Full text
PDF

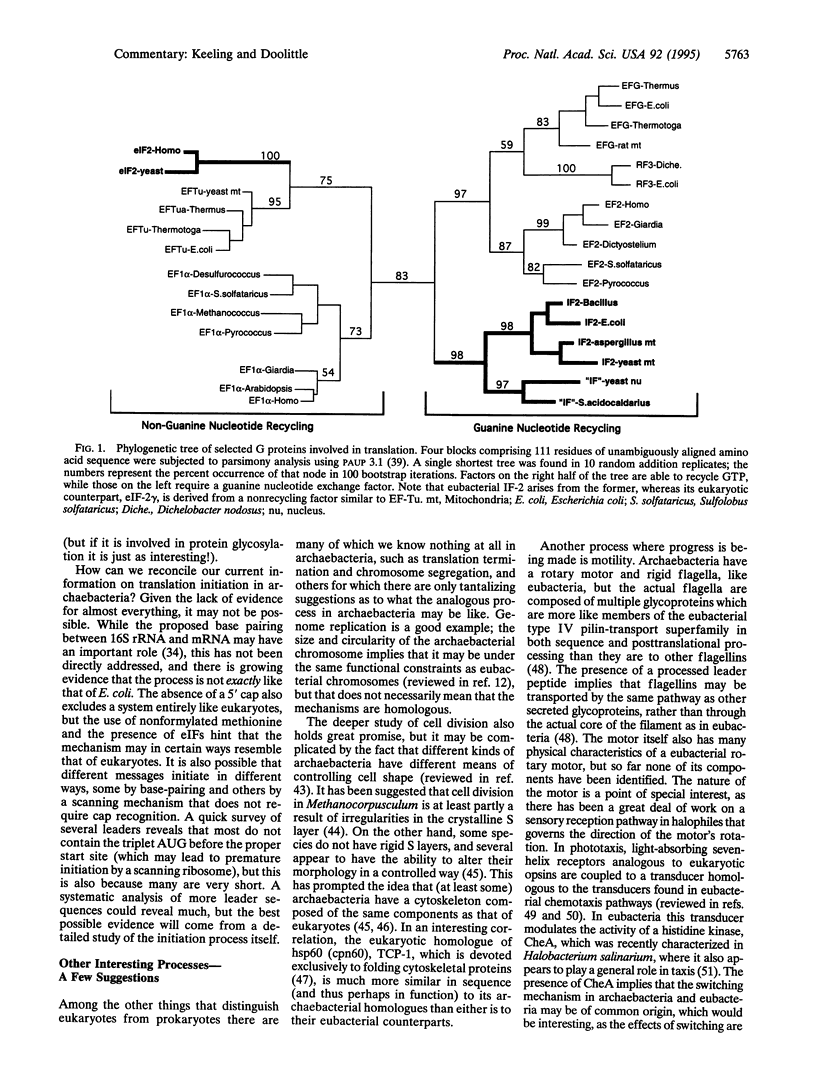

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alam M., Oesterhelt D. Morphology, function and isolation of halobacterial flagella. J Mol Biol. 1984 Jul 15;176(4):459–475. doi: 10.1016/0022-2836(84)90172-4. [DOI] [PubMed] [Google Scholar]
- Bartig D., Lemkemeier K., Frank J., Lottspeich F., Klink F. The archaebacterial hypusine-containing protein. Structural features suggest common ancestry with eukaryotic translation initiation factor 5A. Eur J Biochem. 1992 Mar 1;204(2):751–758. doi: 10.1111/j.1432-1033.1992.tb16690.x. [DOI] [PubMed] [Google Scholar]
- Bork P., Sander C., Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. R., Doolittle W. F. Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc Natl Acad Sci U S A. 1995 Mar 28;92(7):2441–2445. doi: 10.1073/pnas.92.7.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. W., Daniels C. J., Reeve J. N. Gene structure, organization, and expression in archaebacteria. Crit Rev Microbiol. 1989;16(4):287–338. doi: 10.3109/10408418909105479. [DOI] [PubMed] [Google Scholar]
- Brown J. W., Reeve J. N. Polyadenylated, noncapped RNA from the archaebacterium Methanococcus vannielii. J Bacteriol. 1985 Jun;162(3):909–917. doi: 10.1128/jb.162.3.909-917.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman J. L., Asuru A. I., Matts R. L., Hinnebusch A. G. Evidence that GCD6 and GCD7, translational regulators of GCN4, are subunits of the guanine nucleotide exchange factor for eIF-2 in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Mar;13(3):1920–1932. doi: 10.1128/mcb.13.3.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-tsmith T. The origin of nuclei and of eukaryotic cells. Nature. 1975 Aug 7;256(5517):463–468. doi: 10.1038/256463a0. [DOI] [PubMed] [Google Scholar]
- Dunn R., McCoy J., Simsek M., Majumdar A., Chang S. H., Rajbhandary U. L., Khorana H. G. The bacteriorhodopsin gene. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6744–6748. doi: 10.1073/pnas.78.11.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durner J., Böger P. Ubiquitin in the prokaryote Anabaena variabilis. J Biol Chem. 1995 Feb 24;270(8):3720–3725. doi: 10.1074/jbc.270.8.3720. [DOI] [PubMed] [Google Scholar]
- Durovic P., Dennis P. P. Separate pathways for excision and processing of 16S and 23S rRNA from the primary rRNA operon transcript from the hyperthermophilic archaebacterium Sulfolobus acidocaldarius: similarities to eukaryotic rRNA processing. Mol Microbiol. 1994 Jul;13(2):229–242. doi: 10.1111/j.1365-2958.1994.tb00418.x. [DOI] [PubMed] [Google Scholar]
- Erickson H. P. FtsZ, a prokaryotic homolog of tubulin? Cell. 1995 Feb 10;80(3):367–370. doi: 10.1016/0092-8674(95)90486-7. [DOI] [PubMed] [Google Scholar]
- Faguy D. M., Jarrell K. F., Kuzio J., Kalmokoff M. L. Molecular analysis of archael flagellins: similarity to the type IV pilin-transport superfamily widespread in bacteria. Can J Microbiol. 1994 Jan;40(1):67–71. doi: 10.1139/m94-011. [DOI] [PubMed] [Google Scholar]
- Frey G., Thomm M., Brüdigam B., Gohl H. P., Hausner W. An archaebacterial cell-free transcription system. The expression of tRNA genes from Methanococcus vannielii is mediated by a transcription factor. Nucleic Acids Res. 1990 Mar 25;18(6):1361–1367. doi: 10.1093/nar/18.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten J. P., Kibak H., Dittrich P., Taiz L., Bowman E. J., Bowman B. J., Manolson M. F., Poole R. J., Date T., Oshima T. Evolution of the vacuolar H+-ATPase: implications for the origin of eukaryotes. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6661–6665. doi: 10.1073/pnas.86.17.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausner W., Frey G., Thomm M. Control regions of an archaeal gene. A TATA box and an initiator element promote cell-free transcription of the tRNA(Val) gene of Methanococcus vannielii. J Mol Biol. 1991 Dec 5;222(3):495–508. doi: 10.1016/0022-2836(91)90492-o. [DOI] [PubMed] [Google Scholar]
- Holliday R. Ambidextrous RNA. Nature. 1990 Dec 6;348(6301):491–492. doi: 10.1038/348491b0. [DOI] [PubMed] [Google Scholar]
- Huet J., Schnabel R., Sentenac A., Zillig W. Archaebacteria and eukaryotes possess DNA-dependent RNA polymerases of a common type. EMBO J. 1983;2(8):1291–1294. doi: 10.1002/j.1460-2075.1983.tb01583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüdepohl U., Reiter W. D., Zillig W. In vitro transcription of two rRNA genes of the archaebacterium Sulfolobus sp. B12 indicates a factor requirement for specific initiation. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5851–5855. doi: 10.1073/pnas.87.15.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabe N., Kuma K., Hasegawa M., Osawa S., Miyata T. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F. Evolution and tinkering. Science. 1977 Jun 10;196(4295):1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- Kang H. A., Hershey J. W. Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. J Biol Chem. 1994 Feb 11;269(6):3934–3940. [PubMed] [Google Scholar]
- Keeling P. J., Charlebois R. L., Doolittle W. F. Archaebacterial genomes: eubacterial form and eukaryotic content. Curr Opin Genet Dev. 1994 Dec;4(6):816–822. doi: 10.1016/0959-437x(94)90065-5. [DOI] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Zwickl P., Baumeister W. Proteasome sequences in eubacteria. Trends Biochem Sci. 1994 Dec;19(12):533–534. doi: 10.1016/0968-0004(94)90054-x. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. FtsZ ring in bacterial cytokinesis. Mol Microbiol. 1993 Aug;9(3):403–409. doi: 10.1111/j.1365-2958.1993.tb01701.x. [DOI] [PubMed] [Google Scholar]
- Margulis L., Guerrero R. Kingdoms in turmoil. New Sci. 1991 Mar 23;1761:46–50. [PubMed] [Google Scholar]
- Marsh T. L., Reich C. I., Whitelock R. B., Olsen G. J. Transcription factor IID in the Archaea: sequences in the Thermococcus celer genome would encode a product closely related to the TATA-binding protein of eukaryotes. Proc Natl Acad Sci U S A. 1994 May 10;91(10):4180–4184. doi: 10.1073/pnas.91.10.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J. E., Gualerzi C. Translational control of prokaryotic gene expression. Trends Genet. 1990 Mar;6(3):78–85. doi: 10.1016/0168-9525(90)90098-q. [DOI] [PubMed] [Google Scholar]
- Merrick W. C. Mechanism and regulation of eukaryotic protein synthesis. Microbiol Rev. 1992 Jun;56(2):291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mescher M. F., Strominger J. L. Structural (shape-maintaining) role of the cell surface glycoprotein of Halobacterium salinarium. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2687–2691. doi: 10.1073/pnas.73.8.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara E. B., Chekanova J. A., Ingle C. A., Kushner Z. R., Peters E., Kushner S. R. Polyadenylylation helps regulate mRNA decay in Escherichia coli. Proc Natl Acad Sci U S A. 1995 Mar 14;92(6):1807–1811. doi: 10.1073/pnas.92.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota I. M., Varshavsky A. A yeast protein similar to bacterial two-component regulators. Science. 1993 Oct 22;262(5133):566–569. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- Ouzounis C., Sander C. TFIIB, an evolutionary link between the transcription machineries of archaebacteria and eukaryotes. Cell. 1992 Oct 16;71(2):189–190. doi: 10.1016/0092-8674(92)90347-f. [DOI] [PubMed] [Google Scholar]
- Palmer J. R., Daniels C. J. In vivo definition of an archaeal promoter. J Bacteriol. 1995 Apr;177(7):1844–1849. doi: 10.1128/jb.177.7.1844-1849.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pum D., Messner P., Sleytr U. B. Role of the S layer in morphogenesis and cell division of the archaebacterium Methanocorpusculum sinense. J Bacteriol. 1991 Nov;173(21):6865–6873. doi: 10.1128/jb.173.21.6865-6873.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter W. D., Hüdepohl U., Zillig W. Mutational analysis of an archaebacterial promoter: essential role of a TATA box for transcription efficiency and start-site selection in vitro. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9509–9513. doi: 10.1073/pnas.87.24.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlands T., Baumann P., Jackson S. P. The TATA-binding protein: a general transcription factor in eukaryotes and archaebacteria. Science. 1994 May 27;264(5163):1326–1329. doi: 10.1126/science.8191287. [DOI] [PubMed] [Google Scholar]
- Rudolph J., Oesterhelt D. Chemotaxis and phototaxis require a CheA histidine kinase in the archaeon Halobacterium salinarium. EMBO J. 1995 Feb 15;14(4):667–673. doi: 10.1002/j.1460-2075.1995.tb07045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STANIER R. Y., VAN NIEL C. B. The concept of a bacterium. Arch Mikrobiol. 1962;42:17–35. doi: 10.1007/BF00425185. [DOI] [PubMed] [Google Scholar]
- Searcy D. G., Hixon W. G. Cytoskeletal origins in sulfur-metabolizing archaebacteria. Biosystems. 1991;25(1-2):1–11. doi: 10.1016/0303-2647(91)90008-9. [DOI] [PubMed] [Google Scholar]
- Sioud M., Baldacci G., Forterre P., de Recondo A. M. Antitumor drugs inhibit the growth of halophilic archaebacteria. Eur J Biochem. 1987 Dec 1;169(2):231–236. doi: 10.1111/j.1432-1033.1987.tb13602.x. [DOI] [PubMed] [Google Scholar]
- Spudich J. L. Color sensing in the Archaea: a eukaryotic-like receptor coupled to a prokaryotic transducer. J Bacteriol. 1993 Dec;175(24):7755–7761. doi: 10.1128/jb.175.24.7755-7761.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. L. Protein-protein interaction converts a proton pump into a sensory receptor. Cell. 1994 Dec 2;79(5):747–750. doi: 10.1016/0092-8674(94)90064-7. [DOI] [PubMed] [Google Scholar]
- Sutrave P., Shafer B. K., Strathern J. N., Hughes S. H. Isolation, identification and characterization of the FUN12 gene of Saccharomyces cerevisiae. Gene. 1994 Sep 2;146(2):209–213. doi: 10.1016/0378-1119(94)90294-1. [DOI] [PubMed] [Google Scholar]
- Sánchez M., Valencia A., Ferrándiz M. J., Sander C., Vicente M. Correlation between the structure and biochemical activities of FtsA, an essential cell division protein of the actin family. EMBO J. 1994 Oct 17;13(20):4919–4925. doi: 10.1002/j.1460-2075.1994.tb06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A., Goumans H., Voorma H. O., Benne R. The mechanism of action of eukaryotic initiation factor 4C in protein synthesis. Eur J Biochem. 1980;107(1):39–45. doi: 10.1111/j.1432-1033.1980.tb04621.x. [DOI] [PubMed] [Google Scholar]
- Wettach J., Gohl H. P., Tschochner H., Thomm M. Functional interaction of yeast and human TATA-binding proteins with an archaeal RNA polymerase and promoter. Proc Natl Acad Sci U S A. 1995 Jan 17;92(2):472–476. doi: 10.1073/pnas.92.2.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White B. N., Bayley S. T. Methionine transfer RNAs from the extreme halophile, Halobacterium cutirubrum. Biochim Biophys Acta. 1972 Jul 31;272(4):583–587. doi: 10.1016/0005-2787(72)90513-8. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Fox G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillig W. Eukaryotic traits in Archaebacteria. Could the eukaryotic cytoplasm have arisen from archaebacterial origin? Ann N Y Acad Sci. 1987;503:78–82. doi: 10.1111/j.1749-6632.1987.tb40599.x. [DOI] [PubMed] [Google Scholar]


