Abstract
Betaretroviruses exist in endogenous and exogenous forms in hosts that are widely distributed and evolutionarily distantly related. Here we report the discovery and characterization of several previously unknown betaretrovirus groups in the genomes of Mus musculus and Rattus norvegicus. Each group contains both mouse and rat elements, and several of the groups are more closely related to previously known betaretroviruses from nonmurine hosts. Some of the groups also include members from hosts which were not previously known to harbor betaretroviruses, such as the gray mouse lemur (Microcebus murinus) and Seba's short-tailed bat (Carollia perspicillata). Some of the mouse and rat elements possess intact open reading frames for gag, pro, pol, and/or env genes and display characteristics of having retrotransposed recently. We propose a model whereby betaretroviruses have been evolving within the genomes of murid rodents for at least the last 20 million years and, subsequent to (or concomitant with) the global spread of their murid hosts, have occasionally been transmitted to other species.
Endogenous retroviruses are present in the genomes of all vertebrates (5). They are presumed to arise from germ line infection by exogenous retroviruses,although factors controlling endogenization are poorly understood. Endogenous proviruses accumulate mutations while in the germ line but can occasionally escape the germ line and infect other hosts, sometimes following recombination with other endogenous or exogenous retroviruses (5, 20).
The Betaretrovirus genus includes the viruses formerly known as type B and type D retroviruses (33). Betaretroviruses have been discovered in mammalian hosts of wide geographical and evolutionary diversity (Table 1). Mouse mammary tumor virus (MMTV), the prototype type B retrovirus, exists in closely related endogenous and exogenous forms, with variable distribution in both laboratory strains and wild species of mice (6, 7, 11, 14). Jaagsiekte sheep retrovirus (JSRV), enzootic nasal tumor virus (ENTV), and endogenous sheep retrovirus are closely related endogenous and exogenous retroviruses of sheep and goats (9, 12, 35). Type D retroviruses were first discovered in Old World monkeys and include exogenous (simian retrovirus type 1 [SRV-1], SRV-2, and Mason-Pfizer monkey virus [MPMV]) (23, 25, 28) and endogenous (simian endogenous retrovirus [SERV]) (32) forms. Endogenous type D retroviruses have also been discovered in a New World monkey (squirrel monkey retrovirus [SMRV]) (8), mice (Mus musculus type D retrovirus [MusD]) (18), and a metatherian (marsupial) mammal, the Australian common brushtail possum (Trichosurus vulpecula endogenous retrovirus type D [TvERV-D]) (1). PCR approaches, using degenerate primers based on conserved regions of the retroviral pro and/or pol genes, have also been used to detect betaretrovirus-related elements in the genomes of pigs (10, 22), the bower bird, and the stripe-faced dunnart (13), although these elements have not been completely characterized. Many of the endogenous betaretroviruses appear to have entered the genomes of their hosts relatively recently (within the last ∼10 million years) (Table 1). However, no satisfactory explanation as to how betaretroviruses could have become so widely distributed has been presented.
TABLE 1.
Distribution of known betaretroviruses
| Betaretrovirus(es) | Endogenous (En) and/or exogenous (Ex) | Known host(s) | Prehistoric distribution of host(s) | Time of entry into genome of hosta |
|---|---|---|---|---|
| MMTV | En, Ex | Rodents within Mus genus | Africa, Europe, Asia, Southeast Asia | Recent (7,14) |
| JSRV and ENTV | En, Ex | Sheep, goats | Northeast Africa, Southern Europe, Asia | >4-10 MYA (12) |
| SMRV | En | Squirrel monkey (New World) | South America | Recent? (8) |
| TvERV-D | En | Common brushtail possum | Australia | ? (1) |
| MusD | En | Rodents within Mus genus | Africa, Europe, Asia, Southeast Asia | >1-2 MYA? (18) |
| SERV | En | Old World monkeys | Africa, Asia | <9 MYA (32) |
| MPMV, SRV-1, and SRV-2 | Ex | Old World monkeys | Africa, Asia | N/A |
?, time of entry into genome is unknown; N/A, not applicable because retrovirus is exogenous; MYA, million years ago. References are shown in parentheses.
The Muridae family of rodents comprises over 1,300 species and contains approximately one-quarter of all the known mammalian species (21). The family arose 20 to 30 million years ago and rapidly diverged into several (∼17) subfamilies, which are now almost globally distributed (21). Several species of murid rodents are invaluable subjects for laboratory experiments, and the genomes of two murine species—Mus musculus and Rattus norvegicus—have been almost entirely sequenced (34; http://hgsc.bcm.tmc.edu/projects/rat/ [Rat Genome Sequencing Consortium]).
A previous study has investigated the origins of MusD, the type D retrovirus present in the genomes of Mus musculus and closely related members of the Murinae subfamily (18). Here we describe the discovery of multiple groups of betaretroviruses present in the genomes of Mus musculus and Rattus norvegicus. We discuss the possible evolutionary origins of these groups of retroviruses and present the hypothesis that murid rodents are responsible for the current global distribution of betaretroviruses.
MATERIALS AND METHODS
Genome databases.
Initial genome searches were performed with Mus musculus and Rattus norvegicus high-throughput genomic sequences (HTGS) at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov). Subsequent searches and enumeration of copy numbers were performed with the February 2003 version of the MGSCv3 assembly of the C57BL/6J mouse genome and the January and June 2003 assemblies of the rat genome.
BLAST searches.
All searches of genomic DNA databases were performed using either the NCBI BLAST Web server (http://www.ncbi.nlm.nih.gov) or the Network BLAST client server, also available from NCBI. BLAST searches were also performed locally using the Standalone BLAST application.
Pol searches.
We searched the translated mouse genome assemblies, using the tBLASTn program, with the amino acid sequences of a highly conserved region of the Pol proteins of all known betaretroviruses and several class II endogenous retroviruses (the mouse, Chinese hamster, and Syrian hamster intracisternal A-type particles [MIAP, CHIAP, and SHIAP], human endogenous retrovirus K10 [HERV-K10], HERV-HML5, HERV-HML6, and rabbit endogenous retrovirus). The region of the Pol protein used in the searches corresponds to the 246-amino-acid sequence spanning the QWPLTNDKLAAAQQL and FQKLLGDINWLRPYLK motifs of the reverse transcriptase (RT) domain (15) of the MPMV Pol protein (amino acids 940 to 1185 of the sequence corresponding to GenBank accession number NP_056891). Results were retrieved in the hit table format and were parsed using a series of Perl scripts to eliminate partial and redundant matches. The remaining nonredundant nucleotide sequences were used to conduct an all-against-all BLASTn comparison. A single element was selected from any group containing members with >95% identity over their entire lengths.
Sequence retrieval.
Sequences were retrieved either manually using the Entrez server at NCBI or electronically using the EFetch Perl script provided by NCBI (http://www.ncbi.nlm.nih.gov/entrez/query/static/efetch_help.html).
pol nucleotide and Pol amino acid sequence alignment and tree construction.
pol nucleotide and Pol amino acid sequence alignments were performed using ClustalX V1.83 (30) and default parameters. The amino acid sequences of elements containing frameshift mutations were manually reconstructed by comparison with the most closely related intact Pol protein. Phylogenetic trees were constructed from alignments by using the neighbor-joining method within ClustalX and were viewed using Tree Explorer (Koichiro Tamura; http://evolgen.biol.metro-u.ac.jp/TE/TE_man.html).
TM tree.
Where present, transmembrane (TM) sequences were derived from DNA sequences by conceptual translation. The TM region corresponded to the ∼150- to 160-amino-acid region spanning from the cleavage site (RAKR) to the TM domain (LLGPLLCLLLVLSFGPIIF) of the MPMV Env protein (amino acids 391 to 547 of the sequence corresponding to GenBank accession number AAC82575), as described by Bénit et al. (4). Alignment and tree construction were performed as described above.
Primer binding site identification.
For those elements that possess long terminal repeats (LTRs), we attempted to identify the tRNA species used to prime reverse transcription. The 25 nucleotides (nt) immediately adjacent to the 5′ LTR were compared against a database of tRNA sequences (26) by using the BLASTn program of Standalone BLAST, a word size of 7 nt, and a reduced penalty for mismatches (−1). In most cases, the highest-scoring match was assumed to be the priming tRNA.
pol percent identity range and average.
Each subgroup of pol sequences was aligned using ClustalX (see above) and output as a percent identity matrix. The percent identity range gives the lowest and highest percent identities from this matrix, whereas the percent identity average is the average of all the percent identities.
pol and LTR copy numbers.
pol copy numbers were taken from the initial pol nucleotide tree. LTR copy numbers were estimated by conducting BLASTn searches of the mouse and rat genomes with each LTR sequence. Segmented matches were joined if the gap between matching segments was less than 100 nt, and only those matches of greater than 90% of the length of the original LTR were included in the subsequent analysis. Results of all matches to all LTRs were parsed to eliminate redundant matches, and the copy number of each LTR was tallied.
Repeat annotation.
Each pol or LTR sequence was compared with the February 2003 assembly of the mouse genome or the June 2003 assembly of the rat genome by using the BLAT search tool at http://genome.ucsc.edu. The coordinates of the best (and in most cases identical) match were used to parse the repeat annotation (chromOut) files, which were generated using the RepeatMasker program (http://www.repeatmasker.org), for the repeat annotation at that location in the relevant genome and chromosome.
PipMaker alignments and dot plots.
Long alignments were performed using PipMaker (24). Dot plots were generated from the blastz output file returned by PipMaker by using the Perl GD module.
Additional Web-based tools.
Open reading frame (ORF) structures were identified using NCBI's ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/), and translations of nucleotide sequences were performed using the translate tool on the ExPASy molecular biology server (http://ca.expasy.org/tools/dna.html). In cases in which ORFs were interrupted by frameshift mutations or insertions or deletions, relevant ORFs were identified using the tBLASTn and BLASTx functions of the BLAST 2 sequences server (27).
Sequences.
FASTA files of sequences are available upon request.
Accession numbers of retroviral sequences used in BLAST searches and alignments.
Accession numbers of retroviral sequences used in BLAST searches and alignments are as follows: MPMV, AF033815; SRV-1, M11841; SRV-2, M16605; SERV231, U85505; SERV252, U85506; SMRV, M23385; TvERV-D, AF224725 and AF284693 (Env); JSRV, M80216; ENTV, Y16627; MMTV, M15122; MIAP, M17551; MIAP-related element with an envelope gene (MIAPE), M73818;HERV-K10 HML2, M14123; Rous sarcoma virus, AF033808; reticuloendotheliosis virus (REV), X01455;spleen necrosis virus (SNV) Env, M87666; feline retrovirus RD114 (Env), X87829; baboon endogenous virus (BaEV), D10032; gibbon ape leukemia virus, M26927; koala retrovirus, AF151794; Mus musculus endogenous retrovirus (MmERV), AC005743 (nt 112341 to 121005); Mus dunni endogenous virus, AF053745; porcine endogenous retrovirus type A 463H12, AF435966; Moloney murine leukemia virus, AF033811; Mus cervicolor popaeus endogenous virus (McpEV), AF327437;feline leukemia virus, M18247; python endogenous retrovirus, AF500296; murine endogenous retrovirus U1 (Env), AC079043 (nt 96983 to 97459); HERV-H (Env), CAB94192; and HERV-W (Env), AAD14546.2.
Nucleotide sequence accession numbers.
Sequences of new elements from mouse, rat, and other species are located in GenBank under the accession numbers given in Table 2.
TABLE 2.
Features of representative betaretroviruses
| Elementa | Start positionb | Orientationc | Structured | Total length (nt)e | LTR length (nt)f | % Identity of LTRsg | PBSh |
pol copy no.i
|
Identity of pol nucleotidesj
|
LTR copy no.k
|
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | Rat | Range | Avg | Mouse | Rat | ||||||||
| β1 | |||||||||||||
| MmERV-β1_NT_039714 | 479070 | + | Δpol | N/A | N/F | N/F | N/A | 1 | 0 | N/A | N/A | N/A | N/A |
| RnERV-β1_NW_043030 | 1380957 | − | Δgag-Δpro-Δpol | N/A | N/F | N/F | N/A | 0 | 28 | 81-100 | 88 | N/A | N/A |
| RnERV-β1_NW_043429 | 1479228 | + | LTR-Δgag-Δpro-Δpol-Δenv-LTR | 7,544 | 376 | 97.9 | Gln | 0 | 9 | 90-98 | 94 | 0 | 322 |
| RnERV-β1_NW_044437 | 9876667 | + | LTR-Δgag-Δpro-Δpol-Δenv-LTR | 6,301 | 311 | 87.8 | Gln | 0 | 2 | 94 | 94 | 0 | 4 |
| RnERV-β1_NW_044440 | 1868014 | − | LTR-Δgag-Δpro-Δpol-Δenv-LTR | 6,891 | 328 | 90.0 | ? | 0 | 2 | 87-87 | 87 | 0 | 2 |
| β2 | |||||||||||||
| MmERV-β2_AC113463 | 171595 | − | LTR-Δgag-Δpro-Δpol-Δenv-LTR | 9,019 | 1,017 | 91.0 | Lys | 2 | 0 | 83-100 | 90 | 13 | 0 |
| MmERV-β2_AC131667 | 203344 | + | LTR-Δgag-Δpro-pol-env-LTR | 8,983 | 973 | 99.0 | Lys | 5 | 0 | 53 | 0 | ||
| MmERV-β2_NT_039761 | 144979 | + | Δpol | N/A | N/F | N/F | N/A | 6 | 2 | 53-100 | 67 | N/A | N/A |
| RnERV-β2_AC127663 | 166446 | + | LTR-gag-pro-Δpol-Δenv-Δsag-LTR | 9,566 | 1,235 | 98.2 | Lys | 0 | 5 | 97-99 | 98 | 0 | 27 |
| RnERV-β2_NW_043520 | 1658604 | + | Δgag-Δpro-Δpol-Δenv | N/A | N/F | N/F | N/A | 6 | 2 | 71-100 | 83 | N/A | N/A |
| RnERV-β2_NW_043524 | 2808577 | − | Δgag-Δpro-Δpol | N/A | N/F | N/F | N/A | 0 | 19 | 87-100 | 97 | N/A | N/A |
| MMTV (M15122) | N/A | N/A | LTR-gag-pro-pol-env-sag-LTR | 9,901 | 1,328 | 100.0 | Lys | 4 | 0 | 94-99 | 96 | 5 | 0 |
| β3 | |||||||||||||
| MmERV-β3_AC111097 | 134591 | − | LTR-Δgag-Δpro-Δpol-Δenv-LTR | 6,455 | 327 | 98.0 | ? | 2 | 1 | 10 | 2 | ||
| MmERV-β3_AC122238 | 42955 | + | Δgag-Δpro-Δpol-Δenv | N/A | N/F | N/F | N/A | 1 | 2 | 75-89 | 81 | N/A | N/A |
| MmERV-β3_NT_039307 | 16578161 | + | Δgag-Δpro-Δpol-Δenv | N/A | N/F | N/F | N/A | 3 | 0 | N/A | N/A | ||
| RnERV-β3_AC120757 | 9795 | − | LTR-Δgag-Δpro-Δpol-Δenv-LTR | 8,153 | 371 | 84.3 | Arg | 0 | 2 | 20 | 24 | ||
| MmERV-β3_NT_039467 | 12797681 | − | Δpol-Δenv | N/A | N/F | N/F | N/A | 1 | 0 | 80-82 | 81 | N/A | N/A |
| RnERV-β3_AC125695 | 144063 | + | Δgag-Δpro-Δpol-Δenv | N/A | N/F | N/F | N/A | 0 | 2 | N/A | N/A | ||
| ENTV (Y16627) | N/A | N/A | LTR-gag-pro-pol-env-LTR | 7,794 | 373 | N/A | Lys | N/A | N/A | N/A | N/A | N/A | N/A |
| JSRV (M80216) | N/A | N/A | LTR-gag-pro-pol-env-LTR | 7,844 | 395 | N/A | Lys | N/A | N/A | N/A | N/A | N/A | N/A |
| β4 | |||||||||||||
| MmERV-β4_AC102561 | 26199 | − | LTR-Δgag-Δpro-Δpol-Δenv-LTR | 9,358 | 521 | 87.0 | Lys | 2 | 0 | 10 | 0 | ||
| MmERV-β4_AL683829 | 44088 | − | Part Δgag-pro-Δpol-Δenv | N/A | N/F | N/F | N/A | 7 | 0 | 87-100 | 92 | N/A | N/A |
| MmERV-β4_AL805955 | 122014 | + | LTR-gag-pro-pol-env-LTR | 9,338 | 524 | 100.0 | Lys | 1 | 0 | 458 | 0 | ||
| MmERV-β4_AC110500 | 85533 | − | LTR-gag-pro-pol-Δenv-LTR | 9,481 | 558 | 99.0 | Lys | 8 | 0 | 91-97 | 94 | 93 | 0 |
| MmERV-β4_AC124523 | 166212 | − | LTR-Δgag-Δpro-Δpol-Δenv-LTR | 9,453 | 509 | 93.4 | Lys | 14 | 0 | 86-97 | 92 | 56 | 0 |
| MmERV-β4_NT_039539 | 5644121 | − | Δgag-Δpro-Δpol-Δenv | N/A | N/F | N/F | N/A | 10 | 0 | 90-94 | 92 | N/A | N/A |
| MmERV-β4_NT_039643 | 1494486 | + | LTR-Δgag-Δpro-Δpol-Δenv-LTR | 8,980 | 377 | 90.0 | ? | 6 | 0 | 87-100 | 91 | 33 | 0 |
| RnERV-β4_AC106444 | 188975 | + | LTR-Δgag-Δpro-Δpol-Δenv-LTR | 9,137 | 483 | 98.1 | Lys | 0 | 14 | 81-100 | 94 | 0 | 613 |
| RnERV-β4_AC119089 | 33482 | + | LTR-Δgag-pro-Δpol-Δenv-LTR | 9,783 | 500 | 93.6 | Lys | 0 | 4 | 0 | 184 | ||
| RnERV-β4_NW_042829 | 1691663 | − | LTR-Δgag-Δpro-Δpol-Δenv-LTR | 7,968 | 369 | 89.6 | Lys | 0 | 1 | 93-100 | 95 | 0 | 40 |
| RnERV-β4_NW_043168 | 1232374 | + | Δpro-Δpol-Δenv | N/A | N/F | N/F | N/A | 0 | 3 | N/A | N/A | ||
| M murinus_ERV-β4_AC145758 | 225147 | + | LTR-Δgag-Δpro-Δpol-del-Δenv-LTR | 6,703 | 407 | 97.0 | Lys | N/A | N/A | N/A | N/A | N/A | N/A |
| TvERV-D (AF224725) | N/A | N/A | LTR-gag-pro-pol-env-LTR | 8,654 | 376 | N/A | Lys | N/A | N/A | N/A | N/A | N/A | N/A |
| β5 | |||||||||||||
| MmERV-β5_AC098708 | 46744 | − | LTR-Δgag-Δpro-Δpol-Δenv-LTR | 8,805 | 389 | 88.3 | ? | 2 | 0 | 28 | 0 | ||
| MmERV-β5_AC125328 | 58636 | − | LTR-Δgag-Δpro-Δpol-Δenv-LTR | 8,976 | 450 | 91.0 | Lys | 2 | 0 | 76-91 | 86 | 92 | 0 |
| MmERV-β5_NT_039649 | 2846578 | − | LTR-Δgag-Δpro-Δpol-Δenv-LTR | 9,057 | 419 | 94.0 | ? | 2 | 0 | 41 | 0 | ||
| MmERV-β5_NT_039553 | 351307 | − | Δpro-Δpol | N/A | N/F | N/F | N/A | 2 | 0 | 91 | 91 | N/A | N/A |
| RnERV-β5_AC127785 | 12568 | − | LTR-gag-pro-pol-env-LTR | 9,583 | 516 | 100.0 | ? | 0 | 18 | 87-100 | 95 | 0 | 58 |
| RnERV-β5_NW_043324 | 75089 | − | Δpol | N/A | N/F | N/F | N/A | 0 | 4 | 98-100 | 99 | N/A | N/A |
| RnERV-β5_NW_043350 | 1428661 | − | Δgag-Δpro-Δpol-Δenv | N/A | N/F | N/F | N/A | 0 | 2 | 88-91 | 90 | N/A | N/A |
| RnERV-β5_NW_043369 | 1440668 | + | LTR-Δgag-Δpro-Δpol-Δenv-LTR | 9,214 | 502 | 89.7 | Lys | 0 | 2 | 0 | 530 | ||
| RnERV-β5_NW_043819 | 7601422 | + | LTR-Δgag-Δpro-Δpol-Δenv-LTR | 8,698 | 287 | 83.5 | ? | 0 | 6 | 85-90 | 87 | 0 | 14 |
| RnERV-β5_NW_044400 | 1359385 | + | LTR-Δgag-Δpro-?-Δpol-Δenv-LTR | 11,045 | 470 | 85.1 | ? | 0 | 4 | 87-90 | 88 | 0 | 111 |
| SMRV (M23385) | LTR-gag-pro-pol-env-LTR | 8,785 | 456 | N/A | Lys | N/A | N/A | N/A | N/A | N/A | N/A | ||
| CpERV-β5_AC138156 | 36762 | + | LTR-Δgag-del-Δenv-LTR | 4,270 | 363 | 98.9 | Lys | N/A | N/A | N/A | N/A | N/A | N/A |
| β6 | |||||||||||||
| MmERV-β6_NT_039167 | 7343376 | + | LTR-Δgag-Δpro-Δpol-Δenv-LTR | 9,688 | 405 | 94.1 | ? | 1 | 0 | 89-89 | 89 | 3 | |
| MmERV-β6_NT_039210 | 2702259 | + | LTR-Δgag-Δpro-Δpol-Δenv-LTR | 7,461 | 390 | 93.1 | Lys | 1 | 0 | 6 | |||
| MmERV-β6_NT_039424 | 1044685 | − | Δgag-Δpro-Δpol | N/A | N/F | N/F | N/A | 2 | 0 | 88 | 88 | N/A | N/A |
| RnERV-β6_NW_043087 | 9466787 | + | LTR-Δgag-Δpro-Δpol-Δenv-LTR | 8,257 | 434 | 87.8 | ? | 0 | 12 | 83-100 | 88 | 0 | 72 |
| MPMV (AF033815) | N/A | N/A | LTR-gag-pro-pol-env-LTR | 8,155 | 345 | N/A | Lys | N/A | N/A | N/A | N/A | N/A | N/A |
| SERV231 (U85505) | N/A | N/A | LTR-gag-pro-Δpol-Δenv-LTR | 8,393 | 484 | N/A | Lys | N/A | N/A | N/A | N/A | N/A | N/A |
| SERV252 (U85506) | N/A | N/A | Part Δgag-pro-Δpol-Δenv-LTR | 7,113 | 484 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| SRV-1 (M11841) | N/A | N/A | LTR-gag-pro-pol-env-LTR | 8,169 | 346 | N/A | Lys | N/A | N/A | N/A | N/A | N/A | N/A |
| SRV-2 (M16605) | N/A | N/A | LTR-gag-pro-pol-env-LTR | 8,169 | 346 | N/A | Lys | N/A | N/A | N/A | N/A | N/A | N/A |
| β7 | |||||||||||||
| MmERV-β7_BK001485 | N/A | N/A | LTR-gag-pro-pol-LTR | 7,477 | 319 | 100.0 | ? | 60 | 0 | 153 | 0 | ||
| MmERV-β7_AC124426 | 12290 | + | LTR-gag-pro-pol-LTR | 7,492 | 319 | 100.0 | ? | 35 | 0 | 83-100 | 95 | 384 | 0 |
| MmERV-β7_AC124426 | 12290 | + | LTR-gag-pro-pol-LTR | 7,492 | 319 | 100.0 | ? | 35 | 0 | 83-100 | 95 | 384 | 0 |
| MmERV-β7_AC140222 | 58455 | + | LTR-Δgag-Δpro-Δpol-LTR | 7,981 | 377 | 89.0 | Lys | 13 | 0 | 46 | 0 | ||
| MmERV-β7_AC087840 | 3747 | + | LTR-Δgag-Δpro-Δpol-LTR | 6,807 | 359 | 90.0 | ? | 19 | 0 | 60 | 0 | ||
| MmERV-β7_AC091771 | 168351 | − | LTR-Δgag-pro-Δpol-LTR | 6,919 | 460 | 88.3 | ? | 15 | 0 | 19 | 0 | ||
| MmERV-β7_AC114619 | 50293 | + | Part Δgag-Δpro-Δpol | N/A | N/F | N/F | N/A | 1 | 0 | N/A | N/A | ||
| MmERV-β7_AC123949 | 150341 | + | LTR-Δgag-Δpro-Δpol-LTR | 7,204 | 383 | 96.0 | ? | 1 | 0 | 82-100 | 91 | 69 | 0 |
| MmERV-β7_AL772201 | 121304 | + | LTR-Δgag-Δpro-Δpol-LTR | 8,116 | 360 | 92.0 | Lys | 1 | 0 | 46 | 0 | ||
| MmERV-β7-AL807786 | 60768 | + | LTR-Δgag-Δpro-Δpol-LTR | 7,922 | 324 | 92.0 | ? | 1 | 0 | 5 | 0 | ||
| MmERV-β7_NT_039170 | 9539914 | − | LTR-Δgag-Δpro-Δpol-LTR | 8,418 | 394 | 93.3 | ? | 1 | 0 | 67 | 0 | ||
| MmERV-β7_AC125045 | 144547 | + | LTR-Δgag-Δpro-Δpol-LTR | 7,294 | 381 | 88.0 | Asn | 1 | 0 | N/A | N/A | 17 | 0 |
| MmERV-β7_AC130218 | 136369 | − | Part Δgag-Δpro-Δpol | N/A | N/F | N/F | N/A | 31 | 0 | N/A | N/A | ||
| MmERV-β7-AL683829 | 28723 | − | LTR-Δgag-Δpro-Δpol-LTR | 7,237 | 377 | 80.0 | ? | 1 | 0 | 110 | 0 | ||
| MmERV-β7_NT_039185 | 2816314 | − | LTR-Δgag-Δpro-Δpol-LTR | 10,468 | 364 | 91.5 | ? | 2 | 0 | N/A | N/A | 2 | 0 |
| MmERV-β7_NT_039589 | 19547446 | + | Δgag-Δpro-Δpol | N/A | N/F | N/F | N/A | 1 | 0 | N/A | N/A | ||
| MmERV-β7_NT_039170b | 36544295 | + | LTR-Δgag-Δpro-Δpol-LTR | 6,224 | 431 | 88.9 | Lys | 9 | 0 | 21 | 0 | ||
| MmERV-β7_NT_039472 | 8865881 | − | Δgag-Δpro-Δpol-Δenv | N/A | N/F | N/F | N/A | 8 | 0 | 74-100 | 85 | N/A | N/A |
| MmERV-β7_NT_039674 | 5956081 | − | Part Δgag-Δpro-Δpol | N/A | N/F | N/F | N/A | 9 | 0 | N/A | N/A | ||
| MmERV-β7_NT_039684 | 1121143 | + | Part Δgag-Δpro-part Δpol | N/A | N/F | N/F | N/A | 6 | 0 | N/A | N/A | ||
| MmERV-β7_NT_039618 | 1868196 | + | Δpro-Δpol-Δenv | N/A | N/F | N/F | N/A | 1 | 0 | N/A | N/A | N/A | N/A |
| MmERV-β7_NT_039641 | 2392770 | + | Δgag-Δpro-Δpol | N/A | N/F | N/F | N/A | 2 | 0 | N/A | N/A | ||
| MmERV-β7_NT_039719 | 3803644 | − | Part Δgag-Δpol | N/A | N/F | N/F | N/A | 12 | 0 | N/A | N/A | ||
| RnERV-β7_NW_043514 | 810072 | + | Δgag-Δpro-Δpol | N/A | N/F | N/F | N/A | 0 | 1 | N/A | N/A | N/A | N/A |
| RnERV-β7_NW_043214 | 264534 | − | Δgag-Δpro-Δpol | N/A | N/F | N/F | N/A | 0 | 1 | N/A | N/A | N/A | N/A |
| ETnl (M16478) | N/A | N/A | LTR-?-Δpol-?-LTR | 5,528 | 322 | 100.0 | Lys | N/A | N/A | N/A | N/A | 1046 | 0 |
| ETnll (Y17107) | N/A | N/A | LTR-?-Δpol-?-LTR | 5,537 | 319 | 100.0 | ? | N/A | N/A | N/A | N/A | 197 | 0 |
Accession numbers of new elements from mouse, rat, and other species are included in the name of the element; accession numbers of previously known betaretroviruses are in parentheses.
Position of 5′ nucleotide of the pol nucleotide sequence used in the pol alignment. N/A, not applicable.
Orientation of the pol sequence relative to the clone or contig in which it lies. +, the pol gene and the clone or contig are in the same orientation; −, the pol gene and the clone or contig are in opposite orientations.
Part, partial or truncated gene; del, deletion; Δ, gene contains premature termination or frameshift mutations; ?, noncoding region of unknown origin.
Length of full-length element, including complete 5′ and 3′ LTRs. N/A, element lacks LTRs; length could not be determined.
Length of 5′ LTR, N/F, LTRs not found.
Percent identity of the 5′ and 3′ LTRs of the indicated element. Note that percent identity alone cannot be used to infer provirus age due to the possibility of recombination and gene conversion.
tRNA species used to prime reverse transcription. ?, priming tRNA could not be determined; N/A, element lacks LTRs; PBS, primer binding site.
Number of pol elements in the mouse and rat genome assemblies that are most closely related to the indicated element. N/A, nonmouse, nonrat element.
Range and average percent identities of elements that group with the indicated element. N/A, nonmouse, nonrat element or element is sole member of group.
Number of LTRs in the mouse and rat genome assemblies that are most closely related to the indicated element. N/A, nonmouse nonrat element or element lacks LTRs.
RESULTS
Detection of multiple groups of murid betaretroviruses.
We began this study by searching for elements within the mouse genome that were related to the mouse endogenous type D retrovirus, MusD. We soon discovered that numerous groups of retroviruses, which differ in their degree of relatedness to MusD, are present in the mouse genome. Some of the groups we initially discovered were more closely related to nonmouse betaretroviruses and were also found to have relatives in high-throughput sequences of the rat genome. Hence, we embarked on a more thorough investigation of the betaretroviruses in the mouse and rat genomes.
We searched for betaretroviruses in the mouse and rat genomes by conducting tBLASTn searches (i.e., comparing a protein query sequence with the genome translated in all six reading frames) using a ∼246-amino-acid sequence from the RT domains of the Pol proteins of all known betaretroviruses and several class II primate and rodent endogenous retroviruses (see Materials and Methods). Matching sequences were aligned and used to construct a neighbor-joining tree. This initial tree, based on nucleotide sequences, indicated that multiple groups of endogenous betaretroviruses and class II elements are present in the mouse and rat genomes. In this paper, we will focus on those elements which clustered with the betaretroviruses.
All elements grouping with the betaretroviruses were analyzed in more detail. For each element, the sequence of a ∼15.7-kb region,spanning 7.5 kb on either side of the Pol-related sequence, was extracted from GenBank. The resulting sequences were analyzed in terms of their gene contents (the presence or absence of genes for retroviral proteins and the integrity of those genes) and the presence of identifiable LTRs and primer binding sites. Selected elements containing intact or reconstructible ORFs were used in phylogenetic analyses which were performed using pol nucleotide and deduced Pol amino acid sequences. Those elements which were chosen to represent groups of several elements were usually the most intact—in terms of the presence and integrity of ORFs and the presence of LTRs—of their group, although we strived to ensure complete coverage of the betaretrovirus section of the original pol tree. In many cases, this required extensive manual reconstruction of Pol ORFs which contained numerous frameshift mutations. As a consequence, we consider the Pol amino acid tree to be less reliable than the pol nucleotide tree. Trees were also constructed using deduced amino acid sequences corresponding to gag (data not shown) and env (see below) where present.
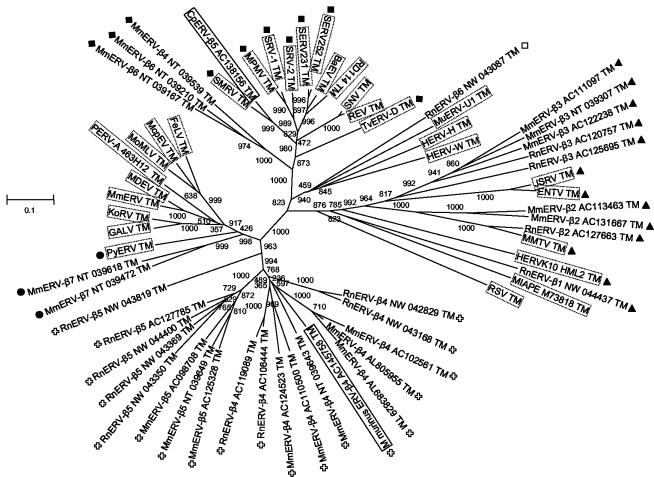
As shown in Fig. 1, multiple groups of betaretrovirus-related pol sequences are present in the mouse and rat genomes. We designated these groups β1 to β7 according to their pol-based phylogenetic relationships to one another and to known betaretroviruses from other species. The branching orders of several of the groups differ between the pol nucleotide and Pol amino acid trees, possibly due to errors introduced during manual reconstruction of mutated Pol ORFs (see above). However, all of the groups contain the same members in both trees and (in most cases) are supported by high bootstrap values (Fig. 1). Two sets of sister groups (namely, β4-β5 and β6-β7) are apparent in both the pol and Pol trees and could arguably be combined. However, we made the arbitrary decision to designate these as separate groups based on the presence of previously known betaretroviruses.
FIG. 1.
Neighbor-joining trees based on alignments of betaretrovirus pol nucleotide (a) and Pol amino acid (b) sequences. The aligned sequences are from the same ∼738-bp or 246-amino-acid region spanning the eight conserved regions of retroviral RT proteins described by Jacobo-Molina and Arnold (15). The trees are rooted on the β1 group of retroviruses. Names of previously known betaretroviruses are surrounded by dashed boxes, and those of newly discovered nonmurine retroviruses are surrounded by solid boxes. Bootstrap values (out of 1,000 replicates) are shown at the nodes. Shapes correspond to those in Fig. 3 and indicate groups determined based on the Env tree.
All of the groups contain both mouse and rat members, and many of the groups are more closely related to previously described betaretroviruses than they are to one another. The majority of the elements in the mouse and rat genomes possess premature termination or frameshift mutations in at least one gene and in most cases in all genes. Characteristics of the members of each group are summarized in Table 2. Notable features of each group are discussed below.
Descriptions of groups β1 to β7. (i) β1.
The β1 group falls outside the large group containing all of the other betaretroviruses in the pol nucleotide tree (Fig. 1a) but lies within a larger group including groups β3 to β7 and excluding group β2, with high bootstrap support, in the Pol amino acid tree (Fig. 1b). We favor the grouping based on nucleotide sequences because of the subjectivity involved in manually determining amino acid sequences from nucleotide sequences which contain frameshift mutations (see above).
The majority of the elements in group β1 fall into four clusters of rat-specific elements, represented by the Rattus norvegicus endogenous retrovirus group β1 element corresponding to accession number NW_043030 (designated RnERV-β1_NW_043030) (28 elements),RnERV-β1_NW_043429 (9 elements), RnERV-β1_NW_044437 (2 elements), and RnERV-β1_NW_044440 (2 elements). The majority of the elements in these clusters possess mutated gag, pro, and pol genes. Although the majority of the members of this group possess remnants of an env gene, few were sufficiently intact to enable inclusion in the TM tree.
A single β1 element, MmERV-β1_NT_039714, is present in the draft mouse genome, and only a mutated pol gene of that element could be detected.
(ii) β2.
The β2 group comprises mouse and rat elements and includes the previously known betaretrovirus MMTV. Several clusters within group β2 are apparent; some of these are species specific, while others contain both mouse and rat elements.
RnERV-β2_NW_043524 represents a cluster of 19 rat-specific elements which are all highly similar and initially appeared to have arisen through a recent replicative burst. However, all of these elements lack LTRs and intact ORFs for gag, pro, or pol, and closer inspection revealed that this group arose through duplication of genomic DNA rather than retrotransposition (data not shown).
MmERV-β2_AC113463a and MmERV-β2_AC131667 belong to a group of seven mouse-specific elements. MmERV-β2_AC131667 possesses intact pol and env ORFs, but its gag and pro genes are interrupted by a small number of premature stop codons and frameshift mutations. Both MmERV-β2_AC113463a and MmERV-β2_AC131667 possess relatively long LTRs (Table 2).
Four endogenous MMTV elements were identified in the C57BL/6J mouse genome. Three of these were full-length, possessing two identical or near-identical LTRs and (largely) intact ORFs. The fourth was an incomplete MMTV from a short contiguous DNA sequence (contig).
The elements most closely related to MMTV are a group of rat elements represented by RnERV-β2_AC127663. This group contains five closely related members, the most intact of which is RnERV-β2_AC127663, with intact gag and pro ORFs, a single terminating mutation in the pol gene, and a frameshift in the env gene (Fig. 2). Not only do these elements group with MMTV based on pol sequences, but they also possess long LTRs (∼1,200 bp) and sag genes, features they share with MMTV. Seventeen solitary LTRs derived from this group of elements are present in the rat genome, and they are all highly (94 to 100%) related to those of RnERV-β2_AC127663 (data not shown). Thus, these elements also appear to have entered the rat genome recently (see Discussion).
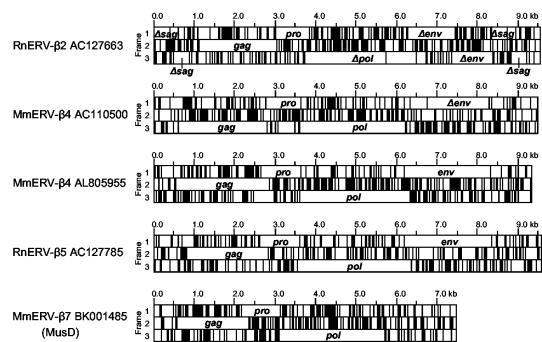
FIG. 2.
ORF structures of selected murine betaretroviruses. The three forward reading frames are shown for each element, with vertical black lines indicating stop codons. The gag, pro, pol, and/or env ORFs from each element are indicated, with Δ indicating the presence of premature termination and/or frameshift mutations.
Searches of nonmouse, nonrat, nonhuman genome survey sequences revealed a β2-related element in a clone (accession number CC563924) from the cow (Bos taurus) genome. This 747-bp clone includes only sequences from the pol gene, which is uninterrupted by mutations. We have named the corresponding provirus BtERV-β2_CC563924, but more of the cow genomic sequence will be required before this endogenous retrovirus can be characterized further.
(iii) β3.
The β3 group comprises two murine clusters, both of which contain mouse and rat elements, as well as the previously known betaretroviruses JSRV and ENTV of sheep and goats. It is a sister group to the β2 elements in the pol nucleotide tree (Fig. 1a) but groups with the β6 and β7 elements (albeit with very low bootstrap support) in the Pol amino acid tree (Fig. 1b). Again, we consider the position in the pol nucleotide tree to be the most likely.
One β3 cluster contains 11 elements and is represented by MmERV-β3_AC111097, MmERV-β3_AC122238, MmERV-β3_NT_039307, and RnERV-β3_AC120757. All of the members of this group have numerous premature stop codons and frameshift mutations. MmERV-β3_AC111097 and RnERV-β3_AC120757 possess identifiable LTRs, and numerous solitary LTRs related to each are present in the mouse and rat genomes (Table 2).
JSRV and ENTV fall inside the β3 group with high bootstrap support in the pol nucleotide tree and moderate bootstrap support in the Pol amino acid tree, although in both cases JSRV and ENTV lie outside the group of mouse and rat elements.
(iv) β4.
Group β4 contains five mouse-specific clusters, two rat-specific clusters, the type D retrovirus TvERV-D from the Australian brushtail possum, and a gray mouse lemur (Microcebus murinus) endogenous retrovirus which we describe for the first time. Several of the members of this group possess intact gag, pro, pol, and/or env ORFs.
RnERV-β4_AC106444 and RnERV-β4_AC119089 belong to a cluster of 18 closely related rat elements—their pol genes are 94% identical, on average (Table 2). All possess LTRs and identifiable retroviral ORFs. Several of the elements have one or more intact ORFs, and the 5′ and 3′ LTRs of many of the elements are highly similar (>97%), a testament to their recent expansion. These elements are also accompanied by a vast excess of solitary LTRs (Table 2).
MmERV-β4_AC110500 belongs to a cluster of eight mouse elements which also appear to have expanded relatively recently. MmERV-β4_AC110500 possesses near-identical LTRs, intact gag, pro, and pol ORFs, and an env gene with three premature stop codons (Fig. 2).
A cluster of 10 mouse-specific elements includes MmERV-β4_AC102561, MmERV-β4_AL683829b, and MmERV-β4_AL805955. The latter possesses intact gag, pro, pol, and env ORFs (Fig. 2) and identical LTRs, suggesting recent retrotransposition. In addition, almost 500 MmERV-β4_AL805955-related LTRs are present in the mouse genome (Table 2), with identity to those of MmERV-β4_AL805955 ranging from 100% down to ∼80%. Most of these are solitary LTRs, although some are associated with a family of LTR retrotransposons present in 15 copies and possessing only remnants of the original MmERV-β4_AL805955 ORFs. Despite the number of MmERV-β4_AL805955-related LTRs in the mouse genome, they are only partially recognized as repeats by the RepeatMasker program (see below and Table 3).
TABLE 3.
Repeat annotation of pol and LTRsa
| Elementb | Repeat annotationc of:
|
|
|---|---|---|
| pol | LTR | |
| β1 | ||
| MmERV-β1_NT_039714 | LTR1_RN-int (1-743, 29.5) | N/A |
| RnERV-β1_NW_043030 | RNERVK9 (1-735, 20.6) | N/A |
| RnERV-β1_NW_043429 | RNERVK9 (1-307, 30.8), RNLTR14-int (295-738, 30.9) | RNLTR14 (1-376, 7) |
| RnERV-β1_NW_044437 | LTR1_RN-int (1-738, 28.9) | N/A |
| RnERV-β1_NW_044440 | RNLTR14-int (1-738, 30.3) | RNLTR14 (38-282, 27.4) |
| β2 | ||
| MmERV-β2_AC113463a | MMTV-int (1-731, 33.5) | RLTR13D2 (1-1017, 7.3) |
| MmERV-β2_AC131667 | ETnERV3 (1-738, 32.5) | RLTR13A3 (1-973, 5.5) |
| MmERV-β2_NT_039761 | RMER16-int (1-736, 31.8) | N/A |
| RnERV-β2_AC127663 | RMER16-int (1-738, 29.6) | RNLTR13 (596-749, 30.9) |
| RnERV-β2_NW_043520 | SRV_RN-int (1-727, 29.3) | N/A |
| RnERV-β2_NW_043524 | RMER16-int (1-736, 29.8) | N/A |
| MMTV (M15122) | N/A | RLTR3_Mm (125-1326, 5.9) |
| β3 | ||
| MmERV-β3_AC111097 | RMER16-int (1-754, 15.4) | RMER16 (1-327, 18.5) |
| MmERV-β3_AC122238 | RMER16-int (1-737, 17.4) | N/A |
| MmERV-β3_NT_039307 | RMER16-int (1-720, 21.9) | N/A |
| RnERV-β3_AC120757 | RMER16-int (1-732, 18.8) | RMER16 (1-371, 11.6) |
| MmERV-β3_NT_039467 | RMER16-int (1-745, 29.6) | N/A |
| RnERV-β3_AC125695 | RMER19B-int (1-707, 33) | N/A |
| ENTV (Y16627) | N/A | N/A |
| JSRV (M80216) | N/A | N/A |
| β4 | ||
| MmERV-β4_AC102561 | MYSERV (1-733, 32.5) | RNLTR3c (5-239, 29.1) |
| MmERV-β4_AL683829b | ETnERV2 (1-734, 31.9) | ETnERV2 (1-159, 15.7), RNLTR10-int (256-319, 23.4) |
| MmERV-β4_AL805955 | RNLTR3c-int (1-738, 33.1) | ETnERV2 (2-131, 26.8), RNLTR3c (253-522, 25.7) |
| MmERV-β4_AC110500 | RLTR13C1-int (1-738, 32.4) | RNLTR3c (471-553, 23.2) |
| MmERV-β4_AC124523 | RNLTR3c-int (1-737, 32.3) | RNLTR3c (1-310, 26.8) |
| MmERV-β4_NT_039539 | RNLTR3b-int (1-733, 33) | N/A |
| MmERV-β4_NT_039643 | SRV_MM-int (1-736, 32.6) | RNLTR3b (66-378, 27.7) |
| RnERV-β4_AC106444 | SRV_RN-int (1-737, 29.9) | RNLTR3a (1-483, 1.4) |
| RnERV-β4_AC119089 | SRV_RN-int (1-735, 31.1) | RNLTR3b (1-500, 6.5) |
| RnERV-β4_NW_042829 | SRV_RN-int (1-738, 31.9) | RNLTR3b (216-369, 25) |
| RnERV-β4_NW_043168 | RNLTR3b-int (1-738, 31.1) | N/A |
| M murinus_ERV-β4_AC145758 | N/A | N/A |
| TvERV-D (AF224725) | N/A | N/A |
| β5 | ||
| MmERV-β5_AC098708 | RNIAP1a (250-381, 23.6), SRV_MM-int (410-752, 29) | RNLTR5 (3-150, 21.3) |
| MmERV-β5_AC125328 | SRV_MM-int (1-733, 33.2) | RLTR8 (4-103, 21.4), RNLTR5 (28-183, 21.7) |
| MmERV-β5_NT_039649 | RLTR19-int (1-730, 34.3) | RNLTR5 (27-159, 18.2) |
| MmERV-β5_NT_039553 | SRV_MM-int (1-736, 33.8) | N/A |
| RnERV-β5_AC127785 | SRV_RN-int (1-738, 31.5) | RNLTR5 (28-168, 19.1) |
| RnERV-β5_NW_043324 | SRV_RN-int (1-615, 30.5) | N/A |
| RnERV-β5_NW_043350 | SRV_RN-int (1-738, 30.5) | N/A |
| RnERV-β5_NW_043369 | SRV_RN-int (1-730, 32.4) | RNLTR5 (1-477, 10.5) |
| RnERV-β5_NW_043819 | RLTR10-int (1-734, 33.3) | SRV_RN-LTR (143-287, 28) |
| RnERV-β5_NW_044400 | SRV_RN-int (1-735, 32.4) | RNLTR5 (27-119, 17.2), RNLTR5 (100-346, 12.7), RNLTR5 (395-470, 13.9) |
| SMRV (M23385) | N/A | N/A |
| CpERV-β5_AC138156 | N/A | N/A |
| β6 | ||
| MmERV-β6_NT_039167 | SRV_MM-int (1-749, 33.1) | RLTR8 (263-396, 25.2) |
| MmERV-β6_NT_039210 | SRV_MM-int (1-754, 33.1) | RLTR8 (277-390, 27) |
| MmERV-β6_NT_039424 | SRV_MM-int (1-732, 7.5) | N/A |
| RnERV-β6_NW_043087 | SRV_RN-LTR-int (1-726, 6) | SRV_RN-LTR (1-434, 7.5) |
| MPMV (AF033815) | N/A | N/A |
| SERV231 (U85505) | N/A | N/A |
| SERV252 (U85506) | N/A | N/A |
| SRV-1 (M11841) | N/A | N/A |
| SRV-2 (M16605) | N/A | N/A |
| β7 | ||
| MmERV-β7_BK001485 | ETnERV2 (1-738, 16.5) | RLTRETN_Mm (1-319, 10.6) |
| MmERV-β7_AC124426 | ETnERV2 (1-738, 14.8) | RLTRETN_Mm (1-319, 9.9) |
| MmERV-β7_AC140222 | ETnERV (1-737, 31.2) | RLTR9E (1-362, 22.9) |
| MmERV-β7_AC087840 | ETnERV2 (1-738, 20.5) | RLTR9E (1-359, 11.4) |
| MmERV-β7_AC091771 | ETnERV2 (1-738, 19.2) | RLTR9E (1-457, 13.6) |
| MmERV-β7_AC114619 | ETnERV2 (1-731, 18.7) | N/A |
| MmERV-β7_AC123949 | ETnERV2 (1-739, 19.2) | RLTR9E (1-381, 8.2) |
| MmERV-β7_AL772201 | ETnERV2 (1-715, 19.1) | RLTR9E (1-360, 21.6) |
| MmERV-β7_AL807786 | ETnERV2 (1-735, 19) | RLTR9E (1-324, 18.6) |
| MmERV-β7_NT_039170 | ETnERV2 (1-739, 19.2) | RLTR9E (1-393, 11.6) |
| MmERV-β7_AC125045 | ETnERV2 (1-739, 19.4) | RLTR9C (1-381, 16.9) |
| MmERV-β7_AC130218 | ETnERV2 (1-740, 20.5) | N/A |
| MmERV-β7_AL683829a | ETnERV2 (1-738, 23.4) | RLTR9D (2-377, 16.4) |
| MmERV-β7_NT_039185 | ETnERV (1-740, 31.2) | RLTR9E (1-177, 34.1) |
| MmERV-β7_NT_039589 | SRV_MM-int (1-759, 30.8) | N/A |
| MmERV-β7_NT_039170b | ETnERV2 (1-729, 21.9) | RLTR9B2 (1-431, 18.2) |
| MmERV-β7_NT_039472 | ETnERV (1-738, 32.2) | N/A |
| MmERV-β7_NT_039674 | ETnERV2 (1-730, 25.9) | N/A |
| MmERV-β7_NT_039684 | SRV_MM-int (1-724, 32.9) | N/A |
| MmERV-β7_NT_039618 | ETnERV2 (1-738, 28.5) | N/A |
| MmERV-β7_NT_039641 | ETnERV2 (1-728, 23) | N/A |
| MmERV-β7_NT_039719 | ETnERV2 (1-737, 18.4) | N/A |
| RnERV-β7_NW_043514 | SRV_RN-int (1-744, 31.9) | N/A |
| RnERV-β7_NW_043214 | SRV_RN-int (1-701, 32.8) | N/A |
| ETnl (M16478) | N/A | RLTRETN_Mm (1-322, 0.9) |
| ETnll (Y17107) | N/A | RLTRETN_Mm (1-319, 10.6) |
Annotation was performed as described in Materials and Methods.
Designations are as described in Table 2, footnote a.
Repeat Masker annotation of pol and LTR sequences. Repeat names are as used by Repbase Update. Sequence ranges (in nucleotides) and percentages of divergence from consensus are shown in parentheses. N/A, not applicable because element is nonmouse, nonrat or lacks LTRs.
Searches of nonmouse, nonrat, nonhuman HTGS revealed a β4-related element in a bacterial artificial chromosome (BAC) (accession number AC145758) from the gray mouse lemur (Microcebus murinus);this BAC is being sequenced as part of the National Institutes of Health Intramural Sequencing Center (NISC) Comparative Vertebrate Sequencing Initiative (http://www.nisc.nih.gov) (29). The provirus of this element (which we have named M_murinus_ERV-β4_AC145758) possesses LTRs which are 97% identical and gag and pro ORFs which are interrupted by only a few frameshift and premature termination mutations. A deletion has removed the 3′ half of the pol gene and the 5′ half of the env gene. The remaining region of pol contains four premature stop codons, whereas the remaining env is uninterrupted. M_murinus_ERV-β4_AC145758 lies within the β4 group with good bootstrap support in both the pol nucleotide and Pol amino acid trees (Fig. 1).
TvERV-D is placed within group β4 with moderate to high bootstrap support in both the pol nucleotide and Pol amino acid trees. However, its deep branching position within the β4 group and its long branch reflect its distant relationship to the mouse and rat β4 elements.
(v) β5.
Group β5 comprises several rat- and mouse-specific clusters, as well as SMRV.
RnERV-β5_AC127785, RnERV-β5_NW_043324, and RnERV-β5_NW_044400 represent smaller clusters within a larger cluster of 26 rat elements. The RnERV-β5_AC127785 cluster of 18 elements has an average pol identity of 95%. Several members of the RnERV-β5_AC127785 group, including RnERV-β5_AC127785 itself, have intact gag, pro, pol, and env ORFs (Fig. 2) and identical or near-identical LTRs, which suggests autonomous and recent replication. In contrast, the members of the RnERV-β5_NW_044400 (four elements) and RnERV-β5_NW_043324 (four elements) clusters possess highly mutated gag, pro, pol, and env ORFs and many lack identifiable LTRs, suggesting ancient retrotransposition events.
The clusters of mouse and rat elements represented by MmERV-β5_NT_039553 and RnERV-β5_NW_043819 appear to be sister groups. These groups fall within the larger β5 group with moderate bootstrap support (65.0%) in the pol nucleotide tree (Fig. 1a) but with only weak support (24.2%) in the Pol amino acid tree (Fig. 1b).
SMRV lies within β5 with moderate to strong bootstrap support in the pol nucleotide and Pol amino acid trees.
(vi) β6.
Group β6 is a relatively small group comprising one cluster of 12 rat elements (represented by RnERV-β6_NW_043087) and two clusters of two mouse elements each (one cluster includes MmERV-β6_NT_039167 and MmERV-β6_NT_039210; the other is represented by MmERV-β6_NT_039424), as well as the exogenous and endogenous type D retroviruses of Old World monkeys. The gag, pro, pol, and env genes of all β6 elements are mutated. The Old World monkey type D retroviruses fall within group β6 with very strong bootstrap support based on both pol nucleotide and Pol amino acid sequences.
(vii) β7.
The β7 elements form the largest murine betaretrovirus group, comprising 229 elements. Only two of these elements are rat elements, and the rest are from the mouse genome. No known betaretroviruses from other species belong to the β7 group.
The only two rat elements belonging to group β7, RnERV-β7_NW_043514 and RnERV-β7_NW_043214, appear to be old insertions. They have mutated gag, pro, and pol genes but no identifiable LTRs.
In contrast, the mouse elements in group β7 have been retrotranspositionally active for some time, and some are still active. Some of the older members of this group (MmERV-β7_NT_039472 and MmERV-β7_NT_039618), which do not have identifiable LTRs and possess highly mutated (and often barely distinguishable) gag, pro, and/or pol ORFs, also possess mutated env genes. However, the majority of β7 elements possess only vestiges of an env gene or lack it completely. Although some clusters are apparent within the β7 group, the clusters are generally poorly defined.
MmERV-β7_BK001485 and MmERV-β7_AC124426 represent a cluster of 78 elements that have retrotransposed recently, as previously reported (3); this group includes the previously identified type D retrovirus MusD (18). The pol genes of this group have an average sequence identity of 95% (Table 2). Both MmERV-β7_BK001485 and MmERV-β7_AC124426 have identical LTRs and intact gag, pro, and pol ORFs. An additional six elements have completely intact gag, pro, and pol ORFs and identical (or nearly identical) LTRs. More than 500 MmERV-β7_BK001485- and MmERV-β7_AC124426-related LTRs reside in the mouse genome (Table 2). Approximately 40% of these are associated with full-length proviruses, another ∼15% are associated with the ETnII family of MusD-derived retroelements (3), and the remainder are solitary LTRs.
The remaining elements in the β7 group all have mutated gag, pro, and pol genes. Generally, those elements which diverge closer to the base of the β7 group (Fig. 1) appear to be older: they contain more mutations in their gag, pro, and pol genes, and their 5′ and 3′ LTRs are either distantly related or unidentifiable (Table 2).
Relationships of env genes.
A neighbor-joining tree was constructed using sequences from a conserved region of the TM domain of the env ORF, where present and/or reconstructible (see Materials and Methods). The TM tree is shown in Fig. 3. In contrast to the trees based on pol nucleotide and Pol amino acid sequences, which appear to represent primarily gradual evolution, the Env tree shows three distinct groups of murine betaretroviruses. One of these groups includes the β1, β2, and β3 groups, as well as the class II endogenous retroviruses MIAPE and HERV-K. A second group comprises the majority of the β4, β5, and β7 elements, as well as the Env proteins of the mammalian type C (gamma) retroviruses. The third group includes individual β4 and β6 elements, as well as all of the type D (and related) retroviruses and an endogenous retrovirus that we discovered in the genomic sequence of Seba's short-tailed bat (Carollia perspicillata; see below). The Env sequences of the rat β6 elements, along with those of HERV-W, HERV-H, and murine endogenous retrovirus U1, are distantly related to this group.
FIG. 3.
Neighbor-joining tree based on alignment of TM amino acid sequences. The aligned sequences are from the region spanning the cleavage site and the TM domain of the Env protein, as described by Bénit et al. (4). Names of previously known retroviruses are surrounded by dashed boxes, and those of newly discovered nonmurine retroviruses are surrounded by solid boxes. Shapes correspond to those in Fig. 1 and indicate groups determined based on the Env tree.
The β2 mouse and rat elements, which cluster with MMTV based on their pol sequences, also group with MMTV based on their Env sequences. Similarly, the β3 group of elements cluster with JSRV and ENTV based on both their pol and Pol and Env sequences.
In the case of groups β4 to β6, it appears that recombination has occurred between the pol and env genes, giving rise to new pol-env combinations. The β4 and β5 groups of murine endogenous retroviruses are sister clades in the Env tree as they are in the pol and Pol trees. This suggests that a single recombination event gave rise to the pol-env combination of the common ancestor of the β4 and β5 groups. One β4 element, MmERV-β4_NT_039539, has undergone an additional recombination event, during which it has acquired a β6-related env gene (Fig. 3).
The env genes of the β6 elements appear to have been acquired through two independent recombination events. The mouse elements MmERV-β6_NT_039167 and MmERV-β6_NT_039210 have env genes which cluster with those of the type D retroviruses, as they do in the pol and Pol trees. The rat β6 elements are the murine betaretroviruses that are most closely related to the type D retroviruses of Old World monkeys on the basis of their pol genes, whereas the mouse β6 elements are more closely related to these retroviruses on the basis of their env genes (Fig. 3).
The only group β7 elements with sufficient Env sequences to include in alignments, MmERV-β7_NT_039472 and MmERV-β7_NT_039618, cluster with the gammaretroviruses (gibbon ape leukemia virus, koala retrovirus, MmERV, Mus dunni endogenous virus, porcine endogenous retrovirus type A, Moloney murine leukemia virus, McpEV, and feline leukemia virus) and an unclassified python retrovirus.
One of the most interesting features of the Env tree is the relationship of the type D group members (MPMV, SRV-1 and SRV-2, SERV251, SMRV, TvERV-D, BaEV, RD114, SNV, and REV) to one another and to the murine betaretroviruses. Whereas the type D retroviruses are placed within or interspersed with groups β4, β5, and β6 based on their pol sequences (Fig. 1), they form a tight cluster with one another, to the exclusion of all murine retroviruses, based on their Env sequences. This suggests that the type D envelope may have been acquired from a nonmurine, and possibly nonmurid, host (see Discussion).
In an attempt to identify the origin of the type D group env gene, we conducted tBLASTn searches using the amino acid sequences of several members of this group against the nonmouse, nonrat, nonhuman genome survey sequence and HTGS databases at NCBI. The highest-scoring match was with a BAC sequence from Seba's short-tailed bat (Carollia perspicillata), which is being sequenced as part of the NISC Comparative Vertebrate Sequencing Initiative. This env gene, which belongs to an endogenous retrovirus which we have named CpERV-β5_AC138156, is most closely related to that of SMRV (Fig. 2). CpERV-β5_AC138156 is an incomplete provirus which possesses almost (98%) identical 363-bp LTRs but has a large deletion which removes approximately one-third of the gag gene (at the 3′ end), the entire pro and pol genes, and approximately 1/10 of the env gene (at the 5′ end). What remains of the gag ORF corresponds to 485 amino acids and has the highest identity to the Gag protein of SMRV (49% identity). The 514 amino acids of the Env protein of CpERV-β5 are 68% identical to the corresponding sequence of the SMRV Env protein. Thus, it appears that SMRV and CpERV-β5_AC138156 share a recent common ancestor (see Discussion).
Common insertions in the mouse and rat genomes.
We attempted to identify insertions of betaretrovirus elements at the same positions in the mouse and rat genomes. In general, the betaretroviruses we discovered formed species-specific clusters, suggesting expansion after the mouse-rat split. Two exceptions to this general rule were two β2 elements and a group of β3 elements.
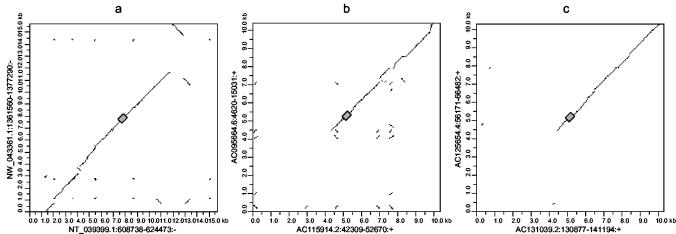
The two β2 elements (MmERV-β2_NT_039339 andRnERV-β2_NW_0433361) are not represented in the pol and Pol trees in Fig. 1, but they group with MmERV-β2_NT_039761. It was apparent from the initial pol tree derived from all mouse and rat pol sequences that these two elements were relatively closely related and that they grouped together to the exclusion of all other mouse and rat elements. Both elements include only a pol-related region and a short region with similarity to the gag gene, but we were unable to detect any homology to other retroviral genes or identify LTRs. However, alignment of the pol regions and flanking sequences showed that these loci display similarity over a large range in the mouse and rat genomes (Fig. 4a), suggesting that these elements represent remnants of a β2 retroviral insertion which occurred prior to the mouse-rat split.
FIG. 4.
Dot plots of aligned common insertions in mouse and rat betaretroviral elements. Alignments and dot plots were generated as described in Materials and Methods. In all cases, the mouse and rat elements are on the horizontal and vertical axes, respectively. The grey boxes represent the retroviral regions. (a) Flanking sequences of 7.5 kb on either side of the pol region of MmERV-β2_NT_039339 and RnERV-β2_NW_0433361. (b) Flanking sequences of 5 kb on either side of RMER16 LTRs in mouse (accession number AC115914; horizontal axis) and rat (accession number AC095664; vertical axis) genomes. (c) Data in this panel are the same as described for panel b with mouse (accession number AC131039; horizontal axis) and rat (accession number AC125654; vertical axis) RMER16 LTRs. Notations along axes are accession numbers followed by the coordinates of the first and last nucleotides of the aligned sequence in the contig or clone and the orientation of the sequence relative to the contig or clone. +, element and contig or clone are in the same orientation; −, element and contig or clone are in opposite orientations.
The β3 group represented by MmERV-β3_AC111097, MmERV-β3_AC122238, MmERV-β3_NT_039307, andRnERV-β3_AC120757 is a cluster of six mouse and five rat elements which are interspersed with one another. We were unable to identify any common β3 insertions based on the locations of their pol genes in the genomes of mouse and rat. However, one mouse β3 element (MmERV-β3_AC111097) and one rat element (RnERV-β3_AC120757) possessed identifiable LTRs, both of which bore closest similarity to the RMER16 LTR in Repbase (16). Although only 30 and 26 RMER16-related LTRs could be identified in the mouse and rat genomes, respectively, by using BLASTn with default parameters, we were able to detect 190 and 168 RMER16 LTRs in the mouse and rat HTGS by using discontiguous MegaBLAST, which is designed to detect more-diverged sequences. We constructed a neighbor-joining tree based on the alignment of these sequences, and it resembled that of the pol sequences of this group in that mouse and rat LTRs were interspersed with one another and few species-specific clusters were observed (data not shown). Comparison of the mouse RMER16 LTRs and their flanking regions with their rat counterparts (see Materials and Methods) revealed several apparent common insertions. Two of these are shown in Fig. 4b and c. In both of the cases shown in Fig. 4b and c, the mouse and rat LTRs diverged by 12% (gaps were ignored), a figure which corresponds to that observed by others (34) for mouse-rat sequence divergence. That so few common insertions were found among so many insertions in each genome and the interspersed nature of the elements in both the pol and LTR trees suggest that these elements were active just prior to, during, and after the mouse-rat split.
Repeat annotation.
We determined the RepeatMasker annotation of the pol and LTR regions of the mouse and rat betaretroviruses as described in Materials and Methods (Table 3). In general, those groups with large numbers of closely related members are well annotated, presumably because they are more readily detected by repeat-seeking programs. Such elements match, over their entire lengths and with low levels of divergence from consensus, repeats in the Repbase Update database. Examples include the LTRs of RnERV-β1_NW_043429, MmERV-β2_AC113463, MmERV-β2_AC131667, MmERV-β3_AC111097, RnERV-β3_AC120757, RnERV-β4_AC106444, RnERV-β4_AC119089, RnERV-β6_NW_043087, and many members of the β7 group and the pol regions of MmERV-β3_AC111097, MmERV-β3_AC122238, RnERV-β3_AC120757, MmERV-β6_NT_039424, RnERV-β6_NW_043087, and many members of the β7 group. Those groups that contain few and/or distantly related members are less well annotated.
Although the majority of the pol elements in the genomes of mice and rats are annotated as repeats, most of them show high levels of divergence from consensus. This suggests that although the murine betaretroviruses are recognized as being of retroviral origin, the annotation of mouse and rat betaretroviruses is currently incomplete. Consequently, some elements are assigned to groups to which they are only distantly related.
Many of the LTRs are only partially annotated. The most striking example of this is the LTRs of MmERV-β4_AL805955. The mouse genome assembly contains almost 500 MmERV-β4_AL805955 LTRs (Table 2) with an average sequence identity of 87%, and yet these LTRs are not assigned their own name in Repbase Update and instead are annotated as having a section with 26.8% divergence from the ETnERV2 consensus, a section with 25.7% divergence from the RNLTR3c consensus, and an intervening section which is a nonrepeat (Table 3). Other examples of numerous yet incompletely annotated LTRs are those of MmERV-β4_AC110500, MmERV-β4_AC124523, MmERV-β5_AC125328, MmERV-β5_NT_039649, and RnERV-β5_AC127785 (Tables 2 and 3). Clearly, the completeness of the repeat databases has implications for both the annotation of genomic sequences and evolutionary deductions (see Discussion).
DISCUSSION
Discovery of new murine betaretroviruses.
We have described the discovery of several groups of betaretroviruses residing in the mouse and rat genomes. These groups, which we named β1 to β7, were defined in terms of their relationships to one another and to previously known betaretroviruses from mice and other species. All of the groups contain mouse and rat elements, and some of the groups also contain previously known betaretroviruses and/or newly discovered betaretroviruses from nonmouse, nonrat hosts. A phylogenetic tree based on sequences from the TM domain of the Env protein suggested that multiple recombination events have occurred during the evolution of murine betaretroviruses.
Four of the murine betaretrovirus groups (β2, β4, β5, and β7) possess coding-competent members, with fully intact ORFs for Gag, Pro, Pol, and/or Env proteins (Fig. 2). Most of the elements with intact ORFs also possess identical or near-identical 5′ and 3′ LTRs. These two features combined suggest recent and autonomous retrotransposition or infection.
Previous reports suggest that the β2 (MMTV) elements of mice have variable distribution in wild mice and inbred strains and appear to have entered the genomes of their hosts recently (7, 14). Our results support these observations and suggest that the closely related β2 viruses in the rat genome (represented by RnERV-β2_AC127663) were also recently acquired. Both groups of elements contain few members, all of which are fully (or almost fully) intact and have highly similar 5′ and 3′ LTRs. In addition, no MMTV solitary LTRs and only 17 solitary LTRs from the RnERV-β2_AC127663 group are observed in the mouse and rat assemblies, respectively. More distantly related β2 elements reside in the mouse (MmERV-β2_AC113463 and MmERV-β2_AC131667) and rat (RnERV-β2_NW_043520) genomes. These may correspond to the MMTV-related elements previously detected by Southern hybridization (6). It is interesting that MMTV and RnERV-β2_AC127663 both possess sag genes, whereas the more distantly related β2 elements do not—acquisition of the sag gene by these viruses may have been crucial in enabling the cross-species transmission back to mice and rats.
The β7 (MusD) elements display insertional polymorphisms in mice (2), suggesting that they are still active retrotransposons. These elements also display elevated embryonal expression in some laboratory strains of mice, which may contribute to (or enable) their retrotranspositional activity (4). The activity of β4 and β5 coding-competent elements is unknown. However, that these elements have retained intact ORFs despite their presence in the genomes of their hosts for such presumed long periods of time suggests that betaretroviruses from these and/or the other three groups may have retained coding competency and, therefore, the ability to undergo cross-species transmission to other murid species.
We have identified some β2 and β3 elements that likely integrated prior to the mouse-rat split—as evidenced by proviruses and solitary LTRs, respectively, at the same positions within the genomes of both species—but we have been unable to do so for the other beta groups. Although this may be because such common integrants do not exist, it is more likely that we have simply missed those integrants because of the nature of our search criteria, because the elements have been mutated or deleted over time, or because the genome sequences are incomplete. For many older elements of some of the groups, only incomplete proviruses (i.e., those lacking LTRs) could be found, and these groups may contain common integrants which we have not detected. It is also likely that many solitary LTRs reside in the mouse and rat genomes that are not represented by their original internal sequences, and these would not be detected by our approach.
Although the majority of the pol elements we have discovered here have already been annotated as repeats, this is the first time the phylogenetic relationships of these elements have been described. The most recently expanded and numerous elements have been identified by repeat-seeking programs and have been well annotated, but older and less numerous elements are poorly annotated. Generally, the pol regions of these elements are recognized as being retroviral, but they are highly diverged from the consensus sequences of the groups to which they have been assigned. In addition, LTRs of these older elements are usually only partially recognized as being repeats. The completeness of annotation obviously has important implications for determining ages of elements and dates of expansion of groups. Divergence from consensus is commonly used to estimate the age of a given element or group of elements (17, 29, 34). However, incomplete identification of repeat groups can lead to the assignment of some repeats to groups to which they are only distantly related, giving high divergences from consensus and skewing measurements of repeat age. Thorough identification and annotation of repeats is therefore of crucial importance for studies of the evolution of repeats and their hosts.
Increase in the known host range of betaretroviruses.
In addition to the newly described mouse and rat betaretroviruses, we have discovered three previously unknown betaretroviruses from other species. CpERV-β5_AC138156 is present in the genome sequence of Carollia perspicillata, a short-tailed leaf-nosed bat of Central and South America, and M_murinus_ERV-β4_AC145758 resides in the genome of the gray mouse lemur (Microcebus murinus) of Madagascar. CpERV-β5_AC138156 is, as far as we are aware, the first known bat retrovirus and is most closely related to the endogenous type D retrovirus of the squirrel monkey (Saimiri sciureus), which also inhabits South America. It is possible that transmission occurred directly between Carollia perspicillata and Saimiri sciureus or between these hosts via an intermediate host or that the retroviruses were transmitted to both hosts from an unknown (perhaps murid) host. M_murinus_ERV-β4_AC145758 possesses sufficient sequence to be included in both pol and Pol and env trees, and in both cases it groups with the murine β4 elements (Fig. 1 and 3). The third novel betaretrovirus sequence was that of BtERV-β2_CC563924, which was discovered in a clone from the cow (Bos taurus) genome. The significance of these newly discovered proviruses to the evolution of betaretroviruses is unknown, but they extend the biological and geographical ranges of known betaretrovirus hosts and suggest that further investigation of betaretroviruses in these and other species is warranted.
Several groups have recently reported the detection of betaretroviruses in the genomes of pigs (10, 22), the bower bird, and the stripe-faced dunnart (13). These elements were detected by PCR using degenerate primers, and the sequences were too short to include in our pol and Pol alignments. We constructed trees using shorter pol and Pol sequences, including two pig elements (PMSN-1 and PMSN-4) (10) and the bower bird and stripe-faced dunnart elements (13), and all of these elements fell outside of the seven groups of betaretroviruses described here (data not shown), suggesting that an even greater diversity of betaretroviruses exists and awaits thorough characterization.
Murid rodents as hosts for evolution and distribution of betaretroviruses.
It is clear from our results that a diverse range of betaretroviruses is present in the genomes of murine rodents. We have also obtained evidence of the presence of several betaretroviruses in the genomes of two North American sigmodontine rodents (our unpublished results), suggesting that betaretroviruses are broadly distributed in the Muridae family. Thus, murid rodents, with their global distribution, appear to have played a major role in the evolution and spread of betaretroviruses. It is also clear that betaretroviruses are present in the genomes of a wide variety of nonmurid hosts—some known, some currently unknown—and that numerous interspecies transmission events must have occurred. At this stage, however, it is unclear whether the majority of betaretrovirus evolution occurred in a murid rodent context, with occasional transmission to other species, or whether other hosts have played an equal or greater role in betaretrovirus evolution.
Transmission between murid and nonmurid hosts, regardless of the direction of transmission, has sometimes involved recombination within the retroviral genome to create new pol-env combinations, as exemplified by the type D retroviruses. These viruses are found within different groups of murine betaretroviruses (β4, β5, and β6) based on their pol genes (Fig. 1). However, they do not group with their murine counterparts in the TM tree and are instead grouped together, to the exclusion of all murine sequences (Fig. 3). This suggests that several different betaretroviruses have acquired the same env gene during transmission between hosts. Several viruses from other (non-beta) retroviral genera—namely, SNV and REV of anseriform and gallinaceous birds, BaEV of baboons, and the feline retrovirus RD114—also possess type D env genes, and these viruses all appear to have arisen relatively recently through recombination and cross-species transmission (19, 20, 31). The type D env gene has thus proven itself to recombine readily and enable infection of a wide range of hosts and may confer a selective advantage on viruses which possess it.
Our results show that the diversity of endogenous betaretroviruses within the genomes of mice and rats (and other mammals) is much greater than was previously appreciated. Studies of other murid rodents, other mammals, and perhaps nonmammalian vertebrates will surely reveal an even greater diversity.
Acknowledgments
This work was supported by New Zealand Foundation for Research, Science and Technology postdoctoral fellowship number TFBC0001 (G.J.B.), a studentship from the National Sciences and Engineering Research Council of Canada (L.N.V.D.L.), and a grant from the Canadian Institute of Health Research (D.L.M.).
We thank the NISC Comparative Sequencing Program for the use of their sequence data.
REFERENCES
- 1.Baillie, G. J., and R. J. Wilkins. 2001. Endogenous type D retrovirus in a marsupial, the common brushtail possum (Trichosurus vulpecula). J. Virol. 75:2499-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baust, C., G. J. Baillie, and D. L. Mager. 2002. Insertional polymorphisms of ETn retrotransposons include a disruption of the wiz gene in C57BL/6 mice. Mamm. Genome 13:423-428. [DOI] [PubMed] [Google Scholar]
- 3.Baust, C., L. Gagnier, G. J. Baillie, M. J. Harris, D. M. Juriloff, and D. L. Mager. 2003. Structure and expression of mobile ETnII retroelements and their coding-competent MusD relatives in mouse. J. Virol. 77:11448-11458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bénit, L., P. Dessen, and T. Heidmann. 2001. Identification, phylogeny, and evolution of retroviral elements based on their envelope genes. J. Virol. 75:11709-11719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeke, J. D., and J. P. Stoye. 1997. Retrotransposons, endogenous retroviruses, and the evolution of retroelements, p. 343-435. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 6.Callahan, R., W. Drohan, D. Gallahan, L. D'Hoostelaere, and M. Potter. 1982. Novel class of mouse mammary tumor virus-related DNA sequences found in all species of Mus, including mice lacking the virus proviral genome. Proc. Natl. Acad. Sci. USA 79:4113-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, J. C., and H. E. Varmus. 1979. Endogenous mammary tumour virus DNA varies among wild mice and segregates during inbreeding. Nature 278:418-423. [DOI] [PubMed] [Google Scholar]
- 8.Colcher, D., R. L. Heberling, S. S. Kalter, and J. Schlom. 1977. Squirrel monkey retrovirus: an endogenous virus of a new world primate. J. Virol. 23:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cousens, C., E. Minguijon, R. G. Dalziel, A. Ortin, M. Garcia, J. Park, L. Gonzalez, J. M. Sharp, and M. de las Heras. 1999. Complete sequence of enzootic nasal tumor virus, a retrovirus associated with transmissible intranasal tumors of sheep. J. Virol. 73:3986-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ericsson, T., B. Oldmixon, J. Blomberg, M. Rosa, C. Patience, and G. Andersson. 2001. Identification of novel porcine endogenous betaretrovirus sequences in miniature swine. J. Virol. 75:2765-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Escot, C., E. Hogg, and R. Callahan. 1986. Mammary tumorigenesis in feral Mus cervicolor popaeus. J. Virol. 58:619-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hecht, S. J., K. E. Stedman, J. O. Carlson, and J. C. DeMartini. 1996. Distribution of endogenous type B and type D sheep retrovirus sequences in ungulates and other mammals. Proc. Natl. Acad. Sci. USA 93:3297-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herniou, E., J. Martin, K. Miller, J. Cook, M. Wilkinson, and M. Tristem. 1998. Retroviral diversity and distribution in vertebrates. J. Virol. 72:5955-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai, S., M. Okumoto, M. Iwai, S. Haga, N. Mori, N. Miyashita, K. Moriwaki, J. Hilgers, and N. H. Sarkar. 1994. Distribution of mouse mammary tumor virus in Asian wild mice. J. Virol. 68:3437-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobo-Molina, A., and E. Arnold. 1991. HIV reverse transcriptase structure-function relationships. Biochemistry 30:6351-6361. [DOI] [PubMed] [Google Scholar]
- 16.Jurka, J. 2000. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 16:418-420. [DOI] [PubMed] [Google Scholar]
- 17.Lander, E. S., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 18.Mager, D. L., and J. D. Freeman. 2000. Novel mouse type D endogenous proviruses and ETn elements share long terminal repeat and internal sequences. J. Virol. 74:7221-7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mang, R., J. Goudsmit, and A. C. van der Kuyl. 1999. Novel endogenous type C retrovirus in baboons: complete sequence, providing evidence for baboon endogenous virus gag-pol ancestry. J. Virol. 73:7021-7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin, J., E. Herniou, J. Cook, R. W. O'Neill, and M. Tristem. 1999. Interclass transmission and phyletic host tracking in murine leukemia virus-related retroviruses. J. Virol. 73:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Musser, G. G., and M. D. Carlton. 1993. Family Muridae, p. 501-755. In D. E. Wilson and D. M. Reeder (ed.), Mammal species of the world. A taxonomic and geographic reference, 2nd ed. Smithsonian Institution Press, Washington, D.C.
- 22.Patience, C., W. M. Switzer, Y. Takeuchi, D. J. Griffiths, M. E. Goward, W. Heneine, J. P. Stoye, and R. A. Weiss. 2001. Multiple groups of novel retroviral genomes in pigs and related species. J. Virol. 75:2771-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Power, M. D., P. A. Marx, M. L. Bryant, M. B. Gardner, P. J. Barr, and P. A. Luciw. 1986. Nucleotide sequence of SRV-1, a type D simian acquired immune deficiency syndrome virus. Science 231:1567-1572. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz, S., Z. Zhang, K. A. Frazer, A. Smit, C. Riemer, J. Bouck, R. Gibbs, R. Hardison, and W. Miller. 2000. PipMaker-a web server for aligning two genomic DNA sequences. Genome Res. 10:577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonigo, P., C. Barker, E. Hunter, and S. Wain-Hobson. 1986. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell 45:375-385. [DOI] [PubMed] [Google Scholar]
- 26.Sprinzl, M., C. Horn, M. Brown, A. Ioudovitch, and S. Steinberg. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26:148-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247-250. [DOI] [PubMed] [Google Scholar]
- 28.Thayer, R. M., M. D. Power, M. L. Bryant, M. B. Gardner, P. J. Barr, and P. A. Luciw. 1987. Sequence relationships of type D retroviruses which cause simian acquired immunodeficiency syndrome. Virology 157:317-329. [DOI] [PubMed] [Google Scholar]
- 29.Thomas, J. W., J. W. Touchman, R. W. Blakesley, G. G. Bouffard, S. M. Beckstrom-Sternberg, E. H. Margulies, M. Blanchette, A. C. Siepel, P. J. Thomas, J. C. McDowell, B. Maskeri, N. F. Hansen, M. S. Schwartz, R. J. Weber, W. J. Kent, D. Karolchik, T. C. Bruen, R. Bevan, D. J. Cutler, S. Schwartz, L. Elnitski, J. R. Idol, A. B. Prasad, S.-Q. Lee-Lin, V. V. B. Maduro, T. J. Summers, M. E. Portnoy, N. L. Dietrich, N. Akhter, K. Ayele, B. Benjamin, K. Cariaga, C. P. Brinkley, S. Y. Brooks, S. Granite, X. Guan, J. Gupta, P. Haghighi, S.-L. Ho, M. C. Huang, E. Karlins, P. L. Laric, R. Legaspi, M. J. Lim, Q. L. Maduro, C. A. Masiello, S. D. Mastrian, J. C. McCloskey, R. Pearson, S. Stantripop, E. E. Tiongson, J. T. Tran, C. Tsurgeon, J. L. Vogt, M. A. Walker, K. D. Wetherby, L. S. Wiggins, A. C. Young, L.-H. Zhang, K. Osoegawa, B. Zhu, B. Zhao, C. L. Shu, P. J. De Jong, C. E. Lawrence, A. F. Smit, A. Chakravarti, C. Haussler, P. Green, W. Miller, and E. D. Green. 2003. Comparative analyses of multi-species sequences from targeted genomic regions. Nature 424:788-793. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Kuyl, A. C., J. T. Dekker, and J. Goudsmit. 1999. Discovery of a new endogenous type C retrovirus (FcEV) in cats: evidence for RD-114 being an FcEVGag-Pol/baboon endogenous virus BaEVEnv recombinant. J. Virol. 73:7994-8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Kuyl, A. C., R. Mang, J. T. Dekker, and J. Goudsmit. 1997. Complete nucleotide sequence of simian endogenous type D retrovirus with intact genome organization: evidence for ancestry to simian retrovirus and baboon endogenous virus. J. Virol. 71:3666-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.). 2000. Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses, 1st ed. Academic Press, San Diego, Calif.
- 34.Waterston, R. H., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520-562. [DOI] [PubMed] [Google Scholar]
- 35.York, D. F., R. Vigne, D. W. Verwoerd, and G. Querat. 1992. Nucleotide sequence of the Jaagsiekte retrovirus, an exogenous and endogenous type D and B retrovirus of sheep and goats. J. Virol. 66:4930-4939. [DOI] [PMC free article] [PubMed] [Google Scholar]