Abstract
Gonadotropin-releasing hormone (GnRH) neurons originate outside the CNS in the olfactory placode and migrate into the CNS, where they become integral components of the hypothalamic-pituitary-gonadal (HPG) axis. Disruption of this migration results in Kallmann syndrome (KS), which is characterized by anosmia and pubertal failure due to hypogonadotropic hypogonadism. Using candidate-gene screening, autozygosity mapping, and whole-exome sequencing in a cohort of 30 individuals with KS, we searched for genes newly associated with KS. We identified homozygous loss-of-function mutations in FEZF1 in two independent consanguineous families each with two affected siblings. The FEZF1 product is known to enable axons of olfactory receptor neurons (ORNs) to penetrate the CNS basal lamina in mice. Because a subset of axons in these tracks is the migratory pathway for GnRH neurons, in FEZF1 deficiency, GnRH neurons also fail to enter the brain. These results indicate that FEZF1 is required for establishment of the central component of the HPG axis in humans.
Main Text
Kallmann syndrome (KS [MIM 308700, 147950, 244200, 610628, 612370, and 612702]) is characterized by a combination of hypogonadotropic hypogonadism and anosmia. This pathophysiological association results from a defect in the shared development of gonadotropin-releasing hormone (GnRH) neurons and olfactory neurons.1,2 Both types of neurons originate in the nasal placode, and GnRH neurons migrate in association with olfactory axon bundles to the CNS. Upon entering the CNS, olfactory axons synapse in the olfactory bulb, whereas GnRH neurons migrate caudally on a subset of nonsensory olfactory axons to reach the hypothalamus. Once within the hypothalamus, GnRH neurons form a functional network, producing pulsatile GnRH secretion. Disruption of this migration results in KS. A number of genes have been implicated to cause KS when mutated. These include KAL13,4 (MIM 308700), genes involved in the fibroblast growth factor (FGF) signaling pathway (FGF8 [MIM 600483], FGFR1 [MIM 136350], FGF17, IL17RD, DUSP6, SPRY4, and FLRT3),5–7 genes associated with the PROK2 signaling pathway (PROK2 [MIM 607002] and PROKR2 [MIM 607123]),8,9 CHD710 (MIM 608892), WDR1111 (MIM 614858), SEMA3,12,13 SOX10,14 and HS6ST115 (MIM 614880). Mutations in these genes account for less than half of all familial cases, and thus identification of newly associated genes is highly likely, which could provide insight into the biology of GnRH neuronal migration.16 Here, we describe two independent consanguineous families in which the KS phenotype is associated with loss-of-function mutations in FEZF1.
The ethics committee of the Cukurova University Faculty of Medicine approved this study, and written informed consent was obtained for each participant. A cohort of 30 individuals was studied in an effort to identify a gene newly associated with KS. In this prospective, multicenter study, the diagnosis of KS was based on the presence of anosmia and the absence of any signs of spontaneous puberty by 13 years of age in girls (Tanner breast stage 1) and by 14 years of age in boys (testicular volume < 4 ml), as well as concentrations of sex steroids at inappropriately normal hypogonadal levels or low gonadotropin levels. There were five consanguineous multiplex families with more than one affected individual. The rest of the cohort consisted of single affected persons. Those multiplex families were prioritized and screened first for mutations in the known KS-associated genes. In one of the multiplex families (family 1), a 19-year-old male (II-3) presented first with absent pubertal development at age 14 years. He received testosterone and human chorionic gonadotropin (HCG) treatments and underwent surgery for undescended testicles. His penis developed to normal adult size with testosterone treatment, but his testicles remained small. His 24-year-old sister (II-1) suffered from absent breast development and primary amenorrhea. Only after starting estrogen replacement at age 18 did her breast development and subsequent menstrual periods begin. In a second multiplex family (family 2), a 14-year-old male (II-2) presented first with micropenis and undescended testicles at age 2 years. For undescended testicles, he received repeated HCG treatments; upon failure, he was subsequently operated on. He has not had any spontaneous pubertal development, for which he was given testosterone injections. Currently, he has 1 ml of testicles bilaterally and 4 cm of phallus with stage 2 axillary and pubic hair. A recent GnRH stimulation test elicited a prepubertal response with a maximum luteinizing hormone response of 0.2 mIU/ml. His 8-year-and-7-month-old male sibling (II-4) also has micropenis and undescended testicles, for which he received an operation at 2 years of age after failure of repeated HGC treatment. Currently, he is prepubertal with 1 ml of testicles bilaterally and 3.6 cm of phallus with stage 1 axillary and pubic hair. Both families are ethnically Kurdish from distant provinces and are not knowingly related to each other. None of the parents or other siblings has experienced problems in pubertal development or fertility. All four individuals from both families are otherwise healthy. They do not have mental disorders, dysmorphic facial features (such as cleft lip, cleft palate, or hypodontia), bone anomalies, deafness, renal agenesis, or bimanual synkinesia, which are sometimes associated with KS.16 They all have anosmia, documented by a quantitative smell-identification test. Olfactory function of both the proband and his unaffected brother (II-2) in family 1 was evaluated with the 40-item University of Pennsylvania smell-identification test (UPSIT). The UPSIT is a validated microencapsulated odor scratch-and-sniff test that correlates with other olfactory tests, including odor-detection thresholds.17 To control for cross-cultural variation of smell identification, we also administered a culturally appropriate 20-item smell test. Results of this test were found to be consistent with the UPSIT results. The culturally appropriate 20-item smell test was also administered to the proband in family 2. The hormonal profiles of all four individuals when they were off treatment are shown in Table 1. Brain-MRI studies of the probands showed bilateral aplasia of the olfactory bulbs (Figure 1B).
Table 1.
Hormonal Results of the Affected Siblings when They Were off Hormonal Treatment
|
Family 1 |
Family 2 |
Normal Range | |||
|---|---|---|---|---|---|
| II-1 | II-3 | II-2 | II-4 | ||
| Sex | F | M | M | M | – |
| Age (years) | 24 | 19 | 14 | 8 | – |
| FSH (mIU/ml) | 0.6 | 0.1 | 0.1 | 0.6 | M: 1.4–18.1 F: 2.5–10.2 |
| LH (mIU/ml) | 0.1 | 0.1 | 0.1 | 0.1 | M: 1.5–9.3 F: 1.9–12.5 |
| Estradiol (ng/dl) | 0.7 | ND | ND | ND | M: 0.8–3.5 F: 6.3–16.5 |
| Testosterone (ng/dl) | ND | 69.0 | 20.0 | 10.0 | M: 175–781 F: 20–38 |
| Prolactin (ng/ml) | 8.9 | 6.6 | ND | ND | M: 2.1–17.7 F: 2.8–29.2 |
| TSH (mIU/ml) | 1.1 | 2.8 | 2.1 | 2.0 | 0.35–4.2 |
| Free T4 (ng/dl) | 0.8 | 1.1 | 1.0 | 1.1 | 0.89–1.8 |
| ACTH (pg/ml) | 34.8 | 9.1 | ND | ND | 5–55 |
| Cortisol (mcg/dl) | 21.2 | 12.9 | ND | ND | 3–25 |
| DHEAS (mcg/dl) | 139.0 | ND | 155.4 | 61.6 | M: 35–430 F: 76–255 |
| LHRH stimulation test (maximum LH) | – | – | 0.2 | – | M: 1.5–9.3 F: 1.9–12.5 |
| LHRH stimulation test (maximum FSH) | – | – | 1.3 | – | M: 1.4–18.1 F: 2.5–10.2 |
The coefficients of variation within assays and between assays were less than 5%. Abbreviations are as follows: ACTH, adrenocorticotropic hormone; DHEAS, dehydroepiandrosterone; F, female; FSH, follicle-stimulating hormone; LH, luteinizing hormone; LHRH, luteinizing-hormone-releasing hormone; M, male; ND, not determined; T4, thyroxine; and TSH, thyroid-stimulating hormone.
Figure 1.
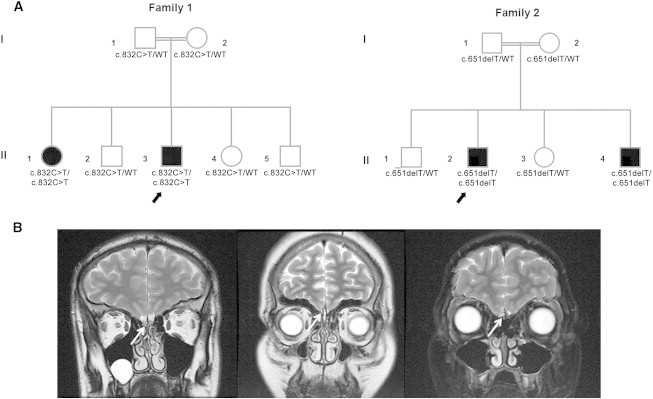
Pedigree and Olfactory MRI of the Probands
(A) Pedigrees of the families; arrows point to probands. Solid symbols indicate affected family members, open symbols indicate unaffected family members, squares indicate male family members, circles indicate female family members, and the double line indicates consanguinity. Under each symbol is the FEZF1 genotype.
(B) Brain MRI at T2 coronal sections. The left panel is from a healthy young adult; the arrow indicates normal olfactory bulbs. The middle panel shows the absence of olfactory bulbs (aplasia) in the proband from family 1. The right panel demonstrates the absence of olfactory bulbs (aplasia) in the proband from family 2.
We first ruled out mutations in known KS-associated genes, including KAL1, FGFR1, FGF8, PROK2, PROKR2, CHD7, WDR11, and HS6ST1, by Sanger sequencing on an Applied Biosystems PRISM 3130 autosequencer. Subsequently, we combined autozygosity mapping and exome sequencing. A genome-wide SNP analysis used 250K NspI SNP microarrays (Affymetrix), and the data were analyzed with AutoSNPa software. For exome sequencing, samples were prepared as an Illumina sequencing library. Sequencing libraries were enriched with the desired target according to the Illumina Exome Enrichment protocol. The captured libraries were sequenced with the Illumina HiSeq 2000 Sequencer. The reads were mapped against the UCSC Genome Browser (hg19). In family 1, autozygosity mapping identified three regions of homozygosity: a 5.8 Mb region from 165.7 to 171.5 Mb on chromosome 1, a 7.7 Mb region from 172.4 to 180.1 Mb on chromosome 2, and a 12 Mb region from 110.7 to 122.4 Mb on chromosome 7. Whole-exome sequencing data, which were filtered with the “rare recessive Mendelian disease” mode in wANNOVAR, indicated two homozygous mutations each located within these homozygous regions. The FEZF1 (FEZ family zinc finger 1 [MIM 613301]) mutation, which was located in the homozygous region on chromosome 7, involved a C-to-T change (c.832C>T [RefSeq accession number NM_001024613.2]) leading to p.His278Tyr (RefSeq NP_001019784.2). Both affected siblings were homozygous for this mutation, and the parents and unaffected siblings (except for II-4, who was homozygous for the wild-type) were heterozygous (Figure S1, available online). This mutation changes the first histidine in the well-known C2H2 motif, which is required for the stability of the central zinc in zinc-finger proteins.18 Therefore, this residue is extremely evolutionarily conserved, and all variants are predicted to be deleterious.
The other variant was a nonsense mutation (c.2270C>T [p.Arg724∗]) in CCDC141 and was located in the homozygous region on chromosome 2. Both individuals were homozygous for the change, and the parents and unaffected siblings were all heterozygous.
The four multiplex families in the KS cohort were prioritized for exome sequencing. In one of the families (family 2), we found an additional FEZF1 mutation (c.652del) resulting in a frameshift and premature stop (p.Ala217fs∗13) (Figure S1). Transcripts harboring this frameshift mutation might be sensitive to nonsense-mediated mRNA decay.19 Whereas all affected siblings were homozygous for c.652del, both parents and unaffected siblings were heterozygous. None of the individuals from family 2 carried mutations in CCDC141. No additional probands from the KS cohort had mutations in FEZF1 or CCDC141. The inheritance pattern in both families was consistent with autosomal-recessive inheritance. In view of the oligogenic inheritance repeatedly reported in KS,8,12,20 we attempted to ensure that all variants were identified to account for the KS phenotype by combining a variety of genetics methods, including candidate-gene screening, autozygosity mapping, and whole-exome sequencing. FEZF1 mutations in this study were not found in public SNP databases (dbSNP136, 1000 Genomes, or the NHLBI Exome Sequencing Project Exome Variant Server). Additionally, neither of the mutations was seen in 100 ethnically matched healthy adult control individuals or in 36 in-house whole exomes.
To determine the predicted consequences of the mutations on FEZF1 function, we used in silico prediction methods and 3D molecular modeling. PolyPhen-2, SIFT, and MutationTaster indicated that both mutations would be harmful. Using the SWISS-MODEL protein-structure homology-modeling server to evaluate the domains surrounding the altered residues indicated that changes to the secondary structure are not predicted, although the stability of the zinc binding structure could be affected.
To assess the transcriptional activity of FEZF1, we transfected human embryonic kidney 293T cells with wild-type and mutant (c.832C>T) plasmids (GeneArt) together with a Hes5p-DsRed reporter. This reporter is known to be repressed by FEZF1 during early cortical neurogenesis.21 Wild-type FEZF1 reduced DsRed accumulation, as measured by immunoblot (Figures 2C and 2D); however, the FEZF1 mutant accumulated intermediate levels of DsRed, indicating a partial loss of function.
Figure 2.
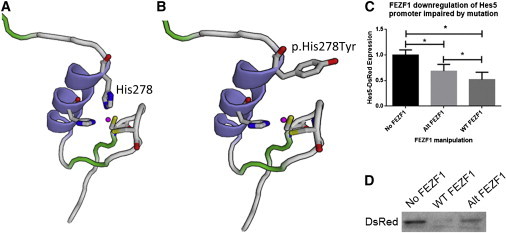
Functional Analysis of Mutated FEZF1
(A) Predicted 3D model of the zinc-finger domain containing histidine 278. Histidine residues (blue) are shown alongside cysteine (yellow) and bound zinc.
(B) Predicted 3D model of the altered FEZF1, in which histidine 278 changed to a tyrosine, from family 1.
(C) Analysis of DsRed band optical density (normalized to tubulin) with either no FEZF1, altered FEZF1, or wild-type FEZF1. Histograms show the mean, and error bars represent the SE (∗p < 0.05, repeated-measures one-way ANOVA with the Holm-Sidak posttest).
(D) Example of an immunoblot showing Hes5-DsRed levels when cotransfected with no FEZF1 (left), altered FEZF1 (middle), or wild-type FEZF1 (right). Immunoblot analysis was performed according to standard protocols. Protein was extracted 24 hr after transfection. Fifty micrograms of total protein was denatured, subsequently loaded into a 7.5% gel, and then separated. Protein was then transferred to a polyvinylidene fluoride membrane, blocked, and incubated in primary antibody overnight. Then the membranes were washed, subsequently incubated with horseradish-peroxidase-conjugated secondary antibody for 2 hr, and visualized with an enhanced chemiluminescence reaction. Band density was quantified with ImageJ. DsRed band density was normalized to β-tubulin levels. Rabbit anti-human ZNF312B (FEZF1, Abcam, catalog no. ab81251, 1:1,000), anti-DsRed (Clontech, catalog no. 632496, 1:1,000), mouse anti-β-tubulin (Sigma, catalog no. T4026, 1:1,000), anti-rabbit HRP (Cell Signaling, catalog no. 7074, 1:5,000), anti-mouse HRP (Cell Signaling, catalog no. 7076, 1:5,000) antibodies were used in these experiments. Statistical significance was determined with Prism (GraphPad).
All together, in a cohort of 30 probands with KS, we found deleterious homozygous FEZF1 variants in two independent consanguineous families each with two siblings afflicted with severe KS, including complete hypogonadotropic hypogonadism and anosmia and absent olfactory bulbs. KS is genetically heterogeneous and has various modes of transmission and oligogenic inheritance.22 In contrast to previously described KS-associated gene defects, such as mutations in FGFR15 or PROKR2,8,9 the phenotypical features in our subjects were invariably severe, and heterozygous parents and siblings had normal reproductive and olfactory functions; thus, the mode of inheritance is solely autosomal-recessive in families affected by FEZF1 mutations. The presence of micropenis and cryptorchidism in the male individuals reflects complete hypogonadotropic hypogonadism since intrauterine life.23 The absence of additional phenotypical features (such as cleft palate, deafness, and synkinesia, which can be seen in other KS-associated gene mutations5,8,9) suggests that FEZF1 is spatially and temporally restricted in expression and functionally specific. Consistency among these phenotypical features of FEZF1-associated KS might not hold in additional future mutations, given that the affected individuals in this study probably represent the most severe cases. In one of the families (family 1), we found a second homozygous mutation in another gene, CCDC141, whose gene product has been implicated in cortical neuronal migration,24 and this mutation might cause additional detrimental effects.
FEZF1 is a zinc-finger gene encoding a transcriptional repressor that is highly and selectively present during embryogenesis in the olfactory epithelium, amygdala, and hypothalamus. Interestingly, these tissues and organs mark the pheromone pathway from the nose to the GnRH pulse generator in the hypothalamus.25–28 Two independent studies have shown that Fezf1-deficient mice have impaired axonal projection of pioneer olfactory receptor neurons (ORNs) that cross the cribriform plate and subsequently innervate the olfactory bulb. This defect was attributed to the inability of ORN axons to penetrate the basal lamina of the CNS. These mice have smaller olfactory bulbs and an absence of GnRH neurons in the brain.27,29 When these experiments were repeated with the basal lamina surgically removed, GnRH neurons were found to be in their expected locations in the hypothalamus.27 Thus, it seems likely that the FEZF1 product promotes the presence of a protease to enable ORNs, and thus accompanying GnRH neurons, to enter the brain.27,30 Upon entering the CNS, ORN axons make synapses with olfactory bulb neurons, which is believed to promote the development of the olfactory bulbs.31 Thus, the absence of olfactory bulbs on MRI in humans with KS indicates a strong parallelism with the rodent data in that ORNs fail to penetrate the basal lamina and subsequently induce the growth of the olfactory bulbs. Fezf1-knockout mice in both studies died early postnatally with abdominal distention, so their puberty and fertility could not be observed. Our findings in humans suggest that hypogonadism associated with a GnRH-migration deficit due to FEZF1 mutations cannot be compensated by some other mechanism, although, again, it should be noted that the affected individuals in this study are likely to be the most severe cases.
This study highlights a step in deciphering the molecular mechanisms involved in the development of the GnRH neuronal system. The nasal-forebrain junction is a crucial point in the migration of GnRH neurons, and crossing of this junction is a finely regulated step in which different players act together in a coordinated manner to guide the entrance of these neurons into the forebrain.32 However, it should be kept in mind that other sites of embryonic expression (i.e., the amygdala and hypothalamus)25–27 might also be involved in the pathogenesis of FEZF1-associated KS. Also, in this regard, it is important to note that the potential genes up- or downregulated by FEZF1 remain to be identified.
The potential mechanism of KS due to FEZF1 mutations appears to be different from those of previously described causes of KS. Fibroblast growth factor signaling, prokineticin signaling, and anosmin-1, defects of which constitute the great majority of KS cases, appear to interact with heparin sulfate glycosominoglycan compounds within an extracellular signaling complex in promoting GnRH neuronal migration.5,22 WDR11 mutations could be the only previously implicated cause of KS that can be in any way likened to FEZF1 deficiency. The WDR11 product is known to interact with transcription factor emx1,11 whose paralog (emx2) is involved in innervation of the olfactory bulb by ORNs in mice.33
In conclusion, the FEZF1 mutations reported here are examples of genetic aberrations accounting for KS. These studies strongly suggest that in humans, FEZF1 plays a role in embryonic migration of GnRH neurons into the brain to eventually form a hypothalamic neuronal network essential for pulsatile GnRH secretion and thus puberty and fertility.
Acknowledgments
This work was supported by the Scientific and Technological Research Council of Turkey (TÜBİTAK, project no. 109S455), by the Cukurova University Scientific Research Projects (BAP), and by International Centre for Genetic Engineering and Biotechnology (CRP/TUR10-01). B.I.H., P.J.C., H.S., and S.W. received support from the Intramural Research Program of the NIH National Institute of Neurological Disorders and Stroke (NS002824-13).
Contributor Information
Susan Wray, Email: wrays@ninds.nih.gov.
A. Kemal Topaloglu, Email: ktopaloglu@cu.edu.tr.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://browser.1000genomes.org
Autozygosity.org, http://autozygosity.org
HUGO Gene Nomenclature Committee, http://www.genenames.org/
MutationTaster, http://www.mutationtaster.org
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
SWISS-MODEL, http://swissmodel.expasy.org/
UCSC Genome Browser, http://genome.ucsc.edu
References
- 1.Schwanzel-Fukuda M., Pfaff D.W. Origin of luteinizing hormone-releasing hormone neurons. Nature. 1989;338:161–164. doi: 10.1038/338161a0. [DOI] [PubMed] [Google Scholar]
- 2.Wray S., Grant P., Gainer H. Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc. Natl. Acad. Sci. USA. 1989;86:8132–8136. doi: 10.1073/pnas.86.20.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Legouis R., Hardelin J.P., Levilliers J., Claverie J.M., Compain S., Wunderle V., Millasseau P., Le Paslier D., Cohen D., Caterina D. The candidate gene for the X-linked Kallmann syndrome encodes a protein related to adhesion molecules. Cell. 1991;67:423–435. doi: 10.1016/0092-8674(91)90193-3. [DOI] [PubMed] [Google Scholar]
- 4.Franco B., Guioli S., Pragliola A., Incerti B., Bardoni B., Tonlorenzi R., Carrozzo R., Maestrini E., Pieretti M., Taillon-Miller P. A gene deleted in Kallmann’s syndrome shares homology with neural cell adhesion and axonal path-finding molecules. Nature. 1991;353:529–536. doi: 10.1038/353529a0. [DOI] [PubMed] [Google Scholar]
- 5.Dodé C., Levilliers J., Dupont J.M., De Paepe A., Le Dû N., Soussi-Yanicostas N., Coimbra R.S., Delmaghani S., Compain-Nouaille S., Baverel F. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat. Genet. 2003;33:463–465. doi: 10.1038/ng1122. [DOI] [PubMed] [Google Scholar]
- 6.Falardeau J., Chung W.C., Beenken A., Raivio T., Plummer L., Sidis Y., Jacobson-Dickman E.E., Eliseenkova A.V., Ma J., Dwyer A. Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J. Clin. Invest. 2008;118:2822–2831. doi: 10.1172/JCI34538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miraoui H., Dwyer A.A., Sykiotis G.P., Plummer L., Chung W., Feng B., Beenken A., Clarke J., Pers T.H., Dworzynski P. Mutations in FGF17, IL17RD, DUSP6, SPRY4, and FLRT3 are identified in individuals with congenital hypogonadotropic hypogonadism. Am. J. Hum. Genet. 2013;92:725–743. doi: 10.1016/j.ajhg.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dodé C., Teixeira L., Levilliers J., Fouveaut C., Bouchard P., Kottler M.L., Lespinasse J., Lienhardt-Roussie A., Mathieu M., Moerman A. Kallmann syndrome: mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet. 2006;2:e175. doi: 10.1371/journal.pgen.0020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pitteloud N., Zhang C., Pignatelli D., Li J.D., Raivio T., Cole L.W., Plummer L., Jacobson-Dickman E.E., Mellon P.L., Zhou Q.Y., Crowley W.F., Jr. Loss-of-function mutation in the prokineticin 2 gene causes Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc. Natl. Acad. Sci. USA. 2007;104:17447–17452. doi: 10.1073/pnas.0707173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim H.G., Kurth I., Lan F., Meliciani I., Wenzel W., Eom S.H., Kang G.B., Rosenberger G., Tekin M., Ozata M. Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am. J. Hum. Genet. 2008;83:511–519. doi: 10.1016/j.ajhg.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim H.G., Ahn J.W., Kurth I., Ullmann R., Kim H.T., Kulharya A., Ha K.S., Itokawa Y., Meliciani I., Wenzel W. WDR11, a WD protein that interacts with transcription factor EMX1, is mutated in idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am. J. Hum. Genet. 2010;87:465–479. doi: 10.1016/j.ajhg.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanchate N.K., Giacobini P., Lhuillier P., Parkash J., Espy C., Fouveaut C., Leroy C., Baron S., Campagne C., Vanacker C. SEMA3A, a gene involved in axonal pathfinding, is mutated in patients with Kallmann syndrome. PLoS Genet. 2012;8:e1002896. doi: 10.1371/journal.pgen.1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young J., Metay C., Bouligand J., Tou B., Francou B., Maione L., Tosca L., Sarfati J., Brioude F., Esteva B. SEMA3A deletion in a family with Kallmann syndrome validates the role of semaphorin 3A in human puberty and olfactory system development. Hum. Reprod. 2012;27:1460–1465. doi: 10.1093/humrep/des022. [DOI] [PubMed] [Google Scholar]
- 14.Pingault V., Bodereau V., Baral V., Marcos S., Watanabe Y., Chaoui A., Fouveaut C., Leroy C., Vérier-Mine O., Francannet C. Loss-of-function mutations in SOX10 cause Kallmann syndrome with deafness. Am. J. Hum. Genet. 2013;92:707–724. doi: 10.1016/j.ajhg.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tornberg J., Sykiotis G.P., Keefe K., Plummer L., Hoang X., Hall J.E., Quinton R., Seminara S.B., Hughes V., Van Vliet G. Heparan sulfate 6-O-sulfotransferase 1, a gene involved in extracellular sugar modifications, is mutated in patients with idiopathic hypogonadotrophic hypogonadism. Proc. Natl. Acad. Sci. USA. 2011;108:11524–11529. doi: 10.1073/pnas.1102284108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Topaloglu A.K., Kotan L.D. Molecular causes of hypogonadotropic hypogonadism. Curr. Opin. Obstet. Gynecol. 2010;22:264–270. doi: 10.1097/GCO.0b013e32833bb425. [DOI] [PubMed] [Google Scholar]
- 17.Doty R.L., Shaman P., Dann M. Development of the University of Pennsylvania Smell Identification Test: a standardized microencapsulated test of olfactory function. Physiol. Behav. 1984;32:489–502. doi: 10.1016/0031-9384(84)90269-5. [DOI] [PubMed] [Google Scholar]
- 18.Brown R.S. Zinc finger proteins: getting a grip on RNA. Curr. Opin. Struct. Biol. 2005;15:94–98. doi: 10.1016/j.sbi.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 19.Chang Y.F., Imam J.S., Wilkinson M.F. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 20.Pitteloud N., Quinton R., Pearce S., Raivio T., Acierno J., Dwyer A., Plummer L., Hughes V., Seminara S., Cheng Y.Z. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J. Clin. Invest. 2007;117:457–463. doi: 10.1172/JCI29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimizu T., Nakazawa M., Kani S., Bae Y.K., Shimizu T., Kageyama R., Hibi M. Zinc finger genes Fezf1 and Fezf2 control neuronal differentiation by repressing Hes5 expression in the forebrain. Development. 2010;137:1875–1885. doi: 10.1242/dev.047167. [DOI] [PubMed] [Google Scholar]
- 22.Hardelin J.P., Dodé C. The complex genetics of Kallmann syndrome: KAL1, FGFR1, FGF8, PROKR2, PROK2, et al. Sex Dev. 2008;2:181–193. doi: 10.1159/000152034. [DOI] [PubMed] [Google Scholar]
- 23.Grumbach M.M. A window of opportunity: the diagnosis of gonadotropin deficiency in the male infant. J. Clin. Endocrinol. Metab. 2005;90:3122–3127. doi: 10.1210/jc.2004-2465. [DOI] [PubMed] [Google Scholar]
- 24.Fukuda T., Sugita S., Inatome R., Yanagi S. CAMDI, a novel disrupted in schizophrenia 1 (DISC1)-binding protein, is required for radial migration. J. Biol. Chem. 2010;285:40554–40561. doi: 10.1074/jbc.M110.179481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimizu T., Hibi M. Formation and patterning of the forebrain and olfactory system by zinc-finger genes Fezf1 and Fezf2. Dev. Growth Differ. 2009;51:221–231. doi: 10.1111/j.1440-169X.2009.01088.x. [DOI] [PubMed] [Google Scholar]
- 26.Eckler M.J., McKenna W.L., Taghvaei S., McConnell S.K., Chen B. Fezf1 and Fezf2 are required for olfactory development and sensory neuron identity. J. Comp. Neurol. 2011;519:1829–1846. doi: 10.1002/cne.22596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe Y., Inoue K., Okuyama-Yamamoto A., Nakai N., Nakatani J., Nibu K., Sato N., Iiboshi Y., Yusa K., Kondoh G. Fezf1 is required for penetration of the basal lamina by olfactory axons to promote olfactory development. J. Comp. Neurol. 2009;515:565–584. doi: 10.1002/cne.22074. [DOI] [PubMed] [Google Scholar]
- 28.Murata K., Tamogami S., Itou M., Ohkubo Y., Wakabayashi Y., Watanabe H., Okamura H., Takeuchi Y., Mori Y. Identification of an olfactory signal molecule that activates the central regulator of reproduction in goats. Curr. Biol. 2014;24:681–686. doi: 10.1016/j.cub.2014.01.073. [DOI] [PubMed] [Google Scholar]
- 29.Hirata T., Nakazawa M., Yoshihara S., Miyachi H., Kitamura K., Yoshihara Y., Hibi M. Zinc-finger gene Fez in the olfactory sensory neurons regulates development of the olfactory bulb non-cell-autonomously. Development. 2006;133:1433–1443. doi: 10.1242/dev.02329. [DOI] [PubMed] [Google Scholar]
- 30.Dodd J., Jessell T.M. Axon guidance and the patterning of neuronal projections in vertebrates. Science. 1988;242:692–699. doi: 10.1126/science.3055291. [DOI] [PubMed] [Google Scholar]
- 31.Engle E.C. Human genetic disorders of axon guidance. Cold Spring Harb. Perspect. Biol. 2010;2:a001784. doi: 10.1101/cshperspect.a001784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cariboni A., Maggi R., Parnavelas J.G. From nose to fertility: the long migratory journey of gonadotropin-releasing hormone neurons. Trends Neurosci. 2007;30:638–644. doi: 10.1016/j.tins.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida M., Suda Y., Matsuo I., Miyamoto N., Takeda N., Kuratani S., Aizawa S. Emx1 and Emx2 functions in development of dorsal telencephalon. Development. 1997;124:101–111. doi: 10.1242/dev.124.1.101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


