Summary
The Mcm2-7 replicative helicase is central to all steps of eukaryotic DNA replication. The hexameric ring of Mcm subunits forms six essential ATPases whose contributions to replication initiation remain unclear. Mcm2-7 complexes containing ATPase-motif mutations showed Mcm2-7 ATP binding and hydrolysis are required for helicase loading. Loading-defective Mcm2-7 mutant complexes were defective in initial Mcm2-7 recruitment or Cdt1 release. Comparison with Cdc6 ATPase mutants showed that Cdc6 ATP hydrolysis is not required for helicase loading but instead drives removal of Mcm2-7 complexes that cannot complete loading. A subset of Mcm2-7 ATPase-site mutants completed helicase loading but could not initiate replication. Individual mutants were defective in distinct events during helicase activation including maintenance of DNA association, recruitment of the GINS helicase activator and DNA unwinding. Consistent with its heterohexameric structure, our findings show that the six Mcm2-7 ATPase active sites are specialized for different functions during helicase loading and activation.
Introduction
Eukaryotic DNA replication initiation is a multi-step process that is tightly regulated to ensure the complete and timely replication of the genome. Eukaryotic helicase loading, also known as pre-replicative complex formation or origin licensing, is the first event of eukaryotic DNA replication initiation and is restricted to the G1 phase of the cell cycle. In contrast, helicase activation, replisome assembly and DNA synthesis require S phase entry. The temporal separation of these events ensures that no origin of replication initiates more than once per cell cycle (reviewed in Costa et al., 2013). The only protein that participates in all of these events is the eukaryotic replicative DNA helicase, the Mcm2-7 complex.
The Mcm2-7 complex is a ring-shaped heterohexamer of related AAA+ ATPases arranged in a defined order (Fig. S1, reviewed in Bochman and Schwacha, 2009). The interface between each pair of Mcm subunits forms an ATP binding and hydrolysis domain. One subunit provides the primary ATP-binding domain including the Walker-A and Walker-B motifs. The adjacent subunit provides an ATPase-activating arginine (the arginine finger). In isolation, Mcm2-7 complexes with mutations in one Walker-A or arginine-finger motif show reduced Mcm2-7 ATP hydrolysis, suggesting all six ATPase motifs are functional (Bochman et al., 2008; Schwacha and Bell, 2001).
Loading the Mcm2-7 helicase at origins of replication requires three additional proteins: the origin recognition complex (ORC), Cdc6 and Cdt1 (reviewed in Yardimci and Walter, 2014). ORC binds origin DNA and localizes helicase loading to these sites (Aparicio et al., 1997; Bell and Stillman, 1992; Donovan et al., 1997). Cdc6 associates with origin-bound ORC during entry into G1 phase (Weinreich et al., 1999). In contrast, Cdt1 is associated with Mcm2-7 before helicase loading (Remus et al., 2009; Tanaka and Diffley, 2002). During G1, these proteins load two Mcm2-7 complexes in the form of an inactive, head-to-head double hexamer that encircles double-stranded, origin DNA (Evrin et al., 2009; Remus et al., 2009). DNA access to the central channel is thought to be mediated by a gap in the Mcm2-7 ring between Mcm2 and Mcm5 (the Mcm2/5 gate, Bochman and Schwacha, 2008; Costa et al., 2011).
ATP binding and hydrolysis regulate eukaryotic helicase loading. In addition to Mcm2-7, Orc1 and Cdc6 are active ATPases (Klemm et al., 1997; Perkins and Diffley, 1998; Randell et al., 2006; Schepers and Diffley, 2001). ATP binding by ORC and Cdc6 is important for their initial origin DNA association (Bell and Stillman, 1992; Perkins and Diffley, 1998; Speck et al., 2005; Weinreich et al., 1999). Mcm2-7/Cdt1 binding to ORC/Cdc6 forms the ORC-Cdc6-Cdt1-Mcm2-7 or OCCM complex. Cdc6 ATP hydrolysis is implicated in the transition between the OCCM complex and loaded Mcm2-7 (Fernández-Cid et al., 2013; Randell et al., 2006). In contrast, ORC ATP hydrolysis is dispensable for double-hexamer formation but required for repeated Mcm2-7 loading events (Bowers et al., 2004; Evrin et al., 2013). ATP hydrolysis by ORC and/or Cdc6 also facilitates release of Mcm2-7 complexes that fail to load (Frigola et al., 2013). Importantly, mutations that interfere with ATP binding and hydrolysis by ORC and Cdc6 are lethal or temperature-sensitive and interfere with helicase loading in vivo (Bowers et al., 2004; Klemm et al., 1997; Perkins and Diffley, 1998; Schepers and Diffley, 2001).
Helicase activation and replisome assembly require additional proteins (reviewed in Tanaka and Araki, 2013). Two helicase activators, Cdc45 and GINS, bind and activate Mcm2-7, forming the Cdc45/Mcm2-7/GINS (CMG) complex (Ilves et al., 2010; Moyer et al., 2006). Cdc45 recruitment requires phosphorylation of Mcm2-7 by the Dbf4-dependent Cdc7 kinase (DDK) and two loading proteins, Sld3 and Sld7 (Heller et al., 2011; Tanaka et al., 2011). S-phase cyclin-dependent kinases (S-CDKs) phosphorylate Sld3 and Sld2, stimulating their binding to the TopBP1 analog, Dpb11 (Tanaka et al., 2007; Zegerman and Diffley, 2007). These interactions, as well as interactions between Sld2, Dpb11, GINS and DNA Pol ε(Muramatsu et al., 2010; Sengupta et al., 2013; Tanaka et al., 2013) drive CMG complex formation. Finally, Mcm10 has been implicated in both CMG activation and recruitment of the remaining DNA polymerases, DNA Pol α/primase and DNA Pol δ (reviewed in Thu and Bielinsky, 2013).
The role of ATP binding and hydrolysis by Mcm2-7 during DNA replication remains unclear. Mutation of any of the six Mcm2-7 ATPase active sites is lethal (Bochman and Schwacha, 2009). Yet several of the lethal ATPase mutations retain helicase activity in vitro (Bochman and Schwacha, 2008; 2010), suggesting roles for Mcm2-7 ATP binding and hydrolysis beyond DNA unwinding. Similar mutations in the Drosophila CMG complex also retain helicase activity (Ilves et al., 2010). Previous studies of several Mcm2-7 complexes with Walker-A or arginine-finger mutations found no helicase loading defects, suggesting these motifs are dedicated to DNA unwinding (Fernández-Cid et al., 2013; Ying and Gautier, 2005).
Here we analyze the effects of individual Mcm2-7 Walker-A and arginine-finger mutations on replication initiation in vitro. These mutations do not alter the assembly of Mcm2-7/Cdt1 complexes. In contrast, all of the mutant complexes show defects in helicase loading with five being fully defective. Analysis of the ATP-hydrolysis-dependent release of Cdt1 showed that a combination of Cdc6 and Mcm2-7 ATP activities stimulates this event. In addition, we found Cdc6 ATP hydrolysis is required for the release of unsuccessfully loaded Mcm2-7 complexes but dispensable for efficient Mcm2-7 loading. Analysis of the loading-competent mutant Mcm2-7 complexes found that only one mutant, which is viable in vivo, initiated replication in vitro. The remaining mutant Mcm2-7 complexes showed defects in Mcm2-7 origin-DNA retention, CMG complex formation or DNA unwinding. Our findings indicate that Mcm2-7 complexes are active participants in helicase loading and activation and that the different Mcm2-7 ATPase sites have distinct roles during these events.
Results
To assess the role of Mcm2-7 ATP binding and hydrolysis during replication initiation, we purified twelve mutant Mcm2-7/Cdt1 complexes. Each contained one subunit with a mutation in its Walker A (K→A) or arginine-finger (R→A) motif (Fig. S1 and Table S1 and S2). We refer to the mutant Mcm2-7 complexes by ‘MCM’ followed by the subunit mutated and the type of mutation (e.g. MCM2KA has a Walker-A mutation in Mcm2). Only one of the twelve mutations tested (mcm4-R701A) complements a deletion of the corresponding wild-type gene (Fig. S1A, Bochman et al., 2008). All of the mutant Mcm2-7/Cdt1 complexes eluted in the same position as wild-type during gel filtration (data not shown) and showed equivalent subunit stoichiometry (Fig. S1B). We conclude that assembly of the mutant complexes is not grossly defective.
Mcm2-7 ATP Binding and Hydrolysis is Required for Helicase Loading
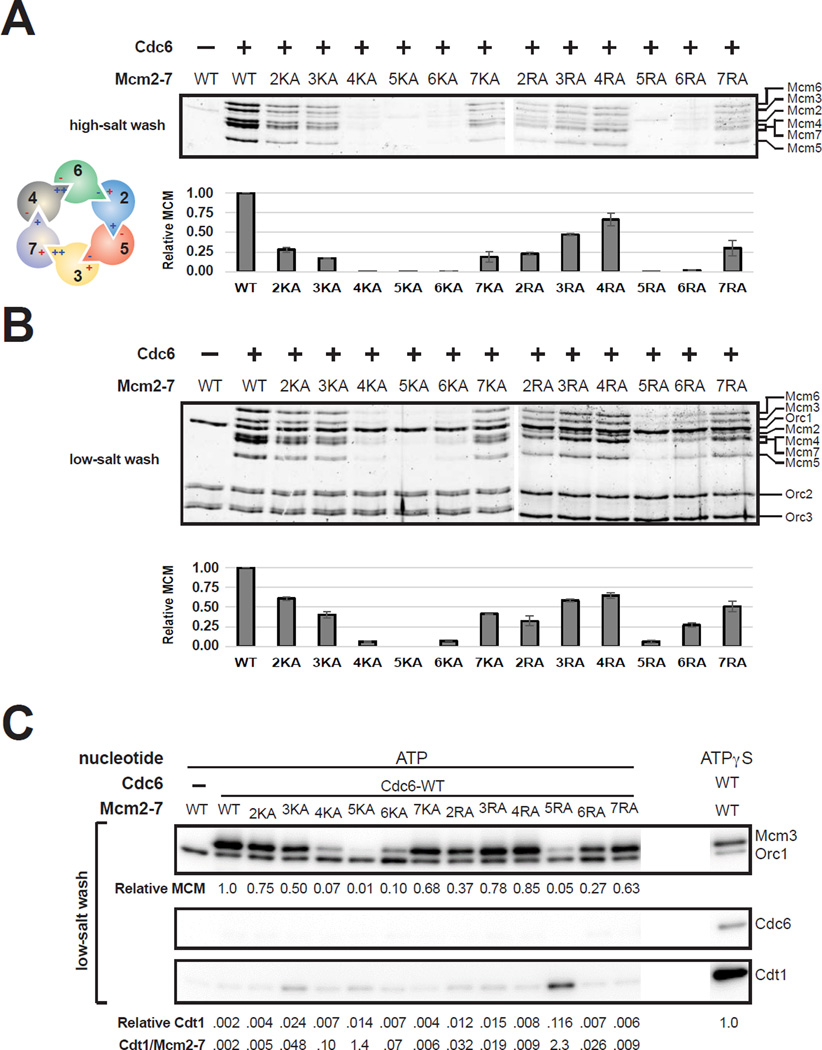
We tested the ability of each mutant Mcm2-7 complex to be loaded onto origin DNA using a reconstituted assay (Remus et al., 2009). Purified ORC, Cdc6 and the indicated Mcm2-7/Cdt1 complex were added to bead-attached, origin-containing DNA. To identify loaded Mcm2-7, DNA-associated proteins were washed under high-salt conditions. Only Mcm2-7 complexes that have completed loading are retained after a high-salt wash (Donovan et al., 1997; Randell et al., 2006).
All twelve Mcm2-7 ATPase motif mutations showed helicase-loading defects (Fig. 1A and S2C) but the extent of defect varied depending on the site and subunit mutated. The strongest defects were observed for mutations in Mcm5 and Mcm6. Both Walker-A and arginine-finger mutations in these subunits eliminated helicase loading. In contrast, mutations in Mcm2, 3 and 7 showed intermediate phenotypes for both types of mutations (2- to 5-fold reductions, Fig. 1A). Only Mcm4 showed strongly different phenotypes for the Walker-A (fully defective) and arginine-finger (2-fold defect) mutants. If we asked which ATPase motifs are involved in helicase loading, we found that only mutations in the Mcm3/7 ATPase interface do not show severe loading defects. For the remaining ATPase sites, one of the two mutations eliminated helicase loading. Interestingly, the type of mutation (K→A or R→A) that eliminates loading varies between the different ATPase interfaces (Fig. 1A).
Figure 1.
Mcm2-7 ATPase motif mutants are defective in helicase loading. (A) Helicase loading of MCM ATPase motif mutants was monitored by incubating loading proteins with ATP and origin-DNA-beads followed by a high-salt wash. The associated graph shows the relative loading of the MCM mutants compared to wild-type Mcm2-7 based on two independent loading experiments. Proteins were detected by fluorescent protein staining. The WT/K→A and R→A samples were analyzed on separate gels. Error bars indicate the standard deviation. Helicase loading of Walker-A (red) and arginine-finger (blue) mutant Mcm2-7 complexes are summarized in the Mcm2-7 illustration (−: less than 3%, +: 3 ~ 33%, ++: more than 33% of wild-type loading, respectively). (B) Origin DNA association of Mcm2-7 ATPase motif mutants and helicase loading proteins. Helicase-loading assays were performed followed by a low-salt wash. DNA-associated proteins were detected by fluorescent protein staining. The WT/K→A and R→A samples were analyzed on separate gels. The associated graph shows the relative amounts of DNA bound Mcm2-7 complexes based on two independent experiments as described for (A). (C) Release of Cdc6 and Cdt1 by Mcm2-7 ATPase motif mutants during helicase loading. Helicase-loading assays were performed followed by a low-salt wash. Bound Mcm2-7 (Mcm3) and helicase loading proteins were monitored by immunoblot (anti-Flag Ab for Mcm3, Orc1, and Cdc6; Strep-Tactin-HRP for Cdt1-StrepII). An ATPγS reaction, which arrests loading after initial association of helicase and helicase loading proteins, was included as a control (two adjacent irrelevant lanes were eliminated). The relative amount of Mcm3 compared to a wild-type Mcm2-7 reaction is indicated below the top panel. The relative amount of Cdt1 was determined by comparing to a wild-type Mcm2-7 ATPγS reaction (last lane) and is reported below the third panel. The ratio of retained Cdt1 and Mcm3 is also reported below the third panel. See also Figure S1 and Tables S1 and S2.
To investigate what step in helicase loading is defective for the Mcm2-7 ATPase motif mutations, we performed the helicase-loading assay followed by a low-salt wash. Under these conditions, stable association of ORC, Cdc6 and Cdt1 can be observed. The pattern of mutant Mcm2-7 DNA association (after low-salt wash) was similar to the pattern of Mcm2-7 loading (after high-salt wash) with one exception (compare Fig. 1A and 1B). MCM6RA showed much higher levels of DNA association than loading (Fig. S2C; 4- vs. 50-fold defect, respectively), suggesting this mutant arrests helicase loading at a stage when Mcm2-7 is associated but not loaded. To increase sensitivity, we used western blotting to monitor Cdc6 and Cdt1 association after the low-salt wash. Under these conditions, wild-type Mcm2-7 showed full release of Cdc6 and Cdt1 (Fig. 1C). None of the mutations in the Mcm2-7 ATPase sites led to Cdc6 retention. In contrast, four of the five loading defective mutants (MCM3KA, MCM4KA, MCM5KA and MCM5RA) showed the most Cdt1 retention as determined by comparing the Cdt1/Mcm2-7 ratio for the different Mcm2-7 complexes (this ratio compensates for the different loading defects of the mutant complexes, see bottom of Fig. 1C). These findings suggest that defective Cdt1 release contributes to or is a symptom of Mcm2-7 loading defects.
To ask if the Mcm2-7 mutations altered the initial recruitment of Mcm2-7/Cdt1, we performed helicase-loading assays in the presence of ATPγS (Fig. 2). In this condition, helicase loading is arrested with incompletely-loaded Mcm2-7 and all three helicase-loading proteins associated with origin DNA (Randell et al., 2006). The majority of the mutant Mcm2-7/Cdt1 complexes were recruited to the origin DNA at levels similar to wild-type (Fig. S2C). In contrast, MCM6RA showed a 4-fold defect in Mcm2-7 recruitment and a 10-fold defect in Cdt1 recruitment. Interestingly, MCM2KA shows the second strongest Cdt1-recruitment defect (as measured by the Cdt1/Mcm2-7 ratio, see bottom of Fig. 2) and alters the same ATPase active site (Mcm6/2) as MCM6RA. These data indicate that mutations in the Mcm6/2 ATPase interfere with the initial recruitment of Mcm2-7/Cdt1 to origin DNA.
Figure 2.
Initial association of ATPase motif mutant Mcm2-7/Cdt1 complexes. Helicaseloading assays were performed in the presence of ATPγS followed by a low-salt wash allowing detection of the initial DNA association of Mcm2-7 and helicase loading proteins (ORC, Cdc6 and Cdt1). DNA bound proteins were detected after SDS-PAGE by fluorescent protein staining (top panel) or immunoblot (middle and bottom panel) as described in Figure 1. The relative association of Mcm2-7 and Cdt1 was measured based on two independent ATPγS experiments and are shown as graphs. Error bars indicate the standard deviation. The ratio of retained Cdt1 and Mcm2-7 is reported below the Cdt1 graph. See also Figure S2.
Two experiments addressed the possibility that helicase loading resulted in the disassembly of mutant Mcm2-7 rings. First, we immunoprecipitated Mcm2-7 from the supernatants of helicase loading reactions using anti-Flag agarose (only the Mcm3 subunit of Mcm2-7 is Flag-tagged) and examined their subunit composition (Fig. S2). The immunoprecipitated Mcm2-7 subunit pattern was similar between wild-type and mutant complexes, although MCM6KA showed moderately lower Mcm2 and Mcm6 levels compared to other subunits. Second, the ATPγS experiment described above showed consistent Mcm2-7 stoichiometry. If the Mcm2-7 ring was disrupted at this stage, it is likely that subunits between the open Mcm2/5 gate and the disrupted interface would be released, since the primary association with the helicase loading proteins at this stage is mediated by one Mcm subunit (Mcm3, Fernández-Cid et al., 2013; Frigola et al., 2013). Thus, the consistent subunit stoichiometry in the ATPγS experiment also suggests that the Mcm2-7 interfaces (with the exception of the Mcm2/5 gate) are intact.
Cdc6 ATP Hydrolysis Is Not Required for Helicase Loading
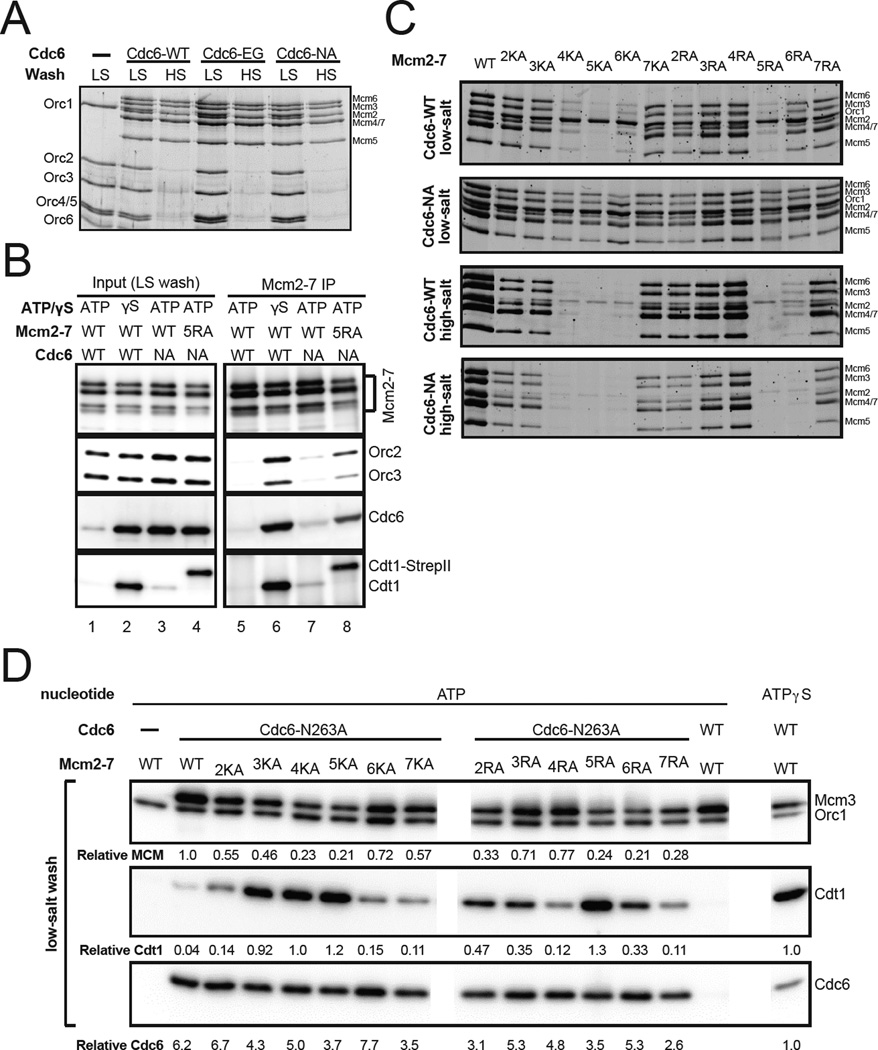
Previous in vivo and in vitro studies found that Cdc6 ATPase mutations were defective for Cdt1 release and helicase loading (Fernández-Cid et al., 2013; Perkins and Diffley, 1998; Randell et al., 2006; Schepers and Diffley, 2001). To compare the contribution of Cdc6 and Mcm2-7 ATPase motifs to helicase loading, we addressed the function of two Cdc6 ATPase-defective mutants (Cdc6-E224G and Cdc6-N263A, Schepers and Diffley, 2001; Speck and Stillman, 2007) in the reconstituted helicase-loading assay (Fig. 3). In contrast to previous findings (Fernandez-Cid et al., 2013; Randell et al., 2006), both Cdc6 mutants supported the assembly of high-salt-resistant Mcm2-7 loading at levels similar to wild-type Cdc6 (Fig. 3A). Importantly, the Mcm2-7 complexes loaded by mutant Cdc6 directed replication initiation and elongation (Fig. S3A). Unlike wild-type Cdc6, however, low-salt wash experiments showed that Cdc6 ATPase mutations resulted in sustained DNA association of Cdc6 and, to a lesser extent, Cdt1 (Fig. 3B, compare lanes 1 and 3).
Figure 3.
Cdc6 ATP hydrolysis is required for release of unsuccesfully loaded helicases. (A) Helicase loading of Cdc6 ATPase mutants. A helicase loading reaction was performed with two Cdc6 ATPase mutants, Cdc6-E224G (Cdc6-EG) and Cdc6-N263A (Cdc6-NA). After low- (LS) or high-salt wash (HS), DNA bound proteins were detected by fluorescent protein staining. (B) Combining a Cdc6 ATPase and Mcm2-7 ATPase-motif mutant halts the loading reaction at the OCCM stage. After loading and low-salt wash with wild-type or the indicated mutant proteins, DNA-associated proteins were released by DNase I and either detected by immunoblotting (input, lanes 1–4) or subjected to immunoprecipitation using anti-Mcm2-7 antibodies coupled to gammabind G sepharose (GE Healthcare). After immunoprecipitation, Mcm2-7 and any associated helicase loading proteins were detected by immunoblotting (Mcm2-7 IP, lanes 5–8). An ATPγS reaction was included as a control. (C) Loading defective Mcm2-7 ATPase motif mutant complexes are not released from DNA in the presence of a Cdc6 ATPase mutant. Helicase loading reactions were performed with wild-type or Cdc6-NA and either wild-type or the indicated Mcm2-7 ATPase motif mutant complex. After loading, complexes were washed with either high- or low-salt buffer as indicated. Loaded (high-salt wash) or associated (low-salt wash) Mcm2-7 was detected by fluorescent proteins staining. (D) Cdc6 and Mcm2-7 ATPases contribute to Cdt1 release. Helicase loading reactions were performed with the indicated proteins followed by a low-salt wash. DNA-associated Mcm2-7 and helicase-loading proteins were monitored by immunoblot as described in Fig. 1C. An ATPγS reaction, which represents initial association of helicase and helicase loading proteins, was included as a control (an irrelevant adjacent lane was eliminated). The relative amount of Mcm3 compared to a wild-type ATP reaction, and the relative amount of Cdt1 and Cdc6 compared to a wild-type ATPγS reaction are shown below each panel. See also Figure S3.
To investigate the differences between Cdc6 function in the reconstituted assay and previous studies, we tested the function of the same Cdc6 ATPase mutants in a G1 extract-based assay. As seen previously (Randell et al., 2006), in the presence of G1 extract, the mutant Cdc6 proteins showed retention of Cdc6 and Cdt1 and a clear defect in helicase loading (Fig. S3B). To ask if this difference was due to the lower efficiency of the extract-based assay, we tested the Cdc6 ATPase mutants in reconstituted reactions under limiting Cdc6 levels (Fig. S3C). Strong Mcm2-7 loading defects were observed in the mutant but not the wild-type reactions at reduced Cdc6 concentrations. Interestingly, these defects were more pronounced for high- vs low-salt washed reactions (Fig. S3D), suggesting that lower-efficiency reactions accumulate incompletely-loaded Mcm2-7 complexes that are removed by the high-salt wash or Cdc6 ATP hydrolysis (see below). These findings indicate that Cdc6 ATPase activity is not intrinsically required for helicase loading but becomes important when helicase loading is inefficient.
Release of Unsuccessfully-loaded Mcm2-7 Requires Cdc6 ATP Hydrolysis
We performed assays with both Mcm2-7 and Cdc6 ATPase motif mutants to investigate their combined effects. In contrast to wild-type Cdc6, in the presence of Cdc6-N263A we observed similar levels of DNA association for wild-type and mutant Mcm2-7 complexes after a low-salt wash (Fig. 3C, compare top two panels). This difference is most notable for the loading-defective Mcm2-7 mutants (e.g. MCM5RA). Cdc6-N263A does not rescue Mcm2-7 loading defects since the pattern of Mcm2-7 mutant loading (after high-salt wash) is similar for wild-type and mutant Cdc6 (Fig. 3C, compare bottom two panels). Consistent with less efficient reactions requiring Cdc6 ATP hydrolysis (Fig. S3C), we observed reduced loading for the Mcm2-7 mutants in the presence of Cdc6-N263A (Fig. 3C, compare bottom two panels). Thus, in the absence of Cdc6 ATP hydrolysis, incompletely-loaded complexes are retained on DNA by the helicase loading proteins.
We also asked if Cdc6 ATPase mutants are defective in the release of successfully loaded Mcm2-7 complexes from helicase loading proteins. To this end, we performed helicase-loading reactions using a low-salt wash to retain associated ORC, Cdc6 and Cdt1. Following DNase I treatment to release proteins from the DNA beads, we immunoprecipitated Mcm2-7 complexes and detected Mcm2-7 and the helicase-loading proteins. In the presence of all wild-type proteins, Cdc6 and Cdt1 were released from DNA during loading (Fig. 3B, lane 1) and ORC did not co-immunoprecipitate with Mcm2-7 (Fig. 3B, lane 5). In the presence of Cdc6-N263A, although the mutant Cdc6 was strongly retained on DNA after the low-salt wash (Fig. 3B, lane 3), the Mcm2-7 immunoprecipitation showed only a slight increase in Mcm2-7-associated ORC, Cdc6 and Cdt1 (Fig. 3B, lane 7). Consistent with ATPγS arresting helicase loading at the OCCM stage, an ATPγS control experiment showed strong retention of all three helicase-loading proteins in the Mcm2-7 immunoprecipitation (Fig. 3B, lane 2 and 6). Thus, Cdc6 ATP hydrolysis is not required to release successfully-loaded Mcm2-7 from helicase-loading proteins.
Further analysis of the Mcm2-7 and Cdc6 ATPase mutants showed ATP hydrolysis by Mcm2-7 and Cdc6 contributed to OCCM complex disassembly. In the presence of Cdc6-N263A and ATP, all of the Mcm2-7 ATPase motif mutants showed increased levels of Cdc6 and Cdt1 DNA retention after low-salt wash (Fig. 3D and S2C). The increase in Cdt1 retention is likely due to retention of incompletely-loaded mutant Mcm2-7 complexes that are still bound to Cdt1, which would be released if Cdc6 could hydrolyze ATP. When only Mcm2-7 or Cdc6 was mutated, we observed much less Cdt1 DNA retention (Fig. 1C and Fig. 3D - lane 2, respectively). Also consistent with both Cdc6 and Mcm2-7 ATPases driving OCCM disassembly, in the presence of ATP, the combination of Cdc6-N263A and MCM5RA resulted in co-immunoprecipitation of Cdc6, ORC and Cdt1 with Mcm2-7 at levels comparable to an ATPγS reaction (Fig. 3B, compare lanes 6 and 8). Together, our findings indicate a combination of Cdc6 and Mcm2-7 ATP hydrolysis is required for Cdt1 and Cdc6 release and the transition from initial Mcm2-7 recruitment to later steps in helicase loading (summarized in Fig. S2C and Fig. S5). However, Cdc6 release is not required to complete helicase loading.
Loaded Mcm2-7 ATPase-motif mutants Form Double Hexamers that are DDK Substrates
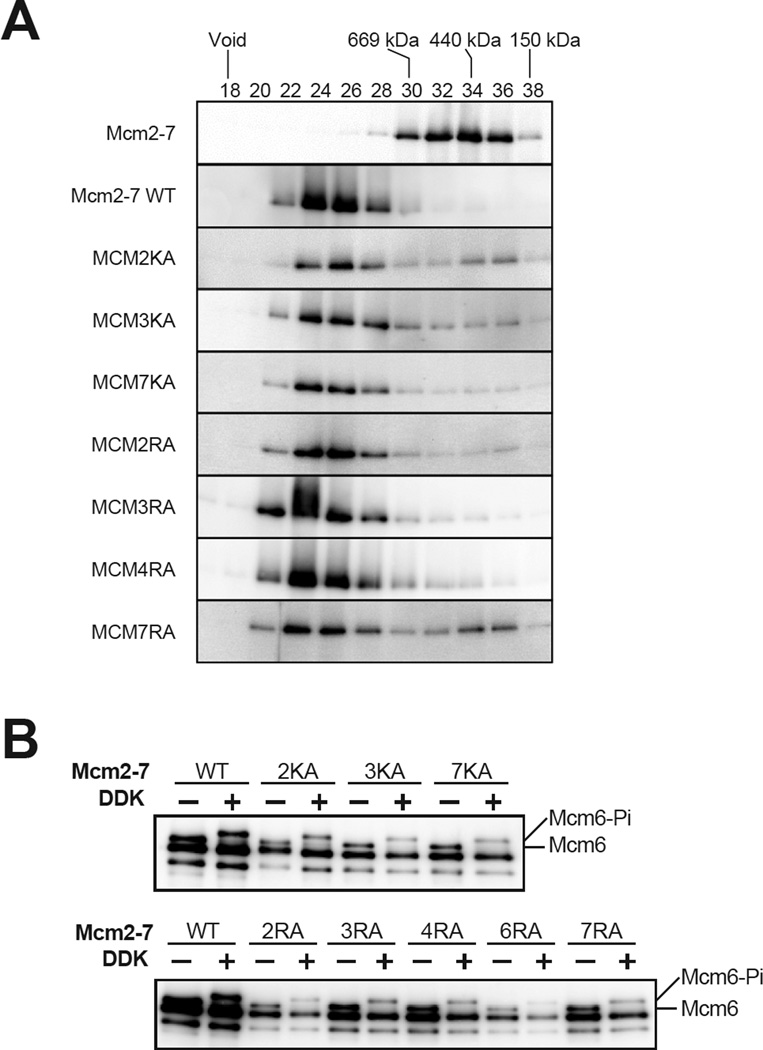
Although multiple Mcm2-7 ATPase mutants formed salt-resistant complexes, it was unclear if they formed double hexamers. To address this issue, we used a previously described gel-filtration assay for double-hexamer formation (Fig. 4A, Evrin et al., 2009). Consistent with previous studies, loaded wild-type Mcm2-7 complexes eluted at a position consistent with double-hexamer formation (Fig. 4A, compare first two rows). Each of the Mcm2-7 ATPase mutant complexes capable of salt-resistant loading primarily eluted as double-hexamers, although each showed increased evidence of single hexamer or smaller Mcm2-7 sub-assemblies (particularly, MCM2KA and MCM7RA).
Figure 4.
Loaded Mcm2-7 ATPase motif mutants form double hexamers that are DDK substrates. (A) Double hexamer formation by ATPase motif mutant Mcm2-7 complexes. After a helicase-loading reaction and high-salt wash, loaded wild-type or mutant Mcm2-7 complexes were released from the bead-bound DNA by DNase I treatment. Released Mcm2-7 complexes were applied to Superose 6 gel-filtration column (AKTA Micro, GE Healthcare). The Flag-Mcm3 subunit of the Mcm2-7 complexes in the indicated fractions was detected by immunoblot. Mcm2-7 double hexamers eluted earlier (fraction 24~26) compared to purified Mcm2-7 (fraction 32~36). Elution peaks of protein standards (thyroglobulin (669 kDa), apoferritin (440 kDa) and alcohol dehydrogenase (150 kDa) are indicated. (B) Phosphorylation of loaded wild-type or ATPase motif mutant Mcm2-7 complexes by DDK. After helicase loading, Mcm2-7 complexes were phosphorylated by addition of purified Cdc7-Dbf4 complex (DDK). Mcm2-7 complexes were detected by SDS-PAGE and immunoblot (anti-Mcm polyclonal). Phosphorylation was monitored by the reduced migration of Mcm6. Mcm6 and the slower-migrating phospho-Mcm6 (Mcm6-Pi) are indicated.
As an orthogonal means to assess double-hexamer formation, we tested DDK phosphorylation of the loaded mutant complexes. Previous studies showed that DDK preferentially phosphorylates loaded (and presumably double-hexameric) Mcm2-7 complexes (Francis et al., 2009). Thus, we asked if the loaded mutant Mcm2-7 complexes were DDK-phosphorylated by monitoring the DDK-dependent reduction in Mcm6 electrophoretic mobility (the largest Mcm subunit, Fig. 4B). Both the wild-type and mutant Mcm2-7 complexes showed complete shifting of the Mcm6 subunit (note: the antibody used detects phosphorylated Mcm6 less efficiently than unmodified Mcm6). Thus, the loaded Mcm2-7 ATPase-motif mutants are DDK substrates, further supporting the conclusion that the loaded mutant complexes are predominantly double hexamers.
Replication Initiation by Mcm2-7 ATPase motif mutants
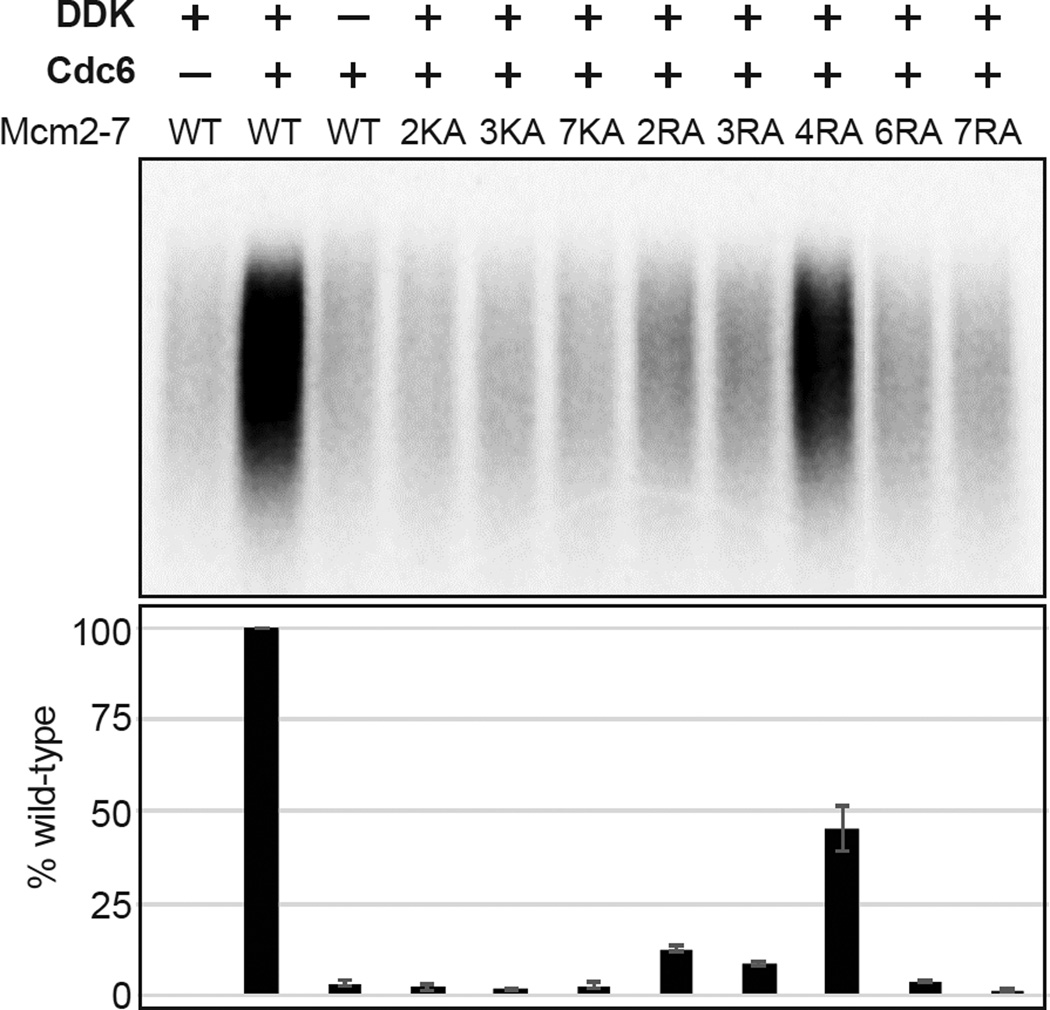
To investigate the role of the Mcm2-7 ATPase motifs in events after helicase loading, we assessed the ability of the mutant Mcm2-7 complexes that showed significant helicase loading (MCM2KA, MCM3KA, MCM7KA, MCM2RA, MCM3RA, MCM4RA and MCM7RA) to participate in downstream events of replication initiation. We included MCM6RA as a control for a mutant complex that displays strong helicase loading defects. Initially, we tested the ability of these mutant Mcm2-7 complexes to direct replication initiation. For these experiments, we assembled Mcm2-7 proteins onto the origin DNA using the reconstituted helicase-loading assay. We then sequentially incubated the loaded Mcm2-7 complexes with purified DDK and an S-phase extract made from cdc7-4 cells arrested at the non-permissive temperature (Heller et al., 2011). We monitored DNA synthesis by including radiolabeled dCTP in the S-phase incubation and separated the resulting replication products on a denaturing gel. Replication with wild-type Mcm2-7 showed that DNA replication required prior helicase loading (-Cdc6) and DDK phosphorylation (Fig. 5, compare lanes 1–3).
Figure 5.
Mcm2-7 ATPase motif mutants are defective for in vitro DNA replication. Replication assays were performed with purified wild-type or Mcm2-7 ATPase motif mutant complexes. Radiolabeled DNA replication products were analyzed by alkaline agarose electrophoresis (top panel). The relative DNA synthesis compared to wild-type Mcm2-7 (based on three independent experiments) is plotted in the bottom panel. Error bars indicate the standard deviation.
Consistent with genetic analysis of the Mcm2-7 ATPase mutations, only the complex containing the viable Mcm4 arginine-finger mutation (MCM4RA) showed robust DNA synthesis (Fig. 5). The MCM4RA complex supported ~45% of wild-type Mcm2-7 nucleotide incorporation. Of the remaining complexes, only MCM2RA and MCM3RA showed nucleotide incorporation over background, each showing less than 15% of wild-type levels.
Non-replicating Mcm2-7 ATPase mutants are defective in distinct initiation steps
To investigate the cause of the replication defects, we first determined the retention of the mutant Mcm2-7 complexes on the origin DNA after DDK-treatment and incubation in the S phase extract (Fig. 6A and S4A). Four of the Mcm2-7 mutant complexes (MCM3KA, MCM7KA, MCM2RA and MCM7RA) showed strong defects in DNA retention. Importantly, the strongest DNA-retention defects required both DDK treatment and S-phase extract addition. We observed intermediate levels of DNA release with S-phase extract alone (Fig. S4A). We suspect that this is due to the S-phase extract inducing release of associated but not loaded Mcm2-7, as the Mcm2-7 release seen with S-phase extract alone generally correlates with the change in Mcm2-7 DNA retention between the high- and low-salt washed helicase loading assays (compare Figs. S2C and S4A). These data indicate that several of the Mcm2-7 ATPase motifs are required to withstand or catalyze changes in the Mcm2-7 complex that are required for DNA retention during helicase activation.
Figure 6.
Mcm2-7 ATPase motif mutants exhibit defects in multiple steps during helicase activation. (A) Mcm2-7 ATPase mutants are defective in DNA retention during helicase activation. The amount of the indicated Mcm2-7 complexes (Flag-Mcm3) retained on DNA was monitored before and after the addition of S-phase extract and/or DDK treatment as indicated. (B) Mcm2-7 ATPase mutants are defective in distinct steps during helicase activation. Mcm2-7 loading before treatment of DDK and S phase extract (top panel) and replication proteins associated with the DNA template during DNA replication (bottom panel) were analyzed by immunoblot. Helicase activation was monitored by association of helicase activating proteins (Cdc45 and the Psf2 subunit of the GINS complex) and RPA binding to DNA. Quantification of these assays is shown in Figure S4.
To assess the defects in helicase activation further, we monitored the recruitment Cdc45, GINS and RPA to the DNA. Each of these proteins showed Cdc6- and DDK-dependent association with replicating DNA in the presence of wild-type Mcm2-7 (Fig. 6B, lanes 1–3). Recruitment of Cdc45 generally paralleled the amount of Mcm2-7 retained on the DNA (Fig. 6B and Fig. S4B), suggesting that, once loaded and DDK-phosphorylated, mutations in the Mcm2-7 ATPase motifs did not effect Cdc45 recruitment. In contrast, the level of GINS recruitment was distinct from the retention of the Mcm2-7 proteins. Although MCM3RA and MCM4RA showed levels of GINS recruitment similar to wild-type Mcm2-7, MCM2KA and MCM3KA showed GINS recruitment defects (Fig. 6B and S4C). Although able to recruit GINS, MCM3RA showed defects in RPA recruitment, strongly suggesting that this mutant is defective for replication-dependent DNA unwinding (although we cannot eliminate the possibility that RPA is associating with a replication protein rather than ssDNA). Consistent with RPA binding reflecting successful DNA unwinding and initiation, only the replication-competent MCM4RA showed strong RPA recruitment. We conclude that different Mcm2-7 ATPase motifs facilitate distinct steps in helicase activation (summarized in Fig. S4C and S5).
Discussion
The ring-shaped, heterohexameric Mcm2-7 complex forms an ATPase at each subunit interface but the function of these enzymes has remained obscure. Here we show that specific Mcm2-7 ATPase motifs contribute to distinct events throughout the initiation of DNA replication (Fig. 7 and S5). Five of the Mcm2-7 ATPase motif mutations show mutant-specific defects that eliminate helicase loading. Combining Mcm2-7 and Cdc6 mutations revealed that the Cdc6 ATPase is required to release incompletely-loaded Mcm2-7/Cdt1. Finally, all but one of the Mcm2-7 ATPase motif mutants capable of helicase loading showed specific defects during helicase activation.
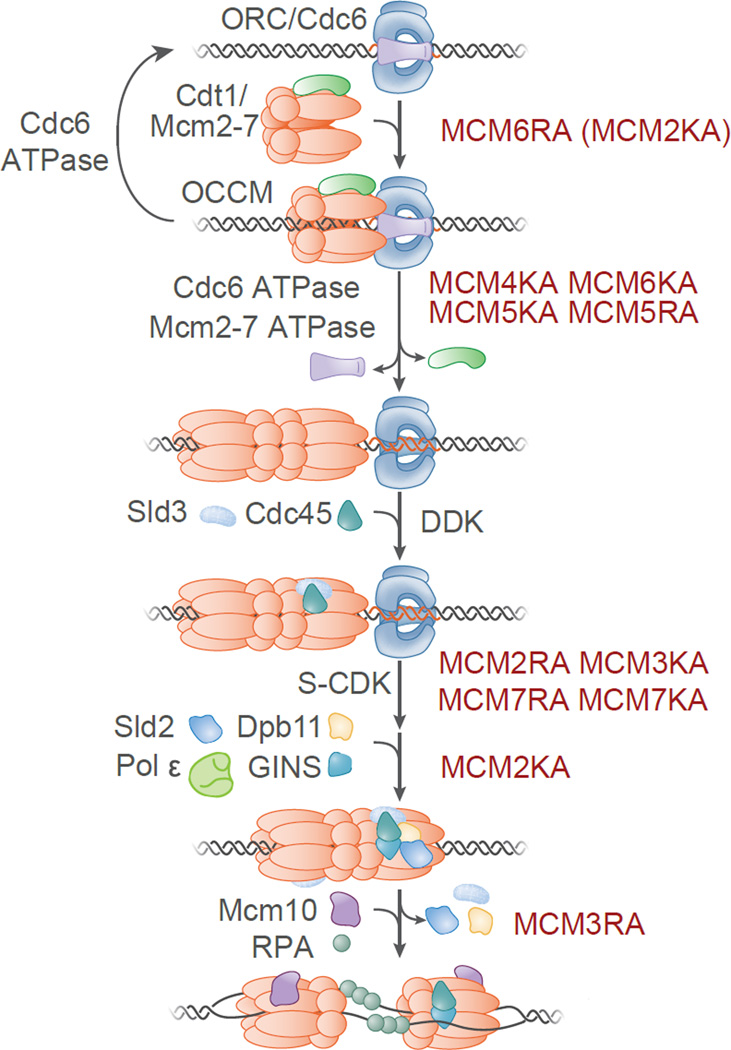
Figure 7.
Mcm2-7 ATPase active sites are specialized for different functions during helicase loading and activation. Helicase loading and activation events are illustrated and the step at which each Mcm2-7 ATPase motif mutant is defective indicated. Origin-DNA-bound ORC/Cdc6 initially recruits Mcm2-7/Cdt1 to form the OCCM. Following a poorly understood additional round of Mcm2-7/Cdt1 recruitment and loading (not shown), a head-to-head Mcm2-7 double hexamer is formed, Cdc6 and Cdt1 are released from DNA and ORC is released from Mcm2-7. Cdc6 ATP hydrolysis releases imcompletely-loaded Mcm2-7/Cdt1 complexes. DDK phosphorylation of loaded Mcm2-7 drives binding of Sld3 and Cdc45 to Mcm2-7. S-CDK phosphorylates Sld2 and Sld3 driving their binding to Dpb11 and the recruitment of GINS (and DNA Pol ε) to facilitate CMG complex formation. Sld2, Sld3 and Dpb11 are released and Mcm10 further activates DNA unwinding driving RPA association with the resulting ssDNA. See also Figure S5.
The Mcm2-7 mutations tested target key ATPase residues. There are two ways in which mutants in these domains could affect Mcm2-7 function: (1) inhibition of ATP binding (typical for Walker-A mutation) or hydrolysis (typical for arginine-finger mutation); or (2) disrupting the integrity of a Mcm2-7 interface. Several observations suggest that under our reaction conditions any effects on integrity are transient and only revealed during a specific initiation event. First, the assembly and integrity of the mutant Mcm2-7/Cdt1 complexes is maintained during purification. Second, although there were minor defects for MCM6KA (Fig. S2), the subunit stoichiometry of the mutant Mcm2-7 complexes is maintained throughout helicase loading (Figs. 1, 2, S2 and 3). Mutations that completely disrupt a Mcm2-7 interface would be expected to separate one or more subunits from the DNA during loading (Frigola et al., 2013). Third, many of the mutant complexes can form double hexamers that are maintained after DNase I treatment (Fig. 4), consistent with robust subunit interactions. Regardless of their primary cause, the diversity of replication initiation defects observed for the different Mcm2-7 ATPase motif mutants (including different phenotypes for mutations in the same interface) shows that these ATP-regulated interfaces contribute to Mcm2-7 function in distinct ways.
Mcm2-7 ATPase motifs are required for helicase loading
Mutations in five out of the six Mcm2-7 ATPase sites prevented helicase loading (Fig. 1 and 7). Interestingly, in all instances, only one of the two mutants in the interface showed a complete loss of loading (Fig. S5A). For the Mcm7/4, Mcm4/6 and Mcm2/5 interfaces, Walker A mutants prevented loading suggesting a requirement for ATP binding at these sites. The lesser effect of the corresponding arginine-finger mutants suggests ATP hydrolysis at these sites does not drive loading. In contrast, the Mcm6/2 and Mcm3/5 interfaces were fully defective when the associated arginine finger was mutated but the corresponding Walker-A mutants retained significant loading. Assuming that the arginine finger mutants allow ATP binding but not hydrolysis, it is likely that these ATPase interfaces must undergo conformational changes during helicase loading that are prevented by continuous ATP binding but are allowed when no ATP is present.
The helicase-loading-defective mutants disrupted this event at distinct steps (Fig. 7). Consistent with direct interactions between Mcm6 and Cdt1 (Wu et al., 2012), MCM6RA (and to a lesser extent its ATPase site counterpart, MCM2KA) showed reduced recruitment of Cdt1 and Mcm2-7 during OCCM complex formation (Fig. 2 and S2C). The remaining loading-defective Mcm2-7 mutants (MCM4KA, MCM5KA, MCM5RA and MCM6KA) failed after OCCM formation and showed Cdt1-release defects (Fig. 1C, 3D; summarized in Fig. S2C), suggesting a connection between successful loading and Cdt1 release. The Mcm5 ATPase motif mutations showed the strongest Cdt1-release defects. Interestingly, the EM structure of the OCCM complex shows that the AAA+ portion of the Mcm2/5 gate is open and that Mcm5 is proximal to Cdt1 (Sun et al., 2013). Perhaps ATP-controlled changes in Mcm5 facilitate gate closing and Cdt1 displacement. Importantly, any or all of these mutations could also inhibit additional steps involved in double-hexamer formation, an event that remains poorly understood.
The finding that most Mcm2-7 ATPase motif mutations form the OCCM complex suggests that Mcm2-7 is a passive participant in this event. Consistent with this view, recent studies suggest a small number of interactions are required for this initial recruitment (Frigola et al., 2013). Formation of the OCCM independent of the Mcm2-7 ATPase function could explain the chromatin-bound forms of Xenopus MCM6KA and MCM6KA/7KA mutant complexes (Ying and Gautier, 2005). Alternatively, the roles of the Mcm2-7 ATPase sites may not be conserved between S. cerevisiae and Xenopus.
In contrast to previous studies, we found that Cdc6 ATP hydrolysis is not required for helicase loading (Fig. 3). Combined with previous findings showing that ORC ATP hydrolysis is not required for helicase loading (Bowers et al., 2004; Evrin et al., 2013), these data lead to the conclusion that Mcm2-7 is the only ATPase required for this event. Our data also suggest that release of successfully loaded Mcm2-7 from the helicase-loading proteins requires Mcm2-7 but not ORC or Cdc6 ATP hydrolysis (Fig. 3B).
It is interesting to compare the role of ATP hydrolysis during helicase loading and the analogous event of sliding clamp loading. Sliding clamp loaders are structurally related to ORC and Cdc6 (Sun et al., 2013) and must hydrolyze ATP to complete loading (Hedglin et al., 2013). Clamp loader ATP hydrolysis is driven by primer-template junction binding to ensure that the clamp is loaded around this DNA structure. In contrast, ATP hydrolysis by ORC and Cdc6 is not required for helicase loading. Instead, our studies suggest that it is Mcm2-7 ATP hydrolysis that couples successful loading to the release of Mcm2-7 from ORC/Cdc6/Cdt1. Although it is the substrate rather than the loader that is hydrolyzing ATP to complete loading, such a model is analogous to sliding clamp loading in coupling ATP hydrolysis and substrate release to completion of the loading event.
Cdc6 ATPase Facilitates Release of Cdc6 and Incompletely Loaded Mcm2-7
Although not required for loading, the Cdc6 ATPase is required for release of Cdc6 and incompletely loaded Mcm2-7 from DNA (Fig. 3). It is likely that these functions are responsible for the helicase-loading defects observed for Cdc6 ATPase mutants (Fernández-Cid et al., 2013; Perkins and Diffley, 1998; Randell et al., 2006; Schepers and Diffley, 2001). Our findings show that Cdc6 ATP hydrolysis becomes important when helicase loading is less efficient (Fig. S3C and S3D). It is possible that ATPase-dependent Cdc6 release/recycling is required to maintain effective Cdc6 levels in less efficient reactions. Alternatively, less efficient reactions could increase the frequency of incomplete Mcm2-7 loading events, creating a need for Cdc6 ATP hydrolysis to release Mcm2-7 and allow a new attempt at loading. In support of the latter hypothesis, when loading is inefficient, Cdc6 ATPase mutations show elevated levels of associated Mcm2-7 (i.e. incompletely-loaded Mcm2-7, Fig. S3D).
Why is ORC and Cdc6 ATP hydrolysis required for helicase loading in vivo? It is possible that frequent incomplete Mcm2-7 loading events in vivo create a need for Cdc6 ATPase activity. Alternatively, the ATP-hydrolysis-dependent release of Cdc6 from DNA may be more important in vivo. Cdc6 is continuously required to maintain loaded Mcm2-7 during G1 (Aparicio et al., 1997), suggesting repeated rounds of Mcm2-7 loading and unloading are occurring. If Cdc6 is used repeatedly at the origin, the inability of Cdc6 ATPase mutants to release from the DNA could deplete Cdc6 or interfere with repeated helicase loading. Similarly, the lethality of ATPase-defective ORC-4R likely reflects the importance of repeated loading of helicases which is inhibited by this mutation (Bowers et al., 2004; Evrin et al., 2013).
Mcm2-7 ATPase Motifs Are Required for Helicase Activation
Seven of the MCM ATPase motif mutations were loaded as salt-resistant double hexamers (Fig. 4), allowing us to investigate their contribution to later steps in replication initiation (Fig. 7). None of the mutants prevented DDK phosphorylation or Cdc45 recruitment, suggesting that, after helicase loading, Mcm2-7 ATP binding and hydrolysis does not contribute to these events. Multiple mutants showed dramatic losses of DNA association at some point during helicase activation (Fig. S5C). Importantly, these defects require DDK-treatment and S-phase extract addition, implicating helicase activation in this release. The DNA release of these mutant Mcm2-7 complexes could be caused by either loss of ring integrity during helicase activation or export of both DNA strands instead of one during the transition from encircling double-stranded to single-stranded DNA (reviewed in Tanaka and Araki, 2013). MCM2KA showed a strong defect in GINS recruitment or maintenance. Because Mcm2 is proximal to the predicted Cdc45 binding site on Mcm2-7 (Costa et al., 2011), it is likely that ATP regulates the ability of this subunit to position Cdc45 for proper GINS binding during CMG complex formation. Alternatively, stable GINS binding could require initial ssDNA formation and this mutant is unable to melt origin DNA. Finally, MCM3RA bound Cdc45 and GINS but was defective in initial DNA unwinding, presumably due to a defect in either CMG helicase activity or activation. The only viable mutant tested, MCM4RA, was also the only mutant that supported DNA synthesis. Interestingly, MCM3RA does and MCM4RA does not unwind DNA on its own (Fig. S1A, Bochman and Schwacha, 2008), indicating that the contribution of the Mcm2-7 ATPase motifs changes upon CMG complex formation.
The events of eukaryotic replication require multiple changes in both Mcm2-7 interactions with DNA and other replication proteins. During loading, the OCCM complex transiently forms and the Mcm2-7 ring has to encircle double stranded DNA. During helicase activation, DDK and S-CDKs phosphorylation drives new Mcm2-7 interactions with Cdc45, GINS and DNA Pol εvia transient interactions of Sld2, Sld3 and Dpb11 (Fig. 7). In addition, the translocating DNA strand must be engaged and the non-translocating strand ejected from the Mcm2-7 central channel. Our findings show that, in addition to their function in DNA unwinding, the six Mcm2-7 ATPase motifs have evolved distinct functions to facilitate and potentially coordinate many of the steps required for DNA replication initiation.
Experimental Procedures
Protein purification
Wild-type and ATPase motif mutant Mcm2-7/Cdt1 complexes were purified from 2 L cultures of the strains listed in Supplementary Table 1. See Supplementary Experimental Procedures for detailed purification protocols. ORC was purified as described previously (Tsakraklides and Bell, 2010) and Cdc6 was purified as described in (Frigola et al., 2013).
Helicase-loading assay
2 pmole ORC, 3 pmole Cdc6 and 6 pmole Mcm2-7/Cdt1 were sequentially added to a 40 µl reaction solution containing 1 pmole of bead-coupled 1.3 Kbps ARS1 DNA in helicase loading buffer (25 mM HEPES-KOH (pH7.6), 12.5 mM magnesium acetate (MgAc), 0.1 mM zinc acetate (ZnAc), 300 mM potassium glutamate (KGlu), 20 µM creatine phosphate, 0.02 % NP40, 10 % glycerol, 3 mM ATP, 1 mM dithiothreitol (DTT) and 2 µg creatine kinase). The reaction mix was incubated at 25°C at 1200 rpm for 30 minutes in a Thermomixer (Eppendorf). Beads were washed three times with Buffer H (25 mM HEPES-KOH (pH 7.6), 1 mM EDTA, 1 mM EGTA, 5 mM MgAc, 10 % glycerol, 0.02% NP40) containing 0.3 M KGlu and DNA bound proteins were eluted from the beads using DNase I. Eluted proteins were separated by SDS-PAGE and stained with a fluorescent protein stain (Krypton, Thermo Scientific). For high-salt wash experiments, Buffer H containing 0.5 M NaCl was used for the second wash step. In ATPγS experiments, 6 mM ATPγS replaced added ATP.
Mcm2-7 IP after helicase loading
After a helicase-loading reaction and low-salt wash, DNA associated proteins were released from beads using DNase I treatment (5 U DNase I, 10 min). Eluted proteins were incubated with 30 µl anti-Mcm2-7 Ab (UM174, rabbit polyclonal) bound to Gammabind G Sepharose (GE) in 400 µl Buffer H containing 0.3 M KGlu for 90 minutes. Beads were washed three times with Buffer H containing 0.3 M KGlu and immunoprecipitated proteins were detected by SDS-PAGE and immunoblotting. ATP or ATPγS was included as indicated throughout the helicase loading and immunoprecipitation process (1 mM).
In vitro replication assay
Helicase loading reactions were performed using 0.5 pmol ORC, 0.75 pmol Cdc6, 2 pmol Mcm2-7/Cdt1 and 250 fmol bead-coupled 7.4 Kbps linear pARS1-Nco-Nco plasmid DNA (Fig. 5) or 3.6 Kbps circular pUC19-ARS1 (Fig. 6). Origin-loaded MCM complexes were phosphorylated with 450 ng purified DDK in DDK reaction buffer (50 mM HEPES-KOH (pH7.6), 3.5 mM MgAc, 0.1 mM ZnAc, 150 mM KGlu, 0.02 % NP40, 10 % glycerol, 1 mM spermine, 1 mM ATP and 1 mM DTT, 30 µl). Phosphorylated MCM complexes were activated by the addition of 750 µg of S phase extract in replication reaction buffer (25 mM HEPES-KOH (pH7.6), 12.5 mM MgAc, 0.1 mM ZnAc, 300 mM KGlu, 20 µM creatine phosphate, 0.02 % NP40, 10 % Glycerol, 3 mM ATP, 40 µM dNTPs, 200 µM CTP/UTP/GTP, 1 mM DTT, 10 µCi [α-P32] dCTP and 2 µg creatine kinase, 40 µl) for 1 hour at 25 °C at 1200 RPM in a Thermomixer (Eppendorf). DNA synthesis was monitored using alkaline agarose gel electrophoresis followed by detection of incorporated [P32] dCMP. DNA bound proteins were released from the beads by DNase I treatment (5 U for 10 minutes) and analyzed by immunoblotting. S phase extracts were prepared from ySKS12 as described previously (Heller et al., 2011).
Mcm2-7 double-hexamer-formation assay
Helicase loading reactions were performed using 1 pmol ORC, 1.5 pmol Cdc6 and 4 pmol Mcm2-7/Cdt1 and 500 fmol bead-coupled 7.4 Kbps linear pARS1-Nco-Nco plasmid DNA in a 160 µL reaction. Following loading, the DNA beads were washed with high salt (as described above). DNA bound proteins were released by DNase I treatment (25 U for 10 minutes) and loaded onto a Superose 6 PC 3.2/30 gel-filtration column (GE Healthcare). 40 µL fractions were collected. Adjacent fractions were pooled, TCA precipitated and analyzed by immunoblotting.
Supplementary Material
Highlights.
Mcm2-7 ATP binding and hydrolysis is required for helicase loading and activation.
Mcm2-7 ATPase mutations inhibit Mcm2-7 recruitment and Cdt1-release during loading.
Cdc6 ATP hydrolysis releases loading-defective Mcm2-7 but is not required for loading.
Loading-competent Mcm2-7 ATPase mutants show defects in CMG formation and DNA retention.
Acknowledgements
We thank John Diffley for communicating results prior to publication, Eric Greene, Daniel Duzdevitch, Marko Looke and Andrea Rothballer for helpful comments on the manuscript and Adam Shoemaker for protein purification. This work was supported by the Howard Hughes Medical Institute and an NIH grant awarded to SPB (GM052339). MDW was supported in part by a NIH Pre-Doctoral Training Grant (GM007287) and a NSF Graduate Fellowship (1122374).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol Cell. 2008;31:287–293. doi: 10.1016/j.molcel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- Bochman ML, Schwacha A. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol Mol Biol Rev. 2009;73:652–683. doi: 10.1128/MMBR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Schwacha A. The Saccharomyces cerevisiae Mcm6/2 and Mcm5/3 ATPase active sites contribute to the function of the putative Mcm2-7 'gate'. Nuc Acids Res. 2010;38:6078–6088. doi: 10.1093/nar/gkq422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Bell SP, Schwacha A. Subunit organization of Mcm2-7 and the unequal role of active sites in ATP hydrolysis and viability. Mol Cell Biol. 2008;28:5865–5873. doi: 10.1128/MCB.00161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JL, Randell JCW, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol Cell. 2004;16:967–978. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Costa A, Hood IV, Berger JM. Mechanisms for initiating cellular DNA replication. Annu Rev Biochem. 2013;82:25–54. doi: 10.1146/annurev-biochem-052610-094414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18:471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci USA. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Fernández-Cid A, Zech J, Herrera MC, Riera A, Clarke P, Brill S, Lurz R, Speck C. In the absence of ATPase activity, pre-RC formation is blocked prior to MCM2-7 hexamer dimerization. Nuc Acids Res. 2013;41:3162–3172. doi: 10.1093/nar/gkt043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Cid A, Riera A, Tognetti S, Herrera MC, Samel S, Evrin C, Winkler C, Gardenal E, Uhle S, Speck C. An ORC/Cdc6/MCM2-7 complex is formed in a multistep reaction to serve as a platform for MCM double-hexamer assembly. Mol Cell. 2013;50:577–588. doi: 10.1016/j.molcel.2013.03.026. [DOI] [PubMed] [Google Scholar]
- Francis LI, Randell JCW, Takara TJ, Uchima L, Bell SP. Incorporation into the prereplicative complex activates the Mcm2-7 helicase for Cdc7-Dbf4 phosphorylation. Genes Dev. 2009;23:643–654. doi: 10.1101/gad.1759609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigola J, Remus D, Mehanna A, Diffley JFX. ATPase-dependent quality control of DNA replication origin licensing. Nature. 2013;495:339–343. doi: 10.1038/nature11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedglin M, Kumar R, Benkovic SJ. Replication Clamps and Clamp Loaders. Cold Spring Harb Perspect Biol. 2013;5:a010165–a010165. doi: 10.1101/cshperspect.a010165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146:80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm RD, Austin RJ, Bell SP. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu S, Hirai K, Tak Y-S, Kamimura Y, Araki H. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol (epsilon}, and GINS in budding yeast. Genes Dev. 2010;24:602–612. doi: 10.1101/gad.1883410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins G, Diffley JF. Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Mol Cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]
- Randell JCW, Bowers JL, Rodríguez HK, Bell SP. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Mol Cell. 2006;21:29– 39. doi: 10.1016/j.molcel.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JFX. Concerted Loading of Mcm2-7 Double Hexamers around DNA during DNA Replication Origin Licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers A, Diffley JF. Mutational analysis of conserved sequence motifs in the budding yeast Cdc6 protein. J Mol Biol. 2001;308:597–608. doi: 10.1006/jmbi.2001.4637. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Bell SP. Interactions between two catalytically distinct MCM subgroups are essential for coordinated ATP hydrolysis and DNA replication. Mol Cell. 2001;8:1093–1104. doi: 10.1016/s1097-2765(01)00389-6. [DOI] [PubMed] [Google Scholar]
- Sengupta S, van Deursen F, De Piccoli G, Labib K. Dpb2 integrates the leading-strand DNA polymerase into the eukaryotic replisome. Curr Biol. 2013;23:543–552. doi: 10.1016/j.cub.2013.02.011. [DOI] [PubMed] [Google Scholar]
- Speck C, Stillman B. Cdc6 ATPase activity regulates ORC•Cdc6 stability and the selection of specific DNA sequences as origins of DNA replication. J Biol Chem. 2007;282:11705–11714. doi: 10.1074/jbc.M700399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol. 2005;12:965–971. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Evrin C, Samel SA, Fernández-Cid A, Riera A, Kawakami H, Stillman B, Speck C, Li H. Cryo-EM structure of a helicase loading intermediate containing ORC-Cdc6-Cdt1-MCM2-7 bound to DNA. Nat Struct Mol Biol. 2013;20:944–951. doi: 10.1038/nsmb.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Araki H. Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb Perspect Biol. 2013;5:a010371. doi: 10.1101/cshperspect.a010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Diffley JFX. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2-7 during G1 phase. Nat Cell Biol. 2002;4:198–207. doi: 10.1038/ncb757. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Komeda Y, Umemori T, Kubota Y, Takisawa H, Araki H. Efficient initiation of DNA replication in eukaryotes requires Dpb11/TopBP1-GINS interaction. Mol Cell Biol. 2013;33:2614–2622. doi: 10.1128/MCB.00431-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Nakato R, Katou Y, Shirahige K, Araki H. Origin association of sld3, sld7, and cdc45 proteins is a key step for determination of origin-firing timing. Curr Biol. 2011;21:2055–2063. doi: 10.1016/j.cub.2011.11.038. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- Thu YM, Bielinsky A-K. Enigmatic roles of Mcm10 in DNA replication. Trends Biochem Sci. 2013;38:184–194. doi: 10.1016/j.tibs.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakraklides V, Bell SP. Dynamics of pre-replicative complex assembly. J Biol Chem. 2010;285:9437–9443. doi: 10.1074/jbc.M109.072504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M, Liang C, Stillman B. The Cdc6p nucleotide-binding motif is required for loading mcm proteins onto chromatin. Proc Natl Acad Sci USA. 1999;96:441–446. doi: 10.1073/pnas.96.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Wang J, Liang C. Cdt1p, through its interaction with Mcm6p, is required for the formation, nuclear accumulation and chromatin loading of the MCM complex. J Cell Sci. 2012;125:209–219. doi: 10.1242/jcs.094169. [DOI] [PubMed] [Google Scholar]
- Yardimci H, Walter JC. Prereplication-complex formation: a molecular double take? Nat Struct Mol Biol. 2014;21:20–25. doi: 10.1038/nsmb.2738. [DOI] [PubMed] [Google Scholar]
- Ying CY, Gautier J. The ATPase activity of MCM2-7 is dispensable for pre-RC assembly but is required for DNA unwinding. Embo J. 2005;24:4334–4344. doi: 10.1038/sj.emboj.7600892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JFX. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.