Abstract
The conserved eukaryotic Pan2–Pan3 deadenylation complex shortens cytoplasmic mRNA 3′ polyA tails to regulate mRNA stability. Although the exonuclease activity resides in Pan2, efficient deadenylation requires Pan3. The mechanistic role of Pan3 is unclear. Here, we show that Pan3 binds RNA directly both through its pseudokinase/C-terminal domain and via an N-terminal zinc finger that binds polyA RNA specifically. In contrast, isolated Pan2 is unable to bind RNA. Pan3 binds to the region of Pan2 that links its N-terminal WD40 domain to the C-terminal part that contains the exonuclease, with a 2:1 stoichiometry. The crystal structure of the Pan2 linker region bound to a Pan3 homodimer shows how the unusual structural asymmetry of the Pan3 dimer is used to form an extensive high-affinity interaction. This binding allows Pan3 to supply Pan2 with substrate polyA RNA, facilitating efficient mRNA deadenylation by the intact Pan2–Pan3 complex.
Keywords: gene expression, mRNA deadenylation, Pan2–Pan3, polyA tail, protein structure
See also: MT Stubbs & E Wahle (July 2014)
Introduction
In eukaryotic cells, mature mRNAs contain a 3′ polyadenosine (polyA) tail that is important for both mRNA stability and translation. The polyA tail is added co-transcriptionally to a length of approximately 70–90 nucleotides in Saccharomyces cerevisiae or approximately 200–250 nucleotides in humans (Zhao et al, 1999). Once mRNA is exported to the cytoplasm, the length of the polyA tail is regulated, which influences translation in some situations (Kapp & Lorsch, 2004; Goldstrohm & Wickens, 2008; Subtelny et al, 2014). For example, polyA tail length plays a key role in translational regulation of mRNAs in inflammation, neuronal processes, and early development (Weill et al, 2012). Deadenylation is also the first and rate-limiting step in cytoplasmic mRNA decay which involves one of two distinct pathways: (i) mRNA degradation by the exosome in a 3′–5′ direction or (ii) degradation from the 5′-end by decapping and the Xrn1 nuclease (Meyer et al, 2004; Parker & Song, 2004; Garneau et al, 2007).
PolyA tails are regulated at a mRNA-specific level (Lowell et al, 1992; Decker & Parker, 1993; Beilharz & Preiss, 2007; Lackner et al, 2007). Two conserved complexes carry out the majority of deadenylation in the cytoplasm: Pan2–Pan3 and Ccr4–Not (Wahle & Winkler, 2013). The catalytic activities are found in exonuclease-containing subunits Pan2 in Pan2–Pan3 and Ccr4 and Caf1/Pop2 in Ccr4–Not (Boeck et al, 1996; Tucker et al, 2001). Although these complexes appear to be at least partially functionally redundant, Pan2–Pan3 may be more efficient at initiating deadenylation, whereas Ccr4–Not may be more efficient at removing the final approximately 20–25 adenosines (Brown & Sachs, 1998; Tucker et al, 2001; Yamashita et al, 2005). This suggests that Pan2–Pan3 and Ccr4–Not could act sequentially. Severe phenotypes in yeast are only seen upon simultaneous deletion of both PAN2 and CCR4 (Tucker et al, 2001). Still, the molecular mechanisms of these complexes, and in particular the roles of non-enzymatic protein subunits, are unclear (Wiederhold & Passmore, 2010; Wolf & Passmore, 2014).
Deadenylase complexes can be recruited to mRNA by binding RNA directly or indirectly through RNA binding proteins (Wahle & Winkler, 2013). The C-terminal domain of polyA-binding protein (PABP/Pab1) interacts with a short peptide motif (PABP interacting motif 2, PAM2) in Pan3 (Siddiqui et al, 2007) (Fig1A). PABP stimulates Pan2–Pan3 activity, both in yeast and human (Sachs & Deardorff, 1992; Uchida et al, 2004), and is assumed to be required for recruitment of Pan2–Pan3 to mRNA (Mangus et al, 2004b; Funakoshi et al, 2007; Siddiqui et al, 2007). Indeed, Pan2–Pan3 inefficiently removes the final 20–25 adenosines, the approximate length required to bind PABP (Baer & Kornberg, 1980; Sachs et al, 1987; Lowell et al, 1992). This suggests that Pan2–Pan3 activity stops or slows after the last PABP has been removed from the mRNA. A function of PABP to stimulate Pan2–Pan3 could play a role in the sequential model of deadenylation.
Figure 1. The deadenylation activity of the Pan2–Pan3 complex does not require PABP/Pab1.
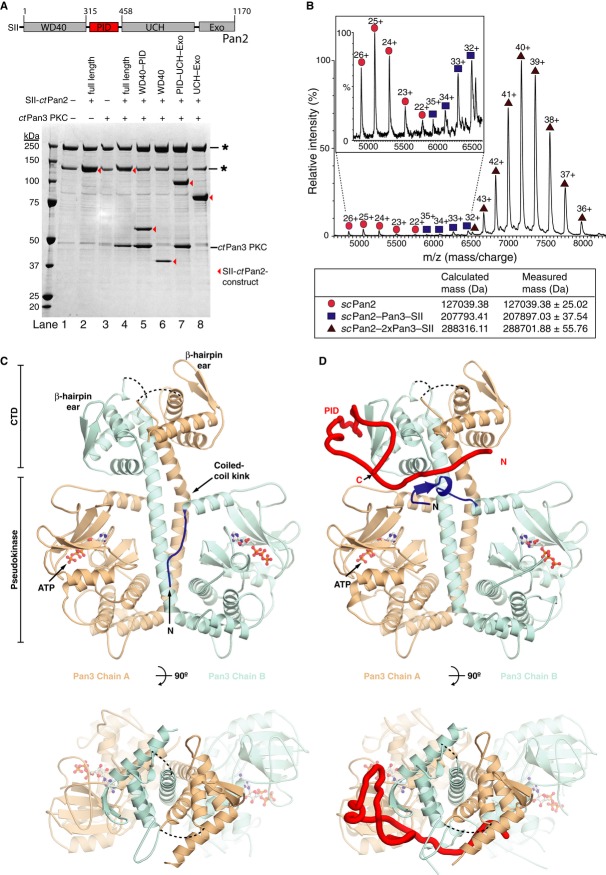
- Schematic representation of Pan3 and Pan2 proteins. The N-terminus of Pan3 contains a zinc finger and a PABP interacting motif 2 (PAM2). The C-terminal part contains a pseudokinase, a coiled coil, and a C-terminal domain (CTD), which form a structural unit (PKC). Pan2 contains WD40, inactive ubiquitin C-terminal hydrolase (UCH), and exonuclease domains. A long linker region (Pan3 interacting domain, PID) connects the WD40 and UCH domains. Sizes are shown for Saccharomyces cerevisiae proteins and constructs used for crystallization are indicated below.
- Recombinant Pan2–Pan3 removes mRNA polyA tails. Deadenylation of a RNA substrate (CYC1 3′ UTR with A80 tail) by recombinant full-length Saccharomyces cerevisiae (sc) Pan2–Pan3 complex was analyzed by denaturing polyacrylamide gel electrophoresis (see Supplementary Fig S1). The lane profiles of various time points are shown here. The sizes of CYC1 3′ UTR RNA with (252 nt) and without (172 nt) an A80 tail are represented by black and gray dotted lines, respectively.
- Pab1 stimulates but is not required for Pan2–Pan3 deadenylation activity. Deadenylation assays using the same substrate as in (B) with scPan2–Pan3 in the absence (black) and presence (green) of the polyA-binding protein Pab1. The average length of the polyA tail is plotted for each time point where a single RNA band was visible on the gel (see Supplementary Fig S1). Dotted lines indicate the sizes of the CYC1 3′ UTR RNA with (black, 252 nt) and without (gray, 172 nt) a polyA80 tail.
Pan3 has also been implicated in direct interactions with GW182 proteins in miRNA-mediated degradation, and with Dun1 kinase in regulation of Rad5 mRNA (Hammet et al, 2002; Braun et al, 2011; Chekulaeva et al, 2011; Fabian et al, 2011). In contrast, Pan2 is reported to interact only with Pan3 (Mangus et al, 2004a; Uchida et al, 2004). This led to the proposal that Pan3 acts as a regulator or adaptor within the complex, facilitating recruitment to mRNA via PABP (Uchida et al, 2004).
The mechanism of Pan2–Pan3 complex formation and substrate binding remain elusive. Recent work showed that Pan3 forms a homodimer in solution (Christie et al, 2013) raising the question of whether the Pan2–Pan3 complex contains one or two exonuclease subunits. Here, we use biochemical and structural approaches to define the mechanism of RNA recruitment by Pan2–Pan3 and demonstrate that the complex contains two Pan3 subunits and one Pan2 subunit. We show that Pan3 uses at least three different mechanisms to bind RNA, explaining its contribution to mRNA deadenylation. Further, we identify the region of Pan2 responsible for binding Pan3 and determine a crystal structure of their complex. This reveals an extensive interface underscoring the importance of a stable interaction between these two proteins to allow Pan3 to supply Pan2 with substrate RNA for efficient mRNA deadenylation.
Results
The deadenylation activity of Pan2–Pan3 does not require Pab1
To investigate the molecular mechanism of mRNA deadenylation by Pan2–Pan3, we purified recombinant S. cerevisiae Pan2–Pan3 complex (scPan2–Pan3) and polyA-binding protein Pab1 (Supplementary Fig S1A). We tested the ability of scPan2–Pan3 to remove a 3′ polyA tail from an in vitro transcribed model mRNA comprising the 3′ untranslated region (UTR) from the CYC1 gene followed by 80 adenosines. The CYC1 3′ UTR has been used extensively to study the addition of polyA tails during mRNA processing (Viphakone et al, 2008). In our assays, the length of the RNA substrate was monitored by gel electrophoresis. We observed robust activity of scPan2–Pan3, which removed the polyA80 tail, even in the absence of Pab1 (Fig1B and C and Supplementary Fig S1). In contrast, Pan2–Pan3 complex containing a catalytic mutation in Pan2 (E912A) did not have nuclease activity (Supplementary Fig S2). Thus, the purified Pan2–Pan3 complex is an active deadenylase in vitro and appears to have an intrinsic ability to bind polyA RNA.
Addition of Pab1 to scPan2-Pan3 stimulated the rate of the reaction by approximately fourfold (Fig1C and Supplementary Figs S1 and S2). Nuclease activity continued beyond the polyA tail, presumably due to binding of Pab1 to A-rich regions of the 3′ UTR. In contrast, in the absence of Pab1, scPan2–Pan3 did not degrade the 3′ UTR region of the substrate and stopped after the 80 adenosines had been removed (Fig1C and Supplementary Fig S1).
In the absence of Pab1, it appears that removal of the final approximately 25 adenosines occurs at a slower rate than removal of the first 50 adenosines (Fig1C and Supplementary Fig S1C). This is similar to a previous study where removal of the last 10–25 adenosines by Pan2–Pan3 was inefficient (Lowell et al, 1992). Our results suggest that inefficient removal of short polyA tails is not necessarily due to dissociation of the last Pab1/PABP and the polyA specificity is not solely determined by polyA-binding proteins. Instead, this behavior is determined primarily by the interaction of Pan2–Pan3 with mRNA itself.
The Pan3 CCCH zinc finger binds polyA RNA
Since Pab1/PABP was not necessary for Pan2–Pan3 to deadenylate RNA in the in vitro assay, we analyzed the role of either Pan3 or Pan2 alone in RNA binding. As illustrated in Fig1A, Pan3 contains a zinc finger at its N-terminus, a pseudokinase domain (PK), a coiled coil and a highly conserved C-terminal domain (CTD) (Brown et al, 1996; Christie et al, 2013; Wolf & Passmore, 2014). Pan2 contains three functional domains: an N-terminal WD40 domain, an inactive ubiquitin C-terminal hydrolase (UCH) domain and a C-terminal exonuclease (Fig1A) (Boeck et al, 1996; Wolf & Passmore, 2014). A linker region separates the WD40 and UCH domains. Although zinc fingers often act as interaction modules with DNA, RNA or protein (Font & Mackay, 2010), none of the domains in either Pan3 or Pan2 have previously been shown to bind directly to RNA.
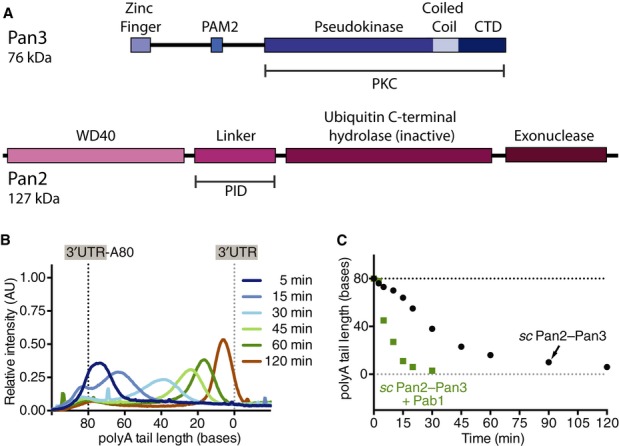
We determined the solution NMR structure of the zinc finger in scPan3 (residues 1–41) and used this to investigate its role in binding RNA. Complete 1H/13C/15N assignments and structure determination were carried out using conventional methods (Supplementary Table S1). The N-terminus of scPan3 has a compact CCCH-type zinc finger with a well-defined core, held in place by the zinc ion (Fig2A–C). It also has an N-terminal extension that is largely ordered. The zinc-coordinating residues Cys14, Cys23, Cys30, and His34 (CCCH) of scPan3 are universally conserved in Pan3 zinc fingers (Supplementary Fig S3A). The overall structure is similar to the CCCH zinc fingers in MBNL1 (muscleblind-like protein), TIS11d (Tristetraprolin family), and Nab2 (Hudson et al, 2004; Teplova & Patel, 2008; Brockmann et al, 2012; Kuhlmann et al, 2014) (Supplementary Fig S3). The TIS11d zinc finger is strikingly similar, including an ordered N-terminal extension, a short α-helix between the first two zinc-coordinating cysteines and a 310 helix between the second and third cysteines. Interestingly, MBNL1, TIS11d and Nab2 all bind RNA.
Figure 2. The zinc finger domain of scPan3 binds polyA RNA.
- Overlay of the 20 best NMR models of the CCCH zinc finger of Saccharomyces cerevisiae Pan3 (residues 1–41) showing the protein backbone colored in rainbow from N- to C-terminus.
- Cartoon representation of the zinc finger of scPan3 with a modeled zinc ion (red sphere). The disordered termini have been omitted.
- Close-up of the zinc finger of scPan3 (ribbon) showing functional residues in stick representation. The Zn atom (red sphere) is coordinated to His34 and the three cysteines.
- The scPan3 zinc finger binds polyA RNA. Plot of the magnitude of chemical shift perturbation of each residue, calculated from 15N-1H HSQC spectra of 150 μM scPan3 zinc finger after addition of 150 μM 15-mer polyA RNA. Many of the zinc finger residues experience large changes in their chemical environment in the presence of polyA RNA. Residues discussed in the text are labeled. The peak for Arg15 (red) disappears upon RNA binding. (Also see Supplementary Fig S4).
To investigate whether the scPan3 zinc finger can also bind RNA, we performed chemical shift perturbation experiments to measure changes in its environment in the presence of polyA15, C15, G15, or U15 RNA. Large shifts of the peaks of many zinc finger residues were observed in the presence of polyA, whereas polyG generated small changes and polyC or polyU generated negligible changes (Fig2D and Supplementary Fig S4). By titrating the amount of RNA in the NMR experiment, we estimate the binding affinity of this zinc finger to the polyA15 RNA to be 60–100 μM (Supplementary Fig S4). These data indicate that the zinc finger of scPan3 binds polyA RNA in preference to other polyribonucleotides.
Pan3 pseudokinase/CTD interacts directly with RNA
To assess whether the zinc finger is the only component in the Pan2–Pan3 complex that binds directly to RNA, we deleted it from scPan3 and investigated the ability of the truncated scPan2–Pan3 complex to bind RNA. We used an active site mutant in scPan2 (E912A) to prevent degradation of the substrate CYC1 3′ UTR model RNA (with or without an A80 tail) in electrophoretic mobility shift assays (EMSAs). The EMSA experiments in Fig3 and Supplementary Fig S5A and B show that wild-type scPan2–Pan3 and scPan2–Pan3 lacking the zinc finger both bind CYC1 RNA, irrespective of whether it has a polyA tail. Wild-type scPan2–Pan3 has a slightly higher affinity for polyadenylated (A80) RNA compared to the non-adenylated 3′ UTR (Fig3A). In contrast, scPan2–Pan3 lacking the zinc finger domain showed the same binding affinity for CYC1 RNA with and without polyA tail (Fig3B). Together, these data show that although the scPan3 zinc finger binds RNA and may convey polyA specificity, other parts of Pan2–Pan3 contribute to RNA binding. In agreement with this, scPan2–Pan3 lacking a zinc finger has a slightly reduced rate of deadenylation compared to wild-type complex (Supplementary Fig S5C).
Figure 3. The zinc finger domain of scPan3 increases the affinity of the Pan2–Pan3 complex to polyA RNA.
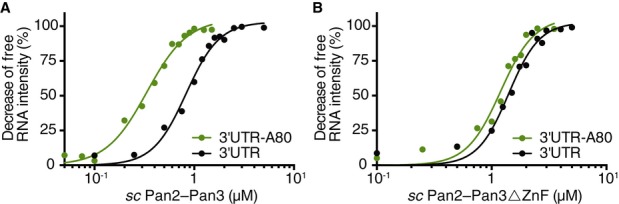
A, B Electrophoretic mobility shift assay (EMSA) using the CYC1 3′ UTR RNA with (green) or without (black) a polyA80 tail. RNA was incubated with (A) scPan2–Pan3 or (B) scPan2–Pan3 with a deletion of the Pan3 zinc finger (scPan2–Pan3ΔZnF). Both complexes contain an active site mutation in scPan2 (E912A). Binding was analyzed by native polyacrylamide gel electrophoresis, and the decrease in the intensity of free RNA was quantified and plotted against protein concentration for a representative experiment. Also see Supplementary Fig S5A and B.
To evaluate whether other domains of Pan3 contribute to RNA binding, we used a short synthetic RNA that could be more readily quantitated. We measured the changes in the fluorescence polarization signal of 5′ Cy3-labeled polyA15 RNA upon addition of a truncated Pan3 protein containing the pseudokinase and C-terminal domains (PKC). Separately expressed Pan2 and Pan3 proteins from budding yeast had a tendency to aggregate in isolation and so we instead used proteins from the thermophilic fungus Chaetomium thermophilum (ct) for these experiments (Supplementary Fig S1A). We observed an increase in polarization of the labeled RNA showing that ctPan3 PKC can bind polyA (Fig4A). Similar binding affinities could be observed when polyC15 or U15 RNA was used, indicating that the interaction of this region of ctPan3 with RNA is not polyA specific (Fig4A). The interaction with polyG15 appeared tighter but interpretation of this result is complicated by an increased magnitude of fluorescence polarization, possibly due to the well-known potential of polyG to form G-quadruplexes (Joachimi et al, 2009). Thus, the PKC region of ctPan3 also binds RNA but is not specific for polyA.
Figure 4. Pan3 functions within the Pan2–Pan3 complex to bind RNA.
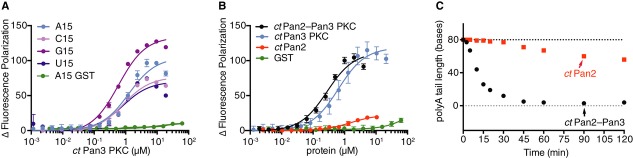
- Pan3 pseudokinase/C-terminal domain (PKC) from Chaetomium thermophilum (ct) binds RNA but has no preference for polyA. The plot shows a comparison of the change in the fluorescence polarization signal of 20 nM 5′ Cy3-labeled polyA, polyC, polyG, and polyU RNA upon addition of ctPan3 pseudokinase/C-terminal domain (PKC) or purified GST, for a representative experiment. Error bars are the standard deviations of triplicate measurements from one experiment. The Kds for RNA binding are: 2.1 ± 1.4 μM (A15); 2.2 ± 1.7 μM (C15); 0.55 ± 0.05 μM (G15); and 1.6 ± 0.6 μM (U15) (average of three independent experiments, errors are the standard deviations).
- Isolated ctPan2 has a low affinity for RNA compared with ctPan3 PKC. The change in the fluorescence polarization signal of 20 nM 5′ Cy3-labeled polyA RNA upon addition of ctPan2 (red), ctPan3 PKC (blue), ctPan2–Pan3 PKC (black), or purified GST (green) is shown as the average of three independent experiments. Error bars are standard deviations. The Kds for A25 RNA binding are: 0.62 ± 0.06 μM (ctPan3 PKC) and 0.27 ± 0.02 μM (ctPan2–Pan3 PKC) (average of three independent experiments, errors are the standard deviations).
- Intact ctPan2–Pan3 complex (black) has much higher activity than isolated ctPan2 (red). Deadenylation assays using CYC1 3′ UTR with A80 tail (Supplementary Fig S1). The average length of the polyA tail is plotted for each time point. Dotted lines indicate the sizes of the CYC1 3′ UTR RNA with (black, 252 nt) and without (gray, 172 nt) a polyA80 tail.
Pan2 does not bind RNA and requires Pan3 for efficient deadenylation
To determine whether Pan2 also binds RNA, we again measured the changes in fluorescence polarization of a labeled polyA RNA after incubation with protein. Changes in the polarization signal of the RNA were substantially reduced in the presence of catalytically inactive ctPan2 compared with ctPan3 PKC (Fig4B). As a control, GST (which is not expected to interact with RNA) did not substantially influence the fluorescence polarization signal (Fig4B). Thus, the interaction of ctPan2 with polyA RNA is much weaker than the interaction of ctPan3 PKC with the same RNA. When we tested the interaction of the ctPan2–Pan3 PKC complex with polyA RNA, the binding was comparable to ctPan3 PKC alone (Fig4B).
The data in Fig4B suggest that interaction of ctPan2 with polyA RNA is negligible. Consequently, we predicted that the activity of isolated Pan2 would be very weak. To test this, we used isolated ctPan2 in the in vitro deadenylation assay described above. As shown in Fig4C and Supplementary Fig S1, ctPan2 alone has greatly reduced activity compared to the Pan2–Pan3 complex and only removes approximately 30 As in 2 h. In comparison, full-length ctPan2–Pan3 removes almost the entire polyA80 tail in 30 min. Like the Saccharomyces cerevisiae proteins, the complex from Chaetomium exhibits robust activity in the absence of Pab1, whereas ctPan2–Pan3 containing a catalytic mutation in Pan2 (E899A) is not active (Supplementary Fig S2C). These results show that Pan2 does not deadenylate mRNA efficiently in the absence of Pan3.
One Pan2 linker region mediates binding to a Pan3 homodimer
The experiments described above highlight the importance of association of Pan2 and Pan3 into a complex for their function in mRNA deadenylation, so we characterized how these two proteins interact at the molecular level in greater detail. Yeast-two-hybrid and mutational studies suggest that the CTD of Pan3 is involved in the interaction with Pan2 (Mangus et al, 2004a; Christie et al, 2013). Furthermore, the WD40 or exonuclease domains of Pan2 can be deleted without losing the interaction with Pan3 (Mangus et al, 2004a; Uchida et al, 2004).
To map the Pan3 binding site on Pan2, we over-expressed a series of StrepII-tagged ctPan2 truncations together with ctPan3 PKC and used pull-down experiments to test for their interaction. Removal of either the C-terminal UCH and exonuclease domains or the N-terminal WD40 domain did not impair the ability of ctPan2 to bind ctPan3 PKC (Fig5A, lanes 5 and 7). However, additional removal of the linker that connects the WD40 and UCH domains resulted in loss of the interaction with ctPan3 PKC (Fig5A, lanes 6 and 8). Together, these data indicate that the linker region of Pan2 (residues 316–458 for ctPan2) mediates the interaction with Pan3. Hence, we refer to this region as the ‘Pan3 interaction domain’ (PID).
Figure 5. The Pan2–Pan3 complex forms a heterotrimer.
A ctPan3 interacts with the linker region (Pan3 interacting domain, PID) of ctPan2. Various truncations of StrepII (SII)-tagged ctPan2 (full length, 1–1170; WD40-PID, 1–458; WD40, 1–315; PID-UCH-Exo, 316–1170; UCH-Exo, 459–1170) were co-expressed with ctPan3 pseudokinase/C-terminal domain (PKC, 205–640) in Saccharomyces cerevisiae, and pull-downs were performed using StrepTactin beads. ctPan2 proteins are indicated by a red arrowhead. Asterisks indicate contaminating bands that bind to StrepTactin beads.
B Intact Pan2–Pan3 has a stoichiometry of 1:2. NanoESI-MS spectrum of recombinant scPan2-Pan3, showing the major peak series (< 95%) of a heterotrimeric complex of one Pan2 and two Pan3 molecules at 6,500–8,000 m/z (dark red triangles).
C, D The ctPan2 linker region wraps around a homodimer of ctPan3. Crystal structures of (C) ctPan3 PKC homodimer and (D) ctPan3 PKC bound to the Pan3 interacting domain (PID) of ctPan2. Pan3 chains A and B are shown in wheat and cyan, respectively, and ctPan2 PID is shown in red. The N-terminal residues of Pan3 chain B that change conformation on Pan2 binding are shown in dark blue. ATP is shown in ball and stick representation. Dashed lines connect loops not visible in the electron density. The two views are related by a rotation of 90° around the horizontal axis.
A recent crystal structure of Pan3 revealed that it forms a homodimer (Christie et al, 2013). Accompanying biochemical data suggested that mutants that were unable to dimerize could still bind Pan2. These data indicated that a Pan3 dimer might bind two copies of Pan2 with an overall stoichiometry of 2:2. To establish the stoichiometry of the Pan2–Pan3 complex, we used non-covalent nanoelectrospray ionization mass spectrometry (nanoESI-MS). This technique allows the determination of composition and stoichiometry of native protein complexes (Hernández & Robinson, 2007). NanoESI-MS of scPan2–Pan3 revealed that the most prominent species (> 95%) corresponds to a heterotrimeric complex containing one scPan2 and two scPan3 molecules (Fig5B). These data are consistent with formation of a Pan2–Pan3 1:2 heterotrimer in solution.
Structural basis of Chaetomium thermophilum Pan2–Pan3 complex formation
To understand the basis of subunit association in the Pan2–Pan3 complex, we determined the crystal structure of the pseudokinase/C-terminal domains of Pan3 alone and when in complex with the Pan3 interaction domain of Pan2. Proteins obtained from C. thermophilum generated crystals that diffracted to 2.4 Å resolution, substantially higher than that obtained from Drosophila melanogaster or Neurospora crassa Pan3 crystals (Christie et al, 2013), and so these were used for structural characterization. We obtained crystals of Se-Met-labeled ctPan3 PKC (residues 205–640) with P1 symmetry and four Pan3 homodimers in the asymmetric unit that were solved using single-wavelength anomalous dispersion (SAD) methods (Supplementary Table S2).
The ctPan3 structure consists of an N-terminal pseudokinase domain (residues 242–497) to which MgATP is bound, a central asymmetric coiled-coil domain (residues 499–538) and a C-terminal domain (residues 555–640) (Fig5C). Although ATP was bound to both pseudokinase domains, ATP was not added to crystallization conditions and is present in all copies in the asymmetric unit of the crystal. The pseudokinase lacks almost all catalytic residues and is predicted to be inactive (Christie et al, 2013). Two distinctive beta-hairpin ‘ears’ protrude from each C-terminal domain. A kink is formed in the coiled-coil region as a result of one of the CTDs (chain B) interacting with the pseudokinase from the other chain, introducing asymmetry into the Pan3 dimer. The pseudokinase and CTD domains of C. thermophilum Pan3 are similar to the recently reported structures for Drosophila (RMSDs of 1.13 Å and 0.87 Å, respectively) and Neurospora (RMSDs of 1.24 Å and 0.93 Å, respectively) proteins (Christie et al, 2013), indicating high structural conservation of Pan3 (Supplementary Fig S6A and B). The principal difference between these structures is that the kink in the coiled coil of ctPan3 is more pronounced, resulting in twists of approximately 28° and 31°, respectively, of the CTD relative to the pseudokinase domain.
We also crystallized ctPan3 PKC bound to the PID of ctPan2. The best diffracting crystals were obtained with a construct that lacks the first 27 residues of the PID region, ctPan2343–458, had P21 symmetry and diffracted to 2.6 Å resolution (Supplementary Table S3) with two copies of the complex in the asymmetric unit. Molecular replacement using the structure of ctPan3 PKC showed clear difference density corresponding to a single ctPan2 chain (comprising residues 355–406) on each ctPan3 dimer. The overall structure of the ctPan3 dimer in the complex closely resembled that obtained for ctPan3 alone (Cα RMSD 0.94 Å), although there was a slight (˜6°) rotation of the C-terminal domain relative to the coiled-coil and pseudokinase domain (Fig5C and D and Supplementary Fig S6C). ATP was bound to the pseudokinase domains. The major difference between free and Pan2-associated ctPan3 was a substantial rotation of the N-terminus of chain B (residues 207–224) by approximately 90° toward the Pan3 CTD (Fig5C and D). This region had been removed from the Neurospora and Drosophila constructs used in previous crystallization studies (Christie et al, 2013).
The PID of ctPan2 exploits the asymmetry of the ctPan3 dimer by wrapping around it at the interface between the pseudokinase and C-terminal domain (Fig6). The N-terminus of the ctPan2 PID (residues 355–360) interacts with chain A of the ctPan3 dimer. In particular, ctPan2 Phe357 and Trp360 are inserted into a pocket of ctPan3 formed by the C-terminal domain of chain A and the coiled coil (Fig6C). Phe357 and Trp360 are conserved hydrophobic residues among Pan2 orthologs (Supplementary Fig S6D).
Figure 6. Pan2 exploits the asymmetry and forms an extensive interface with the Pan3 dimer.
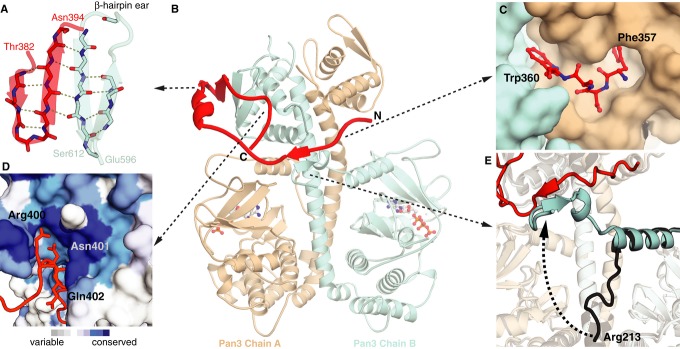
Structural details of the complex of ctPan3 PKC bound to the Pan3 interacting domain (PID) of ctPan2. ctPan2 is shown in red, and ctPan3 PKC chains A and B are depicted in wheat and cyan, respectively.
- Close-up view of the four-stranded intermolecular antiparallel β-sheet formed between a β-hairpin in ctPan2 (residues 384–393) and a β-hairpin ear in ctPan3 chain B (residues 597–612).
- Overall structure of the ctPan3 PKC dimer bound to ctPan2 PID (cartoon). ATP is in ball and stick representation.
- The N-terminal region of ctPan2 PID interacts with the ctPan3 dimer by inserting aromatic residues Phe357 and Trp360 (stick representation) into two pockets formed between the coiled coil and the CTD of ctPan3 chain A.
- Surface representation of the CTD of ctPan3 chain B colored by sequence conservation. ctPan2 PID (residues 400–402) binds into a conserved groove in the CTD.
- Superposition of ctPan3 from the structures of isolated ctPan3 (black) and bound to ctPan2 PID (Pan3 in cyan and wheat, Pan2 in red) in cartoon representation. The N-terminus of chain B of the ctPan3 dimer rotates by approximately 90° upon binding of ctPan2 to form an antiparallel β-sheet with ctPan2.
The PID of ctPan2 next passes over the coiled coil and interacts with the N-terminus of ctPan3 chain B (Gln212–Ala214) through an intermolecular antiparallel β-sheet (Fig6E). This new β-sheet is located between the pseudokinase domain of chain A and the CTD of chain B. The PID continues around Pan3 to form extensive interactions with the CTD of chain B that include an extended 4-strand antiparallel β-sheet between a β-hairpin of the ctPan2-PID (residues 384–393) and a β-hairpin ‘ear’ in ctPan3 (residues 597–612) (Fig6A). Finally, residues 399–403 of the PID of ctPan2 bind in a highly conserved groove formed by residues 599–603 and 562–573 in the CTD of chain B of the Pan3 dimer (Fig6D). Interestingly, binding of ctPan2 to ctPan3 is mediated mainly by interactions between main chain atoms, not side chains.
The extensive Pan2–Pan3 interface generates high affinity
Collectively, the interface between the PID of ctPan2 and the ctPan3 PKC dimer buries a surface area of approximately 2,230 Å2, and the ctPan3 PKC dimer interface buries approximately 3,740 Å2. The extensive surface area buried between these two proteins, together with the hydrogen bonding associated with new secondary structural elements formed on binding, is consistent with a tight association. We characterized the interaction of ctPan3 PKC with ctPan2 PID thermodynamically and kinetically and found a Kd of 10 nM using surface plasmon resonance and a Kd approximately 50 nM with isothermal titration calorimetry, though the necessity to use low protein concentrations for this range of affinities in the latter places a limit on the accuracy of this value (Supplementary Fig S7A–C). A large favorable change in enthalpy (ΔH ˜−30 kcal/mol) was observed during the interaction that was opposed by an unfavorable change in the entropy of the system (−TΔS ˜20 kcal/mol). These values reflect the extensive interactions between ctPan2 and ctPan3 and a potential loss in conformational flexibility upon binding. The stoichiometry of interaction confirmed that only a single Pan2 chain was bound to each Pan3 dimer in solution.
The Pan2 PID observed in the crystal structure is important for Pan3 binding in solution
As we observed only 52 (residues 355–406) of the 142 residues of the Pan2 linker that connects the WD40 and UCH domains in the electron density, we analyzed the contributions made by different regions of the PID to the interaction. Pull-down assays using ctPan3 PKC and N- or C-terminal truncations of ctPan2 (Fig5A and Supplementary Fig S7D and E) suggest that the major Pan3 binding region on ctPan2 includes the segment seen in the crystal structure. Since amino acids 1–342 and 1–352 also bind weakly to ctPan3, there may be additional contributions from these N-terminal residues. Notably, no region is absolutely required for binding, consistent with the extensive interface observed in the crystal structure.
We further assayed the ability of ctPan2 mutants to bind ctPan3 PKC by mixing separately purified proteins and analyzing whether they co-elute on size exclusion chromatography. Because of the extensive nature of the interaction interface, point mutations even of deeply buried residues such as Phe357 and Trp360 were not able to impair the interaction (Supplementary Fig S7F). We therefore assayed the effect of internal deletions of the PID in the context of full-length ctPan2 (Fig7A–E). We assessed the contributions of: (i) residues 343–352 for which electron density was not visible in our structure, (ii) the N-terminal region of the PID visible in our structure (residues 353–375), and (iii) the C-terminal half of the PID observed in the crystal structure, residues 376–406 (Supplementary Fig S6D). Only simultaneous deletion of all three regions (Δ343–406) abrogated the co-elution of ctPan2 and ctPan3 PKC on size exclusion chromatography, resulting in the appearance of separate peaks (Fig7A and 7E). Consistent with these data, when we tested the deadenylation activity of ctPan2–Pan3 PKC complexes formed with PID deletion mutants, larger deletions resulted in decreased activity (Fig7F–J). For example, full-length ctPan2–Pan3 PKC deadenylated all the RNA after approximately 10 min (Fig7F), whereas the Δ343–406 mutant required approximately 90 min for full deadenylation (Fig7J). Because the Δ343–406 mutant retained significant activity compared to isolated ctPan2, there may be additional sites of Pan2–Pan3 interaction, either within the PID (such as residues 316–342, Fig5A and S7D) or outside the PID.
Figure 7. The ctPan2 PID observed in the crystal structure interacts with ctPan3 PKC in solution.
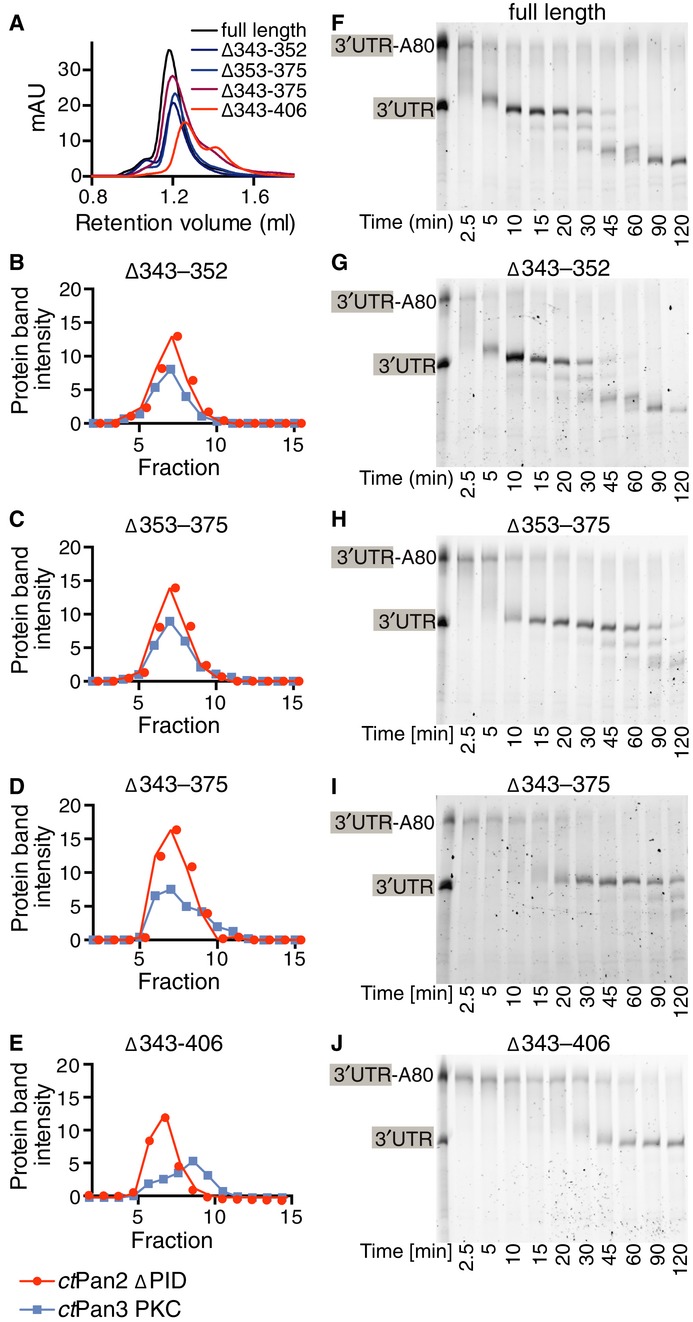
A–E Deletion of 63 residues of the PID is required to disrupt the interaction between ctPan2 and ctPan3 in vitro. Separately purified ctPan2 PID deletion constructs and ctPan3 PKC were mixed, and binding was analyzed by size exclusion chromatography. (A) Elution profiles measuring A280 nm are shown for ctPan3 PKC with full-length ctPan2 (black) and containing the following deletions in the PID: Δ343–352 (dark blue), Δ353–375 (light blue), Δ343–375 (dark red), and Δ343–406 (red). (B–E) Fractions of the size exclusion chromatography runs from (A) were analyzed by SDS–PAGE, and intensities of individual protein bands were quantified and plotted against fraction number. ctPan2 and ctPan3 PKC are depicted in red and blue, respectively.
F–J A similar pattern was observed when the activity of the deletion constructs was assessed using deadenylation of an RNA substrate (CYC1 3′ UTR with A80 tail) by ctPan3 PKC mixed with (F) full-length ctPan2 or (G–J) PID deletion mutants. Time points were analyzed by denaturing polyacrylamide gel electrophoresis. The first lane shows CYC1 3′ UTR with and without A80 tail as a marker. Although the Δ353–375 and Δ343–375 constructs showed a slight decrease in deadenylation rate, a major decrease was only observed with the Δ343–406 construct that also dissociated during size exclusion chromatography (E).
In summary, we found that it was difficult to disrupt the Pan2–Pan3 interaction by deletion of substantial regions of ctPan2 PID and show that a core region (residues 343–406) makes a major contribution to the association of ctPan2 and ctPan3. These results are consistent with the extensive interaction interface observed in the crystal structure of the complex and the low nanomolar binding affinity of ctPan2 to ctPan3.
Discussion
Although Pan2 provides the catalytic activity of the Pan2–Pan3 deadenylation complex and defects in Pan3 lead to loss of Pan2 function (Brown et al, 1996; Boeck et al, 1998; Uchida et al, 2004), the mechanism by which Pan3 facilitates deadenylation was unclear. Together, our data suggest that the role of Pan3 in the Pan2–Pan3 complex is to supply Pan2 with substrate polyA RNA. We show that the reduced deadenylase activity of isolated Pan2 is likely due to an inability to bind RNA. In the complex, Pan3 functions to bind RNA through at least three different mechanisms, facilitating recruitment of Pan2 to the polyA tail. The Pan3 homodimer binds a single Pan2 chain, and the crystal structure of ctPan3 bound to the interacting linker region of ctPan2 (residues 355–406) shows an extensive interface between the two subunits consistent with the integral role of Pan3 in the overall function of the complex.
Pan3 supplies Pan2 with substrate RNA through complementary mechanisms
Although an interaction between the PAM2 motif of Pan3 and PABP/Pab1 can mediate indirect recruitment of Pan2–Pan3 to the polyA tail (Siddiqui et al, 2007), our data demonstrate that Pan3 can function through at least two additional complementary mechanisms to bind RNA directly. First, the N-terminal zinc finger of Pan3 conveys sequence specificity by binding to polyA RNA preferentially over other polyribonucleotides (Supplementary Fig S4). The NMR solution structure of the scPan3 zinc finger (Fig2) revealed a similarity to other zinc fingers that bind RNA (Hudson et al, 2004; Teplova & Patel, 2008; Kuhlmann et al, 2014). Each of these other proteins contains tandem zinc fingers within the same polypeptide chain (four in MBNL1, two in TIS11d, and seven in Nab2), probably because the affinity of a single finger is insufficient to provide binding specificity. In Pan2–Pan3, there are two zinc fingers (one in each Pan3 chain) that may contribute to recognition in an analogous manner.
Second, the highly conserved Pan3 pseudokinase/C-terminal domain also binds RNA but in a manner that is not specific for polyA (Fig4). This region has a theoretical pI of approximately 8.8, and examination of the surface potential reveals a substantial positive charge (Supplementary Fig S8) that could mediate electrostatic interactions with negatively charged RNA backbone phosphates. In contrast, the ability of isolated Pan2 to bind RNA is negligible, and it has very low deadenylation activity in the absence of Pan3 (Fig4).
The importance of correct substrate selection is highlighted by the three distinct ways that Pan3 can use to recognize polyA RNA. Each individual interaction is weak: Kd of approximately 40–150 μM for the PAM2:PABP interaction (Siddiqui et al, 2007), apparent Kd of approximately 60–100 μM for the zinc finger:polyA interaction (Supplementary Fig S4), and Kd of approximately 0.5–2 μM for the PKC:RNA interaction (Fig4A). Because Pan3 is a dimer, there are two zinc fingers and two PAM2 motifs in each Pan2–Pan3 complex. Consequently, avidity effects due to these interactions acting in concert likely increase the overall affinity substantially, thereby facilitating recruitment to mRNA.
The extensive Pan2–Pan3 interface links deadenylation to mRNA binding
We identified the linker region between the Pan2 WD40 and UCH domains as a major binding site for Pan3. NanoESI mass spectrometry, isothermal calorimetry, and a crystal structure of the interacting regions of ctPan2 and ctPan3 show that only one Pan2 molecule binds to a Pan3 dimer to form a heterotrimeric complex. This unusual stoichiometry derives from the asymmetry of the Pan3 dimer itself so that the interface to which Pan2 binds is not replicated on the opposite face of Pan3 (Fig5D). The ctPan2 linker wraps around the Pan3 dimer with interactions dominated by the formation of new secondary structural elements (intermolecular β-sheets) and the insertion of Pan2 into surface grooves on Pan3 (Fig6). The extensive interface is consistent with the high enthalpic change observed on binding, and there being a tight interaction (10–50 nM Kd) that is difficult to disrupt.
A previous study using Pan3 mutants defective in dimerization suggested that a single Pan3 chain could bind Pan2 (Christie et al, 2013). Although some interaction between the Pan2 PID with a single Pan3 chain may be possible, in our crystal structure, we observed that Pan2 interacts extensively with both chains in the Pan3 dimer. Moreover, conserved solvent-exposed residues in the Pan3 dimer proposed to be involved in Pan2 interactions (Christie et al, 2013) do not make contact with the ctPan2 PID in our crystal structure. These regions could participate in additional interactions with Pan2 outside of the PID, in stabilizing the Pan3 structure, or in mediating interactions with other proteins. Consistent with additional Pan2–Pan3 interactions, Pan2 Δ343–406 retained deadenylation activity (Fig7). Furthermore, we found that the conserved N-terminus of the Pan3 pseudokinase, which was truncated in previous studies, changes conformation to form a β-sheet with Pan2.
Pan2–Pan3 alone shows robust deadenylation activity
Regulation of deadenylation is crucial since it can trigger translational repression and/or rapid mRNA decay. Deadenylation in vivo appears to have two phases: an initial phase removes the distal part of the polyA tail until approximately 12–25 As remain (Brown & Sachs, 1998). The proximal polyA tail is then removed during ‘terminal deadenylation’ which triggers mRNA decay (Tucker et al, 2001). The recent observation that the median polyA tail length in yeast is approximately 27 nucleotides (Subtelny et al, 2014) could imply that at steady state, many mRNAs have undergone distal but not proximal deadenylation. In yeast, deletion of PAN2 results in long polyA tails (> 50 As), and deletion of CCR4 impairs removal of the final approximately 20 As (Brown & Sachs, 1998; Tucker et al, 2001). Thus, although the complexes appear to be functionally redundant, in the sequential model of deadenylation, Pan2–Pan3 stimulated by PABP performs the initial phase and Ccr4–Not takes over (Brown & Sachs, 1998; Tucker et al, 2001; Yamashita et al, 2005; Bönisch et al, 2007).
In our in vitro deadenylation assay, the Pan2–Pan3 complex alone completely deadenylates RNA (Fig1C). PolyA-binding protein stimulates the reaction but is not required, suggesting that this function is intrinsic to the Pan2–Pan3 complex. It is conceivable that Pan2–Pan3 could function independently of Pab1 in vivo. Pab1-independent activity of Pan2–Pan3 was observed previously in very low ionic strength conditions and in the presence of spermidine (Lowell et al, 1992). Importantly, Pan2–Pan3 retains polyA specificity in the absence of Pab1 (Fig1B and C; Lowell et al, 1992). Thus, complete deadenylation can be performed by Pan2–Pan3 alone. This is consistent with suggestions that Pan2–Pan3 and Ccr4–Not may have independent roles (Lowell et al, 1992; Decker & Parker, 1993; Fadda et al, 2013; Sun et al, 2013).
At a molecular level, we propose that the Pan2–Pan3 complex, probably through RNA recognition by Pan3, can distinguish between the polyA tail and the 3′ UTR causing a decrease in the deadenylase activity as the polyA tail shortens. One way in which this could be achieved would be for the distance between the RNA binding regions in the Pan3 dimer to sense when the polyA tail is truncated to approximately < 25 nt. For example, because they are on separate Pan3 chains, it may not be possible for both zinc fingers as well as the PKC to bind and position short polyA stretches for efficient deadenylation by Pan2 (Fig8). In this context, positioning of protein domains within the complex is likely to be important. The formation of a β-sheet between ctPan2 and the N-terminal residues of chain B of the ctPan3 dimer that changes its position dramatically on complex formation could influence the position of the two zinc fingers and the two PAM2 motifs within the complex. The N- and C-termini of the ctPan2 PID are in close proximity in our structure, and this could also have implications for the positioning of the N-terminal WD40 and C-terminal UCH and exonuclease domains of Pan2 on the Pan3 dimer. Other components, including the active site of Pan2, may also contribute to polyA specificity.
Figure 8. Model for Pan2–Pan3.
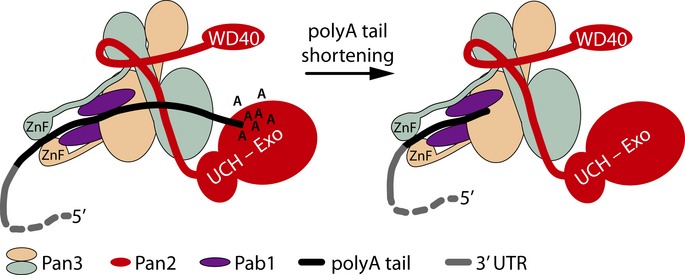
Schematic model for Pan2–Pan3 function, whereby Pan3 channels RNA to the Pan2 exonuclease domain.
In conclusion, through combined biochemical and structural approaches, we have shown that Pan3 is required to couple RNA binding to the Pan2 exonuclease activity. Pan3 binds RNA through multiple mechanisms and forms an extensive interface with Pan2 to ensure a stable interaction for efficient mRNA deadenylation. The zinc fingers, PAM2 motifs, and positively charged surface of Pan3 could channel polyA RNA into the Pan2 exonuclease active site (Fig8). This mechanism of RNA binding could provide Pan2–Pan3 with an intrinsic preference for the polyA tail over the 3′ UTR that would be crucial for its function in the regulation of gene expression.
Materials and Methods
Protein expression and purification
Detailed methods of protein expression and purification can be found in Supplementary Methods. In brief, Pan2–Pan3 complexes, Pan2, and Pab1 were expressed in S. cerevisiae as described (Galej et al, 2013) with a Strep-II affinity tag for purification using StrepTactin affinity chromatography. His-tagged Pan3 zinc finger and GST-Pan3 PKC were expressed in E. coli and purified using Ni2+ affinity chromatography and glutathione Sepharose, respectively.
Deadenylation activity assay
A construct containing a T7 promoter, the CYC1 3′ UTR (169 nucleotides downstream of the stop codon) followed by 80 adenosines, and a BsaI restriction site were created by gene synthesis and cloned into pUC57 plasmid (GenScript). Following restriction digest with BsaI and mung bean nuclease treatment, the linearized plasmid was used in a run-off in vitro transcription reaction. The RNA was eluted from denaturing polyacrylamide gels in 0.5 M ammonium acetate, 1 mM EDTA, 0.2% SDS (w/v) at 37°C overnight and stored in 10 mM ammonium acetate pH 5.4, 50 mM KCl. The CYC1 3′ UTR RNA was obtained in the same way from a plasmid lacking the adenosine stretch. Deadenylation reactions were performed in 10 mM HEPES pH 8.0, 50 mM NaCl, 1 mM MgCl2, and 5 mM β-mercaptoethanol at 30°C. Reactions contained 5 nM Pan2–Pan3 complex or Pan2, 180 nM (1.7 μg) CYC1-A80 RNA, and optional 600 nM Pab1 in a total volume of 115 μl and were started by the addition of RNA. 10 μl samples were taken at different time points, and the reaction stopped by addition of 1 mg/ml proteinase K and 25 mM EDTA and incubated at 37°C for 15 min. Reaction products were separated using 6% (w/v) urea polyacrylamide gel electrophoresis and visualized by RNA staining with SYBR Green II (Life Technologies). For Fig1B, the intensity of the RNA down each lane was measured using ImageJ (Schneider et al, 2012). For Figs1C and 4C and Supplementary Fig S5C, the average polyA tail length was defined as the maximum intensity in the lane.
NMR spectroscopy
For the solution NMR structure of scPan3 zinc finger, complete 1H/13C/15N assignments were obtained and structure determination was carried out using conventional methods. Statistics are summarized in Supplementary Table S1.
Electrophoretic mobility shift assay
The catalytic activity of scPan2 was destroyed by mutating Glu912 to Ala using a QuikChange kit (Agilent Technologies). Various concentrations (0.1–3 μM) of inactive scPan2–Pan3 complex with and without the zinc finger domain were incubated with 40 ng CYC1 3′ UTR or CYC1-A80 in deadenylation assay buffer in a total volume of 10 μl for 15 min at RT. Complex formation was analyzed on 6% NuPage TBE precast gels (Life Technologies), and the RNA stained with SYBR Green II (Life Technologies). The intensity of the free RNA band was measured using ImageJ (Schneider et al, 2012).
Fluorescence polarization
The binding of a 15-mer 5′ Cy3-labeled RNA (polyA, polyC, polyG and polyU), at a final concentration of 20 nM, to a dilution series of ctPan3 PKC in 10 mM Tris pH 8.0, 50 mM NaCl, 1 mM MgCl2, 5 mM β-mercaptoethanol was measured, in triplicate, by fluorescence polarization in a PHERAstar Plus instrument (BMG Labtech). Purified GST was used as a negative control. The background (protein without RNA) was subtracted.
To compare RNA binding of ctPan2, ctPan3 PKC, and the ctPan2–Pan3 PKC complex, a catalytically inactive ctPan2 mutant was made by QuikChange (Agilent Technologies) mutating Glu 899 to Ala. The assay described above was performed using a 25-mer of polyA RNA.
Crystallization and structure determination methods
Crystals of ctPan3 and ctPan2–Pan3 were obtained by vapor diffusion, and data were collected, processed, and refined as described in Supplementary Methods. The refinement statistics are summarized in Supplementary Tables S2 and S3.
Analytical size exclusion
Separately purified ctPan2 constructs containing deletions of parts of the PID domain and ctPan3 PKC dimer were mixed in a 1:1 ratio at 1 μM concentration and incubated at room temperature for 15 min. Fifty microliter samples were injected into a Superdex 200 Increase 3.2/300 size exclusion column (GE Healthcare) pre-equilibrated in 20 mM Tris pH 8.0, 500 mM NaCl, 2 mM TCEP. 65 μl fractions were collected in all experiments and analyzed on 4–12% NuPage Bis–Tris precast gels (Life Technologies). Gels were stained with Coomassie brilliant blue, and protein band intensities measured using ImageJ (Schneider et al, 2012).
Nanoelectrospray ionization (nanoESI) mass spectrometry (MS)
NanoESI mass spectrometry experiments were performed on a high mass Q-TOF-type instrument (Chernushevich & Thomson, 2004) adapted for a QSTAR XL platform (MDS Sciex) (Chernushevich & Thomson, 2004). Prior to MS analysis, 50 μl of 5.5 mg/ml scPan2–Pan3 complex was buffer-exchanged into 200 mM ammonium acetate solution, pH 8.0 twice using Bio-Rad Micro Bio-Spin columns (P-6). Protein solution was loaded for sampling via gold-plated borosilicate glass capillaries made in-house as described previously (Nettleton et al, 1998). The following experimental parameters were used: capillary voltage 1.3 kV, declustering potential 100 V, focusing potential 150 V, second declustering potential 15 V, and focusing rod offset varied from 80 to 100 V, MCP 2,550. Argon was used as a collision gas at maximum pressure. All spectra were calibrated externally using a solution of cesium iodide (100 mg/ml).
Accession codes
Sequences of two Pan3 isoforms cloned from a Chaetomium thermophilum cDNA library have been deposited in GenBank with Accession codes KJ657770 (full-length Pan3) and KJ657771 (splice variant without N-terminal zinc finger). For the three structures reported, the final coordinates have been deposited in the Protein Data Bank with PDB accession codes 4CYI (ctPan3 PKC), 4CYJ (ctPan3 PKC–ctPan2 PID), and 4CYK (scPan3 zinc finger).
Acknowledgments
We thank Minmin Yu and the staff of the Diamond Light Source (Didcot, UK; BAG proposal MX8547) and European Synchrotron Radiation Facility (Grenoble, France; BAG proposal MX1538) for assistance during data collection. We thank Katrin Wiederhold and N. Amy Yewdall for support in the early stages of the project, Helgo Schmidt and Pietro Roversi for many helpful discussions on crystallography, Shintaro Aibara for assistance with RNA purification and fluorescence polarization assays, and Andrew Carter, David Barford, Wojtech Galej and members of the Passmore laboratory for helpful comments. The research leading to these results has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013)/ERC Starting grant agreement no. 261151 to L.A.P., Wellcome Trust Programme Grants 088150/B/09/Z to C.V.R. and 080522/Z/60/Z to M.S., and Medical Research Council grants MC_U105192715 (L.A.P) and MC_U105178939 (M.S.).
Author contributions
JW and LAP designed experiments and led the project. JW, EV, MDA, BM, YG, SHM, and TMO performed experiments. LAP, MS, CVR, and MB supervised research. LAP conceived and initiated the project. JW, LAP, MS, and EV wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Baer BW, Kornberg RD. Repeating structure of cytoplasmic poly(A)-ribonucleoprotein. Proc Natl Acad Sci USA. 1980;77:1890–1892. doi: 10.1073/pnas.77.4.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz TH, Preiss T. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA. 2007;13:982–997. doi: 10.1261/rna.569407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeck R, Tarun S, Rieger M, Deardorff JA, Müller-Auer S, Sachs AB. The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J Biol Chem. 1996;271:432–438. doi: 10.1074/jbc.271.1.432. [DOI] [PubMed] [Google Scholar]
- Boeck R, Lapeyre B, Brown CE, Sachs AB. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol Cell Biol. 1998;18:5062–5072. doi: 10.1128/mcb.18.9.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bönisch C, Temme C, Moritz B, Wahle E. Degradation of hsp70 and other mRNAs in Drosophila via the 5′ 3′ pathway and its regulation by heat shock. J Biol Chem. 2007;282:21818–21828. doi: 10.1074/jbc.M702998200. [DOI] [PubMed] [Google Scholar]
- Braun JE, Huntzinger E, Fauser M, Izaurralde E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell. 2011;44:120–133. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Brockmann C, Soucek S, Kuhlmann SI, Mills-Lujan K, Kelly SM, Yang J-C, Iglesias N, Stutz F, Corbett AH, Neuhaus D, Stewart M. Structural basis for polyadenosine-RNA binding by Nab2 Zn fingers and its function in mRNA nuclear export. Structure. 2012;20:1007–1018. doi: 10.1016/j.str.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CE, Tarun SZ, Boeck R, Sachs AB. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5744–5753. doi: 10.1128/mcb.16.10.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CE, Sachs AB. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol Cell Biol. 1998;18:6548–6559. doi: 10.1128/mcb.18.11.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol. 2011;18:1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernushevich IV, Thomson BA. Collisional cooling of large ions in electrospray mass spectrometry. Anal Chem. 2004;76:1754–1760. doi: 10.1021/ac035406j. [DOI] [PubMed] [Google Scholar]
- Christie M, Boland A, Huntzinger E, Weichenrieder O, Izaurralde E. Structure of the PAN3 pseudokinase reveals the basis for interactions with the PAN2 deadenylase and the GW182 proteins. Mol Cell. 2013;51:360–373. doi: 10.1016/j.molcel.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, Nagar B, Yamamoto T, Raught B, Duchaine TF, Sonenberg N. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol. 2011;18:1211–1217. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- Fadda A, Färber V, Droll D, Clayton C. The roles of 3′-exoribonucleases and the exosome in trypanosome mRNA degradation. RNA. 2013;19:937–947. doi: 10.1261/rna.038430.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Font J, Mackay JP. Beyond DNA: zinc finger domains as RNA-binding modules. Methods Mol Biol. 2010;649:479–491. doi: 10.1007/978-1-60761-753-2_29. [DOI] [PubMed] [Google Scholar]
- Funakoshi Y, Doi Y, Hosoda N, Uchida N, Osawa M, Shimada I, Tsujimoto M, Suzuki T, Katada T, Hoshino S-I. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 2007;21:3135–3148. doi: 10.1101/gad.1597707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galej WP, Oubridge C, Newman AJ, Nagai K. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature. 2013;493:638–643. doi: 10.1038/nature11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- Hammet A, Pike BL, Heierhorst J. Posttranscriptional regulation of the RAD5 DNA repair gene by the Dun1 kinase and the Pan2-Pan3 poly(A)-nuclease complex contributes to survival of replication blocks. J Biol Chem. 2002;277:22469–22474. doi: 10.1074/jbc.M202473200. [DOI] [PubMed] [Google Scholar]
- Hernández H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- Joachimi A, Benz A, Hartig JS. A comparison of DNA and RNA quadruplex structures and stabilities. Bioorg Med Chem. 2009;17:6811–6815. doi: 10.1016/j.bmc.2009.08.043. [DOI] [PubMed] [Google Scholar]
- Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- Kuhlmann SI, Valkov E, Stewart M. Structural basis for the molecular recognition of polyadenosine RNA by Nab2 Zn fingers. Nucleic Acids Res. 2014;42:672–680. doi: 10.1093/nar/gkt876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner DH, Beilharz TH, Marguerat S, Mata J, Watt S, Schubert F, Preiss T, Bähler J. A network of multiple regulatory layers shapes gene expression in fission yeast. Mol Cell. 2007;26:145–155. doi: 10.1016/j.molcel.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell JE, Rudner DZ, Sachs AB. 3′-UTR-dependent deadenylation by the yeast poly(A) nuclease. Genes Dev. 1992;6:2088–2099. doi: 10.1101/gad.6.11.2088. [DOI] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Agrin NS, Smith M, Gongidi P, Jacobson A. Positive and negative regulation of poly(A) nuclease. Mol Cell Biol. 2004a;24:5521–5533. doi: 10.1128/MCB.24.12.5521-5533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Smith MM, McSweeney JM, Jacobson A. Identification of factors regulating poly(A) tail synthesis and maturation. Mol Cell Biol. 2004b;24:4196–4206. doi: 10.1128/MCB.24.10.4196-4206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit Rev Biochem Mol Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- Nettleton EJ, Sunde M, Lai Z, Kelly JW, Dobson CM, Robinson CV. Protein subunit interactions and structural integrity of amyloidogenic transthyretins: evidence from electrospray mass spectrometry. J Mol Biol. 1998;281:553–564. doi: 10.1006/jmbi.1998.1937. [DOI] [PubMed] [Google Scholar]
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Sachs AB, Davis RW, Kornberg RD. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AB, Deardorff JA. Translation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell. 1992;70:961–973. doi: 10.1016/0092-8674(92)90246-9. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui N, Mangus DA, Chang T-C, Palermino J-M, Shyu A-B, Gehring K. Poly(A) nuclease interacts with the C-terminal domain of polyadenylate-binding protein domain from poly(A)-binding protein. J Biol Chem. 2007;282:25067–25075. doi: 10.1074/jbc.M701256200. [DOI] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Schwalb B, Pirkl N, Maier KC, Schenk A, Failmezger H, Tresch A, Cramer P. Global analysis of eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Mol Cell. 2013;52:52–62. doi: 10.1016/j.molcel.2013.09.010. [DOI] [PubMed] [Google Scholar]
- Teplova M, Patel DJ. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat Struct Mol Biol. 2008;15:1343–1351. doi: 10.1038/nsmb.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- Uchida N, Hoshino S-I, Katada T. Identification of a human cytoplasmic poly(A) nuclease complex stimulated by poly(A)-binding protein. J Biol Chem. 2004;279:1383–1391. doi: 10.1074/jbc.M309125200. [DOI] [PubMed] [Google Scholar]
- Viphakone N, Voisinet-Hakil F, Minvielle-Sebastia L. Molecular dissection of mRNA poly(A) tail length control in yeast. Nucleic Acids Res. 2008;36:2418–2433. doi: 10.1093/nar/gkn080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E, Winkler GS. RNA decay machines: deadenylation by the Ccr4-Not and Pan2-Pan3 complexes. Biochim Biophys Acta. 2013;1829:561–570. doi: 10.1016/j.bbagrm.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Weill L, Belloc E, Bava F-A, Mendez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat Struct Mol Biol. 2012;19:577–585. doi: 10.1038/nsmb.2311. [DOI] [PubMed] [Google Scholar]
- Wiederhold K, Passmore LA. Cytoplasmic deadenylation: regulation of mRNA fate. Biochem Soc Trans. 2010;38:1531–1536. doi: 10.1042/BST0381531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf J, Passmore LA. mRNA deadenylation by Pan2-Pan3. Biochem Soc Trans. 2014;42:184–187. doi: 10.1042/BST20130211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Chang T-C, Yamashita Y, Zhu W, Zhong Z, Chen C-YA, Shyu A-B. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.