Summary
The binding of recombinant AtACBP1 to very-long-chain acyl-CoA esters is related to AtACBP1 function in Arabidopsis stem cuticle metabolism. Loss-of-function mutation adversely affected stem cuticle composition and structure.
Key words: Acyl-CoA-binding protein, Arabidopsis thaliana, cuticle, cuticular wax, cutin, very-long-chain acyl-CoAs.
Abstract
The membrane-anchored Arabidopsis thaliana ACYL-COA-BINDING PROTEIN1 (AtACBP1) plays important roles in embryogenesis and abiotic stress responses, and interacts with long-chain (LC) acyl-CoA esters. Here, AtACBP1 function in stem cuticle formation was investigated. Transgenic Arabidopsis transformed with an AtACBP1pro::GUS construct revealed β-glucuronidase (GUS) expression on the stem (but not leaf) surface, suggesting a specific role in stem cuticle formation. Isothermal titration calorimetry results revealed that (His)6-tagged recombinant AtACBP1 interacts with LC acyl-CoA esters (18:1-, 18:2-, and 18:3-CoAs) and very-long-chain (VLC) acyl-CoA esters (24:0-, 25:0-, and 26:0-CoAs). VLC fatty acids have been previously demonstrated to act as precursors in wax biosynthesis. Gas chromatography (GC)–flame ionization detector (FID) and GC–mass spectrometry (MS) analyses revealed that an acbp1 mutant showed a reduction in stem and leaf cuticular wax and stem cutin monomer composition in comparison with the wild type (Col-0). Consequently, the acbp1 mutant showed fewer wax crystals on the stem surface in scanning electron microscopy and an irregular stem cuticle layer in transmission electron microscopy in comparison with the wild type. Also, the mutant stems consistently showed a decline in expression of cuticular wax and cutin biosynthetic genes in comparison with the wild type, and the mutant leaves were more susceptible to infection by the necrotrophic pathogen Botrytis cinerea. Taken together, these findings suggest that AtACBP1 participates in Arabidopsis stem cuticle formation by trafficking VLC acyl-CoAs.
Introduction
The Arabidopsis cuticle is a lipophilic layer that consists of cutin and wax (Nawrath, 2002; Kunst and Samuels, 2009; Lee and Suh, 2013). Cutin is a polymer derived from hydroxy and epoxy-hydroxy C16 and C18 fatty acids (Nawrath, 2006). Cutin biosynthesis consists of sequential reactions including the activation of acyl chains to coenzyme A by long-chain acyl-CoA synthetase (LACS), hydroxylation and epoxidation catalysed by the cytochrome P450 family, and esterification to glycerol-3-phosphate by glycerol-3-phosphate acyltransferases (GPATs) (Li-Beisson et al., 2013). Waxes, including epicuticular waxes that cover the cuticle membrane and intracuticular waxes embedded in the cuticle membrane, are complex mixtures of alcohols, alkanes, aldehydes, ketones, and esters derived from long-chain fatty acids (Jenks et al., 2002). In plant epidermal cells, saturated very-long-chain fatty acids (VLCFAs), comprising acyl chains exceeding 20 carbons (>C20) generated by the extension of C16 and C18 fatty acids in the endoplasmic reticulum (ER), form precursors for the synthesis of aliphatic components of cuticular waxes (Kunst and Samuels, 2003).
Acyl-CoA-binding proteins (ACBPs) constitute a family of eukaryotic proteins that show conservation in an acyl-CoA-binding (ACB) domain with ability to bind long-chain acyl-CoA esters (Knudsen et al., 2000; Xiao and Chye, 2009, 2011a; Fan et al., 2010; Yurchenko and Weselake, 2011). In the model plants Arabidopsis thaliana and Oryza sativa, six genes designated as AtACBP1–AtACBP6 and OsACBP1–OsACBP6, respectively, encode ACBPs that bind acyl-CoA esters and phospholipids with varying affinities (Engeseth et al., 1996; Chye, 1998; Chye et al., 2000; Leung et al., 2004, 2006; Chen et al., 2008; Gao et al., 2009; Xiao et al., 2009; Meng et al., 2011, 2014; Du et al., 2013b ). Variation in subcellular localization has also been observed (Chye et al., 1999; Li and Chye, 2003; Leung et al., 2006; Xiao et al., 2008b ; Xiao and Chye, 2009; Meng et al., 2014). Thus, given the differences in subcellular localization and substrate preference, it appears that some Arabidopsis ACBPs perform distinct cellular functions in vivo while others with similar subcellular localization and binding affinities for acyl-CoA esters may share overlapping roles (Chye et al., 1999, 2000; Li and Chye, 2003; Leung et al., 2006; Xiao et al., 2008b ; Xiao and Chye, 2009, 2011a). For example, AtACBP1 and AtACBP2 both show functions in embryogenesis (Chen et al., 2010) and seedling development (Du et al., 2013a , b ), while AtACBP3 promotes starvation-induced leaf senescence, age-dependent leaf senescence (Xiao et al., 2010), and plant defence against Pseudomonas syringae (Xiao and Chye, 2011b ; Zheng et al., 2012). AtACBPs are also associated with heavy metal/oxidative (Xiao et al., 2008a ; Gao et al., 2009, 2010), freezing (Chen et al., 2008; Du et al., 2010; Liao et al., 2014), and drought (Du et al., 2013a ) stresses. AtACBP1 (Du et al., 2013b ) and AtACBP2 (Gao et al., 2009, 2010) both mediate protein–protein interactions by binding transcription factors and stress-responsive partners.
AtACBP1 was subcellularly localized to the ER and the plasma membrane (PM) (Li and Chye, 2003), and immunoelectron microscopy using anti-AtACBP1 antibody revealed that the AtACBP1 protein accumulates in developing embryos (Chye et al., 1999). The roles of AtACBP1 in embryo development were confirmed by phenotypic and biochemical studies using the acbp1 T-DNA insertional mutant (Chen et al., 2010). Alterations in membrane lipid composition and acyl-CoA content in the acbp1 siliques were observed. In addition, the observation of arrest of early embryo development in the acbp1acbp2 double mutant suggested that AtACBP1 and AtACBP2 are essential during early embryogenesis in Arabidopsis (Chen et al., 2010), most probably in lipid transfer because (His)6-tagged recombinant AtACBP1 (rACBP1) and AtACBP2 bind acyl-CoA esters and both demonstrated preference for unsaturated over saturated long-chain acyl-CoA esters (Chye, 1998; Chye et al., 2000; Leung et al., 2006; Gao et al., 2009).
AtACBP1 has been observed to accumulate in the outer integument cells of the developing seed coat and has been previously proposed to be involved in the biosynthesis of cutin and cuticular waxes (Chye et al., 1999). A membrane-associated ACBP from Agave americana showing 62% amino acid identity to AtACBP1, AaACBP1, was enriched in the epidermis of mature leaves (Guerrero et al., 2006). Besides the presence of the conserved ACB domain, AaACBP1 contains ankyrin repeats which potentially mediate protein–protein interactions (Michaely and Bennett, 1992) similarly to AtACBP1 (Xiao and Chye, 2011a ). The expression of AaACBP1 in the epidermal cells (Guerrero et al., 2006) and observation of AtACBP1 localization at the endomembranes (Chye et al., 1999) support the feasibility of these ACBPs as candidates involved in the biosynthesis of cuticular lipids (Chye et al, 1999; Kunst and Samuels, 2003; Xiao and Chye, 2011a ; Li-Beisson et al., 2013). Interestingly, in mice, ACBP has been reported to be essential in the formation of an epidermal barrier (Bloksgaard et al., 2012), while Xia et al. (2012) demonstrated that AtACBP3, AtACBP4, and AtACBP6 function in cuticle formation in Arabidopsis. To address the role of AtACBP1 in cuticle formation, it was first demonstrated that (His)6-tagged rACBP1 can bind very-long-chain (VLC) acyl-CoA esters in vitro. Subsequently, investigations on the acbp1 T-DNA insertional mutant showed that it displayed reduction in cuticular wax and cutin monomer composition in Arabidopsis stems, suggesting that AtACBP1 functions in stem cuticle formation.
Materials and methods
Plant materials and growth conditions
Seeds of wild-type A. thaliana (ecotype Col-0), the acbp1 mutant (SAIL-653-B06, ecotype Col-0; Xiao et al., 2008a ), and acbp1-COM (Xiao et al., 2008a ) were surface-sterilized, cold-stratified, and germinated on Murashige and Skoog medium (MS medium) (Murashige and Skoog, 1962) supplemented with 2% sucrose for 10 d under cycles of 8h dark (21 °C) and 16h light (23 °C). Plants were transferred into soil and were grown in a growth chamber under a 16h light/8h dark cycle. Stems were harvested from 6-week-old plants for gas chromatography (GC) analysis following Lee et al. (2009b ).
Expression and purification of rACBP1
(His)6-AtACBP1 recombinant protein was expressed in the soluble fraction of Escherichia coli BL21(DE3), and was purified through Ni-NTA agarose (Qiagen, Valencia, CA, USA) affinity columns as previously described (Chye, 1998).
Isothermal titration calorimetry (ITC) measurements
ITC experiments were performed using an isothermal titration calorimeter (MicroCal iTC200 system) from MicroCal Inc. (USA). Long-chain and VLC acyl-CoA esters used in this study were purchased from Avanti Polar Lipids (http://www.avantilipids.com/). The acyl-CoA concentration (250 μM) in the titration syringe was 25-fold higher than the protein concentration (10 μM) in the cell. Acyl-CoA solutions and rACBP1 protein were degassed under vacuum and stirred immediately before use. The experiments were performed at 30 °C, and injections were initiated after equilibration to baseline stability. Each injection was made up to a volume of 1.5 μl and lasted 10 s, with an interval of 240 s between injections. The syringe was rotated at 1000rpm during the assay to ensure immediate mixing. Raw data were integrated, corrected for non-specific heat, and analysed using the ORIGIN software supplied with the instrument by the General Electric Company. The dissociation constant (K D) was calculated by non-linear regression fitting the isotherm.
β-Glucuronidase (GUS) histochemical assays
GUS histochemical assays were carried out on AtACBP1pro::GUS Arabidopsis transformed with construct pAT352 according to Du et al. (2013b ). The standard 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc) solution (100mM sodium phosphate buffer, pH 7.0, 0.1% Triton X-100, 1mg ml–1 X-Gluc) with the addition of 2mM potassium ferricyanide and 2mM potassium ferrocyanide was used. Leaves and stems from 4-week-old AtACBP1pro::GUS transgenic Arabidopsis were vacuum-infiltrated in X-Gluc solution for 30min and kept at 37 °C until a blue colour developed. Samples were destained in 70% ethanol and photographed. The controls in the GUS assays were samples from Col-0 and transgenic Arabidopsis transformed with vector pBI101.3, and they were not stained blue during the same incubation period. The GUS-stained stems and leaves were embedded in Paraplast for sectioning according to Sin et al. (2006).
Quantitative real-time polymerase chain reactions (qRT-PCRs)
Total RNA was isolated from stems of five 5-week-old Arabidopsis plants using the RNeasy Plant Mini Kit following the protocol provided by QIAGEN. RNA (3.5 μg) was reverse-transcribed into cDNA using the SuperScript First-Strand Synthesis System (Invitrogen). PCR was conducted on a StepOne Plus Real-time PCR system using SYBR Green Mix (Applied Biosystems) in the following steps: 10min at 95 °C followed by 40 cycles of 95 °C (15 s) and 56 °C (1min). For each reaction, three experimental replicates were performed with gene-specific primers, and Arabidopsis ACTIN2 was used as an internal control (Supplementary Table S1 available at JXB online). The relative expression of the targeted gene was normalized using the ACTIN2 control as described (Xiao et al., 2011b ).
Scanning electron microscopy (SEM)
Stems of the first internodes above the rosette from 6-week-old wild-type Arabidopsis and the acbp1 mutant were used. Samples were treated with 1% osmium tetroxide (OsO4) for 24h and then air-dried for 3 d, followed by mounting onto standard aluminium stubs and sputter coating with gold particles using six 30-s bursts according to Chen et al. (2003). The coated samples were viewed with a Hitachi S3400 scanning electron microscope.
Transmission electron microscopy (TEM)
The ultrastructure of 6-week-old stems from wild-type Arabidopsis and the acbp1 mutant were prepared for TEM following Sieber et al. (2000) with some modifications. Samples were fixed using 2.5% glutaraldehyde in cacodylate buffer (0.1M sodium cacodylate-HCl buffer, pH 7.4) for 4h at 4 °C, followed by post-fixation treatment with 1% osmium tetroxide in cacodylate buffer for 4h at 4 °C. After gradient dehydration with ethanol, samples were infiltrated overnight in an epoxy resin/propylene oxide 1:1 mixture. This was followed by infiltration overnight in epoxy resin. Samples were subsequently embedded in epoxy resin and polymerized overnight at 60 °C. Ultrathin (60nm) sections were prepared and stained with 2% uranyl acetate and lead citrate (Li et al., 2008), and subsequently examined using a Phillips CM100 transmission electron microscope.
Wax analysis
Cuticular waxes were extracted by immersing two stem segments (each 10-cm in length) in 5ml of chloroform for 30 s. The internal standards used were C28 alkane (n-octacosane), C22 fatty acid (docosanoic acid), and C23:0 fatty alcohol (1-tricosanol). The solvent was then removed by heating (40 °C) under a mild stream of nitrogen. Derivatization was performed by adding 100 μl of pyridine and 100 μl of bis N,N-(trimethylsilyl) trifluoroacetamide (Sigma) to the dried extract and incubating for 30min at 90 °C. The qualitative composition was then evaluated by capillary GC–mass spectrometry (GC-MS; GCMS-QP2010; Shimadzu; column, 60 m HP-5, 0.32mm id, film thickness=0.25 μm; Agilent) using a helium carrier gas inlet pressure of 1.0ml min–1 and a mass spectrometric detector (GCMS-QP2010; Shimadzu). GC-MS conditions were as follows: injection at 220 °C, maintenance of the temperature at 220 °C for 4.5min, followed by an increase to 290 °C at a rate of 3 °C min–1. The temperature was then maintained at 290 °C for 10min, after which it was raised to 300 °C at a rate of 2 °C min–1 and held for 10min (Lee et al., 2009b ). Analysis of quantitative wax materials was performed using a capillary GC program with a flame ionization detector (FID) using the same conditions as in GC-MS. Compounds were quantified relative to the corresponding internal standards by integrating the peak areas.
Cutin analysis
Five- to six 6-week-old primary stems of Arabidopsis were used for cutin analysis according to Lee et al. (2009a ). The internal standards used were C17:0 methyl ester (methyl heptadecanoate) and C15:0 cycloketone (ω-pentadecalactone) (Sigma). Polyesters from dried solvent-extracted residues of stems (wax-free) were depolymerized by hydrogenolysis with methanolysis using sodium methoxide. The products recovered after hydrogenolysis were dried and derivatized as mentioned above and separated and quantified by GC-MS. The GC-MS protocol was as follows: injection at 110 °C, elevation by 2.5 °C min–1 to 300 °C, and holding for 3min at 300 °C. The mass-to-charge ratios (m/z) used to diagnose the cutin compounds are shown in Supplementary Table S2 at JXB online.
Inoculation of plants with Botrytis cinerea
The necrotrophic fungus B. cinerea was maintained on a potato dextrose agar plate (BD Difco) at room temperature. Collection of conidia and plant infection assays (Botrytis suspension concentration of 2×105 spores ml–1) were carried out as previously described (Li et al., 2008; Xiao and Chye, 2011b ). Photographs were taken at 0 and 6 days after infection (DAI).
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: At5g53470 (ACBP1), At3g18780 (ACTIN2), At2g47240 (CER8), At1g67730 (KCR1), At3g55360 (ECR), At1g68530 (CUT1/KCS6), At1g49430 (LACS2), At4g00360 (CYP86A2), At1g01600 (CYP86A4), At4g00400 (GPAT8), At2g19450 (DGAT1), At3g51520 (DGAT2), At1g48300 (DGAT3), At5g13640 (PDAT1), At1g20440 (COR47), At5g52310 (LTI78), At4g25490 (CBF1), and At3g26744 (ICE1).
Results
Recombinant AtACBP1 binds VLC acyl-CoA esters (C24:0-, C25:0-, and C26:0-CoA) in vitro
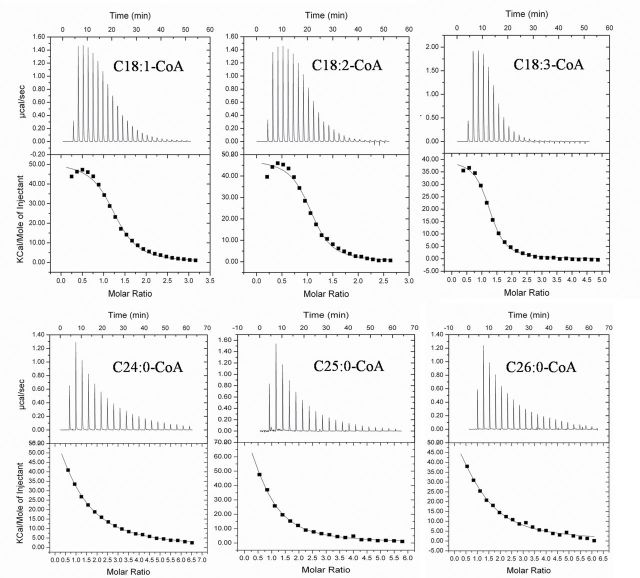
(His)6-tagged rACBP1 was shown to bind long-chain acyl-CoA esters (C18:1-, C18:2-, and C18:3-CoA) in Lipidex assays (Leung et al., 2006) and gel-binding assays (Chye, 1998). As rACBPs have not been reported to bind VLC acyl-CoA esters, rACBP1 was tested using commercially available VLC acyl-CoA esters (C24:0-, C25:0-, and C26:0-CoA) by ITC to determine the K D values. As controls, long-chain acyl-CoA esters (C18:1-, C18:2-, and C18:3-CoA) were included. Analysis of calorimetric data by the ORIGIN software (General Electric Company, USA) indicated that the binding isotherms fitted well with a model of a single binding site (Fig. 1). Consistent with the results from Lipidex assays (Leung et al., 2006) and gel-binding assays (Chye, 1998), rACBP1 interacted with long-chain acyl-CoA esters including C18:1-, C18:2-, and C18:3-CoAs with high affinities in ITC measurements (Fig. 1; Table 1). Furthermore, rACBP1 was also shown to bind to VLC acyl-CoA esters (C24:0-, C25:0-, and C26:0-CoA), although with lower affinities (Fig. 1; Table 1). The affinities as reflected by the K D values of rACBP1 for VLC acyl-CoA esters supported their participation in faty acid elongation during the biosynthesis of VLCFAs.
Fig. 1.
Binding isotherms of recombinant AtACBP1 titrated with C18:1-, C18:2-, C18:3-, C24:0-, C25:0-, and C26:0-CoA esters at 30 °C using isothermal titration calorimetry. Top panels show raw data of 300 μl of 10 μM recombinant AtACBP1 titrated with 20 injections of 1.5 μl of 250 μM acyl-CoA ester solution. Bottom panels show the integrated area of each injection and the plotted graph. Parameters of the dissociation constant (K D) are given in Table 1.
Table 1.
The dissociation constants (K D) of recombinant AtACBP1 (rACBP1) binding to acyl-CoA esters of different acyl chain lengthsThe values are means ±SD (n=3).
| Acyl-CoA esters | K D (μM) |
|---|---|
| C18:1 | 0.76±0.15 |
| C18:2 | 0.83±0.04 |
| C18:3 | 0.44±0.01 |
| C24:0 | 2.14±0.13 |
| C25:0 | 1.69±0.11 |
| C26:0 | 1.94±0.12 |
AtACBP1 is expressed in stem epidermis
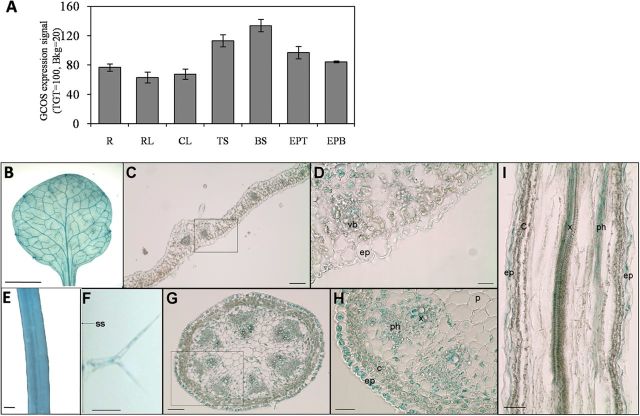
AtACBP1 mRNA has been previously reported to be expressed in all plant organs (Chye, 1998; Chen et al., 2010). The microarray database e-FP Browser (Winter et al., 2007; http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) revealed that AtACBP1 was highly expressed in the top and bottom of stems, in comparison with roots and rosette and cauline leaves, and that this expression localized in the epidermal peels (Fig. 2A).
Fig. 2.
Microarray data and GUS staining for the expression of AtACBP1. (A) Expression pattern of AtACBP1 in vegetative tissues of Arabidopsis including roots (R), rosette leaves (RL), cauline leaves (CL), top of stems (TS), bottom of stems (BS), and epidermal peels from the top (EPT) and bottom (EPB) of stems. The data were retrieved from the microarray database e-FP Browser (Winter et al., 2007; http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). GCOS, gene chip operating software, the method used by Affymetrix MAS5.0 to normalize the microarray data. TGT (target) and Bkg (background) are parameters used in the normalization. (B–I) GUS expression of AtACBP1pro::GUS in 4-week-old transgenic Arabidopsis. Three independent transgenic lines were tested by staining with 1mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide with consistent results. (B) Rosette leaf; (C, D) cross-section of a rosette leaf; (E) stem; (F) trichome on the side stem; (G, H) cross-section of a stem; (I) longitudinal section of a stem. Scale bar=10mm (B); 100 μm (C); 25 μm (D, H); 2mm (E); 10 μm (F); 50 μm (G,I). vb, vascular bundle; ep, epidermis; ss, stem surface; c, cortex; ph, phloem; x, xylem; p, pith.
The expression of AtACBP1 in leaves and stems was investigated in transgenic Arabidopsis lines expressing AtACBP1pro::GUS (Fig. 2B–I). AtACBP1pro::GUS was expressed in the leaf vasculature (Fig. 2B–D) and on the stem surface (Fig. 2E) including the trichomes (Fig. 2F). The cross- and longitudinal-sections of the stem showed AtACBP1pro::GUS expression in the epidermis, the cortex, and the vascular bundles (Fig. 2G–I). Stem and leaf sections from control Arabidopsis transformed with pBI101.3 were not stained.
The acbp1 mutant shows defects in epicuticular wax crystallization and cuticle membrane structure
When SEM was used to investigate wax crystallization patterns on the stem surface of the acbp1 mutant, the occurrence of epicuticular wax crystals was significantly reduced (Fig. 3A) in comparison with the wild type (Fig. 3B). Upright rod-, tube-, and umbrella-shaped wax crystals were arrayed in an orderly manner on the wild type, but not the acbp1 mutant stem. In contrast, the mutant had fewer crystals (Fig. 3).
Fig. 3.
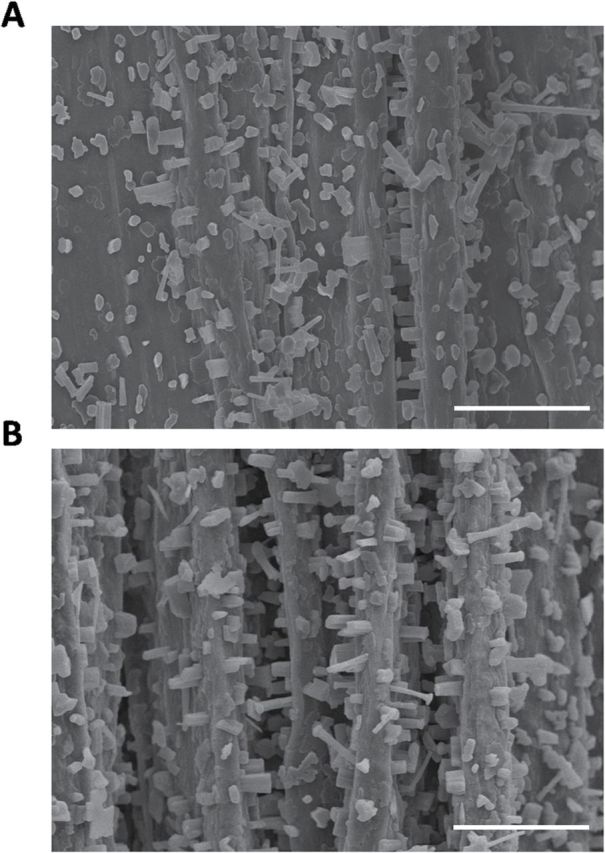
Scanning electron microscopy (SEM) analysis of epicuticular wax crystal patterns on stem surfaces of acbp1 (A) and Col-0 (B). Scale bars=15 μm.
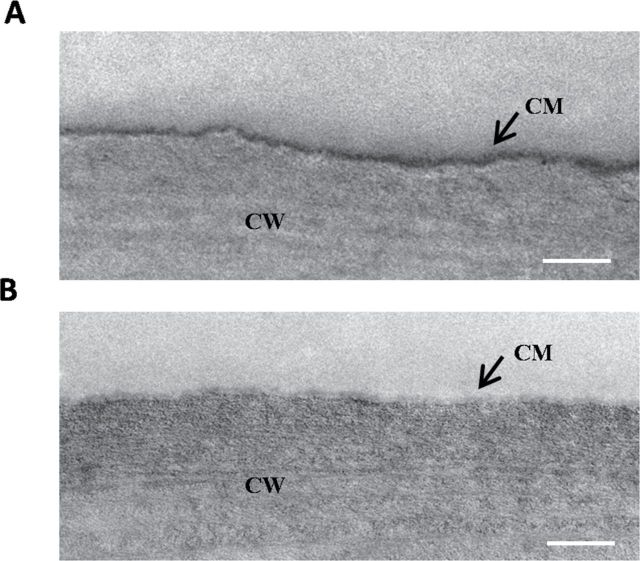
When TEM was used to examine the fine structural changes of the stem cuticle, the cuticle membrane was intact in Col-0 (Fig. 4A) but not in the acbp1 mutant (Fig. 4B). Instead, in the mutant, a ruptured and discontinuous cuticle membrane was observed (Fig. 4B). Absence of expression of AtACBP1 culminated in an aberrant cuticle membrane, suggesting that AtACBP1 is involved in stem cuticle formation.
Fig. 4.
Transmission electron microscopy (TEM) of the cuticle membrane from stem epidermal cells of Col-0 (A) and the acbp1 mutant (B). Scale bars=0.5 μm. CM, cuticle membrane; CW, cell wall.
The acbp1 mutant shows reduction in stem cuticular wax constituents and cutin monomers and down-regulation of cuticular biosynthetic genes
To evaluate further the roles of AtACBP1 in cuticle formation, GC-FID and GC-MS were employed to determine the amount and composition of cuticular waxes from stems of the wild type (Col-0), the acbp1 mutant, and the acbp1-complemented (acbp1-COM; Xiao et al., 2008a ) lines.
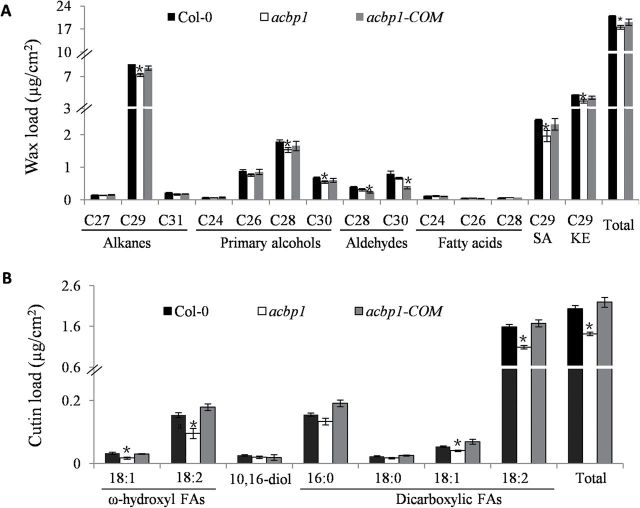
In stems, a 16% reduction of total wax was observed in acbp1 in comparison with the wild type (Fig. 5A). In particular, the levels of C29 alkane, C28 and C30 primary alcohols, and C29 secondary alcohol and ketone were significantly reduced in the acbp1 mutant (Fig. 5A). Their percentage reductions were 16% (C29 alkane), 15% (C28 primary alcohol), 21% (C30 primary alcohol), 21% (C29 secondary alcohol), and 16% (C29 ketone). The normal amounts of total wax and various wax species were recovered in stems of acbp1-COM (Fig. 5A), confirming that the decrease in stem cuticular wax in the acbp1 mutant resulted from knockout of AtACBP1 expression. Subsequently, when the expression of wax biosynthetic genes was determined by qRT-PCR analysis (Fig. 6A), CER8, KCR1, ECR, and CUT1/KCS6 significantly decreased in the acbp1 stem in comparison with the wild type (Fig. 6A), and these decreases in gene expression were recovered in the acbp1-COM plants (Supplementary Fig. S1A at JXB online).
Fig. 5.
Cuticular wax (A) and cutin monomer (B) composition and amount in stems of Col-0, acbp1, and acbp1-COM plants. Six-week-old stems were used in wax and cutin analysis by GC-FID and GC-MS. SA, secondary alcohols; KE, ketones; FA, fatty acid; 10,16-diol, C16-10,16-dihydroxyl fatty acids. Asterisks denote significant differences from the wild type (*P<0.05). Values are means ±SE (n=3).
Fig. 6.
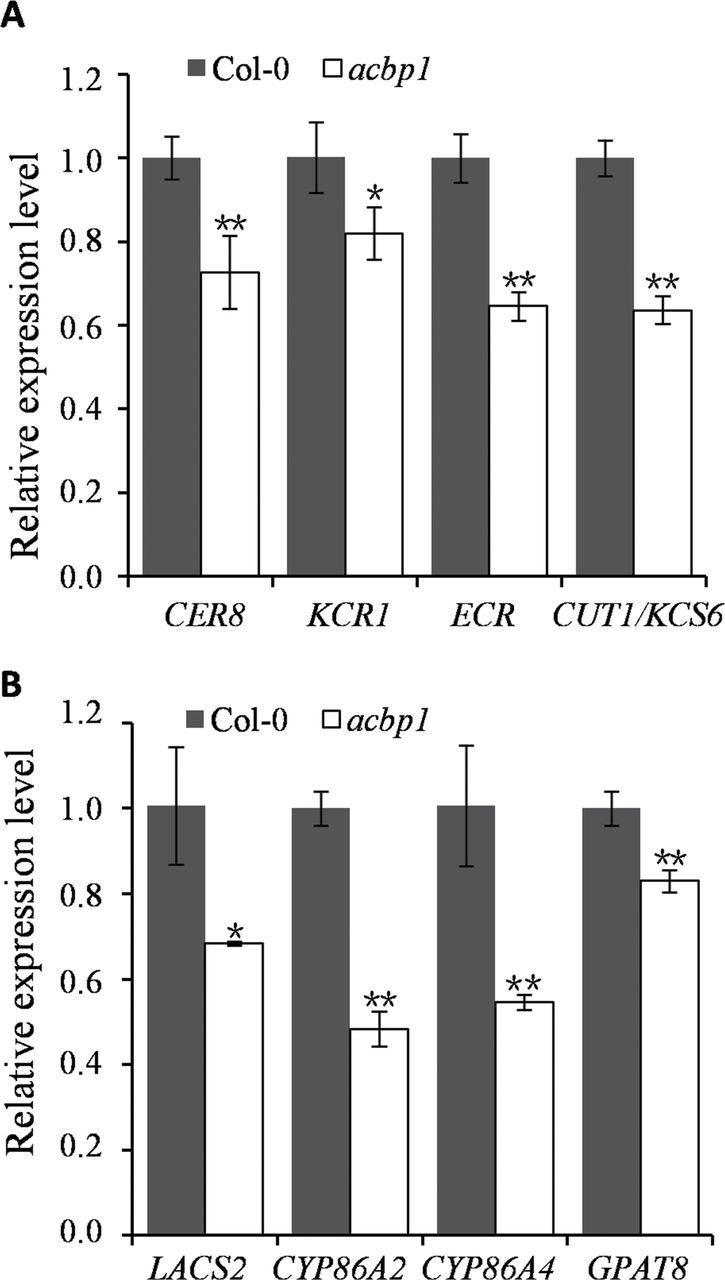
qRT-PCR analysis of wax (A) and cutin (B) biosynthetic genes in stems of the acbp1 mutant and Col-0. Expression of CER8, KCR1, ECR, CUT1/KCS6, LACS2, CYP86A2, CYP86A4, and GPAT8 decreased in stems of acbp1 in comparison with the wild type (Col-0). Asterisks denote significant differences from the wild type (*P<0.05; ** P<0.01). Values are means ±SE (n=3).
GC-MS was also used to determine cutin monomer composition and amount in the stems of the acbp1 mutant, acbp1-COM, and wild type (Fig. 5B). The amounts of total cutin monomer were altered in stems of the acbp1 mutant (Fig. 5B) in comparison with the wild type. Levels of C18:1 and C18:2 ω-hydroxyl fatty acids, as well as C18:1 and C18:2 dicarboxylic fatty acids, were significantly reduced in stems of the acbp1 mutant (Fig. 5B). Their percentage reductions were 45% (C18:1 ω-hydroxyl fatty acid), 38% (C18:2 ω-hydroxyl fatty acid), 24% (C18:1 dicarboxylic fatty acid), and 31% (C18:2 dicarboxylic fatty acid). The chemical change in stems of the acbp1 mutant was recovered in the acbp1-COM line (Fig. 5B), confirming that reduction in the amounts of stem cutin monomer in the mutant resulted from knockout of AtACBP1 expression. Subsequently, on qRT-PCR, expression of some genes involved in cutin synthesis (LACS2, CYP86A2, CYP86A4, and GPAT8) showed a significant decrease in the stems of acbp1 in comparison with wild-type Arabidopsis (Fig. 6B), which could be recovered in acbp1-COM plants (Supplementary Fig. S1B at JXB online). However, the expression of triacylglycerol biosynthetic genes (DGAT1, DGAT2, DGAT3, and PADT1) (Supplementary Fig. S1C) and cold-related genes (COR47, LTI78, CBF1, and ICE1) (Supplementary Fig. S1D), which are not implicated in cuticle formation, was not affected in the acbp1 mutant and acbp1-COM plants in comparison with the wild type.
Seedlings of acbp1 are more susceptible to Botrytis cinerea infection
To examine whether a reduction of wax and cutin in the acbp1 mutant confers an altered response to the necrotrophic fungal pathogen B. cinerea, 3-week-old seedlings of the acbp1 mutant and wild type were inoculated with Botrytis spores. As shown in Fig. 7, the acbp1 mutant seedlings displayed enhanced susceptibility after spraying with Botrytis suspension. At 6 DAI, chlorosis and necrosis were observed in the acbp1 mutant, but not in the wild type (Fig. 7). Measurement of leaf wax and cutin from the acbp1 mutant in comparison with the wild type revealed a significant decrease in wax but not in cutin (Supplementary Fig. S2 at JXB online). In particular, C31 and C33 alkanes, C28 fatty acids, and total wax load significantly declined (Supplementary Fig. S2A). This is not surprising because AtACBP1pro::GUS was expressed more in stem epidermis than in leaf epidermis (Fig. 2). These results suggest that a reduction in wax content in the acbp1 mutant could have caused greater susceptibility to Botrytis infection.
Fig. 7.
Response of acbp1 to Botrytis cinerea infection. Three-week-old wild-type (Col-0) and acbp1 plants were sprayed with B. cinerea (2×105 spores ml–1). Photographs were taken at 0 and 6 days after inoculation (DAI). The experiments were repeated twice with consistent results. Arrows indicate chlorosis and necrosis of leaves.
Discussion
In plants, fatty acids are synthesized in the plastids by the addition of two-carbon units to a growing acyl chain facilitated by the acyl carrier protein (ACP) during de novo fatty acid synthesis (Ohlrogge and Browse, 1995). Subsequently, 16:0-ACP and 18:0-ACP are exported to the ER for the biosynthesis of other lipids including cutin, suberin, and cuticular waxes (Post-Beittenmiller, 1996; Jenks et al., 2002; Nawrath, 2002). Cutin and wax are synthesized exclusively in the epidermis (Nawrath, 2002; Suh et al., 2005; Samuels et al., 2008). AtACBP1pro::GUS is expressed in the embryos, lateral root primordia, vascular bundles, stigmas, and ovaries (Du et al., 2013b ). In this study, transgenic Arabidopsis expressing AtACBP1pro::GUS showed strong GUS expression in stem epidermis, in agreement with the corresponding expression analysis of KCS20 and KCS2/DAISY genes involved in VLCFA elongation (Lee et al., 2009b ). It is noteworthy that the GUS stain was not detected in leaf epidermis (Fig. 2D), suggesting a putative function for AtACBP1 in stem cuticle formation.
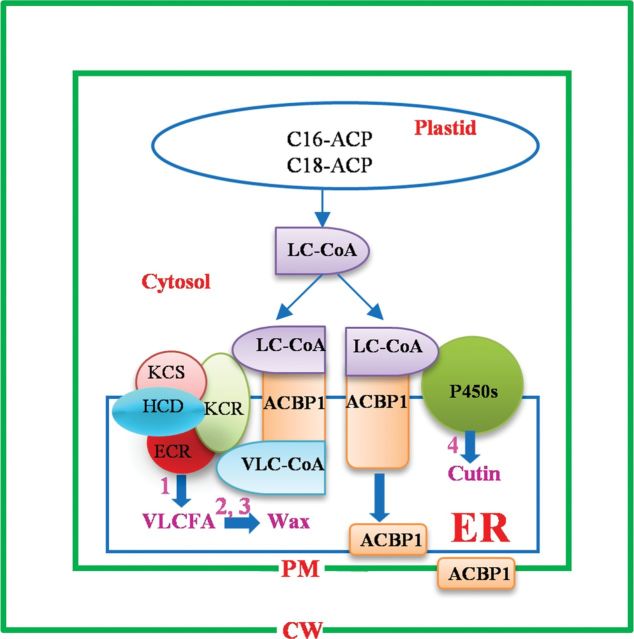
In wax biosynthesis, C18 fatty acyl-CoAs are the predominant precursors for production of VLCFAs in four sequential reactions catalysed by a membrane-bound multiple enzyme system consisting of KCS, β-ketoacyl-CoA reductase (KCR), β-hydroxyacyl-CoA dehydratase (HCD), and enoyl-CoA reductase (ECR) (Kunst and Samuels, 2009). After several cycles of condensation of malonyl-CoA with long-chain acyl-CoAs, reduction to β-hydroxyacyl-CoA, dehydration to an enoyl-CoA, and reduction of the enoyl-CoA, VLCFAs with different acyl chains ranging from C20 to C34 are generated and subsequently converted to various wax components through decarbonylation and acyl reduction (Kunst and Samuels, 2003; Samuels et al., 2008). Through multiple steps of hydroxylation and epoxidation, 16:0 and 18:X fatty acyl-CoAs are converted into cutin monomers (Schnurr et al., 2004). ITC analysis from the present study revealed that rACBP1 binds not only long-chain acyl-CoA esters (C18:1-, C18:2-, and C18:3-CoA) but also saturated VLC acyl-CoA esters (C24:0-, C25:0-, and C26:0-CoA). The reduced binding affinity of rACBP1 for VLC acyl-CoA esters may be attributed to either the longer acyl chain length or unsaturation of the acyl chain. In measurements of rACBP1 interaction with VLC acyl-CoA esters, only C24:0 to C26:0 were tested because acyl-CoAs with acyl chains longer than C27:0 are not commercially available (http://www.avantilipids.com/). The present analysis demonstrated that AtACBP1 is able to bind C18 fatty acyl-CoAs and VLC acyl-CoAs, and can potentially transport these precursors for cutin and wax biosynthesis during stem cuticle formation (Fig. 8).
Fig. 8.
Proposed function of AtACBP1 in the wax and cutin synthesis pathway. The broad range of wax species altered in the acbp1 mutant stem implies that the function of AtACBP1 in wax biosynthesis lies upstream in the pathway, involving VLCFA elongation (1). AtACBP1 may participate in general stem wax biosynthesis and could affect both the decarbonylation and acyl reduction pathways (2, 3). Changes in stem cutin monomer content in the acbp1 mutant suggest that AtACBP1 participates in stem cutin synthesis (4). ACP, acyl carrier protein; LC-CoA, long-chain acyl-CoA; VLC-CoA, very-long-chain acyl-CoA; KCS, β-ketoacyl-CoA synthase; KCR, β-ketoacyl-CoA reductase; HCD, β-hydroxyacyl-CoA dehydratase; ECR, enoyl-CoA reductase; VLCFA, very-long-chain fatty acid; ER, endoplasmic reticulum; PM, plasma membrane; CW, cell wall.
A reduction in wax crystal density and observations on the irregularity of the cuticle membrane on the stems of the acbp1 mutant suggest that a defective cuticle had resulted from functional loss of AtACBP1. Interestingly, the T-DNA insertional mutants of AtACBP3 also showed a highly irregular outermost cell wall surface (Xia et al., 2012). These phenotypes are not only evident in AtACBP mutants but have also been observed in other mutants in cuticle development. Wax crystals were also absent on the stem surfaces of the cer1, cer2, and cer6 mutants (Millar et al., 1999). In addition, the lacs2 mutant showed a reduction in plant size, seed set, and seedling establishment (Schnurr et al., 2004). Furthermore, in the wax2, fdh, and lcr mutants, severe organ fusions occurred and pollen fertility was affected (Lolle et al., 1998; Wellesen et al., 2001; Chen et al., 2003). These drastic phenotypic changes accompanied by severe reduction in wax or cutin load were caused by mutation in the genes of the wax and cutin pathways (Millar et al., 1999; Wellesen et al., 2001; Chen et al., 2003; Lü et al., 2009).
The wax species that were significantly lower in the acbp1 mutant stem included not only alkanes, but also primary alcohols, C29 secondary alcohol, and ketones (Fig. 5A). This is in agreement with the down-regulation in the acbp1 mutant stem of wax biosynthetic genes CER8, KCR1, ECR, and CUT1/KCS6 (Fig. 6A), all of which are associated with VLCFA elongation. CER8 modifies VLCFAs in both wax and cutin syntheses (Lü et al., 2009); ECR (Zheng et al., 2005) and KCR1 (Beaudoin et al., 2009) participate in VLCFA elongation; and CUT1/KCS6 is involved in the production of VLCFA precursors of stem wax (Millar et al., 1999). The broad range of wax species altered in the acbp1 mutant stem implied that the function of AtACBP1 in wax biosynthesis lies upstream in the pathway, involving VLCFA elongation (Fig. 8). It is proposed that AtACBP1 probably participates in general stem wax biosynthesis rather similar to the effect of CUT1/KCS6 in the pathway. Similarly, the decrease in cutin load in the stem of the acbp1 mutant correlated well with the down-regulation of the cutin biosynthetic genes LACS2, CYP86A2, CYP86A4, and GPAT8 (Fig. 6B). LACS2 is required for the correct assembly of the cuticular barrier (Schnurr et al., 2004). CYP86A2 is a fatty acid ω-hydroxylase in the synthesis of hydroxy fatty acids (Xiao et al., 2004), while CYP86A4 and GPAT8 catalyse ω-hydroxylation and esterification to glycerol, respectively, during cutin synthesis (Li et al., 2007; Li-Beisson et al., 2009). Knockout of AtACBP1 probably adversely affected the accumulation of long-chain and VLC acyl-CoAs essential for stem wax and cutin biosyntheses (Fig. 8). Furthermore, the lack of substrates for wax and cutin biosyntheses led to a decrease in the expression of both stem wax and cutin biosynthesis genes, which will reduce wax or cutin production.
Waxes are known to be synthesized in the epidermis (Samuels et al., 2008), and the leaf wax content was lower in the acbp1 mutant in comparison with the wild type (Supplementary Fig. S2A at JXB online). However, AtACBP1pro::GUS was not observed to be expressed in the leaf epidermis (Fig. 2D), suggesting that AtACBP1 may not participate directly in leaf wax biosynthesis. Possibly, leaf wax changes may have been affected by the dramatic alterations observed in stem cuticular contents. Although the AtACBP1pro::GUS-transformed plants did not express detectable GUS activity in the leaf epidermal cells (Fig. 2C, D), reductions in several compounds (i.e. C31 and C33 alkanes, C28 fatty acid, and total wax load) in leaf wax but not leaf cutin (Supplementary Fig. S2) in the acbp1 mutant may be attributed to a systemic change in the expression of wax-related genes and/or the activities of their gene products as a result of the defective stem cuticle. As the acbp1 mutant exhibited lesions in stem cuticle formation, this could have potentially affected the status of the plant as a whole (e.g. water loss, susceptibility to pathogens, etc.), and could have culminated in an indirect effect on wax synthesis in the leaf epidermis. It is well documented that cuticular wax biosynthesis is sensitive to diverse environmental cues, and several transcription factors have been identified to play a role in its biosynthesis and accumulation (Aharoni et al., 2004; Zhang et al., 2007; Seo et al., 2011; Cominelli et al., 2008; Lü et al., 2009). Xia et al. (2012) showed that in the leaves of both acbp3 and acbp4 mutants, the cutin monomers were greatly reduced, with pronounced reduction in C16:0, C18:1, and C18:2 dicarboxylic fatty acids, but no change in most cutin monomers was evident in the acbp6 mutant. In comparison, the present analysis revealed that stem cutin monomer levels also declined in the acbp1 mutant, confirming its role in cutin biosynthesis (Fig. 8). In particular, C18 species (C18:1 and C18:2 ω-hydroxyl fatty acids and dicarboxylic fatty acids) of cutin were more affected in the acbp1 mutant. This corresponds well to ITC data that showed that rACBP1 binds long-chain acyl-CoA esters (C18:1-, C18:2-, and C18:3-CoAs) with a greater affinity (i.e. smaller dissociation constant, K D) than VLC acyl-CoAs (C24:0-, C25:0-, and C26:0-CoA). AtACBP1 is localized in the PM and the ER, but AtACBP3 is targeted to the extracellular space, while AtACBP4 is a cytosolic protein. Although they show differential subcellular localization, they all affect cutin biosynthesis, suggesting that the binding and trafficking of precursors in cutin synthesis transverse across subcellular compartments.
The acbp1 mutant in TEM showed an aberrant cuticle membrane in stems and was more susceptible to infection caused by B. cinerea possibly by entry through the aberrant cuticle, suggesting that alteration of cuticle constituents in this mutant impaired its basal defence responses. These results are consistent with the reduction in fungal pathogen resistance observed in the acbp3, acbp4, and acbp6 mutants which were also cuticle-defective (Xia et al., 2012). Previous findings have also revealed that AtACBP3 overexpression constitutively activated salicylic acid accumulation, PR gene expression and cell death, and increased resistance to the virulent bacterial pathogen P. syringae DC3000 (Xiao and Chye, 2011b ).
Leaf susceptibility of the acbp1 mutant to B. cinerea infection arising from a decline in leaf wax (but not leaf cutin) suggested that this decrease affected the leaf cuticle membrane through extrapolating the observations of altered cuticle in stems including significant decreases in both stem wax and cutin loads (Fig. 4). Lee et al. (2009a ) reported that the ltpg1 mutant showed a reduction in the C29 alkane in stems but not leaves, and they did not see any significant changes in total wax in neither stem nor leaf. Although ltpg1 mutant leaves showed increases in three cutin constituents, they were more susceptive to Alternaria brassicicola (Lee et al., 2009a ). These findings support that changes in wax and cutin loads in stem and leaf may affect the cuticle barrier which is known to protect the plant against pathogen infection (Jenks et al., 1994). Perhaps changes in cuticular content may also have altered the leaf surface structure which then enhanced susceptibility of the acbp1 mutant to B. cinerea. Li et al. (2007) have reported that a change in the thickness or the structure of the pavement cells and the guard cells in the gpat4gpat8 double mutant made it more susceptible to A. brassicicola. Similarly, in the ltpg1 mutant which was more susceptible to A. brassicicola, Lee et al. (2009a ) observed alterations in the structure of the cuticular layer, a protrusive cytoplasm, and disorganized grana and stroma lamellae in the chloroplasts.
In summary, using phenotypic and biochemical analyses of the acbp1 mutant, it is demonstrated that AtACBP1 is involved in stem cuticle formation. Previous studies have suggested that plasma membrane-localized glycosylphosphatidylinositol-anchored lipid transfer proteins function in cuticular lipid transport (DeBono et al., 2009; Lee et al., 2009a ; Kim et al., 2012). It is illustrated herein that ER- and PM-associated AtACBP1 also participates in stem wax and cutin biosynthesis, probably as a carrier protein, as supported by ITC data (Fig. 1; Table 1).
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Expression analysis of wax and cutin biosynthetic genes, and genes with no implication on cuticle formation (triacylglycerol biosynthetic genes and cold-related genes) in stems of Col-0, the acbp1 mutant, and the acbp1-COM line.
Figure S2. Cuticular wax and cutin monomer composition and amount in leaves of Col-0 and the acbp1 mutant.
Table S1. Sequences of gene-specific primers for qRT-PCR.
Table S2. Mass-to-charge ratios (m/z) of cutin compounds used in mass spectrometry.
Acknowledgements
We thank F.Y.F. Chan, A.S.L. Wong, and W.S. Lee (Electron Microscope Unit, the University of Hong Kong) for technical assistance in electron microscopy. This work was supported by the Research Grants Council of the Hong Kong Special Administrative Region, China (project no. HKU765511M), the Wilson and Amelia Wong Endowment Fund, and the University of Hong Kong (postgraduate studentship to YX and postdoctoral fellowships to SX and SCL). Funding for research in the Suh Lab was supported by the Next-Generation BioGreen 21 Program (No. PJ008203), Rural Development Administration, Republic of Korea.
Glossary
Abbreviations:
- ACBP
acyl-CoA-binding protein
- ACP
acyl carrier protein
- ECR
enoyl-CoA reductase
- ER
endoplasmic reticulum
- FID
flame ionization detector
- GC
gas chromatography
- GPAT
glycerol-3-phosphate acyltransferase
- GUS
β-glucuronidase
- HCD
β-hydroxyacyl-CoA dehydratase
- ITC
isothermal titration calorimetry
- KCR
β-ketoacyl-CoA reductase
- KCS
β-ketoacyl-CoA synthase
- LACS
long-chain acyl-CoA synthetase
- MS
mass spectrometry
- MS medium
Murashige and Skoog medium
- PM
plasma membrane
- TEM
transmission electron microscopy
- SEM
scanning electron microscopy
- VLCFA
very-long-chain fatty acid.
References
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A. 2004. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. The Plant Cell 16, 2463–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin F, Wu X, Li F, Haslam RP, Markham JE, Zheng H, Napier JA, Kunst L. 2009. Functional characterization of the Arabidopsis β-ketoacyl-coenzyme A reductase candidates of the fatty acid elongase. Plant Physiology 150, 1174–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloksgaard M, Bek S, Marcher AB, et al. 2012. The acyl-CoA binding protein is required for normal epidermal barrier function in mice. Journal of Lipid Research 53, 2162–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QF, Xiao S, Chye ML. 2008. Overexpression of the Arabidopsis 10-kilodalton acyl-coenzyme A-binding protein ACBP6 enhances freezing tolerance. Plant Physiology 148, 304–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QF, Xiao S, Qi W, Mishra G, Ma J, Wang M, Chye ML. 2010. The Arabidopsis acbp1acbp2 double mutant lacking acyl-CoA-binding proteins ACBP1 and ACBP2 is embryo lethal. New Phytologist 186, 843–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Goodwin SM, Boroff VL, Liu X, Jenks MA. 2003. Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. The Plant Cell 15, 1170–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chye ML. 1998. Arabidopsis cDNA encoding a membrane-associated protein with an acyl-CoA binding domain. Plant Molecular Biology 38, 827–838. [DOI] [PubMed] [Google Scholar]
- Chye ML, Huang BQ, Zee SY. 1999. Isolation of a gene encoding Arabidopsis membrane-associated acyl-CoA binding protein and immunolocalization of its gene product. The Plant Journal 18, 205–214. [DOI] [PubMed] [Google Scholar]
- Chye ML, Li HY, Yung MH. 2000. Single amino acid substitutions at the acyl-CoA-binding domain interrupt 14[C]palmitoyl-CoA binding of ACBP2, an Arabidopsis acyl-CoA-binding protein with ankyrin repeats. Plant Molecular Biology 44, 711–721. [DOI] [PubMed] [Google Scholar]
- Cominelli E, Sala T, Calvi D, Gusmaroli G, Tonelli C. 2008. Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. The Plant Journal 53, 53–64. [DOI] [PubMed] [Google Scholar]
- DeBono A, Yeats TH, Rose JKC, Bird D, Jetter R, Kunst L, Samuels L. 2009. Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. The Plant Cell 21, 1230–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du ZY, Chen MX, Chen QF, Xiao S, Chye ML. 2013. a Overexpression of Arabidopsis acyl-CoA-binding protein ACBP2 enhances drought tolerance. Plant, Cell and Environment 36, 300–314. [DOI] [PubMed] [Google Scholar]
- Du ZY, Chen MX, Chen QF, Xiao S, Chye ML. 2013. b Arabidopsis acyl-CoA-binding protein ACBP1 participates in the regulation of seed germination and seedling development. The Plant Journal 74, 294–309. [DOI] [PubMed] [Google Scholar]
- Du ZY, Xiao S, Chen QF, Chye ML. 2010. Depletion of the membrane-associated acyl-coenzyme A-binding protein ACBP1 enhances the ability of cold acclimation in Arabidopsis. Plant Physiology 152, 1585–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engeseth NJ, Pacovsky RS, Newman T, Ohlrogge JB. 1996. Characterization of an acyl-CoA-binding protein from Arabidopsis thaliana. Archives of Biochemistry and Biophysics 331, 55–62. [DOI] [PubMed] [Google Scholar]
- Fan J, Liu J, Culty M, Papadopoulos V. 2010. Acyl-coenzyme A binding domain containing 3 (ACBD3; PAP7; GCP60): an emerging signaling molecule. Progress in Lipid Research 49, 218–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Li HY, Xiao S, Chye ML. 2010. Acyl-CoA-binding protein 2 binds lysophospholipase 2 and lysoPC to promote tolerance to cadmium-induced oxidative stress in transgenic Arabidopsis. The Plant Journal 62, 989–1003. [DOI] [PubMed] [Google Scholar]
- Gao W, Xiao S, Li HY, Tsao SW, Chye ML. 2009. Arabidopsis thaliana acyl-CoA-binding protein ACBP2 interacts with a heavy-metal-binding farnesylated protein AtFP6. New Phytologist 181, 89–102. [DOI] [PubMed] [Google Scholar]
- Guerrero C, Martín-Rufián M, Reina JJ, Heredia A. 2006. Isolation and characterization of a cDNA encoding a membrane bound acyl-CoA binding protein from Agave americana L. epidermis. Plant Physiology and Biochemstry 44, 85–90. [DOI] [PubMed] [Google Scholar]
- Jenks MA, Eigenbrode SD, Lemieux B. 2002. Cuticular waxes of Arabidopsis. The Arabidopsis book 1, e0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks MA, Joly RJ, Peters PJ, Rich PJ, Axtell JD, Ashworth EN. 1994. Chemically induced cuticle mutation affecting epidermal conductance to water vapor and disease susceptibility in Sorghum bicolor (L.) Moench. Plant Physiology 105, 1239–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Lee SB, Kim HJ, Min MK, Hwang I, Suh MC. 2012. Characterization of glycosylphosphatidylinositol-anchored lipid transfer protein 2 (LTPG2) and overlapping function between LTPG/LTPG1 and LTPG2 in cuticular wax export or accumulation in Arabidopsis thaliana . Plant and Cell Physiology 53, 1391–1403. [DOI] [PubMed] [Google Scholar]
- Knudsen J, Neergaard TB, Gaigg B, Jensen MV, Hansen JK. 2000. Role of acyl-CoA binding protein in acyl-CoA metabolism and acyl-CoA-mediated cell signaling. Journal of Nutrition 130, 294–298S. [DOI] [PubMed] [Google Scholar]
- Kunst L, Samuels AL. 2003. Biosynthesis and secretion of plant cuticular wax. Progress in Lipid Research 42, 51–80. [DOI] [PubMed] [Google Scholar]
- Kunst L, Samuels L. 2009. Plant cuticles shine: advances in wax biosynthesis and export. Current Opinion in Plant Biology 12, 721–727. [DOI] [PubMed] [Google Scholar]
- Lee SB, Go YS, Bae HJ, Park JH, Cho SH, Cho HJ, Lee DS, Park OK, Hwang I, Suh MC. 2009. a Disruption of glycosylphosphatidylinositol-anchored lipid transfer protein gene altered cuticular lipid composition, increased plastoglobules, and enhanced susceptibility to infection by the fungal pathogen Alternaria brassicicola. Plant Physiology 150, 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Jung SJ, Go YS, Kim HU, Kim JK, Cho HJ, Park OK, Suh MC. 2009. b Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. The Plant Journal 60, 462–475. [DOI] [PubMed] [Google Scholar]
- Lee SB, Suh MC. 2013. Recent advances in cuticular wax biosynthesis and its regulation in Arabidopsis. Molecular Plant 6, 246–249. [DOI] [PubMed] [Google Scholar]
- Leung KC, Li HY, Mishra G, Chye ML. 2004. ACBP4 and ACBP5, novel Arabidopsis acyl-CoA-binding proteins with kelch motifs that bind oleoyl-CoA. Plant Molecular Biology 55, 297–309. [DOI] [PubMed] [Google Scholar]
- Leung KC, Li HY, Xiao S, Tse MH, Chye ML. 2006. Arabidopsis ACBP3 is an extracellularly targeted acyl-CoA-binding protein. Planta 223, 871–881. [DOI] [PubMed] [Google Scholar]
- Li HY, Chye ML. 2003. Membrane localization of Arabidopsis acyl-CoA binding protein ACBP2. Plant Molecular Biology 51, 483–492. [DOI] [PubMed] [Google Scholar]
- Li HY, Xiao S, Chye ML. 2008. Ethylene- and pathogen-inducible Arabidopsis acyl-CoA binding protein 4 interacts with an ethylene-responsive element binding protein. Journal of Experimental Botany 59, 3997–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Beisson F, Koo AJK, Molina I, Pollard M, Ohlrogge J. 2007. Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proceedings of the National Academy of Sciences, USA 104, 18339–18344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Pollard M, Sauveplane V, Pinot F, Ohlrogge J, Beisson F. 2009. Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proceedings of the National Academy of Sciences, USA 106, 22008–22013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, et al. 2013. Acyl-lipid metabolism. The Arabidopsis book 11, e0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao P, Chen QF, Chye ML. 2014. Transgenic Arabidopsis flowers overexpressing acyl-CoA-binding protein ACBP6 are freezing tolerant. Plant and Cell Physiology 55, 1055–1071. [DOI] [PubMed] [Google Scholar]
- Lolle SJ, Hsu W, Pruitt RE. 1998. Genetic analysis of organ fusion in Arabidopsis thaliana. Genetics 149, 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü S, Song T, Kosma DK, Parsons EP, Rowland O, Jenks MA. 2009. Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. The Plant Journal 59, 553–564. [DOI] [PubMed] [Google Scholar]
- Meng W, Hsiao AS, Gao C, Jiang L, Chye ML. 2014. Subcellular localization of rice acyl-CoA-binding proteins (ACBPs) indicates that OsACBP6::GFP is targeted to the peroxisomes. New Phytologist 203, 469-–482. [DOI] [PubMed] [Google Scholar]
- Meng W, Su YCF, Saunders RMK, Chye ML. 2011. The rice acyl-CoA-binding protein gene family: phylogeny, expression and functional analysis. New Phytologist 189, 1170–1184. [DOI] [PubMed] [Google Scholar]
- Michaely P, Bennett V. 1992. The ANK repeat: a ubiquitous motif involved in macromolecular recognition. Trends in Cell Biology 2, 127–129. [DOI] [PubMed] [Google Scholar]
- Millar AA, Clemens S, Zachgo S, Giblin EM, Taylor DC, Kunst L. 1999. CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. The Plant Cell 11, 825–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum 15, 473–497. [Google Scholar]
- Nawrath C. 2002. The biopolymers cutin and suberin. The Arabidopsis book 1, e0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C. 2006. Unraveling the complex network of cuticular structure and function. Current Opinion in Plant Biology 9, 281–287. [DOI] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J. 1995. Lipid biosynthesis. The Plant Cell 7, 957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post-Beittenmiller D. 1996. Biochemistry and molecular biology of wax production in plants. Annual Review of Plant Physiology and Plant Molecular Biology 47, 405–430. [DOI] [PubMed] [Google Scholar]
- Samuels L, Kunst L, Jetter R. 2008. Sealing plant surfaces: cuticular wax formation by epidermal cells. Annual Review of Plant Biology 59, 683–707. [DOI] [PubMed] [Google Scholar]
- Schnurr J, Shockey J, Browse J. 2004. The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. The Plant Cell 16, 629–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Lee SB, Suh MC, Park MJ, Go YS, Park CM. 2011. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. The Plant Cell 23, 1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber P, Schorderet M, Ryser U, Buchala A, Kolattukudy P, Métraux JP, Nawratha C. 2000. Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. The Plant Cell 12, 721–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin SF, Yeung EC, Chye ML. 2006. Downregulation of Solanum americanum genes encoding proteinase inhibitor II causes defective seed development. The Plant Journal 45, 58–70. [DOI] [PubMed] [Google Scholar]
- Suh MC, Samuels AL, Jetter R, Kunst L, Pollard M, Ohlrogge JB, Beisson F. 2005. Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiology 139, 1649–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellesen K, Durst F, Pinot F, Benveniste I, Nettesheim K, Wisman E, Steiner-Lange S, Saedler H, Yephremov A. 2001. Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid ω-hydroxylation in development. Proceedings of the National Academy of Sciences, USA 98, 9694–9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An ‘Electronic Fluorescent Pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Yu K, Gao QM, Wilson EV, Navarre D, Kachroo P, Kachroo A. 2012. Acyl CoA binding proteins are required for cuticle formation and plant responses to microbes. Frontiers in Plant Science 3, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Goodwin SM, Xiao Y, Sun Z, Baker D, Tang X, Jenks MA, Zhou JM. 2004. Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO Journal 23, 2903–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Chen QF, Chye ML. 2009. Light-regulated Arabidopsis ACBP4 and ACBP5 encode cytosolic acyl-CoA-binding proteins that bind phosphatidylcholine and oleoyl-CoA ester. Plant Physiology and Biochemistry 47, 926–933. [DOI] [PubMed] [Google Scholar]
- Xiao S, Chye ML. 2009. An Arabidopsis family of six acyl-CoA-binding proteins has three cytosolic members. Plant Physiology and Biochemistry 47, 479–484. [DOI] [PubMed] [Google Scholar]
- Xiao S, Chye ML. 2011. a New roles for acyl-CoA-binding proteins (ACBPs) in plant development, stress responses and lipid metabolism. Progress in Lipid Research 50, 141–151. [DOI] [PubMed] [Google Scholar]
- Xiao S, Chye ML. 2011. b Overexpression of Arabidopsis ACBP3 enhances NPR1-dependent plant resistance to Pseudomonas syringe pv tomato DC3000. Plant Physiology 156, 2069–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Gao W, Chen QF, Chan SW, Zheng SX, Ma J, Wang M, Welti R, Chye ML. 2010. Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. The Plant Cell 22, 1463–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Gao W, Chen QF, Ramalingam S, Chye ML. 2008. a Overexpression of membrane-associated acyl-CoA-binding protein ACBP1 enhances lead tolerance in Arabidopsis. The Plant Journal 54, 141–151. [DOI] [PubMed] [Google Scholar]
- Xiao S, Li HY, Zhang JP, Chan SW, Chye ML. 2008. b Arabidopsis acyl-CoA-binding proteins ACBP4 and ACBP5 are subcellularly localized to the cytosol and ACBP4 depletion affects membrane lipid composition. Plant Molecular Biology 68, 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenko OP, Weselake RJ. 2011. Involvement of low molecular mass soluble acyl-CoA-binding protein in seed oil biosynthesis. New Biotechnology 28, 97–109. [DOI] [PubMed] [Google Scholar]
- Zhang JY, Broeckling CD, Sumner LW, Wang ZY. 2007. Heterologous expression of two Medicago truncatula putative ERF transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Molecular Biology 64, 265–278. [DOI] [PubMed] [Google Scholar]
- Zheng H, Rowland O, Kunst L. 2005. Disruptions of the Arabidopsis enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. The Plant Cell 17, 1467–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SX, Xiao S, Chye ML. 2012. The gene encoding Arabidopsis acyl-CoA-binding protein 3 is pathogen inducible and subject to circadian regulation. Journal of Experimental Botany 63, 2985–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.