawater enriched with different organic matter sources. As a model for mixed organic matter, we used a mixture of yeast extract and peptone (i.e., dominance of polypeptides), and as a single organic compound, we used the amino acid alanine. Alanine was chosen because it is one of the most abundant amino acids in seawater, and it is one of few specific compounds known to sustain growth of native bacteria in the sea (27–29). We hypothesized that PR phototrophy is regulated not only by PR gene responses to light, but also by DOC quality and the expression of genes involved in anaplerotic reactions and/or central metabolic pathways.
Results
Growth and Carbon Incorporation of Dokdonia sp. MED134 Under Different Conditions.
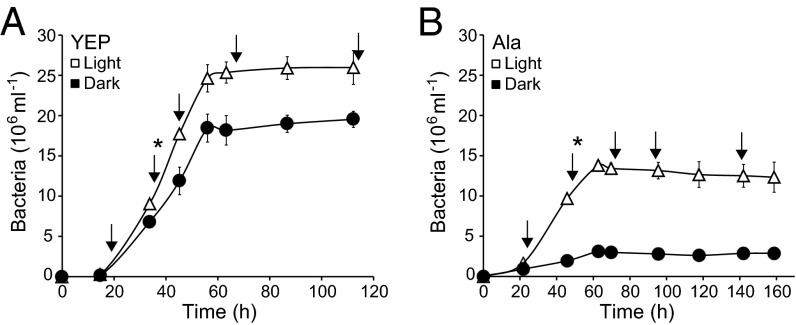
Dokdonia sp. MED134 was incubated in the light and in darkness in seawater enriched with either a mixed [yeast extract and peptone (YEP)] or single (alanine, Ala) dissolved organic carbon source (Fig. 1). In both media, growth rate, final yield, and cell volume were higher in the light than in the dark (Table 1). Differences in specific growth rates and yields were larger in Ala (2.4× and 4.2× higher in light; Fig. 1B) than in YEP (1.2× and 1.3× higher in light; Fig. 1A), whereas cell volumes were 1.2× higher in light in both media (Table 1).
Fig. 1.
Growth response of Dokdonia sp. MED134 in artificial seawater with mixed or single dissolved organic matter compounds at different light conditions. (A) Growth in seawater with yeast extract and peptone (YEP; 0.30 mM carbon). (B) Growth in seawater with alanine (Ala; 0.70 mM carbon). Error bars denote SDs of triplicate cultures; if not visible, error bars are hidden by symbols. Asterisks denote the time points when bicarbonate and leucine incorporation rates were measured, whereas arrows denote the time points for gene expression analyses.
Table 1.
Growth characteristics of Dokdonia sp. MED134 at different light conditions
| Seawater culture enrichment | Treatment | Specific growth rate (µmax, h−1) | Cell yield (106/mL) | Cell volume (μm3)* | Cell production (105 mL−1⋅h−1)† | Carbon production (fgC⋅cell−1⋅h−1)‡ | Bicarbonate incorporation (fgC⋅cell−1⋅h−1) | Bicarbonate incorporation:carbon production (%)§ |
| YEP | Light | 0.059 ± 0.007 | 25.9 ± 1.4 | 0.044 ± 0.005 | 7.59 ± 0.84 | 1.92 ± 0.39 | 0.47 ± 0.06 | 24.5 ± 5.9 |
| Dark | 0.048 ± 0.011 | 19.5 ± 1.1 | 0.036 ± 0.004 | 4.43 ± 1.41 | 1.84 ± 0.28 | 0.10 ± 0.01 | 5.4 ± 1.0 | |
| Ala | Light | 0.074 ± 0.001 | 12.3 ± 0.6 | 0.035 ± 0.004 | 2.42 ± 0.46 | 0.45 ± 0.09 | 0.14 ± 0.02 | 31.1 ± 7.9 |
| Dark | 0.031 ± 0.004 | 2.9 ± 0.2 | 0.029 ± 0.003 | 0.69 ± 0.09 | 0.79 ± 0.12 | 0.11 ± 0.01 | 13.9 ± 2.0 |
Cultures were incubated in the light and in darkness in artificial seawater enriched with either a mixed (YEP) or a single (Ala) dissolved organic carbon source. Also shown is comparison of bicarbonate incorporation in relation to cell carbon production in the different treatments. Values represent mean ± SD for three biological replicates, except for bicarbonate incorporation where values represent mean ± SD for four subsamples of duplicate biological replicates.
Mean cell volume during growth at the time of bicarbonate incorporation measurements (Fig. 1).
Calculated from 33.75 to 45.25 h (YEP) and 45.75 to 62.75 h (Ala) (Fig. 1).
Per cell carbon production calculated from abundance at the time of bicarbonate incorporation measurements, cell production, and carbon per cell. Carbon per cell values were calculated based on measured cell volumes and the experimentally determined strain-specific carbon content for Dokdonia sp. MED134 cells growing in YEP; values for cells growing in the light or in the dark were 515 ± 103 (n = 3) or 778 ± 115 µgC⋅µm−3 (n = 3), respectively (Student t test, P = 0.015). The YEP light and dark carbon content values were applied also to the cells growing in the Ala light and dark cultures, respectively. Strain-specific carbon content errors have been used in error propagation calculations for each biological replicate to determine the SDs for carbon production. Resulting carbon per cell values ranged from 28.2 fgC⋅cell−1 in YEP to 18.2 fgC⋅cell−1 in Ala.
SDs represent the relative cumulative error obtained by error propagation calculations from carbon production and bicarbonate incorporation.
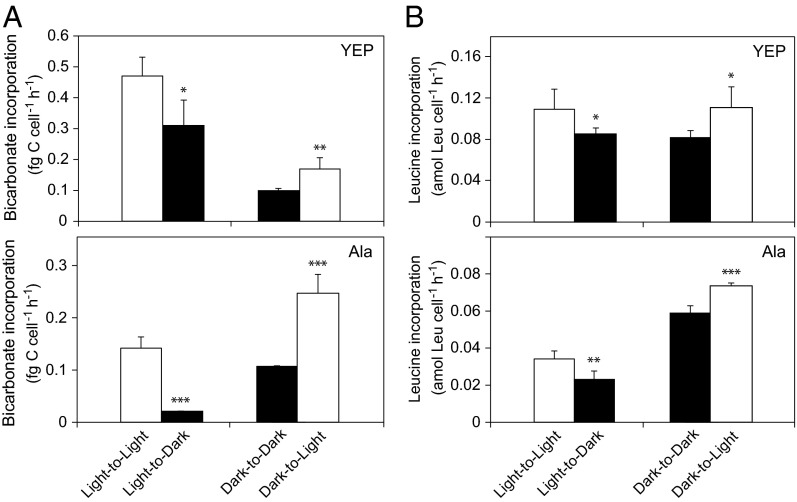
Bicarbonate (a measure of CO2 fixation) and leucine (a measure of protein synthesis/heterotrophic activity) incorporation rates were measured during growth (Fig. 2 and Table 1). In both media, bicarbonate incorporation rates were higher in cultures grown in the light (light-to-light) than in the dark (dark-to-dark): 4.8× and 1.3× higher in YEP and Ala (Fig. 2A), respectively. For leucine incorporation, rates in the light (light-to-light) were 1.4× higher than in the dark in YEP, whereas rates in Ala in the light were 1.8× lower than in the dark (dark-to-dark) (Fig. 2B).
Fig. 2.
Bicarbonate and leucine incorporation rates per cell of Dokdonia sp. MED134 at different light conditions. (A) Bicarbonate incorporation rates in artificial seawater with YEP and Ala. (B) Leucine incorporation rates in artificial seawater with YEP and Ala. Light-to-Light and Light-to-Dark denote subsamples of cultures grown in the light and incubated with radiolabeled bicarbonate or leucine in light and dark, respectively. Dark-to-Dark and Dark-to-Light denote subsamples of cultures grown in darkness and incubated with radiolabeled bicarbonate or leucine in dark and light, respectively. Subsamples for the bicarbonate and leucine incorporation assays were collected at 36 (YEP) and 51 h (Ala) as marked with asterisks in Fig. 1. Error bars denote SDs of four (bicarbonate) or six (leucine) subsamples of duplicate biological replicates. Asterisks denote significance levels (*P < 0.05, **P < 0.01, ***P < 0.001) for the Student t test.
Bicarbonate and leucine incorporation rates in both YEP and Ala media decreased after transfer of cultures from light to dark but increased on transfer from dark to light (Fig. 2). In YEP, bicarbonate and leucine incorporation decreased 1.5× and 1.3×, respectively, in light-to-dark samples, and rates increased in similar magnitude in the dark-to-light samples (Fig. 2A). In Ala, bicarbonate and leucine incorporation decreased 6.8× and 1.5× in light-to-dark samples but increased 2.3× and 1.3× in dark-to-light samples (Fig. 2B). Thus, on transfer between light conditions, stronger responses were found in Ala than in YEP, with the amplitude in response being largest for bicarbonate incorporation in Ala.
We calculated the amount of carbon produced from the increase in cell production, cell volumes, and experimentally determined strain-specific carbon per volume estimates for Dokdonia sp. MED134 cells grown in YEP (Table 1). The bicarbonate incorporation accounted for a substantial part of this produced carbon, and the proportion was significantly higher in the light than in the dark: 24 ± 6% in the light and 5 ± 1% in the dark for YEP (Student t test, P < 0.01) and 31 ± 8% and 14 ± 2% for Ala in the light and the dark (P < 0.05), respectively.
Gene Expression Patterns.
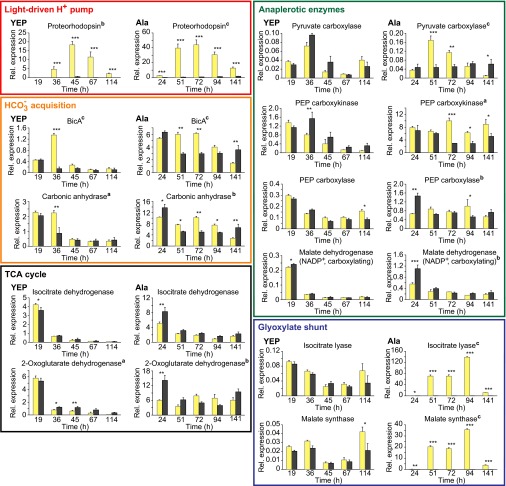
In YEP, the highest relative expression of all quantified genes was that of the PR gene in the light (Fig. 3; see Fig. 4 for summary of expression results). Although PR gene expression was very low early on, it reached a 30-fold higher peak in the light than in the dark at late exponential phase (45 h), and then decreased into stationary phase. Expression levels in the dark remained low throughout the experiment. In Ala, the PR gene expression pattern was similar, with up to 57-fold higher levels in the light during late exponential and early stationary phase. In both YEP and Ala, the difference in PR gene expression between cultures in the light or in darkness changed significantly over time (repeated-measures ANOVA: F > 12.22, df = 4, 16, P < 0.006).
Fig. 3.
Relative gene expression patterns in Dokdonia sp. MED134 of proteorhodopsin, HCO3−-acquisition, TCA cycle, anaplerotic enzyme, and glyoxylate shunt genes during growth in artificial seawater with YEP and Ala. Responses in the light (yellow columns) and in darkness (dark gray columns). qPCR was performed at the time points marked with arrows in Fig. 1. Relative gene-specific expression values were obtained by normalization to the expression levels of the housekeeping genes rpoD and recA. Error bars denote SE of triplicate biological replicates. Superscript letters after gene product names denote significance levels (aP < 0.05, bP < 0.01, cP < 0.001) of differences in gene expression in light or darkness changing over time, as determined by repeated-measures ANOVA. Asterisks denote significance levels (*P < 0.05, **P < 0.01, ***P < 0.001) of differences in gene expression between light and darkness at specific time points, as determined by Fisher’s LSD test. For statistical analysis of changes in expression levels over time within light treatment or in darkness, see Table S1.
Fig. 4.
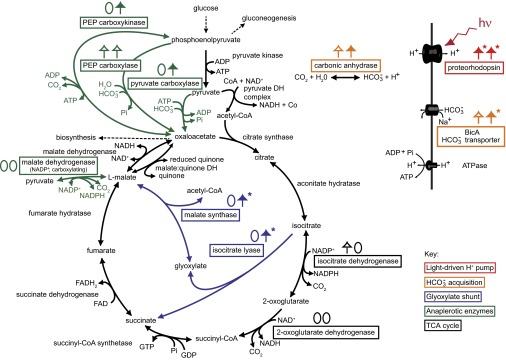
Central metabolic pathways and bicarbonate acquisition enzymes associated with proteorhodopsin phototrophy in Dokdonia sp. MED134. Colored squares denote enzymes for which relative gene expression analyses were carried out on the corresponding genes. For each analyzed gene, light responses in Fig. 3 are summarized as pairs of zeros and/or up-facing arrows in seawater with YEP (left in pair) and Ala (right in pair). Closed arrows denote genes with significantly higher relative expression levels in the light at two or more consecutive time points. Open arrows denote genes with significantly higher expression levels in the light at single or nonconsecutive times only. Asterisks denote gene expression responses reaching greater than fivefold higher levels in the light. Adapted and expanded from González et al. (26).
Gene expression for the bicarbonate membrane transporter BicA and the bicarbonate:carbon dioxide interconvertor carbonic anhydrase generally decreased during the experiment (Fig. 3 and Table S1). Differences in expression levels for these genes between light and dark cultures changed significantly over time in both YEP and Ala (F > 9.72, df = 4, 16, P < 0.015) but were more pronounced in Ala. The relative expression levels of these genes were 5- to 10-fold higher in Ala, where significantly higher expression levels were found in the light from 51 to 94 h (i.e., well into stationary phase; Fig. 3).
For the genes encoding the two TCA cycle enzymes tested, differences in isocitrate dehydrogenase gene expression levels between light and dark cultures did not change significantly over time (Fig. 3; F < 5.28, df = 4, 16, P > 0.05), whereas for the 2-oxoglutarate dehydrogenase (E1) gene there were significant changes in both YEP and Ala (F > 7.73, df = 4, 16, P < 0.023). Gene expression levels for isocitrate dehydrogenase showed marked decreases from initially high levels in both YEP and Ala [Fisher's least significant difference (LSD) test; Table S1; P < 0.01]. The expression pattern was similar for the 2-oxoglutarate dehydrogenase (E1) gene, with the exception of the Ala light cultures where its expression remained stable (Fig. 3).
For the four anaplerotic enzymes [i.e., pyruvate carboxylase, phosphoenolpyruvate (PEP) carboxykinase, PEP carboxylase, and malate dehydrogenase (NADP+, decarboxylating)] (Figs. 3 and 4), there were no consistent differences in expression between light and darkness in YEP over time (F < 3.58, df = 4, 16, P > 0.08). Although pyruvate carboxylase gene expression in YEP peaked in midexponential phase (36 h), the other three genes reached highest levels during early growth and decreased significantly over time (Table S1; P < 0.05). This decrease was especially pronounced for the malate dehydrogenase (NADP+, decarboxylating) gene.
In Ala, the anaplerotic enzyme genes were generally expressed several-fold higher than in YEP, with the exception of the pyruvate carboxylase gene (Fig. 3). In contrast to YEP cultures, differences in expression levels for the four anaplerotic enzymes in Ala light and dark cultures changed significantly over time (F > 6.8, df = 4, 16, P < 0.05). Interestingly, the pyruvate carboxylase gene was significantly up-regulated in the light from 51 to 72 h, whereas it was stably expressed in darkness (Fig. 3). The PEP carboxykinase gene was more highly expressed in the light from 72 h onward.
Gene expression levels for the glyoxylate shunt enzymes isocitrate lyase and malate synthase were very low throughout the experiment in YEP (Figs. 3 and 4). However, unexpectedly, the highest levels of relative gene expression in the present study were recorded for these glyoxylate shunt enzymes in Ala in the light (139 and 36, respectively), whereas values remained very low in the dark (well below 0.4); i.e., up to 300-fold higher levels in the light compared with darkness. Consequently, only in Ala the difference in expression of the glyoxylate shunt genes between cultures in the light or in darkness changed significantly over time (F > 80.67, df = 4, 16, P < 0.001).
Distribution of PR and Glyoxylate Shunt Genes in Seawater.
Integrated across 63 samples of the global ocean survey (GOS) metagenome sequence data, 7,087 PR gene copies were found, and the genes for isocitrate lyase (5,881 copies) and malate synthase (10,727 copies) were similarly abundant (Table S2). Normalization to recA (13,928 copies) indicated that these genes were present in around 54%, 43%, and 78% of marine bacteria, respectively.
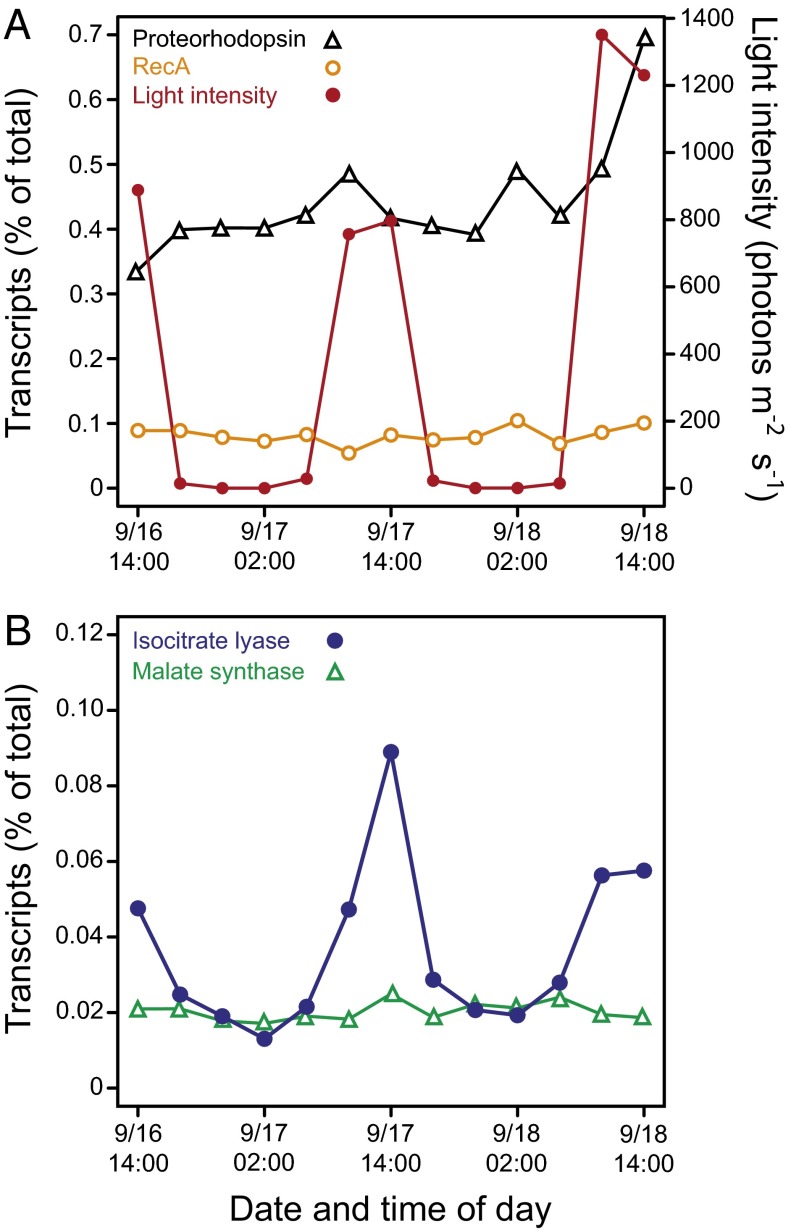
Having determined that glyoxylate shunt genes are widely distributed in marine bacteria, we next analyzed the as yet most exhaustive dataset on variability in gene expression over a 48-h period in Pacific surface water off Monterey Bay (30) to examine potential light-dependent patterns in gene expression (Fig. 5). The relative expression levels of the housekeeping gene recA varied very little, with no clear patterns in relation to light intensity, whereas the PR gene showed a small increase with light the second day and a larger increase on the third day (Fig. 5A). However, for the glyoxylate shunt gene encoding isocitrate lyase, the relative expression peak values coincided with the highest solar irradiance (Fig. 5B). Corresponding patterns were not observed for malate synthase. Diurnal patterns were pronounced also for the isocitrate lyase sequences specifically assigned to the ubiquitous SAR11 clade (Alphaproteobacteria), which was the most abundant taxon of bacteria with PR photoheterotrophic capacity in the dataset (Fig. S1). For the second most abundant bacterial taxon, the SAR86 clade (Gammaproteobacteria), changes in isocitrate lyase sequence abundance were barely visible (Fig. S1), possibly due to the relatively lower number of reads assigned to this taxon.
Fig. 5.
Bacterioplankton community dynamics in selected gene expression patterns in the Pacific off Monterey Bay. Dynamics in relative expression levels of (A) the genes encoding proteorhodopsin and RecA compared with light intensity and (B) genes encoding isocitrate lyase and malate synthase. Figure based on gene-specific analyses of metatranscriptome data obtained from natural seawater and reported by Ottesen et al. (30). Percent of total transcripts in metatranscriptome were obtained by BLASTX against RefSeq.
Discussion
The present study demonstrates that light profoundly affected gene expression patterns, metabolic activities including carbon acquisition, and growth in PR-containing Dokdonia sp. MED134, and that these responses were modulated by the availability of organic matter. Previous work had established that growth of MED134 was stimulated by light in seawater with submillimolar concentrations of DOC (11, 12). Moreover, the net benefit of PR phototrophy was shown to increase with decreasing DOC concentrations. Our experiments showed that, despite lower cell yields with alanine [Ala; consistent with reduced growth of bacteria on single compared with mixed carbon sources (31, 32)], the largest response to light occurred when growing on Ala rather than with YEP—even though the DOC concentration with Ala (0.70 µM C) was twice that used for YEP (0.30 µM C). Thus, the net benefit of PR phototrophy is not only dependent of carbon concentration but is also tightly dependent on DOC quality.
In addition to light stimulation of growth, we also found an increase in anaplerotic CO2 fixation rates in the light. This phenomenon had been noted in the closely related Polaribacter sp. MED152 (25). Together with analyses of the genomes of PR-containing marine Bacteroidetes, this lead to the hypothesis that anaplerotic reactions could be an important component in defining the role of PR phototrophy (25, 26, 33). Although the response observed here was qualitatively similar to that in Polaribacter sp. MED152, the per-cell bicarbonate incorporation rates in MED134 were two orders of magnitude higher than in MED152 (0.02–0.47 compared with <0.001 fgC⋅cell−1⋅h−1, respectively). This difference is likely due to the different culture media used in the two studies; i.e., artificial seawater with submillimolar concentrations of DOC for Dokdonia sp. MED134 and full strength Marine Broth medium diluted only 1:8 with artificial seawater for Polaribacter sp. MED152 (two orders of magnitude higher DOC concentrations). Furthermore, light also stimulated leucine incorporation rates, i.e., an established measure of protein synthesis. Interestingly however, the amplitude in CO2 fixation responses were larger than in leucine incorporation rates, especially in Ala. This difference in response amplitude possibly suggests that anaplerotic reactions are more immediately linked to the energy harvesting process through PR than protein synthesis—i.e., the latter likely represents a more general measure of bacterial activity that integrates the effects of both the heterotrophic metabolism and PR phototrophy. Thus, our findings provide experimental support for the suggestion that anaplerotic reactions provide PR-containing bacteria with an increased flexibility in carbon acquisition pathways to efficiently adjust their biosynthetic machinery to natural variations in light and dissolved organic matter in surface waters (25, 26, 33).
The contribution of anaplerotic CO2 fixation to the cellular carbon demand in marine bacteria remains little studied, particularly in photoheterotrophic bacteria. In model heterotrophic bacteria such as E. coli and Pseudomonas aeruginosa, the contribution of carbon from anaplerotic CO2 fixation to cell carbon varies between 1% and 8% (34). In contrast, a recent experimental study of mixed assemblages of bacteria from Arctic deep waters reported that bicarbonate incorporation, possibly driven by bacterial carboxylases, was equal to bacterial heterotrophic production rates (35). More constrained data on the potential for CO2 fixation in heterotrophic bacteria are provided for the bacteriochlorophyll a-containing Roseobacter denitrificans strain OCh 114. Carbon flux measurements in this strain, using MS analysis of isotopic labeling of amino acids, showed that it obtains 10–15% of its cellular carbon from anaplerotic fixation of CO2 (36), consistent with the presence of anaplerotic enzymes encoded in the genome (37). Light stimulation of CO2 fixation was not observed in this strain (36), although it is observed in several aerobic anoxygenic phototrophic bacteria (38). As a consequence of PR phototrophy, carbon from bicarbonate accounted for 24% and 31% of the carbon incorporated by the MED134 cells in the light in YEP and Ala, respectively, whereas percentages were significantly lower in the dark. This important contribution indicates that MED134 behaves as a facultative double mixotroph in the light: it uses both light and organic matter as energy sources and both bicarbonate and organic matter as carbon sources.
Based on the values in Table 1 and assuming growth efficiencies of 5–35% in our experiments, representative of offshore to coastal waters (39, 40), the CO2 fixation in MED134 could contribute between 0.2% and 5% to the total carbon demand of the cells in the dark and up to 18% in the light. These findings imply that light-induced anaplerotic CO2 fixation contributes substantial portions of cell carbon to PR-containing bacteria and can be more important for marine bacteria than heretofore recognized. Thus, anaplerotic CO2 fixation by this and other photoheterotrophic bacteria may represent an unrecognized entry point for carbon into planktonic biomass, with significant impacts on the biogeochemistry of the oceans.
The significant light-stimulated expression of bicarbonate acquisition genes in both media, and of pyruvate carboxylase in Ala, likely contributed to the observed responsiveness of CO2 fixation rates to light. In addition, the PEP carboxylase and malate dehydrogenase (NADP+, decarboxylating) genes were more highly expressed than the pyruvate carboxylase gene (especially in the early exponential phase), suggesting that these enzymes may also have important roles in anaplerotic CO2 fixation at certain growth stages. These anaplerotic enzymes act to replenish TCA intermediates when they are being used for biosynthesis (41). The TCA cycle is normally used for oxidation of organic matter to produce reducing power, which can subsequently be used for biosynthesis or be oxidized in respiration to generate the proton motive force across the cell membrane. Also, some TCA cycle intermediates are the starting points of central biosynthetic pathways, and during growth, the concentrations of such intermediates decrease and have to be filled in via anaplerotic reactions. In the case of PR-containing bacteria, the proton motive force can be generated by light and, therefore, the TCA cycle can be shifted toward biosynthesis, which in turn would require stimulation of the anaplerotic reactions.
This light-induced adjustment in metabolism appears to be dependent on resource availability. Indeed, the very low expression of the PR gene in early exponential phase coincided with the highest expression of the two classical TCA cycle genes (i.e., isocitrate dehydrogenase and 2-oxoglutarate dehydrogenase) at this time in nearly all treatments. As growth progressed, the expression of these TCA cycle genes decreased, concomitant with a strong increase in PR gene expression in the light cultures. These changes in expression suggest that MED134 cells in both mixed and single carbon media were geared to take advantage of PR photoheterotrophy primarily once the availability of substrates for genuine heterotrophic growth decreased.
Regarding the model carbon sources studied here, YEP provides a very varied combination of substrates, precursors, and vitamins. Likely, the most common situation in nature confronts the cells with a rather unbalanced combination of compounds. Alanine is an extreme case of this situation, because it can only enter the cell metabolism in MED134 by conversion to pyruvate at the step right before the TCA cycle. Growth in seawater with alanine requires the bacterium to synthesize all its precursor metabolites from just one type of molecule, and therefore, it requires the induction of metabolic pathways associated with gluconeogenesis, anaplerotic reactions, and the glyoxylate shunt (41). We infer that the increased dependence on anaplerotic reactions during growth in Ala compared with YEP contributed to explaining the higher proportion of cell carbon obtained through anaplerotic CO2 fixation in Ala compared with YEP dark cultures (14% and 5%, respectively). Consequently, the genes investigated here showed generally higher relative expression levels in Ala compared with YEP, which is in agreement with the more pronounced differences in physiological variables between light and dark found in Ala. Up-regulation of the PEP carboxykinase gene, which typically catalyzes the rate-controlling step in gluconeogenesis (42), suggests that, indeed, light stimulated the gluconeogenic rate in Ala.
Besides the strong light stimulation of PR gene expression (both in seawater with mixed organic carbon and with alanine), light had a strong influence on the expression of the glyoxylate shunt genes in seawater with alanine. The glyoxylate shunt is a long recognized alternative pathway in the TCA cycle that allows cells to use fatty acids or other gluconeogenic compounds, such as acetate and pyruvate, when glucose or mixed (glycolytic) organic matter sources are not available (43–45). It is noted that the glyoxylate shunt pathway in some organisms can be induced when cells have excess ATP (41), but light regulation of this pathway has to our knowledge not been observed previously. The glyoxylate shunt bypasses the TCA cycle steps where two CO2 molecules are released (Fig. 4). In so doing, the steps are also omitted where two NAD(P)H molecules are produced that typically are used for reducing power in biosynthesis or ATP production through oxidative phosphorylation. Thus, in microorganisms with an ATP production potential through PR phototrophy, the glyoxylate shunt, if induced, could have important implications for growth in organic carbon-limited but sunlit marine environments.
We infer that the strong up-regulation of the glyoxylate shunt in MED134 under light exposure in seawater with alanine, and thus the ability to save carbon molecules for biosynthesis, is possible thanks to the generation of ATP through PR phototrophy (Fig. S2). In this scenario, harnessing of light energy in MED134 reduces the dependence of cells on ATP from NADH driven oxidative phosphorylation. Indeed, energy from PR phototrophy reduces the dependence on carbon respiration, as seen from experiments both in marine PR-containing Candidatus Pelagibacter ubique and in a heterologous host PR expression system (17, 24). However, our expression analyses in MED134 showed that the standard TCA cycle genes were expressed in parallel with the glyoxylate shunt genes, consistent with the need of actively growing cells to use TCA cycle intermediates and reducing power for diverse cellular processes. Collectively, these findings suggest that the regulation of central metabolic pathways by light, in parallel with light stimulation of anaplerotic CO2 fixation, account for the strong impact of light on growth of MED134 in seawater.
The high abundance of glyoxylate shunt genes reported here for surface ocean bacterioplankton sampled by the global ocean survey (GOS)–present in roughly half of the bacteria–parallels the widespread distribution of the PR gene (6, 7). When we checked the expression of our genes of interest in natural seawater off Monterey Bay (30), however, only one of the two key enzymes in the glyoxylate shunt, i.e., isocitrate lyase, showed diel cycles (Fig. 5 and Fig. S1). This pattern was similar in MED134, where its expression was up to four times higher than malate synthase. The relatively stable expression of malate synthase in the surface waters may be explained by its involvement in multiple pathways, such as degradation of glycolate, a byproduct of photorespiration in photoautotrophs. It must be considered that things are more complex in nature than in culture and that a very significant fraction of the genes in these waters belonged to the SAR11 and SAR86 clades and not to Flavobacteriia such as MED134. There is as yet no experimental evidence for the ecophysiological importance of PR utilization in members of the SAR86 clade. Further, knowledge on the distribution of genes for the glyoxylate shunt and anaplerotic CO2 fixation among marine Gammaproteobacteria remains scarce, but such genes are found in genera like Vibrio and Marinobacter (25, 45). The use of PR in SAR11 is clearly different from that in Flavobacteriia (11, 24). In particular, although marine Flavobacteriia can use PR phototrophy to improve growth (11, 46), in SAR11 clade bacteria, thus far, PR light harvesting has instead been shown to promote survival during starvation (24). Direct comparative analysis of molecular mechanisms accounting for differences in how light is harnessed by marine bacteria with distinct life strategies remains a future undertaking. Nevertheless, the very well-marked cycle of isocitrate lyase in relation to light suggests that similar shifts between the TCA cycle and the glyoxylate shunt may be occurring in natural bacterioplankton populations dominated by SAR11.
Among the continuum of compounds from low-molecular-weight monomers to complex high-molecular weight polymers constituting the marine dissolved organic matter pool, a number of specific carbon compounds, including, for example, glucose and amino acids like alanine, have been shown to play essential roles in supplying organic carbon for heterotrophic marine bacteria (28, 29, 47). Moreover, such compounds exhibit substantial spatiotemporal variability in both concentrations and turnover times, thus exerting control on the growth of marine bacterial populations (27, 48). Our findings suggest that PR-containing bacteria have a wider than heretofore recognized repertoire of tightly integrated metabolic and physiological responses, including shifts in carbon acquisition pathways, to adapt to natural variations in not only the bulk concentration, but also the quality, of available DOC. Thus, dynamic changes in the dissolved organic matter may have profound impacts on how PR-containing bacteria realize their phototrophic potential.
Materials and Methods
Growth Conditions.
Dokdonia sp. MED134 colonies were grown on marine agar plates (Difco) for 2 d and then inoculated into 20 mL marine broth (Difco) in acid-washed 100-mL glass bottles and cultivated overnight on a shaker (140 rpm; Unimax 2010 orbital shaker, Heidolph) at room temperature. Next, 50 μL of overnight cultures was transferred to 1-L acid-washed polycarbonate bottles containing either marine broth (i.e., essentially a mix of yeast extract and peptone, YEP; Difco) or l-alanine (Ala) diluted in 150 mL artificial seawater (35 practical salinity units, prepared from Sea Salts; Sigma) so that the final concentration of dissolved organic carbon was 0.3 and 0.7 mM, respectively. Note that the higher carbon concentration in seawater with Ala was intentionally adjusted in an attempt to obtain similar levels of cell yields with the two carbon sources, because preliminary experiments had indicated lower yields with Ala as carbon source compared with YEP [as also observed by Kimura et al. (12)]. Yeast extract [0.0005% (wt/vol) final concentration; Difco], NH4Cl, and Na2HPO4 (0.112 and 0.021 mM final concentrations, respectively) were added to the Ala media to provide vitamins, complementary nitrogen, and phosphorus. YEP cultures received final concentrations of 0.048 mM N from NH4Cl and 0.09 mM P from Na2HPO4, respectively. After 2–3 d of cultivation in light conditions (180 μmol photons⋅m−2⋅s−1; Osram, LUMILUX Daylight lamps, L36W/865), 5 × 103 bacteria/mL were transferred to 1-L acid-washed polycarbonate bottles containing 500 mL artificial seawater with YEP or Ala and cultivated in light or dark conditions (triplicate cultures in each condition, i.e., three biological replicates). Cultures were maintained at 21 °C without shaking to reduce aggregation and flock formation. At different time points, bacterial abundance and morphology were determined by epifluorescence microscopy (Zeiss Axiotron) of SYBR Gold (Invitrogen)-stained cells, and sampling for RNA extraction was carried out. Also, subsamples were taken after 36 and 51 h of cultivation in YEP and Ala seawater media, respectively, corresponding to the exponential growth phase and for use in the bicarbonate and leucine incorporation rate measurements.
CO2 Fixation and Heterotrophic Activity.
Bicarbonate incorporation was measured as previously described (25), with some modifications. Briefly, five subsamples from two cultures of each incubation (light/dark) and media (YEP/Ala) condition were transferred to BD Falcon 50-mL tubes (BD Biosciences). From each original incubation condition, two subsamples were placed in transparent tubes, and two subsamples were placed in dark tubes (tubes covered by aluminum foil). The remaining subsample was killed with formaldehyde [3.7% (wt/vol) final concentration] and used as a control. All subsamples were incubated under white light (180 μmol photons⋅m−2⋅s−1) for 4 h with radiolabeled bicarbonate (H14CO3−; 3 μCi final concentration; DHI). That bicarbonate incorporation increased linearly over 4-h incubations was determined experimentally through hourly subsampling. After incubation, the subsamples were filtered through 0.22-μm pore size polycarbonate filters (25 mm diameter, Supor-200; Pall) that were subsequently placed in vials and exposed to 0.7 M HCl overnight. Four milliliters of scintillation mixture (Perkin-Elmer) was added to each filter, and the vials were kept in darkness for at least 24 h. Radioactivity was measured in the scintillation counter (Wallac 1414 Win Spectral). Finally, bicarbonate incorporation rate measurements were calculated based on the standard radioactive carbon assimilation technique (49).
Subsamples from the two seawater media (YEP/Ala) were used for leucine incorporation measurements (50) in the same treatment conditions as bicarbonate incorporation (i.e., light-to-light, light-to-dark, dark-to-light, and dark-to-dark). Triplicate samples of 1.2 mL bacterial culture were incubated with 40 nM [3H] leucine (Perkin-Elmer, specific activity of 170 Ci mmol−1). One hundred twenty microliters of 50% cold TCA was immediately added to duplicate controls. Triplicate samples were incubated for 2 h and fixed with 120 µL of 50% cold TCA. Subsequently, the subsamples and the controls were kept at −20 °C until centrifugation (at ∼12,000 × g) for 20 min, followed by aspiration of the water. Then, 1 mL of scintillation mixture was added to all of the tubes. After 24 h, radioactivity was measured in scintillation liquid counter, and leucine incorporation rates were calculated.
Particulate Organic Carbon.
Strain-specific per cell carbon content in Dokdonia sp. MED134 was determined for cells from midexponential phase cultures in artificial seawater with YEP (for culture growth conditions, see above). Aliquots of 700 mL from each of triplicate 1-L cultures growing in the light or in darkness were filtered onto precombusted 25-mm glass fiber filters (GF/F; Whatman). Epifluorescence microscopy, as described above, confirmed that filtration resulted in efficient harvesting of cells (i.e., no cells were observed in the filtrate). Filters with culture media only and blank precombusted filters were included as controls. Immediately after filtration, filters were dried at 60 °C overnight and then stored in a desiccator until analyzed for carbon content (Costech ECS 4010 Elemental Analyzer; Costech International).
RNA Extraction.
Suspensions from each replicate (2–8 mL) were transferred to individual tubes containing RNAprotect Bacterial Reagent (4–16 mL; Qiagen). After centrifugation (at 15,344 × g) for 10 min, the supernatant was removed and stored at −80 °C. Total RNA was extracted using the RNeasy Mini kit (Qiagen) according to the RNAprotect Bacterial Reagent protocol, and samples were treated with RNase-free DNase I (Qiagen) and TURBO-DNA free (Ambion) to eliminate residual genomic DNA. The NanoDrop 2000 (Thermo Scientific) was used to quantify and check the integrity of the total RNA before it was stored at −80 °C until use.
Quantitative PCR.
Quantitative PCRs (qPCRs) were performed with the StepOnePlus Real-time PCR System (Applied Biosystems) using 11 distinct primer-pairs specific for different genes potentially associated with PR phototrophy (Table S3). The primers were designed using Primer3 (www.bioinfo.ut.ee/primer3/) and then synthesized (Sigma-Aldrich). PCR products were sequenced by Eurofins MWG Operon (Germany). For qPCR, Power SYBR Green RNA-to-CT 1-Step kit (Applied Biosystems) was used according to the manufacturer’s protocol, and reactions were performed in a 10-μL volume containing 0.45 μM of each primer (as determined after optimization), 1 μL RNA, 0.08 μL RT Enzyme Mix (125×), and 5 μL Power SYBR Green RT-PCR Mix (2×). The thermal cycling conditions were as follows: 48 °C for 30 min and 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. The specificity of the primer pairs was checked using the melting curve analysis. All samples were assessed in technical triplicates, with negative control (distilled water) and RT-negative control samples included in each run.
To obtain adequate reference genes for normalization of gene expression, the housekeeping genes rpoB, rpoD, recA, and gyrB were evaluated; qPCR reactions were carried out as mentioned above. The expression stability of potential reference genes was determined with geNorm (51) and BestKeeper (52) software tools. This analysis resulted in the genes encoding RNA polymerase subunit D (rpoD) and recombinase A (recA) being selected as reference genes. The relative expression ratios for each target gene were computed based on the comparative Ct (2−ΔΔCt) method, which combines gene quantification and normalization into a single calculation (53). After normalization to rpoD and recA, average values and SEs of relative expression ratios for our investigated genes were calculated using data from triplicate biological samples.
Statistical Analyses.
Treatment-specific differences in bicarbonate and leucine incorporation rates, cell carbon content, and bicarbonate incorporation:carbon production percentages were analyzed using the Student two-tailed t test (STATISTICA v7.1; StatSoft). For gene expression patterns, repeated-measures ANOVA was first carried out to test whether differences in gene expression levels between light treatment and dark controls changed significantly over time. Thereafter, Fisher’s least significant difference (LSD) tests were done to identify statistically significant differences in gene expression levels: (i) between light and darkness at each individual time point sampled in each of the two media used and (ii) between time points within the light treatment or in darkness (GenStat v16.2; VSN International Ltd.). For the statistical analyses, differences of P < 0.05 were regarded as significant.
Quantification of Genes in Bacterioplankton Community DNA and RNA.
Selected genes were quantified in the metatranscriptome dataset of Ottesen et al. (30), which derives from natural seawater off the Monterey Bay (23 m depth). These authors deployed a robot that drifted in parallel with the Northern California coast in a Lagrangian way and collected samples for metatranscriptomics every 4 h over two day-night cycles. Functional assignment of the mRNA reads was quantified by BLASTX against RefSeq (release 60, July 2013). Processing of the initial sequences and functional annotation was as described in Ottesen et al. (30), except that the annotation of the RefSeq peptide database was confirmed by searching for the protein family corresponding to each gene in the analysis (PF00154, recombinase RecA; PF01036, Bac_rhodopsin; PF00463, isocitrate lyase; PF01274, malate synthase) to rely on a single and robust method of annotation. To this end, hidden Markov models run with HMMER3 (54) were used to determine its identity. A hit was considered valid if its score was equal to or bigger than the gathering score for the model. The label of the peptide in the RefSeq sequence file was modified to accommodate the annotation based on protein family hits.
The RefSeq database was also used to quantify hits to selected genes in the global ocean survey (GOS) metagenome study (6). Before analysis, rRNA sequences were removed by BLASTN against Silva database (55). A sequence was considered part of the rRNA gene if the bitscore was ≥50. The resulting non-rRNA sequences were translated using FragGeneScan (56). The GOS peptides corresponding to the studied genes were identified by BLASTP against the modified RefSeq when the bitscore was ≥50.
Supplementary Material
Acknowledgments
We thank Sabina Arnautovic and Jan Johansson for excellent technical assistance and Brandon Satinsky, Mary Ann Moran, and Saskia Burgers for insightful scientific discussions. The constructive comments and criticism from two reviewers considerably improved the paper. This work was supported by grants from the Göran Gustafsson Foundation for Research in Natural Sciences and Medicine, the Swedish Research Council, the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning, and the Research Programme EcoChange (to J. Pinhassi) and by MarineGems Grant CTM2010-20361 from the Spanish Ministry of Science and Innovation (to J.M.G. and C.P.-A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402617111/-/DCSupplemental.
References
- 1.Béjà O, et al. Bacterial rhodopsin: Evidence for a new type of phototrophy in the sea. Science. 2000;289(5486):1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 2.Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411(6839):786–789. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- 3.de la Torre JR, et al. Proteorhodopsin genes are distributed among divergent marine bacterial taxa. Proc Natl Acad Sci USA. 2003;100(22):12830–12835. doi: 10.1073/pnas.2133554100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabehi G, Béjà O, Suzuki MT, Preston CM, DeLong EF. Different SAR86 subgroups harbour divergent proteorhodopsins. Environ Microbiol. 2004;6(9):903–910. doi: 10.1111/j.1462-2920.2004.00676.x. [DOI] [PubMed] [Google Scholar]
- 5.Sabehi G, et al. Novel Proteorhodopsin variants from the Mediterranean and Red Seas. Environ Microbiol. 2003;5(10):842–849. doi: 10.1046/j.1462-2920.2003.00493.x. [DOI] [PubMed] [Google Scholar]
- 6.Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5(3):e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finkel OM, Béjà O, Belkin S. Global abundance of microbial rhodopsins. ISME J. 2013;7(2):448–451. doi: 10.1038/ismej.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frias-Lopez J, et al. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA. 2008;105(10):3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giovannoni SJ, et al. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature. 2005;438(7064):82–85. doi: 10.1038/nature04032. [DOI] [PubMed] [Google Scholar]
- 10.Riedel T, et al. Constitutive expression of the proteorhodopsin gene by a flavobacterium strain representative of the proteorhodopsin-producing microbial community in the North Sea. Appl Environ Microbiol. 2010;76(10):3187–3197. doi: 10.1128/AEM.02971-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gómez-Consarnau L, et al. Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature. 2007;445(7124):210–213. doi: 10.1038/nature05381. [DOI] [PubMed] [Google Scholar]
- 12.Kimura H, Young CR, Martinez A, Delong EF. Light-induced transcriptional responses associated with proteorhodopsin-enhanced growth in a marine flavobacterium. ISME J. 2011;5(10):1641–1651. doi: 10.1038/ismej.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poretsky RS, et al. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ Microbiol. 2009;11(6):1358–1375. doi: 10.1111/j.1462-2920.2008.01863.x. [DOI] [PubMed] [Google Scholar]
- 14.Lami R, Cottrell MT, Campbell BJ, Kirchman DL. Light-dependent growth and proteorhodopsin expression by Flavobacteria and SAR11 in experiments with Delaware coastal waters. Environ Microbiol. 2009;11(12):3201–3209. doi: 10.1111/j.1462-2920.2009.02028.x. [DOI] [PubMed] [Google Scholar]
- 15.Johnson ET, et al. Enhancement of survival and electricity production in an engineered bacterium by light-driven proton pumping. Appl Environ Microbiol. 2010;76(13):4123–4129. doi: 10.1128/AEM.02425-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Man D, et al. Diversification and spectral tuning in marine proteorhodopsins. EMBO J. 2003;22(8):1725–1731. doi: 10.1093/emboj/cdg183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez A, Bradley AS, Waldbauer JR, Summons RE, DeLong EF. Proteorhodopsin photosystem gene expression enables photophosphorylation in a heterologous host. Proc Natl Acad Sci USA. 2007;104(13):5590–5595. doi: 10.1073/pnas.0611470104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabehi G, et al. New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol. 2005;3(8):e273. doi: 10.1371/journal.pbio.0030273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walter JM, Greenfield D, Bustamante C, Liphardt J. Light-powering Escherichia coli with proteorhodopsin. Proc Natl Acad Sci USA. 2007;104(7):2408–2412. doi: 10.1073/pnas.0611035104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gómez-Consarnau L, et al. Proteorhodopsin phototrophy promotes survival of marine bacteria during starvation. PLoS Biol. 2010;8(4):e1000358. doi: 10.1371/journal.pbio.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akram N, et al. Regulation of proteorhodopsin gene expression by nutrient limitation in the marine bacterium Vibrio sp. AND4. Environ Microbiol. 2013;15(5):1400–1415. doi: 10.1111/1462-2920.12085. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, et al. Function and regulation of Vibrio campbellii proteorhodopsin: Acquired phototrophy in a classical organoheterotroph. PLoS ONE. 2012;7(6):e38749. doi: 10.1371/journal.pone.0038749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stingl U, Desiderio RA, Cho JC, Vergin KL, Giovannoni SJ. The SAR92 clade: An abundant coastal clade of culturable marine bacteria possessing proteorhodopsin. Appl Environ Microbiol. 2007;73(7):2290–2296. doi: 10.1128/AEM.02559-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steindler L, Schwalbach MS, Smith DP, Chan F, Giovannoni SJ. Energy starved Candidatus Pelagibacter ubique substitutes light-mediated ATP production for endogenous carbon respiration. PLoS ONE. 2011;6(5):e19725. doi: 10.1371/journal.pone.0019725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.González JM, et al. Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp. MED152 (Flavobacteria) Proc Natl Acad Sci USA. 2008;105(25):8724–8729. doi: 10.1073/pnas.0712027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.González JM, et al. Genomics of the proteorhodopsin-containing marine flavobacterium Dokdonia sp. strain MED134. Appl Environ Microbiol. 2011;77(24):8676–8686. doi: 10.1128/AEM.06152-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirchman DL. The contribution of monomers and other low-molecular weight compounds to the flux of dissolved organic matter in aquatic ecosystems. In: Findlay SEG, Sinsabaugh RL, editors. Aquatic Ecosystems: Interactivity of Dissolved Organic Matter. New York: Elsevier Science; 2003. pp. 217–241. [Google Scholar]
- 28.Suttle CA, Chan AM, Fuhrman JA. Dissolved free amino acids in the Sargasso Sea: Uptake and respiration rates, turnover times, and concentrations. Mar Ecol Prog Ser. 1991;70:189–199. [Google Scholar]
- 29.Fuhrman J. Close coupling between release and uptake of dissolved free amino acids in seawater studied by an isotope dilution approach. Mar Ecol Prog Ser. 1987;37:42–52. [Google Scholar]
- 30.Ottesen EA, et al. Pattern and synchrony of gene expression among sympatric marine microbial populations. Proc Natl Acad Sci USA. 2013;110(6):E488–E497. doi: 10.1073/pnas.1222099110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Payne WJ, Williams ML. Carbon assimilation from simple and complex media by prototrophic heterotrophic bacteria. Biotechnol Bioeng. 1976;18(11):1653–1655. doi: 10.1002/bit.260181117. [DOI] [PubMed] [Google Scholar]
- 32.Ingraham JL, Maaloe O, Neidhardt FC. Growth of the Bacterial Cell. Sunderland, MA: Sinauer; 1983. [Google Scholar]
- 33.Fernández-Gómez B, et al. Ecology of marine Bacteroidetes: A comparative genomics approach. ISME J. 2013;7(5):1026–1037. doi: 10.1038/ismej.2012.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roslev P, Larsen MB, Jørgensen D, Hesselsoe M. Use of heterotrophic CO2 assimilation as a measure of metabolic activity in planktonic and sessile bacteria. J Microbiol Methods. 2004;59(3):381–393. doi: 10.1016/j.mimet.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 35.Alonso-Sáez L, Galand PE, Casamayor EO, Pedrós-Alió C, Bertilsson S. High bicarbonate assimilation in the dark by Arctic bacteria. ISME J. 2010;4(12):1581–1590. doi: 10.1038/ismej.2010.69. [DOI] [PubMed] [Google Scholar]
- 36.Tang KH, Feng X, Tang YJJ, Blankenship RE. Carbohydrate metabolism and carbon fixation in Roseobacter denitrificans OCh114. PLoS ONE. 2009;4(10):e7233. doi: 10.1371/journal.pone.0007233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swingley WD, et al. The complete genome sequence of Roseobacter denitrificans reveals a mixotrophic rather than photosynthetic metabolism. J Bacteriol. 2007;189(3):683–690. doi: 10.1128/JB.01390-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yurkov VV, Beatty JT. Aerobic anoxygenic phototrophic bacteria. Microbiol Mol Biol Rev. 1998;62(3):695–724. doi: 10.1128/mmbr.62.3.695-724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.del Giorgio PA, Cole JJ. Bacterial growth efficiency in natural aquatic ecosystems. Annu Rev Ecol Syst. 1998;29:503–541. [Google Scholar]
- 40.del Giorgio PA, et al. Coherent patterns in bacterial growth, growth efficiency, and leucine metabolism along a northeastern Pacific inshore–offshore transect. Limnol Oceanogr. 2011;56(1):1–16. [Google Scholar]
- 41.Stryer L. Biochemistry. New York: W. H. Freeman and Company; 1988. [Google Scholar]
- 42.Rognstad R. Rate-limiting steps in metabolic pathways. J Biol Chem. 1979;254(6):1875–1878. [PubMed] [Google Scholar]
- 43.Guest JR, Russell GC. Complexes and complexities of the citric acid cycle in Escherichia coli. Curr Top Cell Regul. 1992;33:231–247. doi: 10.1016/b978-0-12-152833-1.50018-6. [DOI] [PubMed] [Google Scholar]
- 44.Guest JR, et al. Physiological effects of replacing the PDH complex of E. coli by genetically engineered variants or by pyruvate oxidase. In: Gordon F, Patel MS, editors. Thiamine: Catalytic Mechanisms and Role in Normal and Disease States. New York: Marcel Dekker; 2004. [Google Scholar]
- 45.Berdalet E, et al. CO2 production, O2 consumption and isocitrate dehydrogenase in the marine bacterium Vibrio natriegens. Aquat Microb Ecol. 1995;9(3):211–217. [Google Scholar]
- 46.Feng S, Powell SM, Wilson R, Bowman JP. Light-stimulated growth of proteorhodopsin-bearing sea-ice psychrophile Psychroflexus torquis is salinity dependent. ISME J. 2013;7(11):2206–2213. doi: 10.1038/ismej.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rich JH, Ducklow HW, Kirchman DL. Concentrations and uptake of neutral monosaccharides along 140°W in the equatorial Pacific: Contribution of glucose to heterotrophic bacterial activity and the DOM flux. Limnol Oceanogr. 1996;41(4):595–604. [Google Scholar]
- 48.Gómez-Consarnau L, Lindh MV, Gasol JM, Pinhassi J. Structuring of bacterioplankton communities by specific dissolved organic carbon compounds. Environ Microbiol. 2012;14(9):2361–2378. doi: 10.1111/j.1462-2920.2012.02804.x. [DOI] [PubMed] [Google Scholar]
- 49.Parsons TR, Takahashi M, Hargrave B. Biological Oceanographie Processes. 3rd Ed. New York: Pergamon Press; 1984. [Google Scholar]
- 50.Smith DC, Azam F. A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar Microb Food Webs. 1992;6(2):107–114. [Google Scholar]
- 51.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;18(3):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26(6):509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 53.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eddy SR. A probabilistic model of local sequence alignment that simplifies statistical significance estimation. PLOS Comput Biol. 2008;4(5):e1000069. doi: 10.1371/journal.pcbi.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue):D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rho M, Tang H, Ye Y. FragGeneScan: Predicting genes in short and error-prone reads. Nucleic Acids Res. 2010;38(20):e191. doi: 10.1093/nar/gkq747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.