Abstract
We have previously shown that styrylquinolines (SQLs) are integrase inhibitors in vitro. They compete with the long terminal repeat substrate for integrase. Here, we describe the cellular mode of action of these molecules. We show that SQLs do not interfere with virus entry. In fact, concentrations of up to 20 times the 50% inhibitory concentration did not inhibit cell-to-cell fusion or affect the interaction between GP120 and CD4 in vitro. Moreover, the pseudotype of the retrovirus envelope did not affect drug activity. Quantitative reverse transcription PCR experiments showed that SQLs do not inhibit the entry of the genomic RNA. In contrast, the treatment of human immunodeficiency virus type 1-infected cells with SQLs reduced the amount of the late cDNA, suggesting for the first time that integrase targeting molecules may affect the accumulation of DNA during reverse transcription. The cellular target of SQLs was confirmed by the appearance of mutations in the integrase gene when viruses were grown in the presence of increasing concentrations of SQLs. Finally, these mutations led to SQL-resistant viruses when introduced into the wild-type sequence. In contrast, SQLs were fully active against reverse transcriptase inhibitor- and diketo acid-resistant viruses, positioning SQLs as a second group of anti-integrase compounds.
The use of human immunodeficiency virus type 1 (HIV-1) inhibitors such as reverse transcriptase and protease inhibitors has led to the emergence of resistant strains. This highlights the necessity to develop new targeted drugs such as integrase inhibitors.
Retroviral integration, the process that stably insert the DNA copy of the viral genomic RNA into the host cell genome, is an essential step for productive infection (22, 26, 30, 43, 45).
The integration of HIV-1 cDNA is mediated by a complex consisting of both viral and cellular proteins closely associated with the viral DNA. This complex is known as the preintegration complex or PIC (6, 7, 10). Although the molecular organization and composition of the PIC remain to be clarified, the integration of viral DNA is known to be catalyzed by integrase. The integration process comprises three main steps: (i) 3′ end processing, involving the removal of a CA dinucleotide from the 3′ extremity of the U3 and U5 termini of the linear viral genome (4, 28); (ii) strand transfer, in which both 3′ ends of the viral DNA, generated by the first reaction, are covalently linked to host's cellular DNA after importation to the nucleus (21); and (iii) gap repair, where the 5′ ends of the viral DNA are trimmed and then ligated to the host cell DNA following repair of gapped regions generated by the strand transfer reaction (5, 20, 25, 41). Although gap repair is believed to be carried out by cellular proteins (15, 35, 51), both 3′ end processing and strand transfer are primarily mediated by the viral integrase (38).
Although the nonintegration roles of integrase during viral replication are still unclear, mutations in the integrase sequence inhibit different HIV-1 replication steps. Interestingly, some mutations have a particularly dramatically effect on reverse transcription (48). Integrase also promotes reverse transcription through specific interactions (46). In fact, reverse transcription is impaired in viruses lacking integrase or containing mutations in the N-terminal domain (H12A or H16A mutations destroying the zinc finger-like structure), in the C-terminal domain (lacking the 22 amino acids at the C terminus), or in the catalytic core (S81R and F185A for example), but not in a catalytically inactive D116A mutant (31, 44, 50).
A new class of integration inhibitors containing a diketo acid (DKA) moiety was recently described (23, 27). Acute infection in the presence of such compounds (L-731988 and L-708906) not only abolishes productive infection but also results in the accumulation of large amounts of circular DNA forms that cannot undergo integration, indicating that these compounds affect viral DNA integration. In addition, mutations conferring resistance to these drugs in cell culture have been located within the integrase protein (T66I, S153Y, M154I, and/or N155S).
Styrylquinolines (SQLs) are efficient in vitro integrase inhibitors that act both on 3′ processing and strand transfer activities (52), probably interfering with long terminal repeat (LTR)/integrase binding (37) through a competitive inhibition mechanism (17).
We report here a new cellular mode of action for integrase inhibitors that compete for the LTR substrate in vitro. We show that SQLs have antiviral activity against various resistant strains and that they act on the early steps of the viral life cycle. We demonstrate that these molecules do not interfere with virus entry. Indeed, they inhibit neither the GP120/CD4 interaction in vitro nor membrane fusion. Moreover, changing the viral envelope does not modify their activity. Finally, quantitative PCR (Q-PCR) experiments showed that SQLs dramatically diminish the amount of late cDNA without modulating the initial amount of viral RNA. Integrase coding region mutants selected by passaging in the presence of increasing SQL concentrations were resistant to SQLs. This resistance was confirmed with mutations that were genetically introduced into the wild-type sequence. We suggest that these compounds are integrase targeting inhibitors that act at one or several steps prior to integration. Finally, SQLs are active against HIV mutant strains that are resistant to DKAs and to reverse transcriptase inhibitors (RTIs), positioning them as a new means of anti-integrase therapy.
MATERIALS AND METHODS
Cells and viruses.
HeLa, HeLa P4 (encoding a Tat-inducible beta-galactosidase (9), HeLa 243 (encoding the recombinant HIV-1 transactivator Tat and Env proteins), and 293T cells were grown in Dulbecco's modified medium supplemented with 10% heat-inactivated fetal calf serum. MT4 and CEM4fx cells were grown in RPMI medium supplemented with 10% heat-inactivated fetal calf serum.
DKA-resistant virus strains were generated from the wild-type NL43 strain as described by Hazuda et al. by directed mutagenesis (27). Viruses harboring T66I, T66I-S153Y, or T66I-M154I were engineered.
To produce viral stocks, 293T cells were transfected with virus-encoding plasmids by the calcium phosphate method (2).
RTI-resistant strains were generated by use of a recombinant virus assay essentially as described before (42). Briefly, 293T cells were cotransfected with the linear plasmid encoding NL43 lacking reverse transcriptase (ΔRT NL43) and with PCR products corresponding to the reverse transcriptase of primary viruses. Cells were grown for 48 h, and the amount of virus produced was determined by a P24 assay (NEN).
Pseudotyped viruses were produced by a recombinant virus assay. Briefly, 293T cells were cotransfected with a plasmid encoding ΔEnv NL43 and a plasmid encoding the viral envelope of vesicular stomatitis virus G (VSV-G), amphotropic murine leukemia virus (AMLV), or HIV. Cells were grown for 72 h, and the resulting viruses were used to infect P4 cells in the presence of increasing drug concentrations.
IC50 determination.
HeLa P4 cells were infected with viruses and grown in the presence of increasing concentrations of drugs. After 72 h, the infectivity of viruses was determined by measuring beta-galactosidase (β-Gal) production by the chlorophenol red-β-d-galactopyranoside (CPRG) assay (24). The 50% inhibitory concentration (IC50) is the drug concentration at which 50% of β-Gal production was inhibited compared to that by untreated infected cells. Cell survival was also estimated by the use of a standard MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) assay (16).
GP120/CD4 interaction in vitro.
Inhibition of the rGP120/CD4 interaction was measured by a competitive enzyme-linked immunosorbent assay (ELISA) as described previously (49). Briefly, 50 ng of the D7324 anti-GP120 antibody (Aalto Bio Reagents, Dublin, Ireland) was used to coat each well of 96-well plates overnight at 4°C, and 15 ng of GP120LAI/well was then added. This was followed by the addition of 1 ng of full-length soluble CD4/well and different concentrations of soluble competitors, the anti-CD4 monoclonal antibody L120.3, a goat anti-mouse peroxidase-conjugated monoclonal antibody (Jackson ImmunoResearch, West Grove, Pa.), and the 3,3′,5,5′-tetramethylbenzidine substrate (Sigma) for revelation (at 450 nm). The following formula was used to calculate the percent inhibition of CD4 binding: [(OD450 with no competitor − OD450 with a competitor)/OD450 with no competitor] × 100, where OD450 is the optical density at 450 nm. Results are means of triplicate experiments repeated twice. CD4M9, a peptide mimicking CD4, was used as a positive control (49).
Cell-to-cell fusion assay.
HeLa P4 cells that contain the integrated LTR-β-Gal reporter gene (9) were incubated for 30 min in the presence or absence of SQL inhibitors (106 cells/well in 24-well plates). An equal number of HeLa 243 cells expressing Tat and Env were then added, and the concentration of inhibitors was adjusted. The mixed cells were allowed to fuse (48 h at 37°C). The cells were lysed, and transactivated-β-Gal activity was measured by a CPRG colorimetric reaction.
Mutant selection and characterization.
CEM4fx cells were infected with wild-type NL43 virus in the presence of increasing concentrations of FZ41. In fact, the selection of resistance to HIV-1 (NL43) to FZ41 was initiated at a low multiplicity of infection (0.01) in CEM4fx cells and at a drug concentration equal to the IC50, as determined previously. Every 3 to 4 days, the CEM4fx cell culture was monitored for viral production, as measured by p24 protein release. When p24 production was observed, the cell-free culture supernatant was used to infect fresh, uninfected CEM4fx cells in the presence of an equal or higher concentration of the compound. When no virus production was observed, the infected-cell culture was subcultivated in the presence of the same concentration of the compound. The compound concentration was gradually increased. Finally, a selection (two passages) was done with 20 μM drug. RNAs derived from the resistant virus population were extracted, and the integrase-containing fragment was subjected to reverse transcription-PCR (RT-PCR) analysis. These cDNAs were sequenced, and the identified mutations were introduced into NL43 virus by site-directed mutagenesis. Integrated HIV-1 DNAs derived from cells infected with the resistant virus population were extracted, and the integrase-containing fragment was subjected to PCR analysis. These cDNAs were sequenced, and the identified mutations were introduced into NL43 virus by site-directed mutagenesis.
Viruses fitness.
Two days before the infection, 8,000 HeLa P4 cells were inoculated into each well of 96-well plates. Cells were infected with 5 ng of p24 per well, and supernatant was harvested every day from day 3 to day 7 postinfection. Viral production was evaluated by measuring the level of p24 antigen in culture supernatant by ELISA (Perkin-Elmer Life Sciences).
Quantitative PCR experiments.
HeLa cells were transfected with pNL43-encoding wild-type HIV-1 by the calcium phosphate method and grown for 72 h. The supernatant was treated with DNase to remove untransfected plasmid DNA.
To quantify total RNA, HeLa P4 cells were infected with wild-type NL43 in the presence or absence of FZ41 (20 μM) or dextran sulfate (8 μg/ml). After 2 h, RNAs were extracted and used for quantitative RT-PCR experiments. The reverse transcription step was done with a poly(T)12-18 oligonucleotide as the primer, and the amount of cDNA was measured with ARNV+ (5′-CTGTACTGGGTCTCTCTGGTTAGA-3′) and ARNVA2− (5′-TTTTTTTTTTTTTGAGCACTC-3′) as the primers and ARN SONDE (5′-6-carboxyfluorescein [FAM]-TCTGGCTAACTAGGGAACCCACTGCTTAA-6-carboxy tetramethylrhodamine [TAMRA]-3′) as the Q-PCR probe.
To quantify total cDNA, 0.2 × 106 HeLa P4 cells were placed in each well of a six-well plate 24 h before infection and pretreated with 25 μM drug overnight or not pretreated. Cells were washed with phosphate-buffered saline (PBS) and infected with 100 ng of DNase-treated P24 viral stock in the presence or absence of drugs (25 μM). Cells were spinoculated for 2 h at 2,500 rpm at 4°C. The viral supernatant was then replaced by drug-containing (or not) fresh medium, and cells were grown for 5 h at 37°C. Cells were washed twice with PBS and treated with trypsin. DNA was then extracted with the QIAamp DNA blood minikit (Qiagen).
The amount of cDNA present at the end of RT was determined in an ABI7000 thermocycler (Perkin-Elmer) with gag+ (5′-GCGAAAGTAAAGCCAGAGGAGAT-3′) and gag− (5′-TTTTGGCGTACTCACCAGTCG-3′) as the amplification primers and gag-sonde (5′-FAM-CTCGACGCAGGACTCGGTTGCT-TAMRA-3′) as the Q-PCR probe.
To quantify integrated DNA, MT4 cells were infected with DNase-treated viral supernatant for 2 h and then washed twice with PBS. Cells were grown for 3 days in the presence or absence of drugs. Total DNA was extracted using the QIAamp DNA blood minikit (Qiagen). Integrated DNA was then measured by Alu-LTR Q-PCR essentially as described above with MH535 (5′-ACTAGGGAACCCACTGCTTAAG-3′) and SB704 (5′-GCTGGGATTACAGGCGTGAG-3′) as the PCR primers and LTR-sonde (5′-FAM-CGGGCACACACTACTTGAAGCACTC-TAMRA-3′) as the Q-PCR probe.
RESULTS
SQLs are HIV-1 inhibitors that are weakly cytotoxic.
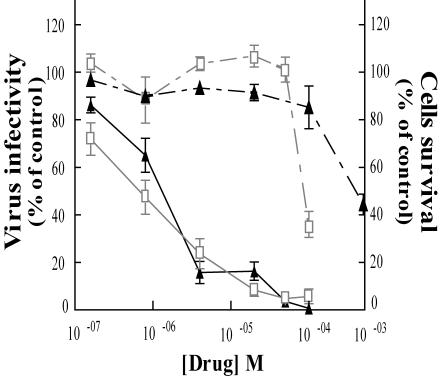
We first tested the antiviral activity of SQLs on the laboratory HIV strain NL43. HeLa P4 cells are HeLa derivatives that express the CD4 membrane receptor and carry the β-Gal gene under the transcriptional control of the HIV-1 LTR. When cells are infected with HIV, the production of Tat results in the trans-activation of the reporter gene. β-Gal activity can then be measured by a colorimetric reaction with CPRG as the substrate. In this test, FZ41 (Fig. 1) was as active as the DKA L-731988, with an IC50 of between 1 and 4 μM in HeLa P4 CD4+ cells (Fig. 2). When using lymphoid CEM4fx cells, the IC50 was about 5 to 10 μM (data not shown). In HeLa and CEM4fx cells, SQL was less cytotoxic than DKA, as the 50% effective concentration was about 300 μM for FZ41 and nearly 100 μM for L-731988, giving a potential therapeutic index of over 250 for FZ41.
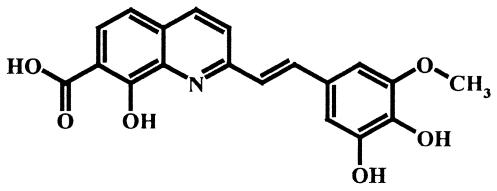
FIG. 1.
Chemical structure of the FZ41 compound.
FIG. 2.
SQL derivatives are potent HIV-1 inhibitors that are weakly cytotoxic. HeLa P4 cells were infected with NL43 in the presence of increasing drug concentrations. First, β-Gal overproduction was quantified by CPRG assay (continuous lines); second, cell survival was measured by MTT assay (dashed lines) 72 h after infection. Triangles, FZ41; squares, L-731988. Curves are the means of at least three experiments, and results are expressed as percentages of the value for the untreated infected cells.
SQLs are active even when added after infection.
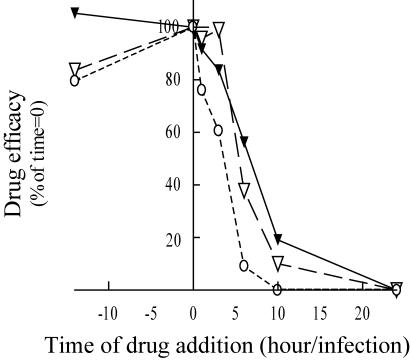
We examined the mode of action of our compounds by a time-of-addition experiment (Fig. 3). Cells were either treated with drugs 14 h before the onset of infection, at the time when infection was initiated (time zero), or after the initiation of infection. Early inhibitors that do not inhibit entry are active when added after infection (entry of the virus) but before the late replication steps of the virus (i.e., before or during integration, which is believed to occur about 12 to 24 h after infection onset).
FIG. 3.
SQLs are active even when added after infection: a time-of-addition experiment. HeLa P4 cells were infected with NL43. The drug was added to the cell culture 14 h before the infection; at the onset of infection; or 2, 3, 5, 10, or 24 h after the onset of infection. FZ41 (▾) was tested, and AZT (○) and nevirapine (▿) were used as controls for inhibition at early replication steps. β-Gal overproduction was quantified by CPRG assay 72 h after infection. Curves are the means of at least three experiments, and IC50s were determined as described in Materials and Methods. The IC50s are expressed as percentages of the IC50 when the drug was added at the same time as the virus.
Zidovudine (AZT) and nevirapine were used as controls for efficient inhibition of early viral cycle steps postentry but before virus integration. As expected, AZT and nevirapine were active when added 14 h before infection and addition at this time was nearly 85% as efficient as addition of the drug at the same time as the virus. The inhibitory effect of AZT rapidly decreased, and only 10% of the activity remained when the drug was added 5 h after infection. This rapid decrease in activity is probably due to the obligatory metabolism of AZT in cells to produce AZT-triphosphate, which is the real RTI. Nevirapine remained active for a longer period, and it had less activity when it was added 3 h or more after infection. FZ41 was also active when added before infection and remained active for at least 5 h (when the drug was added 5 h after infection, its activity was 58% of that when it was added at the same time as the virus). This suggests that entry is not the SQL target. Moreover, when FZ41 was added 24 h after infection, it did not display any antiviral activity, demonstrating that this molecule is not active at postintegration steps. This experiment strongly suggests that SQLs are active on early but postentry viral life cycle steps (i.e., they may have anti-integrase activity).
SQLs do not interfere with virus entry.
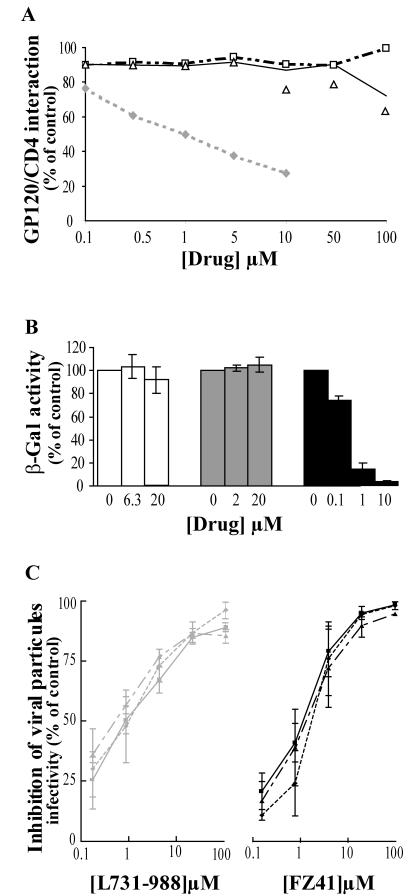
To ensure that SQLs were not entry inhibitors, we analyzed the competition of these molecules with the GP120/CD4 interaction in vitro. Neither FZ41 nor KHD161, another member of the SQL family, inhibited the GP120/CD4 interaction, even at high concentration (100 μM) (Fig. 4A). CD4M9, the positive control, was able to compete for the envelope protein (49). A cell-to-cell fusion experiment showed that FZ41 did not interfere with the membrane fusion mechanism (Fig. 4B). In fact, neither L-731988 nor FZ41 inhibited cell-to-cell fusion, even at concentrations as high as 20 times the IC50, whereas RPR103611, a betulinic acid derivative known to inhibit HIV-1 entry (IC50 = 0.7 μM [29]), was fully active. Moreover, cell fusion was not inhibited when the HIV-1 GP120 protein was replaced by a HIV-2 (NDK) envelope protein (not shown).
FIG. 4.
SQLs do not affect the entry pathway. (A) CD4/GP120 interaction in vitro. A binding competition experiment was performed using the CD4-mimetic peptide CD4M9 (diamonds) as a positive control. Increasing concentrations of SQLs (KHD161 [squares] and FZ41 [triangles]) were tested. (B) Cell-to-cell fusion experiment. HeLa P4 cells were cocultured with HeLa 243 cells for 48 h in the presence of increasing drug concentrations. β-Gal was measured by CPRG colorimetric assay. Results represent the means of three experiments and are expressed as percentages of cell fusion without any treatment. FZ41 (white bars) and L-731988 (gray bars) were used as integrase inhibitors, and RPR103611 (black bars), a betulinic acid derivative, was used as a positive control. (C) SQL activity is not affected by the viral envelope. HeLa P4 cells were infected the NL43 virus carrying the VSV-G (triangles), AMLV (diamonds), or HIV (squares) envelope in the presence of increasing drug concentrations. β-Gal was quantified by the CPRG assay 72 h after infection. Curves are the means of at least three experiments, and results are expressed as percentages of the value obtained for the untreated infected cells.
Finally, we generated ΔEnv NL43 viruses with three different type of envelope to check the effect of the viral entry pathway on drug activity (Fig. 4C). The VSV-G protein binds to phosphatidylserine and other negatively charged lipids (34), leading to an infection by an endocytotic mechanism (1). The AMLV envelope mediates infection via a membrane fusion mechanism involving a specific interaction between the envelope protein and the phosphate symporter membrane receptor Glvr1 (Pit1) (12). The NL43 envelope protein (HIV GP120/GP41 glycoprotein) induces membrane fusion via a GP120/CD4-specific interaction (14). Modulation of the viral cellular infection pathway did not modify drug activity, and IC50s remained near 1 to 2 μM (Fig. 4C).
All these experiments strongly suggest that the cellular target of SQLs is independent from the viral entry pathway.
SQLs decrease the amount of late cDNA present but neither entry of genomic RNA nor reverse transcriptase activity at micromolar concentrations.
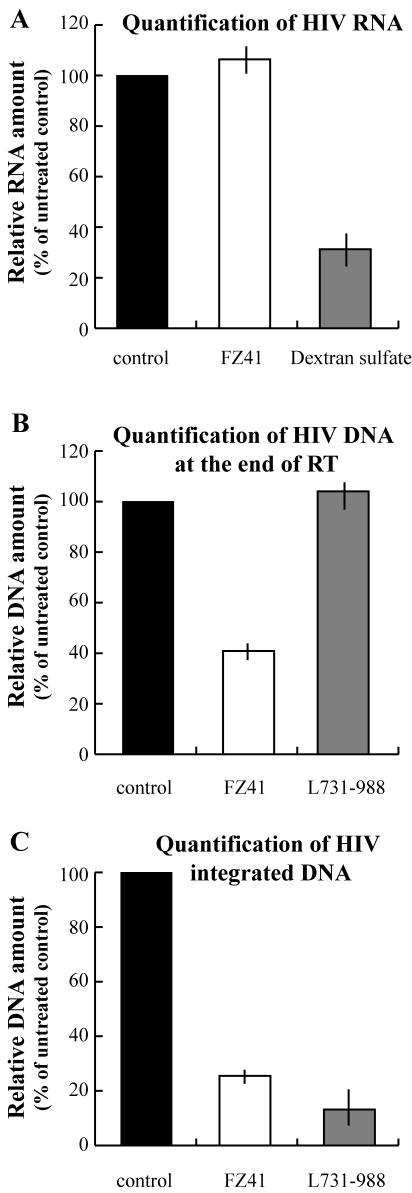
To determine the target of SQLs, we quantified the viral nucleic acid species present in the cells after infection (Fig. 5). To avoid plasmid DNA contamination, HIV viral stocks were treated with DNase before cell infection. Three quantitative PCR assays were developed to quantify entered genomic RNA, total reverse transcribed DNA, and integrated DNA. For the first assay (Fig. 5A), cells were treated with the drug and HIV concomitantly. Two hours after infection, the RNAs were extracted and used as a template for quantitative RT-PCR amplification. For late-reverse-transcription cDNA quantification (Fig. 5B), cells were pretreated with drugs for 24 h and the infection occurred in the presence of drugs. Cells were grown in the presence of drugs for 5 h, and DNA was extracted and subjected to Q-PCR. Finally, the integrated DNA was quantified by an Alu-LTR PCR assay essentially as previously described (8) (Fig. 5C). Cells were infected in the presence of drugs and then grown in the presence of drugs for 3 days. DNA was then extracted and subjected to Q-PCR.
FIG. 5.
SQLs act after virus entry but before integration. For all Q-PCR experiments, cells were treated either with 25 μM FZ41 (white bars) or 25 μM L-731988 (gray bars) inhibitors or not treated (controls; black bars). (A) Quantitative RT-PCR was used to determine the amount of genomic RNA that entered the cells. Dextran sulfate was used as a control inhibitor for unspecific virus entry. Cells were treated with FZ41 or 8 μg of dextran sulfate/ml. (B) Amount of HIV DNApresent at the end of reverse transcription (RT), as determined by R-Gag Q-PCR. (C) Amount of HIV-integrated DNA, as determined by Alu-LTR Q-PCR.
We used dextran sulfate, a polyanion that inhibits the binding of virions to CD4 cells, as a control for nonspecific entry inhibition (36). An 8-μg/ml concentration of dextran sulfate dramatically decreased the amount of viral genomic RNA detected in the cells, whereas 20 μM FZ41 had no effect (Fig. 5A). These results show that SQLs do not inhibit the entry of genomic RNA into the cells. Neither unspecific entry (Fig. 5A) nor the specific CD4/GP120 interaction nor the fusion process (Fig. 4) was inhibited by FZ41.
In contrast, the treatment of infected cells with 25 μM FZ41 led to a decrease of the amount of viral DNA detected compared to untreated or 25 μM L-731988-treated cells (Fig. 5B). In fact, FZ41 treatment resulted in a 59% decrease at the end of reverse transcription. Finally, FZ41 treatment reduced the amount of integrated DNA by 75% compared to that in untreated cells (Fig. 5C). Nevertheless, this decrease can be in part explained by the decreasing amount of late cDNA present in the cells.
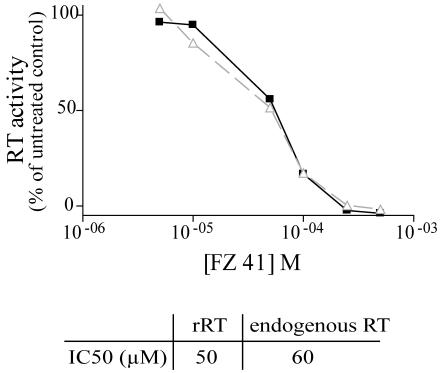
As our results shown a decrease of late cDNA in the presence of SQLs, we tested the activity of SQLs on recombinant and endogenous reverse transcriptase polymerization activity by use of the Quan-T-RT assay system (Amersham Biosciences). This assay is based on the ability of reverse transcriptase to incorporate [3H]dTTP during the extension of an oligo(dT) primer annealed to a poly(rA) template. SQLs weakly inhibited recombinant and endogenous reverse transcriptase activity (Fig. 6). In fact, the IC50 values of FZ41 were 50 μM when 60 pM recombinant reverse transcriptase was used in the assay and 60 μM when 40 pM equivalent endogenous reverse transcriptase was used. These results make it possible to estimate the apparent inhibition constant (Ki) by the following Cheng-Prusoff equation, Ki = Kd · IC50/(s + Kd), where Kd is the apparent dissociation constant of recombinant reverse transcriptase for dTTP and s is the dTTP concentration (11). As s is negligible (5 nM) compared to Kd (6.6 μM) (47), Ki is approximately equal to IC50 (50 μM). As the Ki of FZ41 for integrase is 0.5 μM (17), we can assume that the inhibition of viral particle production in cell cultures is likely due to drug-integrase interactions and not to reverse transcriptase protein inhibition.
FIG. 6.
SQLs weakly inhibit reverse transcriptase (RT) activity. Recombinant RT (60 pM; triangles) or 10 μl of virus equivalent to 40 pM RT activity (squares) was used as the RT source in a Quan-T-RT assay. Relative RT activities (percentages of the activity for untreated sample) are represented as a function of drug concentration in the assay. IC50s are indicated at the bottom.
SQLs are true integrase inhibitors: mutations in the integrase protein lead to SQL resistant viruses.
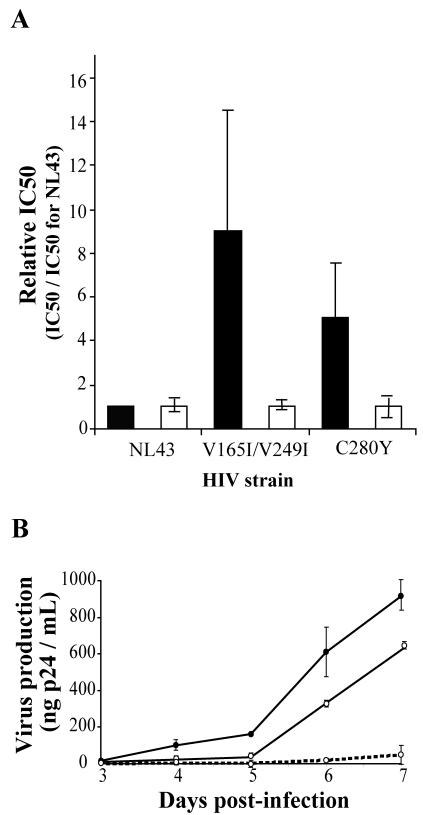
We selected SQL-resistant strains by passaging the virus in the presence of increasing drug concentrations. The resistant viruses obtained in the presence of 20 μM FZ41 were sequenced, and the identified mutations were introduced into the NL43 wild-type strain by site-directed mutagenesis. We further confirmed the effect of the mutations by determining the IC50 of FZ41. Two mutants appeared after incubation with 20 μM FZ41. One of these mutants contained a single mutation (C280Y), and the other contained a double mutation (V165I V249I). The IC50s showed that selected mutations conferred resistance to SQLs, whereas these viruses remained fully sensitive to DKAs (Fig. 7A). The double mutant was the most resistant virus as the IC50 FZ41 for it was nearly ninefold higher than that for the wild-type strain. The IC50 of FZ41 for the C280Y mutant was three- to fivefold higher than that for the wild type. In contrast, the IC50 of the DKA integrase inhibitor was not modified for either of these two mutants.
FIG. 7.
Mutations in the integrase confer resistance to SQLs. (A) Drug efficacy on mutant viruses. The integrase-containing PCR fragments of resistant viruses were sequenced. Detected mutations were then introduced into the wild-type NL43 virus by site-directed mutagenesis. Infected cells were treated with FZ41 (black bars) or L-731988 (white bars), and the IC50s were determined. Results are the means of four independent experiments. (B) Fitness of mutant viruses. HeLa P4 cells were infected with 5 ng of p24 of wild-type (solid circles), C280Y (open circles and solid lines) or V156I V249I (open circles and dashed lines) HIV strains. The p24 release was monitored by ELISA from day 3 to 7 postinfection.
To evaluate whether drug-induced mutations in integrase affect viral fitness, the ability of HIV-1 strains selected in the presence of FZ41 to replicate in HeLa P4 cells was compared to that of the parental HIV-1 (NL43) strain in the absence of drug (Fig. 7B). Replication kinetics of the single (C280Y) mutant and the double (V165I V249I) mutant, as evaluated by p24 measurements, were delayed relative to the those of the replication of the parental HIV-1 (NL43) strain. In fact, 7 days postinfection, only 68 and 5% of wild-type levels of p24 released for the C280Y and V165I V249I mutants, respectively, could be detected.
SQLs are active against HIV-1 strains that are resistant to other antiviral compounds.
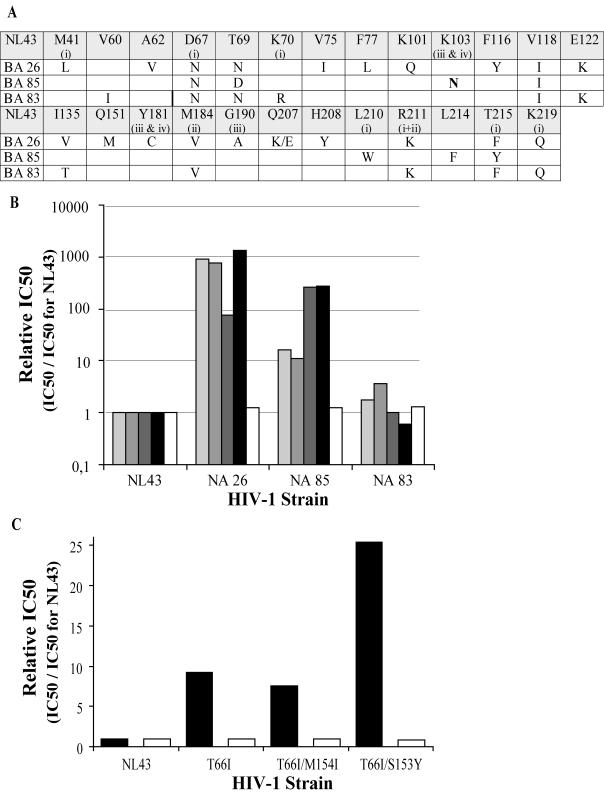
We tested the activity of SQLs against HIV strains that are resistant to RTIs or DKAs. Three virus strains were used as controls for their resistance to RTIs (Fig. 8A and B). These viruses harbored mutations in the reverse transcriptase gene (Fig. 8A). Several of these mutations are specifically associated with the resistance to AZT (M41L, D67N, K70N, L210W, R211K, T215F/Y, and K249Q), lamivudine (3TC) (G190A and R211K), nevirapine (K103N, Y181C, and G190A), or efavirenz (K103N and Y181C) or to combinations of RTIs (for a review, see reference 13). BA26 and BA85 strains harbor mutations conferring resistance to nucleosidic and nonnucleosidic RTIs; BA83 mutations are associated only with resistance to AZT and 3TC. Three other viruses were mutated in the integrase gene to obtain DKA-resistant strains as described previously (27) (Fig. 8C). We genetically engineered the single-mutant T66I strain and the double-mutant T66I M154I and T66I S153Y strains by site-directed mutagenesis.
FIG. 8.
SQLs are active against various HIV-resistant viruses. (A) Mutations in the reverse transcriptase (RT) gene of the three RTI-resistant strains are indicated with respect to RT amino acid numbers. Mutations that are specifically associated with the resistance to AZT, 3TC, nevirapine, or efavirenz are labeled i, ii, iii, and iv, respectively. (B) FZ41 (first bar from the left in each group) was tested on RTI-resistant HIV-1 strains. AZT (second bar) and 3TC (third bar) were used as nucleic RTIs; efavirenz (fourth bar) and nevirapine (fifth bar) were used as nonnucleoside RTIs. (C) FZ41 (white bars) and L-731988 (black bars) were tested against genetically engineered DKA-resistant strains. IC50s were determined as described in Materials and Methods.
As expected, the relative IC50 varied considerably when the drug was an RTI for NA26, NA85, and NA83 strains (Fig. 8B) or L-731988 for integrase mutant strains (Fig. 8C). This demonstrates that the mutants were resistant to inhibitors. In fact, treatment with AZT and 3TC was 1,000-, 10-, and 5-fold less active for BA26, BA85, and BA83, respectively, than for the NL43 strain (Fig. 8B). Nonnucleoside inhibitors were fully active only against the BA83 strain. For BA26 and BA85, the resistance factor was near or greater than 100.
DKA-resistant integrase mutants were treated with the L-731988 DKA inhibitor. As expected, T66I, T66I M154I, and T66I S153Y mutants were 8- to 25-fold less sensitive to DKAs (Fig. 8C). These results are in agreement with previously reported data (27).
In contrast, the relative IC50s, compared with the those for the wild-type NL43 strain, for any mutant strains did not vary when FZ41 was used as an inhibitor. This experiment demonstrates that SQL activity is not impaired by mutations that confer resistance to RTIs or DKAs.
DISCUSSION
To date, only DKA derivatives have been shown to be true integrase inhibitors. These compounds specifically inhibit strand transfer and target the preintegration complex in vivo. Consequently, they are integration inhibitors only and do not interfere with any other viral replication steps (27).
SQLs have been reported to be integrase inhibitors in vitro. It was proposed that in vitro these molecules interfere with integrase DNA binding (17). In fact, these compounds strongly inhibit 3′ processing and also the strand transfer step, even if the inhibition of the second reaction is not as efficient as inhibition of the first one.
Many molecules that inhibit integrase in vitro failed to do so in vivo. For example, chicoric acid derivatives inhibit entry (40). Consequently, we focused on the potential interference of SQLs with virus entry. SQLs remained active when added after virus infection, indicating that they act after this step. Furthermore, SQLs did not inhibit cell-to-cell fusion at concentrations as high as 20 times the IC50 and did not compete with the GP120/CD4 interaction in vitro. Moreover, the pseudotype of the retrovirus envelope did not affect drug activity. These data strongly suggest that SQLs do not interfere with the specific entry pathway. Finally, quantitative RT-PCR experiments demonstrated that SQLs do not inhibit the entry of the genomic RNA.
To determine the step that was targeted by SQLs, we quantified HIV-1 cDNA species present in the cells. We observed that the amount of late cDNA was reduced following the treatment of HIV-1-infected cells with SQLs. In contrast, we did not detect any accumulation of the circular HIV DNA after SQL treatment (not shown), indicating that SQLs are most efficient at steps prior to the nuclear PIC translocation. As micromolar concentrations of SQLs did not inhibit RT activity in vitro, we hypothesize that the decrease of the late cDNA amount is not due to a direct interaction between SQLs and reverse transcriptase. Instead, the inhibition of late cDNA accumulation could be related to a modification of PIC activity. For example, the inability of the integrase to bind to the DNA could destabilize the PIC during or at the end of the RT step and consequently could inhibit DNA synthesis. This hypothesis is reinforced by results for all the integrase mutants previously described, in which a point mutation dramatically reduced the full RT activity (31) and particularly by that in mutants such as the H16V mutant for which in vitro reverse transcriptase activity is not modified despite its inability to synthesize cDNA ex vivo (44, 48, 50).
This hypothesis is reinforced by the fact that SQLs generate resistance mutations in the integrase gene, suggesting that SQLs directly target the integrase. Moreover, SQL-resistant viruses mutated in the integrase gene have been found less fit than the wild type in the absence of drug. Such a loss of viral fitness has been associated with mutations that confer a replicative advantage in the presence of drug (18, 32). Indeed, these mutations could not be due to hazardous replicative error-prone selection but must be related to the selection pressure of SQLs. The replicative kinetics seem to be directly related to the resistance rate for the two mutant viruses. In fact, the double V165I V249I mutant replicated slowly but is more resistant than the single C280Y mutant. Interestingly, the single mutant that harbors the C280Y mutation was impaired in its replicative kinetics whereas the C280S mutant has been shown to replicate similarly to the wild-type strain (3). This result highlights the fact that the C280Y resistant virus is probably not only impaired for disulfide bridge formation. The local conformation of integrase in the vicinity of the C280 residue could be important for viral replication.
It is possible that SQLs target the RNase H activity, given structural similarities between integrase and the RNase H domain of the RT. However, mutations that appeared in the integrase gene after incubation in various concentrations of SQL (that totally abolished wild-type viral replication) led to resistant viruses. The relative IC50s for the single mutant (C280Y) and double (V165I V249I) mutant were five and nine times higher than that of the wild type, respectively. As the C280S mutant was not resistant to SQLs (not shown), the C280Y resistance should not be due to disulfide bridge breaking but more likely is due to a specific integrase conformation that reduces the affinity of SQLs for the integrase. Recent data highlight the importance of the C280 residue for the interaction between drugs and integrase (33). The authors showed that the C280S mutation, in combination with F185K, led to resistance for some DKA derivatives. In the double mutant, one mutation affected the valine 165 of the integrase; it has been proposed that this amino acid participates in the nuclear translocation of the integrase (19). Thus SQLs may not act only on reverse transcription; they may also target integrase nuclear translocation. These mutations did not modify the DKA activity, reinforcing the idea that SQLs and DKAs are two distinct classes of true anti-integrases.
Recently, Pannecouque and coworkers described a new class of anti-integrase molecules (PDPs) that inhibit HIV replication via integration and probably reverse transcription targeting (39). These compounds are more active against 3′ processing activity than against the strand transfer reaction (i.e., V165, the most active compound, exhibited an IC50 of 0.90 μM for the 3′ processing reaction and 16.1 μM for strand transfer) and seem to inhibit the integrase DNA binding in vitro in the same manner as SQLs. For example, these compounds were unable to inhibit the 3′ processing activity of a preformed integrase-DNA complex. Interestingly, Q-PCR experiments showed that HIV DNA could be destroyed after or during reverse transcription. For example, 20 h postinfection, no late cDNA, no circular DNA accumulation, and no integrated DNA were seen. Although the selection of PDP-resistant strains has been unsuccessful, the results obtained by Pannecouque and coworkers are in good agreement with the mode of action of SQLs: targeting integrase can act on steps prior to integration.
Finally, as SQLs are active against various resistant strains, including integrase-mutated DKA-resistant viruses, these compounds represent a new class of antiretroviral compounds that could be used as a complementary means of therapy.
Acknowledgments
We thank F. Clavel and his team for fruitful discussions, and we particularly thank Laurence Jeanson for help in revising the manuscript.
REFERENCES
- 1.Aiken, C. 1997. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J. Virol. 71:5871-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidmen, J. A. Smith, and K. Struhl (ed.). 2003. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 3.Bischerour, J., H. Leh, E. Deprez, J. C. Brochon, and J. F. Mouscadet. 2003. Disulfide-linked integrase oligomers involving C280 residues are formed in vitro and in vivo but are not essential for human immunodeficiency virus replication. J. Virol. 77:135-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, P. O. 1990. Integration of retroviral DNA. Curr. Top. Microbiol. Immunol. 157:19-48. [DOI] [PubMed] [Google Scholar]
- 5.Brown, P. O., B. Bowerman, H. E. Varmus, and J. M. Bishop. 1989. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc. Natl. Acad. Sci. USA 86:2525-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukrinsky, M. I., N. Sharova, M. P. Dempsey, T. L. Stanwick, A. G. Bukrinskaya, S. Haggerty, and M. Stevenson. 1992. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc. Natl. Acad. Sci. USA 89:6580-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 90:6125-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 9.Charneau, P., M. Alizon, and F. Clavel. 1992. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J. Virol. 66:2814-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, H., and A. Engelman. 1998. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc. Natl. Acad. Sci. USA 95:15270-15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, Y., and W. H. Prusoff. 1973. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 percent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22:3099-3108. [DOI] [PubMed] [Google Scholar]
- 12.Chien, M. L., E. O'Neill, and J. V. Garcia. 1998. Phosphate depletion enhances the stability of the amphotropic murine leukemia virus receptor mRNA. Virology 240:109-117. [DOI] [PubMed] [Google Scholar]
- 13.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 14.Dalgleish, A. G., P. C. Beverley, P. R. Clapham, D. H. Crawford, M. F. Greaves, and R. A. Weiss. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763-767. [DOI] [PubMed] [Google Scholar]
- 15.Daniel, R., R. A. Katz, and A. M. Skalka. 1999. A role for DNA-PK in retroviral DNA integration. Science 284:644-647. [DOI] [PubMed] [Google Scholar]
- 16.Denizot, F., and R. Lang. 1986. Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J. Immunol. Methods 89:271-277. [DOI] [PubMed] [Google Scholar]
- 17.Deprez, E., S. Barbe, M. Kolaski, H. Leh, C. Auclair, J. C. Brochon, M. Le Bret, and J. F. Mouscadet. 2004. Mechanism of HIV-1 integrase inhibition by styrylquinoline derivatives in vitro. Mol. Pharmacol. 65:85-98. [DOI] [PubMed] [Google Scholar]
- 18.Domingo, E., and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151-178. [DOI] [PubMed] [Google Scholar]
- 19.Dvorin, J. D., P. Bell, G. G. Maul, M. Yamashita, M. Emerman, and M. H. Malim. 2002. Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J. Virol. 76:12087-12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellison, V., H. Abrams, T. Roe, J. Lifson, and P. Brown. 1990. Human immunodeficiency virus integration in a cell-free system. J. Virol. 64:2711-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engelman, A., K. Mizuuchi, and R. Craigie. 1991. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell 67:1211-1221. [DOI] [PubMed] [Google Scholar]
- 22.Englund, G., T. S. Theodore, E. O. Freed, A. Engleman, and M. A. Martin. 1995. Integration is required for productive infection of monocyte-derived macrophages by human immunodeficiency virus type 1. J. Virol. 69:3216-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Espeseth, A. S., P. Felock, A. Wolfe, M. Witmer, J. Grobler, N. Anthony, M. Egbertson, J. Y. Melamed, S. Young, T. Hamill, J. L. Cole, and D. J. Hazuda. 2000. HIV-1 integrase inhibitors that compete with the target DNA substrate define a unique strand transfer conformation for integrase. Proc. Natl. Acad. Sci. USA 97:11244-11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eustice, D. C., P. A. Feldman, A. M. Colberg-Poley, R. M. Buckery, and R. H. Neubauer. 1991. A sensitive method for the detection of beta-galactosidase in transfected mammalian cells. BioTechniques 11:739-740, 742-743. [PubMed] [Google Scholar]
- 25.Fujiwara, T., and K. Mizuuchi. 1988. Retroviral DNA integration: structure of an integration intermediate. Cell 54:497-504. [DOI] [PubMed] [Google Scholar]
- 26.Hansen, M. S., S. Carteau, C. Hoffmann, L. Li, and F. Bushman. 1998. Retroviral cDNA integration: mechanism, applications and inhibition. Genet. Eng. 20:41-61. [DOI] [PubMed] [Google Scholar]
- 27.Hazuda, D. J., P. Felock, M. Witmer, A. Wolfe, K. Stillmock, J. A. Grobler, A. Espeseth, L. Gabryelski, W. Schleif, C. Blau, and M. D. Miller. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646-650. [DOI] [PubMed] [Google Scholar]
- 28.Katz, R. A., and A. M. Skalka. 1994. The retroviral enzymes. Annu. Rev. Biochem. 63:133-173. [DOI] [PubMed] [Google Scholar]
- 29.Labrosse, B., O. Pleskoff, N. Sol, C. Jones, Y. Henin, and M. Alizon. 1997. Resistance to a drug blocking human immunodeficiency virus type 1 entry (RPR103611) is conferred by mutations in gp41. J. Virol. 71:8230-8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaFemina, R. L., C. L. Schneider, H. L. Robbins, P. L. Callahan, K. LeGrow, E. Roth, W. A. Schleif, and E. A. Emini. 1992. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J. Virol. 66:7414-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leavitt, A. D., G. Robles, N. Alesandro, and H. E. Varmus. 1996. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J. Virol. 70:721-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maeda, Y., D. J. Venzon, and H. Mitsuya. 1998. Altered drug sensitivity, fitness, and evolution of human immunodeficiency virus type 1 with pol gene mutations conferring multi-dideoxynucleoside resistance. J. Infect. Dis. 177:1207-1213. [DOI] [PubMed] [Google Scholar]
- 33.Marchand, C., A. A. Johnson, R. G. Karki, G. C. Pais, X. Zhang, K. Cowansage, T. A. Patel, M. C. Nicklaus, T. R. Burke, Jr., and Y. Pommier. 2003. Metal-dependent inhibition of HIV-1 integrase by beta-diketo acids and resistance of the soluble double-mutant (F185K/C280S). Mol. Pharmacol. 64:600-609. [DOI] [PubMed] [Google Scholar]
- 34.Marsh, M., and A. Helenius. 1989. Virus entry into animal cells. Adv. Virus Res. 36:107-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, M. D., B. Wang, and F. D. Bushman. 1995. Human immunodeficiency virus type 1 preintegration complexes containing discontinuous plus strands are competent to integrate in vitro. J. Virol. 69:3938-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitsuya, H., D. J. Looney, S. Kuno, R. Ueno, F. Wong-Staal, and S. Broder. 1988. Dextran sulfate suppression of viruses in the HIV family: inhibition of virion binding to CD4+ cells. Science 240:646-649. [DOI] [PubMed] [Google Scholar]
- 37.Ouali, M., C. Laboulais, H. Leh, D. Gill, D. Desmaele, K. Mekouar, F. Zouhiri, J. d'Angelo, C. Auclair, J. F. Mouscadet, and M. Le Bret. 2000. Modeling of the inhibition of retroviral integrases by styrylquinoline derivatives. J. Med. Chem. 43:1949-1957. [DOI] [PubMed] [Google Scholar]
- 38.Panganiban, A. T., and H. M. Temin. 1984. The retrovirus pol gene encodes a product required for DNA integration: identification of a retrovirus int locus. Proc. Natl. Acad. Sci. USA 81:7885-7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pannecouque, C., W. Pluymers, B. Van Maele, V. Tetz, P. Cherepanov, E. De Clercq, M. Witvrouw, and Z. Debyser. 2002. New class of HIV integrase inhibitors that block viral replication in cell culture. Curr. Biol. 12:1169-1177. [DOI] [PubMed] [Google Scholar]
- 40.Pluymers, W., N. Neamati, C. Pannecouque, V. Fikkert, C. Marchand, T. R. Burke, Jr., Y. Pommier, D. Schols, E. De Clercq, Z. Debyser, and M. Witvrouw. 2000. Viral entry as the primary target for the anti-HIV activity of chicoric acid and its tetra-acetyl esters. Mol. Pharmacol. 58:641-648. [DOI] [PubMed] [Google Scholar]
- 41.Pommier, Y., and N. Neamati. 1999. Inhibitors of human immunodeficiency virus integrase. Adv. Virus Res. 52:427-458. [DOI] [PubMed] [Google Scholar]
- 42.Race, E., E. Dam, V. Obry, S. Paulous, and F. Clavel. 1999. Analysis of HIV cross-resistance to protease inhibitors using a rapid single-cycle recombinant virus assay for patients failing on combination therapies. AIDS 13:2061-2068. [DOI] [PubMed] [Google Scholar]
- 43.Sakai, H., M. Kawamura, J. Sakuragi, S. Sakuragi, R. Shibata, A. Ishimoto, N. Ono, S. Ueda, and A. Adachi. 1993. Integration is essential for efficient gene expression of human immunodeficiency virus type 1. J. Virol. 67:1169-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shin, C. G., B. Taddeo, W. A. Haseltine, and C. M. Farnet. 1994. Genetic analysis of the human immunodeficiency virus type 1 integrase protein. J. Virol. 68:1633-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stevenson, M., T. L. Stanwick, M. P. Dempsey, and C. A. Lamonica. 1990. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 9:1551-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tasara, T., G. Maga, M. O. Hottiger, and U. Hubscher. 2001. HIV-1 reverse transcriptase and integrase enzymes physically interact and inhibit each other. FEBS Lett. 507:39-44. [DOI] [PubMed] [Google Scholar]
- 47.Thrall, S. H., R. Krebs, B. M. Wohrl, L. Cellai, R. S. Goody, and T. Restle. 1998. Pre-steady-state kinetic characterization of RNA-primed initiation of transcription by HIV-1 reverse transcriptase and analysis of the transition to a processive DNA-primed polymerization mode. Biochemistry 37:13349-13358. [DOI] [PubMed] [Google Scholar]
- 48.Tsurutani, N., M. Kubo, Y. Maeda, T. Ohashi, N. Yamamoto, M. Kannagi, and T. Masuda. 2000. Identification of critical amino acid residues in human immunodeficiency virus type 1 IN required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells. J. Virol. 74:4795-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vita, C., E. Drakopoulou, J. Vizzavona, S. Rochette, L. Martin, A. Menez, C. Roumestand, Y. S. Yang, L. Ylisastigui, A. Benjouad, and J. C. Gluckman. 1999. Rational engineering of a miniprotein that reproduces the core of the CD4 site interacting with HIV-1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA 96:13091-13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu, X., H. Liu, H. Xiao, J. A. Conway, E. Hehl, G. V. Kalpana, V. Prasad, and J. C. Kappes. 1999. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J. Virol. 73:2126-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoder, K. E., and F. D. Bushman. 2000. Repair of gaps in retroviral DNA integration intermediates. J. Virol. 74:11191-11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zouhiri, F., J. F. Mouscadet, K. Mekouar, D. Desmaele, D. Savoure, H. Leh, F. Subra, M. Le Bret, C. Auclair, and J. d'Angelo. 2000. Structure-activity relationships and binding mode of styrylquinolines as potent inhibitors of HIV-1 integrase and replication of HIV-1 in cell culture. J. Med. Chem. 43:1533-1540. [DOI] [PubMed] [Google Scholar]