Abstract
The potential use of the modified vaccinia virus Ankara (MVA) strain as a live recombinant vector to deliver antigens and elicit protective immune responses against infectious diseases demands a comprehensive understanding of the effect of MVA infection on human host gene expression. We used microarrays containing more than 15,000 human cDNAs to identify gene expression changes in human HeLa cell cultures at 2, 6, and 16 h postinfection. Clustering of the 410 differentially regulated genes identified 11 discrete gene clusters with altered expression patterns after MVA infection. Clusters 1 and 2 (accounting for 16.59% [68 of 410] of the genes) contained 68 transcripts showing a robust induction pattern that was maintained during the course of infection. Changes in cellular gene transcription detected by microarrays after MVA infection were confirmed for selected genes by Northern blot analysis and by real-time reverse transcription-PCR. Upregulated transcripts in clusters 1 and 2 included 20 genes implicated in immune responses, including interleukin 1A (IL-1A), IL-6, IL-7, IL-8, and IL-15 genes. MVA infection also stimulated the expression of NF-κB and components of the NF-κB signal transduction pathway, including p50 and TRAF-interacting protein. A marked increase in the expression of histone family members was also induced during MVA infection. Expression of the Wiskott-Aldrich syndrome family members WAS, WASF1, and the small GTP-binding protein RAC-1, which are involved in actin cytoskeleton reorganization, was enhanced after MVA infection. This study demonstrates that MVA infection triggered the induction of groups of genes, some of which may be involved in host resistance and immune modulation during virus infection.
Interaction between mammalian cells and viruses has an impact on a diverse set of cellular processes. Many of these interactions are characterized by antiviral immune responses and changes in cellular transcriptional, translational, and trafficking machinery that in turn depend on the infection stage and the biological condition of the infected cell. The modified vaccinia virus (VV) Ankara (MVA), derived from the Ankara strain, is a highly attenuated virus. MVA has been passaged more than 500 times in chicken embryo fibroblasts. During the course of attenuation, 15% of the parental viral genome was lost (2, 25); the structural genes remained unaltered, but genes involved in immune evasion factors (4) and host range genes (1, 25, 42) have been deleted or fragmented. MVA produces an infectious cycle in chicken embryo fibroblasts and baby hamster kidney (BHK) cells but not in various human cell lines, including the HeLa cell line (7, 11). Although viral replication depends on cell type, blockade of the morphogenetic program in nonpermissive cells occurs in steps after the formation of immature viral forms, with no alteration in early or late viral gene expression (34, 36). In cultured cells, MVA recombinants produced levels of heterologous protein similar to or higher than those of VV-derived vectors (8, 33, 36). In mammals, MVA recombinants induce protective immunity against a wide spectrum of pathogens (7, 18, 23, 24, 35, 37). MVA may be of use in the generation of live vaccines against infectious diseases and in cancer therapy due to its safety and its ability to evoke protection. The generation of such vaccines demands a comprehensive understanding of the effect of MVA infection on human host gene expression. With DNA microarray technology, the expression of several thousand individual genes can be monitored (19), and this technology has been used to identify cellular genes that are differentially expressed in response to infection with several animal viruses (5, 9, 16, 17, 20, 30, 41, 43). Here, we analyzed host gene expression changes in cultures of the human cervical carcinoma cell line HeLa at 2, 6, and 16 h postinfection by using cDNA microarray technology. During MVA infection, we found increased expression of cellular genes associated with the immune response and with a variety of cellular pathways. This study represents the first global analysis of the transcriptional response of HeLa cells to MVA infection.
MATERIALS AND METHODS
Cells, viruses, and infection conditions.
HeLa cells (from the American Type Culture Collection) were cultured in Dulbecco's medium supplemented with 10% newborn bovine serum and antibiotics. MVA was cultured in BHK-21 cells, purified by banding on sucrose gradients, and titrated on BHK-21 cells by immunostaining of fixed infected cultures with a polyclonal anti-VV protein antibody. The VV Western Reserve (WR) strain was grown in monkey BSC-40 cells, purified by sucrose gradient banding, and titrated on BSC-40 cells by plaque assay. MVA and WR infections were carried out at a multiplicity of infection of 5 PFU/cell.
Microarray fabrication.
The Research Genetics 40K sequence-verified clone human cDNA library (http://www.resgen.com/products/SVHcDNA.php3) was used to generate cDNA arrays as described previously (17). Slides contained 15,360 cDNAs, of which 13,295 correspond to known genes and 2,257 correspond to control genes. Printing on CMT-GAPS II slides (Corning) was performed with a Microgrid II (BioRobotics) at 22°C and 40 to 45% relative humidity.
Microarray hybridization.
Total RNA was isolated from MVA-infected (5 PFU/cell) or mock-infected HeLa cells cultured in 10-cm plates with Ultraspect-II RNA (Biotecx) by following the manufacturer's instructions. Uninfected samples were isolated at each infection time point and processed in parallel with infected cells. Two different samples of RNA from MVA-infected cells at 2, 6, and 16 h postinfection and RNA from corresponding mock-infected cells were processed for analysis. Each RNA was used in two different hybridizations. In one hybridization, the mock-infected sample was labeled with dUTP-Cy3 and the MVA-infected sample was labeled with dUTP-Cy5; in the other, the mock-infected sample was labeled with dUTP-Cy5 and the MVA-infected sample was labeled with dUTP-Cy3. Double labeling was used to abolish labeling and hybridization differences due to specific Cy-dUTP characteristics. A mixture containing 40 μg of RNA, 150 pmol of oligo(dT)20, 0.5 mM dATP, 0.5 mM dGTP, 0.5 mM dCTP, 0.1 mM dTTP, 0.05 mM Cy3/Cy5 dUTP (Amersham), 1× first-strand reaction buffer (Invitrogen), and 10 mM dithiothreitol in a volume of 38 μl was heated (65°C, 5 min) and preincubated (42°C, 5 min), after which 400 U of SuperScript II (Invitrogen) and 40 U of RNase Inhibitor (Roche) were added and the mixture was incubated (42°C, 2.5 h). The reaction was terminated with EDTA, and starting RNA template was removed by adding 2 μl of 10 N NaOH, followed by incubation (20 min, 65°C). The reaction was neutralized by adding 4 μl of 5 M acetic acid. Cy5 and Cy3 probes were mixed, and unincorporated dyes were removed by isopropanol precipitation. Probes were resuspended in deionized water; blocking reagents added to increase specificity were poly(A) (20 μg; Sigma), tRNA (20 μg; Sigma), and human Cot-1 DNA (20 μg; Invitrogen). While probes were drying in a Speed-Vac, microarray slides were prehybridized in a mixture containing 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.5% sodium dodecyl sulfate (SDS), and 1% bovine serum albumin (42°C, 1 h), rinsed five times with water, and dried by centrifugation (563 × g, 1 min). Probes were resuspended in 40 μl of hybridization buffer (50% formamide, 6× SSC, 0.5% SDS, 5× Denhardt's solution) and incubated with slides (42°C, 16 h) in hybridization chambers (Array-It) in a water bath in the dark. After incubation, slides were washed twice in 0.1× SSC-0.1% SDS for 5 min each time and three times in 0.1× SSC for 5 min each time. Finally, slides were dried by centrifugation as described above and scanned on a ScanArray 4000 (Packard Biosciences) by using ScanArray 3.1 software. Raw data were obtained from Cy5 and Cy3 images by using QuantArray 3.0 software (Packard Biosciences) and processed by using SOLAR software (BioALMA, Madrid, Spain). Briefly, background is subtracted from the signal, log10(signal) is plotted versus log2(ratio) and, a lowess normalization is done to adjust most spots to log ratio 0. This value is calculated for all four replicates and a table is obtained with mean signal, change (n-fold), log ratio, standard deviation of the log ratio, and z score (a measure of the proximity of one value [log ratio] to other values with similar signals) (32).
Gene expression analysis.
The original data set containing 13,295 clones per slide was prepared for clustering. Genes with an interreplicate standard deviation of >1 were removed from the analysis. The resulting data set was reduced to 9,749 transcripts that showed a consistent expression value among the four replicates. The z score value was used to eliminate genes that did not show significant expression under at least one experimental condition (32). In this way, only genes with z scores of >2 were selected for clustering. A new data set was created with the 410 transcripts that successfully passed through the filter. After the data were preprocessed, genes were clustered by using Kohonen's classical self-organizing map (12, 22, 40). The resulting 7-by-5 map was analyzed by using the Engene software package (15), available at http://www.engene.cnb.uam.es.
Quantitative real-time RT-PCR.
RNA (1 μg) was reverse transcribed by using the Superscript first-strand synthesis system for reverse transcription-PCR (RT-PCR) (Invitrogen). A 1:40 dilution of the RT reaction mixture was used for quantitative PCR. Primers and the probe set used to amplify H2BFB, PCNT2, WASF1, WAS, interleukin 7 (IL-7), IL-6, IFNG, APEXL2, and FLJ20643 were purchased from Applied Biosystems. RT-PCRs were performed by using Assay-on-Demand optimized to work with TaqMan Universal PCR Master Mix, No AmpErase UNG, as previously described (17). All samples were assayed in duplicate. Threshold cycle values were used to plot a standard curve in which the threshold cycle value decreased in linear proportion to the log of the template copy number. The correlation values of standard curves were always >99%.
Western blot.
HeLa cells were infected at 5 PFU/cell with MVA or WR and collected at 2, 6, and 16 h postinfection in lysis buffer (50 mM Tris-HCl [pH 8.0], 0.5 M NaCl, 10% NP-40, 1% SDS). Equal amounts of protein lysates (10 μg) were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on 14 or 8% gels, transferred to nitrocellulose membranes, and incubated with primary antiactin (Sigma), antitubulin (Sigma), anti-NF-κB (Santa Cruz), anti-RAC-1 (kindly provided by J. C. Gallego), anti-WAS (Santa Cruz), and anti-WAVE (Santa Cruz), followed by peroxidase-conjugated mouse and rabbit secondary antibodies. The blots were developed by using the ECL protocol (Amersham).
Analysis of [35S]methionine labeled proteins.
HeLa cell monolayers in 12-well plates were mock infected or infected with WR or MVA at 5 PFU/cell. At the indicated times post infection, cells were washed with methionine-free medium and incubated with methionine-free medium containing 50 μCi of [35S]methionine per well (30 min, 37°C). Proteins in cell extracts prepared in lysis buffer were fractionated by SDS-12% PAGE and developed by autoradiography.
ELISA.
Secreted IL-6 and gamma interferon (IFN-γ) in the medium of MVA- or WR-infected HeLa cells were measured with the quantitative human IL-6 and IFN-γ kit (BD Biosciences). Aliquots (100 μl) of supernatant from uninfected or infected HeLa cells at 2, 6, and 16 h postinfection were used for ELISA according to the manufacturer's instructions. Captured IL-6 and IFN-γ were quantified at 450 nm with a spectrophotometer. Duplicate samples were measured in two independent experiments.
RESULTS
Gene expression analysis.
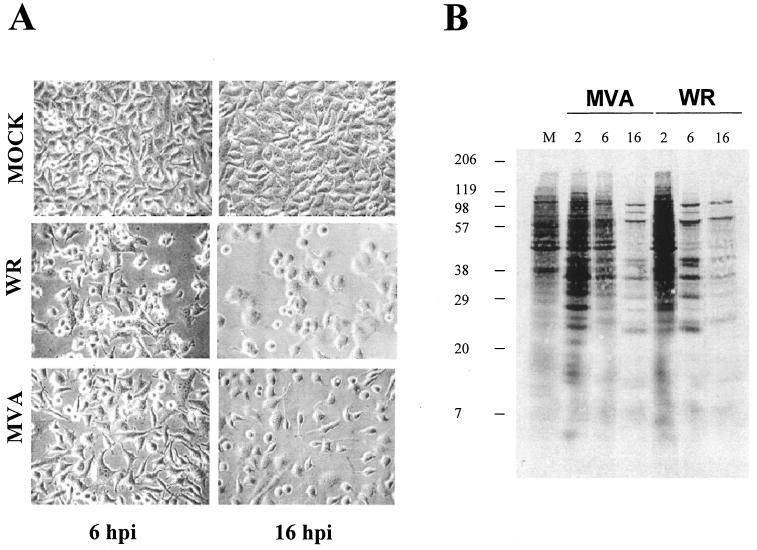
To study the cellular transcriptional response after MVA infection, we first defined the cytopathic effect (CPE) and shutoff of MVA-infected HeLa cells compared to cells infected with the WR strain. WR produced a pronounced CPE at 6 and 16 h postinfection, and this CPE was reduced in MVA-infected cells (Fig. 1A). At a multiplicity of infection of 5 PFU/cell, over 99% of the cells were infected. At 16 h postinfection, more rounded cells were observed after WR infection, whereas MVA produced rounded and bipolar cells, as previously noted (14). The pattern of protein synthesis is shown in Fig. 1B. At 6 h postinfection, WR-infected cells showed a more evident shutoff than MVA-infected cells, while at 16 h postinfection, shutoffs induced by both viruses were similar.
FIG. 1.
Cytopathic effect and protein synthesis pattern of HeLa cells infected with VV WR and MVA strains. (A) Cell morphology after virus infection. Monolayer-cultured HeLa cells were infected with WR or MVA (5 PFU/cell), and CPEs were visualized by phase-contrast microscopy at the indicated times postinfection. hpi, hours postinfection. (B) Pattern of viral protein synthesis. HeLa cells were infected as described above and labeled with [35S]methionine (50 μCi, 30 min) at the times indicated. Cells lysates were analyzed by SDS-12% PAGE and visualized by autoradiography. Based on protein standards, molecular mass (in kDa) is indicated. Uninfected cells (M) served as the control. Postinfection times (in hours) are noted above the gels.
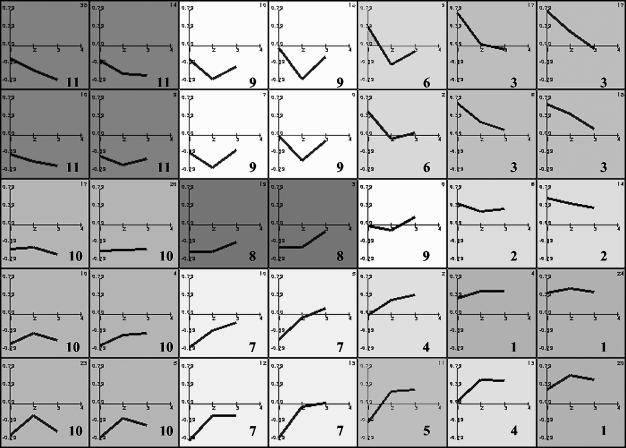
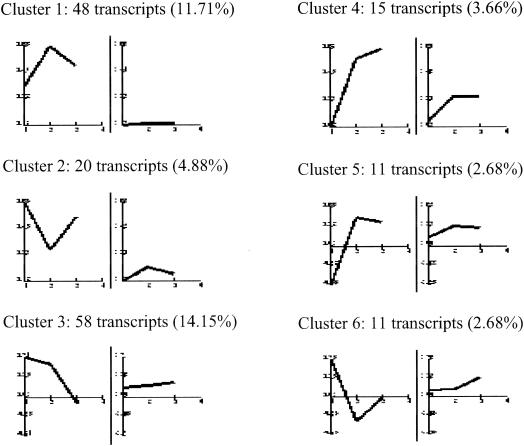
We performed cDNA microarray analysis to determine the relative abundance of specific mRNAs induced in MVA-infected cells compared to that induced in mock-infected cells. The gene expression clusters of the 410 differentially regulated genes are depicted in Fig. 2. Detailed profile analysis led us to group the genes into 11 main clusters according to their behavior at the three time points of MVA infection by using Engene software (15). Clusters 1 and 2 contained 68 transcripts, representing 16.59% of the 410 genes, showing a robust induction pattern that was maintained during the course of infection. Cluster 3 contained 58 transcripts (14.15%), including genes with a generalized induction pattern maintained from 2 to 6 h postinfection that returned to basal levels at 16 h postinfection. Cluster 4 had 15 transcripts (3.66%), with an upregulation pattern maintained from 6 to 16 h postinfection. Cluster 5 contained 11 transcripts (2.68%) that were downregulated at 2 h postinfection and upregulated at 6 and 16 h postinfection. Cluster 6 contained 11 transcripts (2.68%) with upregulated expression only at 2 h postinfection. The average profiles for clusters 1 to 6 are shown in Fig. 3; these six clusters include genes involved in adhesion, cell cycles, immune response, signal transmission, metabolic pathways, and other vital cell processes. Representative human genes upregulated by MVA infection in clusters 1 to 6 are shown in Table 1. Genes whose expression was repressed after MVA infection represented 60.24% of the 410 genes; representative human genes downregulated by MVA in clusters 7 to 11 are detailed in Table 2.
FIG. 2.
Representation of 7-by-5 map obtained by the self-organizing maps algorithm, showing the gene expression cluster for MVA-infected HeLa cells. Each map node represents the average expression profile for a set of similar genes in the data set. Experimental points on the x axis are 1 for 2 h postinfection, 2 for 6 h postinfection, and 3 for 16 h postinfection. The y axis shows normalized expression values. Each cluster is shaded from white to gray and numbered from 1 to 11.
FIG. 3.
Characteristic expression patterns represented in clusters 1 to 6. Shown are mean values (left) and standard deviations (right) of the expression profiles of genes assigned to each cluster. Experimental points on the x axis are 1 for 2 h postinfection, 2 for 6 h postinfection, and 3 for 16 h postinfection. The y axis shows normalized expression values. The values in parentheses are percentages of genes in each cluster with reference to the total of 410 differentially expressed genes.
TABLE 1.
Representative human genes in clusters 1 to 6 (upregulated by MVA)a
| Function or protein description and gene name | Accession no. | Gene symbol | Change (fold) at time (h) postinfection
|
||
|---|---|---|---|---|---|
| 2.00 | 6.00 | 16.00 | |||
| Cluster 1 | |||||
| Adhesion molecules and cytoskeleton | |||||
| Connective tissue growth factor | AA598794 | CTGF | 5.78 | 4.5 | 2.17 |
| Pericentrin | N45326 | PCNT | 3.94 | 3.14 | 2.38 |
| Fibroblast growth factor 9 (glia-activating factor) | AA946776 | FGF9 | 2.05 | 2.58 | 1.41 |
| Attractin | AA683500 | ATRN | 2.16 | 2.6 | 1.41 |
| Adaptin, alpha A | AI018208 | ADTAA | 2.95 | 1.56 | 1.78 |
| Calmegin | AA778675 | CLGN | 2.01 | 2.43 | 1.29 |
| filamin A, alpha (actin-binding protein-280) | AA478436 | FLNA | 3.07 | 3.63 | 2.79 |
| Collagen, type VII, alpha 1 (epidermolysis bullosa, dystrophic, dominant, and recessive) | AA598507 | COL7A1 | 2.35 | 2.71 | 1.47 |
| Cell cycle, DNA damage, apoptosis | |||||
| Immediate early protein | AA496359 | ETR101 | 3.12 | 2.81 | 2.11 |
| CDC7 (cell division cycle 7, Saccharomyces cerevisiae, homolog)-like 1 | N62245 | CDC7L1 | 2.17 | 2.36 | 1.72 |
| Jun B proto-oncogene | N94468 | JUNB | 2.93 | 3.92 | 1.06 |
| Histones | |||||
| H3 histone family, member B | AI399887 | H3FB | 16.34 | 15.67 | 21.71 |
| H4 histone family, member D | AI653010 | H4FD | 12.38 | 24.93 | 24.93 |
| H2B histone family, member B | AA885642 | H2BFB | 9.92 | 10.20 | 17.03 |
| H2A histone family, member N | AI095013 | H2AFN | 7.46 | 4.82 | 11.55 |
| Immune response | |||||
| Interleukin 7 | AI539460 | IL7 | 6.1 | 5.3 | 3.2 |
| Interleukin 15 | N59270 | IL15 | 2.91 | 2.68 | 1.59 |
| Interleukin 8 | AA102526 | IL8 | 2.55 | 2.22 | 1.84 |
| Interleukin 6 (interferon, beta 2) | N98591 | IL6 | 2.65 | 2.14 | 1.56 |
| CD47 antigen (Rh-related antigen, integrin-associated signal transducer) | AA455448 | CD47 | 5.35 | 4.08 | 3.16 |
| CD80 antigen (CD28 antigen ligand 1, B7-1 antigen) | AA983817 | CD80 | 4.29 | 3.36 | 2.57 |
| Interleukin 1, alpha | AA936768 | IL1A | 2.36 | 2.31 | 2.44 |
| Tumor necrosis factor, alpha-induced protein 3 | AA476272 | TNFAIP3 | 2.65 | 2.23 | 1.96 |
| CD8 antigen, beta polypeptide 1 (p37) | AA293671 | CD8B1 | 2.1 | 2.53 | 2.47 |
| TNF receptor-associated factor 3 | AA504259 | TRAF3 | 2.6 | 2.30 | 1.67 |
| Interleukin 22 receptor | AA132964 | IL22R | 3.32 | 3.94 | 2.2 |
| Tumor necrosis factor receptor superfamily, member 17 | AA987627 | TNFRSF17 | 2.97 | 2.58 | 2.24 |
| Miscellaneous | |||||
| Surfactant, pulmonary-associated protein D | AI289238 | SFTPD | 2.73 | 4.38 | 4.14 |
| Thymosin beta 4 X chromosome | AA634103 | TMSB4X | 3.81 | 5.7 | 2.91 |
| Human erythroid isoform protein 4.1 mRNA, complete cds | AA703141 | 2.87 | 3.23 | 1.93 | |
| Signal transmission | |||||
| Dopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2) | AA478553 | DCT | 3.76 | 19.16 | 1.79 |
| Nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) | AA952897 | NFKB2 | 2.03 | 2.22 | 2.34 |
| Mitogen-activated protein kinase kinase 5 | AA250966 | MAP2K5 | 2.37 | 2.74 | 1.69 |
| Nuclear factor (erythroid-derived 2)-like 3 | W76339 | NFE2L3 | 2.36 | 2.36 | 2.52 |
| Adenosine A2a receptor | N57553 | ADORA2A | 3.89 | 4.14 | 4.11 |
| Ras-related C3 botulinum toxin substrate 1 (Rho family, small GTP-binding protein RAC-1) | AA626787 | RAC1 | 2.5 | 1.85 | 2.9 |
| Pronapsin A | AI369218 | NAP1 | 2.28 | 3.63 | 2.43 |
| Transcription, replication, translation, degradation machinery, protein modification | |||||
| TATA box binding protein-associated factor, RNA polymerase II, F, 55 kDa | AA461518 | TAF2F | 2.87 | 2.75 | 2.82 |
| Transcriptional repressor | H89996 | CTCF | 2.55 | 2.2 | 2.23 |
| Protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), alpha isoform | AA598795 | PPP2R2A | 2.49 | 2.02 | 1.68 |
| WASP family | |||||
| Wiskott-Aldrich syndrome (eczema-thrombocytopenia) | H61193 | WAS | 2.58 | 2.25 | 2.61 |
| WAS protein family, member 1 | N59851 | WASF1 | 3.01 | 2.66 | 2.11 |
| Vasoactive intestinal peptide | AI217172 | VIP | 2.01 | 2.58 | 2.32 |
| Cluster 2 | |||||
| Adhesion molecules and cytoskeleton | |||||
| Inter-alpha (globulin) inhibitor H4 (plasma Kallikrein-sensitive glycoprotein) | N73625 | ITIH4 | 2.99 | 2.43 | 1.16 |
| Flamin C, gamma (actin-binding protein-280) | AI675658 | FLNC | 2.95 | 2.44 | 1.15 |
| Elastase 1, pancreatic | AA845015 | ELA1 | 2.9 | 1.59 | 1.25 |
| Claudin 3 | AA039323 | CLDN3 | 2.54 | 2.48 | 1.32 |
| Cadherin 15, M-cadherin (myotubule) | AI571806 | CDH15 | 2.95 | 2.46 | 1.15 |
| Cell cycle, DNA damage, apoptosis | |||||
| Cell division cycle 25A | H59260 | CDC25A | 2.02 | 1.35 | 2.14 |
| RAB6, member of RAS oncogene family | AA934745 | RAB6 | 2.97 | 1.34 | 1.64 |
| DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 11 (Saccharomyces cerevisiae CHL1-like helicase) | AA402879 | DDX11 | 2.91 | 1.48 | 1.37 |
| Miscellaneous | |||||
| Solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 | AI344386 | SLC28A1 | 2.93 | 1.66 | 1.74 |
| Dual-specificity phosphatase 2 | AA759046 | DUSP2 | 3.92 | 3.58 | 1.00 |
| Immune response | |||||
| Nuclear factor of activated T cells, cytoplasmic 3 | AA179812 | NFATC3 | 3.46 | 2.97 | 1.30 |
| Tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) | AI245559 | TNFRSF14 | 2.84 | 2.28 | 1.68 |
| Tumor necrosis factor (ligand) superfamily, member 7 | AI347622 | TNFSF7 | 2.99 | 1.64 | 1.3 |
| Interferon-stimulated protein, 15 kDa | AA406020 | ISG15 | 3.12 | 2.93 | 1.61 |
| Karyopherin (importin) beta 1 | AA251527 | KPNB1 | 2.49 | 2.1 | 1.26 |
| Cluster 3 | |||||
| Adhesion molecules and cytoskeleton | |||||
| Prefoldin 4 | AA253430 | PFDN4 | 2.08 | 1.49 | 1.17 |
| Coronin, actin-binding protein, 2A | AA983765 | CORO2A | 2.04 | 1.73 | 1.04 |
| Kinesin family member 5A | AA984728 | KIF5A | 2.05 | 1.88 | 1.73 |
| Integrin beta 3 (alternatively spliced, clone beta 3C) | AA037229 | ITGB3 | 2.12 | 1.72 | 1.41 |
| Cell cycle, DNA damage, apoptosis | |||||
| RAB2, member RAS oncogene family-like | AA401972 | RAB2L | 2.33 | 1.78 | 1.38 |
| V-myc avian myelocytomatosis viral oncogene homolog | AA464600 | MYC | 2.22 | 1.53 | 1.05 |
| V-jun avian sarcoma virus 17 oncogene homolog | W96155 | JUN | 2.17 | 2.02 | 1.31 |
| Immune response | |||||
| CD1A antigen, a polypeptide | AI240210 | CD1A | 2.2 | 1.39 | 1.45 |
| CD28 antigen (Tp44) | AI375736 | CD28 | 2.16 | 1.49 | 1.27 |
| Interferon, alpha-inducible protein 27 | AA157813 | IFI27 | 2.15 | 1.37 | 1.92 |
| Transporters | |||||
| Solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen) member 16 | AA411554 | SLC25A16 | 2.11 | 1.71 | 1.57 |
| Solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | AI688443 | SLC6A6 | 2.3 | 1.65 | 1.43 |
| Signal transmission | |||||
| Regulator of nonsense transcripts 1 | AA156342 | RENT1 | 2.48 | 2.38 | 1.56 |
| Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon | AA953975 | NFKBIE | 2.41 | 1.61 | 1.12 |
| Zinc finger protein 173 | AA490855 | ZNF173 | 2.31 | 2.14 | 1.39 |
| Cluster 4 | |||||
| Histones | |||||
| H4 histone family, member G | AA868008 | H4FG | 1.91 | 2.58 | 3.34 |
| Immune response | |||||
| B7 protein | N90281 | B7 | 1.97 | 1.75 | 3.53 |
| Metabolism | |||||
| Protein disulfide isomerase related protein (calcium-binding protein, intestine related) | N59626 | ERP70 | 1.34 | 2.87 | 2.06 |
| Miscellaneous | |||||
| Human putative tumor suppressor (LUCA15) mRNA, complete cds | AA456007 | 1.52 | 2.55 | 2.28 | |
| RAE1 (RNA export 1, Schizosaccharomyces pombe) homolog | AA504128 | RAE1 | 1.91 | 2.04 | 1.95 |
| Cluster 5 | |||||
| Adhesion molecules and cytoskeleton | |||||
| Dynein, cytoplasmic, light intermediate polypeptide 2 | AA490963 | DNCLI2 | 0.71 | 2.47 | 1.82 |
| Cluster 6 | |||||
| Adhesion molecules and cytoskeleton | |||||
| Lamin B1 | AA983462 | LMNB1 | 2.00 | 1.54 | 1.91 |
Genes in each cluster are grouped according to predicted biological function.
TABLE 2.
Representative human genes in clusters 7 to 11 (downregulated by MVA)a
| Function or protein description and gene name | Accession no. | Gene symbol | Change (fold) at time (h) postinfection
|
||
|---|---|---|---|---|---|
| 2.00 | 6.00 | 16.00 | |||
| Cluster 7 | |||||
| Enzymes | |||||
| 3-Oxoacid coenzymeA transferase | R40897 | OXCT | 0.60 | 0.67 | 1.05 |
| Aldehyde dehydrogenase 7 | N93686 | ALDH7 | 0.68 | 0.66 | 1.02 |
| Acetylcholinesterase (YT blood group) | R85241 | ACHE | 0.71 | 0.67 | 1.10 |
| Transcription | |||||
| Adenosine A3 receptor | AA863086 | ADORA3 | 0.97 | 0.79 | 1.73 |
| Adenosine deaminase | AA683578 | ADA | 0.84 | 0.58 | 1.49 |
| Cluster 8 | |||||
| ATP synthase | |||||
| ATP synthase, H+ transporting, mitochondrial Fo complex, subunit c (subunit 9), isoform 2 | AA455126 | ATP5G2 | 0.53 | 0.40 | 1.12 |
| Translation | |||||
| Eukaryotic translation elongation factor 1 alpha 2 | AI368766 | EEF1A2 | 0.93 | 0.66 | 0.91 |
| Eukaryotic translation initiation factor 4 gamma 1 | R37276 | EIF4G1 | 0.93 | 0.69 | 0.90 |
| Polymerase (DNA directed), beta | R44427 | POLB | 0.88 | 0.73 | 0.64 |
| Cluster 9 | |||||
| Cell cycle, DNA damage, apoptosis | |||||
| v-maf musculoaponeurotic fibrosarcoma (avian) oncogene family, protein G | AA045436 | MAFG | 0.85 | 0.5 | 0.66 |
| BCL2-associated athanogene | AI017240 | BAG1 | 0.86 | 0.84 | 0.68 |
| Cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | N72115 | CDKN2C | 0.82 | 0.81 | 0.68 |
| Cytochrome | |||||
| Cytochrome P450, subfamily I (aromatic compound-inducible), polypeptide 1 | AA418907 | CYP1A1 | 0.86 | 0.87 | 0.68 |
| Cytochrome P450, subfamily I (dioxin-inducible), polypeptide 1 (glaucoma 3, primary infantile) | AA448157 | CYP1B1 | 0.84 | 0.80 | 0.66 |
| Cytoskeleton | |||||
| Actin-related protein 2/3 complex, subunit 5 (16 kDa) | W55964 | ARPC5 | 0.82 | 0.82 | 0.6 |
| Actinin, alpha 1 | AA669042 | ACTN1 | 0.97 | 0.67 | 0.75 |
| Actinin, alpha 3 | AA196000 | ACTN3 | 0.83 | 0.76 | 0.51 |
| Annexin A1 | H63077 | ANXA1 | 0.97 | 0.7 | 0.67 |
| Collagen, type III, alpha 1 (Ehlers-Danlos syndrome type IV, autosomal dominant) | T98612 | COL3A1 | 0.85 | 0.41 | 1.25 |
| Collagen, type IX, alpha 1 | N69335 | COL9A1 | 0.84 | 0.65 | 0.66 |
| Collagen, type V, alpha 2 | AA461456 | COL5A2 | 0.84 | 0.67 | 0.51 |
| Destrin (actin depolymerizing factor) | AA424824 | ADF | 0.84 | 0.55 | 0.43 |
| Keratin 7 | AA485959 | KRT7 | 0.83 | 0.63 | 0.53 |
| Moesin | R22977 | MSN | 0.85 | 0.55 | 0.44 |
| Myosin, light polypeptide 4, alkali; atrial, embryonic | AA705225 | MYL4 | 0.87 | 0.81 | 0.35 |
| Myosin, light polypeptide 6, alkali, smooth muscle and nonmuscle | AA488346 | MYL6 | 0.85 | 0.58 | 0.45 |
| Prefoldin 5 | AA446453 | PFDN5 | 0.85 | 0.62 | 0.58 |
| Tubulin, gamma polypeptide | T77733 | TUBG1 | 0.87 | 0.40 | 0.69 |
| Enzymes | |||||
| Acetylserotonin N-methyltransferase-like | AA427398 | ASMTL | 0.84 | 0.84 | 0.67 |
| Casein kinase 1, alpha 1 | AA625758 | SSP29 | 0.98 | 0.72 | 0.58 |
| NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 (12 kDa, B12) | AI675527 | NDUFB3 | 0.86 | 0.6 | 0.62 |
| NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1 (6 kDa, KFYI) | AA460251 | NDUFC1 | 0.81 | 0.58 | 0.54 |
| NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1 (6 kDa, KFYI) | AA460251 | NDUFC1 | 0.81 | 0.58 | 0.54 |
| NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 (18 kDa, B18) | AA428058 | NDUFB7 | 0.86 | 0.43 | 0.46 |
| Ribosomal proteins | |||||
| Ribosomal protein L22 | AI688090 | RPL22 | 0.96 | 0.72 | 0.56 |
| Ribosomal protein L35 | AA625634 | RPL35 | 0.93 | 0.66 | 0.76 |
| Ribosomal protein S10 | AI611010 | RPS10 | 0.96 | 0.72 | 0.65 |
| Ribosomal protein S12 | AI689992 | RPS12 | 0.97 | 0.69 | 0.62 |
| Ribosomal protein S23 | N73091 | RPS23 | 0.94 | 0.66 | 0.71 |
| Ribosomal protein S27a | AA625632 | RPS27A | 0.85 | 0.58 | 0.72 |
| Ribosomal protein S28 | AA856556 | RPS28 | 0.89 | 0.63 | 0.69 |
| Ribosomal protein S5 | AA456616 | RPS5 | 0.95 | 0.71 | 0.65 |
| Degradation machinery | |||||
| Proteasome (prosome, macropain) 26S subunit, ATPase 3 | AA987573 | PSMC3 | 0.98 | 0.67 | 0.69 |
| Proteasome (prosome, macropain) activator subunit 2 (PA28 beta) | H65395 | PSME2 | 0.98 | 0.60 | 0.59 |
| Proteasome (prosome, macropain) subunit, beta type 1 | T68758 | PSMB1 | 0.98 | 0.69 | 0.73 |
| Proteasome (prosome, macropain) subunit, beta type 3 | AA620580 | PSMB3 | 0.98 | 0.69 | 0.71 |
| Ubiquinol-cytochrome c reductase binding protein | AA664284 | UQCRB | 0.81 | 0.64 | 0.56 |
| Ubiquitin-specific protease 4 (proto-oncogene) | AA454143 | USP4 | 0.81 | 0.62 | 0.54 |
| Transcription | |||||
| Transcription factor CP2 | AA488618 | TFCP2 | 0.81 | 0.58 | 0.49 |
| Adenine nucleotide translocator 3 (liver) | AA663439 | SLC25A6 | 0.98 | 0.58 | 0.44 |
| Transcription elongation factor B (SIII), polypeptide 3 (110 kDa, elongin A) | AA133129 | TCEB3 | 0.82 | 0.69 | 0.70 |
| General transcription factor IIH, polypeptide 3 (34 kDa subunit) | AA460838 | GTF2H3 | 0.96 | 0.68 | 0.74 |
| Polymerase (RNA) II (DNA directed) polypeptide G | AA477428 | POLR2G | 0.92 | 0.75 | 0.60 |
| Polymerase (RNA) II (DNA directed) polypeptide I (14.5 kDa) | AA777192 | POLR2I | 0.82 | 0.58 | 0.42 |
| Translation | |||||
| Eukaryotic translation initiation factor 3, subunit 6 (48 kDa) | AA669674 | EIF3S6 | 0.95 | 0.70 | 0.55 |
| Eukaryotic translation initiation factor 3, subunit 2 (beta, 36 kDa) | AA936783 | EIF3S2 | 0.91 | 0.52 | 0.42 |
| Cluster 10 | |||||
| Cytoskeleton | |||||
| Tubulin, beta 2 | AI000256 | TUBB2 | 0.84 | 0.79 | 0.48 |
| Collagen, type V, alpha 1 | R75635 | COL5A1 | 0.56 | 0.96 | 0.67 |
| Enzymes | |||||
| Aldose reductase-like 1 | AI301329 | ALDRLn | 0.63 | 0.84 | 0.79 |
| Carboxypeptidase B2 (plasma) | H47838 | CPB2 | 0.64 | 0.68 | 0.88 |
| Carnitine palmitoyltransferase II | AI369287 | CPT2 | 0.60 | 0.72 | 0.63 |
| Acyl-coenzyme A dehydrogenase, very long chain | AA464163 | ACADVL | 0.70 | 0.64 | 0.74 |
| 1-Acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) | AA458922 | AGPAT1 | 0.62 | 0.65 | 0.72 |
| Casein kinase 2, beta polypeptide | AA994790 | CSNK2B | 0.65 | 0.43 | 0.50 |
| COX11 (yeast) homolog, cytochrome c oxidase assembly protein | AA457644 | COX11 | 0.71 | 0.74 | 0.84 |
| Creatine kinase, mitochondrial 1 (ubiquitous) | AI369378 | CKMT1 | 0.69 | 0.76 | 0.64 |
| Cystathionine-beta-synthase | AA430367 | CBS | 0.68 | 0.90 | 0.71 |
| Dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit | AA004759 | DPM1 | 0.66 | 0.76 | 0.68 |
| Ephrin-A1 | AA857015 | EFNA1 | 0.57 | 0.74 | 0.69 |
| NADH dehydrogenase (ubiquinone) Fe-S protein 8 (23 kDa) (NADH-coenzyme Q reductase) | AA127014 | NDUFS8 | 0.63 | 0.55 | 0.55 |
| 7-Dehydrocholesterol reductase | AI652764 | DHCR7 | 0.72 | 0.63 | 0.64 |
| Immune response | |||||
| Erythrocyte membrane protein band 4.1-like 1 | R71689 | EPB41L1 | 0.84 | 0.41 | 0.45 |
| Interferon gamma receptor 1 | H11482 | IFNGR1 | 0.70 | 0.41 | 0.44 |
| Macrophage stimulating 1 (hepatocyte growth factor-like) | T47813 | MST1 | 0.68 | 0.65 | 0.74 |
| Splicing | |||||
| Splicing factor proline/glutamine rich (polypyrimidine tract-binding protein-associated) | AA418910 | SFPQ | 0.74 | 1.12 | 0.66 |
| Splicing factor, arginine/serine-rich 11 | H56944 | SFRS11 | 0.64 | 0.129 | 0.30 |
| Degradation machinery | |||||
| Ubiquinol-cytochrome c reductase core protein II | AA663058 | UQCRC2 | 0.73 | 0.51 | 0.70 |
| Ubiquitin-conjugating enzyme E2L 3 | AA487058 | UBE2L3 | 0.65 | 0.46 | 0.62 |
| Ubiquitin-specific protease 15 | AA253442 | USP15 | 0.84 | 0.59 | 0.64 |
| Transcription | |||||
| General transcription factor IIIC, polypeptide 1 (alpha subunit, 220 kDa) | AA843718 | GTF3C1 | 0.70 | 0.81 | 0.86 |
| Polymerase (RNA) II (DNA directed) polypeptide K (7.0 kDa) | AA458646 | POLR2K | 0.66 | 0.65 | 0.71 |
| Ribophorin II | AA991856 | RPN2 | 0.52 | 0.60 | 0.54 |
| B-cell CLL/lymphoma 10 | AA456036 | BCL10 | 0.70 | 0.88 | 0.67 |
| Adenine nucleotide translocator 3 (liver) | AA496376 | SLC25A6 | 0.69 | 0.59 | 0.54 |
| Translation | |||||
| Human translation initiation factor elF-2alpha mRNA, 3′ untranslated region | AA424956 | 0.69 | 0.72 | 0.65 | |
| Eukaryotic translation initiation factor 3, subunit 9 (eta, 116 kDa) | AA676471 | EIF3S9 | 0.70 | 0.53 | 0.70 |
| Cluster 11 | |||||
| ATP synthase | |||||
| ATP synthase, H+ transporting, mitochondrial Fo complex, subunit f, isoform 2 | AA453995 | ATP5J2 | 0.46 | 0.25 | 0.25 |
| ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide | AA708298 | ATP5B | 0.42 | 0.17 | 0.16 |
| ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit | AA873577 | ATP5O | 0.66 | 0.38 | 0.19 |
| ATP synthase, H+ transporting, mitochondrial F0 complex, subunit f, isoform 2 | AA453995 | ATP5J2 | 0.52 | 0.61 | 0.25 |
| ATP synthase, H+ transporting, mitochondrial F1F0, subunit g | AA126313 | ATP5JG | 0.46 | 0.25 | 0.36 |
| ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c (subunit 9) Isoform 3 | H47080 | ATP5G3 | 0.25 | 0.31 | 0.36 |
| ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit | AA873577 | ATP5O | 0.36 | 0.53 | 0.23 |
| ATP synthase, H+ transporting, mitochondrial F0 complex, subunit e | AA431433 | ATP5I | 0.44 | 0.39 | 0.10 |
| Cell cycle, DNA damage, apoptosis | |||||
| DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 8 (RNA helicase) | AI190747 | DDX8 | 0.67 | 0.65 | 0.45 |
| DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 9 | AA458801 | DDX9 | 0.78 | 0.79 | 0.55 |
| B-cell CLL/lymphoma 7A | H90147 | BCL7A | 0.74 | 0.5 | 0.52 |
| BCL2-antagonist of cell death | AA460291 | BAD | 0.67 | 0.65 | 0.45 |
| Caspase 3, apoptosis-related cysteine protease | AA011446 | CASP3 | 0.67 | 0.57 | 0.54 |
| CDC37 (cell division cycle 37, Saccharomyces cerevisiae, homolog) | AA458870 | CDC37 | 0.67 | 0.45 | 0.54 |
| Cell division cycle 34 | H20743 | CDC34 | 0.65 | 0.56 | 0.51 |
| Cell division cycle 42 (GTP-binding protein, 25 kDa) | AA630164 | CDC42 | 0.67 | 0.4 | 0.42 |
| Colony-stimulating factor 1 receptor | T46880 | CSF1R | 0.67 | 0.45 | 0.54 |
| Cyclin-dependent kinase inhibitor 1C (p57, Kip2) | AI676118 | CDKN1C | 0.67 | 0.57 | 0.54 |
| Neuroblastoma candidate region, suppression of tumorigenicity 1 | AA598830 | NBL1 | 0.79 | 0.53 | 0.72 |
| Cytochrome | |||||
| Cytochrome c oxidase subunit Vb | AI688757 | COX5B | 0.64 | 0.38 | 0.33 |
| Cytochrome c oxidase subunit VIa polypeptide 1 | AA482243 | COX6A1 | 0.86 | 0.42 | 0.37 |
| Cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) | AA872125 | COX7A1 | 0.73 | 0.53 | 0.58 |
| Cytochrome c oxidase subunit VIIa polypeptide 2 (liver) | AI002403 | COX7A2 | 0.9 | 0.75 | 0.67 |
| Cytochrome c oxidase subunit VIIc | AA629719 | COX7C | 0.3 | 0.53 | 0.52 |
| Cytochrome c-1 | AA447774 | CYC1 | 0.71 | 0.37 | 0.23 |
| Cytoskeleton | |||||
| Acid phosphatase 2, lysosomal | T48864 | ACP2 | 0.77 | 0.65 | 0.4 |
| Actin, alpha 2, smooth muscle, aorta | AA634006 | ACTA2 | 0.76 | 0.47 | 0.44 |
| Actin, gamma 2, smooth muscle, enteric | T60048 | ACTG2 | 0.65 | 0.47 | 0.42 |
| Collagen, type IV, alpha 2 | AA430540 | COL4A2 | 0.74 | 0.26 | 0.34 |
| Collagen, type IV, alpha 5 (Alport syndrome) | AA953254 | COL4A5 | 0.84 | 0.35 | 0.18 |
| Collagen, type VI, alpha 2 | AA633747 | COL6A2 | 0.74 | 0.51 | 0.59 |
| Keratin 16 (focal nonepidermolytic palmoplantar keratoderma) | AA928454 | KRT16 | 0.26 | 0.2 | 0.16 |
| Myosin, heavy polypeptide 9, nonmuscle | T69926 | MYH9 | 0.48 | 0.65 | 0.25 |
| Myosin, light polypeptide 1, alkali; skeletal, fast | T52894 | MYL1 | 0.78 | 0.6 | 0.53 |
| Keratin 19 | AA464250 | KRT19 | 0.25 | 0.32 | 0.2 |
| Profilin 2 | AA040703 | PFN2 | 0.76 | 0.65 | 0.45 |
| Rho GDP dissociation inhibitor alpha | AA459400 | ARHGDIA | 0.35 | 0.24 | 0.39 |
| Tubulin-specific chaperone d | AI668870 | TBCD | 0.85 | 0.57 | 0.49 |
| Tubulin, alpha 2 | AA626698 | TUBA2 | 0.82 | 0.52 | 0.77 |
| Actin-related protein 2/3 complex, subunit 1A (41 kDa) | AA490209 | ARPC1A | 0.73 | 0.46 | 0.36 |
| Enzymes | |||||
| 6-Pyruvoyl-tetrahydropterin synthase/dimerization cofactor of hepatocyte nuclear factor 1 alpha | AA459909 | PCBD | 0.75 | 0.67 | 0.62 |
| 3-Hydroxyanthranilate 3,4-dioxygenase | AI005031 | HAAO | 0.90 | 0.89 | 0.6 |
| COX11 (yeast) homolog, cytochrome c oxidase assembly protein | AA450001 | COX11 | 0.71 | 0.61 | 0.61 |
| Crystallin, beta B2 | AA191518 | CRYBB2 | 0.68 | 0.62 | 0.62 |
| DNA segment, single-copy probe LNS-CAI/LNS-CAII (deleted in polyposis) | H99681 | D5S346 | 0.67 | 0.62 | 0.64 |
| Early growth response 2 (Krox-20 [Drosophila] homolog) | AA446027 | EGR2 | 0.66 | 0.59 | 0.61 |
| NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 (8 kDa, B8) | AI017426 | NDUFA2 | 0.78 | 0.35 | 0.31 |
| NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9 (39 kDa) | AA598884 | NDUFA9 | 0.65 | 0.66 | 0.59 |
| Acetyl-coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-coenzyme A thiolase) | H07926 | ACAA2 | 0.73 | 0.59 | 0.56 |
| Immune response | |||||
| CD39 antigen | H10011 | CD39 | 0.54 | 0.58 | 0.85 |
| CD59 antigen p18-20 | H60549 | CD59 | 0.85 | 0.52 | 0.44 |
| CD6 antigen | AI336940 | CD6 | 0.46 | 0.64 | 0.41 |
| Coagulation factor V (proaccelerin, labile factor) | AA680136 | F5 | 0.59 | 0.44 | 0.40 |
| Interferon inducible | AA464417 | IFITM3 | 0.58 | 0.55 | 0.24 |
| Interleukin 1 receptor antagonist | T72877 | IL1RN | 0.55 | 0.49 | 0.46 |
| Delta sleep-inducing peptide, immunoreactor | AA775091 | DSIPI | 0.65 | 0.5 | 0.26 |
| Ribosomal proteins | |||||
| Ribosomal protein S29 | N93715 | RPS29 | 0.69 | 0.48 | 0.37 |
| Ribosomal protein S4, X-linked | AA888182 | RPS4X | 0.69 | 0.48 | 0.37 |
| Ribosomal protein L6 | AA629808 | RPL6 | 0.75 | 0.61 | 0.48 |
| Ribosomal protein S16 | AA668301 | RPS16 | 0.76 | 0.60 | 0.54 |
| Degradation machinery | |||||
| Ubiquitin carrier protein E2-C | AA430504 | UBCH10 | 0.61 | 0.52 | 0.39 |
| Ubiquitin-specific protease 5 (isopeptidase T) | AA465536 | USP5 | 0.56 | 0.5 | 0.50 |
| Ubiquitin-specific protease 5 (isopeptidase T) | AA465536 | USP5 | 0.56 | 0.5 | 0.50 |
| Ubiquitin-like 1 (sentrin) | AA488626 | UBL1 | 0.68 | 0.58 | 0.5 |
| Transcription | |||||
| General transcription factor IIIC, polypeptide 2 (beta subunit, 110 kDa) | AA922691 | GTF3C2 | 0.78 | 0.57 | 0.52 |
| Transcription factor 12 (HTF4, helix-loop-helix transcription factors 4) | H98856 | TCF12 | 0.69 | 0.80 | 0.84 |
| General transcription factor IIIC, polypeptide 2 (beta subunit, 110 kDa) | AA922691 | GTF3C2 | 0.78 | 0.57 | 0.52 |
| Splicing | |||||
| Splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) | AA425853 | SFPQ | 0.56 | 0.24 | 0.44 |
| Splicing factor (CC1.3) | AA193573 | CC1.3 | 0.65 | 0.62 | 0.23 |
Genes in each cluster are grouped according to predicted biological function.
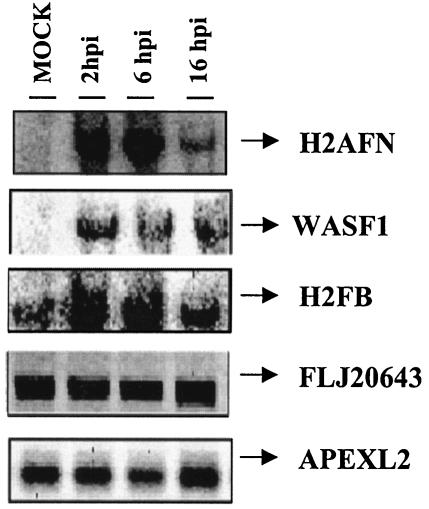
Confirmation of microarray data for selected genes by Northern blot analysis and real-time RT-PCR.
Selected genes with distinct expression patterns after MVA infection, as identified by microarray analysis, were chosen for target verification by Northern blotting. Total RNA was purified from uninfected or MVA-infected cells at 2, 6, and 16 h postinfection, fractionated by gel electrophoresis, blotted, and probed by using 32P-labeled PCR products that were spotted on the microarray. The RNA preparation used for this analysis was the same as that used in the microarray. The amount of RNA on the blot was normalized based on rRNA content. The Northern blot analysis confirmed microarray results in all cases (Fig. 4). Whereas the expression pattern of histone family member N (H2AFN) was weakly detected in mock-infected cells, strong activation compared to that for the uninfected control was observed at 2, 6, and 16 h postinfection. A similar expression pattern was observed for WAS protein family member 1 (WASF1). Peaks of histone F gene H2FB mRNA expression were reached at 2 and 6 h postinfection. A constitutive expression pattern of the apurinic/apyridiminic endonuclease gene (APEXL2) and the EST FLJ20643 was observed in the presence and in the absence of MVA.
FIG. 4.
Validation of microarray data by Northern blotting. Total RNA (20 μg) purified from uninfected or MVA-infected cells at 2, 6, and 16 h postinfection was hybridized with probes derived from the PCR products spotted on the microarray. Genes included in the autoradiogram are H2AFN (histone N), WASF1 (Wiskott-Aldrich syndrome protein), H2FB (histone B), FLJ20643 (EST), and APEXL2 (apurinic/apyridiminic endonuclease). hpi, hours postinfection.
As an alternative to the Northern blot analysis, real-time RT-PCR was used to verify the transcriptional changes in selected genes detected by microarray analysis. Six upregulated genes (H2BFB, PCNT2, WASF1, WAS, IL-7, and IL-6) and three unaltered genes (IFNG, APEXL2, and FLJ20643) were analyzed; hypoxanthine phosphoribosyltransferase was used as an internal control. The RT-PCR data confirmed the microarray results, showing the same relative transcription regulation of the selected genes (Table 3). These findings validate those of the Northern and microarray analyses. Absolute values are not identical when microarray and RT-PCR data are compared, probably due to intrinsic differences in the techniques.
TABLE 3.
Confirmation of microarray data by quantitative real-time RT-PCRa
| Gene product | Change (fold) determined by assay at time (h) postinfection
|
|||||
|---|---|---|---|---|---|---|
| Microarray
|
RT-PCR
|
|||||
| 2 | 6 | 16 | 2 | 6 | 16 | |
| H2BFB | 9.92 | 10.20 | 17.03 | 25 | 18 | 19 |
| PCNT2 | 3.94 | 3.14 | 2.17 | 4.36 | 4.44 | 3.4 |
| WASF1 | 3.01 | 2.66 | 1.11 | 3.73 | 2.72 | 1.53 |
| WAS | 2.58 | 2.25 | 1.61 | 5.08 | 15.2 | 2.3 |
| IL7 | 6.10 | 5.30 | 3.2 | 6.86 | 5.8 | 4.9 |
| IL6 | 2.45 | 3.02 | 1.90 | 1.75 | 2.07 | 2.28 |
| IFNG | 1.27 | 1.36 | 1.45 | 1.35 | 1.15 | 1.19 |
| APEXL2 | 1.10 | 0.98 | 1.11 | 0.96 | 1.2 | 0.84 |
| FLJ | 0.98 | 1.25 | 1.01 | 1.1 | 1.02 | 0.93 |
RT-PCR conditions are described in Materials and Methods.
Target verification by Western blot analysis and ELISA of representative cell proteins.
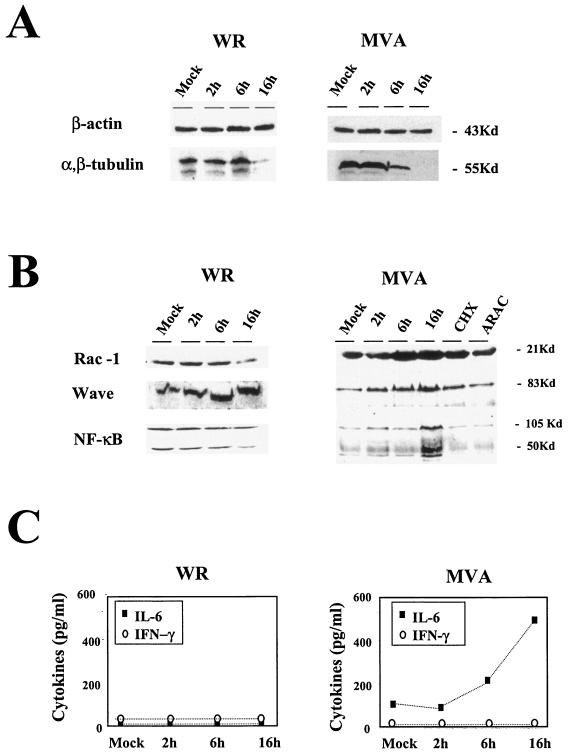
To correlate transcription changes with protein levels, we defined the effects of MVA infection on some cellular components, analyzing protein expression levels by Western blotting. After MVA infection, microarray data indicated that tubulin expression was downregulated (Table 2), a finding confirmed by Western blotting (Fig. 5A). Although actin gene expression was downregulated after 2 h postinfection in microarray analysis (Table 2), the protein was present in equal amounts in mock-infected and MVA-infected cells at 2, 6, and 16 h postinfection (Fig. 5A). This result is probably due to the stability of actin. Similar results were obtained with WR-infected HeLa cells (Fig. 5A). These findings show that the correlation between mRNA and protein levels depends on protein stability.
FIG. 5.
Validation of microarray data by protein level and comparison between MVA and WR infections. Shown are Western blots of different cellular proteins at various times (2, 6, and 16 h postinfection). (A) Actin and tubulin protein levels in HeLa cells (5 PFU/cell). (B) RAC-1, WAVE, and NF-κB protein levels in MVA- or WR-infected HeLa cells (5 PFU/cell) and in cells infected with MVA in the presence of cycloheximide (CHX; 100 μg/ml) or cytosine arabinoside (ARAC; 50 μg/ml) for 16 h. (C) Levels of IL-6 and IFN-γ secreted from HeLa cells after MVA or WR infection (5 PFU/cell), as determined by ELISA. Protein levels of IL-6 and IFN-γ in supernatants of uninfected and MVA- or WR-infected cells were measured at 2, 6, and 16 h postinfection. Duplicate samples were measured in two independent experiments. Kd, kilodaltons
For other proteins, we confirmed microarray data with protein expression patterns. For these analyses, we used the Rho family small GTP-binding protein (RAC-1), the WASP family member WAVE (WASF1), and the nuclear factor kappa light polypeptide (NFKβ1). The RAC-1 protein signal increased in MVA-infected cells, with a peak at 6 h postinfection (Fig. 5B). At this time point, the amount of RAC-1 in MVA-infected cells was about threefold higher than that in control cells. WAVE showed a peak of expression at 16 h postinfection, when the amount of protein was about threefold higher than that in control cells. NF-κB protein expression was more than threefold higher than that in controls at 16 h postinfection, whereas there was no evidence of protein increases at early times postinfection (Fig. 5B). In the case of WR-infected cells, WAVE protein levels were similar for WR- and MVA-infected cells. RAC-1 and NF-κB showed different levels of protein expression at 16 h postinfection; these levels increased with MVA infection and decreased with WR infection (Fig. 5B). Evidence that viral late gene functions are necessary for the increases in RAC-1, WAVE, and NF-κB protein levels was obtained (Fig. 5B) by Western blot analysis of MVA-infected cells cultured in the presence of cytosine-arabinoside (AraC) and an inhibitor of viral DNA replication (21). Moreover, the increase in RAC-1, WAVE, and NF-κB protein expression required de novo protein synthesis, since the accumulation of these proteins was prevented by cycloheximide treatment (Fig. 5B). This result eliminates the possibility that MVA infection might increase protein levels by enhancing protein stability.
We further analyzed the levels of IL-6 and IFN-γ secreted after MVA infection to confirm the data obtained by microarrays and quantitative RT-PCR. The amounts of secreted IL-6 and IFN-γ were determined by ELISA with uninfected and MVA-infected HeLa cells (5 PFU/cell) at 2, 6, and 16 h postinfection. In agreement with data obtained in microarray and quantitative RT-PCR analyses, there was no detectable IFN-γ in supernatants from MVA-infected cells (Fig. 5C). In contrast, we observed a strong increase with time in the amount of secreted IL-6 after MVA infection, in full agreement with the microarray and quantitative RT-PCR data (Fig. 5C). When HeLa cells were WR-infected, no IL-6 or IFN-γ was detected by ELISA in cell-free supernatants (Fig. 5C).
DISCUSSION
The interaction between viruses and the host cell are complex, multifaceted processes. While viruses attempt to take over cellular functions to their advantage, the cell counteracts by mounting a variety of defensive responses that may include induction of interferon, stress response, or apoptotic pathways, all of which are accompanied by changes in gene expression.
In this study, we analyzed the response of the human HeLa cell line to MVA infection by using cDNA microarrays. It was reported previously that MVA undergoes limited replication in HeLa cells (7, 11, 14); virus replication is restricted during infection of HeLa cells after immature virions are formed (7, 14, 34, 36), allowing efficient production of proteins (33, 37, 42). Due to the interest in MVA-based vectors as potential vaccines against pathogens and tumors and current phase I clinical trials with this vector, there is a need to understand the host response to MVA infection. We used microarrays to analyze the changes in host gene expression profiling after MVA infection of cultured human cells.
A total of 410 of 13,295 genes in the array were significantly regulated after MVA infection and assigned to 11 clusters (Fig. 2). Clusters 1 and 2 included 68 genes that were upregulated at 2, 6, and 16 h postinfection. A total of 58 genes were identified in cluster 3; their expression increased at 2 and 6 h postinfection. In cluster 4, we observed 15 genes upregulated at 6 and 16 h postinfection. In cluster 5, 11 genes were downregulated at 2 h but upregulated at 6 and 16 h postinfection. The 11 genes in cluster 6 were upregulated only at 2 h postinfection. The remaining genes, a total of 247, were downregulated with different expression profiling and are represented in clusters 6 to 11 (Table 2). Indeed, these findings differ from the host transcriptional responses observed after WR infection, for which only 37 cellular genes were upregulated in HeLa cells infected at 2, 6, and 16 h postinfection (17). A comparison of gene expression profiles based on data obtained in this study and those from previous work with WR (17) is shown for representative genes in Table 4. In MVA- or WR-infected HeLa cells, there is a group of cellular genes upregulated by both viruses; other genes are upregulated selectively by MVA or WR, and a large number of genes are downregulated by both viruses. These results suggest that MVA and WR use different strategies to regulate cellular transcriptional responses, probably as a consequence of deletions in the MVA genome. This idea is supported by the observed differences in MVA-induced CPE and shutoff of host protein synthesis and those induced by WR (Fig. 1) and is consistent with previous results of analyses of the biology of MVA (14, 33).
TABLE 4.
Representative gene expression profiling of WR and MVA strains in human HeLa cellsa
| Effect and gene name | Gene symbol | Change (fold) with strain at time (h) postinfection
|
|||||
|---|---|---|---|---|---|---|---|
| MVA
|
WR
|
||||||
| 2 | 6 | 16 | 2 | 6 | 16 | ||
| Increased genes in MVA and WR | |||||||
| Pericentrin | PCNT | 3.94 | 3.14 | 2.38 | 3.20 | 7.11 | 1.58 |
| Claudin 3 | CLDN3 | 2.54 | 2.48 | 1.32 | 1.78 | 2.13 | 2.30 |
| H3 histone family, member B | H3FB | 16.34 | 15.67 | 21.71 | 4.92 | 1.12 | 1.11 |
| CD80 antigen | CD80 | 4.29 | 3.36 | 2.57 | 1.75 | 2.58 | 2.06 |
| Thymosin beta 4 X chromosome | TMSB4X | 3.81 | 5.7 | 2.91 | 3.81 | 5.70 | 2.91 |
| Adenosine A2a receptor, | ADORA2A | 3.89 | 4.14 | 4.11 | 3.89 | 4.14 | 4.11 |
| WAS protein family, member 1 | WASF1 | 3.01 | 2.66 | 2.11 | 2.35 | 2.43 | 2.99 |
| H3 histone family, member B | H3FB | 16.34 | 15.67 | 21.71 | 1.17 | 1.12 | 1.11 |
| H4 histone family, member D | H4FD | 12.38 | 24.93 | 24.93 | 2.41 | 0.97 | 1.01 |
| H2B histone family, member B | H2BFB | 9.92 | 10.20 | 17.03 | 5.94 | 0.78 | 0.61 |
| Increased genes in MVA | |||||||
| Interleukin 7 | IL-7 | 6.1 | 5.3 | 3.2 | 1.12 | 0.95 | 0.76 |
| B7 protein | B7 | 1.97 | 1.75 | 3.53 | 1.03 | 0.94 | 0.88 |
| CD47 antigen | CD47 | 5.35 | 4.08 | 3.16 | 1.1 | 0.99 | 1.01 |
| Interleukin 6 | IL-6 | 2.65 | 2.14 | 1.56 | 1.32 | 0.87 | 1.06 |
| Nuclear factor of kappa light polypeptide epsilon | NFKBIE | 2.41 | 1.61 | 1.12 | 1.01 | 1.01 | 0.75 |
| Mitogen-activated protein kinase kinase 5 | MAP2K5 | 2.37 | 2.74 | 1.69 | 1.06 | 1.23 | 0.69 |
| Nuclear factor (erythroid-derived 2)-like 3 | NFE2L3 | 2.36 | 2.36 | 2.52 | 1.12 | 0.89 | 1.01 |
| Nuclear factor of kappa light polypeptide 2 | NFKB2 | 2.03 | 2.22 | 2.34 | 1.2 | 0.54 | 0.59 |
| Kinesin family member 5A | KIF5A | 2.05 | 1.88 | 1.73 | 2.31 | 0.17 | 0.17 |
| Nuclear factor of activated T cells, cytoplasmic 3 | NFATC3 | 3.46 | 2.97 | 1.30 | 1.01 | 0.97 | 1.33 |
| Increased genes in WR | |||||||
| Diacylglycerol kinase delta | DGKD | 1.32 | 1.56 | 1.59 | 2.01 | 1.17 | 0.9 |
| Selenophosphate synthetase | SPS | 1.12 | 1.32 | 0.95 | 3.73 | 7.11 | 2.17 |
| Glutamate decarboxylase 2 (pancreatic islets and brain, 65 kDa) | GAD2 | 1.98 | 1.1 | 0.74 | 3.56 | 2.38 | 3.14 |
| Golgi resident protein | GCP60 | 1.02 | 0.87 | 0.65 | 1.41 | 2.00 | 1.79 |
| Decreased genes in MVA and WR | |||||||
| ATP synthase, subunit c (subunit 9), isoform 2 | ATP9C2 | 0.84 | 0.21 | 0.16 | 1.43 | 0.20 | 0.09 |
| ATP synthase, subunit b, isoform 1 | ATP1B | 0.53 | 0.40 | 1.12 | 1.39 | 0.18 | 0.12 |
| Cytochrome c-1 | CYC1 | 0.71 | 0.37 | 0.23 | 0.30 | 0.21 | 0.04 |
| Tubulin, beta 2 | TUBB2 | 0.84 | 0.79 | 0.48 | 0.73 | 0.22 | 0.26 |
| Actin, alpha 2, smooth muscle, aorta | ACTA2 | 0.76 | 0.47 | 0.44 | 0.84 | 0.56 | 0.12 |
| Interleukin 1 receptor antagonist | IL1RN | 0.55 | 0.49 | 0.46 | 0.15 | 0.21 | 0.18 |
| Macrophage stimulating 1 (hepatocyte growth factor-like) | MST1 | 0.68 | 0.65 | 0.74 | 0.55 | 0.16 | 0.11 |
| Ribosomal protein S27a | RPS27A | 0.85 | 0.58 | 0.72 | 0.84 | 0.34 | 0.43 |
| Ribosomal protein S28 | RPS28 | 0.89 | 0.63 | 0.69 | 0.65 | 0.31 | 0.41 |
| Ubiquitin carrier protein E2-C | UBCH10 | 0.61 | 0.52 | 0.39 | 0.78 | 0.24 | 0.26 |
| Proteasome (prosome, macropain) subunit, beta type 3 | PSMB3 | 0.98 | 0.69 | 0.71 | 0.50 | 0.17 | 0.17 |
Representative groups of genes from the microarray data obtained for MVA (this report) and from previous work with WR (17) were selected according to their up- or downregulation. For comparative purposes, we show cellular genes that are upregulated by both viruses, genes that are upregulated selectively by MVA or WR, and genes that are downregulated by the two viruses. Genes in each cluster were grouped according to predicted biological function.
Examination and analysis of the list of the cellular genes upregulated by MVA indicated several gene families with significant, distinct biological functions (Table 1). Some of these groups include genes involved in adhesion, the cytoskeleton, the cell cycle, apoptosis, histone, and immune modulation. Some of these genes may be involved in processes such as viral replication or cell defense.
Transcription of several genes involved in the immune response (20 transcripts) was activated by MVA infection. At least five cytokines (IL-1A, IL-6, IL-7, IL-8, and IL-15) and five members of the tumor necrosis factor receptor (TNF) family (TNFRSF7, TNFAIP3, TNFRSF14, TNFRSF17, and TRAF3) were upregulated in response to MVA. Genes encoding CD47 and CD80 antigens showed marked inductions at early and late times after MVA infection (Table 1). The immune modulator gene IL-7 upregulation was more than sixfold compared to levels for control cells at early times postinfection (Table 1), a result validated by quantitative RT-PCR (Table 3). The MVA-induced increase in IL-15 gene expression is a characteristic also observed for other viruses (3). At the protein level, we observed elevated IL-6 levels in supernatants from MVA-infected HeLa cells but not in those from cells infected with WR (Fig. 5C). A similar increase in IL-6 and TNF-α levels was found in spleen homogenates from MVA-infected mice but not in those from WR-infected mice (33). These results indicate that MVA and WR induce different proinflammatory cytokine profiles both in vivo and in vitro.
We observed clear upregulation of NF-κB expression during MVA infection, as determined by mRNA (Table 1) and protein levels (Fig. 5B). In the case of WR, NF-κB expression was downregulated at late times postinfection (Table 4). The increase in NF-κB protein required viral DNA replication, as seen with cells infected in the presence of adenosine arabinoside, an inhibitor of virus DNA synthesis. Moreover, the increase in NF-κB protein levels required de novo protein synthesis, as observed after cycloheximide treatment (Fig. 5B). These findings concur with the observation by Oie and Pickup (31) of an increase in NF-κB activity in extracts from MVA-infected 293 cells but not in cells infected with the Copenhagen strain of VV. The NF-κB family has a central role in the transcriptional regulation of inflammatory cytokines and other genes essential for activating immune responses (10). NF-κB also regulates the expression of genes involved in the control of cell proliferation and apoptosis (10, 31). It can be speculated that the increase observed in NF-κB mRNA and protein levels in response to MVA infection corresponds to enhanced NF-κB activity. An increase in NF-κB protein levels, together with the IκB-α degradation previously observed in MVA-infected cells (31), will contribute to the transcription of κB-dependent genes and to the clearance of virus infection as a result of the induction of apoptosis and specific immune responses. Since MVA has large genome deletions, viral genes that interfere with NF-κB, present in orthopoxviruses, might be absent (31).
In accord with previous microarray results for WR-infected cells (17), we observed increased levels of WASF1, claudin 3, CD-80, thymosin-beta-4, and adenosine-A2a receptor during MVA infection (Table 4). Comparison of WR and MVA nonetheless showed differences in expression levels of other genes, including kinesin-5 and histone family members, during infection. In WR-infected cells, these genes were upregulated early but not late in infection, whereas in MVA-infected cells, levels were maintained throughout infection. Among genes with an important role in motility and WR spreading is the Wiskott-Aldrich syndrome protein family member N-WASP (WASL gene) (13). WAS and WASF1 were upregulated in MVA-infected cells, whereas in WR-infected cells, only WASF1 was upregulated. Significantly, the small GTP-binding protein RAC-1 was also induced after MVA infection. Although the role of WAS proteins in the cell is well established (6, 27), the biological meaning of WASP family upregulation by a nonproductive virus is not immediately clear, in contrast to the case for WR, in which it appears to be important for viral spreading (6, 26, 38, 39).
It was reported that MVA has lost functional receptors for TNF, IFN-γ, IFNα/β, and CC chemokines (4, 7, 28, 29). The absence of these viral proteins and the induction of different host immune response molecules such as TNF may be the reason for MVA attenuation and for the potent immune response elicited by MVA recombinants to specific antigens compared with that elicited by VV recombinants (33). Induction of innate and adaptive immune responses during MVA infection could have a beneficial effect when MVA is used as a vaccine. MVA is known to trigger a poor immune response against itself, while it activates a potent immune response to specific antigens delivered by the vector (33). The enhanced expression of genes encoding immunomodulatory molecules (Table 1) during MVA infection might create a microenvironment that allows antigen-presenting cells and activated T cells to help expand CD4+ and CD8+ T cell populations. This possibility may be relevant when immunization protocols that include prime-booster immunization with heterologous vectors are used (33, 35). Maintenance of specific T cells could contribute to MVA-induced expression of cytokines such as IL-15. MVA induction of cellular genes with immunomodulatory functions may be a major contributing factor in the enhanced immunogenicity of MVA-derived vaccines.
In conclusion, through analyses of host cell gene expression profiling after MVA infection of cultured human cells, we found a number of cellular genes whose expression levels are markedly modified by MVA infection and suggest potential roles as regulators of viral infection. Some of these upregulated genes may be involved in the enhanced immune response to specific antigens observed in animals after infection with MVA recombinants. Identification of genes that are differentially regulated and the characterization of their functions is important when the potential benefit of MVA as a vector for vaccination against pathogens and tumors is considered.
Acknowledgments
We are indebted to R. Bablanian, M. Krupa, and C. Mark for critical reviews of the manuscript, to J. C. Gallego for kindly providing the RAC-1 antibody, to J. C. Oliveros for bioinformatic assistance, and to V. Jiménez for expert technical assistance.
This work was supported by grants from the Spanish Ministry of Science and Technology (grants BIO2000-0340P4 and BIO2001-2269), the Spanish Foundation for AIDS Research (grant FIPSE 36344/02), and the EU (grants QLRT-PL-1999-01321 and QLK2-CT-2002-01867) to M.E. A.P.-M was partially supported by Spanish CICYT grant number BIO2001-1237. The Department of Immunology and Oncology was founded and is supported by the Spanish Council for Scientific Research (CSIC) and Pfizer.
REFERENCES
- 1.Altenburger, W., C.-P. Sütter, and J. Altenburger. 1989. Partial deletion of the human host range in the attenuated vaccinia virus MVA. Arch. Virol. 105:15-27. [DOI] [PubMed] [Google Scholar]
- 2.Antonie, G., F., Scheiflinger, F. Dorner, and F. G. Falkner. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology 244:365-396. [DOI] [PubMed] [Google Scholar]
- 3.Azimi, N., K. Brown, R. N. Bamford, Y. Tagaya, U. Siebenlist, and T. A. Waldmann. 1998. Human T cell lymphotropic virus type I Tax protein trans-activates interleukin 15 gene transcription through an NF-κB site. Proc. Natl. Acad. Sci. USA 95:2452-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchard, T. J., A. Alcamí, P. Andrea, and G. L. Smith. 1998. Modified vaccinia virus Ankara undergoes limited replication in human cells and lacks several immunomodulatory proteins: implication for use as a human vaccine. J. Gen. Virol. 79:1159-1167. [DOI] [PubMed] [Google Scholar]
- 5.Bolt, G., K. Berg, and M. Blixenkrone-Moller. 2002. Measles virus-induced modulation of host-cell gene expression. J. Gen. Virol. 83:1157-1165. [DOI] [PubMed] [Google Scholar]
- 6.Caron, E. 2002. Regulation of Wiskott-Aldrich syndrome protein and related molecules. Curr. Opin. Cell Biol. 14:82-87. [DOI] [PubMed] [Google Scholar]
- 7.Carroll, M. W., and B. Moss. 1997. Host range and cytopathogenicity of the highly attenuated MVA strain of vaccinia virus: propagation and generation of recombinant viruses in a nonhuman mammalian cell line. Virology 238:198-211. [DOI] [PubMed] [Google Scholar]
- 8.Carroll, M. W., W. W. Overwijk, R. S. Chamberlain, S. A. Rosenberg, B. Moss, and N. P. Restifo. 1997. Highly attenuated modified vaccinia virus Ankara (MVA) as an effective recombinant vector: a murine tumor model. Vaccine 15:387-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y. E., and L. A. Laimins. 2000. Microarray analysis identifies interferon-inducible genes and stat-1 as major transcriptional target of human papillomavirus type 31. J. Virol. 74:4174-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, F., V. Castranova, X. Shi, and L. M. Demers. 1999. New insights into the role of nuclear factor-κB, a ubiquitous transcription factor in the initiation of diseases. Clin. Chem. 45:7-17. [PubMed] [Google Scholar]
- 11.Drexler, I., K. Heller, B. Wahren, V. Erfle, and G. Sütter. 1998. Highly attenuated modified vaccinia virus Ankara replicates in baby hamster kidney cells, a potential host for virus propagation, but not in various human transformed and primary cells. J. Gen. Virol. 79:347-352. [DOI] [PubMed] [Google Scholar]
- 12.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frischknecht, F., V. Moreau, S. Rottger, I. Reckmann, C. Superti-Furga, and M. Way. 1999. Actin based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature 404:1007-1011. [DOI] [PubMed] [Google Scholar]
- 14.Gallego-Gómez, J. C., C. Risco, D. Rodriguez, P. Cabezas, S. Guerra, J. L. Carrascosa, and M. Esteban. 2003. Differences in virus-induced cell morphology and virus maturation between the WR and MVA strains of vaccinia virus. J. Virol. 77:10606-10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García de la Nava, J., D. Franco Santaella, J. Cuenca Alba, J. M. Carazo, O. Trelles, and A. Pascual-Montano. 2003. Engene: the processing and exploratory analysis of gene expression data. Bioinformatics 19:657-658. [DOI] [PubMed] [Google Scholar]
- 16.Geiss, G. K., R. E. Bumgarner, M. C. An, A. B. Agy, A. B. van't Wout, E. Hammersmark, V. S. Carter, D. Upchurch, J. I. Mullins, and M. G. Katze. 2001. Large-scale monitoring of host gene expression during HIV-1 infection using cDNA microarrays. Virology 266:8-16. [DOI] [PubMed] [Google Scholar]
- 17.Guerra, S., L. A. López-Fernandez, A. Pascual-Montano, M. Muñoz, K. Harshman, and M. Esteban. 2003. Cellular gene expression survey of vaccinia virus infection of human HeLa cells. J. Virol. 77:6493-6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirsch, V. M., T. R. Fuerst, G. Sutter, M. W. Carroll, L. C. Yang, S. Goldstein, M. Piatak, Jr., W. R. Elkins, W. G. Alvord, D. C. Montefiori, B. Moss, and J. D. Lifston. 1996. Patterns of viral replication correlate with outcome in simian immunodeficiency virus (SIV)-infected macaques: effect of prior immunization with a trivalent SIV vaccine in modified vaccinia virus Ankara. J. Virol. 70:3741-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 20.Jones, J. O., and A. M. Arvin. 2003. Microarray analysis of host cell gene transcription in response to Varicella-Zoster virus infection of human T cells and fibroblasts in vitro and SCIDhu skin xenografts in vivo. J. Virol. 77:1268-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keck, J. G., C. J. Baldick, Jr., and B. Moss. 1990. Role of DNA replication in vaccinia virus gene expression: a naked template is required for transcription of three late trans-activator genes. Cell 61:801-809. [DOI] [PubMed] [Google Scholar]
- 22.Kohonen, T. 1997. Self-organizing maps, 2nd ed. Springer-Verlag, Heidelberg, Germany.
- 23.Mahnel, H., and A. Mayr. 2002. Experiences with immunization against orthopox viruses of humans and animals using vaccine strain MVA. Berl. Muench. Tieraerztl. Wochenschr. 107:253-256. [PubMed] [Google Scholar]
- 24.Mayr, A., H. Stickl, H. K. Muller, K. Danner, and H. Singer. 1978. The smallpox vaccination strain MVA: marker, genetic structure, experience gained with parenteral vaccination and behaviour in organism with a debilitated defense mechanism. Zentbl. Bakteriol. B 167:375-390. [PubMed] [Google Scholar]
- 25.Meyer, H., G. Sutter, and A. Mayr. 1991. Mapping of deletions in the genome of highly attenuated vaccinia virus MVA and their influence on virulence. J. Gen. Virol. 72:1031-1038. [DOI] [PubMed] [Google Scholar]
- 26.Miki, H., K. Minura, and T. Takenawa. 1996. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 15:5326-5335. [PMC free article] [PubMed] [Google Scholar]
- 27.Moreau, V., F. Frischknecht, I. Reckmann, G. Superti-Furga, and M. Way. 2000. A complex of N-WASP and WIP integrates signalling cascades that lead to actin polymerization. Nat. Cell Biol. 2:441-448. [DOI] [PubMed] [Google Scholar]
- 28.Moss, B., M. W. Carroll, L. S. Wyatt, J. R. Bennink, V. M. Hirsch, S. Golldstein, W. R. Elkins, W. Overwijk, R. Chamberlain, S. A. Rosenberg, and G. Sutter. 1996. Host range restricted, non-replicating vaccinia virus vectors as vaccine candidates. Adv. Exp. Med. Biol. 397:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moss, B., and J. L. Shisler. 2001. Immunology 101 at poxvirus U: immune evasion genes. Semin. Immunol. 13:59-66. [DOI] [PubMed] [Google Scholar]
- 30.Nees, N., J. M. Geoghegan, T. Hyman, S. Frank, L. Miller, and C. D. Woodworth. 2001. Papillomavirus type 16 oncogenes downregulate expression interferon-responsive genes and upregulate proliferation-associated and NF-κΒ-responsive genes in cervical keratinocytes. J. Virol. 75:4283-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oie, L. K., and D. Pickup. 2001. Cowpox and other members of the orthopoxvirus genus interfere with the regulation of NF-KB activation. Virology 288:175-187. [DOI] [PubMed] [Google Scholar]
- 32.Quackenbush, J. 2002. Microarray data normalization and transformation. Nat. Genet. 32:496-501. [DOI] [PubMed] [Google Scholar]
- 33.Ramirez, J. C., M. M. Gherardi, and M. Esteban. 2000. Biology of attenuated modified vaccinia virus Ankara recombinant vector in mice: virus fate and activation of B- and T-cell immune responses in comparison with the Western Reserve strain and advantages as a vaccine. J. Virol. 74:923-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sancho, M. C., S. Schleich, G. Griffiths, and J. Krijnse-Locker. 2002. The block in assembly of modified vaccinia virus Ankara in HeLa cells reveals new insights into vaccinia virus morphogenesis. J. Virol. 76:8318-8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider, J., S. C. Gilbert, T. J. Blanchard, T. Hanke, K. J. Robson, C. M. Hannan, M. Becker, R. Sinden, G. L. Smith, and A. V. S. Hill. 1998. Enhanced immunogenicity for CD8+ T cell induction and complete protective efficacy of malaria DNA vaccination by boosting with modified vaccinia virus Ankara. Nat. Med. 4:397-402. [DOI] [PubMed] [Google Scholar]
- 36.Sütter, G., and B. Moss. 1992. Nonreplicating vaccinia vector efficiently expresses recombinant genes. Proc. Natl. Acad. Sci. USA 89:10847-10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sütter, G., L. S. Wyatt, P. L. Foley, J. R. Benninnk, and B. Moss. 1994. A recombinant vector derived from the host range-restricted and highly attenuated MVA strain of vaccinia virus stimulates protective immunity in mice to influenza virus. Vaccine 12:1032-1040. [DOI] [PubMed] [Google Scholar]
- 38.Symons, M., J. M. Derry, B. Karlak, S. Jiang, V. Lemahieu, F. McCormick, U. Francke, and A. Abo. 1996. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase Cdc42Hs, is implicated in actin polymerization. Cell 84:723-734. [DOI] [PubMed] [Google Scholar]
- 39.Takenawa, T., and H. Miki. 2001. WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J. Cell Sci. 114:1801-1809. [DOI] [PubMed] [Google Scholar]
- 40.Tamayo, P., D. Slonim, J. Mesirov, Q. Zhu, E. Dmitrovsky, E. S. Lander, and T. R. Golub. 1999. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc. Natl. Acad. Sci USA 96:2907-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van 't Wout, A. B., G. K. Lehrman, S. A. Mikheeva, G. C. O'Keeffe, M. G. Katze, R. E. Bumgarner, G. K. Geiss, and J. I. Mullins. 2003. Cellular gene expression upon human immunodeficiency virus type 1 infection of CD4+-T-cell lines. J. Virol. 77:1392-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wyatt, L. S., M. W. Carroll, C.-P. Czerny, M. Merchlinsky, J. R. Sisler, and B. Moss. 1998. Marker rescue of the host range restrictions defects of modified vaccinia virus Ankara. Virology 251:334-342. [DOI] [PubMed] [Google Scholar]
- 43.Zhu, H., J. P. Cong, G. Mantora, T. Gineras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]