Abstract
Coronavirus discontinuous transcription uses a highly conserved sequence (CS) in the joining of leader and body RNAs. Using a full-length infectious construct of transmissable gastroenteritis virus, the present study demonstrates that subgenomic transcription is heavily influenced by upstream flanking sequences and supports a mechanism of transcription attenuation that is regulated in part by a larger domain composed of primarily upstream flanking sequences which select appropriately positioned CS elements for synthesis of subgenomic RNAs.
Transmissible gastroenteritis virus (TGEV), the causative agent of acute gastroenteritis in swine, is a member of the Coronaviridae family, order Nidovirales (29). TGEV possesses a single-stranded, positive-sense ∼28.5-kb RNA genome that expresses eight large open reading frames (ORFs), which are expressed from full-length or subgenomic-length mRNAs during infection (9, 17). TGEV uses a copy choice discontinuous transcription mechanism for subgenomic mRNA synthesis, resulting in the synthesis of a 3′ coterminal nested set of subgenomic mRNAs that all contain a 5′-proximal leader RNA sequence, which is derived from the 5′ end of the genome (28). Although each mRNA is polycistronic, the 5′-most ORF is preferentially translated, and with the exception of ORF 3b, a distinct mRNA species for each ORF is synthesized (20, 27, 28).
Each of the TGEV ORFs is preceded by a transcription regulatory sequence (TRS) which contains a highly conserved sequence (CS) of six nucleotides (nt) in length (5′-CUAAAC-3′) that functions in the synthesis of each of the subgenomic mRNAs (1, 5, 31). This same CS sequence is located within the genomic 5′ leader RNA sequence, suggesting that base pairing between 5′ leader RNAs and TRS regions plays an important role in coronavirus discontinuous transcription (16, 17). With the equine arterivirus (order Nidovirales) cDNA clone, base pairing between the leader and body CS regions was required for efficient subgenomic transcription (33). In addition, the stability of the leader-body RNA CS interaction was recently shown to be an important factor in the regulation of subgenomic mRNA transcription (23, 24). However, it has become increasingly clear that genomic location and CS flanking sequences are also important and that the simple insertion of CS sequences may not be sufficient in initiating subgenomic transcription (2, 3, 12-14, 18, 19, 22, 32, 34). In addition, a number of noncanonical transcription start sites have been noted within the green fluorescent protein (GFP) gene cloned into the mouse hepatitis virus and TGEV genomes and cannot be explained by a simple base-pairing model (8, 10). Additionally, transcription utilizing noncanonical CS sequences has been demonstrated for mouse hepatitis virus, bovine coronavirus and arteriviruses (13, 22, 26). As a result, the exact sequence requirements for coronavirus subgenomic transcription remain unknown, especially in the context of genome-length RNAs.
Until recently, a number of technical barriers have prevented the generation of a coronavirus infectious cDNA clone, including sequence toxicity and the large genome size (1, 35). Consequently, the major observations concerning the structure and function of coronavirus TRS sequences in discontinuous transcription have been made by using defective interfering (DI) RNAs that are significantly smaller than wild-type genomic RNAs and are assembled from different regions of the viral genome. These RNAs require a helper virus for their replication and replicate faster than full-length genomic RNAs. Because DI RNAs are not authentic with respect to an intact genomic RNA, it is possible that findings using DI RNAs may differ in the context of the complete genome. In this study, we sought to identify the TGEV N gene TRS unit sequence required for driving efficient subgenomic transcription within the context of a full-length genomic RNA of TGEV.
Construction of recombinant TGEV encoding mutated ORF 3a TRS sequences.
We have recently described the construction of a recombinant TGEV that expresses GFP from the ORF 3a locus (TGEV-GFP2PflMI) (8, 35). This virus expresses gfp with the ORF 3a CS and 20 nt from the N gene TRS region (including the CS) upstream of the start codon (3a CS-N CS- gfp start). However, the more upstream ORF 3a CS is preferentially used for the synthesis of ORF 3a mRNA from this virus, indicating that the simple insertion of the 20-bp N gene TRS sequence was not sufficient for directing efficient mRNA synthesis from this location. It is possible either that the inserted 20-nt N gene TRS sequence does not represent a functional N gene TRS element or that the upstream ORF 3a TRS suppresses transcription from proximal TRS elements. Using the TGEV-GFP2PflMI construct as a background, we used recombinant DNA approaches to assemble recombinant viruses that contained an active N gene CS for the synthesis of leader-containing transcripts expressing gfp.
To determine whether the ORF 3a TRS present within TGEV-GFP2PflMI suppressed subgenomic mRNA transcription from the inserted 20- nt N gene TRS sequence, we assembled a construct containing the deletion of the ORF 3a CS and 89 nt of 5′ flanking sequence, TGEVmut1 (Fig. 1A). By 18 h posttransfection, GFP expression was observed by fluorescence microscopy in cultures transfected with full-length TGEVmut1 RNA (data not shown). The supernatant was harvested from the transfected culture, and plaque-purified virus stocks exceeded 107 PFU/ml by 24 h postinfection. At ∼12 h postinfection, total cellular RNA was harvested and used as a template for reverse transcription-PCR (RT-PCR) to detect leader-containing mRNA expressing gfp. A GFP leader containing an amplicon of ∼850 bp was isolated that contained leader-body junctions derived from the 20-nt N gene TRS element (nine of nine independent clones sequenced) (Table 1). These data indicate that the inserted 20-nt N gene TRS is functional in the absence of the upstream ORF 3a CS plus 89 nt of 5′ flanking sequence and that use of this 20-nt N TRS sequence is likely suppressed by the presence of a functional ORF 3a TRS in the TGEV-GFP2PflMI recombinant virus. By Western blot analysis, GFP expression was clearly impaired in the TGEVmut1-infected cells compared with the expression in the TGEV-GFP2PflMI parent (Fig. 2).
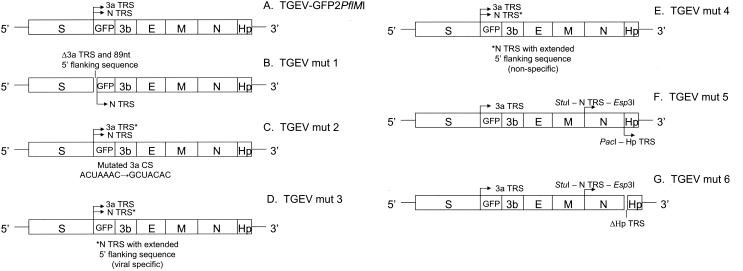
FIG. 1.
TGEV recombinant viruses expressing GFP. A series of TGEV recombinant viruses was generated using the TGEV-GFP2PflMI construct as the backbone for further characterization of the 3a and N gene TRS. (A) The TGEV-GFP2PflMI parent construct contains an ORF 3a deletion and insertion of GFP under the control of the 3a TRS and 20 nt of the N gene TRS. The location of the N and/or ORF 3a TRS sequences upstream of gfp is indicated. Clones were identified by DNA sequencing using an ABI model 377 automated sequencer, and the resulting constructs were subsequently used in the assembly of recombinant TGEV viral cDNAs. (B) TGEVmut1 was constructed by deleting nt 24709 to 24804 of the TGEV genome from TGEV-GFP2PflMI, corresponding to the ORF 3a CS and 89 nt of its 5′ flanking sequence. (C) TGEVmut2 contains the mutation of the upstream ORF 3a CS (ACUAAAC→GCUACAC), such that only the N CS is positioned upstream of gfp. (D) TGEVmut3 contains the 5′ N CS flanking sequence, which has been extended from 8 to 47 nt, (corresponding to nt 26858 to 26904 of the TGEV genome; GenBank accession no. AJ271965) while leaving the upstream ORF 3a CS completely intact. (E) TGEVmut4 contains the insertion of a nonspecific 39-nt sequence just 5′ of the 20 nt N gene TRS. (F) TGEVmut5 was engineered to contain unique StuI and Esp3I restriction sites which flank the CS element upstream of the N gene, and unique BstEII and PacI sites were introduced at the 3′ end of the N gene. The PacI restric-tion site was introduced at nucleotide position 28063 of the TGEV genome by mutagenesis of the Hp gene CS (ACTAAAC→ATTAATTAA). The wild-type Hp CS and gene were then repositioned just downstream of this PacI site, including 10 bp of upstream 5′ flanking sequence (nt 28051 to 28060). (G) To generate TGEVmut6, a ∼500-bp amplicon representing the CS deletion and 5′-truncated Hp gene was generated using the PacI site containing the 5′ primer (5′-TTA ATT AAA CCG GTT CGT CTT CCT CCA TGC TG-3′) and a 3′ primer within the cloning vector (Topo XL TA cloning vector; Invitrogen. Using the unique PacI site, this amplicon was inserted into the TGEVmut5 background to replace the wild-type Hp gene, resulting in a new recombinant TGEV F fragment containing a deletion of the CUAAAC Hp CS and 7 nt of 3′ flanking sequence (nt 28051 to 28074), including the first 4 nt of the Hp ORF. The only two available ATG start codons are out of frame with respect to the Hp ORF at nt positions 28087 and 28186 and would encode a seven- and nine-amino-acid protein, respectively.
TABLE 1.
Origin of leader-containing GFP transcripts
| Recombinant virus | No. of clonesa
|
|
|---|---|---|
| TGEV 3a CS | TGEV N CS | |
| TGEVmut1 | 0/9 | 9/9 |
| TGEVmut2 | 0/10 | 10/10 |
| TGEVmut3 | 1/10 | 9/10 |
| TGEVmut4 | 7/7 | 0/7 |
Number of leader-containing GFP cDNA clones that originated from either the 3a or N CS site/number total.
FIG. 2.
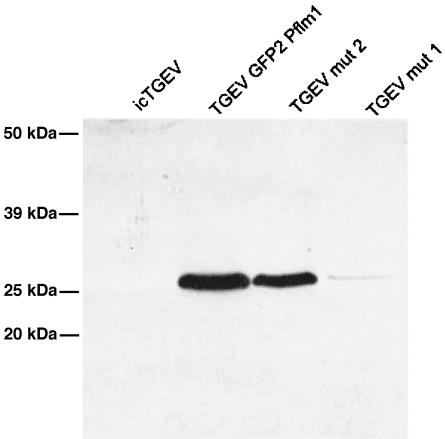
GFP expression from TGEV recombinant viruses. ST cells were infected with wild-type TGEV, TGEVmut2, or TGEVmut3, incubated for 12 h at 37°C, and subsequently lysed. Harvested lysates were subjected to Western blotting with monoclonal antisera directed against GFP (Clontech) or the TGEV N protein. Murine antisera against the TGEV N protein were raised by immunizing mice with Venezuelan equine encephalitis virus replicon vectors encoding the TGEV N gene.
To further examine N gene TRS functionality and to determine whether the ORF 3a TRS was suppressing transcription from the 20-nt N gene TRS, the ORF 3a CS sequence in the TGEVmut2 construct was mutated (ACUAAAC→GCUACAC), such that only a perfect N gene CS sequence was positioned upstream of gfp (Fig. 1B). In this construct, only the ORF 3a CS has been mutated, while the 5′ and 3′ flanking sequences were not modified. GFP expression was evident in TGEVmut2-transfected cultures by fluorescence microscopy at ∼18 h posttransfection (data not shown), and infectious virus was recovered that grew in swine testicular (ST) cells to titers approaching 108 PFU/ml. Total cellular RNA was harvested ∼14 h postinfection and subjected to RT-PCR for detection of subgenomic mRNA expressing gfp. Sequence analysis of leader-containing gfp amplicons revealed that mutation of the ORF 3a CS activated the downstream N gene TRS element (10 of 10 independent clones) (Table 1). These data demonstrate that the 20-bp N TRS sequence is functional, but only in the absence of the upstream ORF 3a CS. Consistent with these studies and following normalization to N protein expression levels (data not shown), deletion of the ORF 3a CS site and 5′ flanking sequence in the TGEVmut1 virus resulted in about 70 to 80% less GFP expression than with TGEV-GFP2PflMI. Under conditions of an intact upstream 3a TRS region but CS knockout, the TGEVmut2 virus expressed about 22% less GFP than TGEV-GFP2-PflMI, as determined by Western blot analysis (Fig. 2).
In a previous study using TGEV-derived minigenomes, transcription levels from the N gene TRS were significantly enhanced by the extension of CS 5′ flanking sequence (2). Therefore, we sought to determine if any suppressing effect from the ORF 3a TRS might be overcome by increasing the N CS 5′ flanking sequence. Consequently, we assembled the recombinant virus TGEVmut3 in which the 5′ N CS flanking sequence has been extended from the 8 nt in TGEV-GFP2-PflMI to 47 nt (26858 to 26904) while leaving the upstream ORF 3a TRS completely intact (Fig. 1C). To control for the increased spacing between the 3a and N CS sequences, we also constructed a TGEVmut4 control, which contained the insertion of a nonspecific 39-nt sequence just 5′ of the 20-nt N gene TRS element (Fig. 1D). By 18 h posttransfection, GFP expression was observed by fluorescence microscopy in the TGEVmut3-transfected cultures but not in the TGEVmut4-transfected cultures (data not shown). Supernatants were harvested from both transfections and passed onto fresh cultures of ST cells, and infectious virus was isolated. At ∼12 to 18 h postinfection, total cellular RNA was harvested and used as a template for RT-PCR to detect leader-containing mRNA expressing gfp. Amplicons of ∼850 bp were isolated from both cultures (data not shown), cloned, and subsequently sequenced to determine which TRS was utilized in these two viruses. Subgenomic mRNA transcription was almost exclusively initiated from the N gene CS in the TGEVmut3 recombinant virus (9 of 10 independent clones), while subgenomic mRNA synthesis was exclusively initiated from the ORF 3a CS of TGEVmut4 (7 of 7 independent clones) (Table 1). In contrast to transcription profiles noted in the TGEV-GFP2PflMI virus, upstream flanking sequences activated the N CS even in the presence of the ORF 3a CS. This N gene TRS element was comprised of the 5′-CUAAAC-3′ consensus sequence, 6 nt of 3′ flanking sequence and 47 nt of 5′ flanking sequence. Furthermore, these data show that the activation of the N CS in TGEVmut3 was not simply a result of increasing the distance between the two TRS elements. Rather, these data are consistent with the hypothesis that CS location in relationship to the flanking sequence context is central to the regulation of subgenomic RNA transcription.
Studies using DI RNAs engineered to encode two closely positioned CS sequences have demonstrated that downstream CS sequences suppress subgenomic mRNA transcription from upstream CS sequences and that upstream CS sequences have little or no effect on downstream CS sequences (12, 14, 15, 34). Additionally, it is apparent that the nature of the flanking sequences themselves and not any secondary structures may be the primary determinant of the site for leader fusion (2, 22). These data are most consistent with transcription attenuation during negative-strand synthesis, where the TRS acts as an attenuator, and also explain why the smaller subgenomic mRNAs are generally produced in larger quantities than the larger subgenomic mRNAs (4, 25). Data from the present study are in partial agreement with these previous DI studies in that they demonstrate the preferential use of a downstream TRS CS site over a closely positioned upstream CS site (TGEVmut3 data). However, data from this study also indicate that these relationships are potentially more complex, as we have demonstrated that a closely positioned upstream CS can attenuate transcription from a downstream CS.
We believe that the ORF 3a CS flanking sequences are optimally positioned to interact with the upstream TRS site. By position context, the 3a CS site is a preferred site for discontinuous transcription compared with the inserted ∼20-nt N TRS, which is only optimally recognized when the normal 3a CS site is disrupted. Consonant with these findings, positional effects were overcome by the extension of the N CS 5′ flanking sequence from 8 to 47 nt, which also has been shown to increase N TRS transcription levels in TGEV minigenomes (2). We believe that this phenomenon was observed because the addition of N CS upstream flanking sequences in TGEVmut3 reconstituted a functional N TRS site, which preferentially recognized the position-proximal N CS for discontinuous transcription. If the model is correct, the upstream TRS sequences likely select the optimal CS sequence that participates in site-assisted discontinuous subgenomic transcription.
Construction of recombinant TGEV encoding mutated Hp TRS sequences.
Our data, as well as those from previous reports (3, 12, 13, 19, 22, 32, 34), demonstrate the importance of flanking sequences in CS function in subgenomic mRNA transcription. We and others have hypothesized that the function of the CS is to serve as an efficient sequence for site-assisted recombination during discontinuous RNA transcription. If the TRS regulates transcription attenuation, then disruption of a normal CS sequence should reveal inefficiently positioned and utilized CS sites that are in close proximity to the TRS. To test this possibility, we altered a second TRS site at another genome location. We reorganized the N and Hp CS sequences located at the 3′ end of the genome by inserting unique restriction sites that flanked the N and Hp TRS elements but did not disrupt the surrounding ORFs. In the wild-type TGEV genome, the N gene stop codon and Hp CS overlap at nucleotide positions 28063 to 28065. In TGEVmut5, the N CS was flanked with unique StuI and Esp3I sites, while a unique PacI site was positioned just downstream of N (Fig. 1E). The N gene stop codon was positioned within the engineered PacI site (nucleotide position 28063), and the Hp CS and flanking sequences (beginning at position 28051) were reengineered downstream of this restriction site. The result is the reengineering of the Hp TRS to a more downstream location within the TGEV genome. Recombinant TGEVmut5 cDNAs were constructed, and full-length capped, polyadenylated RNAs were generated in vitro and transfected into BHK cells as previously described (8, 35). Following electroporation of full-length transcripts, we rapidly identified recombinant TGEVmut5 viruses that expressed GFP and replicated to titers of about 107 by 19 h postinfection (data not shown). These data demonstrate that the TRS reorganization does not necessarily interfere with efficient TGEV replication and will allow for the precise removal of the Hp CS and the mutagenesis of cis-acting sequences required for TGEV replication. TGEVmut5 should also provide a means to test sites around the N and Hp genes as heterologous gene expression sites.
To determine if CS deletion results in the induction of new leader/body junction sites, the Hp CS sequence and 7 nt of 3′ flanking sequence were deleted from TGEVmut5 by primer-mediated mutagenesis, resulting in the construct TGEVmut6 (Fig. 1F). In addition to deletion of the Hp CS, the first 4 nt of the Hp gene were removed, including the ATG start codon, in order to address whether Hp is critical for TGEV replication. Recombinant TGEVmut6 cDNA was constructed, and full-length capped, polyadenylated RNAs were generated in vitro and transfected into BHK cells. Infectious virus was isolated that grew to titers ∼2 logs lower than those of either the parental TGEVmut5 or wild-type TGEV viruses (data not shown), demonstrating that Hp is not required for TGEV replication in vitro but that the absence of this 3′-terminal ORF/transcriptional unit is somewhat detrimental to virus replication. Similar findings have been reported by other laboratories (21). GFP expression was observed by fluorescence microscopy in TGEVmut6-infected ST cell cultures by 18 h postinfection (data not shown). Importantly, while Hp expression was apparent by immunofluorescence assay with rabbit anti-Hp polyclonal antiserum in cultures infected with both wild-type TGEV and TGEVmut5, expression was not evident in cultures infected with TGEVmut6 (data not shown).
Intracellular RNA was isolated from infected cultures at ∼12 h postinfection and analyzed by RT-PCR with a primer pair located within the leader RNA sequence and at the very 3′ end of the TGEV genome. As expected, an amplicon corresponding to leader-containing N gene transcripts was obtained from wild-type TGEV, TGEV-GFP2PflMI, TGEVmut5, and TGEVmut6 virus-infected cultures (Fig. 3A). Interestingly, an amplicon corresponding to leader-containing Hp transcripts was also obtained from each of these cultures, albeit to a lesser quantity in the TGEVmut6-infected cultures, despite the deletion of the Hp CS from the TGEVmut6 genome. To map the precise location of the leader-body junctions of the Hp subgenomic mRNAs, amplicons were cloned and examined by sequence analysis. The leader-body junctions of the transcripts obtained from wild-type TGEV- and TGEVmut5-infected cultures were located at the conserved CS sequence, 5-CUAAAC-3′ (data not shown). However, the leader-body junctions of the transcripts obtained from TGEVmut6-infected cultures varied in their location, ranging from 124 bp upstream to 42 bp downstream of the original Hp CS position (Fig. 3B). Although the consensus Hp CS was not present in the TGEVmut6 genome to allow for complementary base pairing of the subgenomic RNA with the 5′ leader sequence, sequence analysis revealed regions of sequence homology that may have allowed for sufficient base pairing, with as little as a 4-nt overlap (data not shown). These data demonstrate that the conserved CS sequence, 5′-CUAAAC-3′, is not required for Hp subgenomic mRNA synthesis and that Hp CS deletion reveals or activates poorly utilized noncanonical CS sites at nearby locations. Because efficient base pairing between body and leader RNAs is important for efficient production of leader-containing subgenomic mRNAs, the new leader-body junction sites contain various amounts of homology with the leader RNA sequence.
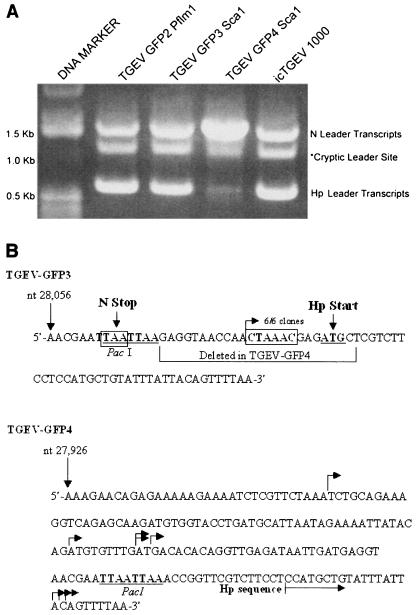
FIG. 3.
RT-PCR and sequence analysis of leader-containing transcripts in TGEV-GFP3- and TGEV-GFP4-infected cells. Cultures of ST cells were infected with TGEV-GFP2, -GFP3, or -GFP4 or wild-type TGEV derived from the infectious construct (icTGEV 1000) at a multiplicity of infection of sim for 1 h at room temperature. Intracellular RNA was harvested ∼12 h postinfection and used as a template for RT-PCR with the 5′ leader-specific primer TGEV-L (5′-CACTATTAGACTTTTAAAGTAAAGTGAGTGTAGC-3′) and a 3′ primer specific to the 3′ terminus of the TGEV genome (TGEV 3′ end; 5′-NNNNNNGCGGCCGCTTTTTTTTTTTTTTTTTTTTTTTTTGGTGTATCACTATCAAAAGG-3′). (A) Agarose gel electrophoresis of RT-PCR amplicons representing leader-containing subgenomic mRNA. RT-PCRs were run on 0.8% agarose gels, and amplicons of ∼1.5 kb and 600 bp were isolated, corresponding to leader-containing transcripts encoding N and Hp, respectively. A third amplicon of ∼1.3 kb was isolated that represents leader-containing transcripts driven from a cryptic start site. (B) Sequence analysis of RT-PCR amplicons from TGEV-GFP3- and TGEV-GFP4-infected cells. The ∼600 bp RT-PCR amplicons from TGEV-GFP3- and GFP4-infected cells shown in panel A were cloned and sequenced to determine the leader-body junctions. Leader-containing transcripts encoding Hp were exclusively initiated from the CS present just upstream of the Hp ORF in TGEV-GFP3 (six of six independent clones sequenced). Subgenomic mRNA synthesis was initiated from a variety of locations in TGEV-GFP4, ranging 124 nt upstream and 42 nt downstream from the original CS location in TGEV-GFP3 (eight independent clones sequence, each indicated by an arrow).
The widely accepted transcription attenuation model hypothesizes that subgenomic RNAs are synthesized during negative-strand synthesis (25). Subgenomic negative-strand synthesis is regulated by a functional TRS, possibly in conjunction with bound transcription factors and 3′-end proximity (6, 25, 27, 30, 36). Data from this study and others suggest that the TGEV TRS is a composite of regulatory domains (RD) and a CS site (Fig. 4) (2, 11, 23). The RD is primarily localized in a ∼50-nt domain upstream of the CS site and displays little obvious homology with 5′ leader RNA flanking sequences. While 3′ flanking sequence also contribute (22), our study is not well designed to measure the contribution of these elements in subgenomic transcription. In this model the CS region serves as a target for site-assisted recombination during discontinuous transcription. Given the number of cryptic CS sites identified following deletion of the HP CS site, transcription attenuation likely generates a gradient pool of truncated incomplete nascent negative-strand RNAs that terminate in and around the TRS body CS site. Those RNAs that most efficiently base pair with the complementary leader RNA CS sequences are positively selected and function as templates for primer extension and synthesis of complete negative-strand templates. Such a mechanism would explain the high variability in the number of cryptic leader-body CS junctions noted during coronavirus transcription, the identification of cryptic CS sites following Hp CS inactivation, and the debilitating effects of CS mutations on discontinuous transcription (7, 8, 26, 37, 38, 39). Importantly, nascent incomplete negative-strand RNAs might require exonuclease processing for proper alignment of complementary CS alignments and discontinuous transcription of the subgenomic negative-strand RNAs, an intriguing possibility since coronavirus phylogenetic analyses suggest that ORF 1b encodes homologues of cellular RNA processing enzymes (29).
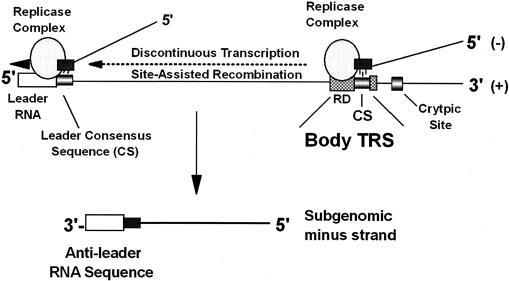
FIG. 4.
Model for the TGEV subgenomic TRS. The genomic RNA is the principle template for the synthesis of full-length and subgenomic-length negative-strand RNAs. Regulation of subgenomic negative-strand synthesis is mediated in part by proximity to the 3′ end of the genome and the presence of a TRS. The TRS is divided into RD and CS, both of which participate in discontinuous transcription. The regulatory domain of the TRS signals transcription attenuation, and the CS site allows for site-assisted recombination between complementary CS sequences located in incomplete nascent negative-strand RNAs and in the leader CS sequences encoded at the 5′ end of the genome. The nascent negative-strand RNAs are extended into complete negative strands by acquisition of anti-leader RNA sequences. In the absence of proximal body CS sites, nearby cryptic sites are inefficiently used in discontinuous transcription of subgenomic negative-strand RNAs.
The nature of the upstream TRS regulatory domain is unknown but may include an undetermined secondary sequence and structure that includes 3′ sequence elements, genome-wide TRS network signaling, and/or regions that bind specific viral or cellular proteins. Little conservation is noted upstream of each body CS site, suggesting that higher-order structures or undescribed protein binding sites may regulate discontinuous transcription. This regulatory domain likely signals transcription attenuation and positively selects among closely positioned CS sequences for those sites that function in efficient site-assisted recombination during the synthesis of complete subgenomic negative strands. Perhaps it is not surprising that a variety of different CS sequence motifs have been noted as sites for subgenomic transcription among the coronaviruses. Given this model, the actual CS sequence may not be so critical, with only efficient networking between homologous leader and body CS sequences needed during discontinuous transcription of subgenomic minus-strand RNAs. Not surprisingly, efficient coronavirus and arterivirus subgenomic transcription is maintained when both leader and body CS sites are coordinately altered, but not singly (39). Proximity to the TRS regulatory domain is also critical for subgenomic transcription. In the case of wild-type virus, the CS site that is proximal to the upstream RD of the TRS and that is positively selected by base pairing is used during subgenomic negative-strand synthesis. In the absence of a specific CS sequence, however, other nearby sequences are utilized, likely resulting in less-efficient subgenomic RNA synthesis. In addition to the present study, this model is supported by previous studies of bovine coronavirus and equine arteritis virus TRS elements (22-24). It also seems likely that regulatory domains in and around the leader RNA sequence at the 5′ end of the genome also contribute to regulation of discontinuous transcription.
Acknowledgments
We are grateful to Nancy Davis, Mark Heise, and Steve Bachenheimer for helpful discussions during the course of this study. In addition, we thank Bob Bagnell and Victoria Madden for assistance with fluorescence microscopy.
This work was supported by research grants from the National Institutes of Health (AI23946, GM63228).
REFERENCES
- 1.Almazan, F., J. M. Gonzalez, Z. Penzes, A. Izeta, E. Calvo, J. Plana-Duran, and L. Enjuanes. 2000. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 97:5516-5521. [DOI] [PMC free article] [PubMed]
- 2.Alonso, S., A. Izeta, I. Sola, and L. Enjuanes. 2002. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J. Virol. 76:1293-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An, S., and S. Makino. 1998. Characterizations of coronavirus cis-acting RNA elements and the transcription step affecting its transcription efficiency. Virology 243:198-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baric, R. S., and B. Yount. 2000. Subgenomic negative-strand RNA function during mouse hepatitis virus infection. J. Virol. 74:4039-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, C. M., D. Cavanagh, and P. Britton. 1995. Cloning and sequencing of a 8.4-kb region from the 3′-end of a Taiwanese virulent isolate of the coronavirus transmissible gastroenteritis virus. Virus Res. 38:83-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi, K. S., P. Huang, and M. M. Lai. 2002. Polypyrimidine-tract-binding protein affects transcription but not translation of mouse hepatitis virus RNA. Virology 303:58-68. [DOI] [PubMed] [Google Scholar]
- 7.Chouljenko, V. N., T. P. Foster, X. Lin, J. Storz, and K. G. Kousoulas. 2001. Elucidation of the genomic nucleotide sequence of bovine coronavirus and analysis of cryptic leader mRNA fusion sites. Adv. Exp. Med. Biol. 494:49-55. [DOI] [PubMed] [Google Scholar]
- 8.Curtis, K. M., B. Yount, and R. S. Baric. 2002. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J. Virol. 76:1422-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enjuanes, L., and B. A. M. van der Zeijst. 1995. Molecular basis of transmissible gastroenteritis coronavirus (TGEV) epidemiology, p. 337-376. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, N.Y.
- 10.Fischer, F., C. F. Stegen, C. A. Koetzner, and P. S. Masters. 1997. Analysis of a recombinant mouse hepatitis virus expressing a foreign gene reveals a novel aspect of coronavirus transcription. J. Virol. 71:5148-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiscox, J. A., K. L. Mawditt, D. Cavanagh, and P. Britton. 1995. Investigation of the control of coronavirus subgenomic mRNA transcription by using T7-generated negative-sense RNA transcripts. J. Virol. 69:6219-6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsue, B., and P. S. Masters. 1999. Insertion of a new transcriptional unit into the genome of mouse hepatitis virus. J. Virol. 73:6128-6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeong, Y. S., J. F. Repass, Y. N. Kim, S. M. Hwang, and S. Makino. 1996. Coronavirus transcription mediated by sequences flanking the transcription consensus sequence. Virology 217:311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joo, M., and S. Makino. 1995. The effect of two closely inserted transcription consensus sequences on coronavirus transcription. J. Virol. 69:272-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishnan, R., R. Y. Chang, and D. A. Brian. 1996. Tandem placement of a coronavirus promoter results in enhanced mRNA synthesis from the downstream-most initiation site. Virology 218:400-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai, M. M., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laude, H., D. Rasschaert, B. Delmas, M. Godet, J. Gelfi, and B. Charley. 1990. Molecular biology of transmissible gastroenteritis virus. Vet. Microbiol. 23:147-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao, C. L., and M. M. Lai. 1994. Requirement of the 5′-end genomic sequence as an upstream cis-acting element for coronavirus subgenomic mRNA transcription. J. Virol. 68:4727-4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makino, S., and M. Joo. 1993. Effect of intergenic consensus sequence flanking sequences on coronavirus transcription. J. Virol. 67:3304-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGoldrick, A., J. P. Lowings, and D. J. Paton. 1999. Characterisation of a recent virulent transmissible gastroenteritis virus from Britain with a deleted ORF 3a. Arch Virol. 144:763-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ortego, J., I. Sola, F. Almazan, J. E. Ceriani, C. Riquelme, M. Balasch, J. Plana, and L. Enjuanes. 2003. Transmissible gastroenteritis coronavirus gene 7 is not essential but influences in vivo virus replication and virulence. Virology 308:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozdarendeli, A., S. Ku, S. Rochat, G. D. Williams, S. D. Senanayake, and D. A. Brian. 2001. Downstream sequences influence the choice between a naturally occurring noncanonical and closely positioned upstream canonical heptameric fusion motif during bovine coronavirus subgenomic mRNA synthesis. J. Virol. 75:7362-7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasternak, A. O., E. van den Born, W. Spaan, and E. J. Snijder. 2003. The stability of the duplex between sense and antisense transcription-regulating sequences is a crucial factor in arterivirus subgenomic mRNA synthesis. J. Virol. 77:1175-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pasternak, A. O., E. van den Born, W. J. Spaan, and E. J. Snijder. 2001. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 20:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawicki, S. G., and D. L. Sawicki. 1990. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 64:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaad, M. C., and R. S. Baric. 1993. Evidence for new transcriptional units encoded at the 3′ end of the mouse hepatitis virus genome. Virology 196:190-198. [DOI] [PubMed] [Google Scholar]
- 27.Sethna, P. B., M. A. Hofmann, and D. A. Brian. 1991. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J. Virol. 65:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sethna, P. B., S. L. Hung, and D. A. Brian. 1989. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc. Natl. Acad. Sci. USA 86:5626-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Zeibuhr, L. L. M. Poon, Y. Guan, M. Rozanov, W. J. M. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spagnolo, J. F., and B. G. Hogue. 2000. Host protein interactions with the 3′ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 74:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tung, F. Y., S. Abraham, M. Sethna, S. L. Hung, P. Sethna, B. G. Hogue, and D. A. Brian. 1992. The 9-kDa hydrophobic protein encoded at the 3′ end of the porcine transmissible gastroenteritis coronavirus genome is membrane-associated. Virology 186:676-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Most, R. G., R. J. de Groot, and W. J. Spaan. 1994. Subgenomic RNA synthesis directed by a synthetic defective interfering RNA of mouse hepatitis virus: a study of coronavirus transcription initiation. J. Virol. 68:3656-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Marle, G., J. C. Dobbe, A. P. Gultyaev, W. Luytjes, W. J. Spaan, and E. J. Snijder. 1999. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. USA 96:12056-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Marle, G., W. Luytjes, R. G. van der Most, T. van der Straaten, and W. J. Spaan. 1995. Regulation of coronavirus mRNA transcription. J. Virol. 69:7851-7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yount, B., K. M. Curtis, and R. S. Baric. 2000. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 74:10600-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, X., and M. M. Lai. 1995. Interactions between the cytoplasmic proteins and the intergenic (promoter) sequence of mouse hepatitis virus RNA: correlation with the amounts of subgenomic mRNA transcribed. J. Virol. 69:1637-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, X., and M. M. Lai. 1994. Unusual heterogeneity of leader-mRNA fusion in a murine coronavirus: implications for the mechanism of RNA transcription and recombination. J. Virol. 68:6626-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, X., and R. Liu. 2000. Identification of a noncanonical signal for transcription of a novel subgenomic mRNA of mouse hepatitis virus: implication for the mechanism of coronavirus RNA transcription. Virology 278:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuniga, S., I. Sola, S. Alonso, and L. Enjuanes. 2004. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 78:980-994. [DOI] [PMC free article] [PubMed] [Google Scholar]