Abstract
Sinorhizobium meliloti is an alpha-proteobacterium able to induce nitrogen-fixing nodules on roots of specific legumes. In order to propagate in the soil and for successful symbiotic interaction the bacterium needs to sequester metals like iron and manganese from its environment. The metal uptake has to be in turn tightly regulated to avoid toxic effects. In this report we describe the characterization of a chromosomal region of S. meliloti encoding the sitABCD operon and the putative regulatory fur gene. It is generally assumed that the sitABCD operon encodes a metal-type transporter and that the fur gene is involved in iron ion uptake regulation. A constructed S. meliloti sitA deletion mutant was found to be growth dependent on Mn(II) and to a lesser degree on Fe(II). The sitA promoter was strongly repressed by Mn(II), with dependence on Fur, and moderately by Fe(II). Applying a genome-wide S. meliloti microarray it was shown that in the fur deletion mutant 23 genes were up-regulated and 10 genes were down-regulated when compared to the wild-type strain. Among the up-regulated genes only the sitABCD operon could be associated with metal uptake. On the other hand, the complete rhbABCDEF operon, which is involved in siderophore synthesis, was identified among the down-regulated genes. Thus, in S. meliloti Fur is not a global repressor of iron uptake. Under symbiotic conditions the sitA promoter was strongly expressed and the S. meliloti sitA mutant exhibited an attenuated nitrogen fixation activity resulting in a decreased fresh weight of the host plant Medicago sativa.
The root nodule bacteria of the genera Rhizobium, Bradyrhizobium, Mesorhizobium, Azorhizobium, and Sinorhizobium, collectively known as rhizobia, are able to establish a nitrogen-fixing symbiosis with their respective leguminous host plants. During this symbiotic interaction the rhizobia first induce the formation of root nodules on their respective hosts, colonize the nodules, and finally, after differentiation into bacteroids, reduce atmospheric nitrogen to ammonia. The fixed nitrogen is then delivered to the host plants in the form of alanine or ammonia. The plant in turn supplies the bacteria with various nutrients. This symbiotic interaction has been described in detail recently (44, 57, 61).
One interesting aspect of the rhizobium-legume symbiosis is the influence of transition metals on nodule formation and nitrogen fixation. A number of these micronutrients are known to be essential for bacterial metabolism (36), yet the mechanisms of metal import and their regulation in rhizobia are largely unknown. One of the most important transition metals is iron, which due to its importance in metabolic electron transport chains is essential for almost all known organisms. Despite the fact that iron is the fourth most abundant element on earth its availability to microorganisms is restricted because of its low solubility under aerobic conditions and physiological pH values. Thus, bacteria have evolved a number of strategies to overcome iron deficiencies (26). Most of these mechanisms involve specialized ABC transporters, which can be classified into three families, namely the siderophore-type, the ferric-type, and the metal-type transporters (37).
The metal-type transporters appear to be less specific for iron than the other transporter types. The best-characterized metal-type iron ABC transporters are the YfeABCD system of Yersinia pestis, the SitABCD system of Salmonella enterica serovar Typhimurium, and the more recently characterized SitABCD transporter of Shigella flexneri. Metal-type ABC transporters were first shown to be involved in the utilization of chelated iron (6, 72), but evidence also exists for their involvement in Mn(II) acquisition (5, 8, 54). The S. flexneri SitABCD and the Y. pestis YfeABCD transporters were repressed by iron and manganese (6, 54, 72), whereas for Salmonella only iron-dependent repression has been investigated so far. In all cases a Fur (ferric uptake regulator) protein is necessary for iron-dependent repression. Additionally, Fur was also shown to be involved in the repression of the Y. pestis yfeABCD operon by Mn(II), whereas in S. flexneri Mn(II)-dependent repression of the sitABCD operon was attributed to the manganese-binding repressor MntR (54).
In many gram-negative bacteria Fur is the main repressor of iron-responsive genes (27). In the classic model of Fur regulation established for Escherichia coli the Fur protein utilizes Fe(II) as a cofactor and binds to specific sequence elements in the promoter regions of these genes, termed Fur boxes, and thereby inhibits gene expression under iron-replete conditions (3, 13). Besides its role as a repressor of iron uptake mechanisms Fur has also been found to be involved in the regulation of alternative sigma factor and activator genes (45, 46), stress response genes (19, 28-31), energy metabolism genes (39, 59, 64), and virulence-associated genes (9, 11, 22, 23, 40, 69). Thus, in many bacterial species the role of Fur extends beyond that of a negative regulator of iron acquisition systems.
In Sinorhizobium meliloti, the microsymbiont of alfalfa, a putative ferric uptake regulator has been identified in the direct vicinity of a metal-type transporter (12); however, the role of Fur in S. meliloti has not been investigated yet. Evidence exists that at least in some rhizobia like Rhizobium leguminosarum the Fur protein is not a global regulator of iron metabolism (70). The adjoining metal-type transporter, designated SitABCD, was found to be involved in manganese utilization of S. meliloti 2042 grown in the presence of a chelator (48). The SitABCD homologues identified in Y. pestis, S. enterica serovar Typhimurium, and S. flexneri contributed to virulence in their animal hosts (5, 35, 54). The relevance of metal-type transporters to bacterium-plant interactions is unclear though. Platero et al. reported that transposon insertions in the sitB and sitD genes of S. meliloti 2042 do not affect the ability of the bacterium to establish nitrogen-fixing symbiosis with alfalfa (48). In contrast, transcriptome and proteome analysis performed with S. meliloti 1021 indicated a possible involvement of the SitABCD transporter in symbiosis (2, 14).
The work of Wexler et al. (70) demonstrated that in rhizobia the Fur protein plays a different role than in other mostly enterobacterial organisms studied so far. Therefore, we applied S. meliloti whole-genome microarrays to investigate the role of this regulator on a genome-wide scale. Furthermore, we examined the regulation of the sitABCD operon of S. meliloti, since previous studies of pathogenic bacteria demonstrated that it is a target of the regulatory Fur protein. Additionally, the roles of the sitABCD operon in metal uptake in the free-living state and during symbiosis were analyzed in order to determine the relevance of the SitABCD transporter for symbiosis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are shown in Table 1. Strains of E. coli were routinely cultured at 37°C in antibiotic medium no. 3 (PA) (Oxoid, Wesel, Germany). S. meliloti strains were cultivated at 30°C either in tryptone yeast (TY) complex medium (7) or in Vincent minimal medium (VMM) (68). For growth assays VMM was prepared without any iron sources and was designated VMM*. Iron and other metals were added after autoclaving as indicated for the respective bioassays. Fe(III) was added as FeCl3, Fe(II) was added as FeSO4, and Mn(II) was added as MnSO4. Where appropriate, antibiotics were added at the following concentrations: neomycin (100 μg ml−1), tetracycline (5 or 10 μg ml−1), kanamycin (50 μg ml−1), and streptomycin (600 μg ml−1). For growth under low-iron conditions glassware was washed with 6 M HCl and rinsed thoroughly with water.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristic(s) | Reference |
|---|---|---|
| S. meliloti | ||
| Rm1021 | Spontaneous mutant of wild-type strain RU47, Smr | 43 |
| Rm1021ΔsitA/Rm1021-02509-01 | Rm1021 derivative, ΔsitA | This study |
| Rm1021Δfur/Rm1021-02510-01 | Rm1021 derivative, Δfur | This study |
| Rm1021Δfur(::pTCB1) | Rm1021Δfur carrying pTCB1 integrated into the chromosome | This study |
| E. coli | ||
| DH5αMCR | F−endA1 supE44 thi-1 λ−recA1 gyrA96 relA1 deoR Δ(lacZYA-argF)U169 φ80dlacZΔM15 mcrA Δ(mrr hsdRMS mcrBC) | 24 |
| S17-1 | E. coli 294::[RP4-2(Tc::Mu)(Km::Tn7)] pro res ΔrecA Tpr | 60 |
| Plasmids | ||
| pK18mobsacB | pUC18 derivative, sacB lacZα Kmr, mobilizable | 56 |
| pK18mob | pUC18 derivative, lacZα Kmr mobilizable | 56 |
| pTCB1 | pK18mob containing a ≈1.5-kb fragment of the S. meliloti chromosome containing the fur gene | This study |
| pTC02510del | pK18mobsacB carrying a Δfur construct | This study |
| pTC02509del | pK18mobsacB carrying a ΔsitA construct | This study |
| pJP2 | pTR102 GUS with artificial multiple cloning site, Apr Tcr | 50 |
| pTCC1 | pJP2 with a 762-bp fragment containing the sitA promoter, Apr Tcr | This study |
DNA manipulations.
The protocols of Sambrook et al. (55) were used for routine manipulations of plasmid and chromosomal DNA. Mutated DNA fragments containing either a 261-bp deletion in the fur gene or a 585-bp deletion in the sitA gene were constructed by gene SOEing (33). In a first PCR step, regions up- and downstream of the desired deletion were amplified and then fused in a second PCR. The obtained deletion constructs were subsequently cloned into the suicide vector pK18mobsacB, which allows sucrose selection for vector loss (56). The resulting plasmids were conjugated into S. meliloti via E. coli S17-1 to introduce deletions by allelic exchange. Mutants were verified by PCR and Southern hybridizations.
In order to analyze the expression of the sitABCD operon and possible regulation by Fur, the sitA promoter region (Fig. 1) was amplified and cloned into the promoter test vector pJP2 (50) in front of a gusA gene. The resulting plasmid, designated pTCC1, was introduced into the S. meliloti 1021 wild-type strain (Rm1021) and the fur mutant (Rm1021Δfur) for expression analyses.
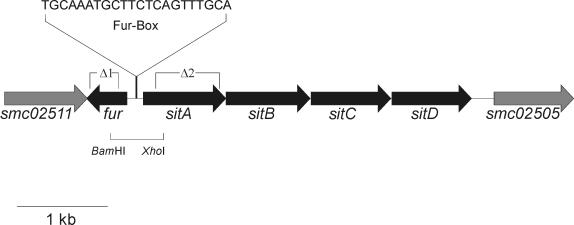
FIG. 1.
Genetic organization of a DNA region of the S. meliloti chromosome containing the fur gene and the sitABCD operon. The DNA region of the S. meliloti genome containing the fur gene, the sitABCD operon, and the two neighboring genes, smc02511 and smc02505, of unknown function, is shown. The putative Fur box sequence as identified by an HMM-based search is indicated. The deletions Δ1 and Δ2 were used to construct mutations in the genes fur and sitA, respectively. The fur deletion Δ1 ranges from bases 140 to 400 of the fur gene, and the S. meliloti strain carrying the deletion Δ1 was designated Rm1021Δfur. The sitA deletion Δ2 ranges from bases 312 to 896 of the sitA gene, and the strain carrying the deletion Δ2 was named Rm1021ΔsitA. In addition, the 760-bp BamHI/XhoI fragment containing most probably the sitA promoter was used to construct the promoter test vector pTCC1 for sitA promoter studies.
Growth inhibition of S. meliloti strains by the compounds H2O2 and MnCl2.
For zone inhibition assays 100-μl portions of overnight cultures of S. meliloti were added to 2 ml of prewarmed 0.7% top agar and layered onto solid TY plates. After the top agar had hardened a sterile filter disk was placed in the middle of the plate. Fifteen microliters of either H2O2 in various concentrations between 0 and 35 mM or MnCl2 with a concentration between 0 and 250 μM was applied to the filter. The inhibition zone was measured after 48 h of incubation at 30°C.
Chromazurol S (CAS) liquid assay to determine siderophore production in S. meliloti cultures.
CAS assay solutions for siderophore detection were prepared as described by Schwyn et al. (58). Supernatants containing the siderophores of S. meliloti cultures grown in TY medium were mixed 1:1 with CAS assay solution. After equilibrium was reached, the absorbance was measured at 630 nm. Relative siderophore activity was determined as optical density ratios of different cultures.
Expression analysis of the S. meliloti sitA promoter, using a gus reporter gene construct.
In order to quantitatively determine the activity of the sitA promoter, exponential-phase cultures (optical density at 580 nm of 0.6 to 0.8) of S. meliloti strains carrying the promoter test vector pTCC1 were centrifuged and diluted into saline buffer (0.89% [wt/vol] NaCl) to 2 × 108 CFU ml−1; 310 μl of the culture was mixed with 1,190 μl of buffer (50 mM sodium phosphate, 50 mM dithiothreitol, 1 mM EDTA [pH 7]). Toluene was added, and the mixture was vortexed and then incubated at 37°C for ca. 30 min to remove the toluene. After a short centrifugation to pellet the cell debris, 290 μl of the mixture was pipetted into each well of a 96-well suspension culture plate. The reaction was started by adding 10 μl of p-nitrophenyl β-glucuronide (10.5 mg/ml; Serva, Heidelberg, Germany) to each well. The plates were incubated at 37°C for 2 h in a FLUOstar Galaxy plate reader (BMG Labtechnologies, Offenburg, Germany). The reader was set to measure the optical density at 405 nm every 5 min after an initial incubation of 45 min. The results from 10 time points for six cultures with eight parallel probes each were used to calculate the mean expression level.
In order to visualize β-glucuronidase activity in the nodules, the nodules were harvested, cut with a Vibratome into slices with a thickness of 60 μm each, and stained with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-Gluc) (Serva) in a final concentration of 2 mM in Tris/NaCl buffer (100 mM Tris/HCl, 50 mM NaCl [pH 7]) for 2 h at 37°C. Digital images were taken with an Olympus C2000Z camera mounted on an Olympus BH-2 microscope.
Assays to determine nitrogen-fixation efficiency in the S. meliloti-M. sativa symbiosis.
Nodulation tests were performed as described by Rolfe et al. (52). Solutions a to c for the plant agar plates (a: 294 g of CaCl2 · 2H2O/liter; b: 50 g of KH2PO4/liter; c: 123 g of MgSO4/liter, 87 g of K2SO4/liter, 0.247 g of H3BO3/liter, 0.288 g of ZnSO4/liter, 0.1 g of CuSO4 · 5H2O/liter, 0.056 g of CoSO4 · 7H2O/liter, 0.048 g of NaMo4 · 2H2O/liter) were autoclaved separately. One-half milliliter of each solution was added to 1 liter of autoclaved water containing 15 g of agar. Alfalfa (M. sativa L. cv. europe) seeds were sterilized with 32% HCl for 30 min and then washed with sterile water. After germination seedlings were placed on nodulation plates and inoculated with equal amounts (around 6 × 107 CFU each) of washed wild-type and mutant cells. Plants were weighed after 30 days of growth, and nitrogen fixation activity was tested by the acetylene reduction assay (52).
S. meliloti transcript profiling using the genome-wide SM6kPCR microarray.
S. meliloti wild-type (Rm1021) and fur mutant (Rm1021Δfur) strains were cultured in VMM in 250-ml Ehrlenmeyer flasks with shaking at 150 rpm to an optical density at 580 nm of 0.9 before harvesting. Cells were centrifuged (10,000 × g, 1 min, 4°C) and afterwards immediately frozen in liquid nitrogen. For total RNA isolation the RNeasy Mini Kit (Qiagen, Hildesheim, Germany) was used. Cells were disrupted in RLT buffer provided in the kit in Fast Protein tubes (Qbiogene, Carlsbad, Calif.), using a Ribolyser (Hybaid, Heidelberg, Germany) (30 s; speed, 6.5) before purification according to the RNeasy Mini Kit RNA purification protocol. Fluorescent labeling of cDNA by amino-allyl dye coupling was performed as described by de Risi et al. (http://www.microarrays.org/protocols.html). For this study the Sm6k microarrays described by Rüberg et al. (53) were used. Each microarray contains 6,046 PCR fragments and 161 70-mer oligonucleotides as open reading frame-specific probes, covering the whole S. meliloti genome. Each probe was spotted in triplicate. Hybridization and image acquisition of the microarrays were performed as described by Rüberg et al. (53). For acquisition of the mean signal and mean local background intensities for each spot of the microarray ImaGene 5.0 software for spot detection, image segmentation, and signal quantification (Biodiscovery Inc., Los Angeles, Calif.) was used. Spots were flagged as “empty” if R was ≤1.5 in both channels, with R = (signal mean−background mean)/background standard deviation). The remaining spots were considered for further analysis. The log2 value of the intensity ratios was calculated for each spot, with Mi = log2(Ri/Gi) (Ri = Ich1i−Bgch1i and Gi = Ich2i−Bgch2i, with Ich1i and Ich2i being the intensity of a spot in channel l or channel 2, respectively, and Bgch1i and Bgch2i being the background intensity of a spot in channel 1 or channel 2, respectively. The mean intensity was calculated for each spot, with Ai = log2(RiGi)0.5 (16). A normalization method based on local regression that accounts for intensity spatial dependence in dye biases was applied. Within a print tip group normalization was performed as described by Yang et al. (71): Mi = log2(Ri/Gi) → log2(Ri/Gi)−cj(A) = log2(Ri/[kj(A)Gi]), where cj(A) is the lowest fit to the MA plot for the jth grid (i.e., for the jth print tip group), j = 1,…, J, with J denoting the number of print tips. A floor value of 20 was introduced before normalization to be able to use logarithmic values. Genes significantly up- or down-regulated were identified by t statistics (16). The expression of a gene was considered significantly different if the P value was ≤0.05, the log2 ratio of the intensities (M value) was ≥1 or ≤−1, and the mean intensity (A value) was ≥9.
Bioinformatics tools.
Database searches for homologues to the S. meliloti SitABCD and Fur proteins were performed with the BLAST program (1). Potential Fur boxes were identified with the HMMer suite (17). Alignments were performed with the ClustalW program (65). Normalization and t statistics of microarray data were carried out using the EMMA 1.0 microarray data analysis software (15).
RESULTS
Genetic organization of the S. meliloti DNA region carrying the fur gene and the sitABCD operon.
The S. meliloti 1021 sitABCD locus (Fig. 1) is located on the chromosome of the tripartite genome of this bacterium. According to BLAST searches of available databases the S. meliloti SitA, -B, -C, and -D proteins exhibit high homologies to metal-type ABC transporter proteins. Most of them were identified in a variety of animal pathogen γ-proteobacteria, with the notable exceptions of the plant pathogen Agrobacterium tumefaciens, an α-proteobacterium, closely related to S. meliloti. Other sequenced rhizobial genomes like Bradyrhizobium japonicum and Mesorhizobium loti do not possess sitABCD homologues. The S. meliloti SitA, -B, -C, and -D proteins showed highest similarities to the proteins of the S. enterica serovar Typhimurium SitABCD transporter (between 65 and 78% identity) (72), to the Y. pestis YfeABCD transporter (between 54 and 66% identity) (6), and to those of an uncharacterized transporter of A. tumefaciens (AGR_L_798, AGR_L_799, AGR_L_801, and AGR_L_803, between 64 and 80% identity). Based on sequence similarities it can be assumed that in S. meliloti sitA encodes the periplasmic binding protein, sitB encodes the ATPase, and sitC and sitD encode the transmembrane domains of the ABC transporter. The genes sitA, -B, -C, and -D in the S. meliloti chromosome appear to be translationally coupled, which is a strong indicator that they represent an operon. Adjacent to the S. meliloti sitABCD operon a fur gene is located in the opposite orientation. The S. meliloti Fur protein showed the highest similarities to the Fur proteins of Bartonella species (60% identity) (47).
To analyze the role of the sitABCD operon in metal acquisition and to investigate the regulatory role of the fur gene in S. meliloti 1021, the marker-free deletion mutants Rm1021ΔsitA and Rm1021Δfur, respectively, were constructed. The construction of the deletion mutants is described in Materials and Methods. The deleted fragments are indicated in Fig. 1.
Roles of the S. meliloti sitABCD operon in Mn(II) and Fe(II) utilization.
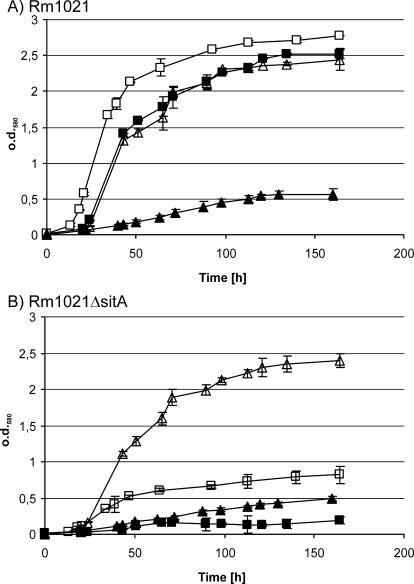
For investigation of the involvement of the S. meliloti sitABCD operon in both manganese and iron utilization, we analyzed the abilities of strain Rm1021ΔsitA to grow under iron- and manganese-limiting conditions. First we used defined VMM* in order to be able to test different metals in defined concentrations (Fig. 2). When grown in VMM* supplemented with Mn(II) but not with any iron source, the wild type and the sitA mutant displayed limited growth with no significant differences between each other. Under these conditions, obviously both strains suffer from iron deficiency. In VMM* supplemented with 30 μM Fe(II) as the sole iron source the sitA mutant exhibited a strongly reduced growth compared to the wild type. Growth was less than during incubation in VMM* without added iron sources. Addition of 1 μM Mn(II) to the medium fully restored growth to wild-type levels, which was not the case when up to 60 μM Fe(II) was added. The manganese-dependent growth defect of Rm1021ΔsitA suggests that at least during growth in VMM the sitABCD operon is needed for the high affinity transport of Mn(II).
FIG. 2.
Growth of the S. meliloti wild type and the S. meliloti sitA mutant strain with dependence on Mn(II) and/or Fe(II) supplementation of the medium. Overnight cultures of the S. meliloti wild-type strain Rm1021 (A) and the S. meliloti sitA mutant Rm1021ΔsitA (B) were washed and diluted in fresh VMM* containing 30 μM Fe(II) (▪), 60 μM Fe(II) (□), 1 μM Mn(II) (▴), or 30 μM Fe(II) + 1 μM Mn(II) (▵). Growth was monitored via optical density at 580 nm (o.d.580). The results are derived from five independent replicates, and the error bars indicate the standard deviations.
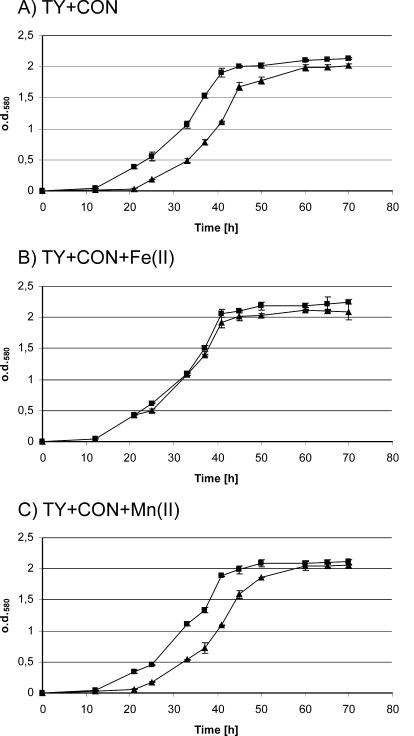
It has been reported that SitABCD homologues are also involved in the acquisition of chelated iron (6). Since the Mn(II) growth deficiency in VMM* might obscure any iron-dependent growth phenotypes, we also tested the growth of the sitA mutant in TY complex medium. Platero et al. demonstrated that the iron chelator EDDHA (ethylenediamine di-o-hydroxyphenylacetic acid) also appears to heavily affect manganese availability in TY medium (48). Therefore, we tested different chelators that might reduce iron availability while not causing manganese deficiency. Under sufficiently high conalbumin (Con) concentrations (>240 mg/liter) the S. meliloti sitA mutant Rm1021ΔsitA exhibited a prolonged lag phase when compared to growth of the wild type. This growth deficit could be alleviated by the addition of Fe(II) but not by the addition of Mn(II) (Fig. 3), indicating that iron deficiency is responsible for this growth phenotype. The prolonged lag phase might be explained by the assumption that the growth of Rm1021ΔsitA is dependent on the expression of alternative iron ion transport systems. The results therefore suggest that the sitABCD system contributes to acquisition of chelated iron during early growth stages.
FIG. 3.
Effects of the iron chelator Con on the growth of the S. meliloti wild-type strain and the S. meliloti sitA mutant strain. Strains Rm1021 (▪) and Rm1021ΔsitA (▴) were grown in TY medium containing 240 mg of Con/liter (A), 240 mg of Con/liter and 25 μM Fe(II) (B), or 240 mg of Con/liter and 25 μM Mn(II) (C). The depicted results were obtained from five independent replicates, and the error bars indicate the standard deviations. o.d.580, optical density at 580 nm.
Phenotypic characterization of the S. meliloti fur mutant Rm1021Δfur.
In many gram-negative bacteria the Fur protein acts as a global repressor, and therefore a defect in Fur often results in pleiotropic phenotypes. Common traits in fur mutants include siderophore overproduction, enhanced manganese resistance, and H2O2 sensitivity (27). Therefore, we tested the inhibitory capacity of Mn(II) and H2O2 on the growth of the S. meliloti wild-type Rm1021 and the fur mutant Rm1021Δfur on TY agar plates as described in Materials and Methods. The inhibition zone was monitored after 48 h of growth, but no difference between the wild-type and mutant strains were observed (data not shown). Additionally, in a CAS assay the amounts of produced siderophores in the supernatants of the wild-type and the Rm1021Δfur culture grown in TY medium were measured, but no differences between the wild type and the fur mutant were found (data not shown).
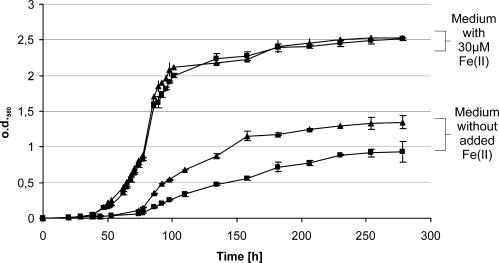
We further tested whether Rm1021Δfur exhibited iron-dependent growth phenotypes. For this purpose overnight cultures of the S. meliloti wild type (Rm1021) and the S. meliloti fur mutant (Rm1021Δfur) were washed and diluted into fresh VMM* containing either no added iron or 30 μM Fe(II). Growth was then monitored by measuring the optical density (Fig. 4). The mutant strain Rm1021Δfur showed no growth differences compared to the wild type when grown in medium containing 30 μM Fe(II). During growth under low-iron conditions in VMM*, both the wild type and fur mutant exhibited a longer lag phase and reduced overall growth than under iron-replete conditions, likely caused by iron deficiency. Surprisingly though, the fur mutant showed a higher initial growth and reached a higher cell density than the wild type.
FIG. 4.
Growth of the S. meliloti wild-type strain Rm1021 and the fur mutant strain Rm1021Δfur under high and low iron concentrations. Overnight cultures of Rm1021 (▪) and Rm1021Δfur (▴) were washed and diluted into fresh VMM* containing 30 μM Fe(II) or no added iron sources and incubated at 30°C for 180 h. The growth was monitored by measuring the optical density at 580 nm (o.d.580). The error bars indicate the standard deviations, which were calculated from six independent experiments.
Regulation of the S. meliloti sitA promoter, with dependence on the fur gene and metal ions.
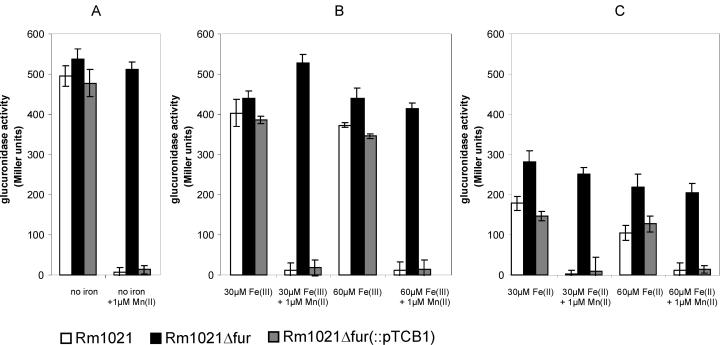
Since the growth experiments suggested that the sitABCD operon is involved in Mn(II) and Fe(II) acquisition, we investigated whether its expression is regulated by those metals. The S. meliloti wild-type strain Rm1021 carrying the reporter plasmid pTCC1 was grown in VMM*, to which various amounts of different metals were added. Since pTCC1 carried the sitA promoter fused to a gusA codon region, sitA expression could be measured by determining the β-glucuronidase activity during logarithmic growth (Fig. 5).
FIG. 5.
Effects of different metal sources on the expression of the S. meliloti sitA promoter. A promoter test plasmid containing a sitA promoter-gusA fusion (pTCC1) was introduced into the S. meliloti wild-type strain Rm1021 (white bars), the S. meliloti fur mutant strain Rm1021Δfur (black bars), and the fur mutant strain Rm1021Δfur complemented with plasmid pTCB1 carrying an intact fur gene (grey bars). The strains were grown in VMM* under the following conditions: (A) no added iron, (B) addition of Fe(III), (C) addition of Fe(II) as the sole iron source. Additionally, Mn(II) was added as indicated. The different strains were grown to an optical density of 0.6 before the assays for glucuronidase activity were carried out. The β-glucuronidase levels are expressed in Miller units, and the error bars indicate the standard deviations. In each case the depicted results were derived from six independent cultures.
The expression of the sitA promoter in wild-type S. meliloti cells grown in VMM* containing 30 μM Fe(III) (Fig. 5B) was only marginally lower than in cells incubated in medium containing no added iron (Fig. 5A). Doubling the amount of Fe(III) to 60 μM did not result in a significant change of sitA expression. In contrast, addition of 30 μM Fe(II) reduced the expression of the sitABCD operon to half of the level (P < 0.01, t test) of that in cells grown in medium with no added iron (Fig. 5C). This indicates that the sitA promoter is only weakly affected by Fe(III) and moderately by Fe(II). Addition of 1 μM Mn(II) on the other hand repressed sitA promoter activity to background levels, regardless of the concentration of other metals. Addition of Zn(II) or Ca2+ to the medium had no effect on the expression of sitA (data not shown), indicating that the moderate repression by Fe(II) and the strong repression by Mn(II) are specific.
We further investigated whether repression by Fe(II) and Mn(II) is mediated by Fur. The sitA promoter test vector pTCC1 was introduced into the fur mutant strain Rm1021Δfur. sitA expression was determined as described above. The sitA promoter activity in Rm1021Δfur was under all tested conditions higher than in the wild type (Fig. 5), indicating that the fur gene regulates the sitABCD operon as a repressor. The sitA promoter activity of the fur mutant was totally unaffected by supplementation with Mn(II); thus, it can be concluded that the fur gene is needed for Mn(II)-mediated repression of the sitA promoter. In contrast, sitA expression in the fur mutant background was still repressed by Fe(II), indicating that Fur is not essential for the repression of the sitA promoter by Fe(II). Since Fe(II)-dependent repression of the sitA promoter appears to be stronger in the wild type than in the fur mutant, it is possible that Fur enhances the repression. Altogether these results strongly suggest that the S. meliloti Fur protein is an Mn(II)-dependent repressor of the sitABCD operon.
To verify that the observations for Rm1021Δfur were indeed solely caused by the mutation in the fur gene, we integrated plasmid pTCB1, carrying the S. meliloti fur gene, into the chromosome of the fur mutant. The sitA promoter activity in the resulting strain, Rm1021Δfur(pTCB1), was under all conditions similar to that of the wild type (Fig. 5). This demonstrates that only the fur gene is needed for the Mn(II)-dependent repression of the sitA promoter.
Micorarray-based analysis of transcription of S. meliloti genes with dependence on the fur gene.
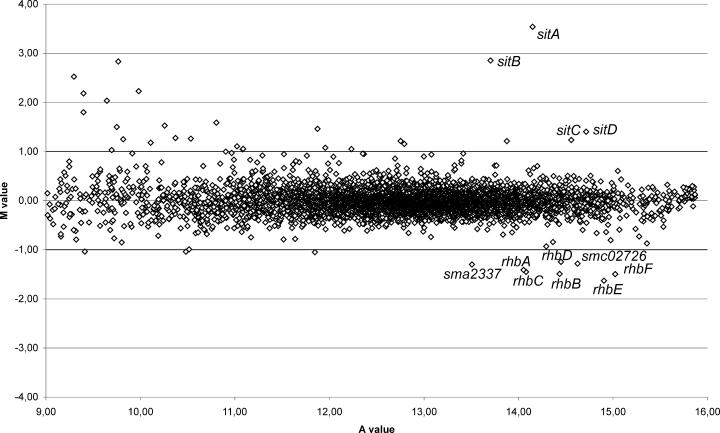
In order to identify which genes beside the sitABCD operon are affected by the fur mutation, a genome-wide expression profiling of the fur-defective S. meliloti strain Rm1021Δfur compared to the wild-type, Rm1021, using the S. meliloti SM6kPCR microarrays (53), was performed. Cultures of both strains were grown in VMM to a cell density of about 8 × 108 CFU ml−1. RNA isolation, labeling, microarray hybridization, and data analysis were performed as described in Materials and Methods. The expression of a gene was considered significantly different if the M value was ≥1 or ≤−1, the A value was ≥9, and P was ≤0.05. The results depicted in the scatter plot (Fig. 6) and summarized in Table 2 were obtained from two independent biological replicates. Altogether, 33 genes displayed an altered expression in the fur mutant when compared to expression in the wild-type. Of these genes, 23 were induced and 10 were repressed in the fur mutant.
FIG. 6.
Scatter plot for the microarray-based analysis of S. meliloti genes affected by deletion of the fur gene. The scatter plot shows the logarithmic mean signal ratios (M values) versus. the logarithmic mean signal intensities (A values) obtained for comparison of the S. meliloti wild-type strain Rm1021 to the fur mutant Rm1021Δfur by microarray hybridization. Indicated are the differentially expressed genes that are involved in metal acquisition. Genes were only regarded as differentially expressed when the M values were ≥1.0 or ≤−1.0, the A values were ≥9.0, and the P values ≤0.05.
TABLE 2.
Genes induced or repressed more than twofold in the S. meliloti fur mutant Rm1021Δfur as compared to the wild type, Rm1021
| Gene | Expression ratio | M value | A value | Function |
|---|---|---|---|---|
| smc04431 | 2.38(+) | 1.25 | 9.82 | Conserved hypothetical protein |
| smc03269 | 3.00(+) | 1.59 | 10.80 | Putative dipeptide periplasmic ABC transporter |
| smc03183 (phaF1) | 2.15(+) | 1.11 | 11.02 | pH adaptation potassium efflux system |
| smc02509 (sitA) | 11.62(+) | 3.54 | 14.15 | Manganese/iron transporter periplasmic binding protein |
| smc02508 (sitB) | 7.22(+) | 2.85 | 13.70 | Manganese/iron transporter ATPase |
| smc02507 (sitC) | 2.35(+) | 1.23 | 14.56 | Manganese/iron transporter membrane protein |
| smc02506 (sitD) | 2.64(+) | 1.40 | 14.72 | Manganese/iron transporter membrane protein |
| smc02474 | 4.69(+) | 2.23 | 9.98 | Putative sugar ATP binding ABC transporter protein |
| smc02416 | 4.54(+) | 2.18 | 9.40 | Putative hydrolase |
| smc02025 | 2.32(+) | 1.21 | 13.88 | Putative dipeptide periplasmic binding protein |
| smc01821 | 2.11(+) | 1.07 | 11.96 | Putative dihydropyrimidinase |
| smc01701 | 2.08(+) | 1.05 | 11.09 | Conserved hypothetical protein |
| smc01666 (mdeA) | 5.75(+) | 2.52 | 9.30 | Putative methionine gamma lyase |
| smc01664 | 7.12(+) | 2.83 | 9.76 | Putative transcription regulator |
| smc01637 | 4.09(+) | 2.03 | 9.64 | Conserved hypothetical protein |
| smc01124 (glnD) | 2.42(+) | 1.28 | 10.37 | Putative uridyltransferase |
| smb21403 | 2.06(+) | 1.04 | 11.08 | Hypothetical protein |
| smb20532 | 2.00(+) | 1.00 | 10.90 | Hypothetical protein |
| smb20335 | 2.26(+) | 1.18 | 10.11 | Conserved hypothetical protein |
| sma2137 | 2.32(+) | 1.21 | 12.75 | Putative glycerate dehydrogenase |
| sma1413 | 2.83(+) | 1.50 | 9.75 | Hypothetical protein |
| sma1016 | 2.23(+) | 1.15 | 12.79 | Putative acetyltransferase |
| sma0875 (nolG) | 2.75(+) | 1.46 | 11.87 | Putative efflux transporter |
| sma0629 | 2.05(−) | −1.04 | 10.48 | Hypothetical protein |
| sma2337 | 2.46(−) | −1.30 | 13.51 | Putative transmembrane transport protein |
| sma2400 (rhbA) | 2.66(−) | −1.41 | 14.05 | Rhizobactin 1021 siderophore biosynthesis |
| sma2402 (rhbB) | 2.81(−) | −1.49 | 14.44 | Rhizobactin 1021 siderophore biosynthesis |
| sma2404 (rhbC) | 2.74(−) | −1.45 | 14.08 | Rhizobactin 1021 siderophore biosynthesis |
| sma2406 (rhbD) | 2.37(−) | −1.25 | 14.45 | Rhizobactin 1021 siderophore biosynthesis |
| sma2408 (rhbE) | 3.09(−) | −1.63 | 14.90 | Rhizobactin 1021 siderophore biosynthesis |
| sma2410 (rhbF) | 2.82(−) | −1.50 | 15.02 | Rhizobactin 1021 siderophore biosynthesis |
| smb20728 | 2.07(−) | −1.05 | 11.85 | Hypothetical protein |
| smc02726 | 2.43(−) | −1.28 | 14.63 | Putative iron transport protein |
In accordance with gus reporter gene analyses, the genes of the sitABCD operon were found to be highly induced in the fur mutant. Surprisingly, none of the other up-regulated genes showed homologies to genes of known metal utilization systems. Instead, the up-regulated genes were found to be involved in the transport of sugar (smc02474) or peptides (smc03269 and smc02025) or to encode efflux transporters (phaF and nolG), a regulator (smc01664), five probable enzymatic proteins (mdeA, glnD, sma2137, smc01821, and smc02416), and seven proteins which have been annotated as hypothetical proteins (smc01637, sma1413, smc04431, smb20335, smc01701, smb21403, and smb20532). An updated annotation of these hypothetical proteins did not give any evidence that they might be involved in metal utilization.
Ten genes were moderately (between 2.05- and 3.09-fold) less expressed in the fur mutant Rm1021Δfur than in the wild-type strain. It is noteworthy that except for the genes smb20728 and sma0629, all of them seem to be involved in iron acquisition. Six of the genes comprise the complete rhbABCDEF operon (Table 2), which is involved in the synthesis of rhizobactin 1021, the siderophore of S. meliloti 1021 (41). Directly upstream of the siderophore synthesis operon a gene encoding a putative transmembrane transporter (sma2337) is located, which is also down-regulated in the fur mutant. Finally, the gene smc02726, which was found to be repressed in the fur mutant Rm1021Δfur, was annotated to encode a putative outer membrane iron receptor.
In order to identify putative Fur boxes in the S. meliloti genome, we performed a genome-wide hidden Markov model-based search (17) for putative Fur binding regions on the basis of the alignments of known Fur boxes. Only in front of the sitABCD operon, but not in front of the other up-regulated genes, a significant Fur-box motif (GCAAATGCTTCTCATTTGC) could be detected. This motif exhibits only moderate homologies to either the classical Fur box motif or the 7-1-7 motif as proposed by Baichoo and Helmann (4), but it could be interpreted as an F-F-x-R motif (GCAAAT GCTTCT C ATTTGC) (18, 38) with two imperfect direct repeats and an inverted repeat.
Roles of the S. meliloti fur gene and the sitABCD operon in nodule symbiosis.
Up to this point we investigated the role of the fur and sitA genes during free-living growth of S. meliloti. To assess whether defects in the sitABCD operon and the fur gene also affect the symbiotic abilities of S. meliloti, we inoculated the S. meliloti mutant strains Rm1021Δfur and Rm1021ΔsitA and the wild-type strain Rm1021 onto M. sativa seedlings. After 4 weeks of growth the fresh weight of the plants was measured and the ability of the nodules to fix nitrogen was assessed by acetylene reduction tests. Both mutants were able to nodulate normally, and the number of nodules varied between 5 and 18 per plant, with no significant difference between the mutants and the wild type. Moreover, M. sativa plants inoculated with the fur-defective S. meliloti strain showed no differences in fresh weight and nitrogen fixation rate compared to plants inoculated with the wild-type strain (Table 3). However, M. sativa plants which were inoculated with the S. meliloti sitA mutant strain showed a slight but significant decrease in wet weight as well as in the acetylene reduction rate (P < 0.001, t test). Thus, the sitA mutation seems to attenuate the nitrogen fixation capability of the microsymbiont. Increasing the Fe(II) concentration of the plant medium to 30 μM abolished the differences between the wild type and sitA mutant, whereas addition of up to 30 μM Mn(II) had no effect (Table 3).
TABLE 3.
Nitrogenase activity and wet weight of M. sativa plants inoculated with the S. meliloti wild type (Rm1021), the fur mutant (Rm1021Δfur), and the sitA mutant (Rm1021ΔsitA) after 4 weeks of growth on plant medium containing the indicated amounts of Fe(II) and Mn(II)
| Strain | Nitrogenase activity (nmol of ethylene plant−1 h−1]
|
Foliage fresh wt (mg plant−1)
|
||||
|---|---|---|---|---|---|---|
| 10 μM Fe(II) | 30 μM Fe(II) | 10 μM Fe(II) + 30 μM Mn(II) | 10 μM Fe(II) | 30 μM Fe(II) | 10 μM Fe(II) + 30 μM Mn(II) | |
| Rm1021 | 3.1 ± 0.8 | 2.4 ± 0.4 | 3.1 ± 0.7 | 152 ± 10 | 134 ± 11 | 141 ± 13 |
| Rm1021Δfur | 3.1 ± 0.2 | 2.1 ± 0.6 | 2.9 ± 0.9 | 132 ± 6 | 138 ± 29 | 148 ± 26 |
| Rm1021ΔsitA | 1.6 ± 0.3 | 2.3 ± 0.3 | 1.4 ± 0.5 | 71 ± 8 | 131 ± 37 | 87 ± 11 |
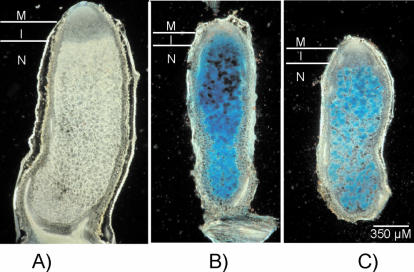
Furthermore, we analyzed the expression of the sitABCD operon during symbiosis by inoculating M. sativa plants with the wild-type strain Rm1021 and the fur mutant Rm1021Δfur, both carrying the sitA promoter gusA reporter construct on the plasmid pTCC1. As a negative control the S. meliloti wild type carrying plasmid pJP2 without inserts was used. By X-Gluc staining of the nodules, the activity of the sitA promoter was visualized. As expected, no β-glucuronidase activity was detected in the negative control (Fig. 7A). Since we demonstrated earlier that the sitABCD operon is derepressed in the fur mutant, it could be expected that this is also the case in the nodule. Indeed, M. sativa nodules that were inoculated with the fur mutant carrying pTCC1 showed a strong blue staining throughout all zones of the nodule with the exception of the meristem (Fig. 7B). This indicates that in the S. meliloti fur mutant background the sitABCD operon is highly expressed in the nodule. Nodules that were induced by the wild-type strain carrying the sitA promoter test plasmid (Fig. 7C) exhibited a β-glucuronidase pattern similar to that of nodules induced by the fur mutant. One difference, however, was that the infection zone of nodules induced by the wild type was generally more weakly colored than the nitrogen fixation zone. This may indicate that in the wild-type strain the sitABCD operon is generally more strongly expressed in the nitrogen-fixing zone than in the infection zone.
FIG. 7.
Visualization of S. meliloti sitA promoter activity in M. sativa nodules. Nodules of M. sativa were harvested, thin sectioned, and stained with the dye X-Gluc. (A) M. sativa nodule induced by the S. meliloti wild-type strain carrying pJP2. (B) M. sativa nodule induced by the S. meliloti fur mutant carrying sitA promoter-gusA fusion plasmid pTCC1. (C) M. sativa nodule induced by the S. meliloti wild-type strain carrying pTCC1. Abbreviations: M, meristem; I, infection zone; N, nitrogen fixation zone.
DISCUSSION
The S. meliloti sitABCD operon is involved in Mn(II) and Fe(II) utilization.
In this study we analyzed S. meliloti mutants to investigate the involvement of the sitABCD operon and the fur gene in metal acquisition and regulation. The manganese-deficient growth of the sitA mutant clearly demonstrated that the sitABCD operon is involved in Mn(II) utilization. Moreover, the strength of the growth inhibition implies that SitABCD is, at least under the tested conditions, the main transporter for Mn(II) in S. meliloti. Since addition of Mn(II) restored the growth of Rm1021ΔsitA, it is probable that S. meliloti possesses alternative transporters with a lower affinity for Mn(II). In the genome of S. meliloti a hitherto uncharacterized, putative manganese transporter is located (sma1115), which is a possible candidate for the low-affinity transporter of Mn(II).
In contrast to the manganese-dependent growth, the iron deficiency of the S. meliloti sitA mutant was far less apparent. Only in the presence of high concentrations of the chelator Con did growth experiments reveal an extended lag phase for the sitA mutant when compared to the wild type. This indicates that other iron transport systems that have been annotated for S. meliloti (12) might be able to compensate for the missing iron transport of SitABCD. Nonetheless, the growth experiment showed that the sitABCD operon does promote growth of S. meliloti in the presence of chelated iron; however, the physiological relevance is not clear. The contribution of metal-type ABC transporters to either iron or manganese uptake varies in different species. In Yersinia and Shigella the Sit homologues appear to be primarily involved in iron uptake (5, 54), while in Salmonella and, according to our results, in S. meliloti the manganese uptake capabilities are of more importance.
The S. meliloti fur gene is required for Mn(II)-dependent repression of the sitABCD operon.
The expression analyses showed that the S. meliloti sitABCD operon is repressed by Mn(II) and Fe(II), but not by Fe(III). These results support the observation that the sitABCD operon is involved in the acquisition of Mn(II) and Fe(II). The repression by Mn(II) was completely eliminated in the S. meliloti fur mutant. However, the fur deletion did not negate the repression of sitA by Fe(II). Thus, the Mn(II)-dependent repression of the S. meliloti sitABCD operon is strongly dependent on the fur gene, while Fe(II)-dependent repression is only weakly affected by the fur mutation. Probably another, hitherto uncharacterized regulator is involved in Fe(II)-dependent repression of the sitABCD operon.
The orthologous yfeABCD operon of Y. pestis and the sitABCD operon of S. enterica serovar Typhimurium were both reported to be repressed by iron, and at least the yfeABCD operon was also repressed in the presence of Mn(II) (6, 72), which is in accordance with our results. One difference is that in S. enterica serovar Typhimurium and Y. pestis a fur gene is needed for the repression of the sitABCD operon by Fe(II), whereas Fur in S. meliloti plays only a subordinate role, if any, in the Fe(II)-dependent repression of the sitA promoter. Our results indicate that Fur is mainly an Mn(II)-dependent repressor of the S. meliloti sitABCD operon.
The S. meliloti fur gene does not encode a global regulator of iron uptake.
In many gram-negative bacteria the fur gene has been well established as a central repressor of iron uptake and other iron-responsive genes (27). Our microarray experiments showed that altogether only 23 genes were significantly up-regulated in a fur mutant. In similar global analyses of iron-responsive genes in E. coli and Neisseria meningitidis (25, 42), a larger number of genes was found to be repressed by Fur. It is of special interest that in our experiments the genes of the sitABCD operon were the only ones known to be involved in metal uptake. The other up-regulated genes have no similarities to those for known iron utilization systems. Additionally, only in front of the sitABCD genes could a significant Fur box motif be found. This supports our reporter fusion-based expression analyses, with which we demonstrated that S. meliloti Fur is a manganese-dependent repressor of the sitABCD system. The lack of putative Fur boxes in front of the other genes identified in the microarray experiment might indicate that those genes are not directly controlled by Fur. On the other hand, in B. japonicum an atypical Fur-binding site was identified (20). Whether this is also the case in S. meliloti remains to be determined.
While we cannot state with absolute certainty whether some of the hypothetical proteins might represent completely novel metal utilization systems, it is intriguing that none of the known iron uptake systems of S. meliloti are upregulated in the fur mutant. It is noteworthy in this context that a putative iron transport gene (smc02726) and the complete rhizobactin 1021 synthesis operon are, in contrast, repressed in the S. meliloti fur mutant, instead of being induced. The simplest explanation is that the derepression of the sitABCD operon led to an increase in intracellular Mn(II) and/or Fe(II) concentration, which in turn caused the observed repression of the iron utilization systems. This would also explain the higher initial growth of the S. meliloti fur mutant compared to the wild type during incubation in iron-limited medium. The enhanced expression of the S. meliloti sitABCD operon caused by the fur mutation might thus improve the ability of S. meliloti to scavenge iron from the medium. The altered intracellular metal content caused by the derepression of the sitABCD operon might also be responsible for the differential expression of other genes identified in our microarray experiments. These results clearly demonstrate that, unlike in other gram-negative bacteria, in S. meliloti the protein encoded by the fur gene does not function as the main repressor of iron uptake. This finding is also supported by the phenotypic analyses of the S. meliloti fur mutant, in which we found that this mutant did not exhibit common phenotypes of other known fur-defective strains like H2O2 sensitivity, manganese resistance, siderophore overproduction (31, 62), and impaired growth in the presence of iron (19, 31, 39, 62, 63, 67). Additionally, the S. meliloti fur mutant nodulated M. sativa plants and fixed nitrogen with the same efficiency as the wild type. These results thus verify that the role of the fur gene in S. meliloti differs substantially from that in other bacterial species. This is in accordance with experiments performed by Wexler et al. in which it was demonstrated that the fur gene in R. leguminosarum is not involved in the regulation of operons involved in iron metabolism and that an R. leguminosarum fur mutant did not exhibit any phenotypes generally associated with a fur mutation (70). In fact, in R. leguminosarum a different protein termed RirA appears to fulfill most functions that are usually attributed to Fur (66).
Given the fact that the Fur homologue in S. meliloti is indeed not a ferric uptake regulator, but rather an Mn(II)-dependent repressor, renaming the S. meliloti fur gene mur (manganese uptake regulator) should be considered. To date only a few regulators specific for manganese uptake systems have been described for bacteria (32). Most of these regulators belong to the group of DtxR-like proteins such as TroR in Treponema pallidum (49), ScaR in Streptococcus gordonii (34), and MntR in Bacillus subtilis (51), none of which exhibits significant homologies to the S. meliloti Fur protein. Another metal-dependent repressor is PerR of Bacillus subtilis (10). PerR represses genes in the presence of either Fe(II) or Mn(II); however, the regulation of fur by PerR in B. subtilis appears to be solely Mn(II) dependent (21). The results of our work now indicate that the S. meliloti Fur homologue is the first characterized member of the Fur family which is mainly involved in manganese regulation in gram-negative bacteria.
The sitABCD operon is induced during nodule symbiosis and contributes to symbiotic nitrogen fixation.
In plant tests we found that the mutation in the S. meliloti sitA gene led to an attenuated nitrogen fixation activity. Additionally, the sitA promoter showed strong expression in the nodule, indicating that the sitABCD operon contributes to successful symbiosis. This is in accordance with macroarray experiments performed by Ampe et al. who found an induction of the S. meliloti sitA gene under symbiotic conditions (2). Additionally, Dvordjevic et al. identified the SitA and SitB protein among proteins isolated from nodules which were induced by S. meliloti (14).
Since the nitrogen fixation efficiency of the sitA mutant could be restored to wild-type levels by supplementing the plant medium with Fe(II), but not with Mn(II), it appears unlikely that the altered symbiotic properties were caused by manganese deficiency. A possible explanation for this observation might be that different sets of transport systems are expressed during free-living growth and in symbiosis. In liquid medium iron transporters other than the SitABCD system (e.g., siderophore-dependent iron transporters) might be the main importers of iron. This would be in agreement with the weak iron-dependent growth phenotype of the S. meliloti sitA mutant in VMM. During symbiosis, however, the genes involved in siderophore synthesis (rhbABCDEF) and transport (rhtA) in S. meliloti are inactive (41). Under these conditions the SitABCD transporter may have an increased importance for iron acquisition. Our data thus suggest that the relevance of the SitABCD transporter for manganese and iron uptake might be different during free-living growth than under symbiotic conditions. It is obvious however that in any case the sitABCD operon is not essential for metal acquisition during symbiosis since the nitrogen fixation rate was only moderately lower in the sitA mutant than in the wild type.
It is interesting that the metal-type ABC transporters encoded by yfeABCD of Y. pestis and sitABCD of S. enterica serovar Typhimurium and S. flexneri contribute significantly to the virulence of the respective pathogens (5, 35, 54). Moreover other (uncharacterized) homologues of these metal-type transporters are mainly found in pathogens like Haemophilus or enteroinvasive E. coli isolates. It thus appears that the highly conserved sitABCD system generally contributes to the supply of the respective bacteria with metals in their animal or plant host organisms but is not always essential.
Acknowledgments
We thank S. Rüberg and E. Schulte-Berndt for performing the microarray experiments and V. Bartelsmeier for the preparations of the nodules. We also thank A. Johnston for insightful discussions.
The work was supported by a scholarship of the Graduate School for Bioinformatics and Genome research, funded by the Ministerium für Wissenschaft und Forschung (MWF), North-Rhine Westphalia, and grants 0311752 and 031U213D from the Bundesministerium für Bildung und Forschung (BMBF), Germany, and grant BIZ 7 from the Deutsche Forschungsgemeinschaft (DFG).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ampe, F., E. Kiss, F. Sabourdy, and J. Batut. 2003. Transcriptome analysis of Sinorhizobium meliloti during symbiosis. Genome Biol. 4:R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagg, A., and J. B. Neilands. 1987. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471-5477. [DOI] [PubMed] [Google Scholar]
- 4.Baichoo, N., and J. D. Helmann. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184:5826-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bearden, S. W., and R. D. Perry. 1999. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol. Microbiol. 32:403-414. [DOI] [PubMed] [Google Scholar]
- 6.Bearden, S. W., T. M. Staggs, and R. D. Perry. 1998. An ABC transporter system of Yersinia pestis allows utilization of chelated iron by Escherichia coli SAB11. J. Bacteriol. 180:1135-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 8.Boyer, E., I. Bergevin, D. Malo, P. Gros, and M. F. Cellier. 2002. Acquisition of Mn(II) in addition to Fe(II) is required for full virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 70:6032-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun, V. 2001. Iron uptake mechanisms and their regulation in pathogenic bacteria. Int. J. Med. Microbiol. 291:67-79. [DOI] [PubMed] [Google Scholar]
- 10.Bsat, N., A. Herbig, L. Casillas-Martinez, P. Setlow, and J. D. Helmann. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29:189-198. [DOI] [PubMed] [Google Scholar]
- 11.Calderwood, S. B., and J. J. Mekalanos. 1987. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J. Bacteriol. 169:4759-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dreano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Puhler, B. Purnelle, U. Ramsperger, C. Renard, P. Thebault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lorenzo, V., S. Wee, M. Herrero, and J. B. Neilands. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 169:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Djordjevic, M. A., H. C. Chen, S. Natera, G. Van Noorden, C. Menzel, S. Taylor, C. Renard, O. Geiger, and G. F. Weiller. 2003. A global analysis of protein expression profiles in Sinorhizobium meliloti: discovery of new genes for nodule occupancy and stress adaptation. Mol. Plant-Microbe Interact. 16:508-524. [DOI] [PubMed] [Google Scholar]
- 15.Dondrup, M., A. Goesmann, D. Bartels, J. Kalinowski, L. Krause, B. Linke, O. Rupp, A. Sczyrba, A. Puhler, and F. Meyer. 2003. EMMA: a platform for consistent storage and efficient analysis of microarray data. J. Biotechnol. 106:135-146. [DOI] [PubMed] [Google Scholar]
- 16.Dudoit, S., Y. H. Yang, M. J. Callow, and T. P. Speed. 2002. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Stat. Sinica 12:111-139. [Google Scholar]
- 17.Eddy, S. R. 1998. Profile hidden Markov models. Bioinformatics 14:755-763. [DOI] [PubMed] [Google Scholar]
- 18.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1998. Binding of the fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J. Mol. Biol. 283:537-547. [DOI] [PubMed] [Google Scholar]
- 19.Foster, J. W., and H. K. Hall. 1992. Effect of Salmonella typhimurium ferric uptake regulator (fur) mutations on iron- and pH-regulated protein synthesis. J. Bacteriol. 174:4317-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman, Y. E., and M. R. O'Brian. 2003. A novel DNA-binding site for the ferric uptake regulator (Fur) protein from Bradyrhizobium japonicum. J. Biol. Chem. 278:38395-38401. [DOI] [PubMed] [Google Scholar]
- 21.Fuangthong, M., A. F. Herbig, N. Bsat, and J. D. Helmann. 2002. Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 184:3276-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldberg, M. B., S. A. Boyko, and S. B. Calderwood. 1991. Positive transcriptional regulation of an iron-regulated virulence gene in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 88:1125-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldberg, M. B., S. A. Boyko, and S. B. Calderwood. 1990. Transcriptional regulation by iron of a Vibrio cholerae virulence gene and homology of the gene to the Escherichia coli fur system. J. Bacteriol. 172:6863-6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grifantini, R., S. Sebastian, E. Frigimelica, M. Draghi, E. Bartolini, A. Muzzi, R. Rappuoli, G. Grandi, and C. A. Genco. 2003. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. USA 100:9542-9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guerinot, M. L. 1994. Microbial iron transport. Annu. Rev. Microbiol. 48:743-772. [DOI] [PubMed] [Google Scholar]
- 27.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 28.Harris, A. G., F. E. Hinds, A. G. Beckhouse, T. Kolesnikow, and S. L. Hazell. 2002. Resistance to hydrogen peroxide in Helicobacter pylori: role of catalase (KatA) and Fur, and functional analysis of a novel gene product designated ‘KatA-associated protein,' KapA (HP0874). Microbiology 148:3813-3825. [DOI] [PubMed] [Google Scholar]
- 29.Hassan, H. M., and H. C. Sun. 1992. Regulatory roles of Fnr, Fur, and Arc in expression of manganese-containing superoxide dismutase in Escherichia coli. Proc. Natl. Acad. Sci. USA 89:3217-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassett, D. J., M. L. Howell, U. A. Ochsner, M. L. Vasil, Z. Johnson, and G. E. Dean. 1997. An operon containing fumC and sodA encoding fumarase C and manganese superoxide dismutase is controlled by the ferric uptake regulator in Pseudomonas aeruginosa: fur mutants produce elevated alginate levels. J. Bacteriol. 179:1452-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hassett, D. J., P. A. Sokol, M. L. Howell, J. F. Ma, H. T. Schweizer, U. Ochsner, and M. L. Vasil. 1996. Ferric uptake regulator (Fur) mutants of Pseudomonas aeruginosa demonstrate defective siderophore-mediated iron uptake, altered aerobic growth, and decreased superoxide dismutase and catalase activities. J. Bacteriol. 178:3996-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horsburgh, M. J., S. J. Wharton, M. Karavolos, and S. J. Foster. 2002. Manganese: elemental defence for a life with oxygen. Trends Microbiol. 10:496-501. [DOI] [PubMed] [Google Scholar]
- 33.Horton, R. M., Z. L. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8:528-535. [PubMed] [Google Scholar]
- 34.Jakubovics, N. S., A. W. Smith, and H. F. Jenkinson. 2000. Expression of the virulence-related Sca (Mn2+) permease in Streptococcus gordonii is regulated by a diphtheria toxin metallorepressor-like protein ScaR. Mol. Microbiol. 38:140-153. [DOI] [PubMed] [Google Scholar]
- 35.Janakiraman, A., and J. M. Slauch. 2000. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol. Microbiol. 35:1146-1155. [DOI] [PubMed] [Google Scholar]
- 36.Johnston, A. W., K. H. Yeoman, and M. Wexler. 2001. Metals and the rhizobial-legume symbiosis—uptake, utilization and signalling. Adv. Microb. Physiol. 45:113-156. [DOI] [PubMed] [Google Scholar]
- 37.Köster, W. 2001. ABC transporter-mediated uptake of iron, siderophores, heme and vitamin B12. Res. Microbiol. 152:291-301. [DOI] [PubMed] [Google Scholar]
- 38.Lavrrar, J. L., and M. A. McIntosh. 2003. Architecture of a fur binding site: a comparative analysis. J. Bacteriol. 185:2194-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Litwin, C. M., and S. B. Calderwood. 1994. Analysis of the complexity of gene regulation by fur in Vibrio cholerae. J. Bacteriol. 176:240-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Litwin, C. M., and S. B. Calderwood. 1993. Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 6:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lynch, D., J. O'Brien, T. Welch, P. Clarke, P. O. Cuiv, J. H. Crosa, and M. O'Connell. 2001. Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J. Bacteriol. 183:2576-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McHugh, J. P., F. Rodriguez-Quinones, H. Abdul-Tehrani, D. A. Svistunenko, R. K. Poole, C. E. Cooper, and S. C. Andrews. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278:29478-29486. [DOI] [PubMed] [Google Scholar]
- 43.Meade, H. M., S. R. Long, G. B. Ruvkum, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetical characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niner, B. M., and A. M. Hirsch. 1998. How many Rhizobium genes, in addition to nod, nif/fix, and exo, are needed for nodule development and function? Symbiosis 24:51-102. [Google Scholar]
- 45.Ochsner, U. A., Z. Johnson, I. L. Lamont, H. E. Cunliffe, and M. L. Vasil. 1996. Exotoxin A production in Pseudomonas aeruginosa requires the iron-regulated pvdS gene encoding an alternative sigma factor. Mol. Microbiol. 21:1019-1028. [DOI] [PubMed] [Google Scholar]
- 46.Ochsner, U. A., and M. L. Vasil. 1996. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc. Natl. Acad. Sci. USA 93:4409-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park, S. Y., K. L. Kelminson, A. K. Lee, P. Zhang, R. E. Warner, D. H. Rehkopf, S. B. Calderwood, and J. E. Koehler. 2001. Identification, characterization, and functional analysis of a gene encoding the ferric uptake regulation protein in Bartonella species. J. Bacteriol. 183:5751-5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Platero, R. A., M. Jaureguy, F. J. Battistoni, and E. R. Fabiano. 2003. Mutations in sitB and sitD genes affect manganese-growth requirements in Sinorhizobium meliloti. FEMS Microbiol. Lett. 218:65-70. [DOI] [PubMed] [Google Scholar]
- 49.Posey, J. E., J. M. Hardham, S. J. Norris, and F. C. Gherardini. 1999. Characterization of a manganese-dependent regulatory protein, TroR, from Treponema pallidum. Proc. Natl. Acad. Sci. USA 96:10887-10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prell, J., B. Boesten, P. Poole, and U. B. Priefer. 2002. The Rhizobium leguminosarum bv. viciae VF39 gamma-aminobutyrate (GABA) aminotransferase gene (gabT) is induced by GABA and highly expressed in bacteroids. Microbiology 148:615-623. [DOI] [PubMed] [Google Scholar]
- 51.Que, Q., and J. D. Helmann. 2000. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol. Microbiol. 35:1454-1468. [DOI] [PubMed] [Google Scholar]
- 52.Rolfe, B. G., P. M. Gresshoff, and J. Shine. 1980. Rapid screening for symbiotic mutants of Rhizobium meliloti and white clover. Plant Sci. Lett. 19:277-284. [Google Scholar]
- 53.Ruberg, S., Z. X. Tian, E. Krol, B. Linke, F. Meyer, Y. Wang, A. Puhler, S. Weidner, and A. Becker. 2003. Construction and validation of a Sinorhizobium meliloti whole genome DNA microarray: genome-wide profiling of osmoadaptive gene expression. J. Biotechnol. 106:255-268. [DOI] [PubMed] [Google Scholar]
- 54.Runyen-Janecky, L. J., S. A. Reeves, E. G. Gonzales, and S. M. Payne. 2003. Contribution of the Shigella flexneri Sit, Iuc, and Feo iron acquisition systems to iron acquisition in vitro and in cultured cells. Infect. Immun. 71:1919-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 56.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 57.Schultze, M., and A. Kondorosi. 1998. Regulation of symbiotic root nodule development. Annu. Rev. Genet. 32:33-57. [DOI] [PubMed] [Google Scholar]
- 58.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 59.Sebastian, S., S. Agarwal, J. R. Murphy, and C. A. Genco. 2002. The gonococcal fur regulon: identification of additional genes involved in major catabolic, recombination, and secretory pathways. J. Bacteriol. 184:3965-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simon, R., U. B. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vitro genetic engineering: Transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 61.Spaink, H. P. 2000. Root nodulation and infection factors produced by rhizobial bacteria. Annu. Rev. Microbiol. 54:257-288. [DOI] [PubMed] [Google Scholar]
- 62.Staggs, T. M., J. D. Fetherston, and R. D. Perry. 1994. Pleiotropic effects of a Yersinia pestis fur mutation. J. Bacteriol. 176:7614-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomas, C. E., and P. F. Sparling. 1996. Isolation and analysis of a fur mutant of Neisseria gonorrhoeae. J. Bacteriol. 178:4224-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thompson, D. K., A. S. Beliaev, C. S. Giometti, S. L. Tollaksen, T. Khare, D. P. Lies, K. H. Nealson, H. Lim, J. Yates, 3rd, C. C. Brandt, J. M. Tiedje, and J. Zhou. 2002. Transcriptional and proteomic analysis of a ferric uptake regulator (fur) mutant of Shewanella oneidensis: possible involvement of fur in energy metabolism, transcriptional regulation, and oxidative stress. Appl. Environ. Microbiol. 68:881-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Todd, J. D., M. Wexler, G. Sawers, K. H. Yeoman, P. S. Poole, and A. W. Johnston. 2002. RirA, an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum. Microbiology 148:4059-4071. [DOI] [PubMed] [Google Scholar]
- 67.Touati, D., M. Jacques, B. Tardat, L. Bouchard, and S. Despied. 1995. Lethal oxidative damage and mutagenesis are generated by iron in Δfur mutants of Escherichia coli: protective role of superoxide dismutase. J. Bacteriol. 177:2305-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vincent, J. M. 1970. A manual for the practical study of root nodule bacteria. IBP handbook no. 15. Blackwell Scientific Publishers, Oxford, United Kingdom.
- 69.Watnick, P. I., J. R. Butterton, and S. B. Calderwood. 1998. The interaction of the Vibrio cholerae transcription factors, Fur and IrgB, with the overlapping promoters of two virulence genes, irgA and irgB. Gene 209:65-70. [DOI] [PubMed] [Google Scholar]
- 70.Wexler, M., J. D. Todd, O. Kolade, D. Bellini, A. M. Hemmings, G. Sawers, and A. W. Johnston. 2003. Fur is not the global regulator of iron uptake genes in Rhizobium leguminosarum. Microbiology 149:1357-1365. [DOI] [PubMed] [Google Scholar]
- 71.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou, D., W. D. Hardt, and J. E. Galan. 1999. Salmonella typhimurium encodes a putative iron transport system within the centisome 63 pathogenicity island. Infect. Immun. 67:1974-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]