Abstract
Mutations in four of the ten human small heat shock proteins (sHSP) are associated with various forms of motor neuropathies and myopathies. In HspB1, HspB3, and HspB8 all known mutations cause motor neuropathies, whereas in HspB5 they cause myopathies. Several features are common to the majority of these mutations: (i) they are missense mutations, (ii) most associated disease phenotypes exhibit a dominant inheritance pattern and late disease onset, (iii) in the primary protein sequences, the sites of most mutations are located in the conserved α-crystallin domain and the variable C-terminal extensions, and (iv) most human mutation sites are highly conserved among the vertebrate orthologs and have been historically exposed to significant purifying selection. In contrast, a minor fraction of these mutations deviate from these rules: they are (i) frame shifting, nonsense, or elongation mutations, (ii) associated with recessive or early onset disease phenotypes, (iii) positioned in the N-terminal domain of the proteins, and (iv) less conserved among the vertebrates and were historically not subject to a strong selective pressure. In several vertebrate sHSPs (including primate sHSPs), homologous sites differ from the human sequence and occasionally even encode the same amino acid residues that cause the disease in humans. Apparently, a number of these mutations sites are not crucial for the protein function in single species or entire taxa, and single species even seem to have adopted mechanisms that compensate for potentially adverse effects of 'mutant-like' sHSPs. The disease-associated dominant sHSP missense mutations have a number of cellular consequences that are consistent with gain-of-function mechanisms of genetic dominance: dominant-negative effects, the formation of cytotoxic amyloid protein oligomers and precipitates, disruption of cytoskeletal networks, and increased downstream enzymatic activities. Future therapeutic concepts should aim for reducing these adverse effects of mutant sHSPs in patients. Indeed, initial experimental results are encouraging.
Keywords: small heat shock protein, mutation, motor neuropathy, myopathy, purifying selection, genetic dominance
1. Introduction
The human genome encodes ten small heat shock proteins (sHSP1), now systematically designated as HspB1 through HspB10 [1–3]. The defining feature of this protein superfamily is the presence of a characteristic stretch of ~85 amino acid residues, the so-called α-crystallin domain, regardless of whether the expression of these genes is regulated by heat or other stress factors [4, 5]. The α-crystallin domain typically is flanked 5' by a variable central region and a less conserved N-terminal domain, and 3' by a variable C-terminal extension (Fig. 1) [1]. Proteins without the α-crystallin domain do not qualify for membership in this protein family, even if overall sequence similarity with sHSPs is detectable [6]. In evolution, sHSPs have been recruited for quite diverse functions resulting in their involvement in many cellular processes including light refraction, apoptosis and growth control (carcinogenesis), protection of tissues and organs from stress, oxidative homeostasis, regulation of the organization of the cytoskeleton, muscle contraction and relaxation, chaperoning, proteolysis, neuron development, and others [5, 7–9]. In spite of this growing body of knowledge, the details of their cellular roles are not completely understood.
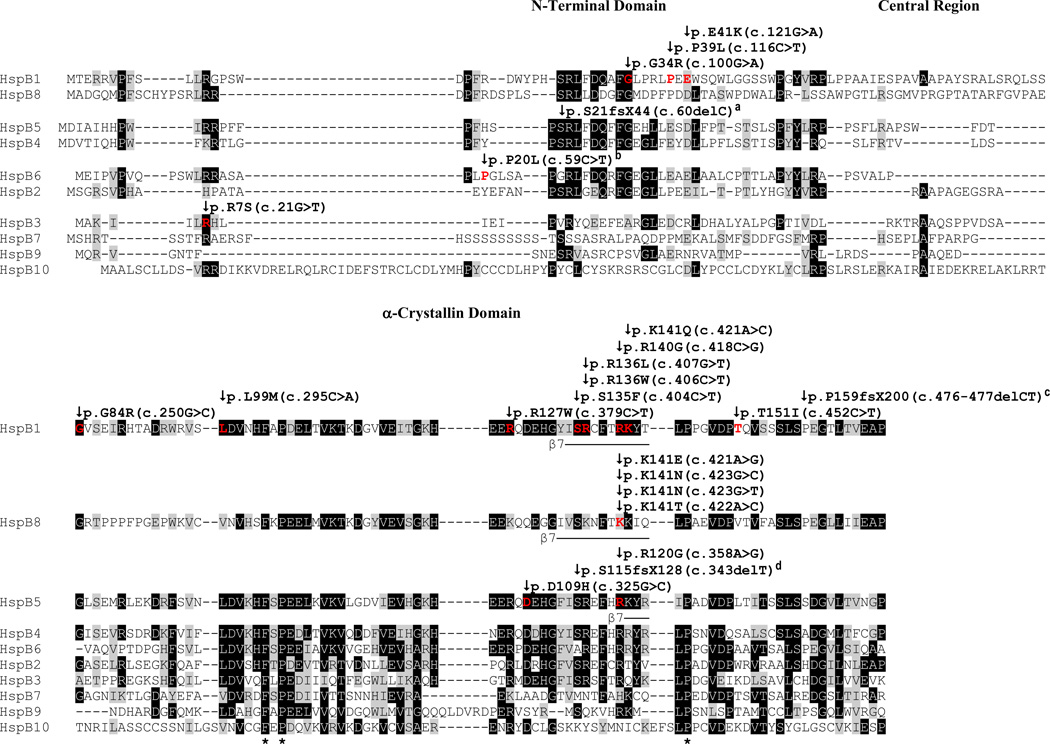
Figure 1. Positions of neuropathy- and myopathy-associated mutations in HspB1, HspB3, HspB5, HspB6, and HspB8 in the aligned ten human sHSPs.
The domain designation is based on the previously published alignment [1]. Identical amino acid residues are highlighted in black if they occur in at least five sequences. Similar amino acid residues (E/D; A/G; H/F/W/Y; S/T; I/L/V; H/R/K) are highlighted in gray if they occur in at least five sequences, or if they occur in at least one sequence in addition to identical amino acid residues in five sequences. The position of the β7-strand secondary structure in HspB1, HspB5, and HspB8 is indicated below these sequences [24]. Positions affected by missense mutations are in red. The asterisk denotes amino acid residues that are conserved in all ten human sHSPs.
Since the first identification of a myopathy-associated missense mutation in HspB5 more than a decade ago [10], the number of reported mutations in sHSPs (HspB1, HspB3, HspB5, HspB8) that affect the functions of muscles or motor neurons has grown to more than 30 (Table 1). Even though the precise molecular and cellular consequences of the sHSP mutations are incompletely understood, they demonstrate the crucial roles of this group of proteins in both muscles and motor neurons. This effect was not entirely unexpected, as data accumulated in the preceding decades on the importance of sHSPs for the physiology of both muscular and neuronal tissues [11–13]. Among the disease-causing mutations, the majority are missense mutations associated with a dominant disease phenotype. In addition to the sHSP mutations with clearly associated diseases, sequence variants with unclear disease association have been found, representing mutations with low penetrance or rare polymorphisms. For example, a variant of HspB6 is suspected to be associated with the impaired ability of the heart to cope with pathological stress [14].
Table 1.
Neuropathy and myopathy-associated mutations in HspB1, HspB3, HspB5, HspB6 and HspB8a
| sHSP | Mutation (protein) |
Mutation (cDNA) |
Type of Mutation |
Zygosity | Inheritance of the Phenotype |
Associated Disease | Onset Age of the Clinical Symptoms (Neuropathy and Myopathy only) |
Comment | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| HSPB1 | p.G34R | c.100G>A | missense | het | isolatedb (D) | dHMNc | >50y | – | 38 |
| p.P39L | c.116C>T | missense | het | D | dHMN2 [37], CMT2 [38]c | 54y [37], 61y [38] | – | 37, 38 | |
| p.E41K | c.121G>A | missense | het | D | dHMNc | <10y | – | 38 | |
| p.G84R | c.250G>C | missense | het | D | dHMN2c | Ø39y [37], 65y [43] | – | 37, 43 | |
| p.L99M | c.295C>A | missense | hom | (R) | dHMN2c | 37 y | – | 37 | |
| p.R127W | c.379C>T | missense | het | D | dHMN2, CMT2c | 18y [40], 35–60y [42] | – | 40, 42, 47 | |
| p.R127R | c.379C>A | synonymous | het | sporadicd | dHMN | – | association with disease unclear | 37 | |
| p.S135F | c.404C>T | missense | het | D | dHMN2, CMT2c | Ø21y [37], 15–25 y [40] | – | 37, 40 | |
| p.R136W | c.406C>T | missense | het | De | CMT2c | – | – | 40 | |
| p.R136L | c.407G>T | missense | het | isolatedb (D) | CMT2, dHMNc | 42y, 60y | – | 38 | |
| p.R140G | c.418C>G | missense | het | D | dHMN2c | Ø33y | – | 37 | |
| p.K141Q | c.421A>C | missense | het | D | dHMN2c | >40y, <60y | possible relationship to diabetic neuropathy | 44 | |
| p.T151I | c.452C>T | missense | het | D | dHMN2c | 25y | – | 40, 47 | |
| p.P159fsX200 | c.476–477delCT | frame shift | het | D | CMT2 [38], dHMN [60] | adulthood [38, 60], 3m [60] | mild form of neuropathy, but severe early onset secondary to diphtheria-tetanus-pertussis vaccination [60] | 38, 60 | |
| p.T180I | c.539C>T | missense | het | (D) | dHMN [38], CMT2L/dHMN [46]c | <18y [38], <10y [46] | – | 38, 46 | |
| p.P182L | c.545C>T | missense | het | parental mosaicism | dHMN2c | 5y | – | 40, 47 | |
| p.P182S | c.544C>T | missense | het | sporadicd (D) | dHMN2c | <10y | wild-type alleles in both parents | 45 | |
| p.R188W | c.562C>T | missense | het | isolatedb (D) | CMT2c | <10y | – | 38 | |
| HspB3 | p.R7S | c.21G>T | missense | het | D | hereditary axonal motor-predominant neuropathy | >20y | – | 48 |
| HspB5 | p.S21fsX44 | c.60delC | frame shift | hom | R | HIMD | shortly after birth | typically fatal within first year of life, respiratory distress | 61 |
| p.D109H | c.325G>C | missense | het | D | MM+C, possibly also CM | 35–45y, Ø40y | multisystemic disease, respiratory distress | 53 | |
| p.S115fsX128 | c.343delT | frame shift | hom | R | MM | 4m | respiratory distress | 62 | |
| p.R120G | c. 358A>G | missense | het | D | MM+CM+C | ~30y | multisystemic disease | 10, 54, 55 | |
| p.Q151X | c.451C>T | nonsense | het | (D) | MM | <40y | – | 63 | |
| p.G154S | c.460G>A | missense | het | (D) | DCM [56], MM [58] | 48y [56], | mild left ventricular dilation [56], mild distal vacuolar myopathy [58] | 56, 58 | |
| p.R157H | c.470G>A | missense | het | (D) | DCM | <70y [58] | – | 57 | |
| p.P155fsX163 | c.464–465delCT | frame shift | het | (D) | MM | <50y | respiratory distress | 63 | |
| p.X176WelX195 | c.527A>G | elongation | het | D | DCM+C | <30y | end stage heart failure | 64 | |
| HspB6 | p.P20L | c.59C>T | missense | het | – | potentially DCM | – | rare polymorphism or mutation with low or delayed penetrance, association with DCM not confirmed | 14 |
| HspB8 | p.K141E | c.421A>G | missense | het | D | dHMN2 | 15–25y | – | 47, 49 |
| p.K141N | c.423G>C | missense | het | D | dHMN2/CMT-M | 14–35y | – | 47, 49 | |
| p.K141N | c.423G>T | missense | het | D | CMT2L | 15–33y | severe symptoms in advanced age | 50 | |
| p.K141T | c.422A>C | missense | het | sporadicd (D) | CMT2L | 13y | 51 | ||
Abbreviations and symbols: Zygosity: het, heterozygous; hom, homozygous; Inheritance: D, dominant; R, recessive (suspected, although not observed, inheritance is given in parentheses); Disorders: C, cataract of the lens of the eye; CM, cardiomyopathy; CMT, Charcot-Marie-Tooth disease; DCM, dilated cardiomyopathy; dHMN, distal hereditary motor neuropathy; HIMD, hypertonic infantile muscular dystrophy; MM, myofibrillar or desmin-related myopathy; Onset age: m, months; y, years; Ø, average.
Positive family history referred, parents not examined.
Missense mutations in HspB1 were assessed to be associated either with CMT subtype 2L or dHMN subtype 2B, depending on the mutations [93]. However, the p.P39L, p.R127W, p.S135F, p.R136L, and p.T180I mutations may be associated with both forms of motor neuropathy.
Parents not affected.
Personal communication by Dr. J. Irobi, Antwerp, Belgium.
The evolutionary history of sHSPs seems to be different from that of other analyzed protein superfamilies in that the various domains and regions of the proteins have evolved independently [15]. sHSPs are found in all domains of life: archaea, bacteria, fungi, and other eukaryotes including metazoa and plants, suggesting their emergence early in evolution. All metazoan sHSPs seem to have evolved from the same single ancestral sequence [15]. In contrast, the shaping of the variable N- and C-terminal regions that border the α-crystallin domain on both sides took place many times in parallel throughout evolution, but independent of the evolution of the α-crystallin domain [15]. Whereas the reason for this peculiarity of sHSP evolution is unknown, this pattern may complicate the analysis of the evolutionary history of the mutation sites in human sHSPs (see Sections 2.3. and 3.1.3.5.). Another specific feature of the evolution of the sHSPs is the absence of recombination events with domains that are common to other protein families, typically resulting in multidomain proteins. This pattern suggests that the functional specification of the sHSPs was achieved by the variation of the terminal sequences without the concomitant diversification of the α-crystallin domain [15, 16]. In this light, it is remarkable that the known mutations in a given sHSP result in similar disease phenotypes, no matter which region of the molecule is affected by the mutation (Fig. 1; Table 1).
In this study, we sought to identify common and disparate features of the myopathy- and neuropathy-associated sHSP mutations, and to elucidate the evolutionary history of the mutation sites. On this basis, conclusions are drawn for future therapeutic approaches to the associated disorders.
2. Methods
2.1. Identification of vertebrate sHSPs with similarity to human HspB1, HspB3, HspB5, HspB6, and HspB8
Gnathostomata (Vertebrata: Gnathostomata) sHSP-like sequences with significant similarity to human HspB1, HspB3, HspB5, HspB6, and HspB8 were identified by a protein Blast (blastp) search at the National Center for Biotechnology Information database at http://blast.ncbi.nlm.nih.gov/Blast.cgi (default settings), using the α-crystallin domains of the human sHSPs as queries [1]. Additionally, the Ensembl databases (http://www.ensembl.org/index.html; http://projects.ensembl.org/neandertal) were queried for Gnathostomata sHSP-like sequences. sHSPs of taxa other than Gnathostomata were excluded. The full-length versions of the identified sequences were retrieved, scrutinized for redundancies and possible artifacts, and used for further analysis. Sequence fragments were included only if they contain at least one site that is homologous to one of the mutation sites in human sHSPs. Sequence variants within a species, whether resulting from within-population polymorphisms, from paralogous genes, or from alternate splicing, are indicated by footnotes, unless they are included in the alignments.
2.2. Multiple alignments of Gnathostomata sHSPs
Gnathostomata sHSPs (protein sequences) were aligned using the full-length sequences and Clustal W (default settings) at Pôle Bioinformatique Lyonnais (http://pbil.univ-lyon1.fr/) with minor manual editing. The various sHSP domains and regions (N-terminal domain, central region, α-crystallin domain, C-terminal extension) were designated as defined previously [1].
Some regions in the Gnathostomata sHSPs that encompass homologous missense mutation sites exhibit considerable variability. In spite of this variability, conserved sequence motifs in these flanking regions can be identified in most orthologs, grouping the amino acid residues into the categories Aliphatic (Al), Aromatic (Ar), Hydrophobic (Hy), Negative (Ne), Polar (Pl), Positive (Ps), Small (Sm), and Tiny (Ti) [17]. For such sites, the conserved motifs are given for the positions -3 to +3 where recognizable, with X indicating any amino acid residue.
Historically, amino acid residue substitutions at homologous positions in orthologs occurred with different frequencies. For example, a Leu/Ile exchange occurred more frequent than a Leu/Gly exchange, as is represented by the BLOSUM62 matrix, derived from a comparison of many homologous mammalian protein families [18]. This matrix was used to classify amino acid residue exchanges among the sHSP orthologs, e.g. as relatively 'rare' (values −4, −3), medium (values −2, −1, 0, +1) or 'frequent' (values +2, +3) compared to what is expected by chance.
2.3. Determination of orthology of the Gnathostomata sHSPs with human sHSPs
Phylogenetic analyses was performed to verify orthology of all Gnathostomata sHSPs included in this study with the corresponding human proteins. However, the reported independent evolution of the different domains and regions of the sHSPs [15] may complicate the determination of the orthology in the more distant Gnathostomata taxa. To address this issue, we conducted phylogenetic analyses using the isolated α-crystallin domains, and in some instances also using the isolated N-terminal domains, in addition to the full-length sequences. The domains of the sHSPs to be analyzed were determined by multiple alignments with human HspB1, HspB3, HspB5, HspB6 and HspB8 [cf. 1]. Each of the sHSPs included in this study were aligned with the 10 human sHSPs in the amino acid space, using the ClustalV and ClustalW algorithms [19] of the DNASTAR Lasergen 8 program package with default settings. Phylogenetic trees were inferred from amino acid p-distances using neighbor-joining, followed by a bootstrap analysis. For most Gnathostomata sHSPs, this approach resulted in consistent phylogenetic trees in which the sHSPs of interest formed well-supported clades with one of the human sHSPs, using the full-length sequences and/or the isolated domains. These trees were used for determining orthology of the Gnathostomata sHSPs. sHSPs with ambiguous phylogenetic placement or with orthology to human HspB2, HspB4, HspB7, HspB9, or HspB10 were excluded from this study.
For some sHSPs, in particular from the more distant taxa, the above procedure provided ambiguous results such that the full-length sHSPs and the isolated α-crystallin and/or N-terminal domains were more closely related to different human sHSPs. In particular, this pattern was found in a number of HspB6-like proteins (cf. Supplemental Information S10). Using the isolated α-crystallin domains of these sequences, the analyses suggested orthology with human HspB6, whereas the analyses of the corresponding full-length sequences, or of the isolated N-terminal domains, suggested orthology with other human sHSPs, or did not resolve well. Several sHSPs with this type of ambiguity were included in the multiple alignments, but excluded from the analysis of selection pressure on the mutation sites (cf. Section 2.4.). In the figures, these sequences are labeled by footnotes. sHSPs with 'atypical' sequence segments were treated in the same way.
2.4. Determination of the evolutionary pressure on the mutations sites
The strength and direction of pervasive natural selection (i.e. purifying versus positive) was assessed at the level of individual codon sites in each of the five sHSPs using a Fixed Effects Likelihood (FEL)[20] method in HYPHY [21], implemented in Datamonkey (www.datamonkey.org) [22]. The analysis was performed using the entire coding region, but we report ω = β/α (the ratio of non-synonymous to synonymous substitution rates - a commonly used measure of selection, expected to be less than one when purifying selection is predominant) only for the missense mutation sites. The detected purifying selection was classified 'significant' or 'marginally significant' if the p-values derived using the asymptotic chi-square test distribution for the likelihood ratio test (LRT) statistic are p<0.05 and 0.05<p<0.1, respectively (note that FEL is generally a conservative test for alignments of the sizes considered here). We also conducted a test for episodic positive selection (acting on only a subset of the lineages in the phylogenetic tree) at the missense mutation sites using MEME [23] but failed to detect evidence for this mode of evolution, possibly due to the lack of power resulting from relatively small numbers of analyzed sequences.
2.5. Species included in this analysis and their taxonomic ranking
Relevant taxonomic ranks are indicated for single organisms or groups according to the National Center Biotechnology Information web site at http://www.ncbi.nlm.nih.gov/taxonomy. This study was limited to sHSP sequences of animals of the taxon Gnathostomata (Vertebrata: Gnathostomata). Among them, all but one are grouped in the taxon Teleostomi (Gnathostomata: Teleostomi) which includes mammals, birds, reptiles, turtles, amphibians and fish, whereas Squalus acanthias (spiny dogfish) is classified in the taxon Chondrichthyes (Gnathostomata: Chondrichthyes). Mammals (Teleostomi: Euteleostomi: Sarcopterygii: Tetrapoda: Amniota: Mammalia) were further grouped into Primates (Mammalia: Theria: Eutheria: Euarchontoglires: Primates), Glires/Scandentia (Mammalia: Theria: Eutheria: Euarchontoglires: Glires/Scandentia), Laurasitheria (Mammalia: Theria: Eutheria: Laurasiatheria), Aftrotheria (Mammalia: Theria: Eutheria: Afrotheria), Xenarthra (Mammalia: Theria: Eutheria: Xenarthra), Metatheria (Mammalia: Theria: Metatheria), and Monotremata (Mammalia: Prototheria: Monotremata). Birds, reptiles and turtles were combined to Sauria/Testudines which includes Archosauria (Sauropsida: Sauria: Archosauria), Lepidosauria (Sauropsida: Sauria: Lepidosauria), and Testudines (Sauropsida: Testudines). All listed amphibians belong to the taxon Anura (Tetrapoda: Amphibia: Batrachia: Anura). The coelacanth (Latimeria chalumnae) is classified in the taxon Coelacanthidae (Teleostomi: Euteleostomi: Sarcopterygii: Coelacanthimorpha: Coelacanthiformes: Coelacanthidae), as opposed to most other fish which belong to the taxon Clupeocephala (Teleostomi: Euteleostomi: Actinopterygii: Actinopteri: Neopterygii: Teleostei: Elopocephala: Clupeocephala). They were further grouped into Holacanthopterygii (Clupeocephala: Euteleostei: Neognathi: Neoteleostei: Eurypterygii: Ctenosquamata: Acanthomorpha: Euacanthomorpha: Holacanthopterygii), Protacanthopterygii (Clupeocephala: Euteleostei: Protacanthopterygii), and Otophysi (Clupeocephala: Otocephala: Otophysi). The spiny dogfish (Squalus acanthias) is classified in the family Squalidae (Chondrichthyes: Elasmobranchii: Neoselachii: Squalea: Hypnosqualea: Squaliformes: Squaloidei: Squalidae). Lower taxonomic ranks are given if the species can be further grouped. If the available sequence of a taxon originates from just one species, the name of the family is given. The complete list of the species included in this study is given in the Supplemental Information S1.
3. Results and Discussion
3.1. Neuropathy- and myopathy-associated mutations in human sHSPs
3.1.1. Common and disparate characteristics
The 33 mutations and sequence variants of sHSPs listed in Table 1 do not share obvious characteristics or consequences that are common to all mutations. However, a few observations are noteworthy with respect to the type and positions of the mutations, the associated disorders, and the inheritance:
Types of mutations
The reported mutations, as listed in Table 1, include missense (26/33), frame shift (4/33), nonsense (1/33), elongation (1/33), and synonymous mutations (1/33). Among the missense mutations, positively charged (basic) arginine and lysine residues are replaced by other amino acid residues in 12/26 cases. Thus, loss of a positive charge seems to be a common feature in a major fraction of these mutations. However, three mutations (p.E41K, p.G84R in HspB1; p.D109H in HspB5) introduce a positive charge.
Positions of mutations
The distribution of the known mutations along the entire length of the sHSPs is uneven, considering either HspB1 alone (the sHSP which is affected by most of the known mutations) or all sHSPs as entity, with 17/33 mutations being sited in the conserved α-crystallin domain (Fig. 1). In HspB1, five mutations (p.S135F, p.R136W, p.R136L, p.R140G, p.K141Q) are clustered in the region of the predicted and conserved β7-strand which is thought to be crucial for the overall structure and function of this and other sHSPs [24]. Mutations in HspB8 (p.K141E, p.K141N, K141T) and HspB5 (p.R120G) are also positioned in the homologous regions of the predicted β7-strands. In fact, the affected positions Arg140/Lys141 in HspB1, Lys141 in HspB8, and Arg120 in HspB5, which have in common the elimination of positively charged amino acid residues, may represent a homologous 'hot spot' position, although this categorization may be premature because of the insufficient number of mutations in human sHSPs that have been reported to date [25].
While the clustering of mutations in the β7-strand region may indicate an important role of this structural element in the sHSP function, this notion should not be overstated since mutations in the adjacent sequence segments (p.R127W in HspB1, p.D109H in HspB5) that result in similar disease phenotypes are not positioned in a region with any recognizable secondary structure [cf. 24].
The N-terminal domains harbored in total only six of the reported mutations (three in HspB1, one in HspB3, HspB5 and HspB6 each). Different from the mutations in the α-crystallin domain, these mutations are not clustered in any recognizable secondary structure [cf. 24]. The relatively short C-terminal extensions harbored nine mutations, all in HspB1 and HspB5. A fraction of these mutations cluster in and around the T-I-P motif that is characteristic for both HspB1 and HspB5 (Fig. 1; cf. also the alignments in the Supplemental Information S5 and S9).
Although a fraction of the missense mutations affect positions that are well conserved among the ten human sHSPs, this notion cannot be generalized as other affected positions are only poorly conserved. For example, Pro39 and Thr151 in HspB1 have no homologous counterparts in six and seven of the other human sHSPs, respectively (Fig. 1).
Numerous studies have addressed the importance of the different domains and regions of the sHSPs for both their structure and function. The conserved α-crystallin domain forms mainly β-strands and seems to be primarily involved in the formation of stable sHSP dimers. These dimers are the building blocks for the larger sHSP complexes, and here the predicted β7-strand may be crucial [24, 26]. The less conserved N-terminal domains and C-terminal extensions, with the latter being flexible and intrinsically disordered, also play a role in the structure of the sHSPs, especially in their self-association into large complexes [27–29]. Differently oriented N-terminal domains interact with each other, or with the α-crystallin domains of the same or neighboring dimers [26]. Both the N-terminal domains and C-terminal extensions were reported to be crucial for the chaperone-like activity of the sHSPs and for binding of the target proteins [29–31]. For example, the ultimate and penultimate lysine residues of the C-terminal extension of HspB5 are required for both its chaperone-like activity [32] and its cardioprotective function [33]. However, these insights into the structural and functional roles of the different parts of the sHSP molecules do not explain the distribution pattern of the mutations along the sHSP sequences: For the associated disease phenotypes, it seems to be of little relevance if the mutations are sited in the β7-strand, other parts of the conserved α-crystallin domain, or in the variable sequence segments. For example, all missense mutations in HspB1 cause the similar disease phenotypes distal hereditary motor neuropathy (dHMN) or Charcot-Marie-Tooth disease (CMT), no matter which structural element of the protein is affected (Table 1).
Association with disorders (motor neuropathy versus myopathy)
A striking observation is that all mutations in HspB1 are associated with motor neuropathy, as opposed to the mutations in HspB5 which are associated with various forms of myopathy, no matter which domain or region of the sHSP sequences is affected or what type mutation is present (Table 1). This observation strongly suggests differential, tissue-specific functions of HspB1 and HspB5. Since for HspB8 and HspB3 only three and one missense mutations, respectively, in the coding amino acid sequences have been reported to date, it is premature to attempt a similar comparison. At least the close phylogenetic relatedness of HspB8 with HspB1 [34] renders it plausible that mutations in HspB8 are also associated with motor neuropathy. Whether HspB8 and HspB3, together with HspB1, eventually will form a 'motor neuropathy group' among the sHSPs remains to be determined as more mutations become identified. On the other hand, HspB6 is most closely related to HspB5, next to HspB4 [34]. The notion that the reported polymorphism in HspB6 is potentially associated with cardiomyopathy [14] suggests that HspB5 and HspB6 may form a 'myopathy group' among the sHSPs. The motor neuropathies associated with mutations in HspB1, HspB3, and HspB8 include distal hereditary motor neuropathy (dHMN) and Charcot-Marie-Tooth disease (CMT), whereas the myopathies associated with mutations in HspB5, and possibly with a polymorphism in HspB6, include various forms of cardiomyopathy (CM), notably desmin-related or myofibrillar myopathy (DRM, MM), dilated cardiomyopathy (DCM), and hypertonic infantile muscular dystrophy (HIMD), partially in combination with cataracts (C) in the lens of the eye (Table 1).
Inheritance
The majority of the missense mutations in sHSPs are associated with disease phenotypes that exhibit a dominant, or possibly semi-dominant, inheritance (Table 1). This prevalence of genetic dominance is noteworthy because wild-type alleles are typically dominant over mutant alleles (see Section 3.2.). In contrast, the frame shift mutations seem to be associated with disease phenotypes that exhibit a higher proportion of recessive inheritance (2/4), although a final conclusion can be drawn only after the identification of more mutations of this type. Most, if not all reported neuropathy- and myopathy-associated mutations in sHSPs exhibit a high degree of penetrance that may approach 100%, even if the disease onset for some mutations occurs late in life.
3.1.2. Polymorphisms
The HspB6 sequence variant p.P20L (Table 1) was found in a study of patients with dilated cardiomyopathy. However, the association with disease is not confirmed as unaffected individuals carry also the heterozygous allele [14]. Thus, p.P20L may represent a rare polymorphism, a mutation with low penetrance, or cause a disease with a late onset. Further examples of polymorphisms in sHSPs are p.S41Y and p.P51L of HspB5 [35], and a number of cardiomyopathy-associated sequence variants of the HSPB7 gene, which, however, do not affect the protein sequence [36].
Similarly, synonymous sequence variants were found for HspB1. Examples are the rare polymorphisms p.R127R (c.379C>A) [37] and p.S187S (c.561G>T) [38], and the frequently occurring substitution p.E3E (c.9G>A) [38, 39]. These synonymous polymorphisms or mutations may indicate that the sHSP mRNAs may also contribute to disease (see Section 3.1.5.).
3.1.3. Missense mutations in human sHSPs and their homologous sites in vertebrate orthologs
3.1.3.1. HspB1
3.1.3.1.1. N-terminal domain
Three missense mutations, p.G34R, p.P39L, and p.E41K, have been reported in the N-terminal domain of HspB1 (Table 1). These mutations were identified in screenings of several families affected with peripheral motor neuropathies. p.G34R and p.P39L caused diseases with a late onset age [37, 38], whereas p.E41K caused a disease within the first decade of life [38]. Family history and sequencing data suggest a dominant inheritance.
Comparison among the human sHSPs
The Gly34 site is conserved in five other human sHSPs: HspB2, HspB4, HspB5, HspB6, and HspB8 (Fig. 1). HspB3 has a biochemically similar alanine residue in homologous position, whereas HspB7, HspB9, and HspB10 have biochemically different amino acid residues. The Pro39 site is conserved in three other human sHSPs (HspB2, HspB7, HspB8), but not in the remaining sequences. The Glu41 site is conserved in seven other human sHSPs (HspB2, HspB4, HspB5, HspB6, HspB7, HspB8, HspB10), if the conservative Glu/Asp substitution is taken into account, leaving only HspB3 and HspB9 with major deviation from HspB1. Interestingly, HspB3 contains a positively charged arginine residue in the homologous position which is similar to the lysine residue in HspB1 that causes the disease. Thus, HspB1 and HspB3 may have different functions in motor neurons.
Comparison with the Teleostomi orthologs
The N-termini of the Sarcopterygii orthologs share a clear similarity with human HspB1 (Supplemental Information S2). Among these sequences, all three mutation sites are well conserved if the conservative Glu/Asp exchange at the positions homologous to Asp41 in a fraction of the Metatheria and all of the Achosauria and Anura orthologs is counted. At the position homologous to Pro39, a few informative exceptions occur: both primates of the taxon Strepsirrhini (G. garnettii, M. murinus) carry an alanine residue in an otherwise similar neighborhood, suggesting that Pro39 in human HspB1 may be dispensable. Of particular interest is HspB1 of T. belangeri that carries a homologous leucine, exactly as does the disease-associated mutant HspB1 in humans, in an otherwise conserved flanking region. Thus, this homologous leucine residue is compatible with life, at least in this Scandentia species, and has not resulted in an apparent evolutionary disadvantage. In this light it remains to be determined why H. sapiens does not tolerate a leucine residue in this position. Historically, such Pro/Leu exchanges were relatively rare events [18], rendering the T. belangeri HspB1 even more interesting. Two explanations for the existence of this leucine residue seem to be plausible: the maximum life span of ~11 years of T. belangeri, as compared to the onset age of 54 years in the reported human patient, may not be sufficient for the manifestation the motor neuropathy. Alternatively, T. belangeri may have adopted a suppressor mechanism, although the specifics are not known.
In C. jacchus with its three HspB1 variants, the deletion of homologous mutation sites in just one sequence, together with the flanking region, may have no consequence as the two other HspB1 forms can be expected to compensate for any functional loss.
Although the N-termini of the Clupeocephala orthologs of HspB1 deviate to a greater extent from human HspB1, the overall similarity still remains noticeable (Supplemental Information S2). The sites homologous to Pro39 and Glu41 are also affected by this variability, although the latter site to a lesser extent not counting the conservative Glu/Asp exchanges. Some of the orthologs retain proline residues in positions homologous to Pro39, although in most sequences the proline is replaced by various other amino acids, including leucine. None of the sequences contain a positively charged amino acid residue in positions homologous to Glu41, suggesting that such residues, as in the disease-causing E41K mutant, may not be tolerated in this position. In contrast to these variable mutations sites, Gly34 and its homologous sites are nearly invariable with only two deviating sequences (T. nigroviridis sequence CAG08169; O. mordax) which, however, exhibit ambiguous orthology.
The unequal degree of variability in this sequence segment is reflected by the minimal amino acid motif Q-Sm-F-G-X-Sm-X-Hy-X-Ne-X-X-X-X (mutation sites underlined) that is common to most Teleostomi sequences (exceptions: T. nigroviridis sequence CAG08169; O. mordax).
Fixed effects likelihood analyses of Gly34, Pro39, Glu41 and their homologous sites reveal that these sites were historically under significant purifying selection (p<0.001; Table 2), in spite of the observed variability in the positions homologous to Pro39 and Glu41. Thus, mutations at these positions in the past were generally associated with negative fitness consequences, although exceptions (e.g. species-level selective sweeps) occurred.
Table 2.
Selection profiles and evidence for purifying selection at human missense mutation sites
| Gene | Codon position |
Amino acids in humans |
α | β | ω | Purifying selection LRTa |
p-value |
|---|---|---|---|---|---|---|---|
| HSPB1 | 34 | G | 0.731 | 0 | 0 | 20.618 | <0.001 |
| 39 | P | 2.748 | 0.399 | 0.145 | 18.484 | <0.001 | |
| 41 | E | 2.732 | 0.179 | 0.065 | 15.130 | <0.001 | |
| 84 | G | 1.492 | 0 | 0 | 37.117 | <0.001 | |
| 99 | L | 0.419 | 0 | 0 | 12.535 | <0.001 | |
| 127 | R | 1.933 | 0.118 | 0.061 | 30.343 | <0.001 | |
| 135 | S | 0.955 | 0 | 0 | 24.648 | <0.001 | |
| 136 | R | 1.219 | 0 | 0 | 36.149 | <0.001 | |
| 140 | R | 1.941 | 0.062 | 0.032 | 36.856 | <0.001 | |
| 141 | K | 0.613 | 0 | 0 | 17.058 | <0.001 | |
| 151 | T | 2.395 | 0.237 | 0.099 | 20.976 | <0.001 | |
| 180 | T | 0.694 | 0.262 | 0.378 | 3.011 | 0.083 | |
| 182 | P | 1.855 | 0 | 0 | 47.288 | <0.001 | |
| 188 | R | 2.562 | 0.142 | 0.055 | 31.402 | <0.001 | |
| HSPB3 | 7 | R | 1.302 | 0.144 | 0.110 | 10.403 | 0.001 |
| HSPB5 | 109 | D | 1.620 | 0 | 0 | 22.963 | <0.001 |
| 120 | R | 2.829 | 0 | 0 | 37.155 | <0.001 | |
| 154 | G | 0.246 | 0.376 | 1.526 | 0.264 | 0.607 | |
| 157 | R | 1.172 | 0 | 0 | 17.843 | <0.001 | |
| HSPB6 | 20 | P | 4.148 | 0.183 | 0.044 | 18.039 | <0.001 |
| HSPB8 | 141 | K | 0.290 | 0 | 0 | 4.599 | 0.032 |
purifying selection likelihood ratio test
3.1.3.1.2. α-crystallin domain
To date, nine missense mutations (affecting eight mutation sites) in the α-crystallin domain of HspB1 have been identified by several groups searching for genetic causes of motor neuropathy (Table 1) [37, 38, 40–44]. Five of these mutations replace positively charged amino acid residues (p.R127W, p.R136W, p.R136L, p.R140G, p.K141Q), whereas one mutation inserts a positively charged residue (p.G84R). One mutation (p.L99M) is recessive as opposed to the other mutations which are dominant. The onset of the motor neuropathies in all patients was in the second decade, or later.
Comparison among the human sHSPs
Most of the affected sites are well conserved among the human sHSPs, albeit none completely (Fig. 1). Leu99 and Arg127 are best conserved with aliphatic and positive amino acid residues, respectively, in the homologous positions across all ten human sHSPs. In comparison, Thr151 is relatively poorly conserved, with HspB5 and HspB8 even carrying aliphatic amino acid residues (leucine and valine, respectively) in the homologous positions, as is found in the disease-associated T151L-HspB1. Arg140/Lys141 may represent a mutational 'hot spot' as the sites homologous to Arg140 in HspB5 and HspB8 are also affected by mutations.
Comparison with the Teleostomi orthologs
All Teleostomi orthologs share a high degree of similarity with human HspB1, in spite of the increasing divergence of the Clupeocephala sequences (the 5'- and 3'-sections of the α-crystallin domain are shown in the Supplemental Information S3 and S4, respectively). Seven of the eight missense mutation sites (Gly84, Leu99, Arg127, Ser135, Arg136, Arg140, Lys141) are perfectly or nearly perfectly conserved among the Teleostomi orthologs. Deviations result from conservative Arg/Lys substitutions in positions homologous to Arg127 (Archosauria, Anura, T. rubripes), or occur in species with multiple HspB1 forms (C. jacchus, T. nigroviridis) where the variant forms may have lost their functional significance. Whether a subpopulation of C. familiaris indeed carries an 'atypical' HspB1 form with a proline residue in the position homologous to Arg140 (sequence NP_001003295) remains to be verified as the other available HspB1 form (sequence ENSCAFP00000019945) perfectly fits into the canonical sequence pattern.
In contrast to these highly conserved sites, the sites homologous to Thr151 are affected by a relatively high degree of variability consistent with the poor conservation among the ten human sHSPs. All Anura, L. chalumnae and Clupeocephala orthologs deviate from the human sequence. Even single Primates (P. abelii) and Laurasitheria (P. vampyrus) orthologs deviate from human HspB1, with the threonine residues being replaced by alanine (Supplemental Information S4). This pattern suggests that the threonine residue in this position may be dispensable for the function of HspB1, including the human protein. Historically, such Thr/Ala substitutions occurred with medium frequency in evolution [18]. The variability in this segment of HspB1 is also reflected by the flanking region with its relatively unspecific minimal amino acid motif Sm-X-X-X-X-Hy-X (mutation site underlined) that is common to all considered sequences. Comparative analyses indicate pervasive purifying selection for all mutation sites in the α-crystallin domain, including for the relatively variable site Thr151 (p<0.001; Table 2).
3.1.3.1.3. C-terminal extension
To date, three missense mutation sites (Thr180, Pro182, Arg188) were identified in this section of the human HspB1 that were affected by four missense mutations (p.T180I, p.P182L, p.P182S, p.R188W) (Table 1) [38, 40, 45, 46]. The motor neuropathies associated with these mutations exhibited a relatively early onset, with the first symptoms presenting in the first decade of life in most cases. The inheritance of the neuropathies associated with the mutations p.T180I, p.P182S, and p.R188W has not been recorded, albeit the heterozygosity of the mutant alleles suggests dominance. The p.P182L mutation was associated with a parental mosaicism with dominant characteristics [47].
Comparison among the human sHSPs
Among the ten human sHSPs, Thr180 and Pro182, together with Ile181, are found only in HspB1 and HspB5, forming the T-I-P motif (Fig. 1). Pro182 alone is also conserved in HspB4, whereas the remaining human sHSPs miss this sequence element. Arg188 is not conserved in any of the other sequences, although HspB4 and HspB5 contain similar lysine residues in homologous positions.
Comparison with the Teleostomi orthologs
In this sequence segment, strong similarity of the human HspB1 with its orthologs is limited to the Mammalia and L. chalumnae sequences, with increasing divergence of the Archosauria, Anura, and Clupeocephala sequences (Supplemental Information S5). Given this high degree of variability that affects the entire length of the C-terminal extension, the nearly perfect conservation of Pro182, together with Ile181, is remarkable, thus suggesting a high selective pressure in favor of maintaining this element. The only apparent exception is the C-terminal extension of A. mississippiensis HspB1 with a major deletion affecting most of the C-terminus, including all three mutation sites. This example, however, demonstrates that the loss of the conserved homologous Pro182 is compatible with life in the Tetrapoda.
In contrast, Thr180 is less conserved with the divergence affecting fractions of the Anura and Clupeocephala sequences. The amino acid residues in the homologous positions do not share common properties [17]. Even isoleucine (O. mordax) or other aliphatic amino acid residues (A. fimbria, S. aurata, S. salar) are found in the homologous position, as is the case in the human disease-associated mutant HspB1. This pattern suggests that Thr180 may not be required for the function of HspB1, and that even an isoleucine residue in this position is compatible with life. Apparently, some Clupeocephala species have developed mechanisms that suppress potential adverse effects of isoleucine or other aliphatic amino acid residues in this position. Similarly, the positions homologous to Arg188 are relatively variable with all of the Archosauria and the Clupeocephala sequences deviating from human HspB1, although in ~50% of the sequences similar lysine residues are found.
The region flanking Thr180 and Pro182 in human HspB1 and the homologous sites in the orthologs represent the best conserved element in the C-terminal extensions in all Teleostomi sequences, as opposed to the variable region flanking Arg188. This situation is reflected by the minimum motif Sm-Ne-Hy-X-Al-P-X-Hy-X-X-X-X-X-X (mutation sites underlined) that is common to all orthologs. Sites Pro182 and Arg188 were historically under significant purifying selection (p<0.001), whereas the evidence for purifying selection at the relatively variable site Thr180 was marginal (p=0.083) (Table 2).
3.1.3.2. HspB3
In a screening of 28 patients with idiopathic peripheral axonal motor neuropathy, covering HSPB1 through HSPB10, the p.R7S missense mutation in HspB3 was identified in two siblings (Table 1) [48]. To date, this is the only known disease-associated mutation affecting HspB3, and p.R7S belongs to the minor fraction of mutations that are positioned in the N-terminal domain (Fig. 1). Like a number of other mutations in human sHSPs, the p.R7S eliminates the positively charged amino acid residue arginine. The onset of clinical symptoms in patients with this mutation was in the early twenties. Sequencing data and familial history suggest genetic dominance of the associated neuropathy.
Comparison among the human sHSPs
Positively charged amino acid residues (arginine, lysine, histidine) at positions homologous to Arg7 are well conserved, with HspB9 being the only exception (Fig. 1). Thus, a positive charge in this position, possibly in conjunction with an additional positive charge in position +1 which is found in 6 human sHSPs, seems to be crucial for the function of most human sHSPs.
Comparison with the Teleostomi orthologs
The N-terminus of human HspB3 shares strong similarity with its Mammalia orthologs, whereas increasing deviation in the Sauria/Testudines, S. tropicales, L. chalumnae, and D. rerio sequences is noted (Supplemental Information S6). Arg7 is perfectly conserved among the Tetrapoda orthologs without exception, and also the L. chalumnae HspB3 carries a similar lysine residue in the homologous position. Such Arg/Lys exchanges were commonly observed in orthologous sequences [18]. Of interest are the two D. rerio sequences that are the only HspB3 orthologs that clearly deviate from human HspB3. Both sequences have serine residues in the homologous position, as is the case in the human disease-associated mutant HspB3. Such Arg/Ser exchanges occurred in protein evolution with medium frequency [18]. In spite of the variability in the more distant Teleostomi species, the vicinity of Arg7 and its homologous sites is recognizably conserved throughout all taxa, fitting into the motif Hy-X-Hy-Pl-H-X-Al (position of the mutation site underlined). This site was under significant purifying selection (p = 0.001; Table 2). Together with the perfect conservation of the Arg7 among the Tetrapoda orthologs, this finding suggests that this residue is crucial for the function of human HspB3 and renders adverse pathological consequences for the R7S mutation plausible.
The two D. rerio sequences demonstrate that serine residues in the homologous positions can be compatible with life, at least in distant taxa. As argued above, one explanation may be the short life span of ~2–4 years of this fish that may not be sufficient for the manifestation of the motor neuropathy, as compared to the onset age of ~20 years in the human patients. Alternatively, D. rerio may have adopted a suppressor mechanism which might be of interest for future therapeutic approaches.
3.1.3.3. HspB8
Three missense mutations in the HspB8 protein (p.K141E, p.K141N, p.K141T), caused by four different mutations in the HSPB8 gene (c.421A>G, c.423G>C, c.423G>T, c.422A>C), were identified in five unrelated families with motor neuropathies, and in a patient without family history (Table 1) [49–51]. These mutations share the elimination of the positively charged Lys141 residue, thus causing an acidic shift in the isoelectric point [52]. The onset age of the disease was in the range from 13 to 35 years. Both family history and sequencing data suggest a dominant inheritance.
Comparison among the human sHSPs
Positively charged amino acid residues (lysine, arginine, histidine) are found in homologous positions in nine of the ten human sHSPs with HspB10 being the only exception (Fig. 1), thus suggesting a crucial role in most of the human sHSPs. Together with the affected Arg140 and Lys141 in HspB1 and Arg120 in HspB5, Lys141 in HspB8 may belong to a mutational 'hot spot'.
Comparison with the Teleostomi orthologs
The α-crystallin domains of all Teleostomi orthologs share a high degree of similarity with human HspB8, and this is also reflected by perfect conservation of this amino acid residue throughout all taxa without exception (Supplemental Information S7). The importance of maintaining Lys141 and its homologous sites is further evidenced by the significant purifying selection detected at this site (p=0.032; Table 2). Both the perfect conservation and the inferred purifying selection render it likely that any substitution historically had negative fitness consequences. Thus, any mutation affecting Lys141 in human HspB8 can be expected to result in malfunction that is associated with disease.
3.1.3.4. HspB5
3.1.3.4.1. α-crystallin domain
The p.R120G mutation in the α-crystallin domain of HspB5 was the first identified sHSP mutation that belongs to the group of the myopathy- or neuropathy-associated mutations (Table 1) [10]. It was identified by positional cloning of a large pedigree of patients with desmin-related myopathy (DRM). More recently, p.D109H was identified as the second mutation in this domain [53]. Both mutations have opposite effects on the charge of this sHSP: p.R120G eliminates a positive charge in HspB5, as opposed to p.D109H which introduces a positive charge. In spite of these opposing molecular features, both mutations result in very similar myopathy phenotypes with onset ages in the third or fourth decades of life [53–55]. Family history and sequencing data suggest a dominant disease inheritance for both mutations.
Comparison among the human sHSPs
The Asp109 and Arg120 mutation sites are largely conserved among the human sHSPs, although exceptions occur: HspB7, HspB8 and HspB9 do not carry aspartate, or another negatively charged residue, in positions homologous to Asp109, whereas HspB10 is the only human sHSP without a positively charged amino acid residue in the position homologous to Arg120 (Fig. 1). Arg120 belongs to a potential mutational hot spot region that is shared with HspB1 and HspB8.
Comparison with the Gnathostomata orthologs
The section of the α-crystallin domain of the HspB5 orthologs of all species, as shown in Supplemental Information S8, shares a high degree of similarity with human HspB1. This similarity is also reflected by the fact that both mutation sites, Asp109 and Arg120, are perfectly conserved in all orthologs. The importance of preserving both Asp109 and Arg120, and their homologous sites, is further supported by the significant purifying selection inferred at both sites (p<0.001; Table 2). As with the mutations in the α-crystallin domain of HspB1 and HspB8, any mutations were likely to have had negative fitness consequences in the past. Thus, mutations affecting Asp109 or Arg120 can be expected to result in malfunctions that are associated with disease.
3.1.3.4.2. C-terminal extension
The variable C-terminal extension harbors two known missense mutations, p.G154S and p.R157H, that were both associated with mild forms of myopathies (Table 1) [56–58]. The onset age of the disorder was after the fourth decade. Family history and the sequencing data suggest a dominant inheritance. Interestingly, the p.R157H mutation replaces the positively charged arginine residue with histidine, another positively charged residue, suggesting that the hydrophobic and aromatic properties of the histidine residue at this position are not tolerated.
Comparison among the human sHSPs
Given the variable nature of the C-terminal extensions of the sHSPs, it is not surprising that both mutation sites are not conserved among the human sHSPs. The only exception is the arginine residue homologous to Arg157 that is found in the closely related HspB4 (Fig. 1).
Comparison with the Gnathostomata orthologs
While all Mammalia orthologs share a high degree of similarity with the human HspB5, increasing deviations are noted in the Sauria, Anura, L. chalumnae, Otophysi and S. acanthias sequences (Supplemental Information S9). In these taxa, this variability affects also the sites homologous to Gly154, featuring the hydrophobic amino acid residues valine, isoleucine, or alanine (exception: D. rerio sequence NP_001002670). In addition, deviations from the human HspB5 are also found in single Eutheria species (C. porcellus, V. pacos). Surprisingly, the test for selection does not indicate any selective force at this position (p=0.607; Table 2). In this respect, site Gly154 is distinguished from all other missense mutation sites in the sHSPs. From this pattern it can be concluded that historically the glycine residue at this position was dispensable, and that the serine residue as found in the disease-associated mutant p.G154S adds an adverse property to human HspB1. Alternatively, Gly154 may have become indispensible only in recent evolutionary history leading to the Primates, including humans. It is also of interest that other sites (e.g. Pro155, Glu156, Ile159) in these variable C-terminal extensions are better conserved than Gly154, however, no disease-associated mutations have been reported for these sites to date.
In contrast, Arg157 is nearly perfectly conserved among all Teleostomi and S. acanthias orthologs, with L. chalumnae HspB5 being the only exception (Supplemental Information S9). Consistent with the observation, there was significant purifying selection at this position (p<0.001; Table 2). Certain features in the vicinity of both mutation sites remain constant in all sequences, fitting into the common motif X-X-X-Hy/R-Sm-E-R/L-X-Al-Sm (mutation sites underlined).
3.1.3.5. HspB6
In a screening of 1347 patients with dilated cardiomyopathy and 744 control individuals, one polymorphism (p.P20L) in the translated sequence of HspB6 was identified in one patient and three control individuals (Table 1)[14]. Thus, the association with cardiomyopathy is not clear at this time. This gene variant may represent a low-frequency polymorphism, or a mutation with a slow or low penetrance. All four reported individuals were heterozygous for this genetic variant.
Comparison among the human sHSPs
Although the corresponding N-terminal regions of the other human sHSPs contain a number of proline residues, none of them seems to be homologous to Pro20 in human HspB6 (Fig. 1). Thus, Pro20 may confer HspB6 a unique property that is not shared by the other human sHSPs. Pro20 may be functionally related to the adjacent phosphorylation site Ser16 [59].
Comparison with the Teleostomi orthologs and HspB6-like sequences
The N-termini of most of the Mammalia orthologs exhibit a high degree of similarity with the human of HspB6 (Supplemental Information S10, part A). Pro20 is conserved in almost all Mammalia orthologs, with two exceptions: The V. pacos and M. domestica sequences exhibit a large deletion and an extended atypical N-terminus, respectively. Both of these non-canonical HspB6 forms have also lost the sites homologous to Pro20. If the non-homologous proline residue (labeled in blue) in this region of M. domestica HspB6 can functionally substitute for the homologous Pro20 is not known.
In the more distant taxa, the N-termini of HspB6-like proteins deviate substantially from that of human HspB6 (Supplemental Information S10, part B). While similarity of the A. carolinensis and X. laevis sequences with human HspB6 (including proline residues homologous to Pro20) is still recognizable, the divergence of the Clupeocephala sequences reaches a degree that obscures similarity with human HspB6. The relatedness of all of these sequences with human HspB6 is suggested by phylogenetic analyses only on the basis of the isolated α-crystallin domain, whereas analyses using the full-length proteins or the isolated N-termini failed to establish orthology with human HspB6 (not shown). This 'chimeric phylogeny' may reflect the independent evolutionary history of the α-crystallin and N-terminal domains reported earlier [15]. Interestingly, even the most diverging Clupeocephala sequences contain probably non-homologous proline residues in their N-termini that might fulfill the function of Pro20 in human HspB6 (labeled in blue). Given the strong divergence in the N-termini between the Mammalia HspB6 orthologs on the one hand and non-Mammalia (Teleostomi) HspB6-like proteins on the other hand, the functional equivalence of both protein groups is unclear at this time and requires further study.
Because of the high degree of sequence divergence in the taxa beyond Mammalia, the natural selection analyses for Pro20 and its homologous sites were restricted to the canonical Mammalia sequences. This site was under significant purifying selection (p<0.001; Table 2), which is consistent with its nearly perfect conservation among the Mammalia sequences. It can be tentatively concluded that a polymorphism or mutation affecting Pro20 in human HspB6 may be associated with some form an adverse phenotype, even if its association with dilated cardiomyopathy was not clear in the available study [14]. However, the absence of this site, together with the flanking regions, in V. pacos and M. domestica also suggests that single Mammalia species developed a strategy to compensate for the loss of whole sequence segments around homologous Pro20.
3.1.4. Frame shift, elongation and nonsense mutations
3.1.4.1. HspB1
One frame shift mutation (p.P159fsX200, c.476–477delCT) in HspB1 has been reported (Table 1) [38, 60]. This mutation results in an aberrant sequence spanning the 3'-end of the α-crystallin domain and the C-terminal extension (Fig. 1). The affected adult male individual presented with a mild form a peripheral motor neuropathy [60]. This, however, is in striking contrast to his 3 month-old child who presented with a severe early onset peripheral neuropathy that developed secondary to a diphtheria-tetanus-pertussis vaccination. This case report suggests an unexpected interplay of pathophysiological and HspB1-related genetic factors, which determine the severity of the motor neuropathy. Sequencing data and family history suggest a genetic dominant inheritance, although environmental factors also play a role.
Surprisingly, alignment of this aberrant C-terminal extension with human wild-type HspB1 reveals a weak, yet detectable, similarity (Supplemental Information S11, part A). One feature of the mutant sequence is the aberrant H-H-P motif exactly at the position of the conserved T-I-P motif, which harbored three missense mutations in HspB1 (cf. Supplemental Information S5). This aberrant H-H-P motif might affect the function of HspB1 by a similar mechanism as the abnormal I-I-P, T-I-S and T-I-L motifs in the p.T180I, p.P182S and p.P182L mutants, respectively, and also as the abnormal C-terminal extensions of some HspB5 mutants (see Section 3.1.4.2.).
3.1.4.2. HspB5
In HspB5, three frame shift mutations (p.S21fsX44, c.60delC; p.S115fsX128, c.343delT; p.P155fsX163, c.464–465delCT), one elongation (p.X176WelX195; c.527A>G) and one nonsense (p.Q151X, c.451C>T) mutation have been reported, all resulting in aberrant proteins which are prematurely terminated (p.S21fsX44, p.S115fsX128, p.Q151X, p.P155fsX163) or C-terminally elongated (p.X176WelX195) (Table 1, Fig. 1).
Those mutations affecting the N-terminal domain (p.S21fsX44) or the α-crystallin domain (p.S115fsX128) caused the most severe phenotypes associated with disease onset within the first months of life, including respiratory distress and early death in most cases [61, 62]. These severe disease forms manifested only in homozygous individuals whom expressed truncated forms of HspB5. Heterozygous siblings, in contrast, exhibited a normal phenotype. These data seem to be consistent with a loss-of-function mechanism causing severe myopathy if both alleles are affected, although the situation may be more complex (see Section 3.2.3.).
The one nonsense (p.Q151X), frame shift (p.P155fsX163) and elongation mutation (p.X176WelX195) each that affect the C-terminal extension cause more benign myopathies with later onsets. The patient with the p.Q151X mutation presented at 53 years of age with leg weakness and foot drop [63]. Similarly, the patient with the p.P155fsX163 mutation presented at 52 years of age with progressive leg weakness and reduced movement of the diaphragm, which however resulted eventually in respiratory failure [63]. The patients carrying the p.X176WelX195 mutation all suffered from cardiomyopathy which may develop in the third decade of life, or later [64]. All individuals affected by these mutations were heterozygous suggesting dominant inheritance.
Interestingly, the p.P155fsX163 mutation results in an aberrant C-terminal extension that shares features with other mutants in both HspB5 and HspB1 (Supplemental Information S11, part B). This aberrant C-terminal extension carries a histidine residue in the same position as the p.R157H mutant of HspB5. Another histidine residue is found at the position of the threonine of the T-I-P motif, as is the case in the p.P159fsX200 mutant of HspB1 (cf. Supplemental Information S11, part A). Additionally, this aberrant C-terminal extension of the p.P155fsX163-HspB5 mutant carries an H-H-P motif in close proximity to the T-I-P motif of the wild-type sequence, similar to the p.P159fsX200 mutant of HspB1.
Taken together, some of the HspB1 and HspB5 frame shift mutations that result in aberrant C-terminal extensions share features with certain missense mutations, and thus may cause similar molecular malfunctions leading to disease.
3.1.5. Synonymous mutations
Several sHSP sequence variants with single nucleotide substitutions that do not affect the protein sequence have been reported both in patients and in control individuals. For example, such synonymous sequence variants were found in HspB1 and HspB7 in patients with motor neuropathies and cardiomyopathies, respectively [36–39]. Although some of these sequence variants seem to correlate with disease, a clear association has not been demonstrated to date. These sequence variants may represent polymorphisms or mutations with low penetrance or late disease onset. Such single synonymous nucleotide substitutions in mRNAs principally can affect the folding structure and processing of the mRNA, and thus can have adverse consequences, without protein involvement [65].
An example of a synonymous sequence variant is p.R127R (c.379C>A) of HspB1 that has been identified in a patient (sporadic case) with distal hereditary motor neuropathy (Table 1) [37]. Interestingly, the p.R127W mutation (c.379C>T) in HspB1 which is clearly associated with disease affects the same position in the mRNA, although this mutation affects also the protein sequence. Thus, cytosine 379 may be a critical position in the HspB1 mRNA, and its mutation may have adverse consequences for the physiology of motor neurons.
Theoretically, this rationale can be also applied to the other sHSP mutations, leading to a provocative hypothesis according to which the sHSP mRNAs would be the damaging agents, with the proteins being irrelevant. This notion is supported by the observation that the mutations c.423G>C and c.423G>T in HspB8 mRNA cause different disease phenotypes (dHMN2 and CMT2L, respectively), in spite of the fact that the resulting mutant protein sequence is identical (p.K141N) [49, 50]. More likely, however, is a scenario in which both mRNA and protein contribute to the disease phenotype.
Before final conclusions can be drawn, more studies on the potential injurious effects of mutated or polymorphic sHSP mRNAs need to be conducted.
3.2. Inheritance of the disease phenotypes associated with the sHSP mutations and their pathomechanisms
3.2.1. Genetic dominance in the context of sHSP evolution
All but one of the disease-associated true missense mutations in sHSPs (p.P20L-HspB6 not counted) exhibit a dominant inheritance of the disease phenotype, or it can be assumed (Table 1). To date, only one recessive missense mutation, p.L99M in HspB1, has been reported. It should be noted, however, that it is not known if the dominant disease phenotypes exhibit true dominant or semi-dominant inheritance as the corresponding mutant/mutant alleles have not been reported.
This prevailing of genetic dominance among the missense mutations in sHSPs is noteworthy as mutagenesis studies in many organisms revealed that ~90% of the wild-type alleles are dominant over mutant alleles. Thus, in the wider picture the dominant phenotypes are the exception [66, 67]. In previous studies it was reported that 'Mendelian disease genes' are under increased purifying selection when the disease-causing alleles are dominant, as compared to more complex disease genes [68, 69]. Table 2 reveals that this observation applies to most of the sHSP missense mutations which belong to the group of the 'Mendelian disease genes', with the mutation sites historically being exposed to detectable significant or marginal purifying selection at the between-species level. The exceptions include p.L99M-HspB1 (associated with a recessive phenotype), p.G154S-HspB5 (no detectable purifying selection), and p.P20L-HspB6 (not a 'Mendelian disease gene').
The term 'genetic dominance' originally referred to the masking of one phenotype by another one in the F1 generation [70]. Since then, different and partially conflicting concepts of genetic dominance were developed: it was considered an inevitable consequence of physiology and metabolism, or, alternatively, a contingent by-product of selection for phenotypic robustness [67, 71]. With regard to the underlying cell biological mechanisms, phenotypic dominance of mutations that affect components of macromolecular complexes (which is a hallmark of sHSPs) was explained by the balanced nature of the relationship of these components (gene balance hypothesis) [72, 73]. Another feature of sHSPs in all studied vertebrates is the existence of paralogous genes. Such an evolutionary retention of paralogs in polyploid species, or species with polyploid ancestry which includes Homo sapiens, was explained by gene dosage effects and dominant-negative effects [72, 74]. Thus, the presence of the 10 paralogous sHSPs [1, 2] in the human genome may well be related to both their receptivity for harboring dominant-negative mutations and their ability to form hetero-oligomeric complexes [75].
3.2.2. Cell-biological and biochemical mechanisms of genetic dominance of the sHSP missense mutations
Apart from these theoretical considerations, a number of cell-biological and biochemical gain-of-function mechanisms of genetic dominance have been identified that seem to accommodate most situations, and which apply also to the phenotypic dominance seen with the neuropathy- and myopathy-associated missense mutations of sHSPs. In addition to the 'classic' dominant-negative effects, these mechanisms include the formation of toxic gene products, altered overall cell architecture, and increased protein or enzyme activities [66]. All these consequences have been observed in cells expressing mutant sHSPs. 'Classic' dominant-negative effects can be readily explained by the oligomer formation of sHSPs, with incorporation of a single mutated sHSP molecule compromising both the structure and function of the entire complex. Indeed, abnormal sHSP-sHSP interactions involving mutant sHSPs may be the underlying mechanism whereby the function of several sHSPs could be impaired by a single mutant sHSP [76–78]. Similarly, toxic properties of the mutant sHSPs are readily explained. Mutant proteins in general are characterized by an increased tendency for misfolding, intracellular accumulation, and mislocalization which is linked to cell injury and death [79–81]. Cells actively sequester these toxic amyloid proteins typically resulting in amorphous protein deposits (inclusion bodies, aggresomes) [82]. This aggresome formation is the hallmark of many 'protein folding diseases' which include neuro- and myodegenerative conditions, although the amyloid precursors seem to be the true injuring agent as opposed to the mature aggresomes which are thought to be the end product of a 'detoxifying' process [81, 83, 84]. All mutant sHSPs studied to date exhibit such an increased tendency to form intracellular aggresomes. Examples are the increased aggresome formation of p.K141E-HspB8, p.K141N-HspB8, p.R120G-HspB5, p.Q151X-HspB5, p.P155fsX163-HspB5, p.S135F-HSPB1, or p.P182L-HspB1 [63, 76, 77, 85, 86]. For p.R120G-HspB5 it was shown that the amyloid protein formation leads to mitochondrial dysfunction, which is suspected to accelerate the amyloid formation in a feed-forward loop [81, 87].
Yet there is another potential mechanism for genetic dominance of disease phenotypes associated with mutant sHSPs. It has been observed that mutated cytoskeletal proteins, or proteins associated with them, cause dominant disease phenotypes. Examples include neuropathies or myopathies that are associated with mutations in the NEFL gene (neurofilament light polypeptide), KIF1B gene (kinesin family member 1B), DNTN1 (dynactin 1), and the DES gene (desmin) [86, 88–90]. All these mutations affect the organization of the cell's cytoskeleton. Similarly, the studied mutant sHSPs exerted negative effects on the cytoskeleton which may be the cause for the observed dominant disease phenotype. Examples include p.S135F-HspB1, p.P182L-HspB1, and p.R120G-HspB5 that resulted in abnormally organized intermediate filaments (neurofilaments, desmin filaments) in neurons or muscles [10, 40, 85, 86]. Three of the HspB1 mutants (p.R120W-HspB1, p.S135F-HspB1, p.R136W-HspB1) exhibited an increased ability to bind both tubulin and the microtubules, resulting in an abnormal dynamics of the microtubules [91].
Finally, increased enzymatic activity is another mechanism of genetic dominance. Such a mechanism has been described for the sHSP mutants p.R120G-HspB5, p.S135F-HspB1, and p.P182L-HspB1. Although HspB5 and HspB1 by themselves do not catalyze 'classic' enzymatic reactions, the expression of these mutant proteins were found to abnormally up-regulate the downstream activities of known enzymes in transgenic mice: i) expression of p.R120G-HspB5 increased the glucose-6-phosphate dehydrogenase (G6PD) activity and other enzymes of the redox metabolism in diseased hearts, thus impairing the redox homeostasis [92], and ii) expression of p.S135F-HspB1 and p.P182L-HspB1 resulted in increased α-tubulin deacetylation, impairing the axonal transport in peripheral neurons [93]. Also, the observed increased chaperone-like activities of several of the HspB1 mutants (p.R127W-HspB1, p.S135F-HspB1, p.R136W-HspB1) fit to this mechanism of genetic dominance [91]. Such abnormally increased enzymatic and functional activities that result from expression of mutant sHSPs are potential targets for pharmacological manipulation (see Section 3.3.5.)
The above considerations suggest that the genetic dominance associated with most of the missense sHSP mutations results from gain-of-function mechanisms but not from haploinsufficiency, which is a loss-of-function mechanism of genetic dominance [66]. This notion is further supported when the phenotypes of HspB5 and HspB1 knock-out mice are compared with mice expressing missense mutations of these sHSPs. HspB5 knock-out mice did not show any overt phenotype up to 40 weeks of development, although in later life some skeletal muscles degenerated [94]. Clearly, deletion of HspB5 did not replicate the myofibrillar type of lesion that was seen after transgenic expression of the p.R120G-HspB5 mutant gene in mice [95]. Similarly, deletion of HspB1 [96] did not replicate the lesion of the motor neurons that was seen after transgenic expression of p.S135F-HspB1 and p.P182L-HspB1 [93]. Further experimental findings tentatively support the notion of loss-of-function mechanisms being not crucial for the effects of the dominant sHSP mutants: some of the studied mutant HspB1 species did not lose key functions such as chaperone-like and anti-apoptotic activities [91, 97].
3.2.3. Recessive sHSP mutations
The two recessive frame shift mutations p.S21fsX44 and p.S115fsX128 in HspB5 are associated with aggressive forms of myofibrillar myopathy in homozygous individuals (see Section 3.1.4.2.). Both mutations result in significantly shortened polypeptides (Fig. 1) which therefore can be assumed to have lost at least part of the HspB5 functions. For both mutations, the heterozygous parents did not show any disease phenotype, in contrast to the homozygous individuals of the F1 generation. Thus, haploinsufficiency can be excluded to cause the disease as the remaining expression of the wild-type HspB5 allele in the parent generation is sufficient for a normal phenotype. The fact that the complete loss of the full-length HspB5 causes such an aggressive disease phenotype contrasts with the HspB5 knock-out mice which did not replicate the myofibrillar type of disease [94]. One possible explanation is that the expression of the truncated mutant forms of HspB5 also contributes to this disease phenotype, thus resulting in a mixed loss/gain-of-function pathogenesis.
The p.L99M missense mutation in HspB1 with a clear autosomal-recessive inheritance of the relatively benign disease phenotype tentatively suggests loss-of-function as the pathomechanism [37]. Also for this mutation, haploinsufficiency can be excluded as the underlying pathomechanism, because the heterozygous parents of the affected patient developed no disease symptoms.
The concept of a loss-of-function for HspB1-associated motor neuropathy is tentatively supported by a case report of a sporadic mutation in the HspB1 promoter in a patient with amyotrophic lateral sclerosis, a related form of motor neuropathy [39]. This mutation decreased the basal HspB1 promoter activity to ~50%, and also affected the stress induction of HspB1. In the patient with this mutation, the reduced expression of wild-type HspB1 apparently was not sufficient for maintaining a healthy phenotype. However, to date the association of this mutation with disease has not been confirmed.
In summary, a loss-of-function type pathogenesis may also play a role for a minority of the mutations in sHSPs. The observed phenotypes and inheritance in these cases can be best explained by assuming mixed loss/gain-of-function mechanisms. Further data on sHSP-related loss-of-function scenarios should be evaluated before final conclusions on this pathogenesis can be drawn.
3.3. Implications for future therapeutic approaches
3.3.1. Reducing the expression of the mutated sHSP species
Given the fact that the disruption of the HspB1 and HspB5 genes in mice did not result in overt motor neuropathy and myopathy phenotypes, respectively [94, 96], a reduction or abrogation of the expression specifically of the affected mutant sHSP seems to be the most straight-forward future therapeutic strategy for the dominant gain-of-function mutations. It can be assumed that this mutant-specific sHSP down-regulation will reduce the cell injury caused by the newly gained, adverse properties of the mutant sHSP, no matter if 'classic' dominant-negative effects, aggresome formation, cytoskeletal effects, or increased enzymatic activities are at the root of the pathogenesis. The advantage of this strategy is that it could be applied to all dominant sHSP mutations, superseding the need for addressing specific downstream consequences for each single mutant sHSP. The disadvantage is that the complete elimination of one particular sHSP may also lead to adverse consequences, although they seem to be less severe.
Whether this approach should be also applied to the recessive mutations remains debatable. If these mutants have exclusively loss-of-function properties, the down-regulation would be futile. However, since it is more likely that diseases result from mixed loss/gain-of-function mechanisms, therapeutic down-regulation of the gain-of-function components of these mutants seems to be desirable.
Whereas this approach of down-regulation specifically of the mutant sHSP in affected individuals bears a great potential, at present the technical impracticality in a clinical setting renders this a task for the future. At least in cell lines, antisense or interfering RNAs have already been successfully used for the reduction of the expression of single sHSPs and could conceivably be specifically targeted for mutant sHSPs [98]. As advances in genome editing progress, these types of precise manipulations may ultimately be very fruitful.
3.3.2. Exploiting suppressor mechanisms adopted by other Gnathostomata species
As shown in the alignments in Supplemental Information S2, S4, S5, S6, and S9, homologous sites in sHSPs of single Gnathostomata species or entire taxa differ from the mutation sites in the human sequences, and occasionally the Gnathostomata sHSP genes even encode the same amino acid residues that cause the disease in humans (examples: T. belangeri and O. mordax orthologs of human HspB1, D. rerio ortholog of human HspB3). This observation suggests that such sHSP variants can be compatible with life and do not necessarily cause disease. These species may have adopted suppressor mechanisms that neutralize the potentially adverse consequences of these sHSP variants. These potential suppressor mechanisms are of interest for future therapeutic approaches, although their existence and nature remain to be ascertained.
3.3.3. Reducing the formation of toxic amyloid oligomers and aggresomes
Given the gain-of-function nature of most dominant mutations, it must be the aim to reduce the adverse downstream consequences resulting from these new or 'enhanced' properties which include the formation of oligomeric amyloids. Thus, the attenuation of this intracellular protein denaturation is desirable. In recent years a number of low-molecular mass compounds have been identified that facilitate protein folding and stabilize their native conformation, thus protecting against denaturation, caused by either external (e.g. oxidative stress) or internal (e.g. mutations) factors. These compounds have the capacity to prevent or reduce the intracellular aggregation and precipitation of proteins. Although these compounds are frequently referred to as low-molecular-mass or chemical chaperones, they are not chaperones in sensu stricto as they do not actively fold proteins. Examples of such chemical chaperones include 4-phenylbutyric acid (PBA)[99], tauroursodeoxycholic acid (TUDCA)[100], trimethylamine oxide (TMAO)[101], and (−)-epigallocatechin-3-gallate (EGCG) [102]. Among them, EGCG is a secondary plant metabolite that is found e.g. in green tea. Although some of these drugs have been studied primarily in the context of endoplasmic reticulum stress [103], it can be assumed that their beneficial effects can be also seen in the cytoplasm which is the primary compartment of mutant sHSP denaturation. Unfortunately, high concentrations of these compounds are typically required to see a beneficial effect, thus increasing the risk of toxic side effects that may hamper their use in humans in near future. However, it can be expected that a new generation of this class of drugs with improved properties will be available in future.
3.3.4. Increasing expression of endogenous wild-type sHSPs
The fact that sHSPs form mixed oligomeric complexes can be potentially exploited for therapeutic approaches. It is thought that increased expression of certain wild-type sHSPs will displace mutant sHSPs in these complexes, thus attenuating the adverse consequences of the mutant sHSPs. Indeed, the amyloid oligomer and aggresome formation by p.R120G-HspB5 was reduced by the presence of HspB1 and HspB8 in cardiomyocytes and also in the hearts of mice [104, 105]. Similarly, the expression of HspB5 or HspB6 markedly reduced the incidence of aggresome formation caused by p.K141N-HspB8 in transfected COS-7 cells [76]. Applying this concept to future therapeutic approaches, however, faces the major challenge of inducing the desired endogenous wild-type sHSPs without inducing the mutant sHSP. Currently no clear strategy for this approach is in sight, although compounds like geranylgeranylacetone (GGA) that induce sHSPs were shown to reduce the formation of amyloid protein oligomers and aggresomes caused by p.R120G-HspB5 in transgenic mice [105]. However, as enhancing overall chaperone levels may also stabilize aggregate formation, this approach must be precisely tailored [106].
3.3.5. Inhibition of abnormally increased downstream enzymatic activities
As outlined in Section 3.2.2, increased downstream enzymatic activities that result from expression of mutant sHSPs have been reported in two cases: i) induction of cardiomyopathy by transgenic expression of p.R120G-HspB5 in mice was associated with reductive stress [92]. The diseased hearts exhibited an increased recycling of oxidized to reduced glutathione which was due to increased expression and activities of glucose-6-dehydrogenase (G6PD) and of other enzymes of the redox metabolism. Interestingly, a reduction of the G6PD expression by cross-breeding with a low-expression G6PD mouse strain rescued the progeny from cardiomyopathy. Thus, pharmacological retention of the redox homeostasis is a strategy that should be pursued in future therapeutic approaches; and ii) induction of motor neuropathy by transgenic expression of p.S135F-HspB1 and p.P182L-HspB1 was associated with a decreased acetylation of the cytoskeletal protein α-tubulin, eventually resulting in an impaired axonal transport in peripheral neurons [93]. The pharmacological inhibition of the enzyme histone deacetylase 6 (HDAC6) with inhibitors such as trichostatin or tubacin increased the motor performance of p.S135F-HspB1 expressing transgenic mice by improving the electrophysiological parameters and restoration of the acetylcholine-receptor clusters at the end plates of the muscles, suggesting that the acetylated α-tubulin is needed to a re-innervation of the peripheral muscles. Thus, inhibition of HDAC6 is a promising therapeutic approach for those mutant sHSP-associated motor-neuropathies that are characterized by abnormally increased HDAC6 activity.
3.3.6. Reducing the adverse effects of mutant sHSPs with peptide aptamers
Peptide aptamers can be used to modulate the interactions of sHSPs, and in consequence also their function. For example, an aptamer designed to stimulate the protective role of HspB1 abolished the adverse effects of p.R120G-HspB5 by disrupting its interactions with HspB1 [107]. Such structure-based interfering strategies are of potential interest for future therapeutic approaches.
4. Conclusions
To date, 33 motor neuropathy- and myopathy-associated mutations have been identified in human HspB1, HspB3, HspB5, and HspB8. The fact that all mutations in HspB1, HspB3, and HspB8 are associated with motor neuropathies, as opposed to the mutations in HspB5, which are associated with various forms of myopathies, suggests tissue-specific functions of these sHSPs. Most mutations are missense mutations that are associated with dominant disease-phenotypes. In the primary protein sequence, most mutations are clustered in the conserved α-crystallin domain and the variable C-terminal extensions. HspB1, HspB5, and HspB8 share an apparent hot spot mutation site, suggesting a crucial feature that is common to these sHSPs. Most of the missense mutation sites are well conserved among vertebrate orthologs, although exceptions occur: in several vertebrate sHSPs, homologous sites differ from the human sequence, and occasionally even encode the same amino acid residues that cause the disease in humans. Apparently, some of these mutations sites are not crucial for the protein function in single species or entire vertebrate groups, and single species seem to have adopted mechanisms that compensates for potentially adverse effects of 'mutant-like' sHSPs.
Mutant sHSP alleles belong to the group of 'Mendelian disease genes'. Like other dominant disease-causing alleles of this type, most missense sHSP mutations were historically under increased purifying selection, suggesting that mutations at these positions in the past were generally associated with negative fitness consequences. The expression of sHSPs with dominant missense mutations results in a number of cellular consequences that are consistent with gain-of-function mechanisms of genetic dominance: (i) 'classic' dominant-negative effects that result from hetero-oligomer complex formation, (ii) formation of cytotoxic amyloid protein oligomers and precipitates, (iii) disruption of cytoskeletal networks, and (iv) increased downstream enzymatic activities.
Future therapeutic approaches for mutant sHSP-associated diseases include the genetic down-regulation of the mutated sHSP species, the exploitation of suppressor mechanisms adopted by other vertebrate species, reducing the formation of toxic amyloid oligomers, increasing the expression of endogenous wild-type sHSPs, inhibition of abnormally increased downstream enzymatic activities, and structure-based interfering strategies of sHSP interactions. Fortunately, initial experimental approaches have provided encouraging results.
Supplementary Material
Acknowledgments
This work was supported by the grant SDG 0235411Z (American Heart Association) to JLM, and by grant AI43638 (National Institutes of Health Training Fellowship) to JOW. The funding sources had no involvement in the study design, in the collection, analysis and interpretations of the data, in writing of the report, and in the decision to submit the article for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: EGCG, (−)-epigallocatechin-3-gallate; G6PD, glucose-6-phosphate dehydrogenase; GGA, geranylgeranylacetone; HDAC6, histone deacetylase 6; TMAO, trimethylamine oxide; TUDCA, tauroursodeoxycholic acid; sHSP, small heat shock protein.
Conflict of interest statement
The authors declare that there are no conflicts of interests.
Contributor Information
Rainer Benndorf, Email: rbenndo@gmx.net.
Jody L. Martin, Email: jmart10@lumc.edu.
Sergei L. Kosakovsky Pond, Email: spond@ucsd.edu.
Joel O. Wertheim, Email: jwertheim@ucsd.edu.
References
- 1.Fontaine J-M, Rest JS, Welsh MJ, Benndorf R. The sperm outer dense fiber protein is the 10th member of the superfamily of mammalian small stress proteins. Cell Stress Chaperones. 2003;8:62–69. doi: 10.1379/1466-1268(2003)8<62:tsodfp>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kappé G, Franck E, Verschuure P, Boelens WC, Leunissen JA, de Jong WW. The human genome encodes 10 alpha-crystallin-related small heat shock proteins: HspB1–10. Cell Stress Chaperones. 2003;8:53–61. doi: 10.1379/1466-1268(2003)8<53:thgecs>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, Bruford EA, Cheetham ME, Chen B, Hightower LE. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones. 2009;14:105–111. doi: 10.1007/s12192-008-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Jong WW, Caspers GJ, Leunissen JA. Genealogy of the alpha-crystallin-small heat shock protein superfamily. Int. J. Biol. Macromol. 1998;22:151–162. doi: 10.1016/s0141-8130(98)00013-0. [DOI] [PubMed] [Google Scholar]
- 5.Mymrikov EV, Seit-Nebi AS, Gusev NB. Large potentials of small heat shock proteins. Physiol. Rev. 2011;91:1123–1159. doi: 10.1152/physrev.00023.2010. [DOI] [PubMed] [Google Scholar]
- 6.Kappé G, Boelens WC, de Jong WW. Why proteins without an alpha-crystallin domain should not be included in the human small heat shock protein family HSPB. Cell Stress Chaperones. 2010;15:457–461. doi: 10.1007/s12192-009-0155-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arrigo A-P. The cellular 'networking' of mammalian Hsp27 and its functions in the control of protein folding, redox state and apoptosis. Adv. Exp. Med. Biol. 2007;594:14–26. doi: 10.1007/978-0-387-39975-1_2. [DOI] [PubMed] [Google Scholar]
- 8.Vos MJ, Hageman J, Carra S, Kampinga HH. Structural and functional diversities between members of the human HSPB, HSPH, HSPA, and DNAJ chaperone families. Biochemistry. 2008;47:7001–7011. doi: 10.1021/bi800639z. [DOI] [PubMed] [Google Scholar]
- 9.Basha E, O'Neill H, Vierling E. Small heat shock proteins and α-crystallins: dynamic proteins with flexible functions. Trends Biochem. Sci. 2012;37:106–117. doi: 10.1016/j.tibs.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vicart P, Caron A, Guicheney P, Li Z, Prévost MC, Faure A, Chateau D, Chapon F, Tomé F, Dupret JM, Paulin D, Fardeau M. A missense mutation in the alphaB-crystallin chaperone gene causes a desmin-related myopathy. Nat. Genet. 1998;20:92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- 11.Wyttenbach A, Hands S, O'Connor V. Small stress proteins in the central nervous system: From neurodegeneration to neuroprotection. In: Simon S, Arrigo A-P, editors. Small stress proteins and human diseases. New York: Nova Science Publishers; 2012. pp. 117–146. [Google Scholar]
- 12.Laskowska E, Matuszewska E, Kuczyńska-Wiśnik D. Small heat shock proteins and protein-misfolding diseases. Curr. Pharm. Biotechnol. 2010;11:146–157. doi: 10.2174/138920110790909669. [DOI] [PubMed] [Google Scholar]
- 13.Benndorf R. HSPB1 and HSPB8 mutations in neuropathies. In: Simon S, Arrigo A-P, editors. Small stress proteins and human diseases. New York: Nova Science Publishers; 2012. pp. 301–324. [Google Scholar]
- 14.Nicolaou P, Knöll R, Haghighi K, Fan GC, Dorn GW, II, Hasenfuss G, Kranias EG. Human mutation in the anti-apoptotic heat shock protein 20 abrogates its cardioprotective effects. J. Biol. Chem. 2008;283:33465–33471. doi: 10.1074/jbc.M802307200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kriehuber T, Rattei T, Weinmaier T, Bepperling A, Haslbeck M, Buchner J. Independent evolution of the core domain and its flanking sequences in small heat shock proteins. FASEB J. 2010;24:3633–3642. doi: 10.1096/fj.10-156992. [DOI] [PubMed] [Google Scholar]
- 16.Han JH, Batey S, Nickson AA, Teichmann SA, Clarke J. The folding and evolution of multidomain proteins. Nat. Rev. Mol. Cell Biol. 2007;8:319–330. doi: 10.1038/nrm2144. [DOI] [PubMed] [Google Scholar]
- 17.Livingstone CD, Barton GJ. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput. Appl. Biosci. 1993;9:745–756. doi: 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
- 18.Berleant D, White M, Pierce E, Tudoreanu E, Boeszoermenyi A, Shtridelman Y, Macosko JC. The genetic code-more than just a table. Cell. Biochem. Biophys. 2009;55:107–116. doi: 10.1007/s12013-009-9060-9. [DOI] [PubMed] [Google Scholar]
- 19.Higgins DG, Sharp PM. Fast and sensitive multiple sequence alignments on a microcomputer. Comput. Appl. Biosci. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 20.Kosakovsky Pond SL, Frost SD. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 21.Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 22.Pond SL, Frost SD. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- 23.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012;8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasakov AS, Bukach OV, Seit-Nebi AS, Marston SB, Gusev NB. Effect of mutations in the beta5-beta7 loop on the structure and properties of human small heat shock protein HSP22 (HspB8, H11) FEBS J. 2007;274:5628–5642. doi: 10.1111/j.1742-4658.2007.06086.x. [DOI] [PubMed] [Google Scholar]
- 25.Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum. Mutat. 2002;19:607–614. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 26.Sudnitsyna MV, Mymrikov EV, Seit-Nebi AS, Gusev NB. The role of intrinsically disordered regions in the structure and functioning of small heat shock proteins. Curr. Protein Pept. Sci. 2012;13:76–85. doi: 10.2174/138920312799277875. [DOI] [PubMed] [Google Scholar]
- 27.Giese KC, Basha E, Catague BY, Vierling E. Evidence for an essential function of the N terminus of a small heat shock protein in vivo, independent of in vitro chaperone activity. Proc. Natl. Acad. Sci. USA. 2005;102:18896–18901. doi: 10.1073/pnas.0506169103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Treweek TM, Ecroyd H, Williams DM, Meehan S, Carver JA, Walker MJ. Site-directed mutations in the C-terminal extension of human alphaB-crystallin affect chaperone function and block amyloid fibril formation. PLoS One. 2007;2:e1046. doi: 10.1371/journal.pone.0001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lelj-Garolla B, Mauk AG. Roles of the N- and C-terminal sequences in Hsp27 self-association and chaperone activity. Protein Sci. 2012;21:122–133. doi: 10.1002/pro.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonald ET, Bortolus M, Koteiche HA, Mchaourab HS. Sequence, structure, and dynamic determinants of Hsp27 (HspB1) equilibrium dissociation are encoded by the N-terminal domain. Biochemistry. 2012;51:1257–1268. doi: 10.1021/bi2017624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asthana A, Raman B, Ramakrishna T, Rao CM. Structural Aspects and Chaperone Activity of Human HspB3: Role of the 'C-Terminal Extension'. Cell. Biochem. Biophys. 2012;64:61–72. doi: 10.1007/s12013-012-9366-x. [DOI] [PubMed] [Google Scholar]
- 32.Plater ML, Goode D, Crabbe MJ. Effects of site-directed mutations on the chaperone-like activity of alphaB-crystallin. J. Biol. Chem. 1996;271:28558–28566. doi: 10.1074/jbc.271.45.28558. [DOI] [PubMed] [Google Scholar]
- 33.Martin JL, Bluhm WF, He H, Mestril R, Dillmann WH. Mutation of COOH-terminal lysines in overexpressed alpha B-crystallin abrogates ischemic protection in cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 2002;283:H85–H91. doi: 10.1152/ajpheart.00512.2001. [DOI] [PubMed] [Google Scholar]
- 34.Sun X, Fontaine J-M, Rest JS, Shelden EA, Welsh MJ, Benndorf R. Interaction of human HSP22 (HSPB8) with other small heat shock proteins. J. Biol. Chem. 2004;279:2394–2402. doi: 10.1074/jbc.M311324200. [DOI] [PubMed] [Google Scholar]
- 35.Hahner A, Erdmann J, Kallisch H, Fleck E, Regitz-Zagrosek V. Five novel genetic variants in the promoter and coding region of the alpha B-crystallin gene (CRYAB): −652G>A, −650C>G, −249G>C, S41Y, P51L. Hum. Mutat. 2000;16:374. doi: 10.1002/1098-1004(200010)16:4<374::AID-HUMU17>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 36.Matkovich SJ, Van Booven DJ, Hindes A, Kang MY, Druley TE, Vallania FL, Mitra RD, Reilly MP, Cappola TP, Dorn GW., II Cardiac signaling genes exhibit unexpected sequence diversity in sporadic cardiomyopathy, revealing HSPB7 polymorphisms associated with disease. J. Clin. Invest. 2010;120:280–289. doi: 10.1172/JCI39085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Houlden H, Laura M, Wavrant-De Vrièze F, Blake J, Wood N, Reilly MM. Mutations in the HSP27 (HSPB1) gene cause dominant, recessive, and sporadic distal HMN/CMT type 2. Neurology. 2008;71:1660–1668. doi: 10.1212/01.wnl.0000319696.14225.67. [DOI] [PubMed] [Google Scholar]
- 38.Capponi S, Geroldi A, Fossa P, Grandis M, Ciotti P, Gulli R, Schenone A, Mandich P, Bellone E. HSPB1 and HSPB8 in inherited neuropathies: study of an Italian cohort of dHMN and CMT2 patients. J. Peripher. Nerv. Syst. 2011;16:287–294. doi: 10.1111/j.1529-8027.2011.00361.x. [DOI] [PubMed] [Google Scholar]
- 39.Dierick I, Irobi J, Janssens S, Theuns J, Lemmens R, Jacobs A, Corsmit E, Hersmus N, Van Den Bosch L, Robberecht W, De Jonghe P, Van Broeckhoven C, Timmerman V. Genetic variant in the HSPB1 promoter region impairs the HSP27 stress response. Hum. Mutat. 2007;28:830. doi: 10.1002/humu.9503. [DOI] [PubMed] [Google Scholar]
- 40.Evgrafov OV, Mersiyanova I, Irobi J, Van Den Bosch L, Dierick I, Leung CL, Schagina O, Verpoorten N, Van Impe K, Fedotov V, Dadali E, Auer-Grumbach M, Windpassinger C, Wagner K, Mitrovic Z, Hilton-Jones D, Talbot K, Martin JJ, Vasserman N, Tverskaya S, Polyakov A, Liem RK, Gettemans J, Robberecht W, De Jonghe P, Timmerman V. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat. Genet. 2004;36:602–606. doi: 10.1038/ng1354. [DOI] [PubMed] [Google Scholar]
- 41.Liu XM, Tang BS, Zhao GH, Xia K, Zhang FF, Pan Q, Cai F, Hu ZM, Zhang C, Chen B, Shen L, Zhang RX, Jiang H. Mutation analysis of small heat shock protein 27 gene in Chinese patients with Charcot-Marie-Tooth disease. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2005;22:510–513. [PubMed] [Google Scholar]
- 42.Tang B, Liu X, Zhao G, Luo W, Xia K, Pan Q, Cai F, Hu Z, Zhang C, Chen B, Zhang F, Shen L, Zhang R, Jiang H. Mutation analysis of the small heat shock protein 27 gene in Chinese patients with Charcot-Marie-Tooth disease. Arch. Neurol. 2005;62:1201–1207. doi: 10.1001/archneur.62.8.1201. [DOI] [PubMed] [Google Scholar]
- 43.James PA, Rankin J, Talbot K. Asymmetrical late onset motor neuropathy associated with a novel mutation in the small heat shock protein HSPB1 (HSP27) J. Neurol. Neurosurg. Psychiatry. 2008;79:461–463. doi: 10.1136/jnnp.2007.125179. [DOI] [PubMed] [Google Scholar]
- 44.Ikeda Y, Abe A, Ishida C, Takahashi K, Hayasaka K, Yamada M. A clinical phenotype of distal hereditary motor neuronopathy type II with a novel HSPB1 mutation. J. Neurol. Sci. 2009;277:9–12. doi: 10.1016/j.jns.2008.09.031. [DOI] [PubMed] [Google Scholar]
- 45.Kijima K, Numakura C, Goto T, Takahashi T, Otagiri T, Umetsu K, Hayasaka K. Small heat shock protein 27 mutation in a Japanese patient with distal hereditary motor neuropathy. J. Hum. Genet. 2005;50:473–476. doi: 10.1007/s10038-005-0280-6. [DOI] [PubMed] [Google Scholar]
- 46.Luigetti M, Fabrizi GM, Madia F, Ferrarini M, Conte A, Del Grande A, Tasca G, Tonali PA, Sabatelli M. A novel HSPB1 mutation in an Italian patient with CMT2/dHMN phenotype. J. Neurol. Sci. 2010;298:114–117. doi: 10.1016/j.jns.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 47.Dierick I, Baets J, Irobi J, Jacobs A, De Vriendt E, Deconinck T, Merlini L, Van den Bergh P, Rasic VM, Robberecht W, Fischer D, Morales RJ, Mitrovic Z, Seeman P, Mazanec R, Kochanski A, Jordanova A, Auer-Grumbach M, Helderman-van den Enden AT, Wokke JH, Nelis E, De Jonghe P, Timmerman V. Relative contribution of mutations in genes for autosomal dominant distal hereditary motor neuropathies: a genotype-phenotype correlation study. Brain. 2008;131:1217–1227. doi: 10.1093/brain/awn029. [DOI] [PubMed] [Google Scholar]
- 48.Kolb SJ, Snyder PJ, Poi EJ, Renard EA, Bartlett A, Gu S, Sutton S, Arnold WD, Freimer ML, Lawson VH, Kissel JT, Prior TW. Mutant small heat shock protein B3 causes motor neuropathy: utility of a candidate gene approach. Neurology. 2010;74:502–506. doi: 10.1212/WNL.0b013e3181cef84a. [DOI] [PubMed] [Google Scholar]
- 49.Irobi J, Van Impe K, Seeman P, Jordanova A, Dierick I, Verpoorten N, Michalik A, De Vriendt E, Jacobs A, Van Gerwen V, Vennekens K, Mazanec R, Tournev I, Hilton-Jones D, Talbot K, Kremensky I, Van Den Bosch L, Robberecht W, Van Vandekerckhove J, Van Broeckhoven C, Gettemans J, De Jonghe P, Timmerman V. Hot-spot residue in small heat-shock protein 22 causes distal motor neuropathy. Nat. Genet. 2004;36:597–601. doi: 10.1038/ng1328. [DOI] [PubMed] [Google Scholar]
- 50.Tang BS, Zhao GH, Luo W, Xia K, Cai F, Pan Q, Zhang RX, Zhang FF, Liu XM, Chen B, Zhang C, Shen L, Jiang H, Long ZG, Dai HP. Small heat-shock protein 22 mutated in autosomal dominant Charcot-Marie-Tooth disease type 2L. Hum. Genet. 2005;116:222–224. doi: 10.1007/s00439-004-1218-3. [DOI] [PubMed] [Google Scholar]
- 51.Nakhro K, Park JM, Kim YJ, Yoon BR, Yoo JH, Koo H, Choi BO, Chung KW. A novel Lys141Thr mutation in small heat shock protein 22 (HSPB8) gene in Charcot-Marie-Tooth disease type 2L. Neuromuscul. Disord. 2013;23:656–663. doi: 10.1016/j.nmd.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 52.Sun X, Fontaine JM, Hoppe AD, Carra S, DeGuzman C, Martin JL, Simon S, Vicart P, Welsh MJ, Landry J, Benndorf R. Abnormal interaction of motor neuropathy-associated mutant HspB8 (Hsp22) forms with the RNA helicase Ddx20 (gemin3) Cell Stress Chaperones. 2010;15:567–582. doi: 10.1007/s12192-010-0169-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sacconi S, Féasson L, Antoine JC, Pécheux C, Bernard R, Cobo AM, Casarin A, Salviati L, Desnuelle C, Urtizberea A. A novel CRYAB mutation resulting in multisystemic disease. Neuromuscul. Disord. 2012;22:66–72. doi: 10.1016/j.nmd.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 54.Fardeau M, Vicart P, Caron A, Chateau D, Chevallay M, Collin H, Chapon F, Duboc D, Eymard B, Tomé FM, Dupret JM, Paulin D, Guicheney P. Myopathie familiale avec surcharge en desmine, sous forme de matériel granulo-filamentaire dense en microscopie électronique, avec mutation dans le gène de l'αB-cristalline. Rev. Neurol. (Paris) 2000;156:497–504. [PubMed] [Google Scholar]
- 55.Fardeau M, Godet-Guillain J, Tome FM, Collin H, Gardeau S, Boffety C, Vernant P. Une nouvelle affection musculaire familiale, definie par l’accumulation intra-sarco-plasmique d’un materiel granulo-filamentaire dense en microscopie electronique. Rev. Neurol. (Paris) 1978;134:411–425. [PubMed] [Google Scholar]
- 56.Pilotto A, Marziliano N, Pasotti M, Grasso M, Costante AM, Arbustini E. alphaB-crystallin mutation in dilated cardiomyopathies: low prevalence in a consecutive series of 200 unrelated probands. Biochem. Biophys. Res. Commun. 2006;346:1115–1117. doi: 10.1016/j.bbrc.2006.05.203. [DOI] [PubMed] [Google Scholar]
- 57.Inagaki N, Hayashi T, Arimura T, Koga Y, Takahashi M, Shibata H, Teraoka K, Chikamori T, Yamashina A, Kimura A. AlphaB-crystallin mutation in dilated cardiomyopathy. Biochem. Biophys. Res. Commun. 2006;342:379–386. doi: 10.1016/j.bbrc.2006.01.154. [DOI] [PubMed] [Google Scholar]
- 58.Reilich P, Schoser B, Schramm N, Krause S, Schessl J, Kress W, Müller-Höcker J, Walter MC, Lochmuller H. The p.G154S mutation of the alpha-B crystallin gene (CRYAB) causes late-onset distal myopathy. Neuromuscul. Disord. 2010;40:255–259. doi: 10.1016/j.nmd.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 59.Dreiza CM, Komalavilas P, Furnish EJ, Flynn CR, Sheller MR, Smoke CC, Lopes LB, Brophy CB. The small heat shock protein, HSPB6, in muscle function and disease. Cell Stress Chaperones. 2010;15:1–11. doi: 10.1007/s12192-009-0127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mandich P, Grandis M, Varese A, Geroldi A, Acquaviva M, Ciotti P, Gulli R, Doria-Lamba L, Fabrizi GM, Giribaldi G, Pizzuti A, Schenone A, Bellone E. Severe neuropathy after diphtheria-tetanus-pertussis vaccination in a child carrying a novel frame-shift mutation in the small heat-shock protein 27 gene. J. Child Neurol. 2010;25:107–109. doi: 10.1177/0883073809334387. [DOI] [PubMed] [Google Scholar]
- 61.Del Bigio MR, Chudley AE, Sarnat HB, Campbell C, Goobie S, Chodirker BN, Selcen D. Infantile muscular dystrophy in Canadian aboriginals is an αB-crystallinopathy. Ann. Neurol. 2011;69:866–871. doi: 10.1002/ana.22331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Forrest KM, Al-Sarraj S, Sewry C, Buk S, Tan SV, Pitt M, Durward A, McDougall M, Irving M, Hanna MG, Matthews E, Sarkozy A, Hudson J, Barresi R, Bushby K, Jungbluth H, Wraige E. Infantile onset myofibrillar myopathy due to recessive CRYAB mutations. Neuromuscul. Disord. 2011;21:37–40. doi: 10.1016/j.nmd.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 63.Selcen D, Engel AG. Myofibrillar myopathy caused by novel dominant negative alpha B-crystallin mutations. Ann. Neurol. 2003;54:804–810. doi: 10.1002/ana.10767. [DOI] [PubMed] [Google Scholar]
- 64.van der Smagt JJ, Vink A, Kirkels JH, Nelen M, Ter Heide H, Molenschot MM, Weger RA, Schellekens PA, Hoogendijk J, Dooijes D. Congenital posterior pole cataract and adult onset dilating cardiomyopathy. Expanding the phenotype of αB-crystallinopathies. Clin. Genet. 2013:1–5. doi: 10.1111/cge.12169. [DOI] [PubMed] [Google Scholar]
- 65.Johnson AD, Wang D, Sadee W. Polymorphisms affecting gene regulation and mRNA processing: broad implications for pharmacogenetics. Pharmacol. Ther. 2005;106:19–38. doi: 10.1016/j.pharmthera.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 66.Wilkie AOM. The molecular basis of genetic dominance. J. Med. Genet. 1994;31:89–98. doi: 10.1136/jmg.31.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Agutter PS. The flux-summation theorem and the 'evolution of dominance'. J. Theor. Biol. 2008;254:821–825. doi: 10.1016/j.jtbi.2008.07.027. [DOI] [PubMed] [Google Scholar]
- 68.Blekhman R, Man O, Herrmann L, Boyko AR, Indap A, Kosiol C, Bustamante CD, Teshima KM, Przeworski M. Natural selection on genes that underlie human disease susceptibility. Curr. Biol. 2008;18:883–889. doi: 10.1016/j.cub.2008.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quintana-Murci L, Barreiro LB. The role played by natural selection on Mendelian traits in humans. Ann. N.Y. Acad. Sci. 2010;1214:1–17. doi: 10.1111/j.1749-6632.2010.05856.x. [DOI] [PubMed] [Google Scholar]
- 70.Mendel G. Versuche über Pflanzenhybriden. Verhandlungen des naturforschenden Vereins Brünn, Volume IV Abhandlungen. 1865:3–47. [Google Scholar]
- 71.Bagheri HC. Unresolved boundaries of evolutionary theory and the question of how inheritance systems evolve: 75 years of debate on the evolution of dominance. J. Exp. Zool. B Mol. Dev. Evol. 2006;306:329–359. doi: 10.1002/jez.b.21069. [DOI] [PubMed] [Google Scholar]
- 72.Veitia RA. A generalized model of gene dosage and dominant negative effects in macromolecular complexes. FASEB J. 2010;24:994–1002. doi: 10.1096/fj.09-146969. [DOI] [PubMed] [Google Scholar]
- 73.Veitia RA, Birchler JA. Dominance and gene dosage balance in health and disease: why levels matter! J. Pathol. 2010;220:174–185. doi: 10.1002/path.2623. [DOI] [PubMed] [Google Scholar]
- 74.Comai L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- 75.Mymrikov EV, Seit-Nebi AS, Gusev NB. Heterooligomeric complexes of human small heat shock proteins. Cell Stress Chaperones. 2012;17:157–169. doi: 10.1007/s12192-011-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fontaine J-M, Sun X, Hoppe AD, Simon S, Vicart P, Welsh MJ, Benndorf R. Abnormal small heat shock protein interactions involving neuropathy-associated HSP22 (HSPB8) mutants. FASEB J. 2006;20:2168–2170. doi: 10.1096/fj.06-5911fje. [DOI] [PubMed] [Google Scholar]
- 77.Simon S, Fontaine J-M, Martin JL, Sun X, Hoppe AD, Welsh MJ, Benndorf R, Vicart P. Myopathy-Associated α B-Crystallin Mutants: Abnormal Phosphorylation, Intracellular Location, and Interactions with other Small Heat Shock Proteins. J. Biol. Chem. 2007;282:34276–34287. doi: 10.1074/jbc.M703267200. [DOI] [PubMed] [Google Scholar]
- 78.Simon S, Dimitrova V, Gibert B, Virot S, Mounier N, Nivon M, Kretz-Remy C, Corset V, Mehlen P, Arrigo A-P. Analysis of the dominant effects mediated by wild type or R120G mutant of αB-crystallin (HspB5) towards Hsp27 (HspB1) PLoS One. 2013;8:e70545. doi: 10.1371/journal.pone.0070545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 80.Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J. Mol. Med. 2003;81:678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 81.Sanbe A. Molecular mechanisms of α-crystallinopathy and its therapeutic strategy. Biol. Pharm. Bull. 2011;34:1653–1658. doi: 10.1248/bpb.34.1653. [DOI] [PubMed] [Google Scholar]
- 82.Ecroyd H, Meehan S, Carver JA. The two-faced nature of small heat shock proteins: Amyloid fibril assembly and the inhibition of fibril formation. Relevance to disease states. In: Simon S, Arrigo A-P, editors. Small stress proteins and human diseases. New York: Nova Science Publishers; 2012. pp. 189–211. [Google Scholar]
- 83.Hung MC, Link W. Protein localization in disease and therapy. J. Cell Sci. 2011;124:3381–3392. doi: 10.1242/jcs.089110. [DOI] [PubMed] [Google Scholar]
- 84.Carra S, Crippa V, Rusmini P, Boncoraglio A, Minoia M, Giorgetti E, Kampinga HH, Poletti A. Alteration of protein folding and degradation in motor neuron diseases: Implications and protective functions of small heat shock proteins. Prog. Neurobiol. 2012;97:83–100. doi: 10.1016/j.pneurobio.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 85.Ackerley S, James PA, Kalli A, French S, Davies KE, Talbot K. A mutation in the small heat-shock protein HSPB1 leading to distal hereditary motor neuronopathy disrupts neurofilament assembly and the axonal transport of specific cellular cargoes. Hum. Mol. Genet. 2006;15:347–354. doi: 10.1093/hmg/ddi452. [DOI] [PubMed] [Google Scholar]
- 86.Zhai J, Lin H, Julien JP, Schlaepfer WW. Disruption of neurofilament network with aggregation of light neurofilament protein: a common pathway leading to motor neuron degeneration due to Charcot-Marie-Tooth disease-linked mutations in NFL and HSPB1. Hum. Mol. Genet. 2007;16:3103–3116. doi: 10.1093/hmg/ddm272. [DOI] [PubMed] [Google Scholar]
- 87.Maloyan A, Sanbe A, Osinska H, Westfall M, Robinson D, Imahashi K, Murphy E, Robbins J. Mitochondrial dysfunction and apoptosis underlie the pathogenic process in alpha-B-crystallin desmin-related cardiomyopathy. Circulation. 2005;112:3451–361. doi: 10.1161/CIRCULATIONAHA.105.572552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goldfarb LG, Vicart P, Goebel HH, Dalakas MC. Desmin myopathy. Brain. 2004;127:23–734. doi: 10.1093/brain/awh033. [DOI] [PubMed] [Google Scholar]
- 89.Irobi J, De Jonghe P, Timmerman V. Molecular genetics of distal hereditary motor neuropathies. Hum. Mol. Genet. 2004;13(Suppl. 2):Rl95–R202. doi: 10.1093/hmg/ddh226. [DOI] [PubMed] [Google Scholar]
- 90.Shy ME. Charcot-Marie-Tooth disease: an update. Curr. Opin. Neural. 2004;17:579–585. doi: 10.1097/00019052-200410000-00008. [DOI] [PubMed] [Google Scholar]
- 91.Almeida-Souza L, Asselbergh B, d'Ydewalle C, Moonens K, Goethals S, de Winter V, Azmi A, Irobi J, Timmermans JP, Gevaert K, Remaut H, Van Den Bosch L, Timmerman V, Janssens S. Small heat-shock protein HSPB1 mutants stabilize microtubules in Charcot-Marie-Tooth neuropathy. J Neurosci. 2011;31:15320–15328. doi: 10.1523/JNEUROSCI.3266-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rajasekaran NS, Connell P, Christians ES, Yan LJ, Taylor RP, Orosz A, Zhang XQ, Stevenson TJ, Peshock RM, Leopold JA, Barry WH, Loscalzo J, Odelberg SJ, Benjamin IJ. Human alpha B-crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell. 2007;130:427–439. doi: 10.1016/j.cell.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.d'Ydewalle C, Krishnan J, Chiheb DM, Van Damme P, Irobi J, Kozikowski AP, Vanden Berghe P, Timmerman V, Robberecht W, Van Den Bosch L. HDAC6 inhibitors reverse axonal loss in a mouse model of mutant HSPB1-induced Charcot-Marie-Tooth disease. Nat. Med. 2011;17:968–974. doi: 10.1038/nm.2396. [DOI] [PubMed] [Google Scholar]
- 94.Brady JP, Garland DL, Green DE, Tamm ER, Giblin FJ, Wawrousek EF. AlphaB-crystallin in lens development and muscle integrity: a gene knockout approach. Invest. Ophthalmol. Vis. Sci. 2001;42:2924–2934. [PubMed] [Google Scholar]
- 95.Sanbe A, Osinska H, Saffitz JE, Glabe CG, Kayed R, Maloyan A, Robbins J. Desmin-related cardiomyopathy in transgenic mice: a cardiac amyloidosis. Proc. Natl. Acad. Sci. USA. 2004;101:10132–10136. doi: 10.1073/pnas.0401900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang L, Min JN, Masters S, Mivechi NF, Moskophidis D. Insights into function and regulation of small heat shock protein 25 (HSPB1) in a mouse model with targeted gene disruption. Genesis. 2007;45:487–501. doi: 10.1002/dvg.20319. [DOI] [PubMed] [Google Scholar]
- 97.Krishnan J, D'YdewaJie C, Dierick I, lrobi J, van den Berghe P, Janssen P, Timmerman V, Robberecht W, van den Bosch L. 38th Annual Meeting of the Society for Neuroscience. Washington, DC: 2008. Nov. Pathogenic mechanisms of HSPB1 mutations associated with distal hereditary motor neuropathies. [Google Scholar]
- 98.Simon S, Arrigo A-P. Beneficial and deleterious, the dual role of small stress proteins in human disease: Implications for the therapeutic strategies. In: Simon S, Arrigo A-P, editors. Small stress proteins and human diseases. New York: Nova Science Publishers; 2012. pp. 457–476. [Google Scholar]
- 99.Liu XL, Doné SC, Yan K, Kilpeläinen P, Pikkarainen T, Tryggvason K. Defective trafficking of nephrin missense mutants rescued by a chemical chaperone. J. Am. Soc. Nephrol. 2004;15:1731–1738. doi: 10.1097/01.asn.0000129826.28932.fd. [DOI] [PubMed] [Google Scholar]
- 100.Neuman MG, Cameron RG, Shear NH, Bellentani S, Tiribelli C. Effect of tauroursodeoxycholic and ursodeoxycholic acid on ethanol-induced cell injuries in the human Hep G2 cell line. Gastroenterology. 1995;109:555–563. doi: 10.1016/0016-5085(95)90345-3. [DOI] [PubMed] [Google Scholar]
- 101.Wei H, Kim SJ, Zhang Z, Tsai PC, Wisniewski KE, Mukherjee AB. ER and oxidative stresses are common mediators of apoptosis in both neurodegenerative and non-neurodegenerative lysosomal storage disorders and are alleviated by chemical chaperones. Hum. Mol. Genet. 2008;17:469–477. doi: 10.1093/hmg/ddm324. [DOI] [PubMed] [Google Scholar]
- 102.Ehrnhoefer DE, Duennwald M, Markovic P, Wacker JL, Engemann S, Roark M, Legleiter J, Marsh JL, Thompson LM, Lindquist S, Muchowski PJ, Wanker EE. Green tea (−)-epigallocatechin-gallate modulates early events in huntingtin misfolding and reduces toxicity in Huntington's disease models. Hum. Mol. Genet. 2006;15:2743–2751. doi: 10.1093/hmg/ddl210. [DOI] [PubMed] [Google Scholar]
- 103.Kraskiewicz H, FitzGerald U. InterfERing with endoplasmic reticulum stress. Trends Pharmacol. Sci. 2012;33:53–63. doi: 10.1016/j.tips.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 104.Sanbe A, Yamauchi J, Miyamoto Y, Fujiwara Y, Murabe M, Tanoue A. Interruption of CryAB-amyloid oligomer formation by HSP22. J. Biol. Chem. 2007;282:555–563. doi: 10.1074/jbc.M605481200. [DOI] [PubMed] [Google Scholar]
- 105.Sanbe A, Daicho T, Mizutani R, Endo T, Miyauchi N, Yamauchi J, Tanonaka K, Glabe C, Tanoue A A. Protective effect of geranylgeranylacetone via enhancement of HSPB8 induction in desmin-related cardiomyopathy. PLoS One. 2009;4:e5351. doi: 10.1371/journal.pone.0005351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bersuker K, Hipp MS, Calamini B, Morimoto RI, Kopito RR. Heat shock response activation exacerbates inclusion body formation in a cellular model of huntington disease. J. Biol. Chem. 2013;288:23633–23638. doi: 10.1074/jbc.C113.481945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gibert B, Simon S, Dimitrova V, Diaz-Latoud C, Arrigo A-P. Peptide aptamers: tools to negatively or positively modulate HSPB1(27) function. Phil. Trans. R. Soc. Lond. B. 2013;368:20120075. doi: 10.1098/rstb.2012.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.