Abstract
Pseudomonas putida KT2440 is a soil microorganism that attaches to seeds and efficiently colonizes the plant's rhizosphere. Lysine is one of the major compounds in root exudates, and P. putida KT2440 uses this amino acid as a source of carbon, nitrogen, and energy. Lysine is channeled to δ-aminovaleric acid and then further degraded to glutaric acid via the action of the davDT gene products. We show that the davDT genes form an operon transcribed from a single σ70-dependent promoter. The relatively high level of basal expression from the davD promoter increased about fourfold in response to the addition of exogenous lysine to the culture medium. However, the true inducer of this operon seems to be δ-aminovaleric acid because in a mutant unable to metabolize lysine to δ-aminovaleric acid, this compound, but not lysine, acted as an effector. Effective induction of the P. putida PdavD promoter by exogenously added lysine requires efficient uptake of this amino acid, which seems to proceed by at least two uptake systems for basic amino acids that belong to the superfamily of ABC transporters. Mutants in these ABC uptake systems retained basal expression from the davD promoter but exhibited lower induction levels in response to exogenous lysine than the wild-type strain.
Fluorescent pseudomonads are good colonizers of the rhizosphere of most crop plants, and it has been established that root exudates play a central role in plant-Pseudomonas sp. interactions (8, 11, 24, 36). The main organic compounds of root exudates are sugars, various organic acids, flavonoids, and a number of amino acids (3, 7). Although sugars account for most of the organic matter in exudates (18), they do not seem to play a major role in plant-bacterium interactions, as deduced from the finding that they do not contribute to tomato root colonization by a well-studied Pseudomonas biocontrol strain (25). Bacterial genes involved in the metabolism of amino acids are differentially expressed in the rhizosphere, or in response to root exudates, and mutants deficient in amino acid utilization are less competitive in rhizosphere colonization than the wild-type strain (4, 5, 12, 44-46). A similar observation was made with mutants of a Rhizobium sp. unable to use proline as the sole carbon and nitrogen source (19).
By means of a transposon carrying a promoterless lux operon to generate transcriptional fusions by insertional mutagenesis, Espinosa-Urgel and Ramos (12) identified the Pseudomonas putida davT gene, whose expression increased in bacteria colonizing the rhizosphere. The davT gene encodes the enzyme δ-aminovalerate aminotransferase, which participates in a key step in lysine catabolism. Expression of the davT::lux fusion was enhanced in response to the addition of lysine in the culture medium. This is consistent with the relatively high concentration of lysine (0.42 mM) estimated to be present in corn root exudates (3, 7, 12, 29, 31). The davT mutant was unable to use lysine or δ-aminovalerate as a carbon source, in consonance with the latter chemical being a point of convergence in the degradation of lysine through two different branches (Fig. 1). Although the davT mutant of P. putida showed a chemoattractive response to root exudates, it was less competitive in root colonization assays than its parental strain (11). Root exudates contain small amounts of different compounds. The ability of rhizobacteria to use different carbon and energy sources simultaneously could be important for the establishment in the rhizosphere.
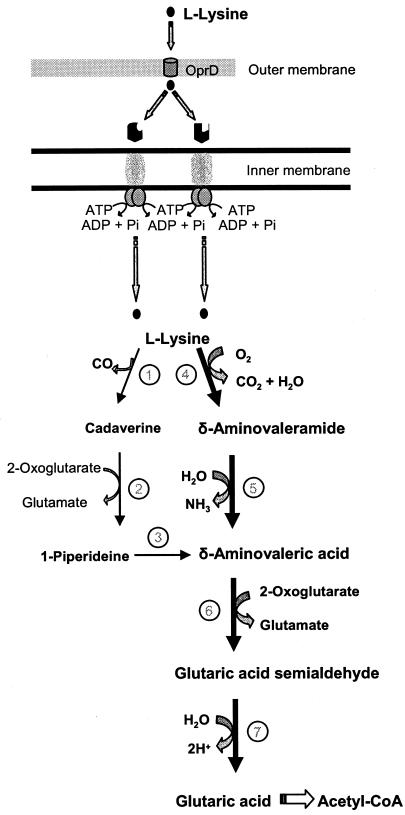
FIG. 1.
l-Lysine catabolism by P. putida. l-Lysine has been proposed to enter into the periplasmic space through the OprD porin (43). Subsequently, it is transported to the cytoplasm (the present study) and is metabolized through two branches that converge at δ-aminovaleric acid (12, 35). Further metabolism of δ-aminovaleric acid yields glutaric acid that is channeled to Krebs cycle intermediates. The thick arrows indicate the main catabolic pathway (Revelles, unpublished). The enzymatic steps are indicated as follows: 1, l-lysine decarboxylase; 2, cadaverine aminotransferase; 3, 1-piperideine dehydrogenase; 4, l-lysine monooxygenase; 5, 5-aminovaleramide amidase; 6, 5-aminovalerate aminotransferase; 7, glutarate semialdehyde dehydrogenase.
In the present study we characterized in detail the davT chromosomal region, and we show that this gene forms an operon with davD, the gene encoding glutaric semialdehyde dehydrogenase. We have identified the promoter of the davDT operon and found that its maximal expression requires efficient transport of lysine into the cell and transformation of this amino acid into δ-aminovaleric acid, which appears to be the true effector for the expression of the davDT operon.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
P. putida KT2440, a derivative of the P. putida soil isolate mt-2, was used (13, 32). P. putida rei2 is a KT2440 derivative that carries the ′luxCDABE genes fused in frame to the davT gene (12). Escherichia coli strain DH5α was used for cloning experiments, and E. coli CC118λpir was used as a host for pUT plasmids (9). These strains were grown at 37°C in Luria-Bertani medium (40). P. putida strains were grown at 30°C either in Luria-Bertani medium or in minimal medium, which was basal M9 medium (40) supplemented with Fe-citrate, MgSO4, and trace metals, as described previously (1), and with benzoate (15 mM), glucose (0.4% [wt/vol]), or sodium citrate (10 mM) as a carbon source. When indicated lysine (10 mM), δ-aminovaleric acid (5 mM) or glutaric acid (10 mM) were added to the culture medium. When appropriate, antibiotics were added at the following concentrations: chloramphenicol, 30 μg/ml; gentamicin (Gm), 30 μg/ml; kanamycin (Km), 50 μg/ml; and tetracycline, 15 μg/ml.
DNA techniques.
Preparation of plasmid and chromosomal DNA, digestion with restriction enzymes, ligation, electrophoresis, and Southern blotting were done by using standard methods (40). P. putida cells were electroporated as described previously (23). Hybridizations were done by using the DIG DNA labeling and detection kit (Roche Laboratories, Mannheim, Germany) according to the manufacturer's instructions. Plasmid sequencing was done with universal or reverse pUC19/M13 oligonucleotides as primers. Sequencing was done on an ABI Prism 310 automated sequencer. Sequences were analyzed with Omiga 2.0 software (Oxford Molecular) and ORF FINDER (available at the National Center for Biotechnology Information [NCBI]) and compared to data from the P. putida KT2440 genome, obtained from NCBI, and with the GenBank database by using basic local alignment search tool (BLAST; www.ncbi.nlm.nih.gov) programs (2).
Preparation of RNA, primer extension analysis, and RT-PCR.
P. putida strains KT2440 and rei2 were grown overnight in M9 minimal medium with benzoate. Cells were then diluted 100-fold in fresh medium, and different aliquots were incubated in duplicate in the absence or in the presence of 5 mM lysine and 5 mM δ-aminovaleric acid until the culture reached a turbidity of ca. 1.0 at 660 nm. Cells (30 ml for primer extension and 1.5 ml for reverse transcription-PCR [RT-PCR]) were harvested by centrifugation (5,000 × g for 10 min) and processed for RNA isolation according to the method of Marqués et al. (27). Extracts were treated with RNase-free DNase I (50 U) in the presence of 3 U of an RNase inhibitor cocktail (Roche Laboratories). For primer extension analysis, we used as a specific primer an oligonucleotide complementary to the davD mRNA (5′-GGTCACCTTGATGGTCTGGCC-3′). The primer was labeled at its 5′ end with [γ-32P]ATP by using T4 polynucleotide kinase as described previously (27). About 105 cpm of the labeled primer was hybridized to 20 μg of total RNA, and extension was carried out by using avian myeloblastosis virus reverse transcriptase as described previously (27). Electrophoresis of cDNA products was done in a urea-polyacrylamide sequencing gel to separate the reaction products.
RT-PCR was done with 1 μg of RNA in a final volume of 20 μl by using the Titan OneTube RT-PCR system according to the manufacturer's instructions (Roche Laboratories). The annealing temperature used for RT-PCR was 60°C, and the cycling conditions were as follows: 94°C for 30 s, 60°C for 30 s, and 68°C for 1 min. Positive and negative controls were included in all assays. The primers used to test contiguity in the mRNA of the davD and davT genes are available upon request.
Construction of a P. putida orf4 mutant strain.
A mutant strain bearing an inactivated chromosomal orf4 was constructed as follows: plasmid pCHESIΩKm is a pUC18 derivative containing the origin of transfer oriT of RP4 and the Ω-Km interposon of plasmid pHP45ΩKm cloned as a HindIII fragment (23). To generate the orf4 mutation, a 593-bp fragment of P. putida orf4 was amplified by PCR with primers provided with EcoRI and BamHI sites and subsequently cloned between the EcoRI and BamHI sites of pCHESIΩKm in the same transcriptional direction as the Plac promoter. The resulting plasmid, pCHESI-ORF4, was mobilized from E. coli CC118λpir into P. putida KT2440 by triparental mating with the E. coli HB101(pRK600) helper strain (15). P. putida transconjugants bearing a cointegrate of the plasmid in the host chromosome were selected on M9 minimal medium with benzoic acid (10 mM) as the sole carbon source and Km. A few Km-resistant clones were chosen to confirm that pCHESI-ORF4 integrated into and disrupted the orf4 gene. All clones analyzed contained an inactivated orf4 gene, and a single random clone was chosen and called P. putida ORF4::ΩKm.
Mini-Tn5 mutagenesis of rei2 mutant and selection of mutants with decreased light emission.
P. putida rei2 (lux+, Kmr) was mated with E. coli CC118λpir(pUT-Gm) (S. Molin, unpublished data) and the helper strain E. coli HB101(pRK600) as described above. Transconjugants of P. putida rei2 were selected on M9 minimal medium with citrate as the C source and with Km and Gm. Approximately 5,000 independent transconjugants were selected, and light emission was examined in the presence of 5 mM lysine added to M9 minimal medium plates with citrate as a C source. We used a charge-coupled device camera to monitor light emission (38) and selected six mutants that did not emit light or emitted significantly lower levels of light than the parental strain. To exclude those that may have resulted from insertion of mini-Tn5 in the lux genes, total DNA of the mutants was hybridized against the entire lux operon labeled with digoxigenin. Four clones were discarded because of inactivation of the lux genes, and the other two were retained for further characterization (see below).
β-Galactosidase assays.
The low-copy-number promoter probe pMP220 (42) is a useful vector for gene expression analysis because it carries a promoterless ′lacZ gene with the appropriate Shine-Dalgarno sequence upstream from the first ATG. We amplified the intergenic regions between davD and davT and between orf3 and davD and cloned them in front of ′lacZ in pMP220 to yield pOR1 and pOR2, respectively. The intergenic region between orf3 and davD, and between davD and davT (Fig. 2) was amplified by PCR with primers incorporating restriction sites, an EcoRI site in the primer designed to meet the 5′ end, and a KpnI site in the primer designed to meet the 3′ end. Upon amplification, DNA was digested with EcoRI and KpnI and ligated to EcoRI-KpnI-digested pMP220 to create the appropriate fusion to ′lacZ to yield pOR1 and pOR2. These plasmids were sequenced to make sure that no mutations were introduced in the amplified fragments. The plasmids were electroporated into P. putida KT2440, and the corresponding transformants were grown overnight on M9 minimal medium with 15 mM benzoate and tetracycline. Then cultures were diluted 100-fold in the same medium. After 1.5 h of incubation at 30°C with shaking, the cultures were supplemented or not with inducer compounds, and 4 h later β-galactosidase activity was assayed in permeabilized whole cells according to the method of Miller (28).
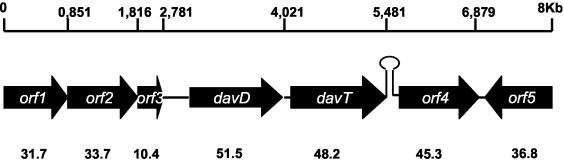
FIG. 2.
Structure of an 8-kb fragment of the chromosome of P. putida KT2440 bearing the davD and davT genes. The davDT genes were sequenced as described in Materials and Methods, and our results agreed with the P. putida genome sequence available at the NCBI (33). Sequence analysis was carried out as described in Materials and Methods. The size of the product of each ORF is given below the corresponding gene in kilodaltons.
14C-labeled lysine uptake assays.
P. putida KT2440 and its mutant derivatives were grown on M9 minimal medium with benzoate until they reached mid-logarithmic phase (0.7 to 0.8 turbidity units at 660 nm). Cells were harvested by centrifugation (3,500 × g for 15 min), washed with M9 medium and benzoate, and incubated with 0.5 μM lysine (7.8 μCi of 14C per assay) for different periods of time. The cells were then filtered and washed with 2 volumes of medium, and the filter was transferred to a scintillation vial. Nonspecific lysine-binding assays were done as described above except that the cells were preincubated for 30 min at 4°C and, after the addition of [14C]lysine, were kept at 4°C.
RESULTS
The davD-davT genes form an operon that is expressed from the PdavD promoter. Espinosa-Urgel and Ramos (12) reported that the davD and davT genes were closely linked in the chromosome of P. putida KT2440 and oriented in the same transcriptional direction (Fig. 2), but it was not determined whether the genes formed an operon. Previous work indicated that davT encodes δ-aminovalerate aminotransferase, and the sequence similarity led us to postulate that the preceding gene, davD, encodes the next enzyme in the lysine catabolic route, glutaric acid semialdehyde dehydrogenase (12). No further analysis of the chromosomal region was done in the original study by Espinosa-Urgel and Ramos (12). We have now identified three open reading frames (ORFs) upstream of davD (orf1, orf2, and orf3) whose translated products correspond to PP0209, PP0210, and PP0211, respectively (33), and one gene (orf4 PP0215) downstream from davT (Fig. 2). All of the genes are predicted to be transcribed in the same direction. Further downstream from orf4 there is another ORF that is transcribed divergently with respect to orf4 (Fig. 2).
orf1 is 852 bp long and its translated gene product exhibits a high degree of similarity (∼75%) to a number of ATP-binding components of ABC transporter systems, such as the nitrate ABC transporter system of Methanococcus janaschii and Synechocystis sp., as well as the sulfonate transporter system of Methanosarcina acetivorans (6, 14, 20). The stop codon of orf1 overlaps with the start codon of orf2, which is 966 bp long and whose translated protein exhibits in some stretches a low degree of similarity to ficobiliproteins. However, the function of this protein is unknown. The putative orf3 presumably encodes a hypothetical protein of unknown function. At 487 bp downstream from the stop codon of orf3 we identified the putative first ATG of davD gene. The intergenic space between davD and the downstream davT gene is 293 bp, whereas the distance between the stop codon of davT and the start codon of orf4 is only 138 bp. The translated product of orf4 exhibits similarity to DNA-binding regulators belonging to the two-component response regulator family.
We decided to test whether all six genes with the same orientation could be transcribed as a single mRNA. To test the potential operon structure of these genes, we grew P. putida KT2440 cells in M9 minimal medium with benzoate as the carbon source and with 5 mM lysine and isolated mRNA from cells in the late-log growth phase. Then we carried out RT-PCRs with primers based on the 3′ end of each of the ORFs and the 5′ end of the next ORFs. We found cDNA products of the expected sizes with the primers corresponding to orf1 and orf2 and davD and davT (not shown) but not with primers corresponding to orf3 and davD or davT and orf4. It should be mentioned that we have not been able to detect mRNA corresponding to orf3 under any growth condition tested in the present study. We therefore considered that the six ORFs could be organized in three transcriptional units, with davD and davT forming a unit that seems to be transcribed independently from orf4 and orf1/orf2.
The similarity of orf4 to genes encoding response regulators led us to test whether the orf4 gene product is involved in the regulation of the davDT operon. To this end, we generated a mutant in orf4 as described in Materials and Methods. The mutant grew as fast as the wild type with lysine as the sole carbon and energy source, and the gene product was therefore considered not to be involved in the regulation of lysine metabolism.
davD promoter region.
Plasmids pOR1 (intergenic davD-davT region′::′lacZ) and pOR2 (PdavD′::′lacZ) were electroporated in P. putida KT2440, and cells were grown as described in Materials and Methods in the absence or in the presence of 5 mM lysine. We found no β-galactosidase activity with pOR1 regardless of the growth conditions. In contrast, with pOR2 we detected basal expression in the absence of lysine, which increased when cells were grown in its presence (Fig. 3). This suggested that a functional promoter could only be located in front of the davD gene. These findings, along with the RT-PCR results, suggest that transcription of davDT genes is driven exclusively from a promoter upstream of davD.
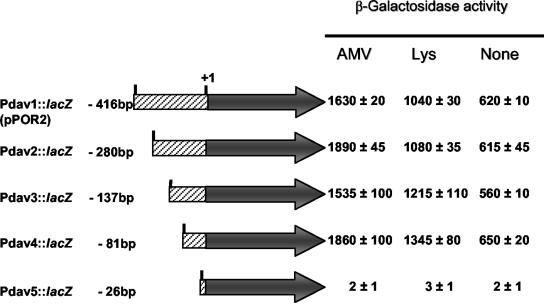
FIG. 3.
Identification of the minimal region of the davD promoter that allowed regulated expression. Intergenic regions of different length between davD and orf3 were amplified by PCR with the same primer to meet the 3′ end (provided with a Kpn site) and different primers at the 5′ end (provided with an EcoRI site). The PCR products, once digested with these restriction enzymes, yielded fragments of 416, 280, 137, 81, and 26 bp upstream from the +1 transcription start point. P. putida KT2440 cells bearing the indicated fusion were grown on M9 minimal medium with benzoate alone or with 5 mM lysine or 5 mM δ-aminovaleric acid. The β-galactosidase activity was determined as described in Materials and Methods, and values are the averages of three independent assays done in duplicate.
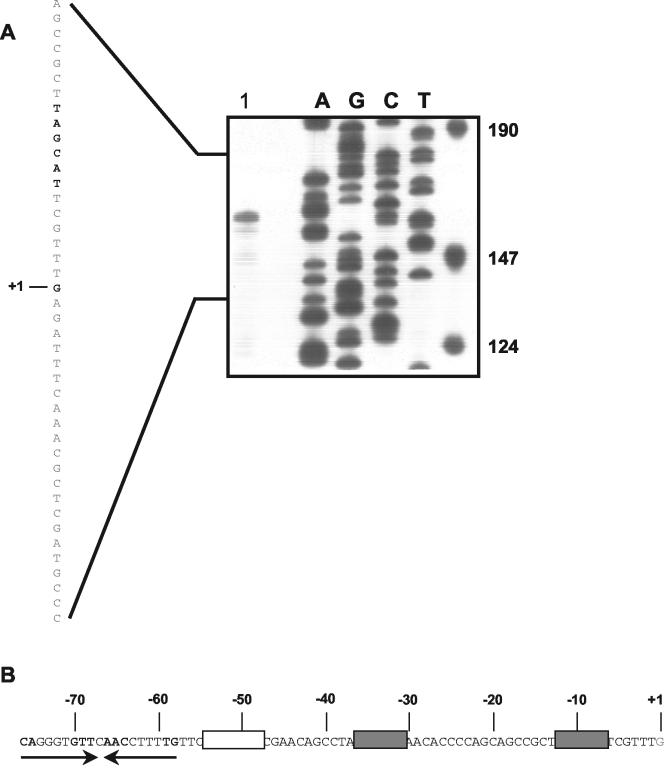
To identify the transcription initiation point of the davDT operon and define the minimum region required for induction to take place, we carried out the following assays. First, we prepared mRNA from KT2440 grown with 5 mM lysine and used a primer located 50 bp within the davD gene for primer extension. Our results revealed a major transcription initiation point (Fig. 4) located 66 bp upstream with respect to the A of the first ATG in davD. Analysis of the sequence upstream revealed a −10 region (5′-TAGCAT-3′) very similar to 5′-TATCAT-3′, which is best recognized by the P. putida sigma-70 factor (10). The promoter also exhibited a relatively well-conserved −35 region (5′-TTGTCC-3′) with 17 bases between the two boxes.
FIG. 4.
Transcription initiation point of the davD promoter and key sequence features in the proximal 5′ region. (A) Transcription initiation mapping of the davD gene. Lane 1, RNA isolated from cells grown on M9 minimal medium with 5 mM lysine. The other lanes correspond to a sequencing ladder. The DNA sequence corresponding to the davD promoter region is shown on the left. The transcription start point is in boldface and is marked +1. The proposed −10 box is in boldface. (B) Organization of the davD promoter region. The −10 and −35 regions are indicated in boldface and by gray boxes. A T-rich region is shown in an open box. Convergent arrows indicate a potential inverted repeat.
To determine the region upstream from the +1 position in davD necessary for the regulated expression of the operon, we amplified different portions of the region by PCR using in all cases the same primer at the 3′ end and different primers at the 5′ end, so that we generated PCR-amplified fragments of different sizes (Fig. 3). These fragments were cloned in pMP220 and β-galactosidase activity was measured. We found high β-galactosidase levels (>1,500 Miller units) when the primer allowed amplification of at least 81 bp upstream from the +1 point.
Sequence analysis of the region between positions −81 and +1 revealed, in addition to the −10/−35 boxes described above, a T-rich region between −48 and −62 that could function as a UP element or a curvature element, and an imperfect inverted repeat that extended from −76 to −58 and that could be the target for a regulator responsible for the induction of this promoter (Fig. 4).
Efficient lysine transport is required for full expression of the davDT operon.
Mutant rei2 carries an in-frame fusion of davT to the ′luxABCDE genes, and the cells show increased light emission upon exposure to lysine. We mutagenized the rei2 mutant with a mini-Tn5-Gm transposon as described in Materials and Methods and searched for mutants with reduced luminescence. These mutants were expected to result from the limited uptake of lysine or from inactivation of a putative regulator. Two mutants called rei2-6 and rei2-7 emitted a basal level of light and only a small increase in light emission was found in response to lysine (Table 1). Southern blotting assays revealed a single insertion of the mini-Tn5-Gm in each of the mutants, and we sequenced the insertion site. In rei2-6 and rei2-7 mutants, the mini-transposon had interrupted ORFs of 753 and 885 bp, respectively (corresponding to PP0283 and PP4486). BLAST analyses against different data banks were carried out with the translated sequence of each ORF. We found that in the rei2-6 and rei2-7 mutants the inactivated genes were related to amino acid uptake systems belonging to the ABC superfamily (17, 30, 39, 41). The gene inactivated in mutant rei2-6 (PP0283) exhibited high similarity to the ATP-binding component of the arginine/ornithine transport system of Pseudomonas aeruginosa (94% similarity), the ATP-binding protein of the histidine ABC transporter of P. syringae pv. tomato strain DC3000 (92% similarity) and E. coli (79% similarity), and the ATP-binding component of the ornithine transport of Salmonella enterica serovar Typhimurium LT2 (79% identity). The insertion in mutant rei2-7 had inactivated a periplasmic binding protein (PP4486) of a transport system for basic amino acids (16, 34). This periplasmic binding protein is a member of the LAO (lysine/arginine/ornithine) group (21). We hypothesized that the lower induction from PdavD in these mutants probably resulted from their limited lysine uptake. To test this hypothesis, we carried out assays with [14C]lysine in cells grown on minimal medium with benzoate as a C source and NH4+ as an N source. We found that uptake of [14C]lysine in the rei2, rei2-6, and rei2-7 mutants was linear in time for about 3 min. [14C]lysine was incorporated by rei2 cells at a rate of 2.5 nmol/mg of protein per min, whereas in the mutants the rate of [14C]lysine incorporation was ca. 30% of that determined for the rei2 strain.
TABLE 1.
Relative level of light emission by rei2 and its mutant derivativesa
| Strain | Relative light units ± SD
|
Increase (fold) | |
|---|---|---|---|
| −Lys | +Lys | ||
| rei2 | 5 ± 0.1 | 20 ± 0.2 | 4 |
| rei2-6 | 5 ± 0.1 | 6 ± 0.1 | 1.2 |
| rei2-7 | 5 ± 0.1 | 7.5 ± 0.1 | 1.5 |
Bacterial cells were grown on M9 minimal medium with citrate as the carbon source without (−) and with (+) 5 mM lysine (Lys) for 3 h at 30°C. Light emission was then determined in a luminometer as described in Materials and Methods.
δ-Aminovaleric acid enhances expression of the davDT operon.
To identify the signal molecule(s) that induces expression from the davD promoter, we cultured P. putida KT2440 bearing pOR2 on M9 minimal medium with 5 mM lysine (the first substrate in the pathway), 5 mM δ-aminovaleric acid (the product in which the two potential lysine degradation pathways converge [Fig. 1]), 2.5 mM cadaverine (a potential intermediate in one of the lysine branches that can yield δ-aminovaleric acid), and 5 mM glutaric acid (the product resulting from δ-aminovaleric acid metabolism). Our results revealed that, whereas glutaric acid was not an inducer (Table 2), the other three compounds induced expression from the PdavD promoter. Whether lysine and cadaverine are true inducers or whether δ-aminovaleric acid resulting from the metabolism of lysine and cadaverine is the true inducer could not be discerned from these results.
TABLE 2.
Induction of the davD promoter in the wild-type P. putida KT2440 and its isogenic mutant KT2440 mut-1 and RpoNa
| Strain | Activity (mean Miller units ± SD) on substrate
|
||||
|---|---|---|---|---|---|
| None | Lysine | Cadaverine | Aminovalerate | Glutarate | |
| KT2440 | 620 ± 10 | 1,280 ± 100 | 930 ± 60 | 1,730 ± 20 | 600 ± 10 |
| mut-1 | 470 ± 10 | 530 ± 30 | 840 ± 10 | 1,700 ± 120 | 580 ± 20 |
| RpoN | 620 ± 40 | 1,450 ± 30 | 1,370 ± 100 | 1,800 ± 50 | 550 ± 10 |
Wild-type, mut-1, and RpoN strains bearing pOR2 were grown in M9 minimal medium with benzoate as the C source. Where indicated, cultures were supplemented with 5 mM lysine, 2.5 cadaverine, 5 mM δ-aminovalerate, and 5 μM glutarate. The β-galactosidase activity was determined 7 h later in cells growing exponentially. The activity is given in Miller units, and values are averages of three to six independent determinations.
To further elucidate the nature of the inducer we repeated the above assays in P. putida KT2440 mut-1, a mutant unable to convert δ-aminovaleramide into δ-aminovaleric acid (O. Revelles, unpublished data). In this mutant, δ-aminovaleric acid induced expression from PdavD at a level similar to that found in the wild type (∼3-fold over the basal level), whereas induction by lysine was null, and induction with cadaverine was low (<2-fold). This suggests that δ-aminovaleric acid resulting from the metabolism of lysine or cadaverine may be the true inducer of the davDT operon (Table 2).
Köhler et al. (22) reported that an rpoN mutant that lacked σ54 was unable to grow on lysine. Although the davDT operon promoter presented σ70-dependent characteristics, its expression could be influenced by σ54 if the operon were part of a regulatory cascade with a master regulator involving σ54, as is the case in 3-methylbenzoate metabolism by the TOL plasmid of P. putida when cells are grown on m-xylene (37). To test this hypothesis, we transformed pOR2 in P. putida strain TK19, an rpoN mutant, and determined the β-galactosidase activity in the presence or in the absence of lysine cadaverine and δ-aminovaleric acid. We found an induction ratio of about 2.5, a level similar to the one obtained in the wild type. Therefore, σ54 is not directly or indirectly involved in expression of the davD gene, and the inability of the RpoN mutant to grow on lysine should therefore be ascribed to the need to express other catabolic segments of the lysine catabolic pathway.
DISCUSSION
Espinosa-Urgel and Ramos (12) showed before that mutagenesis of P. putida KT2440 with a mini-Tn5-lux transposon was useful for identifying genes whose expression increased in cells that colonize plant roots or are exposed to plant root exudates. The inability of the rei2 mutant to use lysine identified a gene, called davT, involved in the use of this amino acid as a C source.
DNA sequencing and the series of RT-PCR assays carried out in the present study have now shown that davT forms an operon with the davD gene. Our in vivo assays revealed light emission by cells of the rei2 strain growing in the absence of lysine, which increased ∼3-fold in response to the exogenous addition of this amino acid.
It has been proposed that in P. putida, the initial metabolism of lysine occurs through two branches that converge at δ-aminovaleric acid (Fig. 1). P. putida mut-1 is blocked at δ-aminovaleramide aminotransferase—a key step in one of the loops (Fig. 1)—and grows slowly on lysine (Revelles, unpublished). This suggests that the δ-aminovaleramide branch is the major lysine assimilation pathway in this strain. In agreement with this hypothesis is the finding that in the mut-1 strain, lysine did not induce the expression of the davDT operon promoter, and cadaverine only induced low levels of expression of the davDT genes. In contrast, δ-aminovaleric acid behaved as the strongest effector. We are therefore tempted to propose that δ-aminovaleric acid is the true inducer of the davDT operon.
The transcription start point of the davD promoter was identified, and deletion analysis of the promoter region revealed the need for at least 80 bp upstream from this point for expression from the davD promoter. Sequence analysis of the promoter region revealed good −10 and −35 boxes for σ70 promoters. Because expression from PdavD can be induced by exogenous lysine, we hypothesized the participation of an as-yet-unidentified regulator, which might possibly bind to an inverted repeat in the −58/−76 region. Although this motif is relatively far from the RNA polymerase binding site, the presence of a putative UP element in the −50/−60 region might facilitate contacts between RNA polymerase and the putative regulator. This type of organization is seen in some E. coli CRP regulated promoters (7, 26) and, to our knowledge, this is the first such case proposed for a pseudomonad.
Downstream of davT we identified orf4, whose translated product showed homology to regulators belonging to the two-component family. An orf4::ΩKm mutant exhibited a pattern of davDT expression similar to that of the wild type, which ruled out this regulator as responsible for the regulated expression of this operon. An alternative approach to search for a potential regulator was designed. We reasoned that the intensity of light emitted by the mutants in the putative regulator of the davDT operon would be diminished. After Tn5 mutagenesis of the rei2 strain we searched for such mutants among 5,000 transconjugants; only two of them exhibited low induction levels. Analysis of the knockout genes suggested that the mutants might exhibit defects in lysine uptake. The results with mutants rei2-6 and rei2-7 showed that efficient induction of the davDT operon by exogenous lysine requires effective uptake systems.
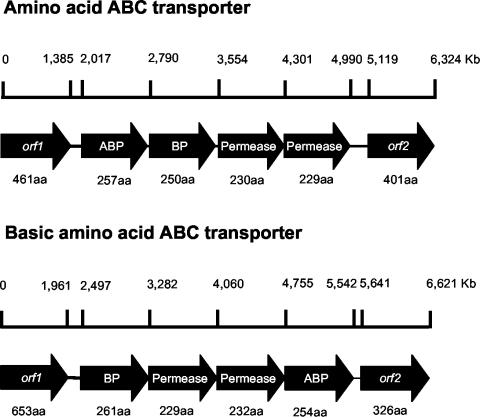
Lysine in P. putida KT2440 seems to be taken up through at least two ABC transporters that appear to consist of four proteins. Figure 5 shows the genomic organization of the two systems. In mutant rei2-6 the gene that encodes the ATP-binding protein was followed by the gene that encodes the solute-binding protein and two integral membrane permeases. In mutant rei2-7 the genes encoding the two permeases lay between the genes that encode the solute-binding protein and the ATP-binding protein. Inactivation of either of the two uptake systems resulted in a significant decrease in the rate of lysine uptake and resulted in reduced induction from the davD promoter.
FIG. 5.
Genetic organization of the two ABC transporter systems for l-lysine uptake in P. putida KT2440. (Top) Organization of the l-lysine transport system inactivated in mutant rei2-6; (bottom) organization of the l-lysine transport system inactivated in rei2-7. Genome organization is shown according to the findings of Nelson et al. (33). The number of amino acids of each translated product is given below the gene. BP stands for periplasmic binding protein, and ABC stands for ATP-binding protein. The function of the products encoded by adjacent ORFs in both loci is unknown.
Köhler et al. (22) reported that an RpoN mutant of P. putida did not grow with lysine. In the RpoN mutant strain both lysine and δ-aminovaleric acid induced expression from PdavD, and therefore the inability to grow could not be ascribed to the prevention of expression of the branches that converge in δ-aminovaleric acid or of the davDT operon itself. Instead, the RpoN mutant seems to be unable to metabolize glutaric acid (Revelles, unpublished). It follows that lysine catabolism in P. putida involves the assemblage of several pathway stretches that are independently regulated: (i) the two loops that convert lysine into δ-aminovaleric acid, (ii) the davD operon for metabolism of δ-aminovaleric acid to glutaric acid, and (iii) a segment that converts glutaric acid into Krebs cycle intermediates. This modular organization endows the strain with high metabolic versatility to respond to different carbon or nitrogen sources supplied exogenously or generated internally during cell metabolism.
Acknowledgments
This study was supported by EU project QLRT-02884 and national grant BIO2003-00515 from the CICYT.
We thank K. Shashok and C. Lorente for checking the English in the manuscript.
REFERENCES
- 1.Abril, M. A., C. Michán, K. N. Timmis, and J. L. Ramos. 1989. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J. Bacteriol. 171:6782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basic, A., S. F. Moody, and A. E. Clarke. 1986. Structural analysis of secreted root slime from maize (Zea mays L.). Plant Physiol. 80:771-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayliss, C., E. Bent, D. E. Culham, S. MacLellan, A. J. Clarke, G. L. Brown, and J. M. Wood. 1997. Bacterial genetic loci implicated in the Pseudomonas putida GR12-2R3-canola mutualism: identification of an exudate-inducible sugar transporter. Can. J. Microbiol. 43:809-818. [DOI] [PubMed] [Google Scholar]
- 5.Bhagwat, A. A., and D. L. Keister. 1992. Identification and cloning of Bradyrhizobium japonicum genes expressed selectively in soil and rhizosphere. Appl. Environ. Microbiol. 58:1490-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bult,C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. Fitzgerald, R. A. Clayton, J. D. Gocyne, A. R. Kerlavage, B. A. Dougherty, J. F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. Geoghagen, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 7.Busby, S., and R. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 8.Dakora, F. D., C. M. Joseph, and D. A. Phillips. 2002. Alfalfa (Medicago sativa L.) root exudates contain isoflavonoids in the presence of Rhizobium meliloti. Plant Physiol. 101:819-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domínguez-Cuevas, P., and S. Marqués. 2004. Compiling sigma-70-dependent promoters, p. 319-344. In J. L. Ramos (ed.), Pseudomonas: virulence and gene regulation, vol. II. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 11.Espinosa-Urgel, M., A. Salido, and J. L. Ramos. 2000. Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J. Bacteriol. 182:2363-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espinosa-Urgel, M., and J. L. Ramos. 2001. Expression of a Pseudomonas putida aminotransferase involved in lysine catabolism is induced in the rhizosphere. Appl. Environ. Microbiol. 67:5219-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franklin, F. C. H., M. Bagdasarian, M. M. Bagdasarian, and K. N. Timmis. 1981. Molecular and functional analysis of the TOL plasmid pWW0 from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta-cleavage pathway. Proc. Natl. Acad. Sci. USA 78:7458-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galagan, J. E., C. Nusbaum, A. Roy, M. G. Endrizzi, P. Macdonald, W. Fitzhugh, S. Calvo, R. Engels, S. Smirnov, D. Atnoor, A. Brown, N. Allen, J. Naylor, N. Stange-Thomann, K. Dea Arellano, R. Johnson, L. Linton, P. McEwan, K. McKernan, J. Talamas, A. Tirrell, W. Ye, A. Zimmer, R. D. Barber, I. Cann, D. E. Graham, D. A. Grahame, A. M. Guss, R. Hedderich, C. Ingram-Smith, H. C. Kuettner, J. A. Krzycki, J. A. Leigh, W. Li, J. Liu, B. Mukhopadhyay, J. N. Reeve, K. Smith, T. A. Springer, L. A. Umayam, O. White, R. H. White, E. Conway de Macario, J. G. Ferry, K. F. Jarrell, H. Jing, A. J. Macario, I. Paulsen, M. Pritchett, K. R. Sowers, R. V. Swanson, S. H. Zinder, E. Lander, W. W. Metcalf, and B. Birren. 2002. The genome of Methanosarcina acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higgins, C. F., and G. F. Ames. 1981. Two periplasmic transport proteins which interact with a common membrane receptor show extensive homology: complete nucleotide sequences. Proc. Natl. Acad. Sci. USA 78:6038-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosie, A. H. F., D. Allaway, D.-S. Galloway, H. A. Dunsby, and P. S. Poole. 2002. Rhizobium leguminosarum has a second general amino acid permease with unusually broad substrate specificity and high similarity to branched-chain amino acid transporters of the ABC family. J. Bacteriol. 184:4071-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaeger, C. H., III, S. E. Lindow, W. Miller, E. Clark, and M. K. Firestone. 1999. Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and tryptophan. Appl. Environ. Microbiol. 65:2685-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiménez-Zurdo, J. I., F. M. García-Rodríguez, and N. Toro. 1997. The Rhizobium meliloti putA gene: its role in established of the symbiotic interaction with alfalfa. Mol. Microbiol. 23:85-93. [DOI] [PubMed] [Google Scholar]
- 20.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsumo, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:185-209. [DOI] [PubMed] [Google Scholar]
- 21.Kang, C. H., W. C. Shin, Y. Yamagata, Goksen, S., G. F. Ames, and S. H. Kim. 1991. Crystal structure of the lysine-, arginine-, ornithine-binding protein (LAO) from Salmonella typhimurium at 2.7-Å resolution. J. Biol. Chem. 266:23893-23899. [PubMed] [Google Scholar]
- 22.Köhler, T., S. Harayama, J. L. Ramos, and K. N. Timmis. 1989. Involvement of Pseudomonas putida RpoN σ-factor in regulation of various metabolic functions. J. Bacteriol. 171:4326-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Llamas, M. A., J. J. Rodríguez-Herva, R. E. W. Hancock, W. Bitter, J. Tommasen, and J. L. Ramos. 2003. Role of the Pseudomonas putida tol-oprL gene products in the uptake of solutes through the cytoplasmic membrane. J. Bacteriol. 185:4707-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lugtenberg, B. J., L. Dekkers, and G. V. Bloemberg. 2001. Molecular determinants of rhizosphere colonization by Pseudomonas. Annu. Rev. Phytopathol. 39:461-490. [DOI] [PubMed] [Google Scholar]
- 25.Lugtenberg, B. J., L. V. Kravchenko, and M. Simons. 1999. Tomato seed and root exudates sugars: composition, utilization by Pseudomonas biocontrol strains and role in rhizosphere colonization. Environ. Microbiol. 1:439-446. [DOI] [PubMed] [Google Scholar]
- 26.Lutz, R., T. Lozinski, T. Ellinger, and H. Bujard. 2001. Dissecting the functional program of Escherichia coli promoters: the combined mode of action of Lac repressor and AraC activator. Nucleic Acids Res. 29:3873-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marqués, S., J. L. Ramos, and K. N. Timmis. 1993. Analysis of the mRNA structure of the Pseudomonas putida TOL meta fission pathway operon around the transcription initiation point, the xylTE and the xylFJ region. Biochim. Biophys. Acta 1216:227-237. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Molina, L., C. Ramos, E. Duque, M. C. Ronchel, J. M. García, L. Wyke, and J. L. Ramos. 2000. Survival of Pseudomonas putida KT2440 in soil and in the rhizosphere of plants under greenhouse and environmental conditions. Soil Biol. Biochem. 32:315-321. [Google Scholar]
- 30.Montesinos, M. L., A. Herrero, and E. Flores. 1997. Amino acid transport in taxonomically diverse cyanobacteria and identification of two genes encoding elements of a neutral amino acid permease putatively involved in recapture of leaked hydrophobic amino acids. J. Bacteriol. 179:853-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mozafar, A. 1992. Effect of Pseudomonas fluorescens on the root exudates of tomato mutants differently sensitive to Fe chlorosis. Plant Soil 144:167-176. [Google Scholar]
- 32.Nakazawa, T. 2002. Travel of a Pseudomonas from Japan around the world. Environ. Microbiol. 4:782-785. [DOI] [PubMed] [Google Scholar]
- 33.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. Chris Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. A. Eisen, K. N. Timmis, A. Dusterhoft, B. Tummler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 34.Oh, B. H., J. Pandit, C. H. Kang, K. Nikaido, S. Gokcen, G. F. Ames, and S. H. Kim. 1993. Three-dimensional structures of the periplasmic lysine/arginine/ornithine-binding protein with and without a ligand. J. Biol. Chem. 268:11348-11355. [PubMed] [Google Scholar]
- 35.Phillips, A. T. 1986. Biosynthetic and catabolic features of amino acid metabolism in Pseudomonas, p. 385-438. In J. R. Sokatch (ed.), The bacteria: a treatise on structure and function, vol. X. Academic Press, Inc., Orlando, Fla. [Google Scholar]
- 36.Rainey, P. B. 1999. Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ. Microbiol. 1:243-257. [DOI] [PubMed] [Google Scholar]
- 37.Ramos, J. L., S. Marqués, and K. N. Timmis. 1997. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid encoded regulators. Annu. Rev. Microbiol. 51:341-373. [DOI] [PubMed] [Google Scholar]
- 38.Ramos, C., L. Molina, L. Molbak, J. L. Ramos, and S. Molin. 2000. A bioluminescent derivative of Pseudomonas putida KT2440 for deliberate release into the environment. FEMS Microbiol. Ecol. 34:91-102. [DOI] [PubMed] [Google Scholar]
- 39.Saier, M. H. 2000. A function-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64:354-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Saurin, W., M. Hofnung, and E. Dassa. 1999. Getting in or out: early segregation between importers and exporters in the evolution of ATP-binding cassette (ABC) transporters. J. Mol. Evol. 48:22-41. [DOI] [PubMed] [Google Scholar]
- 42.Spaink, H. P., R. J. H. Okker, C. A. Wijffelman, E. Pees, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation of the Rhizobium leguminosarum Sym plasmid pRL1J1. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 43.Trias, J., and H. Nikaido. 1990. Protein D2 channel of the Pseudomonas aeruginosa outer membrane has a binding site for basic amino acids and peptides. J. Biol. Chem. 265:15680-15684. [PubMed] [Google Scholar]
- 44.Van Overbeck, L. S., and J. D. van Elsas. 1995. Root exudate-induced promotor activity in Pseudomonas fluorescens mutants in the wheat rhizosphere. Appl. Environ. Microbiol. 62:890-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vílchez, S., M. Manzanera, and J. L. Ramos. 2000. Control of expression of divergent Pseudomonas putida put promoters for proline catabolism. Appl. Environ. Microbiol. 66:5221-5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vílchez, S., L. Molina, C. Ramos, and J. L. Ramos. 2000. Proline catabolism by Pseudomonas putida: cloning, characterization, and expression of the put genes in the presence of root exudates. J. Bacteriol. 182:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]