Abstract
Evolution drives, and is driven by, demography. A genotype moulds its phenotype’s age patterns of mortality and fertility in an environment; these two patterns in turn determine the genotype’s fitness in that environment. Hence, to understand the evolution of ageing, age patterns of mortality and reproduction need to be compared for species across the tree of life. However, few studies have done so and only for a limited range of taxa. Here we contrast standardized patterns over age for 11 mammals, 12 other vertebrates, 10 invertebrates, 12 vascular plants and a green alga. Although it has been predicted that evolution should inevitably lead to increasing mortality and declining fertility with age after maturity, there is great variation among these species, including increasing, constant, decreasing, humped and bowed trajectories for both long- and short-lived species. This diversity challenges theoreticians to develop broader perspectives on the evolution of ageing and empiricists to study the demography of more species.
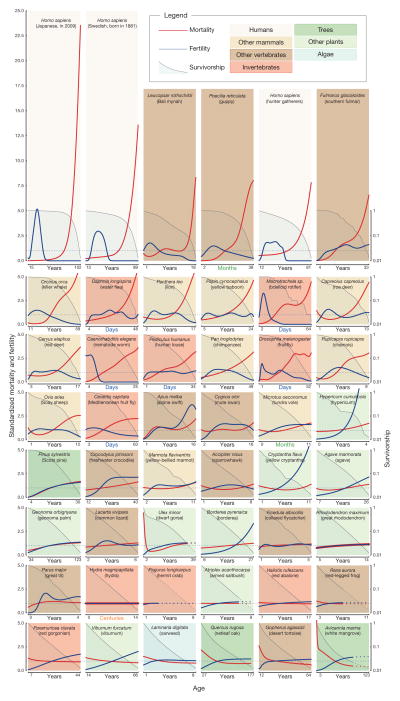
To examine demographic age trajectories across the tree of life, we studied life tables1 (that is, patterns of mortality and fertility over age) and population projection matrices2 for multicellular species from a wide range of taxonomic groups (Fig. 1; see Supplementary Methods for data sources and further rationale). We strived to find species with reliable data and from diverse taxa. From the data for each species we estimated smoothed trajectories of fertility, mortality and survivorship over age. Further research will undoubtedly refine the curves shown for many of the species in Fig. 1 and reveal variation in different environments and for different genotypes, but the general patterns are, we believe, serviceably accurate.
Figure 1. Demographic trajectories.
Relative mortality (red) and fertility (blue) as functions of age, from maturity to the age when only 5% of the adult population is still alive; mortality and fertility are scaled relative to their means. Subplots are arranged in order of decreasing relative mortality at the terminal age. Survivorship (on a log scale) from maturity is depicted by the shaded areas. Broken lines, for trajectories derived from projection matrices, start at the age when cohorts have converged to within 5% of their quasi-stationary distribution (see also Supplementary Methods).
We standardized the demographic trajectories to facilitate comparison. Specifically we standardized the age axis so that it starts at the mean age of reproductive maturity and ends at a terminal age when only 5% of adults are still alive. After this terminal age, sample sizes were usually small and determination of age was often problematic. Fertility and mortality were mean-standardized by dividing age-specific fertility and mortality by the respective weighted average levels of fertility and mortality for all adults alive from maturity to the terminal age (see Methods). We refer to these standardized values as relative fertility and relative mortality. From the highest level of relative mortality at the terminal age (Fig. 1, top left) to the lowest level (Fig. 1, bottom right), species are ordered sequentially, row-by-row and from left-to-right. For the 46 diverse species depicted here, the range of variation in trajectories of fertility and mortality is unexpected. As an indication of variability across species, in modern Japanese women (Fig. 1, top left), mortality at the terminal age (102 years) is more than 20 times higher than the average level of adult mortality, whereas for white mangrove (Avicennia marina; Fig. 1, bottom right) the level of mortality at 123 years is less than half the average adult value.
Such variability is not predicted by the standard evolutionary theories of ageing1,3–6. Such theories provide explanations solely for age patterns of increasing mortality and decreasing fertility from maturity; the disposable soma theory6 does so for species that segregate the germ line from the soma. Furthermore, for those species that show a lifetime increase in mortality, the canonical theory cannot account for the different magnitudes of that increase, although the disposable soma theory points to the crucial importance of trade-offs between the allocation of limited resources to repair and maintenance versus fertility and other imperatives.
Mortality
The most notable pattern is the mortality trajectory for post-industrial humans, exemplified by Japanese women in 2009. The steep rise in relative mortality for the Japanese women is extreme compared with the rise for other species and sharper than that for historical populations such as the Swedish cohort born in 1881 and for hunter-gatherers such as the Aché of Paraguay whose mortality experience may be typical of humans over most of human existence1,7. The increased steepness of the rise of human mortality has largely occurred over the past century, indicating that it was behavioural and environmental change (including advances in health care) and not genetic change that moulded the current pattern7–9. Our close relatives, chimpanzees (Pan troglodytes) and baboons (Papio cynocephalus) also show a rise in mortality with age but far less than that for hunter-gatherers.
In several species mortality declines with age (Fig. 1, bottom row) and, in some cases, notably for the desert tortoise (Gopherus agassizii), the decline persists up to the terminal age. In other cases, an initial decline is followed by more or less constant mortality (for example, netleaf oak, Quercus rugosa). For species for which the underlying data are based on stages, such as dwarf gorse (Ulex minor) or the red-legged frog (Rana aurora), an asymptote is inevitable at older ages8,10. To alert readers to this, the mortality (and fertility and survival) curves derived from stage-classified models are represented by dashed curves in Fig. 1 at ages beyond which a cohort will have converged to within 5% of the quasi-stationary distribution (see Methods).
For most species in Fig. 1 the age pattern of mortality is derived from data on ages rather than stages. For some of these species, mortality levels off at advanced ages (for example, for the collared flycatcher, Ficedula albicollis, the great tit, Parus major, the fruitfly, Drosophila melanogaster) and in others remains constant at all adult ages (for example, for Hydra magnipapillata). For hydra in the laboratory, this risk is so small that we estimate that 5% of adults would still be alive after 1,400 years under those controlled conditions.
Fertility
The fertility trajectories show considerable variation. For humans the trajectories are bell-shaped and concentrated at younger adult ages, but other shapes are apparent in Fig. 1. The patterns for killer whales (Orcinus orca), chimpanzees, chamois (Rupicapra rupicapra) and spar-rowhawks (Accipiter nisus) are also approximately bell-shaped but spread over more of the course of life. Other species show trajectories of gradually increasing fertility (for example, southern fulmars, and the agave, Agave marmorata), asymptotic fertility (for example, tundra voles, Microtus oeconomus), or constant fertility (for example, hydra). In addition to humans and killer whales, bdelloid rotifers (Macrotrachela sp.), nematode worms (Caenorhabditis elegans) and Bali mynah birds (Leucopsar rothschildi) have post-reproductive life spans, which lends further support to the idea that this phenomenon may be widespread3–6,11.
Axes of senescence
Although the demographic trajectories in Fig. 1 vary widely, most of the 46 species can be roughly classified along a continuum of senescence; running from strong deterioration with age, to negligible deterioration, to negative senescence12 and improvement with age. However, there are some deviations, for example, for Soay sheep (Ovis aries) and dwarf gorse, which show mortality reductions with adult age followed by deterioration. Fertility patterns show similar diversity.
A fast–slow continuum has been proposed to order species from those with short lives and intense early reproduction to those with long lives and an extended reproductive period13–16. Figure 1 displays mortality and fertility over the adult lifespan; pre-reproductive mortality trajectories are also of interest but beyond the scope of this article. If distinguished by the length of life, then fast and slow life histories are scattered irregularly across Fig. 1. Lifespans range from 1,400 years for the hydra to just 25 days for nematode worms. Species with fast life histories, such as water fleas (Daphnia longispina), are followed in Fig. 1 by species with slow life histories, such as the lion, and those with slow life histories, such as the chimpanzee, occur adjacent to those with fast life histories, such as the human louse (Pediculus humanus) and the fruitfly (D. melanogaster). Furthermore, species with very different life spans can display similar patterns of mortality, fertility and survivor-ship. For example, the water flea’s trajectories are similar to the fulmar’s, although water fleas reach advanced old age at 48 days, whereas the fulmars do so at 33 years.
If senescence is measured by how long it takes for death rates to rise from some level to a higher level, then long-lived species senesce slowly. It is more interesting to define senescence by the sharpness or abruptness rather than the speed of the increase in mortality. Baudisch8 distinguishes the pace of life; that is, whether reproduction is fast and life spans are short or reproduction is slow and life spans are long, from the shape of mortality and fertility trajectories (whether mortality rises sharply with age and fertility falls sharply or whether mortality and fertility levels are more constant). One measure of pace, the measure that we have used, is the terminal age to which only 5% of adults survive; this measure is in days or years or some other unit of time. One measure of shape, the measure that we have used, is the ratio of mortality at the terminal age to the average level of adult mortality; this time-invariant measure does not change if time is measured in days versus years. More senescent species, with sharper increases in mortality with age, have higher values of this measure of shape.
The measure can be used to explore further the unexpected lack of association between the length of life and the degree of senescence. Among the first 24 graphs, those with the sharpest senescence, 11 species have relatively long life spans and 13 have relatively short life spans. Among the final 24 graphs, those with less senescence, 13 species have relatively long life spans and 11 have relatively short life spans. This weak negative association between the length of life and the degree of senescence is reflected in a weak Spearman rank correlation of –0.13, which is not significantly different from zero (P =0.362). The Spearman correlations are also non-significant when assessed for animals (P =0.414) and for plants (P =0.07) examined separately. If the 12 plants in Fig. 1 are cross-tabulated as longer or shorter lived, and as more or less senescent, then three species fall into each of the four categories. Hence the data support Baudisch’s8 conjecture that pace and shape may be two orthogonal axes of life histories.
A survivorship curve indicates the proportion of individuals that are still alive at a given age. In Fig. 1, we plot survivorship from reproductive maturity on a logarithmic scale. If mortality increases with age, the log-survivorship curve is concave. If mortality is independent of age, log-survivorship is linear (for example, roughly from the hydra to the red abalone (Haliotis rufesens) in Fig. 1). For species with death rates that decline with age, the curve is convex (for example, from the red-legged frog to the white mangrove at the bottom of Fig. 1). The classification of survivorship curves into concave, linear and convex curves is known among biologists as type I, II and III, respectively17,18, but normally the curves are plotted for lifespans starting at birth rather than at maturity. When the evolutionary theory of ageing3–6 was being developed, there was very little empirical evidence for type III survivorship for adults and little evidence for type II survivorship. The widespread recognition that traditional theories of ageing predict adult senescence to be a universal trait led researchers to strive to find evidence for senescence in, for example, the mute swan (Cygnus olor)19. For this species, fertility does decline and mortality does increase at the oldest ages. However, the overall life course is characterized by fertility that increases and then slowly declines and by roughly constant mortality: the log-survivorship curve is nearly straight. It is clear from our analyses that the full spectrum of type I, II and III survivorship curves are found for adults in nature.
Phylogenetic patterns
Phylogenetic relatedness seems to have some role in the order of species in Fig. 1, as shown by taxonomic clustering of mortality, fertility and survivorship patterns. All mammals are clustered in the top part of Fig. 1, whereas birds are somewhat more scattered, from the Bali mynah in the first row to the great tit in the seventh row. Amphibians and reptiles are found in the lower half of the panel, with flat mortality shapes and almost no overlap with mammals. In contrast, invertebrates are scattered across the continuum of senescence, with bdelloid rotifers and water fleas sharing the mammalian mortality pattern. The plants in our sample tend to occur lower in our ordering, with the first being Hypericum cumulicola. Although some angiosperm species seem to senesce20–22, many angiosperm species seem not to23, perhaps as an artefact of the use of stage-based data10. The only alga in our data set, oarweed (Laminaria digitata), falls in the last row.
Such clustering within broad taxonomic levels of kingdom (plants, animals), or class (mammals, birds), suggests that primitive traits related to the bauplan of species may have a pivotal role in determining patterns of ageing. In fact, the evolutionary conservatism of mechanistic determinants of ageing has been highlighted by genetic studies24 and it has been suggested that asexual reproduction25, modularity26, lack of germ-line sequestration from the soma27,28, the importance of protected niches29, regenerative capacity, and the paucity of diverse cell types30, may facilitate the escape from senescence in some clades. Many of the species in the lower half of Fig. 1—the reptiles, vascular plants, alga, and coral—continue to grow after reproductive maturity to sizes much larger than those at maturity. For these indeterminate growers, mortality is approximately constant or decreases somewhat with age, whereas fertility is more or less constant or increases to some extent. Species with indeterminate growth may exhibit patterns of senescence that are fundamentally different from those of species with determinant growth12,31–33.
Approximately constant mortality and fertility are experienced by vertebrates such as collared flycatchers and red-legged frogs, invertebrates such as hermit crabs (Pagurus longicarpus) and red abalone, and vascular plants such as great rhododendron (Rhododendron maximum) and armed saltbush (Atriplex acanthocarpa), with the age at 5% survivorship ranging from 5 years for the collared flycatcher to 14 centuries for hydra. It remains to be seen whether the similarity of patterns of mortality, fertility and survivorship among disparate groups of species is a coincidence or represents convergent solutions to similar evolutionary challenges.
Continuing the exploration of ageing
Although hundreds of theories have been proposed to explain the proximate mechanisms of ageing34,35, theories to explain the ultimate evolutionary causes of the varieties of ageing, illustrated by the diverse range of trajectories in Fig. 1, are in their infancy. However, scattered studies suggest profitable directions for research. It is only recently that researchers have extended their analyses beyond the traditional age-structured framework36; more complex demographic models show that selection gradients in clonal or stage-structured organisms can be non-monotonic37–40. As recognized in the disposable soma theory6, differences in life-history constraints among species, and the resulting differences in optimal resource allocation among vital processes provide a promising direction for explaining empirical observations about diverse fertility32,37–39,41,43 and mortality32,41–43 trajectories. However, current theoretical approaches do not yet explain in detail why senescence has evolved in some species and not in others. Data sets that are currently available for research on ageing are taxonomically biased: high-quality data on hundreds of mammalian and bird species exist but data on other vertebrate taxa and on invertebrates are sparse. There is very limited knowledge of the age patterns of mortality and fertility in species of algae, fungi and bacteria32,43,44.
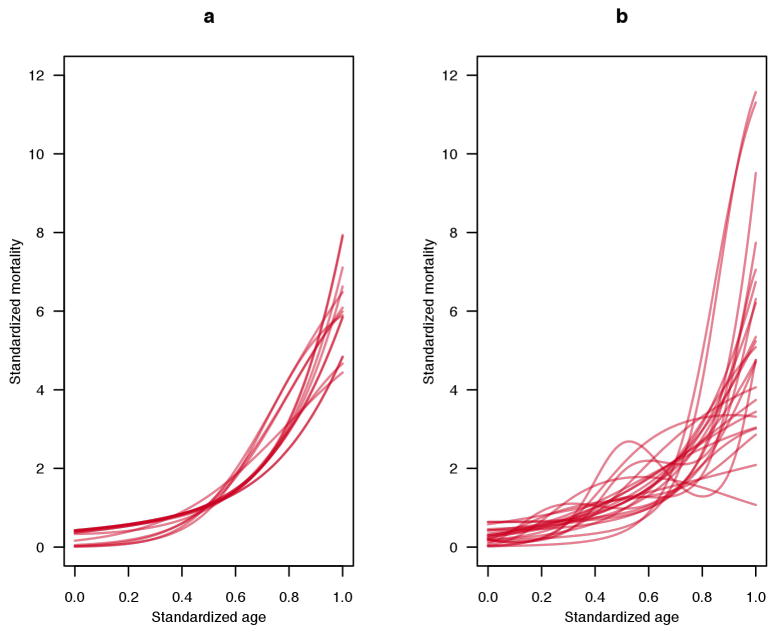
The mortality and fertility trajectories of any species depend on the environment in which they are measured. Most human experience is bounded by the trajectories of modern Japanese and the hunter-gatherers in Fig. 1. Although population ecologists have long studied the responses of mortality and fertility to environmental factors, few studies have focused on the shape of the age trajectories. Environmental and genotypic variation has been documented in laboratory studies of nematode worms, medflies, Drosophila and other model species45, and in a field study of Plantago20. Available evidence suggests that variation can be considerable for a species but that the qualitative shapes of mortality and fertility trajectories are similar, as illustrated by humans in Fig. 1 (see the Supplementary Note and Extended Data Fig. 1, which highlights intraspecific variation in the mortality trajectories of laboratory rats (Rattus norvegicus) and mice (Mus musculus)). In addition to the lack of data for most species, and for variation within a species, little information is available on mortality at advanced ages beyond the age cut-off in Fig. 1. In the species for which such data are available, mortality approaches a plateau at the oldest ages (for example, for humans, fruitflies (D. melanogaster) and nematode worms) or declines (for Mediterranean fruitflies, Ceratitis capitata)45–47. The deceleration of mortality at high ages is more apparent if death rates are plotted on a log scale rather than the linear scale used in Fig. 1 (ref. 45).
Deeper understanding of the evolutionary demography of ageing depends on the compilation of demographic data on diverse species investigated in the wild as well as in laboratories and zoos8, and on the development of more inclusive theories that can account for negligible and negative senescence42,48 as well as for the steepness of deterioration with age in senescent species. In such empirical and theoretical studies, researchers should guard against anthropocentric intuition about ageing: humans, especially modern humans, are extreme outliers in Fig. 1.
METHODS SUMMARY
Selection of examples
We aimed to examine demographic trajectories for organisms across the tree of life. We therefore chose representative data sets compiled from the published literature for the major groups of organisms including vertebrate and invertebrate animals, plants and algae. Within the vertebrates we included exemplars of every major clade, including primates and other mammals, birds, reptiles, amphibians and fish. Representatives for the invertebrates included insects, molluscs, cnidarians and a crustacean. In the plant group we included both gymnosperms and angiosperms and, finally, we included a green alga. We favoured data sets that covered longer time periods, with larger sample sizes and, when possible, we preferred data sets that included information on realized reproduction and recruitment to those that simply recorded reproductive output. In addition, for dioecious species, we favoured data sets for females. See Supplementary Methods 1 and 2 for details.
Calculation of standardised trajectories
We classified the studies as: first, cohort studies; second, period studies with number at risk and numbers dying within a period; third, period studies depicting an age structure at a single point in time; or fourth, stage-structured population projection matrices (see Supplementary Methods 2 for details). We considered mortality and fertility trajectories from the age at maturity to the age at which 5% survivorship from maturity occurs. The trajectories of all data types, except the projection matrix data, were smoothed using P-splines49. We then calculated the force of mortality (μx) and fertility rate (mx) before standardizing them by dividing them by the respective averages, weighted by survivorship from maturity (lx).
Extended Data
Extended Data Figure 1. Standardized mortality trajectories.
a, Trajectories for laboratory rats. b, Trajectories for laboratory mice. Each line represents a different strain, sex or population (see Supplementary Methods for sources). We standardized the age axis to consider the trajectories from age at maturity to the age at which 5% survivorship from maturity occurs. The trajectories were smoothed using P-splines. We then calculated the force of mortality (μx) and standardized it by dividing by the average value, weighted by survivorship from maturity (lx). Note that the sample sizes in most cases were small (approximately 50 to 60 individuals) and thus random fluctuations may lead to erratic curves in some cases.
Supplementary Material
Acknowledgments
We thank S. Alberts for data on baboon demography, J. Curtsinger for data on Drosophila demography and O. Burger, D. Levitis, B. Pietrzak, F. Quade, F. Ringelhan and L. Vinicius for contributing published data about various species. J.W.V. and A.S. acknowledge support from NIH grant PO1 AG-031719. H.C. acknowledges a Research Award from the Alexander von Humboldt Foundation and Advanced Grant 322989 from the European Research Council. R.S.-G. acknowledges support from ARC DP110100727. A.B. acknowledges funding from the Max Planck Society to establish the Max Planck Research Group ‘Modeling the Evolution of Aging’.
Footnotes
Online Content Any additional Methods, Extended Data display items and Source Data are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Information is available in the online version of the paper.
Author Contributions
This research project was initiated by J.W.V. A.S. wrote the first draft; O.R.J., with help from A.S., R.S.-G., H.C., A.B. and J.W.V., wrote subsequent drafts; J.W.V. and O.R.J. completed the final draft. The Figure was produced by O.R.J. with suggestions from J.W.V., A.S., A.B. and H.C. A.B. suggested the method of standardization and the distinction between shape and pace. C.G.C. developed methods to smooth mortality and fertility trajectories. H.C. and R.S.-G. contributed to the analysis of stage-classified species. A.S., R.S.-G., O.R.J. and H.C. each provided data, derived from the literature, for several species. R.S. contributed unpublished data for hydra; J.E., J.D. and M.B.G. for Borderea; R.S.-G. and B.B.C. for Cryptantha; and E.M. and P.F.Q.-A. for Hypericum. O.R.J., A.S., R.S.-G. and H.C. screened the species for data quality.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Chiang CL. The life table and its applications. Krieger Publishing; 1984. [Google Scholar]
- 2.Caswell H. Matrix population models. Sinauer Associates; 2001. [Google Scholar]
- 3.Medawar PB. An unsolved problem of biology. H. K. Lewis; 1952. [Google Scholar]
- 4.Williams G. Pleiotropy, natural selection, and the evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]
- 5.Hamilton WD. The moulding of senescence by natural selection. J Theor Biol. 1966;12:12–45. doi: 10.1016/0022-5193(66)90184-6. [DOI] [PubMed] [Google Scholar]
- 6.Kirkwood TBL. Evolution of ageing. Nature. 1977;270:301–304. doi: 10.1038/270301a0. [DOI] [PubMed] [Google Scholar]
- 7.Burger O, Baudisch A, Vaupel JW. Human mortality improvement in evolutionary context. [15 October 2012];Proc Natl Acad Sci USA. doi: 10.1073/pnas.1215627109. http://dx.doi.org/10.1073/pnas.1215627109. [DOI] [PMC free article] [PubMed]
- 8.Baudisch A. The pace and shape of ageing. Methods Ecol Evol. 2011;2:375–382. [Google Scholar]
- 9.Oeppen J, Vaupel JW. Broken limits to life expectancy. Science. 2002;296:1029–1031. doi: 10.1126/science.1069675. [DOI] [PubMed] [Google Scholar]
- 10.Horvitz CC, Tuljapurkar S. Stage dynamics, period survival, and mortality plateaus. Am Nat. 2008;172:203–215. doi: 10.1086/589453. [DOI] [PubMed] [Google Scholar]
- 11.Cohen AA. Female post-reproductive lifespan: a general mammalian trait. Biol Rev Camb Philos Soc. 2004;79:733–750. doi: 10.1017/s1464793103006432. [DOI] [PubMed] [Google Scholar]
- 12.Vaupel JW, Baudisch A, Dölling M, Roach DA, Gampe J. The case for negative senescence. Theor Popul Biol. 2004;65:339–351. doi: 10.1016/j.tpb.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Gaillard JM, et al. An analysis of demographic tactics in birds and mammals. Oikos. 1989;56:56–76. [Google Scholar]
- 14.Promislow DEL, Harvey PH. Living fast and dying young: a comparative analysis of life-history variation among mammals. J Zool (Lond ) 1990;220:417–437. [Google Scholar]
- 15.Stearns SC. The Evolution of Life Histories. Oxford Univ. Press, USA; 1992. [Google Scholar]
- 16.Jones OR, et al. Senescence rates are determined by ranking on the fast-slow life-history continuum. Ecol Lett. 2008;11:664–673. doi: 10.1111/j.1461-0248.2008.01187.x. [DOI] [PubMed] [Google Scholar]
- 17.Pearl R, Miner JR. Experimental studies on the duration of life. XIV. The comparative mortality of certain lower organisms. Q Rev Biol. 1935;10:60–79. [Google Scholar]
- 18.Deevey ES. Life tables for natural populations of animals. Q Rev Biol. 1947;22:283–314. doi: 10.1086/395888. [DOI] [PubMed] [Google Scholar]
- 19.Charmantier A, Perrins C, McCleery RH, Sheldon BC. Quantitative genetics of age at reproduction in wild swans: Support for antagonistic pleiotropymodels of senescence. Proc Natl Acad Sci USA. 2006;103:6587–6592. doi: 10.1073/pnas.0511123103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shefferson RP, Roach DA. Longitudinal analysis in Plantago: strength of selection and reverse age analysis reveal age-indeterminate senescence. J Ecol. 2013;101:577–584. doi: 10.1111/1365-2745.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tuomi J, et al. Prolonged dormancy interacts with senescence for two perennial herbs. J Ecol. 2013;101:566–576. [Google Scholar]
- 22.Salguero-Gómez R, Shefferson RP, Hutchings MJ. Plants do not count… or do they? New perspectives on the universality of senescence. J Ecol. 2013;101:545–554. doi: 10.1111/1365-2745.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baudisch A, et al. The pace and shape of senescence in angiosperms. J Ecol. 2013;101:596–606. [Google Scholar]
- 24.McElwee JJ, et al. Evolutionary conservation of regulated longevity assurance mechanisms. Genome Biol. 2007;8:R132. doi: 10.1186/gb-2007-8-7-r132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bell G. Measuring the cost of reproduction. I. The correlation structure of the life table of a plank rotifer. Evolution. 1984;38:300–313. doi: 10.1111/j.1558-5646.1984.tb00289.x. [DOI] [PubMed] [Google Scholar]
- 26.Franco M, Silvertown J. Life history variation in plants: an exploration of the fast-slow continuum hypothesis. Phil Trans R Soc B. 1996:1341–1348. [Google Scholar]
- 27.Buss LW. Diversification and germ-line determination. Paleobiology. 1988;14:313–321. [Google Scholar]
- 28.Martínez DE, Levinton JS. Asexual metazoans undergo senescence. Proc Natl Acad Sci USA. 1992;89:9920–9923. doi: 10.1073/pnas.89.20.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baudisch A, Vaupel J. Senescence vs. sustenance: evolutionary-demographic models of aging. Demogr Res. 2010;23:655–668. [Google Scholar]
- 30.Martínez DE. Mortality patterns suggest lack of senescence in hydra. Exp Gerontol. 1998;33:217–225. doi: 10.1016/s0531-5565(97)00113-7. [DOI] [PubMed] [Google Scholar]
- 31.Finch CE. Longevity, Senescence and the Genome. Univ. Chicago Press; 1994. [Google Scholar]
- 32.Baudisch A. Inevitable Aging? Contributions to Evolutionary-Demographic Theory. Springer; 2008. [Google Scholar]
- 33.Charnov EL. Reproductive constraints and the evolution of life histories with indeterminate growth. Proc Natl Acad Sci USA. 2001;98:9460–9464. doi: 10.1073/pnas.161294498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medvedev ZA. An attempt at a rational classification of theories of ageing. Biol Rev Camb Philos Soc. 1990;65:375–398. doi: 10.1111/j.1469-185x.1990.tb01428.x. [DOI] [PubMed] [Google Scholar]
- 35.Kirkwood TBL. Systems biology of ageing and longevity. Phil Trans R Soc B. 2010;366:64–70. doi: 10.1098/rstb.2010.0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Charlesworth B. Evolutionin Age-structured Populations. Cambridge Univ. Press; 1994. [Google Scholar]
- 37.Caswell H. Matrix models and sensitivity analysis of populations classified by age and stage: a vec-permutation matrix approach. Theor Ecol. 2012;5:403–417. [Google Scholar]
- 38.Pedersen B. An evolutionary theory of clonal senescence. Theor Popul Biol. 1995;47:292–320. [Google Scholar]
- 39.Caswell H. In: Population Biology and Evolution of Clonal Organisms. Jackson JBC, Bus LW, Cook RE, editors. Yale Univ. Press; 1985. pp. 187–224. [Google Scholar]
- 40.Caswell H, Salguero-Gomez R. Age, stage and senescence in plants. J Ecol. 2013;101:585–595. doi: 10.1111/1365-2745.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orive ME. Senescence in organisms with clonal reproduction and complex life histories. Am Nat. 1995;145:90–108. [Google Scholar]
- 42.Baudisch A, Vaupel JW. Getting to the root of aging. Science. 2012;338:618–619. doi: 10.1126/science.1226467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gadgil M, Bossert WH. Life historical consequences of natural selection. Am Nat. 1970;104:1–24. [Google Scholar]
- 44.Schaffer WM. Selection for optimal life histories — effects of age structure. Ecology. 1974;55:291–303. [Google Scholar]
- 45.Vaupel JW, et al. Biodemographic trajectories of longevity. Science. 1998;280:855–860. doi: 10.1126/science.280.5365.855. [DOI] [PubMed] [Google Scholar]
- 46.Chen J, et al. A demographic analysis of the fitness cost of extended longevity in Caenorhabditis elegans. J Gerontol A Biol Sci Med Sci. 2007;62:126–135. doi: 10.1093/gerona/62.2.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaupel JW. Biodemography of human ageing. Nature. 2010;464:536–542. doi: 10.1038/nature08984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finch CE. Update on slow aging and negligible senescence - A mini-review. Gerontology. 2009;55:307–313. doi: 10.1159/000215589. [DOI] [PubMed] [Google Scholar]
- 49.Eilers PHC, Marx BD. Flexible smoothing with B-splines and penalties. Stat Sci. 1996;11:89–121. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.