SUMMARY
Cell type-specific master transcription factors (TFs) play vital roles in defining cell identity and function. However, the roles ubiquitous factors play in the specification of cell identity remain underappreciated. Here we show that the ubiquitous CCAAT-binding NF-Y complex is required for the maintenance of embryonic stem cell (ESC) identity and is an essential component of the core pluripotency network. Genome-wide studies in ESCs and neurons reveal that not only does NF-Y regulate genes with housekeeping functions through cell type-invariant promoter-proximal binding, but also genes required for cell identity by binding to cell type-specific enhancers with master TFs. Mechanistically, NF-Y’s distinct DNA-binding mode promotes master/pioneer TF binding at enhancers by facilitating a permissive chromatin conformation. Our studies unearth a conceptually unique function for histone-fold domain (HFD) protein NF-Y in promoting chromatin accessibility and suggest that other HFD proteins with analogous structural and DNA-binding properties may function in similar ways.
INTRODUCTION
Master transcription factors (TFs) establish and/or maintain cellular identity by orchestrating distinct profiles of gene expression that are faithfully transmitted through cell division. The simplified paradigm for master TFs relies on the premise that their expression is spatiotemporally restricted to one or few cell types or lineages that depend on their activity (Oestreich and Weinmann, 2012). This assumption may have unintentionally led to the underappreciation of the roles “housekeeping” or other essential TFs, expressed ubiquitously or in a multitude of cell types, might play in cell specification. Recent studies, however, have shown that many ubiquitous factors, previously thought to have an exclusive housekeeping function, have additional cell type-specific roles (Chia et al., 2010; Cinghu et al., 2014; Golan-Mashiach et al., 2012; Kagey et al., 2010; Kim et al., 2010a; Pijnappel et al., 2013). This emerging body of evidence indicates that we are yet to fully appreciate the significance of ubiquitously expressed proteins in many settings, and argues for a comprehensive reassessment of the roles other housekeeping proteins may play in cell specification.
NF-Y, also known as the CCAAT-binding factor CBF, is a ubiquitously expressed heterotrimeric TF composed of NF-YA, NF-YB, and NF-YC subunits, all of which are conserved from yeast to human (Maity and de Crombrugghe, 1998). NF-Y binds to the CCAAT box, which occurs at ~30% of all eukaryotic promoters (Dolfini et al., 2012a). NF-YB and NF-YC dimerize via their histone-fold domains (HFDs) before associating with NF-YA (Romier et al., 2003), which harbors both DNA-binding and transactivation domains. The crystal structure of NF-Y bound to DNA shows that while NF-YA makes sequence specific DNA contacts, NF-YB/NF-YC interacts with DNA via non-specific HFD-DNA contacts (Nardini et al., 2013). The key structural feature of the NF-Y/DNA complex is the minor-groove interaction of NF-YA, which induces an approximately 80° bend in the DNA. The structure and DNA-binding mode of NF-YB/NF-YC HFDs are similar to those of the core histones H2A/H2B, TATA-binding protein (TBP)-associated factors (TAFs), the TBP/TATA-binding negative cofactor 2 (NC2α/β), and the CHRAC15/CHRAC17 subunits of the nucleosome remodeling complex CHRAC (Nardini et al., 2013). Yet, unlike H2A/H2B which lack sequence specificity, NF-YB/NF-YC interaction with NF-YA provides the NF-Y complex with sequence-specific targeting capability as well as nucleosome-like properties of non-specific DNA binding, a combination that allows for stable DNA binding.
NF-Y, largely described as a transcription activator via its promoter-proximal binding, is a key regulator of cell cycle progression in proliferating cells (Benatti et al., 2011; Bungartz et al., 2012; Hu and Maity, 2000), with its activity often downregulated during cellular differentiation and senescence (Bungartz et al., 2012; Farina et al., 1999). In addition to binding core promoters, NF-Y has also been shown to bind enhancer elements away from TSSs (Fleming et al., 2013; Testa et al., 2005), but its function and mechanism of action at these distal regulatory elements remain to be elucidated.
Consistent with its role in cell cycle regulation, NF-Y is required for ESC and hematopoietic stem cell proliferation (Bungartz et al., 2012; Dolfini et al., 2012b; Grskovic et al., 2007). While NF-YA heterozygous mice are normal and fertile, NF-YA null mice die prior to 8.5 d.p.c (Bhattacharya et al., 2003), suggesting an essential role for NF-Y in early mouse embryonic development. Interestingly, conditional deletion of NF-YA in postmitotic mouse neurons induces progressive neurodegeneration (Yamanaka et al., 2014), which suggests a role for NF-Y that is independent of its role in cell cycle regulation as has also been shown in hepatocytes (Luo et al., 2011).
Given the relatively high expression of one or more NF-Y subunits in mouse oocytes (Su et al., 2004) and the inner cell mass (ICM) of the mouse blastocyst (Yoshikawa et al., 2006), we set out to determine NF-Y’s role in ESCs, derivatives of the ICM. Although NF-Y has been predominantly studied in the context of its promoter-proximal binding at key cell cycle genes, given the prevalence of NF-Y targeting sites at greater distances from TSSs (Fleming et al., 2013), we investigated NF-Y’s function and mechanism of action at distal regulatory elements. We demonstrate a requirement for all three NF-Y subunits in the expression of core ESC self-renewal and pluripotency genes, and in the maintenance of ESC identity. Through genome-wide occupancy and transcriptomic analyses in ESCs and neurons, we show that not only does NF-Y regulate genes with housekeeping functions through cell type-invariant promoter-proximal binding, but also genes required for cell identity by binding to cell type-specific enhancers with master TFs. We present evidence that NF-Y’s distinct DNA-binding mode facilitates a permissive chromatin conformation and promotes enhanced binding of master ESC TFs at enhancers. Our studies unearth, to the best of our knowledge, a novel function for NF-Y in promoting chromatin accessibility and specification of cell identity.
RESULTS
Genomic profiling of NF-YA, NF-YB, and NF-YC binding sites in mouse ESCs
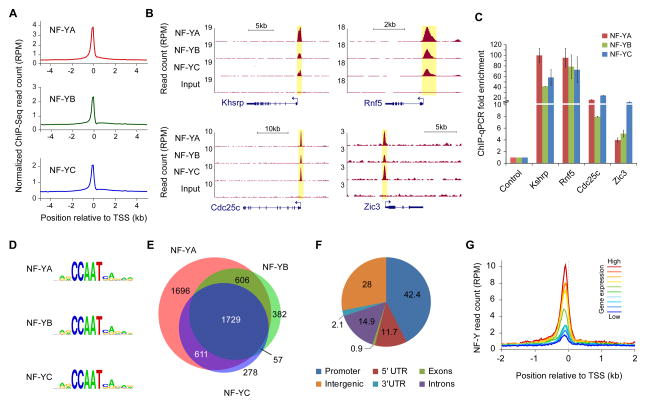
To gain insight into NF-Y-mediated transcriptional regulation in early embryogenesis, we investigated the genome-wide occupancy of all three subunits of the NF-Y complex in mouse ESCs using chromatin immunoprecipitation (ChIP) followed by sequencing (ChIP-Seq). ChIP-Seq analyses revealed enrichment for NF-Y occupancy near TSSs of annotated genes (Figure 1A), consistent with NF-Y’s preference for binding and recruiting RNA Polymerase II and general TFs to various CCAAT motif-containing promoters (Kabe et al., 2005). Binding of all three subunits to the promoters of known NF-Y targets including Cdc25c, Rnf5, and Zic3 (Grskovic et al., 2007; Yamanaka et al., 2014) attested to the sensitivity of the ChIP-Seq data (Figure 1B). ChIP followed by quantitative polymerase chain reaction (qPCR) analysis of certain sites, either previously demonstrated as NF-Y bound regions (Cdc25c, Rnf5, Zic3) or highly enriched for NF-Y binding (Khsrp) further validated the quality and the reproducibility of the ChIP-Seq data (Figure 1C). Analysis of ChIP-Seq datasets using the SISSRs peak calling algorithm (Jothi et al., 2008; Narlikar and Jothi, 2012) identified a total of 4,642, 2,774, and 2,675 binding sites (peaks) for NF-YA, NF-YB, and NF-YC, respectively, with at least 7-fold or more ChIP over input enrichment (P < 10−3). As expected, de novo sequence motif analysis identified the known RRCCAATVR consensus motif within the binding sites for all three subunits (Figure 1D).
Figure 1. Genomic profiling of NF-YA, NF-YB, and NF-YC binding sites in mouse ESCs.
(A) ChIP-Seq read density plot showing NF-YA, NF-YB, and NF-YC occupancy near TSSs of all mouse RefSeq genes.
(B) Genome browser shots showing NF-YA, NF-YB, and NF-YC occupancy at gene promoters.
(C) ChIP-qPCR validation of the NF-Y occupancy at sites highlighted in (B). Error bars represent S.E.M. of three experiments.
(D) Consensus sequence motifs enriched within NF-YA, NF-YB, and NF-YC sites using de novo motif analysis.
(E) Overlap among NF-YA, NF-YB, and NF-YC sites (peaks).
(F) Genome-wide distribution of the NF-Y sites (union of the NF-YA, NF-YB, and NF-YC sites). Promoter, 500 bp upstream of TSS.
(G) Correlation between gene expression levels and NF-Y occupancy near TSSs. See also Figure S1.
NF-Y binding requires all three subunits and correlates with gene expression
Although a vast majority of the binding sites for NF-YA, NF-YB, or NF-YC overlap with those for one or both of the other two subunits (Figure 1E), about one-third of the NF-YA binding sites did not show statistically significant evidence for the co-binding of the other two subunits. To determine whether NF-YA alone binds certain genomic loci, we knocked down NF-YA or NF-YC using siRNAs for 48 hours (Figures S1A and S1B) and analyzed their binding patterns at sites bound by all three NF-Y subunits (Figure 1B) as well as those bound only by NF-YA (Figure S1C). ChIP-qPCR analyses show that upon NF-YC knockdown (KD), NF-YA occupancy on DNA is compromised even at sites that seem to bind only NF-YA (Figure S1D), indicating that NF-YC is essential for NF-YA binding. Low NF-YB and NF-YC occupancy at sites defined as bound only by NF-YA compared to those bound by all three subunits (Figure S1E) suggest that the failure to call peaks for NF-YB/NF-YC at these sites is likely due to weaker ChIP-Seq signals that did not pass the statistical threshold for peak-calling, and/or less immuno-efficient antibodies for NF-YB /NF-YC compared to that of NF-YA. Together with the fact that nearly all of the NF-YB-only and NF-YC-only sites contain the CCAAT motif, we conclude that NF-Y binding requires all three subunits. Although we cannot completely rule out that a very low and/or transient occupancy of NF-YA alone or NF-YB/NF-YC heterodimer may exist, our data show that such events are highly unlikely if not sparse, consistent with in vitro biochemical studies showing NF-Y binding requiring all three subunits (Sinha et al., 1995). Therefore, hereafter, we will refer to the union of all NF-YA, NF-YB and NF-YC binding sites (5,359 in total) as NF-Y binding sites.
Relative to annotated RefSeq genes, a vast majority of all NF-Y binding sites are located near genic regions (Figure 1F), suggesting a role for NF-Y in regulation of gene expression. Indeed, the levels of NF-Y binding at gene promoters correlate positively with gene expression (Figure 1G), which is consistent with NF-Y’s established role as a transcription activator (Dolfini et al., 2012a).
NF-Y co-occupies enhancers with ESC-specific master TFs
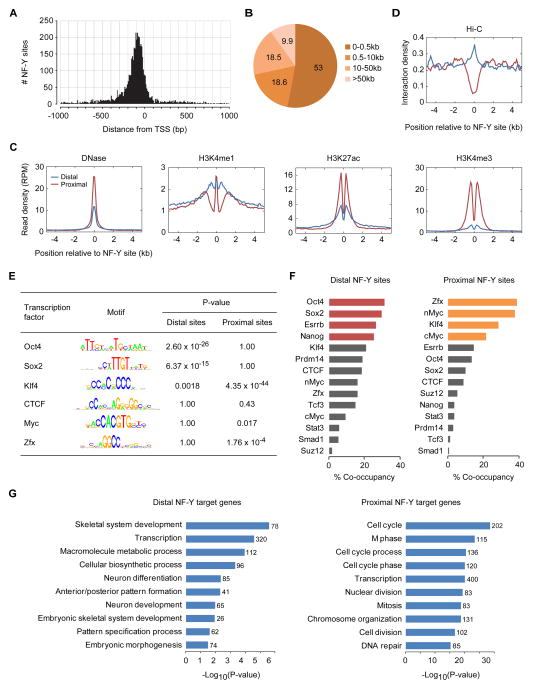
NF-Y has predominantly been reported to bind in close proximity to TSSs. While we confirm a strong enrichment for NF-Y binding sites around TSSs (within 500bp of TSSs), with a clear preference for binding to the region immediately upstream of the TSS (Figure 2A), nearly half of all NF-Y binding sites are located at distances greater than 500bp from the TSSs (Figure 2B). These data suggest a role for NF-Y in ESC transcription regulation by binding to sites distal to TSSs. To investigate the role distal sites may play in the regulation of gene expression, and how they might differ in function compared to that of promoter-proximal NF-Y sites, we examined published ChIP-Seq datasets for histone modifications, DNase I hypersensitivity, Hi-C interaction profiles, 14 different TFs, and other genomic features. As one would expect from their proximity to TSSs, nearly 80% of all promoter-proximal NF-Y sites overlap with CpG islands (Figure S1F), consistent with ~85% of all promoters overlapping CpG islands. In contrast, only about a tenth of distal NF-Y sites overlap with CpG islands (Figure S1F), a fraction similar to that observed for enhancers. Furthermore, ChromHMM annotation of chromatin states (Ernst and Kellis, 2012) show a majority of distal NF-Y sites with chromatin features reminiscent of enhancers (Figure S1G). Chromatin marks at distal NF-Y sites revealed open chromatin architecture marked by high DNase I activity, accompanied by enhancer marks H3K4me1 and H3K27ac but not the promoter mark H3K4me3 (Figure 2C). Additionally, enrichment of Hi-C interaction density at distal but not proximal NF-Y strongly support the notion that distal NF-Y sites are within enhancers (Figure 2D).
Figure 2. NF-Y co-occupies enhancers with ESC-specific master TFs.
(A) Frequency distribution of NF-Y sites within 1 kb of TSSs.
(B) Proximity of NF-Y sites to TSSs.
(C) Levels of DNase I hypersensitivity, H3K4me1, H3K27ac, and H3K4me3 near proximal and distal NF-Y sites.
(D) Hi-C interaction density near proximal and distal NF-Y sites.
(E) TF binding motifs enriched within distal vs proximal NF-Y sites. CTCF is not enriched at either set of NF-Y sites.
(F) Co-occupancy between ESC TFs (Chen et al., 2008; Ho et al., 2011; Ma et al., 2011; Marson et al., 2008) and NF-Y at distal and proximal sites.
(G) Gene ontology categories enriched among distal and proximal NF-Y target genes.
To gain insight into the mechanism underlying NF-Y’s function at enhancers, we studied the frequency of known TF binding motifs within distal and proximal NF-Y sites. We found that distal NF-Y sites are significantly enriched for sequence motifs bound by master ESC TFs known to bind distal enhancers, including Oct4 and Sox2, but not for motifs bound by ubiquitous TFs, such as CTCF (Figure 2E). In contrast, promoter-proximal NF-Y sites are significantly enriched for sequence motifs bound by Klf4, cMyc and Zfx, all known to preferentially bind to promoters (Figure 2E) (Chen et al., 2008). Furthermore, analysis of NF-Y binding in conjunction with other TF binding data revealed an extensive colocalization of NF-Y with ESC-specific master TFs Oct4, Sox2, Nanog, and Esrrb at distal NF-Y sites, and with TFs Zfx, cMyc, and Klf4 at promoter-proximal NF-Y sites (Figure 2F). Consistent with NF-Y’s co-occupancy with master ESC TFs at distal NF-Y sites, gene ontology analysis revealed enrichment for genes with roles in early embryonic development among those targeted by distal NF-Y sites (Figure 2G). Interestingly, genes with NF-Y sites at proximal-promoters are enriched for housekeeping functions including cell cycle and proliferation. Taken together, these data suggest a potential role for NF-Y in the regulation of the core pluripotency transcription network through distal enhancer binding.
NF-Y is required for the expression of core ESC self-renewal and pluripotency genes
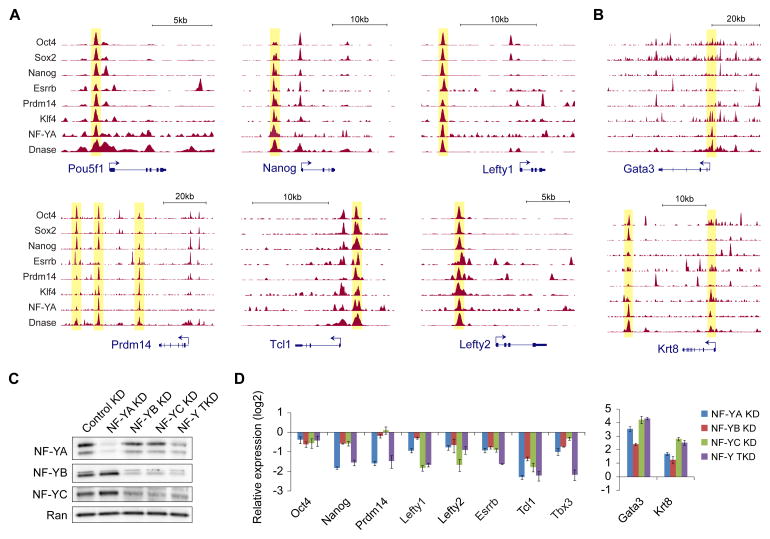
Genes targeted by NF-Y include many key pluripotency-associated genes that have been implicated in the control of ESC identity (Figure 3A). These include genes encoding the master ESC TFs Oct4, Nanog, Prdm14, Esrrb, Tcl1, Tbx3, Rex1, Lefty1, and Lefty2. More importantly, in a majority of cases, NF-Y colocalizes with master ESC TFs at distal enhancers previously demonstrated (Kagey et al., 2010; Zhang et al., 2013) to interact with promoters of these ESC identity genes (Figures 3A and S2A). This suggests that NF-Y might be regulating these genes via enhancer-promoter interactions. In addition to its binding at the enhancers of self-renewal and pluripotency genes, NF-Y binds the promoters of genes with known roles in cell cycle (Figure S2B) as well as development (including Gata3, Krt18, Krt8, Srf, Jun, Prom1, and Lef1) (Figures 3B and S2C).
Figure 3. NF-Y binds to and regulates core ESC self-renewal and pluripotency genes.
(A–B) Genome browser shots showing NF-Y co-occupancy with master ESC TFs (Chen et al., 2008; Ma et al., 2011; Marson et al., 2008) at the enhancers of ESC identity genes (A), and at the promoters/enhancers of differentiation/developmental genes (B).
(C) Western blot analysis of NF-YA, NF-YB, and NF-YC in NF-YA knockdown (KD), NF-YB KD, NF-YC KD, or NF-Y triple KD (TKD; KD of all three NF-Y subunits) ESCs 96h after siRNA transfection. Ran is used as a loading control.
(D) RT-qPCR analysis of relative mRNA levels of ESC identity genes (left), and differentiation markers (right) in NF-YA KD, NF-YB KD, NF-YC KD, and NF-Y TKD ESCs compared to control KD ESCs 96h after siRNA transfection. Data normalized to Actin, HAZ, and TBP. Error bars, SEM. See also Figure S2.
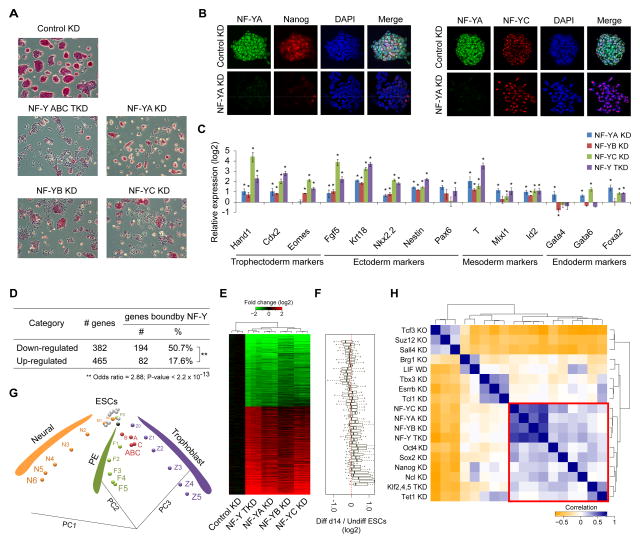
To establish a functional role for NF-Y in the regulation of these genes, we used RNAi to knockdown subunits of the NF-Y complex individually or in combination (Figures 3C and S2D) and profiled gene expression changes using qPCR following reverse-transcription (qRT-PCR). As a positive control, we observed significant changes in the expression of key cell cycle genes in cells depleted of individual NF-Y subunits (Figure S2E) accompanied by severe proliferation defects and growth arrest (Figures S2F and S2G), corroborating NF-Y’s established role in proliferation (Dolfini et al., 2012a). Depletion of NF-Y subunits decreased the expression of pluripotency-associated factors by 2- to 5-fold, and increased the expression of differentiation genes by up to an order of magnitude (Figure 3D). Together, these findings show that the NF-Y complex is required for the expression of key ESC self-renewal and pluripotency genes, and support the conclusion that not only does NF-Y regulate genes with housekeeping functions through its canonical promoter-proximal binding, but also genes required for ESC identity by co-binding distal enhancers with cell type-specific master regulators.
All three NF-Y subunits are required for ESC self-renewal and pluripotency
To investigate whether NF-Y is essential for the maintenance of ESCs in an undifferentiated state, we examined colony morphology and alkaline phosphatase (AP) staining 96 h after RNAi-mediated silencing of individual or all subunits of the NF-Y complex (Figure S2D). We observed notable self-renewal defects accompanied by loss of characteristic ESC colony morphology and AP staining (ESC marker), all consistent with ESC differentiation (Figure 4A). NF-YA KD ESCs showed profound proliferation defects compared to NF-YB/NF-YC KD ESCs, perhaps due to NF-YA being the limiting subunit of the NF-Y complex with relatively low expression compared to the other two subunits (Figure S2H). Nevertheless, these cellular changes are consistent with NF-Y’s control over both housekeeping and ESC-specific functions through distinct modes of regulation (Figures 3 and S2). At the molecular level, immunostaining experiments after NF-YA silencing revealed a drastic loss of Nanog protein levels, while NF-YC protein levels remain unchanged (Figure 4B). Furthermore, RT-qPCR analysis of ESCs depleted of NF-Y subunits revealed a significant increase in the expression of differentiation and lineage markers (Figure 4C) in addition to a significant decrease in the expression of pluripotency genes including Oct4, Nanog, Prdm14, Esrrb, Tcl1, Tbx3, and nodal antagonists Lefty1 and Lefty2 (Figure 3D), which are among the earliest to be downregulated during ESC differentiation. Based on these data, we conclude that the NF-Y complex is essential to maintain ESC identity, and that the depletion of even one of the three NF-Y subunits leads to the loss of the pluripotent state.
Figure 4. NF-Y is required for ESC self-renewal and is an essential component of the core pluripotency network.
(A) Morphology and alkaline phosphatase staining of NF-YA KD, NF-YB KD, NF-YC KD, and NF-Y TKD ESCs 96h after siRNA transfection.
(B) Co-immunostaining of NF-YA and Nanog (left)/NF- YC (right) in control and NF-YA KD ESCs 96h after siRNA transfection. Nuclei counterstained by DAPI.
(C) RT-qPCR analysis of relative mRNA levels of differentiation markers in NF-YA KD, NF-YB KD, NF-YC KD, and NF-Y TKD ESCs compared to control KD ESCs 96h after siRNA transfection. Data normalized to Actin, HAZ, and TBP. Error bars, SEM.
(D) Number of genes up- and down-regulated in NF-Y TKD ESCs 96h after siRNA transfection, and percent of those that are also bound by NF-Y.
(E) Gene expression fold changes upon NF-YA KD, NF-YB KD, NF-YC KD, or NF-Y TKD, measured 96h after siRNA transfection. Experiments performed in triplicates. Only genes that were differentially expressed (FDR ≤ 0.05 and fold change ≥ 2) in NF-Y TKD are shown.
(F) Relative gene expression changes of NF-Y regulated genes during the normal course of embryoid body formation (day 14) compared to undifferentiated ESCs. NF-Y regulated genes, ordered as in Figure 4E, were grouped into 50 bins.
(G) Principal component analysis of gene expression profiles showing NF-YA KD, NF-YB KD, NF-YC KD, and NF-Y TKD ESCs (all in red) alongside differentiation of ESCs into three different lineages (Nishiyama et al., 2009). Z0–Z5, trophoblast lineage (ESCs differentiating into trophoblast cells from day 0 to day 5; purple); N0–N6, neural lineage (ESCs differentiating into neural lineage from day 0 to day 6; orange); F0–F5, primitive endoderm (PE; embryonal carcinoma cells differentiating into PE from day 0 to day 5; green); ESCs, white; Control KD ESCs, black; A, NF-YA KD; B, NF-YB KD; C, NF-YC KD; ABC, NF-Y TKD.
(H) Correlation between global gene expression changes upon NF-YA KD, NF-YB KD, NF-YC KD, and NF-Y TKD and those observed after KD or knockout (KO) of other ESC-associated factors, as previously reported. Rows/columns ordered based on unsupervised hierarchical clustering. TKD, Triple KD; WD, Withdrawal. See also Figures S3 and S4.
NF-Y is an essential component of the core pluripotency network
To determine the extent to which NF-Y regulates gene expression programs influencing ESC self-renewal and pluripotency, we used microarrays to profile global gene expression in ESCs transfected with a control siRNA or siRNA(s) targeting individual or all of the NF-Y subunits (Figures 3C and S3A). Consistent with gene expression changes observed in the RT-qPCR analysis (Figures 3D and 4C), global gene expression changes upon silencing of all three NF-Y subunits correlated highly with those after individual knockdowns (Figure S3B). Notably, many of the NF-Y targets lie at a ‘crossroads’ of cell-fate determination during embryonic development (Figure S4). Using a stringent criteria (FDR < 0.05; fold change ≥ 2.0), we identified 847 genes that were differentially expressed in cells depleted of all three NF-Y subunits (NF-Y TKD) (Figure 4D), a vast majority of which were also differentially expressed in individual knockdowns (Figure 4E). Notably, while a majority of the down-regulated genes harbor NF-Y binding sites either within 5 kb upstream of the TSS or the gene body, only ~18% of the up-regulated genes have NF-Y binding sites (Figure 4D). With much of the up-regulation attributable to indirect effects, this is a clear indication that NF-Y plays a predomintantly activating role in ESCs.
We observed a strong positive correlation between gene expression changes upon NF-Y TKD ESCs and during the normal course of embryoid body differentiation (Fig. 4F). Consistent with this observation, principle component analysis of gene expression profiles during differentiation of ESCs into three different lineages showed ESCs depleted of NF-Y clustering away from undifferentiated ESCs (Figure 4G). Analysis of published gene expression microarray data from ESCs depleted of key pluripotency-associated factors revealed a strong correlation between the global gene expression changes upon loss of individual or all NF-Y subunits and those observed after the loss of Oct4, Sox2, Nanog, Ncl, Tet1, or Klfs (Figure 4H). These data, together with NF-Y’s co-occupancy with and regulation of master ESC TFs (Figures 2F, 3A, and 3D), places NF-Y as a member of the core pluripotency network.
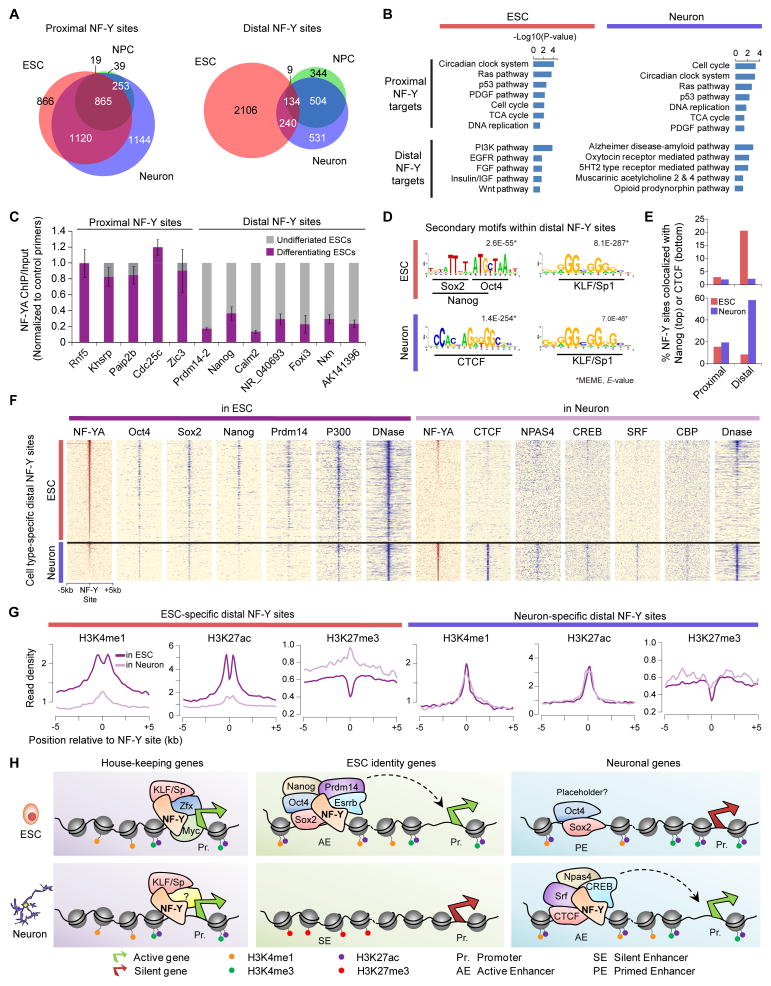
NF-Y binding at enhancers is cell type-specific, whereas NF-Y binding at promoters is cell type-invariant
To investigate whether NF-Y’s function at enhancers is broadly conserved in other cell types, we compared NF-Y binding sites in ESCs with previously published NF-Y ChIP-Seq data from ESC-derived neuronal progenitors (NPCs) and terminal neurons (Tiwari et al., 2012). Remarkably, while promoter-proximal NF-Y binding is largely conserved across the three cell types, distal NF-Y binding is cell type-specific (Figure 5A). Panther pathway analysis of genes associated with either proximal or distal NF-Y binding sites in ESCs or neurons revealed that while genes associated with proximal NF-Y binding are enriched for housekeeping functions in both cell types, genes associated with distal NF-Y binding in ESCs or neurons are highly enriched for pathways known to have critical roles in ESC maintenance or neuronal function, respectively (Figure 5B). This suggests that while promoter-proximal NF-Y binding is largely cell-type invariant and regulates several housekeeping functions, distal NF-Y binding is largely cell type-specific and controls pathways characteristic of cell identity. Hence, we reasoned that we should observe loss of ESC-specific NF-Y binding at distal sites immediately after ESCs exit from the pluripotent state. As predicted, NF-YA ChIP followed by qPCR in differentiating ESCs revealed a significant loss of NF-Y binding at distal but not cell type-invariant proximal sites (Figure 5C).
Figure 5. NF-Y binding at enhancers is cell type-specific, whereas NF-Y binding at promoters is cell type-invariant.
(A) Overlap among NF-Y sites in ESCs (red), neural progenitor cells (NPCs; green), and neurons (blue).
(B) Panther pathway enrichment analysis of proximal and distal NF-Y target genes in ESCs and neurons.
(C) ChIP-qPCR analysis of NF-YA sites in undifferentiated ESCs and differentiating ESCs (induced by retinoic acid, 96h). Error bars, SEM.
(D) Sequence motifs, other than that of NF-Y’s, enriched within distal NF-Y sites in ESCs or neurons, identified using de novo motif analysis.
(E) Nanog and CTCF (ENCODE, GSE49847) co-occupancy at promoter-proximal and ESC-specific (red) and neuron-specific (purple) distal NF-Y binding sites.
(F) TF occupancy (ChIP-Seq read density) near ESC-specific and neuron-specific distal NF-Y sites in ESCs and neurons.
(G) Levels of active enhancer marks H3K4me1 and H3K27ac, and repressive mark H3K27me3 near ESC-specific and neuron-specific distal NF-Y binding sites in ESCs (Creyghton et al., 2010; Ho et al., 2011) and neurons (ENCODE, GSE49847).
(H) Model for NF-Y mediated transcriptional regulation of housekeeping and cell identity genes. See also Figures S5 and S6.
NF-Y co-occupies active enhancers with cell-type specific master TFs
To gain additional insight into mechanisms underlying NF-Y’s cell type-specific binding, we performed de novo motif analysis of 200 bp regions centered on distal NF-Y binding sites in ESCs and neurons, in search of sequence motifs for potential cell type-specific cofactors. We found that distal NF-Y binding sites in ESCs are significantly enriched for sequence motifs known to bind Oct4, Sox2, and Nanog (Figure 5D), consistent with their colocalization at distal NF-Y sites (Figure 2F). In contrast, distal NF-Y binding sites in neurons are enriched for sequence motif known to bind CTCF, an essential master regulator of neural development (Golan-Mashiach et al., 2012). In good agreement, Oct4, Sox2, Nanog, and Prdm14 colocalize specifically at distal but not proximal NF-Y binding sites in ESCs (Figure 5E, S5A, and S5B), whereas these factors are not enriched at distal or proximal sites bound by NF-Y in neurons. In contrast, CTCF colocalizes specifically at distal NF-Y binding sites in neurons but not in ESCs (Figure 5E and S5B), and CTCF is not enriched at proximal NF-Y sites in neurons or ESCs (Figure 5E). Examination of published ChIP-Seq data for various TFs in ESCs and neurons further confirmed NF-Y binding distal enhancer sites with cell type-specific master TFs: NF-Y colocalizes with master ESC TFs Oct4, Sox2, Nanog, and Prdm14 in ESCs, and with master neuronal regulators CTCF, NPAS4, CBP, and CREB in neurons (Kim et al., 2010b) (Figure 5F). Notably, in neurons, NF-Y co-binds with CTCF, NPAS4, CBP, and CREB at distant enhancer elements controlling stochastic promoter choice and alternative isoform expression of clustered protocadherin genes (Pcdhα, -β, and -γ) (Figure S6A and S6B), which are essential for neuronal diversity (Hirayama et al., 2012).
Examination of chromatin features at ESC-specific distal NF-Y binding sites revealed an open chromatin architecture marked by high levels of H3K4me1 and H3K27ac but lacking H3K27me3 in ESCs, all hallmarks of active enhancers, as opposed to high levels of H3K27me3 and no H3K4me1 and H3K27ac in neurons (Figure 5G, left). Interestingly, however, neuron-specific distal NF-Y binding sites, despite their not binding NF-Y in ESCs, had surprisingly high levels of H3K4me1 and H3K27ac but no H3K27me3 in ESCs, comparable to those observed in neurons, indicating that neuron-specific NF-Y binding sites are “primed” in ESCs (Figure 5G, right). This, in combination with low to moderate levels of Oct4/Sox2 occupancy (Figures 5F and S5A) suggests a potential role for Oct4/Sox2 in priming lineage-specific enhancers in addition to regulating ESC specific gene expression programs. Together, these data support a model whereby NF-Y uses distinct mechanisms to regulate housekeeping and cell identity genes: NF-Y regulates housekeeping genes by binding to their proximal promoters, and promotes genes required for cell identity by binding to active enhancers along with cell type-specific master TFs (Figure 5H).
NF-Y enhances Oct4, Sox2, Nanog, and Prdm14 binding on DNA, but not vice versa
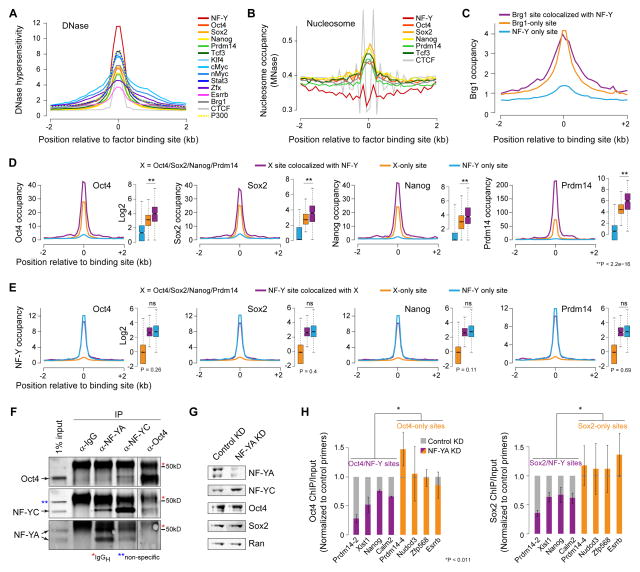
To gain insight into the mechanisms underlying NF-Y’s function at active enhancers, we examined DNase I hypersensitivity at sites bound by NF-Y and various TFs in ESCs. Intriguingly, NF-Y binding sites are the most hypersensitive to DNase I digestion compared to other TF sites analyzed, and are about two-fold more hypersensitive than those bound by master ESC TFs Oct4, Sox2, Nanog, and Prdm14 (OSNP) (Figure 6A). In agreement with this observation, NF-Y binds to sites that are relatively nucleosome-depleted (Figure 6B). In contrast OSNP bind to sites within/near nucleosomal DNA, which is consistent with previous reports showing their penchant for binding “closed chromatin” (Soufi et al., 2012; Teif et al., 2012).
Figure 6. Oct4/Sox2 binding is dependent on NF-Y.
(A) DNase I hypersensitivity, as measured by DNase-Seq (ENCODE, GSE37074), at distal binding sites for various TFs in ESCs.
(B) Nucleosome occupancy, as measured by MNase-Seq (Teif et al., 2012), at distal binding sites for various TFs in ESCs.
(C) Brg1 occupancy at distal Brg1 sites colocalized with NF-Y (purple), distal Brg1-only sites (orange), distal NF-Y sites colocalized with Brg1 (cyan).
(D) Oct4/Sox2/Nanog/Prdm14 (X) occupancy (Ma et al., 2011; Marson et al., 2008) at distal X sites colocalized with NF-Y (purple), distal X-only sites (orange), and distal NF-Y only sites (blue).
(E) NF-Y occupancy at distal NF-Y sites colocalized with X (purple), distal NF-Y only sites (blue), and distal X-only sites (orange).
(F) Co-immunoprecipitation and western blot analysis showing NF-YC and Oct4 co-immunoprecipitating with Oct4 and NF-YC, respectively. As a positive control, NF-YC and NF-YA, two subunits of NF-Y complex, co-immunoprecipitates with NF-YA and NF-YC, respectively.
(G) Western blot analysis of NF-YA, NF-YC, Oct4, and Sox2 in NF-YA KD ESCs 48h after siRNA transfection. Ran used as a loading control.
(H) ChIP-qPCR analysis of sites co-bound by Oct4/Sox2 and NF-Y (purple) or bound by Oct4/Sox2 but not NF-Y (orange) in control or NF-YA KD ESCs. Error bars, SEM. See also Figure S7.
To investigate whether the increased accessibility at NF-Y binding sites is due to NF-Y potentially recruiting a chromatin remodeler to facilitate permissive chromatin, we analyzed our previously published ChIP-Seq data for Brg1 and Ino80 (Wang et al., 2014), the ATPase subunits of the chromatin remodeling complexes esBAF and INO80, respectively. We found that the occupancy levels of chromatin remodeling factors at the NF-Y binding sites are relatively low compared to those observed at many other TF binding sites (Figures S7A and S7B), suggesting that increased hypersensitivity at NF-Y sites cannot be simply explained by higher levels of chromatin remodeling activity. Examination of NF-Y and Brg1 (esBAF) occupancy at their sites of colocalization and at sites where they bind without the other reveals that the levels of esBAF occupancy at NF-Y/esBAF colocalized sites are no higher than those at esBAF-only sites (Figure 6C), suggesting that it is less likely that NF-Y recruits esBAF. Conversely, the levels of NF-Y occupancy are the same whether or not NF-Y colocalizes with esBAF (Figure S7C), suggesting that NF-Y occupancy is independent of esBAF. Taken together, these data argue against interdependency between NF-Y and esBAF, and for a direct role for NF-Y in promoting enhanced accessibility.
NF-Y interacts with DNA through insertion of the NF-YA A2 helix into the DNA minor groove, which induces a significant 78°–80° bend in the DNA. Because NF-YA utilizes such a unique DNA-binding mode, and because the resulting bend in DNA may promote binding of other TFs in the adjacent major grooves, we hypothesized that NF-Y might foster binding of master ESC TFs at their sites of colocalization. Towards testing this hypothesis, we examined NF-Y and OSNP occupancy at their sites of colocalization and at sites where they bind without the other. Strikingly, the levels of OSNP occupancy are significantly higher at sites where they colocalize with NF-Y compared to those at sites where they do not (Figure 6D). In contrast, levels of NF-Y occupancy are the same whether or not NF-Y colocalizes with OSNP (Figure 6E), indicating that NF-Y promotes enhanced binding of OSNP at their sites of colocalization, but not vice versa. A similar phenomenon was also observed in neurons at sites colocalized by NF-Y and CTCF (Figure S7D). Consistent with this observation, target genes of Oct4/Sox2 with NF-Y co-occupancy have significantly higher expression compared to those of Oct4/Sox2 without NF-Y co-occupancy, and are significantly downregulated in NF-Y TKD cells (Figure S7E). Interestingly, de novo motif analysis of Oct4/Sox2 sites with or without NF-Y co-occupancy revealed identical Oct4/Sox2 motifs (Figure S7F), suggesting that enhanced Oct4/Sox2 binding at sites of NF-Y co-occupancy is likely due to NF-Y and not the underlying sequence motifs.
NF-Y interacts with Oct4, and Oct4/Sox2 binding is dependent on NF-Y
To establish the mode of functional interaction between NF-Y and TFs whose binding it might promote, we performed co-immunoprecipitation experiments using protein extracts from ESCs and antibodies against NF-YA or NF-YC subunits of the NF-Y complex. As expected, NF-YA co-immunoprecipitated with NF-YC and vice versa (Figure 6F). More importantly, we found that Oct4 co-immunoprecipitated with NF-YC but not NF-YA (Figure 6F). Conversely, NF-YC, but not NF-YA, co-immunoprecipitated with Oct4. Given that Oct4 interacts with the NF-YC subunit of the NF-Y complex and that Oct4 binding is enhanced at sites of NF-Y co-binding (Figure 6D), we next asked whether Oct4 binding is dependent on NF-Y. To explore this connection, we studied NF-YA KD ESCs 48h after siRNA transfection, when NF-YA is depleted but the cells appear normal with no obvious reduction in Oct4 or Sox2 levels compared to control KD ESCs (Figures 6G, S7G, and S7H). ChIP using antibodies against Oct4 or Sox2 followed by qPCR revealed a significant loss of Oct4/Sox2 binding specifically at Oct4/Sox2 sites co-bound by NF-Y but not at sites where Oct4/Sox2 bind without NF-Y (Figure 6H). These data indicate that Oct4/Sox2 binding is NF-Y dependent at sites where Oct4/Sox2 colocalize with NF-Y.
NF-Y promotes Oct4/Sox2 binding by facilitating a favorable chromatin conformation
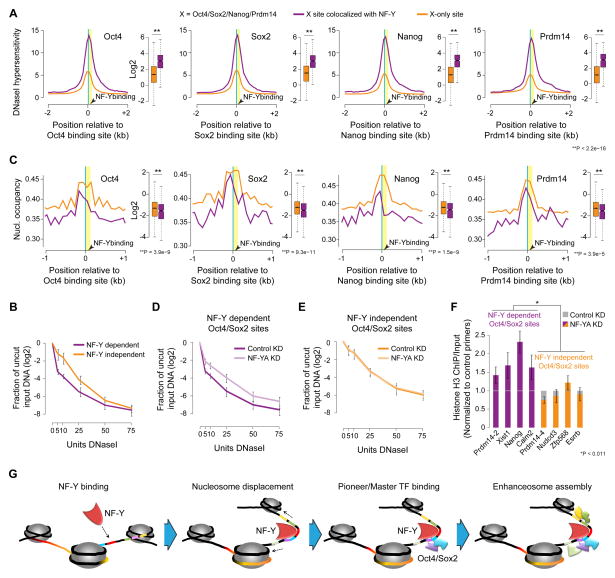
To probe the link between NF-Y deficiency and diminished Oct4/Sox2 binding at their sites of colocalization, we next examined the DNase I hypersensitivity at sites bound by OSNP either with or without NF-Y. We found that those sites co-bound by NF-Y and OSNP are two- to three-fold more hypersensitive to DNase I digestion than those that are bound only by OSNP (Figure 7A). DNase I hypersensitivity assay followed by qPCR amplification confirmed that NF-Y independent Oct4/Sox2 sites in ESCs are less accessible compared with NF-Y dependent Oct4/Sox2 sites, as shown by their greater resistance to digestion by DNase I (Figure 7B). Consistent with the higher DNase I hypersensitivity at NF-Y dependent OSNP sites (Figures 7A and 7B), we find lower nucleosome occupancy at sites where OSNP colocalize with NF-Y (Figure 7C), which is especially pronounced on the side of NF-Y occupancy. Hence, we reasoned that diminished Oct4/Sox2 binding upon NF-Y deficiency might be due to loss of intrinsic chromatin accessibility.
Figure 7. NF-Y facilitates Oct4/Sox2 binding by promoting chromatin accessibility.
(A) DNase I hypersensitivity (ENCODE, GSE37074) at NF-Y dependent (purple) and independent (orange) Oct4/Sox2/Nanog/Prdm14 sites in ESCs.
(B) DNase I and qPCR analysis of NF-Y dependent and NF-Y independent Oct4/Sox2 sites in ESCs. Error bars, SEM.
(C) Nucleosome occupancy (Teif et al., 2012) at NF-Y dependent (purple) and independent (orange) Oct4/Sox2/Nanog/Prdm14 sites in ESCs.
(D–E) DNase I and qPCR analysis of NF-Y dependent (D) and NF-Y independent (E) Oct4/Sox2 sites (n = 3) in control and NF-YA KD ESCs. Error bars, SEM.
(F) ChIP-qPCR analysis of NF-Y dependent (purple) and NF-Y independent (orange) Oct4/Sox2 sites in Control and NF-YA KD ESCs. Error bars, SEM.
(G) Proposed model for NF-Y mediated chromatin accessibility. See also Figure S7.
To investigate this possibility, we depleted NF-Y and examined the accessibility of NF-Y dependent versus NF-Y independent Oct4/Sox2 sites using a DNase I hypersensitivity assay followed by qPCR amplification of the target region to quantitatively assess the relative ‘openness’ of the region. Depletion of NF-Y results in a significant reduction in the accessibility at NF-Y dependent but not NF-Y independent Oct4/Sox2 sites (Figures 7D, 7E, S7I, and S7J). Because sites that only bind NF-Y (and not Oct4/Sox2) also lose accessibility (Figure S7K) but not control sites devoid of both NF-Y and Oct4/Sox2 binding (Figure S7L), we conclude that the observed reduction in accessibility is a direct consequence of NF-Y deficiency.
The observation that NF-Y is required to induce enhanced chromatin accessibility for Oct4/Sox2 binding prompted us to examine whether increased nucleosome occupancy accompanies reduced chromatin accessibility upon NF-Y depletion. Examination of nucleosome occupancy, as assessed using histone H3 ChIP followed by qPCR, reveals elevated levels of nucleosome presence at NF-Y dependent but not NF-Y independent Oct4/Sox2 sites upon NF-Y KD (Figure 7F). Together, these data support the conclusion that NF-Y promotes Oct4/Sox2 binding by facilitating enhanced chromatin accessibility.
DISCUSSION
NF-Y’s function has almost exclusively been studied in relation to its promoter-proximal binding (Dolfini et al., 2012a), likely driven by the prevalence of CCAAT motif within core promoters and promoter-focused arrays. In this study, by focusing on the surprisingly high proportion of NF-Y sites at distal regulatory elements, we report a cell type-specific function for NF-Y, whereby NF-Y promotes chromatin accessibility for cell type-specific master TFs at active enhancers.
NF-Y’s distinct modes of binding
Through comprehensive genome-wide occupancy analysis of NF-YA, NF-YB, and NF-YC subunits of the NF-Y complex in mouse ESCs, we find that nearly half of all NF-Y binding sites are within enhancers located at least 500 bp from TSSs. At these distal sites, NF-Y preferentially colocalizes with master ESC TFs known to bind active enhancers. In addition to confirming its regulation of key cell cycle genes via promoter-proximal binding, we show that NF-Y regulates many key pluripotency-associated genes including Oct4, Nanog, and Prdm14 by binding to their enhancers, where it colocalizes with master ESC TFs. Overall, genes targeted by distal NF-Y binding are enriched for processes related to early embryonic development, whereas those bound by NF-Y at the proximal promoter are enriched for housekeeping functions, including cell cycle.
NF-Y is required for ESC identity
In line with NF-Y’s regulation of master ESC TFs, global gene expression changes upon NF-Y KD are similar to those observed after Oct4, Sox2, Nanog, Ncl, or Tet1 KD. ESCs lacking one or more of the NF-Y subunits exhibit severe self-renewal defects, undergo differentiation, and have global expression profile that is comparable to cells differentiating toward the trophoblast lineage, all consistent with the propensity of ESCs lacking many of these master ESC factors to differentiate towards trophoectoderm (Cinghu et al., 2014; Freudenberg et al., 2012; Li et al., 2007; Nichols et al., 1998). In light of overexpression of NF-YA counteracting ESC differentiation induced by Leukemia inhibitory factor (LIF) withdrawal (Dolfini et al., 2012b), NF-Y’s broad control of key ESC identity genes including master ESC TFs places NF-Y as part of the core pluripotency network. While we have not yet established whether the proclivity of ESCs lacking NF-Y to favor differentiation towards trophectoderm (TE) is a direct effect of NF-Y deficiency, we postulate that this outcome is perhaps indirect due to NF-Y’s positive regulation of genes including Oct4 and Sox2, which are known to inhibit master TE regulator(s). An understanding of NF-Y’s role in pluripotency, lineage specification/commitment, and developmental potential will require analysis of NF-Y deficient mice during early embryonic development.
Cell type-specific function for NF-Y
A comparison of NF-Y occupancy in ESCs, and ESC-derived neuronal progenitors and terminal neurons revealed that while promoter-proximal NF-Y binding is largely conserved across cell types, distal NF-Y binding is cell type-specific. More importantly, as in ESCs where NF-Y colocalizes with master ESC TFs, NF-Y preferentially colocalizes with master neuronal regulators at active enhancers of neuronal genes. These findings, along with those linking NF-Y to neuronal identity (Tiwari et al., 2012), point to a critical role for NF-Y in the specification of neuronal cells that is independent of its role in cell cycle progression. This supposition is in accordance with a recent study showing that conditional deletion of NF-YA in postmitotic mouse neurons induces progressive neurodegeneration (Yamanaka et al., 2014).
Consistent with NF-Y’s role in ESC specification, ESCs lose NF-Y binding at distal but not proximal sites immediately after they exit the pluripotent state, with ESC-specific distal NF-Y sites epigenetically silenced in neurons. Interestingly, however, neuron-specific NF-Y binding sites, despite their not binding NF-Y in ESCs, are within ‘open’ chromatin regions marked by DNase I hypersensitivity, histone marks characteristic of active enhancers, and low to moderate levels of Oct4/Sox2 occupancy. Although further studies are required to ascertain the potential role for Oct4/Sox2 at these sites, we postulate that Oct4/Sox2 binding at lineage-specific enhancers might serve as placeholders for lineage-specific TFs, ‘priming’ these sites for rapid initiation of lineage-specific gene expression programs during differentiation. Such a role for Oct4/Sox2 in priming the chromatin would be different from, but consistent with, their pioneer activity during induced pluripotent stem cell (iPSC) reprogramming (Soufi et al., 2012). Further investigation is needed to precisely characterize the functional significance of these “primed enhancers”.
NF-Y promotes chromatin accessibility for master TFs
The levels of Oct4, Sox2, Nanog, and Prdm14 occupancy in ESCs, and CTCF occupancy in neurons are significantly higher at sites where they colocalize with NF-Y compared to those sites where they bind without NF-Y. This enhancement cannot be attributed exclusively to cooperative binding of TFs because NF-Y occupancy levels are invariant whether or not it colocalizes with other factors. This inequitable augmentation suggests that NF-Y promotes binding of master TFs at their sites of colocalization. Supporting this line of argument, NF-Y depletion in ESCs results in reduced Oct4/Sox2 occupancy at NF-Y dependent but not NF-Y independent Oct4/Sox2 sites, accompanied by reduced chromatin accessibility.
NF-Y’s role in nucleosome displacement, pioneer factor binding, and enhanceosome assembly
As pioneer factors, Oct4 and Sox2 bind nucleosomal DNA in ESCs (Teif et al., 2012), as also observed here and during reprogramming (Soufi et al., 2012). Given the enhanced binding of master TFs at sites of NF-Y colocalization, our observations raise a tantalizing possibility that Oct4/Sox2 and perhaps other pioneer factors with a preference for binding nucleosomal DNA view the NF-Y/DNA complex, with its nucleosome-like properties, as an attractive target. This supposition may not be farfetched given that the structure and DNA-binding mode of the NF-YB/NF-YC dimer are nearly identical to that of histones H2A/H2B. Additionally, Oct4’s interaction with NF-Y, as shown here, would add to a stronger and a more stable Oct4/Sox2 binding, enabling Oct4/Sox2 to recruit other TFs for an ordered enhanceosome assembly (Chen et al., 2014), consistent with the proposed role for pioneer factors (Zaret and Carroll, 2011). Given that in vitro studies have previously demonstrated NF-Y to displace nucleosomes (Coustry et al., 2001), it will be interesting to explore whether and how NF-Y mediates pioneer activity or even how NF-Y itself could play the role of a pioneer factor.
Based on our findings, we propose a general model wherein NF-Y facilitates enhanced chromatin accessibility via two modes. First, NF-Y’s nucleosome-like properties provide for a stable and a stronger binding of pioneer factors Oct4/Sox2. Second, NF-Y’s unique DNA-binding mode, which induces ~80° bend in the DNA, imposes sufficient spatial constraints to induce local nucleosome repositioning (such as flanking nucleosomes sliding down), thus allowing and/or promoting binding of other TFs, whose recognition sequences become more accessible (Figure 7F). While we could not find evidence supporting increased chromatin remodeling activity at NF-Y binding sites by known chromatin remodelers, we cannot rule out the possibility that the increased accessibility at NF-Y binding sites could be due to NF-Y potentially recruiting a bona fide chromatin remodeler to facilitate permissive chromatin. Given that CHRAC15 and CHRAC17, two small subunits of the nucleosome remodeling complex CHRAC with HFDs structurally similar to that of NF-YB/NF-YC and histones H2A/H2B, have previously been shown to facilitate nucleosome sliding (Kukimoto et al., 2004), it is tempting to speculate that NF-Y, with its sequence specific binding ability, may be a ‘pseudo chromatin remodeler’ mimicking the role of a bona fide ATP-dependent chromatin remodeler, albeit at a very small and a limited scale. As in the case of chromatin remodeling enzymes, a formal demonstration of NF-Y’s role in chromatin remodeling, direct or indirect, will require in vitro biochemical experiments involving reconstituted nucleosomes.
Although NF-Y’s role in transcription is widely accepted to be through transactivation, mechanistic details remain undetermined. Based on our findings, it is conceivable that NF-Y’s primary mode of action, both at enhancers and promoters, is to promote binding of other TFs including basal transcription machinery by facilitating an accessible chromatin. This supposition is consistent with a strong positive correlation between NF-Y binding and gene expression.
Our findings establish NF-Y as an essential regulator of the pluripotent state in ESCs, and suggest a role for NF-Y in cell specification. It remains to be seen how NF-Y selectively targets cell type-specific enhancers. Perhaps, this is accomplished through direct recruitment by a cell type-specific TF, alternative isoform usage, and/or post-translational modifications. In summary, our studies unearth a cell type-specific function for NF-Y in promoting chromatin accessibility. Our data suggest that other HFD proteins, with structural and DNA-binding properties analogous to NF-Y, may function in similar ways.
EXPERIMENTAL PROCEDURES
See Supplemental Experimental Procedures.
Mouse ES Cell Culture, RNAi, and AP staining
Mouse ESC culture, siRNA transfection, and alkaline phosphatase (AP) staining were performed as previously described (Freudenberg et al., 2012), and as detailed in the Supplemental Experimental Procedures.
Chromatin Immunoprecipitation (ChIP)
Mouse ESCs (1×107) were cross-linked with 1% formaldehyde, and the reaction was quenched with glycine for 5 min. Cells were lysed and then sonicated for 16 cycles (30sec ON; 50sec OFF) to obtain ~200–500 bp fragments. ChIP was peformed as described in Supplemental Experimental Procedure. See Table S1 for a list of ChIP primers used.
DNase I Hypersensitivity
Mouse ESCs treated with non-targeting control siRNA or NF-YA siRNA were collected 48 hours post-transfection. Nuclei isolation and DNase I digestion were performed as previously described (Burch and Weintraub, 1983), with minor modifications as detailed in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank K Adelman, G Hu, and PA Wade for critical comments on the manuscript, and RS Williams and TK Archer for useful discussion. We thank G Hu and X Zheng for guidance on RNAi experiments, S Peddada for guidance on statistical tests, J Hoffman for guidance on DNase experiments. We thank NIEHS Microarray, Next-Generation Sequencing, Confocal Microscopy, and Flow Cytometry cores for their support. This work was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (R.J.: 1ZIAES102625).
Footnotes
ACCESSION NUMBER
ChIP-Seq and microarray data generated for this study have been deposited in the GEO repository under the accession number GSE56840.
Supplemental information includes seven figures, two tables, and Supplemental Experimental Procedures.
AUTHOR CONTRIBUTIONS
A.J.O. and R.J. conceived the study. A.J.O., A.E.C., and S.C., performed the experiments. P.Y., J.E.M., S.Y., and R.J. analyzed the data. A.J.O., P.Y., and R.J. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Benatti P, Dolfini D, Vigano A, Ravo M, Weisz A, Imbriano C. Specific inhibition of NF-Y subunits triggers different cell proliferation defects. Nucleic Acids Res. 2011;39:5356–5368. doi: 10.1093/nar/gkr128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Deng JM, Zhang Z, Behringer R, de Crombrugghe B, Maity SN. The B subunit of the CCAAT box binding transcription factor complex (CBF/NF-Y) is essential for early mouse development and cell proliferation. Cancer Res. 2003;63:8167–8172. [PubMed] [Google Scholar]
- Bungartz G, Land H, Scadden DT, Emerson SG. NF-Y is necessary for hematopoietic stem cell proliferation and survival. Blood. 2012;119:1380–1389. doi: 10.1182/blood-2011-06-359406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch JB, Weintraub H. Temporal order of chromatin structural changes associated with activation of the major chicken vitellogenin gene. Cell. 1983;33:65–76. doi: 10.1016/0092-8674(83)90335-5. [DOI] [PubMed] [Google Scholar]
- Chen J, Zhang Z, Li L, Chen BC, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156:1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Chia NY, Chan YS, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau MS, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- Cinghu S, Yellaboina S, Freudenberg JM, Ghosh S, Zheng X, Oldfield AJ, Lackford BL, Zaykin DV, Hu G, Jothi R. Integrative framework for identification of key cell identity genes uncovers determinants of ES cell identity and homeostasis. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1318598111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustry F, Hu Q, de Crombrugghe B, Maity SN. CBF/NF-Y functions both in nucleosomal disruption and transcription activation of the chromatin-assembled topoisomerase IIalpha promoter. Transcription activation by CBF/NF-Y in chromatin is dependent on the promoter structure. J Biol Chem. 2001;276:40621–40630. doi: 10.1074/jbc.M106918200. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolfini D, Gatta R, Mantovani R. NF-Y and the transcriptional activation of CCAAT promoters. Crit Rev Biochem Mol Biol. 2012a;47:29–49. doi: 10.3109/10409238.2011.628970. [DOI] [PubMed] [Google Scholar]
- Dolfini D, Minuzzo M, Pavesi G, Mantovani R. The short isoform of NF-YA belongs to the embryonic stem cell transcription factor circuitry. Stem Cells. 2012b;30:2450–2459. doi: 10.1002/stem.1232. [DOI] [PubMed] [Google Scholar]
- Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina A, Manni I, Fontemaggi G, Tiainen M, Cenciarelli C, Bellorini M, Mantovani R, Sacchi A, Piaggio G. Down-regulation of cyclin B1 gene transcription in terminally differentiated skeletal muscle cells is associated with loss of functional CCAAT-binding NF-Y complex. Oncogene. 1999;18:2818–2827. doi: 10.1038/sj.onc.1202472. [DOI] [PubMed] [Google Scholar]
- Fleming JD, Pavesi G, Benatti P, Imbriano C, Mantovani R, Struhl K. NF-Y coassociates with FOS at promoters, enhancers, repetitive elements, and inactive chromatin regions, and is stereo-positioned with growth-controlling transcription factors. Genome Res. 2013;23:1195–1209. doi: 10.1101/gr.148080.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenberg JM, Ghosh S, Lackford BL, Yellaboina S, Zheng X, Li R, Cuddapah S, Wade PA, Hu G, Jothi R. Acute depletion of Tet1-dependent 5-hydroxymethylcytosine levels impairs LIF/Stat3 signaling and results in loss of embryonic stem cell identity. Nucleic Acids Res. 2012;40:3364–3377. doi: 10.1093/nar/gkr1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan-Mashiach M, Grunspan M, Emmanuel R, Gibbs-Bar L, Dikstein R, Shapiro E. Identification of CTCF as a master regulator of the clustered protocadherin genes. Nucleic Acids Res. 2012;40:3378–3391. doi: 10.1093/nar/gkr1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grskovic M, Chaivorapol C, Gaspar-Maia A, Li H, Ramalho-Santos M. Systematic identification of cis-regulatory sequences active in mouse and human embryonic stem cells. PLoS Genet. 2007;3:e145. doi: 10.1371/journal.pgen.0030145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Tarusawa E, Yoshimura Y, Galjart N, Yagi T. CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep. 2012;2:345–357. doi: 10.1016/j.celrep.2012.06.014. [DOI] [PubMed] [Google Scholar]
- Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol. 2011;13:903–913. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Maity SN. Stable expression of a dominant negative mutant of CCAAT binding factor/NF-Y in mouse fibroblast cells resulting in retardation of cell growth and inhibition of transcription of various cellular genes. J Biol Chem. 2000;275:4435–4444. doi: 10.1074/jbc.275.6.4435. [DOI] [PubMed] [Google Scholar]
- Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabe Y, Yamada J, Uga H, Yamaguchi Y, Wada T, Handa H. NF-Y is essential for the recruitment of RNA polymerase II and inducible transcription of several CCAAT box-containing genes. Mol Cell Biol. 2005;25:512–522. doi: 10.1128/MCB.25.1.512-522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010a;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010b;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukimoto I, Elderkin S, Grimaldi M, Oelgeschlager T, Varga-Weisz PD. The histone-fold protein complex CHRAC-15/17 enhances nucleosome sliding and assembly mediated by ACF. Mol Cell. 2004;13:265–277. doi: 10.1016/s1097-2765(03)00523-9. [DOI] [PubMed] [Google Scholar]
- Li J, Pan G, Cui K, Liu Y, Xu S, Pei D. A dominant-negative form of mouse SOX2 induces trophectoderm differentiation and progressive polyploidy in mouse embryonic stem cells. J Biol Chem. 2007;282:19481–19492. doi: 10.1074/jbc.M702056200. [DOI] [PubMed] [Google Scholar]
- Luo R, Klumpp SA, Finegold MJ, Maity SN. Inactivation of CBF/NF-Y in postnatal liver causes hepatocellular degeneration, lipid deposition, and endoplasmic reticulum stress. Sci Rep. 2011;1:136. doi: 10.1038/srep00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Swigut T, Valouev A, Rada-Iglesias A, Wysocka J. Sequence-specific regulator Prdm14 safeguards mouse ESCs from entering extraembryonic endoderm fates. Nat Struct Mol Biol. 2011;18:120–127. doi: 10.1038/nsmb.2000. [DOI] [PubMed] [Google Scholar]
- Maity SN, de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci. 1998;23:174–178. doi: 10.1016/s0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardini M, Gnesutta N, Donati G, Gatta R, Forni C, Fossati A, Vonrhein C, Moras D, Romier C, Bolognesi M, et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell. 2013;152:132–143. doi: 10.1016/j.cell.2012.11.047. [DOI] [PubMed] [Google Scholar]
- Narlikar L, Jothi R. ChIP-Seq data analysis: identification of protein-DNA binding sites with SISSRs peak-finder. Methods Mol Biol. 2012;802:305–322. doi: 10.1007/978-1-61779-400-1_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Nishiyama A, Xin L, Sharov AA, Thomas M, Mowrer G, Meyers E, Piao Y, Mehta S, Yee S, Nakatake Y, et al. Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors. Cell Stem Cell. 2009;5:420–433. doi: 10.1016/j.stem.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich KJ, Weinmann AS. Master regulators or lineage-specifying? Changing views on CD4+ T cell transcription factors. Nat Rev Immunol. 2012;12:799–804. doi: 10.1038/nri3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijnappel WW, Esch D, Baltissen MP, Wu G, Mischerikow N, Bergsma AJ, van der Wal E, Han DW, Bruch H, Moritz S, et al. A central role for TFIID in the pluripotent transcription circuitry. Nature. 2013;495:516–519. doi: 10.1038/nature11970. [DOI] [PubMed] [Google Scholar]
- Romier C, Cocchiarella F, Mantovani R, Moras D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J Biol Chem. 2003;278:1336–1345. doi: 10.1074/jbc.M209635200. [DOI] [PubMed] [Google Scholar]
- Sinha S, Maity SN, Lu J, de Crombrugghe B. Recombinant rat CBF-C, the third subunit of CBF/NFY, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc Natl Acad Sci U S A. 1995;92:1624–1628. doi: 10.1073/pnas.92.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teif VB, Vainshtein Y, Caudron-Herger M, Mallm JP, Marth C, Hofer T, Rippe K. Genome-wide nucleosome positioning during embryonic stem cell development. Nat Struct Mol Biol. 2012;19:1185–1192. doi: 10.1038/nsmb.2419. [DOI] [PubMed] [Google Scholar]
- Testa A, Donati G, Yan P, Romani F, Huang TH, Vigano MA, Mantovani R. Chromatin immunoprecipitation (ChIP) on chip experiments uncover a widespread distribution of NF-Y binding CCAAT sites outside of core promoters. J Biol Chem. 2005;280:13606–13615. doi: 10.1074/jbc.M414039200. [DOI] [PubMed] [Google Scholar]
- Tiwari VK, Stadler MB, Wirbelauer C, Paro R, Schubeler D, Beisel C. A chromatin-modifying function of JNK during stem cell differentiation. Nat Genet. 2012;44:94–100. doi: 10.1038/ng.1036. [DOI] [PubMed] [Google Scholar]
- Wang L, Du Y, Ward JM, Shimbo T, Lackford B, Zheng X, Miao Y-l, Zhou B, Han L, Fargo DC, et al. INO80 Facilitates Pluripotency Gene Activation in Embryonic Stem Cell Self-Renewal, Reprogramming, and Blastocyst Development. Cell Stem Cell. 2014;14:575–591. doi: 10.1016/j.stem.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka T, Tosaki A, Kurosawa M, Matsumoto G, Koike M, Uchiyama Y, Maity SN, Shimogori T, Hattori N, Nukina N. NF-Y inactivation causes atypical neurodegeneration characterized by ubiquitin and p62 accumulation and endoplasmic reticulum disorganization. Nat Commun. 2014;5:3354. doi: 10.1038/ncomms4354. [DOI] [PubMed] [Google Scholar]
- Yoshikawa T, Piao Y, Zhong J, Matoba R, Carter MG, Wang Y, Goldberg I, Ko MS. High-throughput screen for genes predominantly expressed in the ICM of mouse blastocysts by whole mount in situ hybridization. Gene Expr Patterns. 2006;6:213–224. doi: 10.1016/j.modgep.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wong CH, Birnbaum RY, Li G, Favaro R, Ngan CY, Lim J, Tai E, Poh HM, Wong E, et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504:306–310. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.