Abstract
Methionine sulfoxide reductase A (MsrA) is an antioxidant repair enzyme which reduces oxidized methionine to methionine. Since oxidation of methionine in proteins impairs their function, an absence of MsrA leads to abnormalities in different organisms, including alterations in the adherence patterns and in vivo survival of certain pathogenic bacteria. To understand the role of MsrA in intracellular survival of bacteria, we disrupted the gene encoding MsrA in Mycobacterium smegmatis through homologous recombination. The msrA mutant strain of M. smegmatis exhibited significantly reduced intracellular survival in murine J774A.1 macrophages compared to the survival of its wild-type counterpart. Furthermore, immunofluorescence and immnunoblotting of phagosomes containing M. smegmatis strains revealed that the phagosomes with the msrA mutant strain acquired both p67phox of phagocyte NADPH oxidase and inducible nitric oxide synthase much earlier than the phagosomes with the wild-type strain. In addition, the msrA mutant strain of M. smegmatis was observed to be more sensitive to hydroperoxides than the wild-type strain was in vitro. These results suggest that MsrA plays an important role in both extracellular and intracellular survival of M. smegmatis.
Although mononuclear phagocytes are specially equipped to produce superoxide (O2−˙) radicals through the NADPH oxidase (phox) complex (32, 35), most living cells also generate superoxide as a by-product of normal aerobic metabolism. This occurs due to incomplete reduction of oxygen at the level of the cytochrome oxidase system, which accounts for approximately 4% of the oxygen that enters into this system (50). Superoxide reacts with a variety of organic and inorganic compounds and generates toxic reactive oxygen intermediates (ROI), such as H2O2, HO ˙ −, HOCl, and OONO ˙ − (25, 52). Toxic ROI oxidize cellular macromolecules like proteins, lipids, and nucleic acids, which accelerates aging processes and disease in living organisms (4).
In proteins, oxidation occurs mainly at the sulfur-containing amino acids cysteine and methionine (40). Oxidation of methionine leads to the formation of methionine sulfoxide, and methionine sulfoxide reductase A (MsrA), which was first identified in Escherichia coli (5), has long been implicated in the reduction of methionine sulfoxide in proteins. However, recent studies have demonstrated that oxidation of methionine leads to two stereomeric forms of methionine sulfoxide, methionione S-sulfoxide and methionine R-sulfoxide (21, 50); the reduction of these compounds is catalyzed by MsrA and MsrB, respectively, (21, 30). Analysis of the sequences of different genomes has revealed that the genes coding for both MsrA and MsrB are present in almost all living organisms, from prokaryotes to humans (11, 18, 21). Interestingly, however, MsrB from different organisms shows no sequence identity with MsrA (17). Also, in contrast to MsrA, MsrB requires the metal cofactor selenium or zinc (18) for reactivity.
Since the oxidation of methionine in proteins impairs the function of proteins, the absence of MsrA has been implicated in a variety of abnormalities in different organisms. Mice lacking MsrA have been reported to develop atypical walking patterns (tip-toe) and have reduced life spans (26). In addition, prokaryotes like Streptococcus pneumoniae, Neisseria gonorrhoeae, and Escherichia coli deficient in MsrA show altered adherence patterns (51). An absence of MsrA also affects the adherence of the urogenital pathogen Mycoplasma genitalium to sheep red blood cells (8). Furthermore, the yeast Saccharomyces cerevisiae (27) and the prokaryotes E. coli (29), Erwinia chrysanthemi (13), M. genitalium (8), and Staphylococcus aureus (37) lacking the msrA gene show increased sensitivity to H2O2 stress. A recent study demonstrated that an msrA mutant strain of E. coli is hypersensitive to nitric oxide stress, and introduction of a plasmid containing msrA of Mycobacterium tuberculosis could correct this defect (41). Moreover, human T cells (28) and yeast cells (28) overexpressing MsrA and transgenic Drosophila overexpressing MsrA are more resistant to oxidative stress (34). Finally, MsrA has been shown to be important for the survival of E. chrysanthemi (13) and M. genitalium (8) inside the host.
This study was undertaken to determine whether MsrA had any effect on the intracellular survival of bacteria. We used the fast-growing nonpathogenic bacterium Mycobacterium smegmatis in this study. M. smegmatis can survive inside mononuclear phagocytes, including adherent human peripheral blood monocytes (3), for several days, although it cannot replicate inside macrophages like its pathogenic counterparts, such as M. tuberculosis and Mycobacterium leprae. This property of M. smegmatis has been exploited to identify genes of M. tuberculosis that are important for intracellular survival (23, 24, 31, 49). We show here that an M. smegmatis msrA mutant (strain MSΔmsrA) has a decreased ability to survive in macrophages compared to wild-type M. smegmatis (strain MSWt). We also demonstrate that phagosomes containing the MSΔmsrA strain acquire oxidative markers of macrophages earlier than phagosomes containing the MSWt strain acquire them.
MATERIALS AND METHODS
Materials and reagents.
Restriction enzymes and DNA modification enzymes used in this study were purchased from either New England Biolabs (Beverly, Mass.) or GIBCO-BRL (Rockville, Md.). Taq polymerase and deoxynucleoside triphosphates were purchased from Perkin-Elmer (Foster City, Calif.). Qiaprep columns were obtained from Qiagen Inc. (Valencia, Calif.). Hydrogen peroxide, methyl viologen, t-butyl hydroperoxide, and cumene hydroperoxide were purchased from Sigma Chemical Co. (St. Louis, Mo.). S-Nitrosoglutathione (GSNO) and papanonate were purchased from Alexis (San Diego, Calif.). Nitrocellulose membranes were obtained from Schleicher & Schuell (Keene, N.H.).
Bacterial strains, media, and growth conditions.
M. smegmatis mc2155 (= MSWt) was grown either in Middlebrook 7H9 broth containing 0.2% glycerol, albumin dextrose complex (ADC) (5 g of bovine serum albumin per liter, 0.85 g of NaCl per liter), and 0.05% Tween 80 (7H9-TW-ADC) or on Middlebrook 7H10 agar containing 0.2% glycerol, ADC, and 0.05% Tween 80 (7H10-TW-ADC). M. smegmatis harboring a plasmid was grown in 7H9-TW-ADC or 7H10-TW-ADC containing the antibiotic kanamycin (25 μg/ml) or hygromycin (50 μg/ml) or both. M. tuberculosis H37Rv was grown in 7H9 broth containing 0.05% Tween 80 and the oleic acid-albumin-dextrose-catalase supplement. All subcloning procedures were performed in E. coli strain Inv-α (Invitrogen, Carlsbad, Calif.). Plasmid-containing E. coli was grown in Luria-Bertani broth or agar with the appropriate antibiotic (100 μg of ampicillin per ml, 25 μg of kanamycin per ml, or 100 μg of hygromycin per ml). Unless indicated otherwise, all bacteria were grown at 37°C.
DNA manipulations.
Chromosomal DNA of mycobacteria was isolated by using cetyltrimethylammonium bromide as described previously (43). Plasmid DNA from E. coli was isolated by using a Qiaperp kit (Qiagen Inc.). Southern hybridization and PCR were performed as described by Ausubel et al. (1). Oligonucleotide primers were synthesized at the Center for DNA Technology, University of Texas Health Science Center at San Antonio.
Disruption plasmid for M. smegmatis msrA.
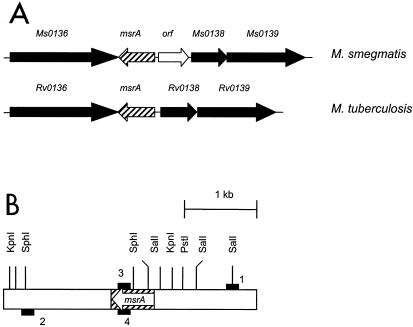
In order to construct an msrA disruption plasmid for M. smegmatis, we first cloned the msrA gene of M. smegmatis. We used The Institute for Genome Research database to obtain the M. smegmatis genome sequence for this. We analyzed the M. smegmatis genome sequence by BLAST analysis using the msrA gene of M. tuberculosis, and we identified the sequence containing the M. smegmatis msrA gene. The information was downloaded from the database, and based on the sequence, primers to amplify an approximately 2.7-kb region were synthesized. In fact, four primers were synthesized, as shown in Fig. 1B. Primers MSMSRA1 (5′-ACGTGCGATCCCAGGAATCC-3′) and MSMSRA4 (5′-CGTCGGATGGTCGTTCAGGCCTCCGGTGTAGCC-3′) were used to amplify the 5′ portion of msrA and its upstream region (1,097 bp), and primers MSMSRA3 (5′-GGCTACACCGGAGGCCTGAACGACCATCCGACG-3′) and MSMSRA2 (5′-AATGTCCAGCCGAACCTCAGCC-3′) were used to amplify the 3′ portion of msrA and its downstream region (1575 bp). The sequences of primers MSMSRA3 and MSMSRA4 were modified (underlined nucleotides) to create an StuI site within the msrA gene. The amplified 1,097- and 1,575-bp fragments were separately cloned in pCR2.1 to create plasmids pMSMSRA1 and pMSMSRA2. Plasmid pMSMSRA2 was cut with StuI and BamHI to release the 1,575-bp fragment, and this fragment was ligated to StuI- and BamHI-cut pMSMSRA1 to create plasmid pMSMSRA3. This step was necessary to create a unique StuI site within the msrA gene. Plasmid pMSMSRA3 was cut with EcoRI to release the 2.6-kb fragment, and this fragment was cloned into EcoRI-cut pUC18 to create plasmid pMSMSRA4. Finally, plasmid pMSMSRA4 was cut with StuI and ligated to the 1.2-kb kanamycin resistance gene from pMV206 (42) to create plasmid pMSMSRA5. This plasmid served as the M. smegmatis msrA disruption construct, which was electroporated into wild-type M. smegmatis to obtain M. smegmatis msrA mutant strains.
FIG. 1.
(A) Organization of msrA in the genomes of M. smegmatis and M. tuberculosis. M. smegmatis open reading frames showing similarity to M. tuberculosis open reading frames, other than the MsrA open reading frame, are designated Ms0136, Ms0138, and Ms0139. (B) Schematic representation of the msrA locus in the chromosome of M. smegmatis. The open boxes represent the flanking regions, the striped box represents the msrA gene, and the solid boxes represent the primers. 1, 2, 3, and 4 indicate primers MSMSRA1, MSMSRA2, MSMSRA3, and MSMSRA4, respectively. The arrow indicates the direction of msrA transcription. KpnI, PstI, SalI, and SphI indicate the restriction sites in and around msrA.
Construction of other plasmids.
Several other plasmids were created to enable complementation of MsrA activity in the M. smegmatis msrA mutant strains or expression of green fluorescent protein in M. smegmatis or for other purposes. Plasmid pHG361 was constructed by deleting the 1.2-kb aph gene (kanamycin resistance gene) in pMV261 (42) by cutting with NheI and SpeI and ligating the remaining fragment (blunt ended with the Klenow fragment) with the 1.7-kb hygromycin resistance gene (cut with BamHI and PstI and blunt ended with the Klenow fragment) from pIJ963 (10). Similarly, plasmid pHG206 was constructed by deleting the 1.2-kb aph gene (kanamycin resistance gene) in pMV206 by cutting the sequence with NheI and SpeI and ligating the remaining fragment (blunt ended with the Klenow fragment) with the 1.7-kb hygromycin resistance gene (cut with BamHI and PstI and blunt ended with the Klenow fragment) from pIJ963 (10). Plasmid pMSMSRA6 was generated by cloning the 1.2-kb M. smegmatis msrA-containing PCR fragment (blunt ended) into pHG361. Plasmid pMTSIGHGFP3 was generated by cloning the DNA containing the M. tuberculosis sigH promoter and the gfpuv gene region (sigH::gpuv) in the EcoRV site of pOLYG. The sigH::gpuv region was obtained by cutting plasmid pMTSIGHGFPUV1 (unpublished data) with EcoRI and NotI.
Electroporation.
Transformation of M. smegmatis with plasmids containing the mycobacterial origin of replication or plasmids without the mycobacterial origin of replication was achieved by electroporation (16).
Intracellular survival of M. smegmatis.
The adherent mouse macrophage cell line J774A.1 grown in Dulbecco modified Eagle medium with 10% fetal bovine serum was used to determine the intracellular survival of M. smegmatis strains. Freshly grown suspensions of M. smegmatis strains (viability, more than 90%) were gently sonicated at 4 W for 15 s and matched to a McFarland no. 1 standard suspension. J774A.1 macrophages were infected at various multiplicities of infection (MOI) ranging from 1 to 10 during phagocytosis. After 4 h of phagocytosis, macrophages were washed to remove nonphagocytosed bacteria and stained with fluorescent dyes to determine the number of bacteria per macrophage. Triplicate wells containing macrophages (106 macrophages per well) were examined for each MOI in three independent experiments to determine the optimal uptake. Strains MSWt, MSΔmsrA, and MSΔmsrA/c (a mutant strain complemented for msrA) were taken up equally well by macrophages, and there was no significant difference in their abilities to phagocytose into macrophages. An MOI of 1 was chosen for all further experiments since most macrophages contained 1 CFU and this value correlated with the natural mode of infection of macrophages. For viability experiments, monolayer cultures of J774A.1 cells were established in 24-well plates and then were infected with bacteria. After nonphagocytosed bacteria were washed with warm Dulbecco modified Eagle medium, 1 ml of fresh medium was added to each well, and the plates were incubated at 37°C in the presence of 5% CO2. Macrophages in quadruplicate wells for each strain for each time point were harvested at zero time and at 1 and 3 days and lysed with 0.05% sodium dodecyl sulfate (SDS) for 15 min at room temperature. The lysed suspensions were diluted 10-fold, and 100 μl of each dilution was plated in triplicate on 7H10-TW-ADC plates. The plates were incubated at 37°C for 3 days, and colonies were counted. The viability assays were repeated three times.
Immunofluorescent localization of phagosomal oxidative markers.
For this study, we used sonicated and well-dispersed green fluorescent protein-expressing msrA mutant (MSΔmsrA-gfp) and wild-type (MSWt-gfp) M. smegmatis strains. These strains were produced by transforming the msrA mutant (MSΔmsrA) and wild-type (MSWt) M. smegmatis strains with plasmid pMTSIGHGFP3. Macrophages were infected (MOI, 1) in eight-well slide chambers for 4 h at 37°C in the presence of 5% CO2, washed, and incubated in fresh medium for different times up to 72 h. At 24-h intervals, cells were washed with warm phosphate-buffered saline once, fixed in ice-cold methanol for 15 s, and then fixed for 30 min in 2.7% paraformaldehyde. Cells were permeabilized for 30 min with a staining buffer containing 1% saponin, 0.1% glycine, and 5% heat-inactivated normal mouse or goat antiserum (depending upon the primary antibody) in phosphate-buffered saline, and then they were washed and incubated at 4°C for 18 h with 1/500 to 1/1,000 dilutions of rabbit anti-mouse inducible nitric oxide synthase (iNOS), antibody to nitrotyrosine (NT), and antibody to mouse p67phox (Santa Cruz Biotechnology, Santa Cruz, Calif.). A monoclonal anti-iNOS antibody (Sigma Chemical Co.) was also used. Macrophages were washed four times with the staining buffer and then counterstained with Texas red conjugated with goat anti-rabbit or goat immunoglobulin G (Jackson Immunochemicals, West Grove, Pa.) at room temperature for 2 h. After further washing, cells were mounted in elvanol mountant and examined with a Nikon fluorescence microscope. Green fluorescent and Texas red images were acquired, and the images were merged by using the metaview software to examine colocalization of stains. A positive control consisting of gamma interferon (IFN-γ)-activated and uninfected macrophages and a negative control consisting of untreated and uninfected macrophages were stained with isotype controls, followed by conjugates. At least 100 microscopic fields were scored for each macrophage preparation. Usually, each microscopic field contained one to five macrophages containing fluorescent organisms. Three independent experiments for each time point were performed, and the results are presented below as mean percentages. Colocalization was confirmed by using a deltavision laser scanning microscope.
Immunoblot analysis of phagosomal maturation and trafficking markers.
J774A.1 cells, grown in 75-ml flasks, were infected with M. smegmatis strains or latex bead controls at an MOI of 1, the cells were mixed for 4 h to enable uniform infection, and monolayers were then washed and incubated for another 72 h. Phagosomes were purified by sucrose gradient centrifugation as described previously (9, 46). Phagosome pellets were separated by SDS—10% polyacrylamide gel electrophoresis (PAGE) and transferred to Immobilon membranes. Phagosomal contamination with endoplasmic or Golgi membranes was monitored by staining for the Golgi-derived grp28 protein or calnexin (Santa Cruz Biotechnology) and was found to be less than 2%. The membranes were then probed with antibodies to free radical markers like p67phox and iNOS. The membranes were also probed with Rab5b antibodies to determine the maturation of phagosomes. The blots were developed with an ECL kit (Amersham, Piscataway, N.J.).
SDS-PAGE and immunoblotting.
SDS-PAGE and immunoblotting for M. smegmatis proteins were performed as described previously (1). Protein extracts were separated by SDS—12% PAGE. Protein concentrations in extracts were determined by using the bicinchoninic acid reagent (38), and 75 μg of protein was loaded in each lane. Separated proteins were transferred to nitrocellulose membranes and probed with the appropriate antiserum. The blots with anti-MsrA antiserum were developed with the 5-bromo-4-chloro-3-indolylphosphosphate (BCIP)—nitroblue tetrazolium substrate.
Survival under oxidative stress conditions.
Survival of M. smegmatis strains after exposure to oxidative stress was examined as follows. M. smegmatis strains grown in 7H9-TW-ADC were diluted to obtain an optical density at 595 nm of 0.300 in the same medium. Aliquots (1 ml) of each diluted culture were exposed to different oxidants (H2O2, t-butyl hydroperoxide, cumene hydroperoxide, and methyl viologen) for 1 h at 37°C. Control cultures were treated similarly but without oxidants. For experiments with the nitric oxide (NO) donors GSNO and sodium papanonoate, the pH of 7H9-TW-ADC was kept at pH 5.0. Cultures were serially diluted to 10-fold, and 100-μl portions of the diluted cultures were plated on 7H10-TW-ADC plates. Colonies were counted after 3 days of incubation at 37°C, and the results are reported below as log10 values.
RESULTS
msrA gene and its organization in the genome of M. smegmatis.
The sequence encoding MsrA of M. smegmatis and its flanking regions was obtained from The Institute for Genome Research database for the M. smegmatis genome sequence. Although in some species of bacteria multiple genes code for MsrA (36), the M. smegmatis genome has only one gene encoding MsrA. The amino acid sequence of M. smegmatis MsrA (170 amino acids) exhibited 80 and 77% identity with the sequences of MsrAs of M. tuberculosis (182 amino acids) and M. leprae (176 amino acids), respectively. It also showed significant identity with several prokaryotic and eukaryotic MsrAs. Furthermore, MsrA of M. smegmatis also possesses the critical N-terminal GCFWG residues at positions 12 to 16 which are a prerequisite for MsrA activity (20). Analysis of the flanking regions (4 kb) of M. smegmatis msrA revealed that the organization is more or less similar to that of msrA (Rv0137c) of M. tuberculosis. The flanking regions of M. smegmatis msrA have homologues for the Rv0136, Rv0138, and Rv0139 genes of M. tuberculosis, which are located in the flanking regions of M. tuberculosis msrA (Fig. 1A). We designated these genes Ms0136, Ms0138, and Ms0139, respectively. However, the intergenic region between msrA and Ms0138 in the M. smegmatis genome is longer (511 bp) than the intergenic region between msrA and Rv0138 in the M. tuberculosis genome (69 bp). In fact, this region of M. smegmatis has a putative open reading frame with a coding capacity of 130 amino acids. However, the function of the protein encoded by this open reading frame is not known, although it has some identity with the C-terminal region of a putative Streptomyces coelicolor large membrane protein (SC6G10.33).
Disruption of M. smegmatis msrA.
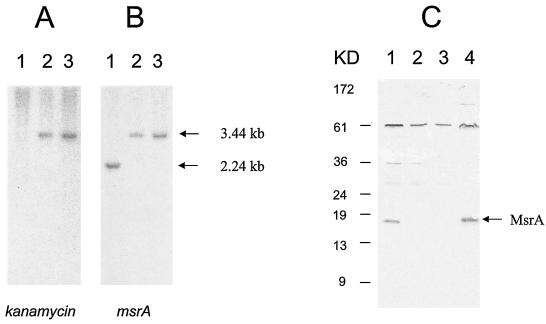
To examine the role of MsrA in intracellular survival, we first disrupted the msrA gene in M. smegmatis through homologous recombination. We electroporated plasmid pMSMSRA5 into wild-type M. smegmatis and plated the organisms onto 7H10-TW-ADC plates containing 25 μg of kanamycin per ml. Several electroporations resulted in approximately 200 kanamycin-resistant colonies. Genomic DNA isolated from these colonies was cut with KpnI and probed in Southern blots with a 1.2-kb kanamycin resistance gene (aph), a 2.2-kb M. smegmatis msrA gene, and 2. 7-kb plasmid pUC18. We predicted that an msrA mutant resulting from a double-crossover event should produce a single signal band (3.4 kb) with the msrA and aph probes and no signal with the pUC18 probe. Transformants MS97 and MS105 exhibited the expected results (Fig. 2A and B); hence, the msrA gene in these transformants was considered disrupted. To confirm the disruption of msrA expression in strains MS97 and MS105, we probed the protein extracts of these strains by Western blotting by using anti-MsrA antiserum produced against the overexpressed and purified MsrA protein of M. tuberculosis (41). As expected, an msrA mutant strain of M. smegmatis (MSΔmsrA) produced no band for MsrA, although wild-type M. smegmatis and M. tuberculosis produced strong bands (Fig. 2C). This indicated that MsrA expression was completely disrupted in the MSΔmsrA strain. In addition to the cross-reactivity with the MsrA protein, the anti-MsrA antiserum showed strong cross-reactivity with some other proteins of both M. tuberculosis and M. smegmatis, which is a common problem for many polyclonal antisera. However, the cross-reactivity did not prevent use of this antiserum against MsrA in immunoblotting since the MsrA band was very distinct (Fig. 2C). The MSWt and MSΔmsrA strains showed similar in vitro growth patterns (data not shown).
FIG. 2.
Southern and Western analysis of M. smegmatis strains. (A and B) Genomic DNA of M. smegmatis strains probed with the 1.2-kb kanamycin resistance gene (A) and the 2.2-kb M. smegmatis msrA gene (B). The results of a Southern analysis with the pUC18 probe are not shown as there were no signals in any of the lanes. Lane 1, wild-type M. smegmatis strain; lane 2, M. smegmatis msrA mutant 97; lane 3, M. smegmatis msrA mutant 105. The arrows on the right indicate the positions of the DNA fragments. (C) Immunoblot showing the reactivity of mycobacterial proteins with anti-MsrA antiserum. Lane 1, M. smegmatis wild-type strain; lane 2, M. smegmatis msrA mutant 97; lane 3, M. smegmatis msrA mutant 105; lane 4, M. tuberculosis H37RV (wild-type strain). The arrow indicates the position of the MsrA protein.
Intracellular survival of the msrA mutant.
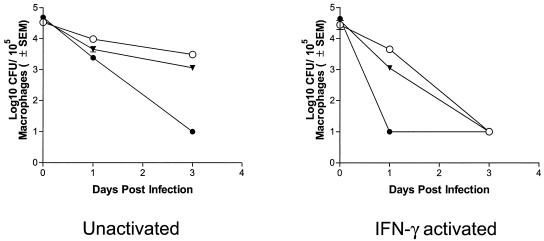
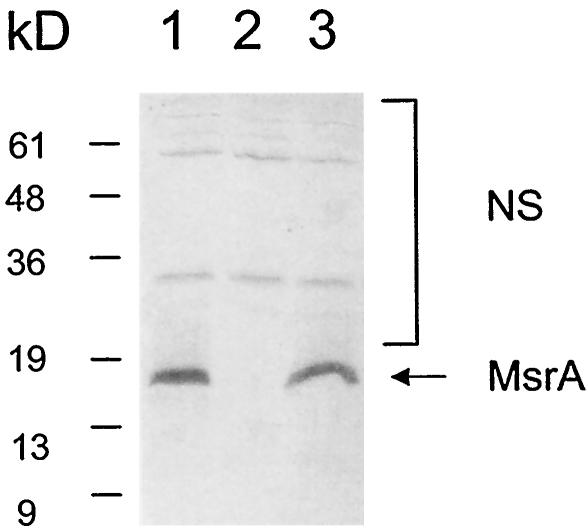
We infected unactivated (naïve) and IFN-γ-activated macrophage cell line J774A.1 with the MSWt and MSΔmsrA (MS97) strains of M. smegmatis and monitored the survival of the strains within the macrophages for 3 days (Fig. 3). Naïve and activated macrophages were deliberately used because phagosome maturation can occur independent of activation, although oxidative killing is regulated normally by IFN-γ in macrophages and IFN-γ is known to enhance certain stages of phagosome maturation. In the unactivated macrophages the survival of the MSΔmsrA strain decreased slightly 1 day postinfection, and this strain was almost completely nonviable by 3 days postinfection. In contrast, the survival of the MSWt strain was only moderately decreased even on day 3 postinfection. In the activated macrophages, both the MSΔmsrA and MSWt strains showed different profiles. While the survival of the MSWt strain was only slightly decreased on day 1 postinfection, the MSΔmsrA strain exhibited no survival on day 1 postinfection, which is similar to the survival profiles in naïve cells on day 3 postinfection. Furthermore, by 3 days postinfection, the MSWt strain was no longer viable in the activated macrophages. These results indicated that MsrA protected against killing in macrophages. To determine whether the relatively lower level of survival of the MSΔmsrA strain in macrophages was really due to the lack of MsrA, we performed complementation experiments. We transformed the MSΔmsrA strain (MS97) with plasmid pMSMSRA6, which contained the msrA gene of M. smegmatis in the integration vector pHG306, and the resulting strain (MSΔmsrA/c) was examined for MsrA expression in immunoblots (Fig. 4) and was used to infect unactivated and IFN-γ-activated macrophages. As Fig. 3 shows, the survival rate of this strain was more or less similar to that of wild-type strain, confirming that the lower level of survival of the MSΔmsrA strain inside macrophages was due to the lack of MsrA. Although deletion of the msrA gene affects the adherence patterns in certain bacterial species (51), microscopic analysis revealed that the binding of the MSΔmsrA and MSΔmsrA/c strains to macrophages and the subsequent phagocytosis by macrophages were similar to the binding and phagocytosis of the MSWt strain. This may have been due to the availability of multiple receptors for entry into macrophages. Consequently, the reduced survival the MSΔmsrA strain was not due to defects in its entry into macrophages.
FIG. 3.
Survival of M. smegmatis wild-type strain MSWt, msrA mutant 97 (= MSΔmsrA), and a complemented msrA mutant (MSΔmsrA/c) in murine macrophage-like J774A.1 cells. Naïve or IFN-γ-stimulated macrophages were infected at an MOI of 1 for 4 h, washed, lysed at intervals, and plated onto Middlebrook 7H10 agar plates to determine the number of CFU. ○, strain MSWt; •, mutant MSΔmsrA; ▾, complemented strain MSΔmsrA/c. There are no error bars for some data points because of the small standard errors.
FIG. 4.
Immunoblot showing expression of MsrA in an msrA mutant complemented with an integration plasmid-borne msrA gene. Lane 1, M. smegmatis wild-type strain MSWt; lane 2, M. smegmatis msrA mutant MSΔmsrA; lane 3, M. smegmatis msrA mutant complemented with the integration plasmid-borne msrA gene (MSΔmsrA/c). The arrow indicates the position of the MsrA protein. NS, nonspecific bands.
Immunofluorescence analysis of phagosomal oxidative stress markers.
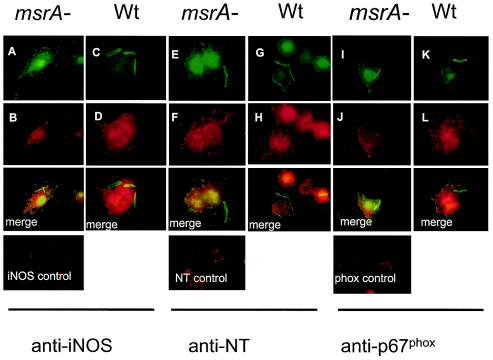
Since the level of intracellular survival of the MSΔmsrA strain of M. smegmatis was low, we hypothesized that this mutant was more susceptible to oxidative killing by macrophages than the MSWt strain was. Macrophages generate oxidative radicals, like superoxide ((O2−˙) and nitric oxide (NO ˙ −), by using the phox complex (NADPH oxidase) of proteins and iNOS, respectively (35). O2−˙ in turn combines with NO ˙ − to form the bactericidal radical peroxynitrite (ONOO ˙ −). Peroxynitrite, an effective agent for Met oxidation in proteins, also reacts with the tyrosine residue of proteins and forms NT. Recently, colocalization of phagosomes with iNOS, p67phox, and NT stains (45) has been used to determine the role of the oxidative mechanisms of macrophages in killing bacteria. We therefore infected J774A.1 macrophages with the MSWt and MSΔmsrA strains and labeled the phagosomes for iNOS, NT, and p67phox (Fig. 5). After 24 h of infection phagosomes containing the MSΔmsrA strain exhibited 76, 72, and 89% staining for iNOS, NT, and p67phox, respectively. In contrast, phagosomes containing the MSWt strain showed only 11, 14, and 11% staining for iNOS, NT, and p67phox, respectively, at the same time. The percentages of staining for iNOS and NT were greater in both the MSΔmsrA strain (100% for iNOS and 100% for NT) and the MSWt strain (90% for iNOS and 78% for NT) containing phagosomes after 72 h of infection. However, the staining for p67phox was greater in the phagosomes containing the MSΔmsrA strain (100%) and not in the phagosomes containing the MSWt strain (20%) (Table 1).
FIG. 5.
Colocalization of iNOS, NT, and phagocyte oxidase protein (p67phox) with the strain MSWt and MSΔmsrA phagosomes. J774A.1 macrophages were infected with gfp-expressing strains, washed, incubated for 24 h, fixed, and stained. (A and B) Strain MSΔmsrA (msrA−) in macrophages stained with iNOS-specific antibodies; (E and F) strain MSΔmsrA in macrophages stained with NT-specific antibodies; (I and J) strain MSΔmsrA in macrophages stained with p67phox-specific antibodies; (C and D) strain MSWt (Wt) in macrophages stained with iNOS-specific antibodies; (G and H) strain MSWt in macrophages stained with NT-specific antibodies; (K and L) strain MSWt in macrophages stained with p67phox-specific antibodies. Fluorescein isothiocyanate and Texas red images are followed by merged images that showed colocalization of the mutant phagosomes with free radical-specific markers. The controls panels show naïve macrophages stained with primary antibody, followed by conjugates.
TABLE 1.
Immunofluorescence colocalization of iNOS, NT, and phagocyte oxidase (p67phox) with M. smegmatis phagosomes in murine macrophagesa
| Straina | Colocalization (%)
|
|||||
|---|---|---|---|---|---|---|
| iNOS
|
NT
|
p67phox
|
||||
| 24 h | 72 h | 24 h | 72 h | 24 h | 72 h | |
| MSWt-gfp | 11 ± 4 | 90 ± 5 | 14 ± 4 | 78 ± 6 | 11 ± 4 | 20 ± 4 |
| MSΔmsrA-gfp | 76 ± 3b | 100 | 72 ± 7b | 100 | 89 ± 6b | 100 |
J774A.1 macrophages were phagocytosed with green fluorescent protein-expressing strains MSWt (MSWt-gfp) and MSΔmsrA (MSΔmsrA-gfp) for 4 h, washed, incubated for 24 to 72 h, fixed, stained with primary antibodies against iNOS, NT, and p67phox, and counterstained with Texas red conjugates. Colocalization of iNOS, NT, and the p67phox component of NADPH oxidase with M. smegmatis wild-type strain MSWt and msrA mutant strain MSΔmsrA in macrophages is expressed as a mean percentage ± standard error for three independent experiments. Colocalization was confirmed by laser confocal microscopy. At least 100 microscopic fields were scored for each quadruplicate mouse macrophage preparation per experiment. Each microscopic field contained 5 to 10 macrophages, and at least 8 macrophages contained at least one fluorescent organism.
P < 0.05 as determined by a t test.
Immunoblot analysis of free radical markers of phagosomes.
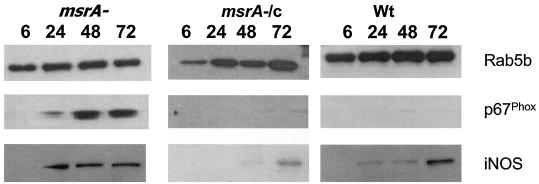
To validate the confocal findings, purified phagosomes were also probed with anti-iNOS and anti-p67phox antibodies in immunoblots (Fig. 6), and the results largely confirmed the results of the phagosome colocalization experiments. Phagosomes with the MSΔmsrA strain indeed acquired iNOS and p67phox markers earlier than phagosomes with the MSWt strain acquired these markers. The fact that phagosomes containing the MSWt strain acquired less p67phox even after 72 h of infection was also evident. The MSΔmsrA/c strain also behaved like the MSWt strain with respect to p67phox and iNOS acquisition. These results together suggest that the MSWt strain has some components that naturally slow down the acquisition of phox components and iNOS by the phagosomal membrane. Nevertheless, the presence of the Rab5b phagosmal maturation marker indicated that initiation of phagosomal maturation started even before 6 h postinfection. It should be mentioned that assembly of phox and assembly of iNOS in the phagosomes occur independent of each other and probably independent of the phagosome maturation process.
FIG. 6.
Immunoblot analysis of M. smegmatis phagosomes with anti-p67phox and anti-iNOS. Unactivated J774A.1 mouse macrophages were infected with MSΔmsrA (msrA−), MSΔmsrA/c (msrA−/c), and MSWt (Wt), and phagosomes were purified on sucrose gradients at different times (6, 24, 48, and 72 h) after infection. Rab5b, p67phox, and iNOS indicate reactivity of phagosomal proteins with the antibodies in the immunoblots.
Sensitivity of msrA mutants to oxidants.
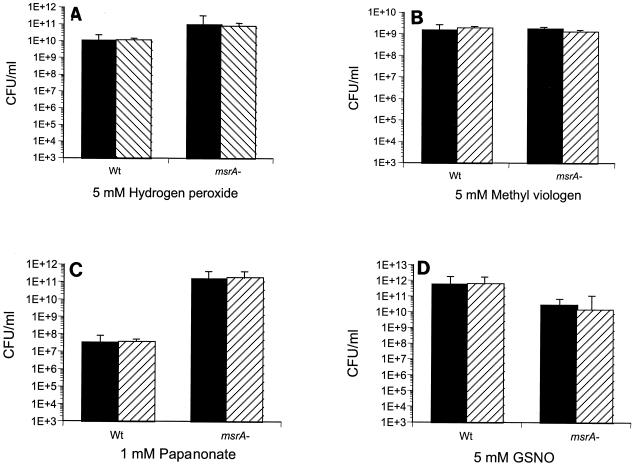
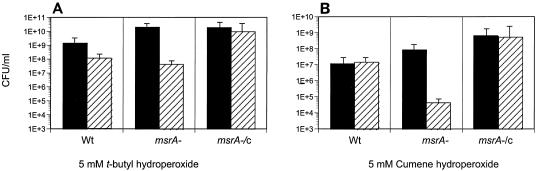
Previous studies indicated that in both eukaryotes and prokaryotes, the absence of MsrA leads to hypersensitivity to oxidative stress (8, 13, 29, 37). To determine if MsrA of mycobacteria plays a similar role, we tested the MSΔmsrA strain with different oxidative stresses. The MSWt and MSΔmsrA strains exhibited similar sensitivities to H2O2, the superoxide generator methyl viologen, and the nitric oxide generators sodium nitrite, sodium papanonoate, and GSNO (Fig. 7). Neither an increase in the concentration of these chemicals nor prolongation of the incubation period (data not shown) had any additional effect. However, the MSΔmsrA strain was more sensitive to cumene hydroperoxide and t-butyl hydroperoxide (Fig. 8), and complementation of the mutant strain (strain MSΔmsrA/c) with msrA of M. smegmatis with the integration plasmid pMSMSRA6 completely reversed this sensitivity.
FIG. 7.
Survival of M. smegmatis after exposure to oxidants. Wt, M. smegmatis wild-type strain MSWt; msrA−, M. smegmatis msrA mutant MSΔmsrA. Solid bars, untreated cultures; striped bars, treated cultures. Aliquots (1 ml) of cultures were exposed to hydrogen peroxide (A), methyl viologen (B), papanonate (C), and GSNO (D) for 1 h at 37°C. The cultures were serially diluted and plated on 7H10-TW-ADC plates.
FIG. 8.
Survival of M. smegmatis after exposure to hydroperoxides. Wt, M. smegmatis wild-type strain MSWt; msrA−, M. smegmatis mutant MSΔmsrA; msrA−/c, M. smegmatis complemented msrA mutant MSΔmsrA/c. Solid bars, untreated cultures; striped bars, treated cultures. Aliquots (1 ml) of cultures were exposed to t-butyl hydroperoxide (A) and cumene hydroperoxide (B) for 1 h at 37°C. The cultures were serially diluted and plated on 7H10-TW-ADC plates.
DISCUSSION
Macrophages and other mononuclear phagocytes are host defense cells that are capable of generating ROI and reactive nitrogen species (RNI) in addition to a variety of hydrolytic enzymes and antimicrobial peptides known as defensins (15, 32). Intracellular pathogens, including M. tuberculosis, are able to resist these responses of phagocytes. In order to understand the molecular mechanism of bacterial resistance to antimicrobial products, antioxidants of bacteria, such as superoxide dismutase (SOD), catalase-peroxidase (KatG), and alkyl hydroperoxide reductase (AhpC), have been investigated intensively. The investigations have been based on the premise that bacterial antioxidants can detoxify the ROI and RNI generated by phagocytes and thereby help the cells resist killing by macrophages. Although the precise role of bacterial antioxidants in the detoxification of phagocyte-derived ROI or RNI is not fully understood yet, the absence of some antioxidants had an effect on the intracellular and/or in vivo survival of bacteria. For example, a Cu,Zn-SOD mutant strain of Salmonella enterica serovar Typhimurium exhibited reduced survival in macrophages and attenuated virulence in mice (7). Likewise, a Cu,Zn-SOD mutant of the M. tuberculosis Erdman strain exhibited decreased survival in activated macrophages (33). A catalase-peroxidase (KatG) mutant of M. tuberculosis has also been implicated in intracellular survival and in survival and persistence in guinea pigs (14, 19, 22). Paradoxically, however, an ahpC mutant of M. tuberculosis showed no defect in intracellular or in vivo survival (39), suggesting that not all antioxidants play similar roles in promoting intracellular or in vivo survival. MsrA is probably different from the antioxidants mentioned above due to its unique specificity for oxidized methionine sulfoxide in proteins, and this study indicates that its role may be vital during intracellular survival. Disruption of the gene encoding MsrA of M. smegmatis led to a defect in survival of this organism inside macrophages, regardless of whether the macrophages were activated or naïve. This is consistent with previous reports which suggested the importance of MsrA in in vivo survival of M. genitalium and E. chrysanthemi (8, 13).
Within macrophages, oxidative damage due to free radicals is dependent upon two independent pathways, phagocyte oxidase (phox complex) and the iNOS enzyme, which produce O ˙ − and NO ˙ −, respectively. The phox complex includes membrane-bound gp91phox and p22phox subunits, which together form the flavocytochrome b and cytosolic p40phox, p47phox, p67phox, Rac1, and Rac2 subunits (47). The compartmentalization of phox proteins prevents production of superoxide in the resting cells. However, upon activation of macrophages, cytosolic p47phox, p67phox, and Rac1 or Rac2 move towards the membrane and assemble with gp91phox and p22phox to form an active complex, and this process is mediated by protein kinase C isoforms. Since there is an invagination of the plasma membrane during phagocytosis, the assembly of the phox complex results in superoxide secretion into the lumen of the phagosome (2, 12). While superoxide is freely diffusible, many of the downstream reactive products of the reactive oxygen species cascade, like the HO · − radicals, are not very stable; hence, localization of the phox complex becomes a critical event in targeting the microbicidal free radicals to the phagosomes (2, 12). Although localization of phox components with some bacterial phagosomes has been reported (45), this is the first report of localization of phox with mycobacterial phagosomes.
In contrast to the well-studied assembly of the phox complex, little is known about the mechanisms by which iNOS is targeted to phagosomes, although it has recently been found that IFN-γ induces the translocation of iNOS onto phagosomes containing either inert beads or mycobacteria through a Rab-dependent pathway (S. Daniel, G. Dai, C. Singh, D. Lindsey, S. Dhandayuthapani, R. Hunter, and C. Jagannath, submitted for publication). iNOS exists as a single 132- to 135-kDa polypeptide and has been reported to be present as a free or vesicle-bound form in the cytosol of IFN-γ-activated mouse macrophages (48). In this study, iNOS was present on MSΔmsrA phagosomes earlier than it was present on wild-type phagosomes. Since the association of NO · − with superoxide to form NT is well known, we suggest that the increased colocalization of phox and iNOS to form NT resulted in a rapid loss of viability of the MSΔmsrA mutant in macrophages compared to the loss of viability of the wild-type strain.
Enhanced colocalization of the phox subunit and iNOS with MSΔmsrA phagosomes led to speculation that wild-type M. smegmatis naturally possesses some molecules that either prevent or slow down the assembly of phox and iNOS in macrophages. Previously, S. enterica serovar Typhimurium pathogenicity island 2 (SPI2) proteins were found to affect the localization of phox and iNOS. Vazquez-Torres et al. (45) reported that salmonellae use SPI2 to exclude NADPH oxidase from Salmonella-containing phagosomes, and Chakravortty et al. (6) observed that SPI2 affects the colocalization of iNOS with Salmonella phagosomal vacuole. In both cases, it was suggested that the SPI2 products, which include host effector proteins (reviewed in reference 44), prevent localization of phox and iNOS to the phagosomes by interfering with the host cytoskeleton. By using the same line of logic, it is tempting to propose that MsrA somehow modulates the trafficking of phox and iNOS at least during the initial 24 h of infection in macrophages, although the mechanism remains unclear.
The disruption of msrA leading to hydroperoxide hypersensitivity in M. smegmatis is consistent with previous reports (8, 13, 29, 37) suggesting that MsrA plays a role in defending cells against oxidative stress (Fig. 8). An obvious difference between an msrA mutant of M. smegmatis and most other microbial msrA mutants is the lack of sensitivity to H2O2. In addition, the MSΔmsrA strain of M. smegmatis showed no sensitivity to NO donors. In contrast, St. John and his colleagues (41) have reported that an msrA mutant of E. coli is sensitive to nitrosative stress and that complementation of the mutant with the msrA gene of M. tuberculosis reverses the sensitivity. This difference in sensitivities between the msrA mutants of E. coli and M. smegmatis may be due to differences in the regulation of oxidative stress in these two species. However, the in vitro sensitivity of an organism to a specific oxidant does not determine its susceptibility to killing by macrophages, because the phagosomal environment is rich in metals, metalloenzymes, cofactors, and various ligands that produce even more toxic species from superoxide as part of the cascade (2, 12).
Overall, our results indicate that the absence of MsrA in M. smegmatis leads to increased sensitivity to certain oxidants in vitro and reduced intracellular survival, probably as a consequence of enhanced localization of phox and iNOS components to phagosomes. At this point, it is not clear whether MsrA plays a similar role in other intracellular bacteria, including M. tuberculosis. However, this possibility cannot be totally excluded, since MsrA is critical to the normal function of proteins in both bacteria and eukaryotic organisms. It is likely, therefore, that studies of MsrA in pathogenic intracellular bacteria, which have an inherent ability to prevent localization of phox and iNOS to the phagosomal membrane, will provide additional insights into the role of MsrA.
Acknowledgments
We thank C. Nathan, Cornell University, Ithaca, N.Y., for providing anti-M. tuberculosis MsrA antiserum and G. Zhong, University of Texas Health Science Center at San Antonio, for critically reading the manuscript.
S.D. is grateful to San Antonio Area Foundation, to the Japan Health Science Foundation, and to the Howard Hughes Medical Institute for grant support. Research in the laboratory of C.J. is supported by NIH/NIAID grant AI49534.
REFERENCES
- 1.Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl. 1989. Current protocols in molecular biology. Wiley, New York, N.Y.
- 2.Babior, B., J. Lamberth, and W. Nauseef. 2002. The neutrophil NADPH oxidase. Arch. Biochem. Biophys. 397:342-344. [DOI] [PubMed] [Google Scholar]
- 3.Barker, K., H. Fan, C. Carroll, G. Kaplan, J. Barker, W. Hellmann, and Z. A. Cohn. 1996. Nonadherent cultures of human monocytes kill Mycobacterium smegmatis, but adherent cultures do not. Infect. Immun. 64:428-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beckman, K. B., and B. N. Ames. 1998. Mitochondrial aging: open questions. Ann. N. Y. Acad. Sci. 854:118-127. [DOI] [PubMed] [Google Scholar]
- 5.Brot, N., L. Weissbach, J. Werth, and H. Weissbach. 1981. Enzymatic reduction of protein bound methionine sulfoxide. Proc. Natl. Acad. Sci. USA 78:2155-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakravortty, D., I. Hansen-Wester, and M. Hensel. 2002. Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J. Exp. Med. 195:1155-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Groote, M. A., U. A. Ochsner, M. U. Shiloh, C. Nathan, J. M. McCord, M. C. Dinauer, S. J. Libby, A. Vazquez-Torres, Y. Xu, and F. C. Fang. 1997. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc. Natl. Acad. Sci. USA 94:13997-14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhandayuthapani, S., M. W. Blaylock, C. M. Bebear, W. G. Rasmussen, and J. B. Baseman. 2001. Peptide methionine sulfoxide reductase (MsrA) is a virulence determinant in Mycoplasma genitalium. J. Bacteriol. 183:5645-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fratti, R. A., J. Chua, and V. Deretic. 2002. Cellubrevin alterations and Mycobacterium tuberculosis phagosome maturation arrest. J. Biol. Chem. 277:17320-17326. [DOI] [PubMed] [Google Scholar]
- 10.Garbe, T. R., J. Barathi, S. Barnini, Y. Zhang, C. Abou-Zeid, D. Tang, R. Mukherjee, and D. B. Young. 1994. Transformation of mycobacterial species using hygromycin resistance as selectable marker. Microbiology 140:133-138. [DOI] [PubMed] [Google Scholar]
- 11.Grimaud, R., B. Ezraty, J. K. Mitchell, D. Lafitte, C. Briand, P. J. Derrick, and F. Barras. 2001. Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J. Biol. Chem. 276:48915-48920. [DOI] [PubMed] [Google Scholar]
- 12.Hampton, M., A. Kettle, and C. Winterbourne. 1998. Inside the neutrophil phagosome: oxidants, myeloperoxidase, and bacterial killing. Blood 92:3007-3017. [PubMed] [Google Scholar]
- 13.Hassouni, M. E., J. P. Chambost, D. Expert, F. Van Gijsegem, and F. Barras. 1999. The minimal gene set member msrA, encoding peptide methionine sulfoxide reductase, is a virulence determinant of the plant pathogen Erwinia chrysanthemi. Proc. Natl. Acad. Sci. USA 96:887-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heym, B., E. Stavropoulos, N. Honore, P. Domenech, B. Saint-Joanis, T. M. Wilson, D. M. Collins, M. J. Colston, and S. T. Cole. 1997. Effects of overexpression of the alkyl hydroperoxide reductase AhpC on the virulence and isoniazid resistance of Mycobacterium tuberculosis. Infect. Immun. 65:1395-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ismail, N., J. P. Olano, H. M. Feng, and D. H. Walker. 2002. Current status of immune mechanisms of killing of intracellular microorganisms. FEMS. Microbiol. Lett. 207:111-120. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs, W. R., Jr., G. V. Kalpana, J. D. Cirillo, L. Pascopella, S. B. Snapper, R. A. Udani, W. Jones, R. G. Barletta, and B. R. Bloom. 1991. Genetic systems for mycobacteria. Methods Enzymol. 204:537-555. [DOI] [PubMed] [Google Scholar]
- 17.Kryukov, G. V., R. A. Kumar, A. Koc, Z. Sun, and V. N. Gladyshev. 2002. Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proc. Natl. Acad. Sci. USA 99:4245-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar, R. A., A. Koc, R. L. Cerny, and V. N. Gladyshev. 2002. Reaction mechanism, evolutionary analysis and role of zinc in Drosophila methionine-R-sulfoxide reductase. J. Biol. Chem. 277:37527-37535. [DOI] [PubMed] [Google Scholar]
- 19.Li, Z., C. Kelley, F. Collins, D. Rouse, and S. Morris. 1998. Expression of katG in Mycobacterium tuberculosis is associated with its growth and persistence in mice and guinea pigs. J. Infect. Dis. 177:1030-1035. [DOI] [PubMed] [Google Scholar]
- 20.Lowther, W. T., N. Brot, H. Weissbach, J. F. Honek, and B. W. Matthews. 2000. Thiol-disulfide exchange is involved in the catalytic mechanism of peptide methionine sulfoxide reductase. Proc. Natl. Acad. Sci. USA 97:6463-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowther, W. T., H. Weissbach, F. Etienne, N. Brot, and B. W. Matthews. 2002. The mirrored methionine sulfoxide reductases of Neisseria gonorrhoeae pilB. Nat. Struct. Biol. 9:348-352. [DOI] [PubMed] [Google Scholar]
- 22.Manca, C., S. Paul, I. C. Barry, V. Freedman, and G. Kaplan. 1999. Mycobacterium tuberculosis catalase and peroxidase activities and resistance to oxidative killing in human monocytes in vitro. Infect. Immun. 67:74-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, B. H., and T. M. Shinnick. 2000. Evaluation of Mycobacterium tuberculosis genes involved in resistance to killing by human macrophages. Infect. Immun. 68:387-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, B. H., and T. M. Shinnick. 2001. Identification of two Mycobacterium tuberculosis H37Rv ORFs involved in resistance to killing by human macrophages. BMC Microbiol. 1:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, R. A., and B. E. Britigan. 1997. Role of oxidants in microbial pathophysiology. Clin. Microbiol. Rev. 10:1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moskovitz, J., S. Bar-Noy, W. M. Williams, J. Requena, B. S. Berlett, and E. R. Stadtman. 2001. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc. Natl. Acad. Sci. USA 98:12920-12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moskovitz, J., B. S. Berlett, J. M. Poston, and E. R. Stadtman. 1997. The yeast peptide-methionine sulfoxide reductase functions as an antioxidant in vivo. Proc. Natl. Acad. Sci. USA 94:9585-9589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moskovitz, J., E. Flescher, B. S. Berlett, J. Azare, J. M. Poston, and E. R. Stadtman. 1998. Overexpression of peptide-methionine sulfoxide reductase in Saccharomyces cerevisiae and human T cells provides them with high resistance to oxidative stress. Proc. Natl. Acad. Sci. USA 95:14071-14075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moskovitz, J., M. A. Rahman, J. Strassman, S. O. Yancey, S. R. Kushner, N. Brot, and H. Weissbach. 1995. Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J. Bacteriol. 177:502-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moskovitz, J., V. K. Singh, J. Requena, B. J. Wilkinson, R. K. Jayaswal, and E. R. Stadtman. 2002. Purification and characterization of methionine sulfoxide reductases from mouse and Staphylococcus aureus and their substrate stereospecificity. Biochem. Biophys. Res. Commun. 290:62-65. [DOI] [PubMed] [Google Scholar]
- 31.Mundayoor, S., and T. M. Shinnick. 1994. Identification of genes involved in the resistance of mycobacteria to killing by macrophages. Ann. N. Y. Acad. Sci. 730:26-36. [DOI] [PubMed] [Google Scholar]
- 32.Nathan, C., and M. U. Shiloh. 2000. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. USA 97:8841-8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piddington, D. L., F. C. Fang, T. Laessig, A. M. Cooper, I. M. Orme, and N. A. Buchmeier. 2001. Cu,Zn superoxide dismutase of Mycobacterium tuberculosis contributes to survival in activated macrophages that are generating an oxidative burst. Infect. Immun. 69:4980-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruan, H., X. D. Tang, M. L. Chen, M. A. Joiner, G. Sun, N. Brot, H. Weissbach, S. H. Heinemann, L. Iverson, C. F. Wu, and T. Hoshi. 2002. High-quality life extension by the enzyme peptide methionine sulfoxide reductase. Proc. Natl. Acad. Sci. USA 99:2748-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiloh, M. U., and C. F. Nathan. 2000. Reactive nitrogen intermediates and the pathogenesis of Salmonella and mycobacteria. Curr. Opin. Microbiol. 3:35-42. [DOI] [PubMed] [Google Scholar]
- 36.Singh, V. K., and J. Moskovitz. 2003. Multiple methionine sulfoxide reductase genes in Staphylococcus aureus: expression of activity and roles in tolerance of oxidative stress. Microbiology 149:2739-2747. [DOI] [PubMed] [Google Scholar]
- 37.Singh, V. K., J. Moskovitz, B. J. Wilkinson, and R. K. Jayaswal. 2001. Molecular characterization of a chromosomal locus in Staphylococcus aureus that contributes to oxidative defence and is highly induced by the cell-wall-active antibiotic oxacillin. Microbiology 147:3037-3045. [DOI] [PubMed] [Google Scholar]
- 38.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem 150:76-85. [DOI] [PubMed]
- 39.Springer, B., S. Master, P. Sander, T. Zahrt, M. McFalone, J. Song, K. G. Papavinasasundaram, M. J. Colston, E. Boettger, and V. Deretic. 2001. Silencing of oxidative stress response in Mycobacterium tuberculosis: expression patterns of ahpC in virulent and avirulent strains and effect of ahpC inactivation. Infect. Immun. 69:5967-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stadtman, E. R., and R. L. Levine. 2000. Protein oxidation. Ann. N. Y. Acad. Sci. 899:191-208. [DOI] [PubMed] [Google Scholar]
- 41.St. John, G., N. Brot, J. Ruan, H. Erdjument-Bromage, P. Tempst, H. Weissbach, and C. Nathan. 2001. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc. Natl. Acad. Sci. USA 98:9901-9906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, et al. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 43.Van Soolingen, D., and P. Hermans. 1995. Epidemiology of tuberculosis by DNA fingerprinting. Eur. Respir. J. 20:649s-656s. [PubMed] [Google Scholar]
- 44.Vazquez-Torres, A., and F. C. Fang. 2001. Salmonella evasion of the NADPH phagocyte oxidase. Microbes Infect. 3:1313-1320. [DOI] [PubMed] [Google Scholar]
- 45.Vazquez-Torres, A., Y. Xu, J. Jones-Carson, D. W. Holden, S. M. Lucia, M. C. Dinauer, P. Mastroeni, and F. C. Fang. 2000. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science 287:1655-1658. [DOI] [PubMed] [Google Scholar]
- 46.Via, L. E., D. Deretic, R. J. Ulmer, N. S. Hibler, L. A. Huber, and V. Deretic. 1997. Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J. Biol. Chem. 272:13326-13331. [DOI] [PubMed] [Google Scholar]
- 47.Vignais, P. 2002. The superoxide-generating NADPH oxidase: structural aspects and activation mechanism. Cell. Mol. Life Sci. 59:1428-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vodovotz, Y., D. Russell, Q. W. Xie, C. Bogdan, and C. Nathan. 1995. Vesicle membrane association of nitric oxide synthase in primary mouse macrophages. J. Immunol. 154:2914-2925. [PubMed] [Google Scholar]
- 49.Wei, J., J. L. Dahl, J. W. Moulder, E. A. Roberts, P. O'Gaora, D. B. Young, and R. L. Friedman. 2000. Identification of a Mycobacterium tuberculosis gene that enhances mycobacterial survival in macrophages. J. Bacteriol. 182:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weissbach, H., F. Etienne, T. Hoshi, S. H. Heinemann, W. T. Lowther, B. Matthews, G. St. John, C. Nathan, and N. Brot. 2002. Peptide methionine sulfoxide reductase: structure, mechanism of action, and biological function. Arch. Biochem. Biophys. 397:172-178. [DOI] [PubMed] [Google Scholar]
- 51.Wizemann, T. M., J. Moskovitz, B. J. Pearce, D. Cundell, C. G. Arvidson, M. So, H. Weissbach, N. Brot, and H. R. Masure. 1996. Peptide methionine sulfoxide reductase contributes to the maintenance of adhesins in three major pathogens. Proc. Natl. Acad. Sci. USA 93:7985-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zahrt, T. C., and V. Deretic. 2002. Reactive nitrogen and oxygen intermediates and bacterial defenses: unusual adaptations in Mycobacterium tuberculosis. Antioxid. Redox Signal. 4:141-159. [DOI] [PubMed] [Google Scholar]