Abstract
Substitution mutagenesis of EBNA2 shows that its interaction with hSNF5/Ini1 involves two sites (286IPP and DQQ313), and a mutation at a CKII phosphorylation site (SS469) is essential for the interaction. An alanine substitution (SS469AA) prevents binding to EBNA2 and diminishes the growth-promotion potential of EBNA2 in the transcomplementation assay.
The ability of Epstein-Barr virus (EBV) to drive resting B lymphocytes into G1 and maintain them in an immortalized state is absolutely dependent on the expression of the viral nuclear protein 2 (EBNA2) (17, 23, 24). The resultant proliferative program, referred to as type III latency, shares phenotypic features with the malignant B lymphocytes that populate EBV-associated lymphomas that occur in immunosuppressed patients whose underlying disease or immunosuppressive therapy results in impairment of the normal cellular immune response that curbs the proliferation of B cells latently infected by EBV (55).
EBNA2 acts as an adaptor molecule that has the potential for recruitment of a large number of interacting proteins that modulate the expression of a subset of crucial viral and cellular genes that are in turn involved in the immortalization program. The EBNA2-responsive genes of the viral genome include those encoding the viral latency-associated nuclear proteins (42) and the regulatory region for the viral transforming protein LMP1 and a membrane protein involved in maintaining the latent state, LMP2A (21, 31, 33, 39, 48, 58). The cellular genes that respond to EBNA2 include the potential B-cell growth factors CD23 (5, 6, 47) and tumor necrosis factor beta (9), the Fgr tyrosine kinase (26), a component of the EBV receptor, CD21 (7, 29), the key proliferative transcription factor, Myc (20, 22), and the transcription factor AML-2 (41).
The mechanism by which EBNA2 influences the expression of its target genes is complex and dependent on the cell context. EBNA2 complexes are directed to their sites of action by binding to the DNA binding proteins CBF1/RBPJκ, PU.1, or ATF/CRE (15, 18, 21, 30, 40, 57). It interacts with basal transcription factors to assemble the preinitiation complex (43, 44) or can recruit transcription coactivators, such as SKIP and BS69 (1, 56), as well as the dual-function proteins, such as the transcription activator and antiapoptosis protein NUR77 (32), the RNA helicase DP103 (16), and the RNA processing complex associated with the survival motor neuron protein (3). Among the proteins that interact with EBNA2 are those involved in chromatin remodeling through covalent modifications of histone tails, PCAF and p300/CBP (49), or through the association with multiprotein chromatin remodeling complexes (51).
The paradigm of these chromatin remodeling machines is the SWI/SNF complex of Saccharomyces cerevisiae and its human homologue, the human SWI/SNF complex (hSWI/SNF) (also called the BAF [BRG1-associated factor] complex). Similar complexes in yeast, Drosophila, and frogs have been studied (4). The SWI/SNF complex can activate or repress transcription of a subset of genes through alteration of chromatin structure, probably by altering the effects on transcription imposed by nucleosomal packaging of DNA. This effect results both from altering the physical association between histones and DNA in an ATP-dependent, catalytic fashion and from recruitment by the complex of other factors that modulate gene expression or modify chromatin structure. SWI/SNF can both catalyze the movement of histones and displace them from DNA (reviewed in references 10, 34, and 36)]. Our work and that of several others have demonstrated that the complex is directed to sites of action by its association with specific transcription factors, EBNA2 providing one example of this activity (2, 27, 52).
We have found that a phosphorylated fraction of lymphocyte EBNA2 associates with a component of the hSWI/SNF complex, hSNF5/Ini1 (51). Using chromatin immunoprecipitation, we showed that EBNA2 directs the complex to an episomal chromatin template containing the LMP2A regulatory segment. We also showed that this is dependent on binding by CBF1/RBPJκ to its recognition sequence, as well as on EBNA2 expression. In addition, we found that EBNA2 directs the hSWI/SNF complex to the cellular CD23 gene regulatory region. These activities were also dependent on the presence of the TATA element of the promoter (52).
In an effort to establish the role that the association between EBNA2 and the hSWI/SNF complex may play in B-cell proliferation, we identified the sites in EBNA2 that mediate its binding to hSNF5/Ini1 by the generation of mutant alleles of EBNA2 that diminish or disrupt its interaction with hSNF5/Ini1. We have incorporated these noninteracting alleles of EBNA2 into retrovirus vectors to determine whether they support the growth of lymphocytes in an established transcomplementation assay (11). We have found that two sites that reside within the divergent region of EBNA2 are important for its binding to hSNF5/Ini1. Phosphorylation of a consensus CKII site residing near the carboxyl terminus of EBNA2, and remote from the region that directly mediates the EBNA2-hSNF5/Ini1 interaction, is essential for binding. Mutant alleles of EBNA2 that are not substrates for CKII do not bind hSNF5/Ini1 and show a reduction in the growth-promoting effects that result from the expression of this mutant form relative to that seen as a result of expression of wild-type EBNA2.
Two sites within the divergent region of EBNA2 are important for binding to hSNF5/Ini1.
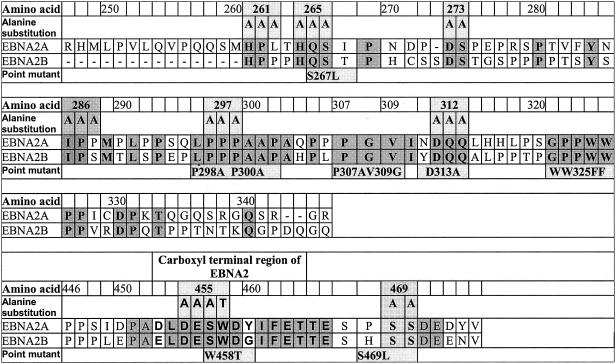
The association between EBNA2 and hSNF5/Ini1 was originally inferred from the results of a two-hybrid screen with yeast cells. The bait construct in that screen spanned amino acids 244 to 349 of EBNA2, which encompasses a region of the molecule that differs significantly between the type A and type B alleles, as depicted in Fig. 1 (8). Type A alleles of EBNA2 function as more potent transcription activators in experimental assays, and virions harboring type A alleles immortalize B cells more efficiently than do those harboring type B alleles (38). We found that hSNF5/Ini1 fusion proteins bound with comparable affinities to both type A and type B alleles of EBNA2 when expressed in yeast cells (data not shown). We also found that phosphorylated EBNA2 coimmunoprecipitated with hSNF5/Ini1 from EBV-immortalized lymphocytes or Burkitt lymphoma cell lines (51). We exploited these results in designing a set of alanine substitution and point mutants of EBNA2 that might disrupt its interaction with hSNF5/Ini1. These are depicted in Fig. 1. We introduced alanine substitution mutations in peptide stretches that are conserved between both type A and type B alleles within the divergent region. Because of the importance of phosphorylation for the interaction with hSNF5/Ini1, we also made a substitution mutant at a presumed CKII phosphorylation site at position 469. In addition, we made substitution mutations in the CBF1/RBPJκ binding site (WW325FF) and in the acidic transactivation domain that mediates interaction with basal transcription factors (DESW455AAAA). Mutant alleles similar to the latter two have been shown to be inactive in viral immortalization assays.
FIG. 1.
Mutant alleles of EBNA2 used in this study. Portions (amino acids 247 to 346 and 446 to 475) of the derived amino acid sequence of the coding sequence of the EBNA2A gene from the M-ABA strain of EBV that encompasses the divergent and carboxyl-terminal regions are depicted. For comparison, the sequence encoded by the EBNA2B allele (8) is indicated. Amino acid identities between A and B types are shaded. Point mutations that were constructed for this study are indicated below the sequence, and alanine substitution mutations are depicted above the sequence. Alanine substitution mutations were generated by standard techniques, using PCR and oligonucleotides that introduced the alanine substitution mutations indicated (19). The modification resulting in the alanine substitutions also generated a new PstI site within the coding region in addition to a preexisting PstI site that resides at its 3′ end. Specific PCR-generated fragments representing the amino and carboxyl portions of these mutations were cleaved with appropriate enzymes and ligated together in the pSG5 expression vector. All mutants were sequenced to verify the presence of the mutation and the absence of additional mutations. pSG5FLAG-Ini1 was constructed by introducing two tandem FLAG-encoding sequences at the amino terminus of the entire hSFN5/Ini1 open reading frame in the pSG5 vector.
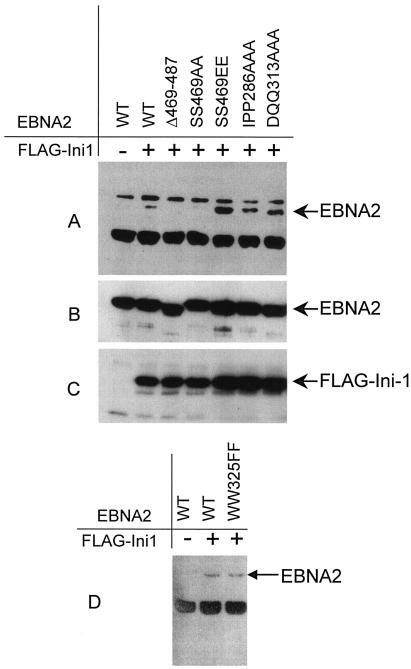
Plasmids expressing these EBNA2 mutations were cotransfected into 293T cells together with plasmids expressing full-length hSNF5/Ini1 fused at its amino terminus to the FLAG epitope. Following transfection, cell lysates were prepared and complexes were immunoprecipitated with anti-FLAG antibody (M2). The washed immunoprecipitates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), blotted, and probed with a monoclonal antibody directed against EBNA2, R3 (28). This assay resulted in variable results that were very dependent on transfection efficiency and antibody preparations, and the experiment required multiple replications in order to arrive at definitive conclusions. In this assay, the observed association of FLAG-Ini1 with EBNA2 was relatively weak (Fig. 2A, lane 2) and was significantly weaker than the interaction that we have consistently observed between endogenous hSNF5/Ini1 and EBNA2 in lymphocyte nuclear extracts (51). Although the results were somewhat variable, we observed a decrease in FLAG-Ini1 binding by EBNA2 mutants IPP286AAA and DQQ313AAA compared with the other mutants and an overall slight decrease in binding compared to wild-type FLAG-EBNA2 in the majority of replication of these experiments. Examples of these results are depicted in Fig. 2A, lanes 6 and 7. By contrast, when the terminal 19 amino acids of EBNA2 were deleted in the Δ469-487 mutant or when the serines at positions 469 to 470 were replaced by alanines, binding to FLAG-Ini1 was abolished (Fig. 2A, lanes 3 and 4). The sequence around amino acid 469 predicts it to be a consensus substrate for CKII phosphorylation, and it has been suggested that this site is a major phosphorylation site in lymphocyte EBNA2 (12). Substitution of phenylalanines for tryptophans at position 325, which abolished binding to RBPJκ, does not affect binding to hSNF5/Ini1 (Fig. 2D). These results suggest, but do not conclusively demonstrate, that these sites are involved in the interaction with hSNF5/Ini1. In cases where several peptides within a protein mediate a protein-protein interaction, it may not be possible to definitively identify the specific amino acid residues that are critical to the interaction.
FIG. 2.
In vivo interactions of mutant EBNA2 and hSNF5/Ini1 as shown by coimmunoprecipitation of EBNA2 and FLAG-Ini1 in 293T cells. 293T cells were transfected by the calcium phosphate technique with 5 μg of indicated plasmid DNA. After 48 h, transfected cells were lysed in NP-40 lysis buffer (0.5% NP-40, 50 mM Tris [pH 7.5], 150 mM NaCl, 5 mM EDTA), the nuclear debris was removed by centrifugation, and the supernatant was used for immunoprecipitation by binding to anti-FLAG agarose (Sigma, Saint Louis, Mo.). (A) Samples were separated by SDS-PAGE, blotted, probed with the EBNA2-specific rat monoclonal antibody (R3) (14), and developed with anti-rat horseradish peroxidase and the ECL substrate kit (Amersham). Panels B and C are immunoblots showing the expression of EBNA2 and FLAG-Ini1, respectively. Panel D shows a coimmunoprecipitation experiment using a mutant of EBNA2, WW325FF, that does not bind to CBF1/RBPJκ.
Phosphorylation of a CKII site in EBNA2 is essential for binding to hSNF5/Ini1.
We have shown that a phosphorylated subfraction of EBNA2 bound hSNF5/Ini1 in lymphocytes. Grasser and his colleagues have shown that EBNA2 is phosphorylated, predominantly at serine residues (12, 13), and they have further shown that the carboxyl-terminal region encompassing residues 469 to 470 is a substrate for CKII in vitro (12). Based on these results, we mutated this site by alanine substitution, as shown in Fig. 1. In the coimmunoprecipitation assay, we found that substitution of the potential phosphorylation sites at serines 469 and 470 abolished detectable hSNF5/Ini1 binding. When we generated the pseudo-revertant mutation by substituting glutamic acid residues for phosphoserines, (SS469EE), binding to hSNF5/Ini1 was restored to a level that consistently exceeded that seen with the wild-type allele. This result suggests that the constitutive negative charge imparted by the substitution of glutamic acids in 293T cells results in enhanced binding of hSNF5/Ini1 compared with that seen with the wild-type allele in this cell line. It is therefore possible that this mutation unmasks all potential interaction sites within the domain and that the 286 and 313 peptides are not directly involved in the hSNF5/Ini1 interaction in B cells, as they appear to be in 293T cells. Nevertheless, these results demonstrate that phosphorylation at the putative CKII site is essential for hSNF5/Ini1 binding.
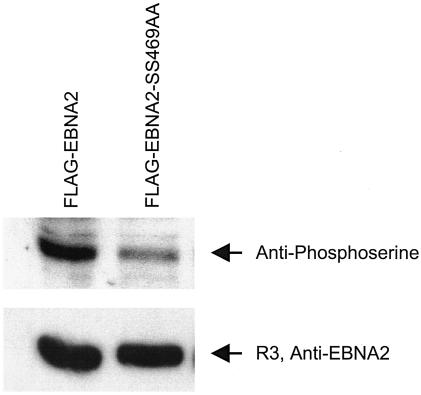
In order to determine whether in fact this site was phosphorylated under these experimental conditions, extracts of 293T cells were transfected with these EBNA2 alleles expressed as FLAG-EBNA2 fusion proteins, immunoprecipitated with anti-FLAG antibody, washed extensively, separated by SDS-PAGE, blotted, and probed with antibodies directed against phosphoserine or EBNA2. These results, shown in Fig. 3, show that substitution of serines at the putative CKII site resulted in a more than 50% reduction in overall EBNA2 serine phosphorylation. This result shows that this is a major phosphorylation site of EBNA2 under these conditions, consistent with previously published results (12).
FIG. 3.
Serine phosphorylation of EBNA2 at the CKII site. 293T cells were transfected by the calcium phosphate technique with pSG5-FLAG-EBNA2 wild type or the SS469AA substitution mutant. Cells were lysed and processed as described in the legend to Fig. 2 in NP-40 lysis buffer supplemented with both protease inhibitor (Roche Diagnostics GmbH, Mannheim, Germany) and phosphatase inhibitor (phosphatase inhibitor cocktail I, P-2850, and phosphatase inhibitor cocktail II, P-5726 [Sigma]). Lysed cell supernatants were immunoprecipitated with anti-FLAG M2 affinity gel (Sigma), washed extensively, blotted, and probed with mouse monoclonal anti-phosphoserine ascites fluid, P-3430, (Sigma), or a rat monoclonal antibody against EBNA2 (R3). Immunoblots were developed by using ECL, and bands were quantitated by using OptiQuant version 4.0 analysis software (Packard Instrument Co.).
The bait construct that was used in the yeast two-hybrid screen that identified hSNF5/Ini1 as a potential EBNA2 binding partner lacked the phosphorylation site that is crucial for its interaction. This suggests that the phosphorylation or its resultant negative charge domain results in some structural alteration that is crucial for binding. This could result from unfolding or exposure of a domain as a result of electrostatic forces engendered by the phosphate (or carboxyl) side chains. We propose that this domain is exposed and accessible for hSNF5/Ini1 binding in the yeast fusion protein and in both the phosphorylated form of EBNA2 and the phenotypic revertant that results from substitution of glutamic acids for serine at the putative CKII. An analogous conformational modification in the intracellular domain of Src has been demonstrated (53, 54). In the latter case, phosphorylation of a crucial tyrosine in the activation loop relieves stearic inhibition of substrate binding and establishes enzymatic activity. A similar conformational change may uncover a region of EBNA2 that is directly involved in hSNF5/Ini1 binding, such as the sequence at 286 or 313.
Binding to hSNF5/Ini1 correlates with EBNA2 growth transformation functions.
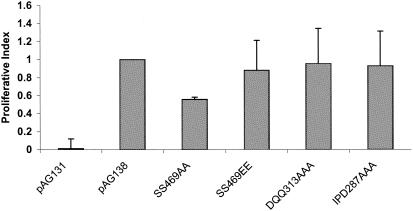
In an effort to determine whether binding to hSNF5/Ini1 correlated with the global effect of EBNA2 on cell growth and survival, we determined the function of the binding and nonbinding alleles of EBNA2 in a modification of the transcomplementation assay (11). In this assay, the EREB2.5 cell line that conditionally expresses an estrogen receptor-EBNA2 fusion protein (ER-EBNA2) molecule is used. These cells grow normally in the presence of estrogen but undergo growth arrest in its absence. The cells can be rescued by retrovirus transduction of a functional allele of EBNA2, permitting proliferation in the absence of exogenous estrogen. This assay serves to determine whether the EBNA2 proteins expressed serve to maintain growth transformation, an established function of EBNA2. Various mutant forms of EBNA2 that either bind or do not bind hSNF5/Ini1 were introduced into the plasmid pAG138 (also referred to as pLIG.EBNA2) (11), which expresses the wild-type allele of type A EBNA2. These were transferred into EREB2.5 cells following a previously described protocol (11). The transduced cells were grown in estrogen-free medium for 4 weeks, and after this period of outgrowth, we performed Alamar blue assays (BioSource) of aliquots of the cells. This assay provides a semiquantitative measure of the ability of the EBNA2 mutants to drive proliferation. These results are shown in Fig. 4. These data show that the mutants that partially disrupt the EBNA2-hSNF5/Ini1 interaction (DQQ313AAA and IPP286AAA) are essentially indistinguishable from wild-type EBNA2 (pAG138) in this assay. By contrast, the SS469AA mutant, which disrupts the interaction, demonstrates about half of the proliferative potential of wild-type EBNA2. We conclude that phosphorylation of the CKII site at residue 469 is important for B-cell growth transformation or immortalization. Interestingly, the SS469EE mutant, which apparently binds hSNF5/Ini1 with enhanced efficiency compared with the wild type, is as active as wild-type EBNA2 in this assay. Cells transduced by virions derived from the empty vector, pAG131, showed no activity in this modification of the transcomplementation assay.
FIG. 4.
Proliferative activities of mutant EBNA2 alleles. Specific mutants of EBNA2 were placed into the pAG138 vector (11) by removal of the appropriate sequences from the various pSG5-EBNA2 alleles. This was done by partial digestion of the pSG5 mutant construct with PstI, blunt-end formation, and subsequent complete digestion with BstEII. Fragments of appropriate size were then ligated into pAG138 previously digested with SmaI and BstEII. The alteration was verified by sequence analysis. Transfection of plasmids and transduction of EREB2.5 cells were performed as previously described (11). The transduced cells were cultured in the presence of 1 μM estradiol for 5 days, after which they were transferred to estrogen-free medium for 4 weeks. Proliferation was determined by reduction of Alamar blue according to the manufacturer's instructions (BioSource). For each transduced culture, a proliferative index was established by determining the least-squares fit of the time course of the measured percentage of Alamar blue reduction for a fixed number of cells (5 × 104 cells/well) in a 96-well plate over a 48-h period. The ordinate indicates the relative proliferation compared with pAG138-transduced cells in this assay. All assays were carried out in triplicate. The complete assay was performed on three separate occasions, using separate plasmid preparations for all of the components of the assay. Each error bar indicates the standard deviation for the three different determinations.
These results are consistent with the conclusion that the EBNA2-hSNF5/Ini1 interaction is important for B-cell immortalization; however, other interpretations are possible. Because the phosphorylation site may be remote from the physical binding site, it is possible that its modification results in exposure of other regions of the molecule that play indispensable roles in B-cell immortalization, such as the Ski interaction region. We have found that recombinant virions harboring a mutation that would be predicted to interrupt this interaction (P307A,V309G) are completely inactive in a standard B-cell immortalization assay (data not shown). Also, because CKII is ubiquitous and confers wide-ranging effects as a result of its ability to phosphorylate a large number of cellular substrates (35), the diminished binding to hSNF5/Ini1 may be only one of a potentially large number of molecular interactions that CKII phosphorylation may effect in EBV-immortalized B cells.
SNF5/Ini1 is essential for murine embryonic development (25), and although hSNF5/Ini1 is not the core catalytic component of the hSWI/SNF complex, all of the SWI/SNF homologues that have been characterized contain a SNF5 component, its presence contributes to the catalytic activity of the complex (37), and antibodies directed against hSNF5/Ini1 can be used to affinity purify the entire complex from human cells (50), suggesting that hSNF5/Ini1 plays a central and critical role in both the structure and function of the whole complex. It has also been suggested that hSNF5/Ini1 acts as a tumor suppressor independently of its chromatin remodeling activity. Homozygous deletion or inactivation of hSNF5/Ini1 is found in a malignant rhabdoid tumor of childhood (46), and ectopic expression hSNF5/Ini1 in cell lines deficient for this protein can induce cell cycle arrest in an Rb-dependent fashion (45). How this function of hSNF5/Ini1 relates to its role in EBV transformation is unclear. It is possible that binding of EBNA2 to hSNF5/Ini1 results in inactivation of the tumor suppressor activity of the latter and contributes to immortalization through removal of a normal block to entry into S phase.
Acknowledgments
We thank Paul Ling for reagents and advice in instituting the transcomplementation assay and Bill Tuttle for technical help.
This work was supported by grants from the NIH (CA82459) and the Department of Veterans Affairs to W.H.S.
REFERENCES
- 1.Ansieau, S., and A. Leutz. 2002. The conserved Mynd domain of BS69 binds cellular and oncoviral proteins through a common PXLXP motif. J. Biol. Chem. 277:4906-4910. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, J. A., J. J. Bieker, and B. M. Emerson. 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95:93-104. [DOI] [PubMed] [Google Scholar]
- 3.Barth, S., M. Liss, M. D. Voss, T. Dobner, U. Fischer, G. Meister, and F. A. Grasser. 2003. Epstein-Barr virus nuclear antigen 2 binds via its methylated arginine-glycine repeat to the survival motor neuron protein. J. Virol. 77:5008-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cairns, B. R. 1998. Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem. Sci. 23:20-25. [DOI] [PubMed] [Google Scholar]
- 5.Calender, A., M. Cordier, M. Billaud, and G. M. Lenoir. 1990. Modulation of cellular gene expression in B lymphoma cells following in vitro infection by Epstein-Barr virus (EBV). Int. J. Cancer. 46:658-663. [DOI] [PubMed] [Google Scholar]
- 6.Cordier, M., A. Calender, M. Billaud, U. Zimber, G. Rousselet, O. Pavlish, J. Banchereau, T. Tursz, G. Bornkamm, and G. M. Lenoir. 1990. Stable transfection of Epstein-Barr virus (EBV) nuclear antigen 2 in lymphoma cells containing the EBV P3HR1 genome induces expression of B-cell activation molecules CD21 and CD23. J. Virol. 64:1002-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cordier-Bussat, M., M. Billaud, A. Calender, and G. M. Lenoir. 1993. Epstein-Barr virus (EBV) nuclear-antigen-2-induced up-regulation of CD21 and CD23 molecules is dependent on a permissive cellular context. Int J. Cancer. 53:153-160. [DOI] [PubMed] [Google Scholar]
- 8.Dambaugh, T., K. Hennessy, L. Chamnankit, and E. Kieff. 1984. U2 region of Epstein-Barr virus DNA may encode Epstein-Barr nuclear antigen 2. Proc. Natl. Acad. Sci. USA 81:7632-7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estrov, Z., R. Kurzrock, E. Pocsik, S. Pathak, H. M. Kantarjian, T. F. Zipf, D. Harris, M. Talpaz, and B. B. Aggarwal. 1993. Lymphotoxin is an autocrine growth factor for Epstein-Barr virus-infected B cell lines. J. Exp. Med. 177:763-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flaus, A., and T. Owen-Hughes. 2001. Mechanisms for ATP-dependent chromatin remodelling. Curr. Opin. Genet. Dev. 11:148-154. [DOI] [PubMed] [Google Scholar]
- 11.Gordadze, A. V., R. Peng, J. Tan, G. Liu, R. Sutton, B. Kempkes, G. W. Bornkamm, and P. D. Ling. 2001. Notch1IC partially replaces EBNA2 function in B cells immortalized by Epstein-Barr virus. J. Virol. 75:5899-5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grasser, F. A., S. Gottel, P. Haiss, B. Boldyreff, O. G. Issinger, and N. Mueller-Lantzsch. 1992. Phosphorylation of the Epstein-Barr virus nuclear antigen 2. Biochem. Biophys. Res. Commun. 186:1694-1701. [DOI] [PubMed] [Google Scholar]
- 13.Grasser, F. A., P. Haiss, S. Gottel, and N. Mueller-Lantzsch. 1991. Biochemical characterization of Epstein-Barr virus nuclear antigen 2A. J. Virol. 65:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grasser, F. A., P. G. Murray, E. Kremmer, K. Klein, K. Remberger, W. Feiden, G. Reynolds, G. Niedobitek, L. S. Young, and N. Mueller-Lantzsch. 1994. Monoclonal antibodies directed against the Epstein-Barr virus-encoded nuclear antigen 1 (EBNA1): immunohistologic detection of EBNA1 in the malignant cells of Hodgkin's disease. Blood 84:3792-3798. [PubMed] [Google Scholar]
- 15.Grossman, S. R., E. Johannsen, X. Tong, R. Yalamanchili, and E. Kieff. 1994. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc. Natl. Acad. Sci. USA 91:7568-7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gundhoff, A. T., E. Kremmer, O. Tureci, A. Glieden, C. Gindorf, J. Artz, N. Mueller-Lantzsch, W. H. Schubach, and F. A. Grasser. 1999. Identification of dp103, a novel member of the DEAD box family which interacts with the Epstein-Barr virus nuclear proteins EBNA2 and EBNA3C. J. Biol. Chem. 274:19136-19144. [DOI] [PubMed] [Google Scholar]
- 17.Hammerschmidt, W., and B. Sugden. 1989. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature 340:393-397. [DOI] [PubMed] [Google Scholar]
- 18.Henkel, T., P. D. Ling, S. D. Hayward, and M. G. Peterson. 1994. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science 265:92-95. [DOI] [PubMed] [Google Scholar]
- 19.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jayachandra, S., K. G. Low, A. E. Thlick, J. Yu, P. D. Ling, Y. Chang, and P. S. Moore. 1999. Three unrelated viral transforming proteins (vIRF, EBNA2, and E1A) induce the MYC oncogene through the interferon-responsive PRF element by using different transcription coadaptors. Proc. Natl. Acad. Sci. USA 96:11566-11571. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Johannsen, E., E. Koh, G. Mosialos, X. Tong, E. Kieff, and S. R. Grossman. 1995. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU.1. J. Virol. 69:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaiser, C., G. Laux, D. Eick, N. Jochner, G. W. Bornkamm, and B. Kempkes. 1999. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J. Virol. 73:4481-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kempkes, B., D. Spitkovsky, P. Jansen-Durr, J. W. Ellwart, E. Kremmer, H. J. Delecluse, C. Rottenberger, G. W. Bornkamm, and W. Hammerschmidt. 1995. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 14:88-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kempkes, B., U. Zimber-Strobl, G. Eissner, M. Pawlita, M. Falk, W. Hammerschmidt, and G. W. Bornkamm. 1996. Epstein-Barr virus nuclear antigen 2 (EBNA2)-oestrogen receptor fusion proteins complement the EBNA2-deficient Epstein-Barr virus strain P3HR1 in transformation of primary B cells but suppress growth of human B cell lymphoma lines. J. Gen. Virol. 77:227-237. [DOI] [PubMed] [Google Scholar]
- 25.Klochendler-Yeivin, A., L. Fiette, J. Barra, C. Muchardt, C. Babinet, and M. Yaniv. 2000. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 1:500-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knutson, J. C. 1990. The level of c-fgr RNA is increased by EBNA-2, an Epstein-Barr virus gene required for B-cell immortalization. J. Virol. 64:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kowenz-Leutz, E., and A. Leutz. 1999. A C/EBP beta isoform recruits the SWI/SNF complex to activate myeloid genes. Mol. Cell 4:735-743. [DOI] [PubMed] [Google Scholar]
- 28.Kremmer, E., B. R. Kranz, A. Hille, K. Klein, M. Eulitz, G. Hoffmann-Fezer, W. Feiden, K. Herrmann, H. J. Delecluse, G. Delsol, G. W. Bornkamm, N. Mueller-Lantzsch, and F. A. Grassert. 1995. Rat monoclonal antibodies differentiating between the Epstein-Barr virus nuclear antigens 2A (EBNA2A) and 2B (EBNA2B). Virology 208:336-342. [DOI] [PubMed] [Google Scholar]
- 29.Larcher, C., B. Kempkes, E. Kremmer, W. M. Prodinger, M. Pawlita, G. W. Bornkamm, and M. P. Dierich. 1995. Expression of Epstein-Barr virus nuclear antigen-2 (EBNA2) induces CD21/CR2 on B and T cell lines and shedding of soluble CD21. Eur. J. Immunol. 25:1713-1719. [DOI] [PubMed] [Google Scholar]
- 30.Laux, G., B. Adam, L. J. Strobl, and G.-F. Moreau. 1994. The Spi-1/PU. 1 and Spi-B ets family transcription factors and the recombination signal binding protein RBP-J kappa interact with an Epstein-Barr virus nuclear antigen 2 responsive cis-element. EMBO J. 13:5624-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laux, G., F. Dugrillon, C. Eckert, B. Adam, U. Zimber-Strobl, and G. W. Bornkamm. 1994. Identification and characterization of an Epstein-Barr virus nuclear antigen 2-responsive cis element in the bidirectional promoter region of latent membrane protein and terminal protein 2 genes. J. Virol. 68:6947-6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, J. M., K. H. Lee, M. Weidner, B. A. Osborne, and S. D. Hayward. 2002. Epstein-Barr virus EBNA2 blocks Nur77-mediated apoptosis. Proc. Natl. Acad. Sci. USA 99:11878-11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Longnecker, R., and C. L. Miller. 1996. Regulation of Epstein-Barr virus latency by latent membrane protein 2. Trends Microbiol. 4:38-42. [DOI] [PubMed] [Google Scholar]
- 34.Martens, J. A., and F. Winston. 2003. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13:136-142. [DOI] [PubMed] [Google Scholar]
- 35.Meggio, F., and L. A. Pinna. 2003. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 17:349-368. [DOI] [PubMed] [Google Scholar]
- 36.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 37.Phelan, M. L., S. Sif, G. J. Narlikar, and R. E. Kingston. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3:247-253. [DOI] [PubMed] [Google Scholar]
- 38.Rickinson, A. B., L. S. Young, and M. Rowe. 1987. Influence of the Epstein-Barr virus nuclear antigen EBNA 2 on the growth phenotype of virus-transformed B cells. J. Virol. 61:1310-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sjoblom, A., A. Jansson, W. Yang, S. Lain, T. Nilsson, and L. Rymo. 1995. PU box-binding transcription factors and a POU domain protein cooperate in the Epstein-Barr virus (EBV) nuclear antigen 2-induced transactivation of the EBV latent membrane protein 1 promoter. J. Gen. Virol. 76:2679-2692. [DOI] [PubMed] [Google Scholar]
- 40.Sjoblom, A., W. Yang, L. Palmqvist, A. Jansson, and L. Rymo. 1998. An ATF/CRE element mediates both EBNA2-dependent and EBNA2-independent activation of the Epstein-Barr virus LMP1 gene promoter. J. Virol. 72:1365-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spender, L. C., G. H. Cornish, A. Sullivan, and P. J. Farrell. 2002. Expression of transcription factor AML-2 (RUNX3, CBFα-3) is induced by Epstein-Barr virus EBNA-2 and correlates with the B-cell activation phenotype. J. Virol. 76:4919-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sung, N. S., S. Kenney, D. Gutsch, and J. S. Pagano. 1991. EBNA-2 transactivates a lymphoid-specific enhancer in the BamHI C promoter of Epstein-Barr virus. J. Virol. 65:2164-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tong, X., R. Drapkin, D. Reinberg, and E. Kieff. 1995. The 62- and 80-kDa subunits of transcription factor IIH mediate the interaction with Epstein-Barr virus nuclear protein 2. Proc. Natl. Acad. Sci. USA 92:3259-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tong, X., F. Wang, C. J. Thut, and E. Kieff. 1995. The Epstein-Barr virus nuclear protein 2 acidic domain can interact with TFIIB, TAF40, and RPA70 but not with TATA-binding protein. J. Virol. 69:585-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Versteege, I., S. Medjkane, D. Rouillard, and O. Delattre. 2002. A key role of the hSNF5/INI1 tumour suppressor in the control of the G1-S transition of the cell cycle. Oncogene 21:6403-6412. [DOI] [PubMed] [Google Scholar]
- 46.Versteege, I., N. Sevenet, J. Lange, M. F. Rousseau-Merck, P. Ambros, R. Handgretinger, A. Aurias, and O. Delattre. 1998. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394:203-206. [DOI] [PubMed] [Google Scholar]
- 47.Wang, F., C. D. Gregory, M. Rowe, A. B. Rickinson, D. Wang, M. Birkenbach, H. Kikutani, T. Kishimoto, and E. Kieff. 1987. Epstein-Barr virus nuclear antigen 2 specifically induces expression of the B-cell activation antigen CD23. Proc. Natl. Acad. Sci. USA 84:3452-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, F., S. F. Tsang, M. G. Kurilla, J. I. Cohen, and E. Kieff. 1990. Epstein-Barr virus nuclear antigen 2 transactivates latent membrane protein LMP1. J. Virol. 64:3407-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, L., S. R. Grossman, and E. Kieff. 2000. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc. Natl. Acad. Sci. USA 97:430-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, W., J. Cote, Y. Xue, S. Zhou, P. A. Khavari, S. R. Biggar, C. Muchardt, G. V. Kalpana, S. P. Goff, M. Yaniv, J. L. Workman, and G. R. Crabtree. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15:5370-5382. [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, D. Y., G. V. Kalpana, S. P. Goff, and W. H. Schubach. 1996. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J. Virol. 70:6020-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, D. Y., A. Krumm, and W. H. Schubach. 2000. Promoter-specific targeting of human SWI-SNF complex by Epstein-Barr virus nuclear protein 2. J. Virol. 74:8893-8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu, W., A. Doshi, M. Lei, M. J. Eck, and S. C. Harrison. 1999. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol. Cell 3:629-638. [DOI] [PubMed] [Google Scholar]
- 54.Xu, W., S. C. Harrison, and M. J. Eck. 1997. Three-dimensional structure of the tyrosine kinase c-Src. Nature 385:595-602. [DOI] [PubMed] [Google Scholar]
- 55.Young, L., C. Alfieri, K. Hennessy, H. Evans, C. O'Hara, K. C. Anderson, J. Ritz, R. S. Shapiro, A. Rickinson, and E. Kieff. 1989. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N. Engl. J. Med. 321:1080-1085. [DOI] [PubMed] [Google Scholar]
- 56.Zhou, S., M. Fujimuro, J. J. Hsieh, L. Chen, and S. D. Hayward. 2000. A role for SKIP in EBNA2 activation of CBF1-repressed promoters. J. Virol. 74:1939-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zimber-Strobl, U., L. J. Strobl, C. Meitinger, R. Hinrichs, T. Sakai, T. Furukawa, T. Honjo, and G. W. Bornkamm. 1994. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-J kappa, the homologue of Drosophila Suppressor of Hairless. EMBO J. 13:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zimber-Strobl, U., K. O. Suentzenich, G. Laux, D. Eick, M. Cordier, A. Calender, M. Billaud, G. M. Lenoir, and G. W. Bornkamm. 1991. Epstein-Barr virus nuclear antigen 2 activates transcription of the terminal protein gene. J. Virol. 65:415-423. [DOI] [PMC free article] [PubMed] [Google Scholar]