Abstract
Artificial minigenomes are powerful tools for studying the replication and transcription of negative-strand RNA viruses. Bunyamwera virus (BUN; genus Orthobunyavirus, family Bunyaviridae) is an arbovirus that shows fundamental biological differences when replicating in mammalian versus mosquito cells. To study BUN RNA synthesis in mosquito cells, we developed a bacteriophage T7 RNA polymerase-based minireplicon system similar to that described previously for mammalian cells. An Aedes albopictus C6/36-derived mosquito cell line stably expressing T7 RNA polymerase was established. Viral proteins and artificial minigenomes (containing Renilla luciferase as a reporter) were transcribed and expressed in these cells from transfected T7 promoter-containing plasmids. Transcription of the minigenome required two viral proteins, the nucleocapsid protein N and the RNA-dependent RNA polymerase L, a situation similar to that in mammalian cells. However, unlike the situation in mammalian cells, the viral polymerase was not inhibited by the viral nonstructural protein NSs. We also report that promoter strength is different for vertebrate versus invertebrate cells. The development of this system opens the way for a detailed comparison of bunyavirus replication in cells of disparate phylogeny.
Bunyamwera virus (BUN; genus Orthobunyavirus) is used as a model with which to study the biology of the Bunyaviridae, a family of mostly arthropod-borne viruses. Its genome consists of three segments of single-stranded RNA of negative polarity. The largest segment, L, codes for an RNA-dependent RNA polymerase (L protein). The M segment codes for a precursor to two virion glycoproteins (Gn and Gc) and for a nonstructural protein (NSm). The smallest segment, S, codes for the nucleoprotein, N, and for a second nonstructural protein called NSs; these two proteins are translated from the same mRNA but in different open reading frames (ORF). The N protein associates with the genomic and antigenomic RNA segments to form helical ribonucleoprotein complexes (RNP) termed nucleocapsids. Genome replication and transcription take place in the cytoplasm, while virus budding generally occurs at the Golgi apparatus (7, 8).
BUN replication and transcription in mammalian cells have been investigated by use of a minireplicon system, based on a Renilla luciferase reporter gene cloned in an antisense orientation between the 3′ and 5′ noncoding terminal sequences of the L, M, or S segment under the control of the φ10 promoter from bacteriophage T7 (22). The BUN noncoding regions contain promoter sequences necessary for transcription and/or replication. The viral L and N proteins, necessary and sufficient for replication and transcription, are expressed from pTM1-based plasmids (13) under the control of a φ10 promoter where expression is enhanced by the presence of the encephalomyocarditis virus (EMCV) internal ribosome entry sequence (IRES). The minigenome mimics a viral genomic RNA, and its encapsidation, transcription, and replication are demonstrated by measuring Renilla luciferase (1, 22). A number of T7 RNA polymerase expression systems have been tested, and at present we use BSR-T7/5 cells, which stably express T7 RNA polymerase (6). Similar systems in mammalian cells have described for other bunyaviruses such as La Crosse, Uukuniemi, Hantaan, and Crimean-Congo hemorrhagic fever viruses (2, 10-12).
In nature BUN is transmitted by mosquitoes, and in the laboratory it is able to infect both mammalian cells and arthropod cells, such as Aedes albopictus C6/36 mosquito cells. Infection of mammalian cells leads to rapid shutoff of host protein synthesis and apoptosis of infected cells in the late stages of infection. Infection of C6/36 mosquito cells by BUN leads to persistent infection (9, 18) without induction of apoptosis (unpublished data), a phenomenon similar to the biology of La Crosse virus in mosquito vectors (3). In order to study the differences in BUN replication between mammalian and mosquito cells, we have established a BUN minireplicon system in A. albopictus C6/36 cells. We show that, as in mammalian cells, only the viral N and L proteins are needed for transcription of the minireplicon, but the pattern of promoter activity of the three genome segments differs in mosquito versus mammalian cells. In contrast to the situation in mammalian cells (22), the NSs protein does not repress the viral polymerase, suggesting that the role of NSs may differ during viral replication in these different cell types.
MATERIALS AND METHODS
Media, cells, and viruses.
BSR-T7/5 cells, which stably express T7 RNA polymerase (6), were a kind gift of K.-K. Conzelmann. BHK-21 and BSR-T7/5 cells were maintained in Glasgow minimal essential medium supplemented with 10% tryptose phosphate broth, 10% fetal calf serum, and, for BSR-T7/5 cells only, 1 mg of Geneticin per ml. A. albopictus C6/36 (mosquito) cells were maintained in Leibovitz's L-15 medium supplemented with 10% fetal calf serum and 8% tryptose phosphate broth. Mosquito cells were incubated at 28°C, their usual growth temperature, unless otherwise indicated. Mammalian cells were grown at 37°C. Working stocks of wild-type BUN (wtBUN) and the mutant BUNdelNSs 9a were grown in BHK-21 cells at 33°C, and titers were determined by plaque assays on BHK-21 cells as previously described (4, 20).
Plasmids.
Plasmids pTM1-BUNN, pTM1-BUNM, pTM1-BUNL, pTM1-BUNNSs, pTM1-FF-Luc, pT7riboBUNLRen(−), pT7riboBUNMREN(−), pT7riboBUNSREN(−), and pT7riboBUNSREN(−)mut16 have been described previously (4, 22), as has plasmid pT7AcCat, which contains a baculovirus translational enhancer element to increase expression in transfected insect cells (19). To generate plasmids capable of expressing viral proteins in T7 RNA polymerase-expressing cells, pT7AcCat was digested with NcoI and BamHI to eliminate the cat gene, and the ORF for N, L, or NSs, or the complete S-segment ORF (i.e., N plus NSs), was inserted as an BsmBI-BamHI fragment to yield pT7AcN, pT7AcL, pT7AcNSs, or pT7AcS, respectively. The firefly luciferase ORF was isolated as an NcoI/BamHI fragment from plasmid pGL3-control (Promega) and cloned in a similar way to yield pT7AcLuc. This strategy could not be used to clone the M ORF, because it contains several BamHI sites. Therefore, pT7AcCat was first cut with BamHI, the overhanging DNA ends were filled by using Pfu Turbo DNA polymerase, and the cat gene was then excised by digestion with NcoI. The M ORF was amplified by PCR using Pfu Turbo DNA polymerase, with pT7riboBUNM (4) as the template. The amplified DNA was digested with BsmBI (the 5′ PCR primer contained a BsmBI restriction site that would generate an NcoI-compatible end following digestion), and the M ORF was cloned as a blunt-NcoI fragment to yield pT7AcM. Details of the oligonucleotides used to amplify the appropriate coding regions and of the cloning strategies described here are available on request. All constructs were confirmed by DNA sequencing.
Dual-reporter constructs containing the RhPV (Rhopalosiphum padi virus) 5′ IRES (Δ1; the complete 5′ untranslated region, nucleotides 1 to 579), the EMCV IRES, or no IRES flanked by cat at the 5′ end and by firefly luc at the 3′ end have been described previously (23). Monocistronic constructs contained the RhPV IRES (Δ1) cloned in the sense (pSP72Δ1luc) or antisense (pSP72Δ1asluc) orientation downstream of a φ10 promoter. Plasmid pT7ribo-cBUNM-REN contains a Renilla luciferase reporter in the positive sense orientation with BUN M cRNA promoter ends. Plasmid phRL-CMV (Promega) contains a Renilla luciferase reporter gene under the control of a φ10 and a cytomegalovirus (CMV) promoter; plasmid pRL-SV40 (Promega) contains a Renilla luciferase gene under the control of a φ10 and a simian virus 40 promoter.
Selection of stable cell lines.
A stable A. albopictus C6/36 cell line expressing T7 RNA polymerase was isolated by the InsectSelect BSD system (Invitrogen), using blasticidin to select transfected cells. The T7 RNA polymerase ORF was amplified by PCR from pSFV-T7 (14), and the amplified product was first digested with HindIII and XbaI and then inserted into the HindIII/XbaI-digested expression vector pIB/V5-His (Invitrogen). A stop codon was inserted at the 3′ end of the T7 RNA polymerase ORF to avoid expression of the T7 RNA polymerase-V5 epitope fusion protein originally present in pIB/V5-His. Stable polyclonal (C6-IBT7/3) or monoclonal (C6-T7/66) cells were selected according to the instructions of the InsectSelect protocol (Invitrogen) by using 100 μg of blasticidin/ml. Expression of T7 RNA polymerase was monitored by measuring luciferase activity after transfection of pT7AcLuc.
Transfection, minireplicon reconstitution, and reporter gene assays.
Approximately 1 × 106 to 1.5 × 106 mosquito cells or 5 × 105 mammalian cells grown in 35-mm-diameter dishes were transfected by using DAC-30 (Eurogentec, Southampton, United Kingdom) as a transfection agent. For minireplicon assays in mammalian cells, 1 μg of pTM1-BUNL, 0.5 μg of pTM1-BUNN, 0.1 μg of pTM1-FF-Luc, and 0.5 μg of either pT7riboBUNLREN(−), pT7riboBUNMREN(−), or pT7riboBUNSREN(−) with 5 μg of DAC-30 were used. For minireplicon assays in T7 RNA polymerase-expressing mosquito cells, 1 μg of pT7AcN, 5 μg of pT7AcL, 0.2 μg of pT7AcLuc, and 0.5 μg of either pT7riboBUNLREN(−), pT7riboBUNMREN(−), or pT7riboBUNSREN(−) with 10 μg of DAC-30 were transfected per dish. pT7Ribo-cBUNM-REN was used in various assays as indicated. In some experiments, pTM1-BUNS or pT7AcS was used instead of N-expressing plasmids. When NSs expression plasmids were cotransfected, the overall DNA amounts were kept equivalent by adding the empty plasmid pTM1 or pT7AcCat as appropriate. Transfection efficiencies were normalized by cotransfection of the luciferase expression plasmid pTM1-FF-Luc or pT7AcLuc as appropriate in order to calculate induction levels of Renilla luciferase in minireplicon reconstitution assays. Luciferase activities were determined by using a Dual-Luciferase or Single Renilla Luciferase assay kit (Promega) as required; cells were lysed in a total volume of 200 μl of lysis buffer at 24 h posttransfection, and luciferase activities were measured in 1 μl (for mammalian cells) or 40 μl (for mosquito cells) of cell extract.
Infection and metabolic labeling.
BHK-21 and A. albopictus C6/36 cells in 35-mm-diameter dishes were infected at a multiplicity of infection of 5 with wtBUN or BUNdelNSs 9a (5) virus. At 18 h postinfection, cells were labeled in methionine-free medium for 2 h with 50 μCi of [35S]methionine. Extracts were prepared by using radioimmunoprecipitation assay (RIPA) buffer and were analyzed by sodium dodecyl sulfate-18% polyacrylamide gel electrophoresis (SDS-18% PAGE).
Western blotting.
Transfected or infected cells were incubated overnight at 37°C and scraped into 100 μl of RIPA buffer containing Complete protease inhibitor cocktail (Roche). Equal amounts of cell extract were separated by SDS-18% PAGE and transferred to a Hybond C-extra membrane (Amersham), followed by overnight incubation in saturation buffer (phosphate-buffered saline-10% dry milk-0.1% Tween 20). The membrane was incubated for 2 h in saturation buffer containing an anti-NSs antibody (T. J. Hart et al., unpublished data) at a dilution of 1:300, followed by incubation with an alkaline phosphatase-labeled anti-rabbit antibody (Cell Signaling Technology) diluted 1:1,000 in saturation buffer. After a wash in saturation buffer, the membrane was rinsed in phosphate-buffered saline, and the signal was revealed by using SuperSignal WestPico chemiluminescent substrate (Pierce).
RESULTS
Establishment of T7 RNA polymerase-expressing mosquito cells.
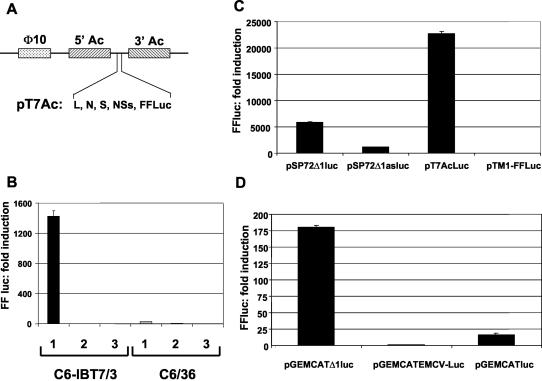
The gene encoding T7 RNA polymerase was amplified by PCR using pSFV-T7 as a template (14) and cloned into the HindIII/XbaI site of expression vector pIB/V5-His (Invitrogen), which also contains a blasticidin resistance gene, to yield pIB/T7-3. A. albopictus C6/36 mosquito cells were transfected with pIB/T7-3, followed by selection with blasticidin (100 μg/ml) as described in Materials and Methods. A blasticidin-resistant population of cells (termed C6-IBT7/3) was grown and analyzed for T7 RNA polymerase activity. Since T7 RNA polymerase would be expressed in the cytoplasm of these cells, translation of mRNA transcribed from transfected plasmid DNA would be initiated in a cap-independent manner. It has previously been shown that translational enhancers routinely used in mammalian cells, such as the EMCV IRES, to allow cap-independent initiation are not functional in mosquito cells but that the translation-enhancing 5′ and 3′ elements of the baculovirus p10 gene can be used as an alternative (14, 19). Therefore, the gene for firefly luciferase was cloned into pT7AcCat from which the cat gene cassette had been removed (Fig. 1A) to yield pT7AcLuc.
FIG. 1.
(A) Components of a T7 RNA polymerase-based BUN-minireplicon system in mosquito cells. The L, N, S, NSs, or firefly luciferase (FFLuc) ORF was cloned into pT7AcCat to produce the pT7Ac constructs used in this study. 5′Ac and 3′Ac, 5′ and 3′ translational enhancers of the baculovirus p10 gene; φ10, T7 RNA polymerase promoter. (B) Induction of firefly luciferase activity in mosquito cells expressing T7 RNA polymerase (C6-IBT7/3) or in parental cells (C6/36). Cells were either transfected with 100 ng of pT7AcLuc (bar 1) or of pT7AcCat (bar 2) or were not transfected (bar 3). (C) Analysis of various IRESs and translational enhancer elements in C6-IBT7/3 mosquito cells expressing T7 RNA polymerase. Monocistronic reporter constructs containing either the RhPV IRES in the sense (pSP72Δ1luc) or antisense (pSP72 Δ1asluc) orientation, the baculovirus p10 gene 5′ and 3′ translational enhancers (pT7AcLuc), or the EMCV IRES (pTM1-FF-Luc) were used. C6-IBT7/3 cells were transfected with 1 μg of the indicated plasmid. Firefly luciferase activities (expressed here as fold induction) were determined at 24 h posttransfection and normalized against an internal standard (the φ10 promoter-containing construct expressing Renilla luciferase). (D) Dual-reporter constructs containing either the RhPV IRES (pGEMCATΔ1luc), the EMCV IRES (pGEMCATEMCV-Luc), or no IRES element (PGEMCATluc), as indicated. C6-IBT7/3 cells were transfected with 1 μg of the indicated plasmid. Firefly luciferase activities (expressed here as fold induction) were determined at 24 h posttransfection and normalized against an internal standard (the φ10 promoter-containing construct expressing Renilla luciferase).
C6-IBT7/3 cells or parental C6/36 cells grown in 35-mm-diameter dishes were transfected with 100 ng of either pT7AcLuc or pT7AcCat as a control. Luciferase activities were determined at 24 h posttransfection. As shown in Fig. 1B, for C6-IBT7/3 cells, firefly luciferase activity was more then 1,500-fold increased over background (or activity in nontransfected cells) in cells transfected with pT7AcCat, while in the parental C6/36 cells, luciferase activity was only just detectable above background. This result shows that functional T7 RNA polymerase is expressed in C6-IBT7/3 cells. Subsequently, a cloned T7 RNA polymerase-expressing cell line, designated C6-T7/66, was isolated and shown to give similar levels of luciferase induction (data not shown). T7 RNA polymerase expression, as determined above, was found to be stable and at similar levels, even after 50 passages (passages still ongoing).
To further test the mosquito cell-T7 RNA polymerase system, we also analyzed an IRES element derived from the aphid-infecting bicistrovirus RhPV (23). Cells were transfected with monocistronic constructs containing the firefly luciferase coding sequence downstream of the RhPV 5′ IRES (Δ1) cloned in the sense (pSP72Δ1luc) or antisense (pSP72Δ1asluc) orientation under the control of the T7 φ10 promoter, and luciferase activity was compared to that generated from pT7AcLuc (Fig. 1C). While the RhPV IRES sense construct produced about 10-fold-higher luciferase activity then its antisense counterpart (5,000-fold induction above background), this activity was still considerably lower than that from pT7AcLuc (about 23,000-fold induction). The EMCV IRES-firefly luciferase construct, pTM1-FF-Luc, gave activity only slightly above background.
We also tested bicistronic reporter constructs (23) containing the RhPV 5′ IRES (Δ1) flanked by the coding sequences for CAT at the 5′ end and firefly luciferase at the 3′ end. As controls, similar bicistronic reporter plasmids with the EMCV IRES or without an IRES were used. Firefly luciferase activity was measured to monitor whether the RhPV IRES was functional in these cells, and as shown in Fig. 1D, about 180-fold induction of activity was observed over background, compared to the relatively low levels obtained in the absence of IRES and the even weaker activity with the EMCV IRES. Thus, for further experiments to establish a minireplicon system, BUN protein coding sequences were cloned into a pT7Ac-background (Fig. 1A).
Minigenome activity in mosquito cells expressing T7 RNA polymerase.
C6-IBT7/3 cells were transfected with various combinations of pT7AcL, pT7AcN, and pT7riboBUNMREN(−) along with pT7AcLuc as an internal transfection control. Cells were incubated at 28°C for 24 h and then lysed, and both Renilla luciferase and firefly luciferase activities were measured. No Renilla luciferase activity was detected in cells transfected with only two of the three BUN plasmids. However, in cells expressing both the BUN N and L proteins together with the minireplicon, about 100-fold induction of Renilla luciferase activity was detected (Fig. 2). Coexpression of the M-segment gene products by transfection of cells with pT7AcM did not enhance minireplicon activity (data not shown). These results indicate that the expressed BUN N and L proteins are necessary and sufficient to transcribe (and, by analogy with the mammalian cell system, probably to replicate) the minireplicon. Experiments with pTM1-based expression plasmids or expression plasmids without translational enhancers gave Renilla luciferase activities only slightly above background, highlighting the need for appropriate translational enhancers (data not shown). However, the minireplicon activity was much lower than that observed in typical mammalian-cell-based experiments. Attempts to optimize the system by trying different amounts of transfected plasmids did not improve activity above that recorded with these amounts (data not shown).
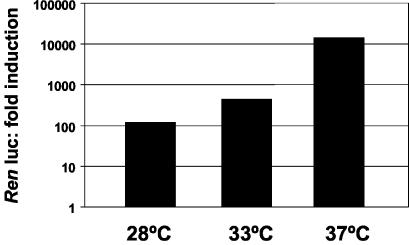
FIG. 2.
Minireplicon activities at 28, 33, or 37°C in mosquito cells expressing T7 RNA polymerase. C6-IBT7/3 cells were transfected with minireplicon components pT7AcL, pT7AcN, pT7riboBUNMREN(−), and pT7AcLuc (internal transfection control). Renilla luciferase (Ren luc) activities were determined at 24 h posttransfection and normalized against the internal transfection control. Activities are expressed as fold induction.
Although mosquito cells are usually maintained at 28°C and BUN replication is quite efficient at that temperature (18), BUN will also replicate at higher temperatures in mammalian cells, e.g., 37°C. Therefore, we determined whether incubating transfected mosquito cells at higher temperatures would improve minireplicon activity. Following transfection with BUN plasmids, C6-IBT7/3 cells were incubated at 33 or 37°C for 24 h, and then Renilla luciferase activity was measured (Fig. 2). The cells survived the higher temperatures for 24 h with no obvious ill effects. Renilla luciferase activity was about five times greater in cells incubated at 33°C than at 28°C (500-fold induction), but at 37°C the activity was much higher, showing 10,000-fold induction over background. These results indicate that we have successfully established a mosquito cell-based BUN minireplicon system.
Transcriptional activities of L, M, and S promoters in mosquito cells.
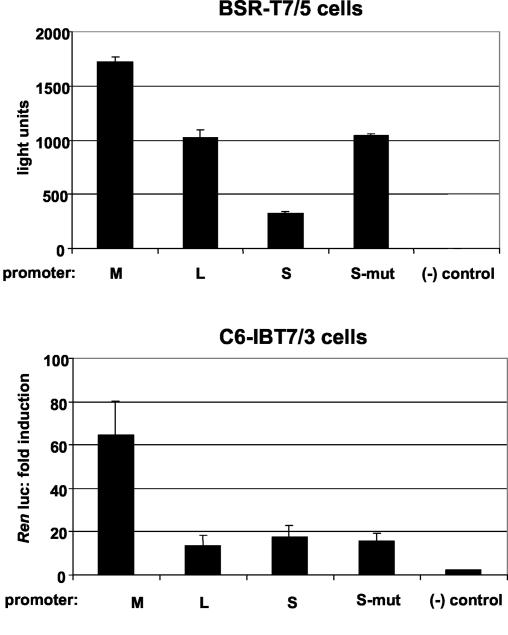
The noncoding regions at the 3′ and 5′ ends of bunyavirus genomes or antigenomes interact to form a panhandle in the RNP, and these noncoding sequences also contain promoter elements that regulate replication levels for each segment. Previous studies with mammalian cells, where minireplicon RNA levels were measured directly, indicated that promoter activity levels differed in the three segments: they were highest in M, intermediate in L, and lowest in S (1). Measurement of Renilla luciferase activity in cell extracts does not allow discrimination between replication and transcription but shows that a biologically active RNP is reconstituted and transcribed in the transfected cell. Therefore, we used this as a means to compare promoter activity in mosquito and mammalian cells. BSR-T7/5 cells were transfected with plasmids pTM1-BUNL (1 μg), pTM1-BUNN (0.5 μg), and pTM1-FF-Luc (100 ng, to normalize transfection efficiencies) and the minigenome-encoding plasmid pT7riboBUNMREN(−), pT7riboBUNLREN(−), pT7riboBUNSREN(−), or pT7riboBUNSREN(−)mut16 (0.5 μg). pT7riboBUNSREN(−)mut16 contains a U-to-G mutation at position 16 of the viral genome end, which results in a strong increase in promoter activity in mammalian cells (15). As shown in Fig. 3 (top), Renilla luciferase activities from the different minigenomes in BSR-T7/5 cells correspond to the replicative abilities of the L, M, and S promoters as deduced by RNA analysis (1); that is, Renilla luciferase activities were strongest in cells transfected with pT7riboBUNMREN(−), followed by pT7riboBUNLREN(−) and pT7riboBUNSREN(−). As previously shown (6), pT7riboBUNSREN(−)mut16 gives about three times higher activity then pT7riboBUNSREN(−), the minigenome containing the wild-type sequence. The same experiment was repeated with C6-IBT7/3 cells by using the optimized amounts of plasmid DNA [5 μg of pT7AcL, 1 μg of pT7AcN, 200 ng of pT7AcLuc, and 0.5 μg of pT7riboBUNMREN(−), pT7riboBUNLREN(−), pT7riboBUNSREN(−), or pT7riboBUNSREN(−)mut16] as described in Materials and Methods. Renilla luciferase activities were measured after 24 h and are displayed in Fig. 3 (bottom). While the strongest activity was mediated by the M-segment-derived minigenome, the activities of the other minigenomes were similar to each other, giving on average four times less Renilla luciferase then the corresponding M sequences. Coexpression of the M-segment gene products (by transfection with pTM1-BUNM in mammalian cells or pT7AcM in mosquito cells) did not affect the relative activities of the different promoters (data not shown). Hence, when a minireplicon system is used, there appears to be a difference in promoter activities between mammalian and mosquito cells.
FIG. 3.
Analysis of promoter strength in mammalian BSR-T7/5 or mosquito C6-IBT7/3 cells. Cells were transfected with pTM1-based (for mammalian cells) or pT7Ac-based (for mosquito cells) L and N expression vectors and the appropriate minigenome plasmid vector pT7riboBUNMREN(−), pT7riboBUNLREN(−), pT7riboBUNSREN(−), or pT7riboBUNSREN(−)mut16. pT7AcLuc was cotransfected as an internal control to standardize luciferase activities. (−) control, negative control, showing background minigenome activity in the absence of functional L protein. Activities are expressed in light units or as fold induction of Renilla luciferase (Ren luc) activity.
The effects of NSs on a reconstituted BUN minireplicon differ in mammalian versus mosquito cells.
It has previously been shown that the NSs protein, whether it is expressed with N from an S-segment-like mRNA or from separate plasmids, strongly inhibits minireplicon activity in mammalian cells (22). Therefore, we investigated whether the same situation held in mosquito cells. Because the baculovirus translation-enhancing elements also work, although to a lesser extent, in mammalian cells, all components were tested first for activity in the mammalian BSR-T7/5 cell-based BUN minireplicon system. As shown in Fig. 4A, expression of both S-segment proteins, N and NSs (pTM1-S and pT7AcS), led to a reduction in Renilla luciferase activity compared to that with expression of N alone (pTM1-N and pT7AcN) in a pT7riboBUNMREN(−)-based assay in BSR-T7/5 cells. The same experiment using pT7riboBUNMREN(−) was repeated with C6-IBT7/3 mosquito cells. As shown in Fig. 4B, expression of both N and NSs from pT7AcS did not result in a reduction in reporter gene activity compared to that with expression of N alone.
FIG. 4.
Effects of NSs on minireplicon activity in mosquito and mammalian cells. Cells were transfected with pTM1-based or pT7Ac-based L expression vectors as appropriate and the minigenome plasmid pT7riboBUNMREN(−), along with plasmids expressing either N and NSs (pTM1-BUNS or pT7AcS) or N alone (pTM1-BUNN or pT7AcN), as indicated. pT7AcLuc was cotransfected as an internal control to standardize luciferase activities. (−) control, negative control without functional L protein. The amounts of DNA transfected were kept constant by adding the corresponding empty plasmid if necessary. Activities are expressed in light units (for BSR-T7/5) or as fold induction of Renilla luciferase (Ren luc) activity (for C6-IBT7/3). (A) Control experiment in BSR-T7/5 cells transfected with N- or N-plus-NSs-expressing pTM1- or pT7Ac-based expression plasmids and minireplicon components as described above. (B) C6-IBT7/3 cells transfected with N- or N-plus-NSs-expressing pT7Ac-based expression plasmids and minireplicon components as described above. (C) Control experiment in BSR-T7/5 cells transfected with NSs-expressing pTM1- or pT7Ac-based plasmids and minireplicon components as described above. (D) C6-IBT7/3 cells transfected or not with the pT7AcNSs plasmid and minireplicon components as described above. (E) Expression of NSs protein in transfected mosquito cells. C6-IBT7/3 cells were either transfected with 1 (lane 1) or 2 (lane 2) μg of pT7AcNSs or left untransfected (lane 3). Cells infected with wtBUN (lane 4) were used as a positive control. Following SDS-PAGE and blotting to a membrane, NSs protein was detected by using an NSs-specific antibody; different exposures were needed to visualize NSs in transfected and infected cells. The positions of NSs and molecular size standards (in kilodaltons) are indicated.
We then tested if NSs expressed from a separate plasmid might have a different effect. As a control, we tested the NSs expression construct by using the minireplicon system in mammalian BSR-T7/5 cells with pT7riboBUNMREN(−) (Fig. 4C). As expected, a decrease in minireplicon activity was observed, even though it was not as strong as that with pTM1-BUNNSs, a difference which is probably due to less-efficient expression from pT7AcNSs in mammalian cells. However, as shown in Fig. 4D, coexpression of NSs with other components of the minireplicon system [pT7AcL, pT7AcN, and pT7riboBUNMREN(−)] in mosquito cells had no major effect on overall minireplicon activity. Expression of NSs proteins in transfected C6-IBT7/3 mosquito cells was confirmed by Western blotting (Fig. 4E).
BUN infection and its effect on protein synthesis in mammalian and mosquito cells.
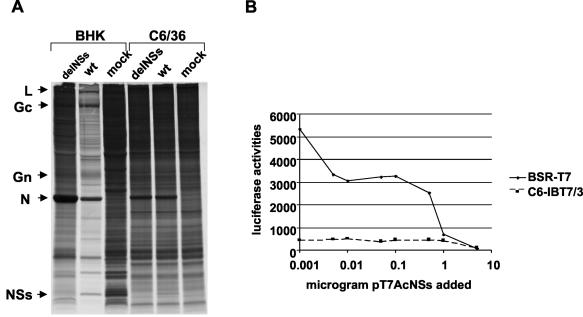
Bridgen and Elliott recently described the production by reverse genetics of a Bunyamwera virus mutant (BUNdelNSs 9a) that does not express NSs (4). As shown in Fig. 5A, infection of mammalian cells with wtBUN leads to shutoff of host protein synthesis, whereas in cells infected with BUNdelNSs 9a, shutoff was markedly reduced. In addition, the N protein was overexpressed in BUNdelNSs 9a-infected cells because of the U-to-G mutation at position 16 of the viral genome (15). In contrast, no shutoff of host protein synthesis was seen in infected C6/36 mosquito cells, and N protein levels were similar in wtBUN- and BUNdelNSs 9a-infected cells (Fig. 5A). This result correlates with the results of minireplicon activity shown in Fig. 3.
FIG. 5.
Effects of NSs expression on protein synthesis. (A) Metabolic labeling of BUN-infected mammalian BHK-21 and mosquito C6/36 cells. Cells were infected at a multiplicity of infection of 5 by wtBUN or BUNdelNSs 9a; at 18 h postinfection, they were labeled with 50 μCi of [35S]methionine for 2 h. Cell extracts were analyzed by SDS-PAGE (18% acrylamide gel). (B) Effects of NSs on transient reporter gene expression in mammalian BSR-T7/5 and mosquito C6-IBT7/3 cells. Cells were cotransfected with phRL-CMV (Promega) (0.5 μg per well) and pT7AcNSs at various concentrations (5, 1, 0.5, 0.1, 0.05, 0.01, 0.005, or 0.001 μg per well). Renilla luciferase reporter gene activities were determined at 16 h posttransfection by using the single Renilla luciferase assay kit (Promega).
The metabolic labeling data suggest that NSs plays a role in the shutoff of host protein synthesis. To test this hypothesis, we compared transient expression of a reporter protein in mammalian BSR-T7/5 cells with that in mosquito C6-IBT7/3 cells also expressing NSs. Cells were transfected with 0.5 μg of plasmid phRL-CMV (Promega), which allows expression of Renilla luciferase from either a T7 promoter or a CMV promoter. (The CMV promoter has been shown previously to be functional in other dipteran cell lines [17].) The cells were cotransfected with various amounts of pT7AcNSs. As shown in Fig. 5B, increasing amounts of the pT7AcNSs expression plasmid led to a strong reduction in Renilla luciferase activity in BSR-T7/5 cells. However, this was not the case in C6-IBT7/3 cells. Although the level of Renilla luciferase was lower than in BSR-T7/5 cells, coexpression of increasing amounts of NSs did not result in a decrease in luciferase activity. This result again indicates that NSs functions differently in mammalian and mosquito cells.
DISCUSSION
In this paper we describe a mosquito cell-based minireplicon system for the study of BUN transcription and replication. We developed polyclonal and monoclonal C6/36 cell-derived cells expressing T7 RNA polymerase and showed that reporter gene activity can be detected after transfection of constructs containing different insect cell translational enhancers, such the RhPV 5′ IRES element (23) or the baculovirus p10 gene 5′ and 3′ translational enhancers (19). The EMCV IRES, which allows high levels of protein expression in mammalian cells, was, as expected (14), ineffective at enhancing protein expression in mosquito cells. The baculovirus translational enhancer was found to be stronger in monocistronic constructs, although the RhPV 5′ IRES functioned efficiently in bicistronic mRNAs to express a second ORF (Fig. 1D). Expression of viral proteins necessary to drive transcription and replication of a BUN-derived minigenome was mediated by transfection of plasmids containing BUN genes flanked by baculovirus p10 gene 5′ and 3′ sequences. Overall, activities were about 1,000- to 2,000-fold lower than the corresponding high reporter gene activities obtained in mammalian BSR-T7/5 cells. We found that reporter gene activities were strongly stimulated at higher temperatures (approximately 10-fold at 33°C relative to activities at 28°C, and approximately 1,000-fold at 37°C), possibly because of higher T7 RNA polymerase activity in the cells. This might be important for experiments where higher levels of protein synthesis are needed, such as rescue of infectious virus from cDNA.
Our system shows fundamental differences between mammalian and mosquito cells. Strikingly, while it has been shown that in mammalian cells, there is a gradation of promoter activity from M to L to S (highest to lowest) (1), this is not the case in mosquito cells. Although the M-segment promoter-based minigenome still gave the strongest reporter gene activity, the L- and S-derived promoters were found to be similar to each other. Moreover, a mutation in the S-segment promoter that has increased activity in mammalian cells (15) did not have the same effect in mosquito cells. Interestingly, this promoter-upregulating mutation was identified in the S segment of BUNdelNSs 9a, which overexpresses N protein in mammalian cells (15) but not, as demonstrated in Fig. 5A, in mosquito cells. The molecular mechanisms behind these biological differences are not clear, but the findings strongly suggest that different host factors might be involved.
Our experiments also show that the function(s) of NSs differs in mammalian and mosquito cells. No significant inhibition of minigenome activity was observed when NSs was expressed either alone or from an intact S-segment mRNA in mosquito cells. In addition, metabolic labeling and reporter gene assays (Fig. 5) indicate that NSs can induce shutoff of host protein synthesis in mammalian cells, but this does not seem to be the case in mosquito cells. Perhaps NSs, which also counteracts the interferon response and early induction of apoptosis in mammalian cells (16, 21), is more important for replication in mammals, whose immune system seems different in many respects from that of arthropods. We are currently analyzing activation of arthropod immune response pathways after infection by wtBUN or BUNdelNSs 9a in order to look for a possible role of NSs.
In summary, we have developed new tools for the study of bunyavirus molecular biology and have shown several differences between replication in mammalian and mosquito cells. This should help us to better understand the fundamental differences between the two host systems.
Acknowledgments
We thank M. van Oers (Utrecht University, Utrecht, The Netherlands) for the gift of plasmid pT7AcCat and K. K. Conzelmann (Max von Pettenkofer Institute, Munich, Germany) for the gift of BSR-T7/5 cells.
T.J.H. was in receipt of a Wellcome Trust Prize Studentship (062679), and work in R.M.E.'s laboratory is supported by Wellcome Trust grants 058356 and 065121.
REFERENCES
- 1.Barr, J. N., R. M. Elliott, E. F. Dunn, and G. W. Wertz. 2003. Segment-specific terminal sequences of Bunyamwera bunyavirus regulate genome replication. Virology 311:326-338. [DOI] [PubMed] [Google Scholar]
- 2.Blakqori, G., G. Kochs, O. Haller, and F. Weber. 2003. Functional L polymerase of La Crosse virus allows in vivo reconstitution of recombinant nucleocapsids. J. Gen. Virol. 84:1207-1214. [DOI] [PubMed] [Google Scholar]
- 3.Borucki, M. K., B. J. Kempf, B. J. Blitvich, C. D. Blair, and B. J. Beaty. 2002. La Crosse virus: replication in vertebrate and invertebrate hosts. Microbes Infect. 4:341-350. [DOI] [PubMed] [Google Scholar]
- 4.Bridgen, A., and R. M. Elliott. 1996. Rescue of a segmented negative-strand RNA virus entirely from cloned complementary DNAs. Proc. Natl. Acad. Sci. USA 93:15400-15404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bridgen, A., F. Weber, J. K. Fazakerley, and R. M. Elliott. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc. Natl. Acad. Sci. USA 98:664-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott, R. M. 1996. The Bunyaviridae: concluding remarks and future prospects, p. 295-332. In R. M. Elliott (ed.), The Bunyaviridae. Plenum Press, New York, N.Y.
- 8.Elliott, R. M. 1997. Emerging viruses: the Bunyaviridae. Mol. Med. 3:572-577. [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott, R. M., and M. L. Wilkie. 1986. Persistent infection of Aedes albopictus C6/36 cells by Bunyamwera virus. Virology 150:21-32. [DOI] [PubMed] [Google Scholar]
- 10.Flick, K., J. W. Hooper, C. S. Schmaljohn, R. F. Pettersson, H. Feldmann, and R. Flick. 2003. Rescue of Hantaan virus minigenomes. Virology 306:219-224. [DOI] [PubMed] [Google Scholar]
- 11.Flick, R., K. Flick, H. Feldmann, and F. Elgh. 2003. Reverse genetics for Crimean-Congo hemorrhagic fever virus. J. Virol. 77:5997-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flick, R., and R. F. Pettersson. 2001. Reverse genetics system for Uukuniemi virus (Bunyaviridae): RNA polymerase I-catalyzed expression of chimeric viral RNAs. J. Virol. 75:1643-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohl, A., A. Billecocq, C. Prehaud, F. Z. Yadani, and M. Bouloy. 1999. Transient gene expression in mammalian and mosquito cells using a recombinant Semliki Forest virus expressing T7 RNA polymerase. Appl. Microbiol. Biotechnol. 53:51-56. [DOI] [PubMed] [Google Scholar]
- 15.Kohl, A., A. Bridgen, E. Dunn, J. N. Barr, and R. M. Elliott. 2003. Effects of a point mutation in the 3′ end of the S genome segment of naturally occurring and engineered Bunyamwera viruses. J. Gen. Virol. 84:789-793. [DOI] [PubMed] [Google Scholar]
- 16.Kohl, A., R. F. Clayton, F. Weber, A. Bridgen, R. E. Randall, and R. M. Elliott. 2003. Bunyamwera virus nonstructural protein NSs counteracts interferon regulatory factor 3-mediated induction of early cell death. J. Virol. 77:7999-8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saraiva, E., P. Fampa, V. Cedeno, M. Bergoin, E. Mialhe, and L. H. Miller. 2000. Expression of heterologous promoters in Lutzomyia longipalpis and Phlebotomus papatasi (Diptera: Psychodidae) cell lines. J. Med. Entomol. 37:802-806. [DOI] [PubMed] [Google Scholar]
- 18.Scallan, M. F., and R. M. Elliott. 1992. Defective RNAs in mosquito cells persistently infected with Bunyamwera virus. J. Gen. Virol. 73:53-60. [DOI] [PubMed] [Google Scholar]
- 19.Scheper, G. C., R. G. Vries, M. Broere, M. Usmany, H. O. Voorma, J. M. Vlak, and A. A. Thomas. 1997. Translational properties of the untranslated regions of the p10 messenger RNA of Autographa californica multicapsid nucleopolyhedrovirus. J. Gen. Virol. 78:687-696. [DOI] [PubMed] [Google Scholar]
- 20.Watret, G. E., C. R. Pringle, and R. M. Elliott. 1985. Synthesis of bunyavirus-specific proteins in a continuous cell line (XTC-2) derived from Xenopus laevis. J. Gen. Virol. 66:473-482. [DOI] [PubMed] [Google Scholar]
- 21.Weber, F., A. Bridgen, J. K. Fazakerley, H. Streitenfeld, N. Kessler, R. E. Randall, and R. M. Elliott. 2002. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J. Virol. 76:7949-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber, F., E. F. Dunn, A. Bridgen, and R. M. Elliott. 2001. The Bunyamwera virus nonstructural protein NSs inhibits viral RNA synthesis in a minireplicon system. Virology 281:67-74. [DOI] [PubMed] [Google Scholar]
- 23.Woolaway, K. E., K. Lazaridis, G. J. Belsham, M. J. Carter, and L. O. Roberts. 2001. The 5′ untranslated region of Rhopalosiphum padi virus contains an internal ribosome entry site which functions efficiently in mammalian, plant, and insect translation systems. J. Virol. 75:10244-10249. [DOI] [PMC free article] [PubMed] [Google Scholar]