Abstract
Coronavirus budding at the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) requires accumulation of the viral envelope proteins at this point in the secretory pathway. Here we demonstrate that the spike (S) protein from the group 3 coronavirus infectious bronchitis virus (IBV) contains a canonical dilysine endoplasmic reticulum retrieval signal (-KKXX-COOH) in its cytoplasmic tail. This signal can retain a chimeric reporter protein in the ERGIC and when mutated allows transport of the full-length S protein as well as the chimera to the plasma membrane. Interestingly, the IBV S protein also contains a tyrosine-based endocytosis signal in its cytoplasmic tail, suggesting that any S protein that escapes the ERGIC will be rapidly endocytosed when it reaches the plasma membrane. We also identified a novel dibasic motif (-KXHXX-COOH) in the cytoplasmic tails of S proteins from group 1 coronaviruses and from the newly identified coronavirus implicated in severe acute respiratory syndrome. This dibasic motif also retained a reporter protein in the ERGIC, similar to the dilysine motif in IBV S. The cytoplasmic tails of S proteins from group 2 coronaviruses lack an intracellular localization signal. The inherent differences in S-protein trafficking could point to interesting variations in pathogenesis of coronaviruses, since increased levels of surface S protein could promote syncytium formation and direct cell-to-cell spread of the infection.
Most enveloped viruses assemble at the cytoplasmic face of the plasma membrane and bud out of the cell (reviewed in reference 12). The envelope proteins of these viruses are synthesized in the secretory pathway and accumulate at the plasma membrane. However, some enveloped viruses assemble intracellularly, obtaining their lipid envelope from intracellular, membrane-bound compartments. These viruses bud into the lumens of intracellular compartments and exit the cell by exocytosis. For example, flaviviruses assemble at the endoplasmic reticulum (ER) (9, 31, 34), coronaviruses assemble at the ER-Golgi intermediate compartment (ERGIC) (27), and bunyaviruses (35) and rubella virus (22) bud into the Golgi. The envelope proteins of viruses that assemble in intracellular compartments possess signals that direct them to the site of viral assembly (reviewed in reference 18). These signals mimic those used by endogenous cellular proteins and utilize cellular machinery for localization.
The first ER localization signal for a membrane protein was identified in the adenovirus E3-19K protein (24, 37). This signal consists of lysine residues at the −3 and −4 (or −5) positions relative to the C terminus (51). Dilysine signals were subsequently shown to direct retrieval of escaped proteins from post-ER compartments back to the ER. Proteins with the dilysine signal bind the coatomer complex (COPI) and are recruited into vesicles that travel in a retrograde direction relative to the ER (8, 13). The efficiency of binding to COPI is influenced by the sequence context surrounding the dilysine signal, which contributes to steady-state localization of proteins bearing this signal to the ER, ERGIC, or Golgi complex (51). The envelope glycoprotein from the retrovirus human foamy virus also contains a dilysine signal (15, 16). This dilysine signal directs budding of this virus into intracellular compartments (14). Other types of targeting signals have been identified in envelope proteins of viruses that assemble at the ERGIC or Golgi, although the mechanism by which they work is not understood (reviewed in reference 21).
Coronaviridae are members of the Nidovirales order and contain a positive-strand RNA genome ranging from 27 to 31 kb in size (47). Coronaviruses are classified into groups 1, 2, or 3 by sequence homology (17) and infect a wide range of vertebrate species. Their cellular tropism also varies, since different coronaviruses infect the gastrointestinal tract, respiratory tract, and nervous system. The recent emergence of the coronavirus that causes severe acute respiratory syndrome (SARS) has focused a great deal of interest on coronaviruses (23).
Coronaviruses contain three envelope proteins: envelope (E), membrane (M), and spike (S). The E protein is present in low levels in the mature virion but plays a critical role in viral assembly (10, 28, 41). M is the most abundant protein in the viral envelope and is important for virus maturation, interacting with E, S, and the nucleocapsid during assembly (40, 49, 53). When expressed together from cDNA, the coronavirus E and M proteins interact and form virus-like particles (5, 6, 53). The S protein is less abundant than M in virions and is responsible for binding and fusion to host cells (reviewed in reference 11).
We study the group 3 coronavirus infectious bronchitis virus (IBV) as a model for intracellular assembly at the ERGIC. IBV E contains a Golgi targeting signal within its cytoplasmic tail (4). IBV M contains a Golgi targeting signal located in its first transmembrane domain (33, 50). Thus, IBV M and E move past the virus assembly site when expressed individually, and it is not yet known how the viral envelope proteins are collected together in the ERGIC in infected cells.
When S proteins from different coronaviruses are exogenously expressed, a large portion remains intracellular (54). Slow folding of the large lumenal domain in the ER could contribute to this localization (39). Here we demonstrate a canonical dilysine ER retrieval signal located at the C terminus of IBV S. This dilysine signal is sufficient to retain IBV S intracellularly and retains a chimeric protein containing the cytoplasmic tail of IBV S in the ERGIC. We also discovered a novel dibasic signal in group 1 coronavirus and SARS S proteins that is similar to the dilysine signal in IBV S. These two types of signals are likely to contribute to the localization of S protein at the virus assembly site. Our observations support the hypothesis first proposed by Vennema et al. (54) suggesting that coronavirus S proteins contain intracellular retention signals.
MATERIALS AND METHODS
Cells and transfection.
HeLa cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum and antibiotics. Transient transfections were performed using 2 μg of pcDNA3.1/Hygro (Invitrogen, Grand Island, N.Y.) encoding the appropriate cDNA and 6 μl of LT-1 transfection reagent (Mirus, Madison, Wis.) following the manufacturer's instructions. Expression was analyzed at 18 h posttransfection, except for pcDNA3.1/S and pcDNA3.1/S2A transfections, which were analyzed at 3 days posttransfection.
Plasmids and expression vectors.
We cloned the cDNA encoding IBV S by reverse transcription-PCR) using RNA from Vero cells infected with the Vero-adapted strain of IBV (33). The full-length cDNA was assembled in pBluescript (Stratagene, La Jolla, Calif.) and sequenced by dideoxy sequencing at the Johns Hopkins DNA Synthesis and Sequencing Facility. The cloned Vero-adapted IBV S gene contained 14 amino acid changes compared to the published Beaudette IBV S sequence (2). Twelve of these amino acid changes (Leu122 to Ile, Leu130 to Phe, Asn261 to Thr, Lys330 to Asn, Ile336 to Val, Lys364 to Ser, Asn421 to His, Ile474 to Val, Asn683 to Thr, Lys689 to Arg, Asn709 to Asp, and Phe723 to Leu) were found in other strains of IBV, while two unique amino acid changes were Asn842 to Asp and Ser1012 to Ile. For transient transfection in HeLa cells, the full-length cDNA encoding IBV S was subcloned into pcDNA3.1/Hygro utilizing the XbaI and ApaI sites. The dilysine located at the C terminus of the S protein was mutagenized using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.), generating the pcDNA3.1/S2A mutant signal (Lys1159 to Ala and Lys1160 to Ala).
The G-S constructs were generated by introducing a BamHI site by QuikChange site-directed mutagenesis at the end of the S transmembrane domain at Gly1119. The resulting protein, STMB, contained one amino acid change (Cys1120 to Ile), but this did not affect normal trafficking of the S protein (data not shown). The S cytoplasmic tail coding sequence was then subcloned in place of the G cytoplasmic tail in pcDNA3.1/GTMB (42), using the BamHI and XhoI sites, generating pcDNA3.1/G-S and pcDNA3.1/G-S2A. Mutation of the putative tyrosine internalization signal (Tyr1143 to Ala and Tyr1144 to Ala) located upstream of the dilysine signal (see Fig. 1A) was performed using the QuikChange site-directed mutagenesis kit in the pcDNA3.1/G-S2A plasmid to generate pcDNA3.1/G-S4A. To generate the pcDNA3.1/G-SBCV construct, sense and antisense 42-mer oligonucleotides encoding the C-terminal 11 amino acids of bovine coronavirus (BCV) S (GenBank accession no. P25193) flanked by AatI and ApaI restriction sites were annealed in a solution containing 100 mM potassium acetate, 30 mM HEPES-KOH (pH 7.4), and 2 mM magnesium acetate and phosphorylated with T4 DNA kinase. A silent AatII site was introduced 11 amino acids before the C terminus of the pcDNA3.1/G-S by QuikChange site-directed mutagenesis to create pcDNA3.1/G-S(AatII). A DNA fragment encoding the last 11 residues of IBV S was excised from pcDNA3.1/G-S (AatII) with AatII and ApaI, and the BCV tail sequence was inserted, generating pcDNA3.1/G-SBCV. The same procedure was used to generate other G-S chimeric proteins containing the last 11 amino acids of coronavirus S proteins from transmissible gastroenteritis virus (TGEV) (GenBank accession no. NP_058424), murine hepatitis virus (MHV) strain A59 (GenBank accession no. VGIH59), and Urbani SARS virus (GenBank accession no. AAP72986).
FIG. 1.
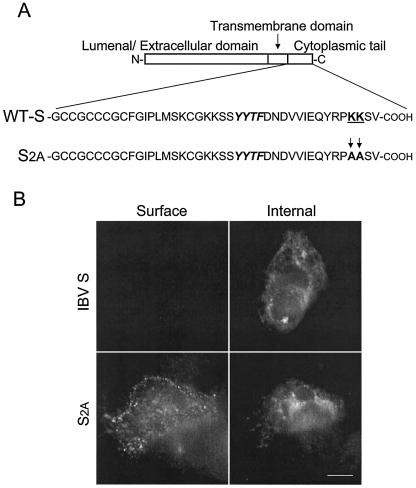
Mutation of the C-terminal dilysine signal promotes transport of IBV S to the cell surface. (A) The sequence of the IBV S cytoplasmic tail, with the KKXX motif at the C terminus. Alanine residues (indicated with arrows) replace the dilysine motif in the S2A mutant. A potential endocytosis signal (YYTF) is indicated in italics. (B) Intact HeLa cells expressing wild-type IBV S or S2A were stained with mouse anti-IBV S at 4°C and then fixed, permeabilized, and stained with rabbit anti-IBV S for internal expression. Secondary antibodies were fluorescein-conjugated donkey anti-rabbit immunoglobulin G (IgG) and Texas Red-conjugated goat anti-mouse IgG. Bar, 10 μm.
The pcDNA3.1/G-SSARS2A construct was generated by mutagenizing the codons for His1158 and Lys1161 of pcDNA3.1/G-SSARS to alanines by QuikChange site-directed mutagenesis. The same procedure was used to construct pcDNA3.1/G-STGEV2A from pcDNA3.1/G-STGEV.
Antibodies.
The mouse monoclonal anti-IBV S antibody (9B1B6) recognizes the lumenal domain and was a kind gift of Ellen Collisson (Texas A&M University, College Station, Tex.) (55). The rabbit polyclonal anti-IBV S antibody was generated from a bacterial fusion protein consisting of a His6 tag fused to the cytoplasmic tail of IBV S. The IBV S-tail sequence was subcloned into pRSET expression vector C (Invitrogen, Grand Island, N.Y.) and transformed into Escherichia coli BL21(DE3)/pLysS (Invitrogen). Expression was induced for 1 h with 100 mM isopropyl-β-d-thiogalactopyranoside. The fusion protein was purified on nickel-agarose beads (Qiagen, Valencia, Calif.), following the manufacturer's instructions. The rabbit anti-IBV S tail antibody was specific for IBV S in immunoprecipitation experiments (data not shown). The mouse anti-vesicular stomatitis virus glycoprotein (VSV-G) monoclonal antibody used in immunofluorescence recognizes the lumenal domain of VSV-G (29). For immunofluorescence, rabbit polyclonal affinity-purified anti-VSV-G was also used (45). For immunoprecipitation of radiolabeled VSV-G and G-S proteins, a rabbit antibody to whole VSV was used (56). The mouse monoclonal anti-ERGIC-53 antibody was a kind gift of H. P. Hauri (Basel, Switzerland). Mouse anti-GM130 was from Transduction Labs (San Jose, Calif.), and rabbit anticalnexin was from StressGen (San Diego, Calif.)
Indirect immunofluorescence microscopy.
HeLa cells were plated on coverslips in 35-mm-diameter dishes 1 day before transfection and transfected as described above. At 18 h posttransfection (or 3 days for IBV S), cells were fixed in 3% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min at room temperature, permeabilized with 0.5% Triton X-100 for 3 min, and stained as previously described (50). For staining with anticalnexin, cells were permeabilized with 0.1% saponin instead of Triton X-100. To label surface IBV S or G-S chimeric proteins, intact cells were washed with ice-cold PBS and incubated with mouse anti-IBV S or anti-VSV-G for 10 min at 4°C. Cells were then washed two times with ice-cold PBS and fixed and permeabilized as described above. Cells were then stained for intracellular proteins with either rabbit affinity-purified anti-VSV-G or rabbit anti-IBV S tail as described above. Live cell antibody uptake was performed by incubating HeLa cells expressing G-S chimeric proteins with mouse anti-VSV-G (0.5 μg/ml in culture medium) for 15 min at 37°C. Cells were then fixed and permeabilized as described above and stained for internal protein with rabbit affinity-purified anti-VSV-G. Images were acquired with a Zeiss (Thornwood, N.Y.) Axioscop microscope equipped for epifluorescence with a Sensys charge-coupled-device camera (Photometrics, Tucson, Ariz.), using IPLab software (Scanalytics, Vienna, Va.).
Metabolic labeling, immunoprecipitation, and glycosidase digestion.
HeLa cells were transfected as described above and at 16 h post transfection were starved for 20 min with methionine- and cysteine-free medium at 37°C. The cells were then labeled with 50 uCi of 35S-Promix (Amersham Pharmacia Biotech, Piscataway, N.J.) per ml in methionine- and cysteine-free medium for 15 min at 37°C. Cells were chased in unlabeled medium for 0, 15, 30, or 60 min. After a rinse in ice-cold PBS, the cells were lysed in a detergent solution (62.5 mM EDTA, 50 mM Tris-HCl [pH 8.0], 0.4% deoxycholate, 1% NP-40) containing protease inhibitors. After removal of nuclei and debris by centrifugation at 16,000 × g for 3 min, the lysate was incubated at 37°C for 20 min with 2 μl of rabbit anti-VSV. Antibody-antigen complexes were collected by incubation with fixed Staphylococcus aureus (Pansorbin; Calbiochem, San Diego, Calif.) for 15 min at room temperature. Immunoprecipitates were washed in RIPA buffer (0.1% sodium dodecyl sulfate [SDS], 50 mM Tris-HCl [pH 8.0], 1% Na-deoxycholate, 150 mM NaCl, 1% Triton X-100) and subsequently eluted in 20 μl of 50 mM Tris-HCl (pH 6.8)-1% SDS by incubation at 100°C for 3 min. An equal volume of 0.15 M sodium citrate, pH 5.5, containing 0.4 mU of recombinant endoglycosidase H (endo H) (New England Biolabs, Beverly, Mass.) was added, and samples were incubated for 16 h at 37°C. The samples were then subjected to SDS-polyacrylamide gel electrophoresis (PAGE) using 10% gels, and oligosaccharide processing was quantified on a Molecular Imager FX phosphorimager (Bio-Rad, Hercules, Calif.) by comparing band densities of endo H-sensitive and -resistant forms, using Quantity One software (Bio-Rad).
RESULTS
Mutation of the putative dilysine signal in IBV results in surface expression.
Vennema et al. (54) showed that coronavirus S proteins expressed exogenously were transported through the secretory pathway more slowly than S proteins in infected cells. They proposed that coronavirus S proteins might contain intracellular retention signals that would be masked after virus assembly. Examination of the Beaudette IBV S protein sequence indicated a putative ER retrieval signal in its cytoplasmic tail, consisting of lysine residues at the −3 and −4 positions from the C terminus (Fig. 1A). These two lysines are conserved in S proteins from eight different strains of IBV whose sequences are available in GenBank, as well as in turkey coronavirus. To determine the contribution of the putative dilysine signal to the localization of IBV S, we expressed the wild-type protein and a mutant where both lysines were replaced by alanines. Transiently transfected HeLa cells were examined by indirect immunofluorescence microscopy (Fig. 1B). Wild-type S was not detected at the plasma membrane, and it accumulated in intracellular compartments resembling the ER and ERGIC (see below). The mutant lacking the dilysine signal (S2A) was readily detected at the cell surface as well as throughout the secretory pathway. Thus, the dilysine signal plays a role in accumulation of the IBV S protein near the virus assembly site. However, the low levels of expression of the IBV S protein that were obtained from transfection of cDNA made further study of this targeting signal difficult in the context of the full-length S protein.
Targeting information in the IBV S cytoplasmic tail retains a reporter protein intracellularly.
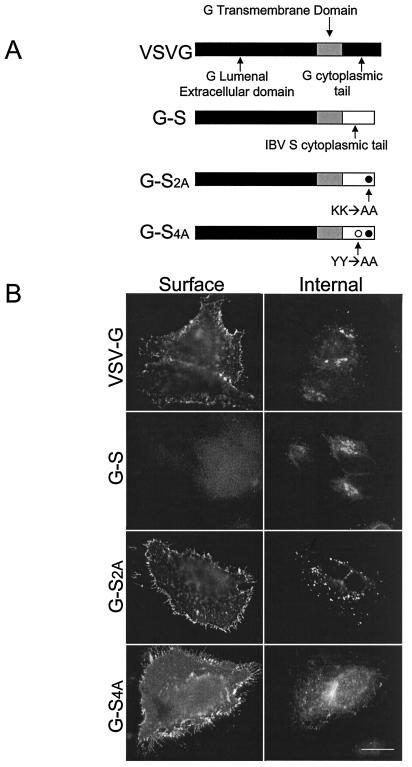
To study the targeting signal in the IBV S cytoplasmic tail, we asked if this domain could retain a reporter protein intracellularly. VSV-G is a well-studied plasma membrane protein that has the same membrane topology as the IBV S protein (a type I membrane protein). We replaced the cytoplasmic tail of VSV-G with that of IBV S to generate the chimeric protein G-S (Fig. 2A). HeLa cells were transiently transfected with plasmids encoding VSV-G, G-S, or G-S2A (with the dilysine signal replaced with alanines) and were examined by indirect immunofluorescence microscopy (Fig. 2B). As expected, VSV-G exhibited significant surface expression with internal staining throughout the secretory pathway. G-S was absent from the plasma membrane in most cells and was instead localized to intracellular compartments. A low percentage of transfected cells (∼13%) expressed G-S on the cell surface (data not shown). These cells appeared to express higher levels of G-S than cells that did not have detectable protein at the cell surface. It is likely that the machinery that recognizes the intracellular localization signal can be saturated (52).
FIG. 2.
The IBV S cytoplasmic tail retains a reporter protein intracellularly. (A) The G-S chimera contains the lumenal head (black) and transmembrane domain (gray) from VSV-G and the cytoplasmic domain from IBV S (white; sequence shown in Fig. 1A). G-S2A has the dilysine signal mutagenized to alanines residues, and G-S4A has both the upstream tyrosine internalization signal and the dilysine signal mutagenized to alanine residues. (B) Intact HeLa cells expressing VSV-G, G-S, G-S2A, or G-S4A were stained with mouse anti-VSV-G at 4°C, fixed, permeabilized, and then stained with rabbit anti-VSV-G for internal expression. Secondary antibodies were fluorescein-conjugated donkey anti-rabbit IgG and Texas Red-conjugated goat anti-mouse IgG. Bar, 10 μm.
Replacing the two lysine residues with alanines (G-S2A) resulted in expression on the cell surface with a patchy distribution. Internal staining of cells expressing G-S2A demonstrated that this protein was largely localized in the Golgi region and numerous small puncta throughout the cytoplasm. Upon closer examination of the IBV S-tail sequence, we observed a potential tyrosine internalization signal (YYTF) (Fig. 1A) (reviewed in reference 3) upstream of the dilysine signal. Replacing the two tyrosine residues in this sequence with alanines (G-S4A) resulted in homogeneous plasma membrane distribution, without the puncta seen with G-S2A (Fig. 2B).
Steady-state localization of G-S chimeric proteins.
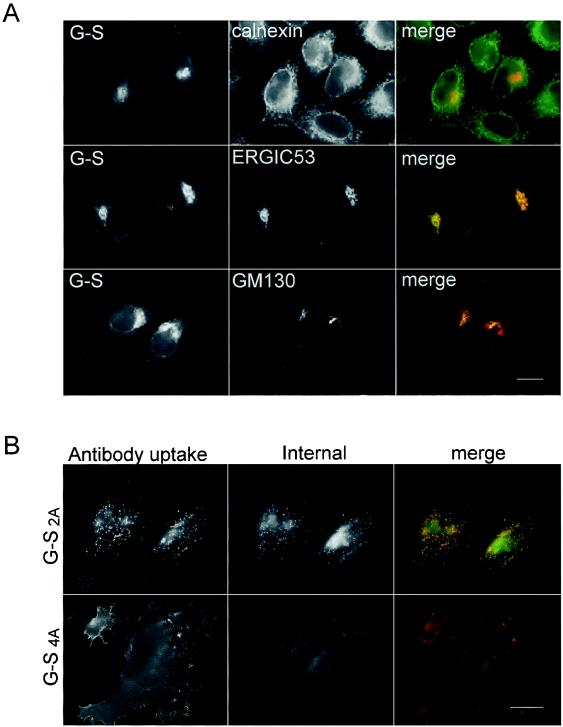
To determine the intracellular localization of G-S, transiently transfected HeLa cells were double labeled with antibodies recognizing various resident proteins localized in different intracellular compartments (Fig. 3A). The localization of G-S partially overlapped those of the ER resident protein calnexin and the Golgi protein GM130. However, the distribution of G-S most strongly overlapped with that of ERGIC-53, indicating that G-S largely accumulated in this intermediate compartment (Fig. 3A, color merge panels). The steady-state localization of G-S in the ERGIC instead of the ER may reflect the contribution of the sequence surrounding the dilysine signal, as shown for the ERGIC-53 protein (1). The full-length IBV S protein, however, showed more ER staining than G-S (compare Fig. 1B and 3A). This difference in steady-state localization probably reflects the slow folding of the large lumenal domain of coronavirus S proteins in the ER (39). By replacing the lumenal domain of IBV S with that of VSV-G, we were able to explore the contribution of the dilysine signal to targeting in the absence of slow folding and exit from the ER.
FIG. 3.
G-S is retained mostly in the ERGIC, and mutation of the dilysine motif results in transport to the plasma membrane and endocytosis. (A) HeLa cells expressing G-S were fixed, permeabilized, stained with either rabbit or mouse anti-VSV-G, and double labeled with either rabbit anticalnexin, mouse anti-ERGIC-53, or mouse anti-GM130. For cells double labeled with calnexin, the secondary antibodies were fluorescein-conjugated donkey anti-rabbit IgG and Texas Red-conjugated goat anti-mouse IgG. For cells double labeled with ERGIC-53 or GM130, secondary antibodies were fluorescein-conjugated goat anti-mouse IgG and Texas Red-conjugated donkey rabbit IgG. (B) Intact HeLa cells expressing G-S2A and G-S4A were incubated with mouse anti-VSV-G at 37°C for 15 min, fixed, permeabilized, and then stained with rabbit anti-VSV-G for internal expression. Secondary antibodies were fluorescein-conjugated donkey anti-rabbit IgG and Texas Red-conjugated goat anti-mouse IgG. Bars, 10 μm.
Unlike G-S, G-S2A was localized in distinct puncta that could represent endocytotic structures following internalization from the plasma membrane (Fig. 2B). To test this idea, live cells expressing G-S2A or G-S4A were incubated for 15 min at 37°C with mouse anti-VSV-G antibodies, as described in Materials and Methods. After fixation and permeabilization, cells were stained with rabbit anti-VSV-G for detection of the total G-S2A pool (Fig. 3B). The intracellular puncta containing G-S2A were labeled by exogenously added antibody (Fig. 3B, color merge), indicating that at least a portion of G-S2A was efficiently endocytosed after reaching the plasma membrane. Mutation of the tyrosine residues in G-S4A demonstrated that the YYTF sequence was the likely internalization signal, since G-S4A was evenly distributed at the cell surface and little anti-VSV-G antibody was internalized. Note that the constructs described below containing the C-terminal residues of other coronavirus S proteins all have the IBV S tyrosine internalization motif.
Mutation of the dilysine signal in G-S results in efficient trafficking through the Golgi complex.
During biosynthesis, VSV-G obtains two N-linked oligosaccharides in the ER. The oligosaccharides are processed in the medial Golgi, where they become resistant to digestion with the enzyme endo H. Thus, the rate of trafficking through the Golgi can be inferred from the rate of acquisition of endo H resistance. HeLa cells expressing VSV-G, G-S, G-S2A, or G-S4A were pulse labeled with [35S]methionine-cysteine for 15 min and chased for various times in medium containing unlabeled amino acids. The expressed proteins were immunoprecipitated and subjected to endo H digestion (Fig. 4A). Note that G-S chimeric proteins are slightly larger than VSV-G and run accordingly on SDS-PAGE. VSV-G, G-S2A, and G-S4A proteins rapidly became resistant to endo H during the chase. However, only a small portion of G-S acquired endo H resistance by 60 min. Quantitation of four similar experiments showed that VSV-G, G-S2A, and G-S4A all move through the Golgi at similar rates, whereas G-S is mostly retained in a compartment prior to the medial Golgi (Fig. 4B). The low rate of processing of G-S could reflect the low percentage of cells that exhibit surface staining for this protein. Given the steady-state localization and slow processing of G-S, we conclude that the dilysine sequence serves as a functional ER retrieval signal for IBV S.
FIG. 4.
G-S is rapidly trafficked through the Golgi after mutation of the dilysine signal. (A) HeLa cells expressing VSV-G, G-S, G-S2A, or G-S4A were pulse labeled with [35S] methionine-cysteine for 15 min, chased for the times indicated, lysed, and immunoprecipitated with anti-VSV-G antibody. The immunoprecipitates were treated with endo H and subjected to SDS-PAGE. The endo H-resistant and -sensitive forms are indicated. (B) Quantitation of oligosaccharide processing rates. These data represent the averages from four independent experiments, with the error bars representing the standard deviations.
Group 2 coronaviruses lack an intracellular localization signal in their cytoplasmic tails.
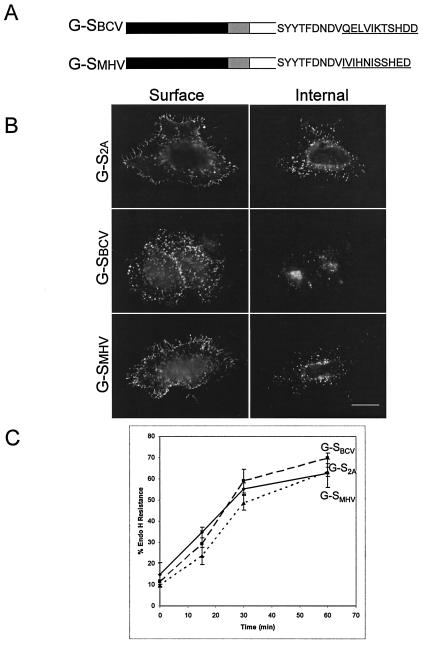
The cytoplasmic tails of group 2 coronaviruses showed no obvious signal or motif that would lead to their sequestration within intracellular compartments. To test this directly, we replaced the last 11 amino acids of the G-S chimera with the final 11 amino acids from BCV or MHV strain A59 to generate G-SBCV and G-SMHV, respectively (Fig. 5A). We chose to swap the last 11 residues in G-S in case a targeting signal existed in this sequence and because a unique restriction site could be introduced at this position with a silent mutation. HeLa cells were transfected with cDNAs encoding G-S2A, G-SBCV, and G-SMHV and assayed by indirect immunofluorescence microscopy. G-SBCV and G-SMHV were expressed on the cell surface, with distinct internal staining resembling the puncta seen with G-S2A (Fig. 5B). Pulse-chase analysis of G-SBCV and G-SMHV demonstrated that oligosaccharide processing in the Golgi occurred at rates comparable to that of G-S2A (Fig. 5C). Thus, unlike the group 3 coronavirus IBV, the C-terminal 11 amino acids of BCV S and MHV S lack signals for localization in the early secretory pathway.
FIG. 5.
The C-terminal 11 amino acids of S proteins from group 2 coronaviruses lack intracellular localization signals. (A) The final 11 amino acids of G-S were swapped with those from the BCV or MHV-A59 S proteins (underlined). Note the lack of any apparent dibasic motif at the −3 and −4 or −5 position from the C terminus in the BCV and MHV sequences. (B) Intact HeLa cells expressing G-S2A, G-SBCV, or G-SMHV were stained with mouse anti-VSV-G at 4°C, fixed, permeabilized, and then stained with rabbit anti-VSV-G for internal expression. Secondary antibodies were fluorescein-conjugated donkey anti-rabbit IgG and Texas Red-conjugated goat anti-mouse IgG. Bar, 10 μm. (C) HeLa cells expressing G-S2A, G-SBCV, or G-SMHV were pulse labeled with [35S]methionine-cysteine for 15 min, chased for 0, 15, 30, or 60 min, lysed, and immunoprecipitated with anti-VSV-G antibody. The immunoprecipitates were treated with endo H and subjected to SDS-PAGE. Endo H resistance was quantitated for three independent experiments (error bars represent standard deviations).
S proteins of group 1 coronaviruses and the SARS coronavirus possess a novel intracellular localization signal.
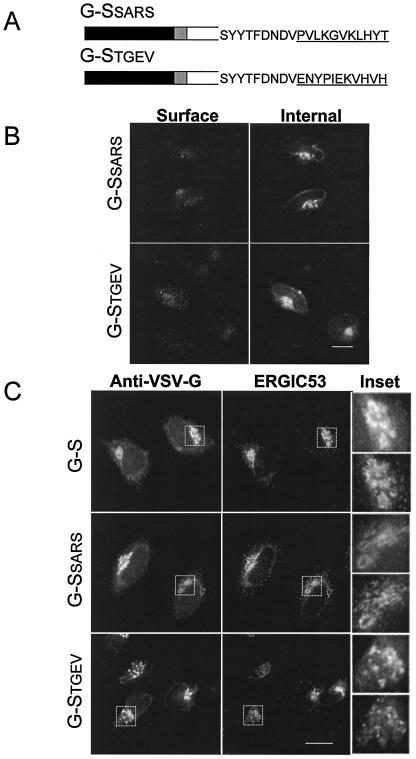
TGEV is a group 1 coronavirus (17), while the recently identified SARS coronavirus has been reported to be distantly related to group 2 coronaviruses (48). Examination of the cytoplasmic tail sequences of the S proteins from TGEV and SARS identified a unique motif, KXHXX, at their C termini (Fig. 6A). S proteins from other group 1 coronavirus (human coronavirus 229E, canine coronavirus, and feline infectious peritonitis virus) also contain this sequence at their C termini. The KXHXX motif fits the criteria for a dibasic motif (51) if the histidine residue is protonated. We postulated that this potential dibasic signal could act similarly to the dilysine signal in IBV S. To test this hypothesis, we replaced the final 11 amino acids of G-S with the corresponding sequence of the SARS or TGEV S proteins, generating the G-SSARS and G-STGEV constructs, respectively.
FIG. 6.
Group 1 coronaviruses and SARS S proteins contain intracellular localization signals in their cytoplasmic tails. (A) The final 11 amino acids of TGEV S or SARS S (underlined), which contain putative dibasic signals, were swapped for the same residues of G-S. (B) Intact HeLa cells expressing G-SSARS or G-STGEV were stained with mouse anti-VSV-G at 4°C, fixed, permeabilized, and then stained with rabbit anti-VSV-G for internal expression. Secondary antibodies were fluorescein-conjugated donkey anti-rabbit IgG and Texas Red-conjugated goat anti-mouse IgG. (C) HeLa cells expressing G-S, G-SSARS, or G-STGEV were fixed, permeabilized, and stained with rabbit anti-VSV-G and mouse anti-ERGIC-53. Secondary antibodies were fluorescein-conjugated goat anti-rabbit IgG and Texas Red-conjugated donkey anti-rabbit IgG. The insets show enlargements of the boxed regions, with the anti-VSV-G panel on top and the anti-ERGIC-53 panel below. Bars, 10 μm.
Transiently transfected HeLa cells expressing G-SSARS and G-STGEV were examined by indirect immunofluorescence microscopy (Fig. 6B). The majority of cells lacked surface expression, with intracellular staining in a post-ER compartment. A low percentage of cells expressing G-SSARS and G-STGEV (∼11 and ∼6%, respectively) had detectable surface expression (data not shown). These were cells with overall high levels of expression, similar to our findings with G-S. The intracellular distribution of G-SSARS and G-STGEV most closely overlapped with that of ERGIC-53 (Fig. 6C). These data suggest that the KXHXX motif is an intracellular localization signal similar to the dilysine signal in IBV S.
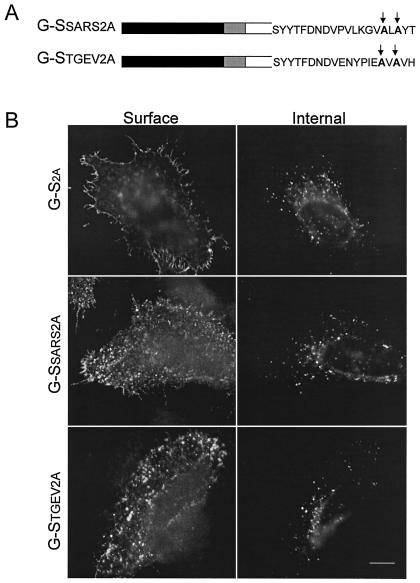
Mutagenesis of the lysine and histidine residues in the KXHXX signal results in loss of ERGIC localization.
To test the hypothesis that the KXHXX motif functions as a genuine localization signal, the histidine and lysine residues in G-SSARS and G-STGEV were mutagenized to alanines to construct the G-SSARS2A and G-STGEV2A chimeras (Fig. 7A). HeLa cells transiently transfected with G-S2A, G-SSARS2A, or G-STGEV2A were examined by indirect immunofluorescence microscopy (Fig. 7B). G-SSARS2A and G-STGEV2A were efficiently transported to the surface, with internal puncta similar to those for G-S2A.
FIG. 7.
Mutagenesis of the histidine and lysine residues in G-SSARS and G-STGEV results in transport to the plasma membrane. (A) The histidine at −3 and the lysine at −5 were replaced by alanines (G-SSARS2A and G-STGEV2A). (B) Intact HeLa cells expressing G-S2A, G-SSARS2A, or G-STGEV2A were stained with mouse anti-VSV-G at 4°C, fixed, permeabilized, and then stained with rabbit anti-VSV-G for internal expression. Secondary antibodies were fluorescein-conjugated donkey anti-rabbit IgG and Texas Red-conjugated goat anti-mouse IgG. Bar, 10 μm.
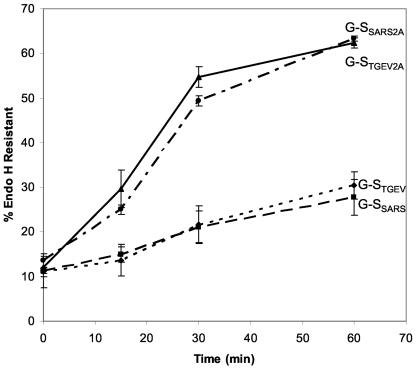
Oligosaccharide processing of G-SSARS, G-STGEV, G-SSARS2A, and G-STGEV2A (Fig. 8) confirmed that mutation of the lysine and histidine residues in the cytoplasmic tails of G-SSARS and G-STGEV abrogated intracellular retention. Both G-SSARS2A and G-STGEV2A obtained endo H resistance at rates similar to those for VSV-G and G-S2A, whereas only low rates of processing occurred with chimeric proteins containing the KXHXX motif.
FIG. 8.
Mutation of the histidine and lysine residues in G-SSARS and G-STGEV results in rapid trafficking through the Golgi complex. HeLa cells expressing G-SSARS, G-STGEV, G-SSARS2A, or G-STGEV2A were pulse labeled with [35S]methionine-cysteine for 15 min, chased for 0, 15, 30, or 60 min, lysed, and immunoprecipitated with anti-VSV-G antibody. The immunoprecipitates were treated with endo H and subjected to SDS-PAGE. Endo H resistance was quantitated (three independent experiments, with the error bars representing the standard deviations).
These results point to a novel intracellular localization signal (KXHXX) at the C terminus of type I membrane proteins. Further experiments are required to determine if proteins that posses this motif use the same ER retrieval pathway followed by proteins with the KKXX signal. Overall, our findings demonstrate that S proteins from groups 1 and 3 and the SARS coronavirus contain signals near their C termini that specify intracellular localization near the virus assembly site.
DISCUSSION
The purpose of this study was to explore the targeting and accumulation of coronavirus S proteins at the site of virus assembly. We report that a dilysine signal (KKXX-COOH) contributes to the localization of IBV S near the virus assembly site. The cytoplasmic tail of IBV S retained a plasma membrane reporter protein (VSV-G) in the ERGIC and reduced the rate of oligosaccharide processing in the Golgi complex. We also studied the cytoplasmic tails of S proteins of other coronaviruses. The C-terminal 11 amino acids of the S protein from the group 1 coronavirus TGEV as well as the SARS coronavirus contained a signal similar to the dilysine motif (KXHXX-COOH). Like the KKXX motif, the KXHXX sequence specified intracellular localization of the G-S chimera in the ERGIC. However, S proteins from group 2 coronaviruses (including BCV and MHV) lacked such signals in this region of their cytoplasmic tails. Here we discuss how these signals may play a role in mediating the localization of coronavirus S proteins to the site of virus assembly.
A canonical dilysine signal is present at the carboxy terminus of IBV S.
We found that when transiently expressed, the IBV S protein was localized to the ER and ERGIC. Mutation of the C-terminal dilysine signal resulted in readily detectable surface expression. The dilysine signal could be responsible for localizing IBV S near the site of virus assembly and could serve to limit surface expression. Both IBV M and E are transported past the virus assembly site when expressed from cDNA (7, 32). Presumably, interactions between IBV S, M, and E result in localization of the three proteins at the ERGIC during virus assembly. It is interesting that all three of the IBV envelope proteins possess targeting signals that maintain their localization near the ERGIC, although all three signals are different. It is perhaps most efficient to retain the three envelope proteins individually near the assembly site.
To fully explore the contribution of the dilysine signal to targeting, we studied it independently of the full-length IBV S protein. Replacing the cytoplasmic tail of a plasma membrane reporter protein (VSV-G) with the cytoplasmic tail of IBV S resulted in ERGIC localization of the G-S chimera. When the two lysine residues were mutagenized to alanines, surface expression of G-S2A was readily detected and trafficking through the Golgi occurred at a rate similar to that of VSV-G. At very high expression levels, G-S was transported to the plasma membrane. It is likely that the retrieval machinery can be saturated, as shown for the H/KDEL signal for soluble ER resident proteins (52).
Recognition of the dilysine signal.
The dilysine signal consists of lysines located at the −3 and −4 or −5 position from the carboxy terminus (24) and is recognized by the COPI coatomer complex (8, 20). Our preliminary experiments suggest that the cytoplasmic tail of IBV S also binds the COPI coatomer complex (data not shown). The coatomer complex is composed of seven different subunits and is the major building block of COPI-coated vesicles. COPI vesicles mediate the retrograde transport of proteins from the Golgi to the ER (reviewed in references 13 and 43) and perhaps other vesicle transport steps. While the COPI complex directly interacts with the dilysine signal, the efficiency of binding is influenced by residues surrounding the two lysines (51). For example, two phenylalanine residues immediately following a dilysine signal contribute to steady-state localization of ERGIC-53 to the ERGIC instead of the ER (1). We have not yet begun to explore the residues surrounding the dilysine signal in IBV S that influence its steady state localization.
The role of other dibasic motifs in intracellular localization.
Other types of signals have been found to mediate ER localization of cellular proteins. The Iip33 form of the invariant chain (a type II membrane protein) has two arginine residues near its N-terminal cytoplasmic tail that contribute to its ER localization (44). More recently, internal ER localization signals (e.g., RXR) have been identified in certain oligomeric ion channels (59). For these dibasic ER localization signals, sequence context contributes to efficiency of localization (58), as does their distance from the membrane (46). In addition, as shown for the dilysine motif (30), oligomerization of diarginine-containing subunits with other proteins can mask the recognition of the localization signal, allowing transport to the plasma membrane in a regulated manner (38, 57, 59). Intracellular localization signals on viral proteins would effectively be masked by the assembly process, allowing exocytosis of budded virions.
A novel intracellular localization motif.
In the work described here, we found that a previously unknown motif (KXHXX) in group 1 coronaviruses and the SARS coronavirus can mimic the dilysine signal in IBV S, resulting in ERGIC localization. If the histidine residue is protonated, the KXHXX motif fits the criteria for a dibasic ER retrieval signal (51). Consistent with this, it was shown that a histidine residue at the −3 position of the oligosaccharide transferase subunit OST48 could substitute for the lysine residue normally present in this position (19). Sequence context is clearly important, since histidine could not replace the lysine at the −3 position in the adenovirus E3-19K protein (25). The lysine-histidine motif located at the carboxy terminus of TGEV is also present in other group 1 coronaviruses, including canine coronavirus, feline infectious peritonitis virus, and human coronavirus 229E. When attached to a plasma membrane reporter protein, the C-terminal 11 amino acids of TGEV or SARS coronavirus S proteins promoted ERGIC localization. Efficient transport to the plasma membrane occurred when the lysine and histidine residues were mutagenized to alanines in the G-SSARS or G-STGEV chimera.
Although dibasic signals are present in S proteins from groups 1 and 3 and the SARS coronavirus, there was no such motif in the final 11 amino acids of group 2 coronaviruses BCV and MHV. Interestingly, group 2 coronaviruses encode a hemagglutinin esterase (HE) protein, although not all express it. HE is a type I membrane protein with arginine at the −5 position and histidine at the −3 position from the C terminus (26). This RXHXX motif can function to sequester OST48 in the ER (19). BCV HE has been shown to oligomerize indirectly with S through its interaction with M (36). Thus, group 2 coronaviruses that express HE might use a similar ER retrieval pathway for localization of the S protein.
Implications of ER retrieval signals for coronavirus S protein function.
One of the hallmarks of coronavirus infection of cultured cells is formation of syncytia, caused by cell-cell fusion induced by S protein expressed at the plasma membrane. How do our results impact this known activity of the coronavirus S protein? We hypothesize that ER retrieval signals in conjunction with slow folding of the lumenal domain in the ER effectively limit the level of surface S protein (39). At high expression levels (as might occur late in coronavirus infection or with use of expression vectors, such as vaccinia virus), the S protein could saturate the ER retrieval machinery and be transported to the plasma membrane. Retention of S near the budding site would thus be regulated by the level of S expressed. In support of the idea that coronaviruses regulate the level of surface S protein, we found that IBV S contains an endocytosis signal upstream of the dilysine signal in its cytoplasmic tail. This internalization signal ensures that IBV S protein that reaches the plasma membrane does not remain there for long. Since S proteins from group 2 coronaviruses lack ER retrieval signals in their cytoplasmic domains, this group may depend on cell-to-cell spread of the infection by syncytium formation to a greater extent than other coronaviruses. Further experiments are needed to explore the importance of dibasic localization signals in assembly and pathogenesis of different coronaviruses.
Acknowledgments
This work was supported by National Institutes of Health grant GM64647.
We thank Andrea Medrano for cloning the IBV S cDNA, Ellen Collisson and Hans-Peter Hauri for antibodies, and the members of the Machamer lab for useful comments on the manuscript.
REFERENCES
- 1.Andersson, H., F. Kappeler, and H. P. Hauri. 1999. Protein targeting to endoplasmic reticulum by dilysine signals involves direct retention in addition to retrieval. J. Biol. Chem. 274:15080-15084. [DOI] [PubMed] [Google Scholar]
- 2.Binns, M. M., M. E. Boursnell, I. J. Foulds, and T. D. Brown. 1985. The use of a random priming procedure to generate cDNA libraries of infectious bronchitis virus, a large RNA virus. J. Virol. Methods 11:265-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonifacino, J. S., and L. M. Traub. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395-447. [DOI] [PubMed] [Google Scholar]
- 4.Corse, E., and C. E. Machamer. 2002. The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting. J. Virol. 76:1273-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corse, E., and C. E. Machamer. 2003. The cytoplasmic tails of infectious bronchitis virus E and M proteins mediate their interaction. Virology 312:25-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corse, E., and C. E. Machamer. 2000. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 74:4319-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corse, E., and C. E. Machamer. 2001. Infectious bronchitis virus envelope protein targeting: implications for virus assembly. Adv. Exp. Med. Biol. 494:571-576. [DOI] [PubMed] [Google Scholar]
- 8.Cosson, P., and F. Letourneur. 1994. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science 263:1629-1631. [DOI] [PubMed] [Google Scholar]
- 9.Dubois-Dalcq, M., K. V. Holmes, and B. Rentier. 1984. Assembly of enveloped RNA viruses. Springer-Verlag, New York, N.Y.
- 10.Fischer, F., C. F. Stegen, P. S. Masters, and W. A. Samsonoff. 1998. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 72:7885-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallagher, T. M., and M. J. Buchmeier. 2001. Coronavirus spike proteins in viral entry and pathogenesis. Virology 279:371-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garoff, H., R. Hewson, and D. J. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaynor, E. C., T. R. Graham, and S. D. Emr. 1998. COPI in ER/Golgi and intra-Golgi transport: do yeast COPI mutants point the way? Biochim. Biophys. Acta 1404:33-51. [DOI] [PubMed] [Google Scholar]
- 14.Goepfert, P. A., K. Shaw, G. Wang, A. Bansal, B. H. Edwards, and M. J. Mulligan. 1999. An endoplasmic reticulum retrieval signal partitions human foamy virus maturation to intracytoplasmic membranes. J. Virol. 73:7210-7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goepfert, P. A., K. L. Shaw, G. D. Ritter, Jr., and M. J. Mulligan. 1997. A sorting motif localizes the foamy virus glycoprotein to the endoplasmic reticulum. J. Virol. 71:778-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goepfert, P. A., G. Wang, and M. J. Mulligan. 1995. Identification of an ER retrieval signal in a retroviral glycoprotein. Cell 82:543-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez, J. M., P. Gomez-Puertas, D. Cavanagh, A. E. Gorbalenya, and L. Enjuanes. 2003. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 148:2207-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffiths, G., and P. Rottier. 1992. Cell biology of viruses that assemble along the biosynthetic pathway. Semin. Cell Biol. 3:367-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardt, B., and E. Bause. 2002. Lysine can be replaced by histidine but not by arginine as the ER retrieval motif for type I membrane proteins. Biochem. Biophys. Res. Commun. 291:751-757. [DOI] [PubMed] [Google Scholar]
- 20.Harter, C., and F. T. Wieland. 1998. A single binding site for dilysine retrieval motifs and p23 within the gamma subunit of coatomer. Proc. Natl. Acad. Sci. USA 95:11649-11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hobman, T. C. 1993. Targeting of viral glycoproteins to the Golgi complex. Trends Microbiol. 1:124-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hobman, T. C., M. L. Lundstrom, C. A. Mauracher, L. Woodward, S. Gillam, and M. G. Farquhar. 1994. Assembly of rubella virus structural proteins into virus-like particles in transfected cells. Virology 202:574-585. [DOI] [PubMed] [Google Scholar]
- 23.Holmes, K. V. 2003. SARS coronavirus: a new challenge for prevention and therapy. J. Clin. Investig. 111:1605-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson, M. R., T. Nilsson, and P. A. Peterson. 1990. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 9:3153-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson, M. R., T. Nilsson, and P. A. Peterson. 1993. Retrieval of transmembrane proteins to the endoplasmic reticulum. J. Cell Biol. 121:317-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kienzle, T. E., S. Abraham, B. G. Hogue, and D. A. Brian. 1990. Structure and orientation of expressed bovine coronavirus hemagglutinin-esterase protein. J. Virol. 64:1834-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klumperman, J., J. K. Locker, A. Meijer, M. C. Horzinek, H. J. Geuze, and P. J. Rottier. 1994. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 68:6523-6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo, L., and P. S. Masters. 2003. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 77:4597-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lefrancois, L., and D. S. Lyles. 1982. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. Virology 121:168-174. [DOI] [PubMed] [Google Scholar]
- 30.Letourneur, F., S. Hennecke, C. Demolliere, and P. Cosson. 1995. Steric masking of a dilysine endoplasmic reticulum retention motif during assembly of the human high affinity receptor for immunoglobulin E. J. Cell Biol. 129:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe, P. M. Howley, R. Griffin, A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 32.Machamer, C. E., S. A. Mentone, J. K. Rose, and M. G. Farquhar. 1990. The E1 glycoprotein of an avian coronavirus is targeted to the cis Golgi complex. Proc. Natl. Acad. Sci. USA 87:6944-6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Machamer, C. E., and J. K. Rose. 1987. A specific transmembrane domain of a coronavirus E1 glycoprotein is required for its retention in the Golgi region. J. Cell Biol. 105:1205-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mackenzie, J. M., and E. G. Westaway. 2001. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 75:10787-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuoka, Y., S. Y. Chen, and R. W. Compans. 1991. Bunyavirus protein transport and assembly. Curr. Top. Microbiol. Immunol. 169:161-179. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen, V. P., and B. G. Hogue. 1997. Protein interactions during coronavirus assembly. J. Virol. 71:9278-9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nilsson, T., M. Jackson, and P. A. Peterson. 1989. Short cytoplasmic sequences serve as retention signals for transmembrane proteins in the endoplasmic reticulum. Cell 58:707-718. [DOI] [PubMed] [Google Scholar]
- 38.O'Kelly, I., M. H. Butler, N. Zilberberg, and S. A. Goldstein. 2002. Forward transport. 14-3-3 binding overcomes retention in endoplasmic reticulum by dibasic signals. Cell 111:577-588. [DOI] [PubMed] [Google Scholar]
- 39.Opstelten, D. J., P. de Groote, M. C. Horzinek, H. Vennema, and P. J. Rottier. 1993. Disulfide bonds in folding and transport of mouse hepatitis coronavirus glycoproteins. J. Virol. 67:7394-7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Opstelten, D. J., M. J. Raamsman, K. Wolfs, M. C. Horzinek, and P. J. Rottier. 1995. Envelope glycoprotein interactions in coronavirus assembly. J. Cell Biol. 131:339-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ortego, J., D. Escors, H. Laude, and L. Enjuanes. 2002. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 76:11518-11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puddington, L., C. E. Machamer, and J. K. Rose. 1986. Cytoplasmic domains of cellular and viral integral membrane proteins substitute for the cytoplasmic domain of the vesicular stomatitis virus glycoprotein in transport to the plasma membrane. J. Cell Biol. 102:2147-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sannerud, R., J. Saraste, and B. Goud. 2003. Retrograde traffic in the biosynthetic-secretory route: pathways and machinery. Curr. Opin. Cell Biol. 15:438-445. [DOI] [PubMed] [Google Scholar]
- 44.Schutze, M. P., P. A. Peterson, and M. R. Jackson. 1994. An N-terminal double-arginine motif maintains type II membrane proteins in the endoplasmic reticulum. EMBO J. 13:1696-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sevier, C. S., O. A. Weisz, M. Davis, and C. E. Machamer. 2000. Efficient export of the vesicular stomatitis virus G protein from the endoplasmic reticulum requires a signal in the cytoplasmic tail that includes both tyrosine-based and di-acidic motifs. Mol. Biol. Cell 11:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shikano, S., and M. Li. 2003. Membrane receptor trafficking: evidence of proximal and distal zones conferred by two independent endoplasmic reticulum localization signals. Proc. Natl. Acad. Sci. USA 100:5783-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siddell, S. G. 1995. The Coronaviridae. Plenum Press, New York, N.Y.
- 48.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sturman, L. S., K. V. Holmes, and J. Behnke. 1980. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J. Virol. 33:449-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swift, A. M., and C. E. Machamer. 1991. A Golgi retention signal in a membrane-spanning domain of coronavirus E1 protein. J. Cell Biol. 115:19-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teasdale, R. D., and M. R. Jackson. 1996. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the golgi apparatus. Annu. Rev. Cell Dev. Biol. 12:27-54. [DOI] [PubMed] [Google Scholar]
- 52.Townsley, F. M., G. Frigerio, and H. R. Pelham. 1994. Retrieval of HDEL proteins is required for growth of yeast cells. J. Cell Biol. 127:21-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vennema, H., G. J. Godeke, J. W. Rossen, W. F. Voorhout, M. C. Horzinek, D. J. Opstelten, and P. J. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vennema, H., L. Heijnen, A. Zijderveld, M. C. Horzinek, and W. J. Spaan. 1990. Intracellular transport of recombinant coronavirus spike proteins: implications for virus assembly. J. Virol. 64:339-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, L., R. L. Parr, D. J. King, and E. W. Collisson. 1995. A highly conserved epitope on the spike protein of infectious bronchitis virus. Arch. Virol. 140:2201-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weisz, O. A., A. M. Swift, and C. E. Machamer. 1993. Oligomerization of a membrane protein correlates with its retention in the Golgi complex. J. Cell Biol. 122:1185-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan, H., K. Michelsen, and B. Schwappach. 2003. 14-3-3 dimers probe the assembly status of multimeric membrane proteins. Curr. Biol. 13:638-646. [DOI] [PubMed] [Google Scholar]
- 58.Zerangue, N., M. J. Malan, S. R. Fried, P. F. Dazin, Y. N. Jan, L. Y. Jan, and B. Schwappach. 2001. Analysis of endoplasmic reticulum trafficking signals by combinatorial screening in mammalian cells. Proc. Natl. Acad. Sci. USA 98:2431-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zerangue, N., B. Schwappach, Y. N. Jan, and L. Y. Jan. 1999. A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron 22:537-548. [DOI] [PubMed] [Google Scholar]