Abstract
Significance: Reduction-oxidation (Redox) status operates as a major integrator of subcellular and extracellular metabolism and is simultaneously itself regulated by metabolic processes. Redox status not only dominates cellular metabolism due to the prominence of NAD(H) and NADP(H) couples in myriad metabolic reactions but also acts as an effective signal that informs the cell of the prevailing environmental conditions. After relay of this information, the cell is able to appropriately respond via a range of mechanisms, including directly affecting cellular functioning and reprogramming nuclear gene expression. Recent Advances: The facile accession of Arabidopsis knockout mutants alongside the adoption of broad-scale post-genomic approaches, which are able to provide transcriptomic-, proteomic-, and metabolomic-level information alongside traditional biochemical and emerging cell biological techniques, has dramatically advanced our understanding of redox status control. This review summarizes redox status control of metabolism and the metabolic control of redox status at both cellular and subcellular levels. Critical Issues: It is becoming apparent that plastid, mitochondria, and peroxisome functions influence a wide range of processes outside of the organelles themselves. While knowledge of the network of metabolic pathways and their intraorganellar redox status regulation has increased in the last years, little is known about the interorganellar redox signals coordinating these networks. A current challenge is, therefore, synthesizing our knowledge and planning experiments that tackle redox status regulation at both inter- and intracellular levels. Future Directions: Emerging tools are enabling ever-increasing spatiotemporal resolution of metabolism and imaging of redox status components. Broader application of these tools will likely greatly enhance our understanding of the interplay of redox status and metabolism as well as elucidating and characterizing signaling features thereof. We propose that such information will enable us to dissect the regulatory hierarchies that mediate the strict coupling of metabolism and redox status which, ultimately, determine plant growth and development. Antioxid. Redox Signal. 21, 1389–1421.
Introduction
Redox chemistry is an intrinsic part of plant metabolism. The cellular redox state is determined by oxidation or reduction of various redox-active species, which are involved in a large number of metabolic reactions (95). In the chloroplast, reductants such as ferredoxin (Fdx) and NADPH are produced by the photosynthetic electron transport chain (Fig. 1), and along with ATP, used to generate sugar-phosphates, amino acids, and many other metabolites that are supplied to the rest of the cell (Fig. 2). In addition to this, NAD(P)H metabolism is involved in central processes such as glycolysis, fermentation, and oxidative pentose phosphate pathway (OPP) in the cytosol, tricarboxylic acid (TCA) cycle, respiratory electron transport, and biosynthetic processes in mitochondria, and photorespiration in plastids, mitochondria, and peroxisomes.
FIG. 1.
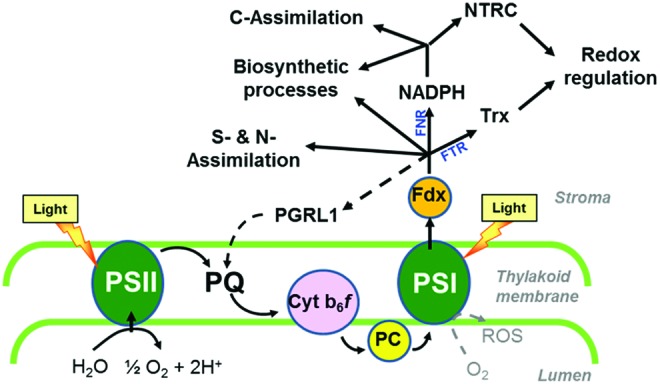
Overview of photosynthetic electron transport pathways in the chloroplast. Linear electron flow requires photosystems (PS) I and II working in series, leading to electron transfer from water to NADP+ to generate NADPH as reducing power. This involves electron transfer from PS II to PS I via plastoquinone (PQ), the cytochrome b6f complex (Cyt b6f), and plastocyanin (PC) as redox carriers. At the stromal side of PS I, electrons are subsequently donated to ferredoxin (Fdx), which functions as a mobile electron carrier distributing electrons to NADP+ via Fdx-NADP-reductase (FNR) to produce NADPH or directly to specific processes located in the stroma, such as S and N assimilation, biosynthetic pathways, and reactions involved in chloroplast redox regulation, catalyzed by Fdx-Trx-reductase (FTR) and thioredoxins (Trxs). NADPH produced by FNR is used by the carbon fixation cycle and various biosynthetic processes as a reductant and by NADPH-dependent Trx-reductase C (NTRC) for redox regulation. In the thyllakoid membrane, proton-gradient-regulation-like protein 1 (PGRL1) acts as an Fdx-PQ reductase in cyclic electron flow, reintroducing electrons from Fdx into the electron transport chain, as indicated by the dotted line (141). A second pathway for cyclic electron flow around PS I involving a NADH oxidase-like complex is not shown for clarity. Under conditions of acceptor limitation of PS I, electrons from PS I will photoreduce oxygen to reactive oxygen species (ROS). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
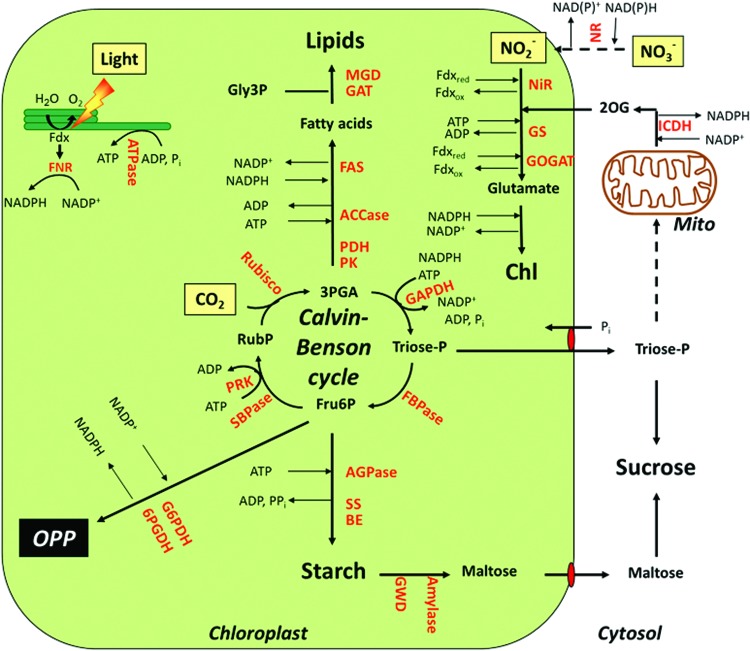
FIG. 2.
Overview of central metabolic pathways in the chloroplast. NADPH, Fdx, and ATP produced by the photosynthetic light reactions at the thylakoid membrane are used by different metabolic pathways in the stroma such as the carbon fixation cycle and the synthesis of starch, lipids, amino acids, and chlorophyll (Chl). In the dark, NADPH is produced by the oxidative pentose phosphate pathway (OPP), while carbon derives from starch degradation. For Abbreviations, see Abbreviations Used section. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
In addition to being an intrinsic part of metabolism, redox status plays an active role in metabolic regulation. In this regard, the redox status operates as a major integrator of cellular metabolism and is simultaneously regulated itself by metabolic processes. This enables readjustment of global metabolic pathways and redox status homeostasis in response to changes in environmental conditions, involving reprogramming of gene expression and post-translational modification of target enzymes by thiol-disulfide modulations (19, 21, 44, 177). The underlying signaling pathways have only partially been discovered in the previous years. While much is known about redox status signals involved in light activation of photosynthetic processes, little is known about redox regulation of other metabolic pathways in the plastid and of extra-plastidial metabolism. While recent studies provide evidence for the existence of redox signals coordinating metabolism and gene expression between different organelles, such as plastid, mitochondrion, and nucleus, their nature has not yet been clarified.
In this review, we will describe the redox status control of metabolism and the metabolic control of redox status at both the cellular and subcellular levels, mainly focusing on post-translational mechanisms. Despite the vastness of literature concerned with redox status-regulated gene expression, we will only describe this in passing given that it is the subject of a couple of excellent recent reviews (114, 309). In the first part, redox status-related metabolic processes will be described within their subcellular context, with regard to redox status-regulatory properties and intra-organellar signals involved in their co-ordination. In this regard, our major focus is placed on organelles such as plastids, mitochondria, and peroxisomes with readers being referred to other comprehensive reviews for details on both cytosolic and apoplastic aspects of redox status (96, 265, 270) with only a broad overview of the most important features in the context of cellular metabolism and function being provided here. In the second part, we will discuss the integration at the cellular level while mainly focusing on inter-organellar signals co-ordinating redox status regulation of metabolism between different subcellular compartments.
Plastidial Redox Status Biology
Chloroplasts are plant-specific organelles with important properties, the most prominent being their ability to carry out oxygenic photosynthesis (51). During this process, light energy is absorbed by photosystems I (PS I) and II (PS II) located in the thylakoid membrane and used to activate photosynthetic electron transport (Fig. 1). Linear electron flow requires both photosystems working in series, leading to electron transfer from water to NADP+ to generate NADPH as a reducing power and a trans-thylakoid proton gradient that drives ATP synthesis via CF0F1ATPase. This involves electron transfer from PS II to PS I via plastoquinone (PQ), the cytochrome b6f complex, and plastocyanin as additional redox carriers (Fig. 1). At the stromal side of PS I, electrons are subsequently donated to Fdx, which functions as a mobile electron carrier distributing electrons to NADP+ via Fdx-NADP-reductase (FNR) to produce NADPH or directly to specific processes located in the stroma, such as S and N assimilation, the synthesis of chlorophyll and fatty acids, and reactions involved in chloroplast redox regulation (128).
In the latter, electrons are transferred from Fdx to thioredoxins (Trxs) via Fdx-Trx-reductase (FTR). Trxs are small regulatory proteins containing a redox-active disulfide group that controls the thiol-disulfide exchange of target proteins (145, 237, 280, 307). In plants, Trxs comprise a medium-sized gene family with 10 different isoforms (f1–2, m1–4, x, y1–2 and z) being located in the chloroplast of Arabidopsis (70), while other isoforms are located in the cytosol and mitochondria (219). In vitro studies using purified proteins indicate Trxs f and m to be involved in the regulation of stromal metabolism, while x-, y-, and z-types serve as reducing substrates for antioxidant enzymes (61) (Table 1). More recently, genetic studies have been used to further dissect the specific roles of different Trxs in vivo, providing evidence for different isoforms of Trxs f and m having different functions in plants (34, 346, 375).
Table 1.
Selected Plastidial Processes Regulated by Thioredoxins and/or NADPH-Dependent Thioredoxin Reductase C and Confirmed Targets
| Process | Target | Trx-types involved | References |
|---|---|---|---|
| Calvin–Benson cycle | NADP-GAPDH | Trx f | 213, 384 |
| FBPase | Trx f | 61, 383 | |
| SBPase | Trx f | 45, 247 | |
| PRK | Trxs f and m | 213, 385 | |
| Rubisco activase | Trx f | 392, 393 | |
| CP12 | Trxs | 213 | |
| Starch synthesis | AGPase (APS1) | Trx f, NTRC | 17, 104, 197, 222, 346 |
| Starch synthase | Trxs | 115 | |
| Starch degradation | GWD | Trxs f and m | 226 |
| SEX4 | Trxs | 313 | |
| TR-BAMY | Trxs f, m and y, NTRC | 313, 321, 356 | |
| Lipid synthesis | ACCase | Trxs f and m | 299 |
| MGD | Trxs f and m | 388 | |
| Amino-acid synthesis | GOGAT | Trx m | 201 |
| GS2 | Trxs | 19, 59 | |
| Chlorophyll synthesis | GluTR | NTRC | 292 |
| Mg chelatase | Trx f | 153, 210 | |
| CHLM | NTRC | 292 | |
| OPP | Glc6PDH | Trxs f and m | 244 |
| ATP synthesis | CF1-ATPase | Trxs f and m | 181, 217, 219, 383 |
| Malate valve | NADP-MDH | Trxs f and m | 61, 159, 225, 303 |
| Protein import | TIC 55 | Trxs f and m | 30 |
| TIC 110 | Trxs f and m | 22 | |
| Gene expression | PEP complex | Trx z | 14, 42 |
| Nac2-RBP40 | NTRC | 308 | |
| Antioxidant system | 2Cys-Prx | Trx x, NTRC | 180, 266 |
For Abbreviations, see Abbreviations Used section.
In Arabidopsis, FNR has been found to be associated with the thylakoid and inner chloroplast envelope membranes, in addition to its location in the soluble stroma (128). Two FNR-binding proteins have been previously identified, Tic62 a component of the chloroplast import machinery (36) and the intrinsic thylakoid protein TROL (thylakoid rhodanese-like protein) (163), leading to binding of FNR to envelope and thylakoid membranes, respectively. Membrane recruiting of FNR has been found to be redox regulated in response to light signals (36). The role of membrane binding in the activity of FNR and the distribution of electrons to stromal reactions has, however, not yet been fully resolved. Recently, a combined knockout of Tic62 and TROL in an Arabidopsis double mutant led to restriction of FNR to the soluble stroma, resulting in decreased FNR level, decreased NADPH/NADP+ ratio, and altered starch metabolism in leaves (204).
In contrast to linear electron transfer, cyclic electron flow is driven by PS I alone to produce ATP without generating NADPH (Fig. 1). Here, electrons from Fdx are reintroduced into the electron transport chain rather than being distributed to stromal components. Two distinct pathways of cyclic electron flow have been identified as involving the proton-gradient-regulation-5/proton-gradient-regulation-like protein 1 (PGRL1) complex or the NADH dehydrogenase-like complex (158). The transmembrane thylakoid protein PGRL1 has recently been found to act as an Fdx-PQ reductase in Arabidopsis plants (141). The enzyme is most likely redox regulated, as its activity requires several redox-active cysteine residues and is controlled by Trx m4, providing a possible mechanism to link cyclic electron transport and stromal redox state (63, 141). Sensitive regulation of cyclic and linear electron flow will be required to rapidly adjust the ratio of NADPH and ATP production in response to fluctuating light conditions and to changes in the activities of stromal reactions with different requirements of ATP and/or NADPH as cofactors (160). In addition to this, excess reducing equivalents can be exported to the cytosol via the malate/oxaloacetate (OAA) shuttle, involving redox-dependent NADP-dependent malate dehydrogenase (MDH) in the chloroplast and NAD-dependent MDH in the cytosol for interconversion (302) (see also Table 1).
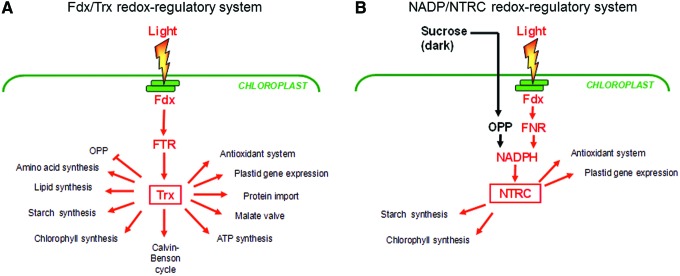
In the next section, we will not only describe the most important metabolic processes in the plastid stroma using NADPH, Fdx, and/or ATP provided by the photosynthetic light reactions, but we will also cover redox signals that are involved in their regulation. Light has been identified as the most important factor that is involved in the redox-status control of stromal metabolism via the Fdx/Trx system (Fig. 3a and Table 1). In addition to this, metabolic, developmental, and abiotic/biotic stress signals have to be integrated into this redox-regulatory network, possibly via alternative redox systems linked to NADPH-dependent thioredoxin reductase C (NTRC) or antioxidant metabolism (Fig. 3b and Table 1).
FIG. 3.
Overview of plastid processes subject to redox regulation via (A) the Fdx/Trx or (B) the NADPH/NTRC system. While the Fdx/Trx system is directly linked to light-driven electron transport, the NADPH/NTRC system is linked to photoreduced Fdx via Fdx-NADP-reductase (FNR) and to sugar metabolism via the OPP in the dark. An overview of the enzymes and proteins that have been found to be subject to redox regulation by these systems as well as the respective references are given in Table 1. NTRC is a bifunctional enzyme that combines both a NADP-Trx reductase and a Trx in a single polypeptide. Compared with Fdx-linked Trxs, there is only little information on possible targets of NTRC. More studies are needed to identify further plastidial processes that are subject to redox regulation by NTRC. For Abbreviations, see Abbreviations Used section. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Carbon fixation cycle
The Calvin–Benson cycle utilizes the products of the light reactions of photosynthesis, ATP and NADPH, to reduce inorganic CO2 to organic sugars, serving as the primary pathway for carbon fixation in higher plants (106, 223). The sequence of reactions of this cycle is well known since its discovery in the years 1946–1954 (35, 50, 246). It can be divided into three stages (Fig. 2). In the initial stage, carbon enters the cycle via carboxylation of ribulose-1,5-bisphosphate (RuBP) by ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) to produce two molecules of glycerate-3-phosphate (3PGA). In the second stage, 3PGA is reduced to triose phosphates via two subsequent reactions catalyzed by phosphoglycerate kinase (PGK) and NADP-dependent glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using most of the ATP and NADPH delivered by the light reactions. In the final stage, the acceptor RuBP is regenerated from triose phosphates via a sequence of different reactions catalyzed by aldolase, fructose-1,6-bisphosphatase (FBPase), transketolase (TK), sedoheptulose bisphosphatase (SBPase), and phosphoribulokinase (PRK), with the latter reaction also requiring ATP.
Studies in the 1960's on subcellular metabolite levels in leaves showed that activation of the Calvin–Benson cycle on illumination is accompanied by a transient decrease in the NADPH/NADP+ and ATP/ADP ratios in the chloroplast stroma (136). This indicated that light activation of the Calvin–Benson cycle is regulated by a mechanism which can override regulation by changes in the stromal NADPH and ATP levels. The mechanism was discovered by Buchanan and coworkers who found that FBPase and other enzymes of the photosynthetic carbon reduction cycle are activated by the Fdx/Trx system, which is directly linked to light-driven photosynthetic electron transport, rather than to stromal metabolism (48). Illumination promotes the reduction of Fdx at the acceptor side of PS I, which, in turn, leads to the sequential reduction of FTR and Trxs, which activate several target enzymes of the Calvin–Benson cycle (GAPDH, FBPase, SBPase, PRK, and Rubisco activase) by reducing intramolecular disulfide bonds (307). This enables the co-ordinated activation of the carbon fixation cycle in the light and its inhibition in the dark, when the Trx system is reoxidized (307). As shown by biochemical studies, the Trx-dependent enzymes of the Calvin–Benson cycle are preferentially or exclusively regulated by f-type Trxs, with m-type Trxs playing a much less prominent role (223, see Table 1). Thiol modulation is combined with additional regulatory mechanisms such as (i) protein complex formation of GAPDH and PRK via CP12 (146) and (ii) metabolic fine control acting on the mid-point redox potential of the regulatory cysteines (i.e., redox activation of FBPase requires fructose-1,6-bisphosphate) (281) or via allosteric effects (101). Light regulation is additionally supported by changes in pH and Mg2+ concentrations in the chloroplast stroma on illumination, which have direct regulatory effects on Rubisco, FBPase, SBPase, and PRK (48, 101). The combination of these mechanisms act to ensure that the Calvin–Benson cycle can be switched on in the light and off in the dark to avoid futile cycling of carbon metabolism. Moreover, they enable balancing of the different reactions of the cycle to avoid its blockage by accumulation or depletion of intermediates. While recent proteomic studies suggest that Calvin–Benson cycle enzymes also undergo other types of post-translational redox modifications, such as nitrosylation and glutathionylation, the importance and function of these additional redox modifications still have to be determined (223).
Rubisco also catalyzes a side reaction with O2, leading to the formation of one molecule of 3PGA and one molecule of 2-phosphoglycolate (2PG). The latter is detoxified and recycled back to 3PGA via photorespiration, a complex energy consuming pathway that liberates fixed carbon and nitrogen as CO2 and NH4+, respectively (31). Moreover, the photorespiratory cycle is a highly compartmentalized process with enzymatic reactions occurring across four compartments. In the chloroplast stroma, 2PG is dephosphorylated to glycolate, which is subsequently exported from the chloroplasts—via the cytosol—to the peroxisomes where it is oxidized to glyoxylate and transaminated to glycine, which—via the cytosol—enters the mitochondria, where two molecules of glycine are deaminated and decarboxylated to form one molecule each of serine, ammonia, and carbon dioxide. Serine is exported from the mitochondria to the peroxisomes, where it is mainly converted to glycerate, which leaves the peroxisomes and is taken up—via the cytosol—into the chloroplast, where it is phosphorylated to yield 3PGA. While the enzymes involved in the photorespiratory cycle have been identified and characterized, little is known about the identity of the transporters catalyzing the exchange of photorespiratory metabolites between the different subcellular compartments. However, first steps in this direction are being taken with a chloroplastidic glycolate/glycerate transporter recently being identified in Arabidopsis that is necessary for the function of the photorespiratory cycle (269). Strategies to decrease photorespiration by CO2-concentrating mechanisms or to improve photorespiratory pathways have been recently reviewed extensively in the literature and will not be covered here (32, 267, 326). The role of the photorespiratory cycle in mitochondria and peroxisomes and its role in the exchange of redox equivalents between organelles will be discussed in greater detail in the respective chapters later.
Starch metabolism
The stoichiometry of the Calvin–Benson cycle requires 5/6th of the triose phosphates to be used to regenerate RuBP as CO2 acceptor, while the remainder can exit the cycle to be used to synthesize sucrose and starch as major end products (Fig. 2), which serve as the ultimate source of carbon for plant growth (326, 338). While sucrose synthesis follows the export of triose phosphates to the cytosol via the triose phosphate/inorganic phosphate (Pi) translocator at the inner membrane of the chloroplast envelope (293), starch is synthesized in the chloroplast stroma, using fructose-6-phosphate (Fru6P) delivered by FBPase in the Calvin–Benson cycle (103, 391). After conversion of Fru6P to glucose-1-phosphate (Glc1P) via the sequential action of phosphoglucose isomerase (PGI) and phosphoglucomutase (PGM), the first committed step of starch synthesis involves the conversion of Glc1P and ATP to ADP-glucose (ADPGlc) and inorganic pyrophosphate (PPi), catalyzed by ADPGlc pyrophosphorylase (AGPase). ADPGlc acts as the glucosyl donor for different classes of starch synthases (SS), which elongate the α-1,4-linked glucan chains of the starch polymers. Five distinct SS classes are known in plants: granule-bound SS, which is responsible for the synthesis of amylose, and soluble SS 1–4, which is responsible for amylopectin synthesis. Branch points are introduced by two classes of starch branching enzymes (SBE 1 and 2), which differ in terms of length of the glucan chains transferred and substrate specificities. Interestingly, starch synthesis also involves two types of debranching enzymes (ISA 1 and 2), which cleave branch points and are probably involved in tailoring the branched glucans into a form that is capable of crystallization within the starch granule.
In Arabidopsis leaves, the majority of control of starch biosynthesis has been found to reside in the reaction catalyzed by AGPase (79). AGPase is rapidly activated on illumination by reduction of an intermolecular disulfide bond between the Cys residues joining the two small subunits (APS1) of this heterotetrameric enzyme (139, 346). Using transgenic Arabidopsis plants expressing a mutated AGPase in which the regulatory Cys 81 of APS1 has been substituted by Ser, genetic evidence has been provided that redox regulation of AGPase contributes significantly to photosynthetic starch turnover during the light/dark cycle in leaves (125). AGPase from potato tubers and pea (Pisum sativum) leaf chloroplasts has been shown to be reduced by Trxs f and m in vitro (17, 104, 346). Using recombinant purified Arabidopsis proteins, it was shown that Trx f1 redox activates AGPase more efficiently than other types of plastidial Trxs, such as Trx m1, x, and y1 (346). Recent studies in Arabidopsis mutants with an insertion in the trx f1 gene provided in planta evidence for the role played by Trx f in the light activation of AGPase and photosynthetic carbon partitioning in plants (346). In these mutants, inactivation of Trx f1 led to decreased light activation of AGPase and decreased starch accumulation in leaves (346), while inactivation of isoforms of Trx m and Trx y in other Arabidopsis mutants had no such effects (I. Thormählen, M. Paul, E. Issakidis-Bourguet, and P. Geigenberger, unpublished results). This indicates a specific role of Trx f in regulating starch synthesis, similar to the established role of Trx f in regulating the Calvin–Benson cycle (see Table 1). The common regulation by Trx f of both metabolic processes enables photosynthesis and end-product synthesis to be co-ordinately regulated in response to light via the same signaling pathway.
Interestingly, Arabidopsis mutants lacking Trx f1 revealed no changes in photosynthetic parameters and growth, although Trx f protein levels were decreased by more than 95% relative to the wild type (346). This is surprising, given the exclusive regulation of individual steps of the Calvin–Benson cycle (i.e., FBPase) by f-type Trxs (see Table 1). While this may point to a more complex regulation of the Calvin–Benson cycle as initially expected, this may also be due to Trx f having much lower affinities for AGPase (17) compared with FBPase (239), indicating that changes in Trx f expression levels in response to transgenesis (346) circadian rhythms (26), thiol status, and sugars (25) or indeed any other stimulus will affect starch synthesis to a greater extent than the Calvin–Benson cycle. Consistently, in transplastomic tobacco plants, very severe over-expression of Trx f led to a strong increase in leaf starch accumulation, without leading to an increase in photosynthesis (290, 298).
In addition to redox, AGPase is also the subject of allosteric regulation, being activated by the first product of the Calvin–Benson cycle (3PGA) and inhibited by Pi (18). Since the concentrations of these effectors will change during light/dark transitions in a characteristic manner, this will contribute to light activation of AGPase. Specifically, allosteric regulation and redox regulation will act synergistically on AGPase to ensure starch synthesis is efficiently activated in the light and inactivated in the dark. While the allosteric activator 3PGA promotes light-dependent redox activation of AGPase by Trxs (139), the sensitivity of AGPase to its allosteric effectors is strongly affected by reversible oxidation of its regulatory cysteine (347).
In addition to light, redox activation of AGPase is also promoted by sugars, in illuminated as well as in darkened leaves and in non-photosynthetic tissues (139, 178, 347). In leaves, this enables starch synthesis to be regulated in response to changes in the balance between carbon supply and growth (111); while in non-photosynthetic storage organs such as potato tubers, this enables starch synthesis to be regulated in response to fluctuations in the supply of sucrose from the leaves (105, 347). Redox activation of AGPase was found to be closely correlated with the sugar content across a range of physiological and genetic manipulations, with light leading to an additional activation in leaves (139, 347). Reductive activation of AGPase in non-photosynthetic tissues or in nocturnal leaves requires alternative systems of electron transfer linked to NADPH generated from sugars, rather than to photoreduced Fdx (Fig. 4). This involves dark operative redox systems that are based on (i) Fdx/Trx systems involving a more oxidizing Fdx receiving electrons from NADPH in amyloplasts (20) or (ii) NTRC, localized in both chloroplasts and amyloplasts (176).
FIG. 4.
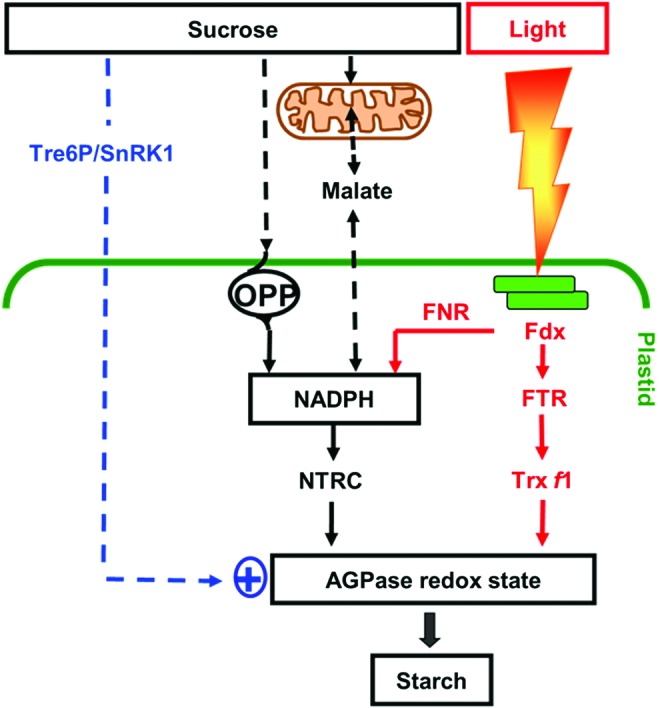
Model of the signaling pathways controlling redox regulation of starch biosynthesis. Light activation of starch synthesis involves post-translational redox activation of AGPase in the chloroplast via the Fdx/Trx f system (139, 346). NTRC, containing both a NADP-Trx reductase and a Trx in a single polypeptide, serves as an alternative system for transferring reducing equivalents from NADPH to AGPase, thereby enhancing storage starch synthesis (197, 222). In the light, NTRC is mainly linked to photoreduced Fdx via Fdx-NADP reductase (FNR) and complements the FTR/Trx system in activating AGPase. In the dark or in non-photosynthetic tissues, NTRC is primarily linked to NADPH provided by sugar oxidation via the initial reactions of the oxidative pentose phosphate pathway (OPP) and in this way, regulates AGPase independently of the Fdx/Trx system. The role of the OPP in regulating starch synthesis has been recently confirmed (322). There is also evidence that alterations in mitochondrial redox status are transmitted to the plastid via the malate valve (302), leading to changes in redox activation of AGPase and starch synthesis (55) (Fig. 7). Redox activation of AGPase is also promoted by sugars, involving cytosolic sugar signaling components such as trehalose-6-phosphate (Tre6P) and SNF1-related protein kinase (SnRK1), although the underlying mechanisms have not yet been resolved (161, 178, 209, 348). For Abbreviations, see Abbreviations Used secton. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
NTRC is a bimodular protein containing both an NADPH-dependent thioredoxin reductase (NTR) and a Trx domain on a single polypeptide (38). By using NADPH as a reducing power, NTRC conjugates both NTR and Trx activities to reductively activate AGPase and concomitant starch synthesis (222). Using an insertional knock-out mutant, NTRC has been found to play a role in the regulation of AGPase and starch synthesis in leaves and non-photosynthetic tissues in response to both light and sugars in the dark (196, 197, 222). In the light, NTRC can use photo-reduced NADPH provided by FNR; while in the dark, NADPH is most likely provided by the initial reactions of the OPP, glucose-6-phosphate dehydrogenase (G6PDH), and 6-phosphogluconate dehydrogenase (6PGDH). The role of the OPP in redox regulation of starch synthesis has been confirmed by recent studies showing chloroplast-localized 6PGDH to be required for starch accumulation in maize mutants (322). The chloroplast isoforms of G6PDH are subject to post-translational redox regulation via Trx f, leading to inactivation of the OPP in the light and activation in the dark (244). In heterotrophic amyloplasts, G6PDH isoforms are less sensitive to redox status control, enabling high levels of NADPH to be produced to drive biosynthetic processes in non-photosynthetic tissues (378). While glucose leads to redox activation of AGPase via an increase in the NADPH/NADP+ ratio, sucrose and trehalose most likely act via additional signaling pathways that have not yet been clarified (178). These disaccharide sugars are linked to trehalose-6-phosphate (Tre6P), a signal metabolite implemented in diverse developmental (371) and metabolic responses, including redox regulation of AGPase and starch metabolism (178, 209). In addition to Tre6P, redox activation of AGPase is also linked to other elements of the cytosolic sucrose signaling cascade, such as the conserved SNF1-related protein kinase (SnRK1) in Arabidopsis leaves (161) and potato tubers (216, 348), although the underlying mechanisms have not yet been resolved.
In addition to AGPase, reductive activation by thiol/disulfide modulation has also been demonstrated for other enzymes involved in the pathway of starch synthesis downstream of AGPase, such as SS 1 and SS 3, SBE 2, and ISA 1 and ISA 2, by using comprehensive in vitro studies (115). This enables a co-ordinated activation of the supply and the use of ADPGlc for starch synthesis by Trx-dependent redox regulation, linking external factors such as light and sucrose to redox activation of the whole pathway. It also implies that redox status regulation may influence starch structure in addition to quantity, depending on the contribution of the different redox-regulated isoforms of SS and (de-)branching enzymes to synthesize the polymers. In addition to this, redox regulation of SS 1, SS 3, and SBE 2 might be important for the involvement of these isoforms in the formation of protein complexes, which have been suggested to play a role in the regulation of starch biosynthesis (140, 345). Interestingly, SS isoforms and NTRC have been implicated in the accumulation of starch in response to microbial volatiles (198).
In leaves, starch is remobilized at night to support chloroplast metabolism and sucrose export when photosynthesis is inactive; while in heterotrophic storage organs, starch is remobilized later in development to support phases of reproductive growth (391). The pathway of starch degradation has been recently established in Arabidopsis leaves (391). After reversible phosphorylation of glucans on the surface of the starch granule via glucan water dikinase (GWD) and glucan phosphatase (SEX4), starch granules are attacked most probably by alpha-amylase and the resulting branched glucans are subsequently converted to unbranched alpha-1,4-glucans via debranching enzymes (isoamylase and pullulanase). Linear glucans are metabolized by the concerted action of β-amylase and disproportionating enzyme to glucose and maltose, which leaves the chloroplast via a maltose transporter in the inner envelope membrane to support cytosolic metabolism (Fig. 2).
Despite our sound knowledge with regard to the pathway, relative little is known about its regulation. There is evidence that starch degradation is controlled by diurnal rhythms (117) and by redox status (115). It has been found that both of the enzymes involved in reversible starch phosphorylation, GWD (226) and SEX4 (313), as well as the more downstream acting enzymes alpha-amylase (115, 381), beta-amylase (321), and limit dextrinase (115, 306), are reductively activated by Trxs f and m. While this shows that redox status regulation also extends to enzymes of starch degradation, reductive activation of these enzymes is counterintuitive in the context of light/dark regulation in the chloroplast, as it would imply that starch degradation is inactivated in the dark, when the Trx system is oxidized. While the roles, in this context, of different Trx isoforms and NTRC have not been clarified, the use of NADPH as an electron donor via NTRC could enable redox status activation of starch degrading enzymes also in the dark. In addition to this, recent studies provide evidence for a role of the redox status in regulating starch degradation under certain stress conditions (165, 166, 356) or in the context of specific tissues, cell types (356), or subcellular compartments (376) having different requirements for regulation of starch degradation.
Lipid synthesis
Similar to the metabolism of starch in most species and tissues, the reactions of de novo fatty acid biosynthesis occur exclusively in the plastid (259). In leaf chloroplasts, carbon used for fatty acid synthesis mainly derives from the Calvin–Benson cycle, using a pathway that converts 3PGA to acetyl-CoA involving pyruvate kinase (PK) and the pyruvate dehydrogenase (PDH) enzyme complex (23). Acetyl-CoA carboxylase (ACCase) catalyzes the first committed step of de novo fatty acid synthesis in the plastid stroma, converting acetyl-CoA and CO2 to malonyl CoA by using ATP as an energy donor (Fig. 2). Malonyl CoA is used as a substrate for fatty acid synthase (FAS), a large multisubunit enzyme complex that performs the cyclic condensation of two carbon units with the extending acyl chains being covalently bound to acyl carrier protein (259). These reactions are a strong sink of ATP and NADPH, which are provided by the light-driven photosynthetic electron transport in leaves, or respiratory processes in non-photosynthetic tissues. Synthesis of one molecule of C16-palmitic acid from eight molecules of acetyl-CoA requires 14 molecules of NADPH and seven molecules of ATP. Conversion of saturated fatty acids to unsaturated forms is catalyzed by fatty acid desaturases, which require Fdx as an electron acceptor in the plastid (259). Fatty acids are used as substrates for the synthesis of glycerolipids, which form the lipid bilayer of all cellular membranes or serve as important carbon reserves in selected seeds (170).
ACCase is a key-regulatory step in the pathway of fatty acid biosynthesis in different organisms, including plants (149). The chloroplast enzyme is a multienzyme complex that consists of four different polypeptides, biotin carboxylase, biotin carboxyl carrier protein, and the carboxyltransferase alpha and beta subunits, encoded in the nucleus and the chloroplast, respectively. In vitro studies revealed that chloroplast ACCase is activated by Trxs f and m (299), catalyzing the reduction of an intermolecular disulfide bond between the carboxyltransferase subunits (183). Light leads to reductive activation of ACCase in isolated pea chloroplasts (183) and Arabidopsis leaves (A. Kolbe, M. Ehrlich, and P. Geigenberger, unpublished results), indicating that fatty acid biosynthesis is regulated by the Fdx/Trx system in a similar manner as the Calvin–Benson cycle (see Fig. 3A). Since fatty acid synthesis is a strong sink of NADPH and ATP (see 51), the use of these compounds has to be strictly coordinated with their supply by photosynthetic light reactions. In addition to light, reductive activation of ACCase has also been shown to be promoted by sucrose, in darkened leaves as well as in non-photosynthetic tissues (A. Kolbe, P. Waldeck, M. Ehrlich, H. Vigeolas, and P. Geigenberger, unpublished results). Similar to the sucrose-dependent redox regulation of AGPase (see Fig. 4), this could involve NTRC as a dark operative redox system in the plastid as well as specific sucrose signals from the cytosol, although direct evidence for their contribution in the regulation of ACCase is lacking at the moment. Recent studies show that in addition to the carboxyltransferases, the other subunits of ACCase are also potentially controlled by redox regulation. There is proteomic evidence for S-thiolation of the biotin carboxyl carrier subunit by glutathione in Chlamydomonas (224) and for glutathionylation of biotin carboxylase in Arabidopsis (71). Moreover, redox status regulation also extends to downstream metabolic reactions of galactolipid biosynthesis, as the envelope bound monogalactosyldiacylglycerol synthase that synthesizes the major lipid component of chloroplast thylakoid membranes has been found to be subject to Trx-dependent redox regulation (388).
Nitrate assimilation, chlorophyll synthesis, and antioxidant metabolism
Key reactions of many other important metabolic pathways, such as N-assimilation, chlorophyll synthesis, and antioxidant metabolism, are also located in the plastid (Fig. 2). The initial step of nitrate assimilation, the conversion of nitrate to nitrite, is catalyzed by nitrate reductase in the cytosol, using NAD(P)H as a reducing power. The highly reactive nitrite is immediately transported into the plastid, where it is converted to ammonium by nitrite reductase (NiR) using Fdx as an electron donor (144, 297). Eight molecules of reduced Fdx are required to reduce one molecule of nitrate to ammonium. The ammonium ions provided by NiR or photorespiration are rapidly assimilated into amino acids via the sequential action of glutamine synthetase (GS) and glutamine:oxoglutarate amino transferase (GOGAT), which catalyze the ATP-dependent conversion of ammonium and glutamate (Glu) to glutamine (Gln) and the conversion of Gln and 2-oxoglutarate (2OG) to two molecules of Glu, respectively (190). In the chloroplast, GOGAT accepts electrons directly from Fdx; while in non-photosynthetic plastids, electrons are provided by NADPH. Provision of 2OG is most likely a function of isocitrate dehydrogenase (ICDH) isoforms in the cytosol and mitochondria catalyzing the conversion of isocitrate and NAD(P)+ to 2OG and NAD(P)H, with 2OG being transported into the chloroplast via a 2OG/malate transporter (143).
Given that the reactions of nitrate assimilation are a strong sink for electrons, they have to be closely coordinated with the electron pressure of the photosynthetic light reactions. This is achieved by direct electron transfer from photoreduced Fdx to NiR and GOGAT in the chloroplast stroma (128). In addition to this, there is in-vitro evidence that NiR (212), GOGAT (201), and GS2 (19, 59, 241) are subject to redox status regulation by Trxs. Moreover, when DTT was fed to Arabidopsis leaves to increase the thiol state of the tissue, the levels of 2OG decreased while amino-acid levels increased, which indicates that redox status regulation of the GOGAT/GS cycle is most likely operational in vivo (177).
Glutamate (Glu), as the first product of ammonium assimilation, is used to synthesize many other amino acids and nitrogenous metabolites in plants. It serves as a precursor for the synthesis of 5-aminolevulinic acid (ALA), which is the starting point for the biosynthesis of chlorophyll (Chl) and heme in the chloroplast (51). Glutamate is converted to ALA by a sequence of reactions involving NADPH-dependent glutamyl-transfer RNA reductase (GluTR), Glu1-semialdehyde aminotransferase, porphobilinogen synthase, and uroporphyrinogen decarboxylase. ALA is subsequently converted in a series of enzymatic steps to protoporphyrin IX, the common branch point for the synthesis of heme and Chl. To make Chl, a polymeric magnesium (Mg) chelatase complex inserts Mg2+ into the protoporphyrin IX ring, which is further modified via the subsequent reactions of a methyltransferase (CHLM), an oxidative cyclase complex, an NADPH:vinyl reductase, and an NADPH:protochlorophyllide oxido-reductase (POR) to synthesize Chl a. The synthesis of Chl b occurs with Chl a as precursor most likely via Fd-dependent chlorophyll a oxygenase (283). After binding to the photosystems and light-harvesting complexes in the thylakoid membranes, both Chl a and Chl b serve as important components in photosynthetic light absorption.
Chlorophyll synthesis has been identified as a light-dependent process in photosynthetic angiosperms (51). The CHL1 subunit of Mg-chelatase was found to be reductively activated by Trx f in vitro (153), while virus-induced gene silencing of Trx f and Trx m in transgenic pea plants showed that this mechanism is also operational in vivo (153). Proteomic approaches also identified other enzymes of Chl biosynthesis as potential Trx targets. These involve enzymes catalyzing the conversion of Glu to ALA, such as of Glu1-semialdehyde aminotransferase, porphobilinogen synthase, and uroporphyrinogen decarboxylase (49). Recent studies provide evidence that Chl synthesis is also regulated by NTRC as a second redox system (292). NTRC uses NADPH to reductively activate GluTR, involved in the synthesis of ALA, and CHLM, involved in the conversion of protoporphyrin IX to Chl a. A physical interaction of NTRC with GluTR and CHLM was confirmed by biomolecular fluorescence complementation assays, while knockout of NTRC led to a decrease in GluTR and CHLM activities and concomitant chlorophyll synthesis in Arabidopsis mutants in vivo (292). This shows that different stages of Chl synthesis are subject to post-translational redox regulation, involving Fdx/Trx and NADPH/NTRC as redox systems (Fig. 3 and Table 1). While the Fdx/Trx system enables Chl synthesis to be regulated in response to light signals, the role of the NADPH/NTRC system is less clear in this context. Similar to the role of NTRC in activation of starch synthesis (222), NTRC could complement the Fdx/Trx system in light activation of Chl synthesis, by acting at different target enzymes in this pathway (Table 1). Moreover, NTRC could Iink the rate of Chl synthesis to the availability of NADPH, which is required as an important reducing equivalent at different steps of the pathway (Fig. 2) (51).
Finally, metabolic reactions are required to scavange reactive oxygen species (ROS) that are produced during photosynthetic electron transport in the chloroplast. Under conditions of acceptor limitation of PS I, electrons from PS I will photoreduce oxygen to superoxide radicals, which subsequently disproportionate to hydrogen peroxide, either spontaneously or in a reaction catalyzed by superoxide dismutase (15). While hydrogen peroxide becomes toxic when it accumulates at a high level (4), it also serves as an important signaling molecule regulating photosynthesis and other processes in plants (268). A sensitive regulation of hydrogen peroxide levels is, therefore, important for its signaling function and detoxification. Hydrogen peroxide is reduced to water by enzymes such as ascorbate peroxidase, converting ascorbate to monodehydroascorbate, with the latter being recycled to ascorbate by using electrons from photoreduced Fdx (15). Hydrogen peroxide detoxification in the chloroplast also involves a 2Cys-peroxyredoxin (Prx). In the light, photoreduced Fdx serves as a source of reducing power to reduce 2Cys-Prx by Trx x via the FTR/Trx x pathway (180). In addition to this, 2Cys-Prx can be reduced by NTRC using electrons provided by NADPH, which is generated via FNR in the light or the OPP in the dark (266). NTRC knockout in Arabidopsis mutants led to increased levels of hydrogen peroxide and lipid peroxidation under conditions of prolonged darkness followed by illumination, while no such effects were observed in a normal diurnal cycle (266). While NTRC is complementing the Fdx/Trx x system in diverting electrons to the reduction of 2Cys-Prx in the light, it may serve as a key detoxification system in the dark, using metabolically produced NADPH as a reducing power. The role of the OPP to provide NADPH for ROS detoxification under these conditions has been recently confirmed (67). The NADPH-dependent hydrogen peroxide-scavenging system via 2Cys-Prx and NTRC has been shown to be required for the protection of the highly ROS-sensitive Mg-protoporphyrin monomethylester cyclase involved in chlorophyll biosynthesis (323, 324). Redox status-sensitive regulation of the cyclase complex will affect accumulation of Mg-protoporphyrin IX, which has been suggested to act as a plastid retrograde signal controlling the expression of photosynthetic genes in the nucleus (24) (see section “Integration at the Cellular Level”).
Interaction of plastid metabolism and gene expression
Due to the endosymbiotic origin of chloroplasts, they contain their own genome and gene expression machinery. This machinery is known to be highly regulated, especially in response to different environmental stimuli (27). Light plays a crucial role, especially for chloroplast protein translation based on findings that synthesis rates of chloroplast encoded photosynthetic proteins increase on illumination despite the fact that their respective mRNA levels stay constant (78). In addition to this, metabolic and developmental signals have to be integrated in this context. Recent studies in Chlamydomonas provide evidence for a cross-talk between chloroplast protein translation and carbon metabolism. In the study of (41), it was shown that a subunit of the plastidial PDH complex (DLA2), which synthesizes acetyl-CoA as a precursor for lipid synthesis, forms ribonucleoprotein particles and influences chloroplast mRNA translation. Conversely, RNA binding affects plastidial PDH metabolic activity. This reciprocal regulation may function in co-ordinating the synthesis of lipids and proteins for the biogenesis of photosynthetic membranes. In a second example, the same group showed that redox status regulation of chloroplast mRNA translation of the psbD gene, encoding the D2 protein of PS II, involves the reduction of a single intermolecular disulfide bridge between two translation-activation proteins in the dark, most likely via NADPH-dependent NTRC, acting as a dark operative chloroplast redox system (308). This provides evidence for a connection between plastid carbon metabolism and protein synthesis via NTRC-mediated redox signaling.
Mitochondrial Redox Biology
Mitochondrial redox biology and, indeed, mitochondrial metabolism is dominated by its major flux-bearing pathways, namely respiration (228) and the pathway of photorespiration that it shares with the peroxisome, mitochondria, chloroplast, and cytosol (31, 351). Mitochondrial respiration provides ATP, reductant, and carbon skeletons in both the light and the dark (Fig. 5). It is, however, considerably downregulated in the light; however, to what extent remains somewhat controversial (256, 343). The mitochondrial proteome is responsive not just to light conditions but also to tissue type and to a range of biotic and abiotic stresses (156, 193, 336), as such it follows that the mitochondria need to harbor a considerable degree of dynamic flexibility in order to maintain both its own function and that of the cell in general (309). In this section, we not only will focus largely on the redox chemistry of the mitochondrial respiratory machinery but will also integrate the mitochondrial steps of photorespiration into this discussion as well as covering relevant signal pathways emanating from the mitochondria. Oxidative phosphorylation in the mitochondria of plants is most commonly fueled by the breakdown of sucrose (Fig. 5). Sucrolysis is the pathway by which sucrose is degraded into pyruvate in the cytosol [although some reactions are additionally localized in the plastid (3) and a small proportion of the enzyme activities of the entire glycolytic pathway is localized to the outer mitochondrial membrane (112, 119)], the reactions of the TCA cycle completely oxidize pyruvate to CO2 (85) with electrons being transferred to NAD+ and FAD yielding NADH and FADH2 while phosphorylating some ADP directly. Complete respiration of a molecule of sucrose will result in the release of 20 NADH and 4 FADH2—these reduced co-enzymes are subsequently oxidized by the mitochondrial electron transport chain (mETC) (Fig. 5). The free energy released by mitochondrial electron transport is partially coupled to the translocation of protons across the inner mitochondrial membrane, creating an electrochemical gradient across the inner membrane, although in plants flexibility exists here due to the presence of non-proton pumping enzymes. The free energy released by the movement of the protons back across the inner membrane through the F0 proton channel of the ATP synthase complex is used by the F1 component to convert ADP and Pi to ATP (85). However, it is important to note that many of the intermediates of this pathway are important precursor molecules in their own right for a plethora of biosynthetic pathways and, as such, these numbers are likely to highly overestimate the true respiratory yield of sucrose breakdown.
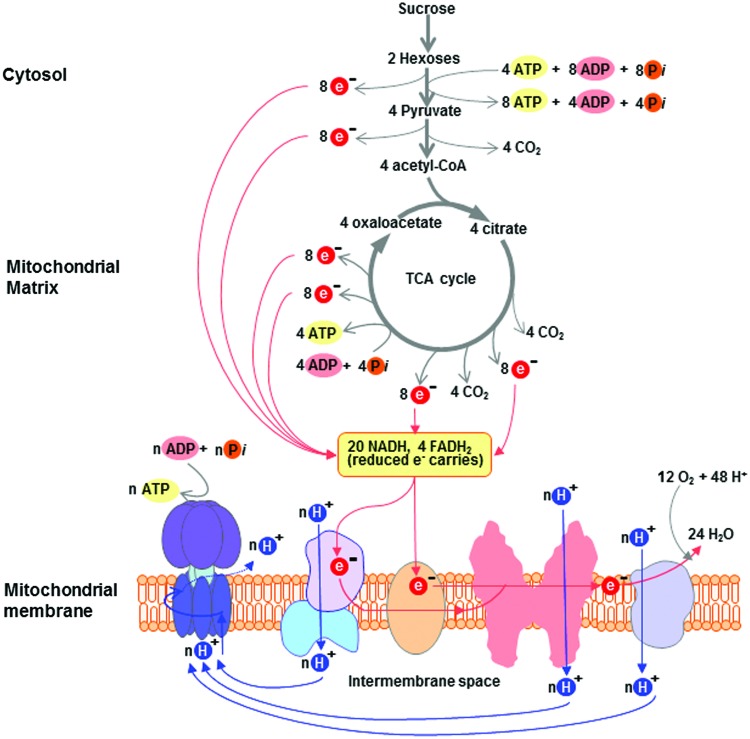
FIG. 5.
The general mechanism of oxidative phosphorylation in plant mitochondria. Electrons released during oxidative reactions of glycolysis and the tricarboxylic acid cycle produce 20 molecules of NADH and 4 molecules of FADH2. These reduced coenzymes are subsequently oxidized by the mitochondrial electron transport chain (mETC). The free energy release during the operation of the mETC is coupled to proton translocation across the inner mitochondrial membrane, concomitantly generating an electrochemical proton gradient across this membrane. This free energy is subsequently released by the movement of protons back across the inner membrane through the F0 channel of the ATP synthase complex and is used by the catalytic site of the F1 component of the ATP synthase complex to convert ADP and inorganic phosphate (Pi) to ATP within the mitochondrial matrix. Modified from Buchanan et al. (51). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Mitochondrial contribution to energy metabolism—respiration
As stated earlier, the mitochondria is dominated by respiration and in photosynthetic cells, the oxidative reactions of photorespiration that reside in this organelle, namely the concerted action of glycine decarboxylase and serine hydroxymethyltransferase (32). Mitochondrial respiratory activity, in turn, is closely connected with NADH production with four of the eight core enzymes of the cycle as well as the intimately related mitochondrial PDH producing this co-factor. To re-capitulate, the TCA cycle is composed by a set of eight enzymes primarily linking the product of the oxidation of pyruvate and malate (generated in the cytosol) to CO2 with the generation of NADH for the oxidation by the mitochondrial respiratory chain (85, 228). Mitochondria are the only organelles in which a full cycle is present, although this does not in itself necessitate that it always functions in a fully cyclic mode (333). The presence of organic acids is known to support numerous and diverse functions within and beyond cellular metabolism; the level of accumulation of the various organic acids is extremely variable between species, developmental stages, and tissue types (7, 86). For this reason, it is likely that the enzymes involved in the inter-conversion of these metabolic intermediates are subject to tight regulatory control. Hints to the regulation of the TCA cycle have been provided by a recent metabolic control analysis which shows that much of the control through this pathway is resident in fumarase, MDH, and 2OG dehydrogenase (8). An important comment needs to be made here. Although the summed control co-efficient values reported exceed 1 and, as such, appear to violate the summation theory, it is critical to note that the summation theory is only valid in instances in which there is no control (which can, of course, be a negative as well as a positive resident outside of the pathway under consideration). This fact, apart from this study, is somewhat in conflict with the finding of modeling studies of Hagedorn et al., which suggested that the rate of oxygen consumption was insensitive to changes in MDH activity (126), suggesting a further study is required to clarify the actual contribution of this enzyme.
Unfortunately, the lack of subcellular information with regard to the levels of intermediates of the cycle (335) currently precludes us from being able to assess the potential of the constituent enzymes to play regulatory roles merely on the basis of disequilibrium ratios. That said, a massive amount of data has, nevertheless, been compiled from both reductionist in vitro studies and more recent holistic, systems-based approaches.
It has long been demonstrated that pyruvate decarboxylase (PDC) as well as TCA cycle dehydrogenases displays product inhibition in vitro by NADH [for a review, see Ref. (238)]. Accordingly, given that the in vivo activities of PDC and other TCA cycle enzymes are responsive to the NADH/NAD+ ratio, this provides a very sensitive mechanism by which it is possible to balance the rate of pyruvate oxidation by PDC and the TCA cycle activity with the rate of oxidative phosphorylation (272). Given that NAD+ is a common co-factor of three enzymes of the TCA cycle proper as well as by the PDC, it is reasonable to assume that mitochondrial NADH/NAD+ ratio has a major impact on the flux through the TCA cycle. However, it is equally important to note, with regard to these enzymes, that elegant studies from the Møller laboratory have revealed that the free NADH concentration is kept constant in plant mitochondria under different metabolic conditions, rendering it crucial to interpret implications of in vitro kinetics with caution (168). In addition to this regulation, the mitochondrial PDC is regulated by product inhibition by acetyl CoA (353) and activated by thiamine pyrophosphate (40).
The subsequent and first true reaction of the TCA cycle that was catalyzed by citrate synthase (CS) is also regulated by the cellular NADH/NAD+ ratio as well as activated by high ATP/ADP ratios and succinyl CoA levels, suggesting that its activity is tightly regulated at the metabolite level (253). The next step of the TCA cycle is catalyzed by aconitase, which catalyzes the reversible hydration of cis aconitate to either citrate or isocitrate. Irrespective of how many genes encode this enzyme, it appears that activities are localized in the mitochondria and cytosol (13, 53) and it is also well documented as being highly sensitive to oxidative stress (194, 258, 312). It has, furthermore, been demonstrated that the lack of manganese superoxide dismutase results in the inhibition of aconitase and the subsequent enzyme of the cycle, NAD-dependent ICDH (240). NAD-dependent ICDH additionally demonstrates product inhibition by NADH, and it is also inhibited by NADPH (192). The next reaction of the cycle that was catalyzed by the 2OG dehydrogenase complex (OGDHC) represents a metabolic branch point connecting 2OG (and the TCA cycle) with nitrogen assimilation with 2OG either being irreversibly degraded by the OGDHC or providing carbon skeletons for nitrogen assimilation. OGDHC is allosterically regulated in response to second messengers and metabolic indicators, such as Ca2+, ATP/ADP, NADH/NAD+, and thiamine pyrophosphate (11). The subsequent enzyme of the cycle, succinyl CoA ligase (ScoAL), is feedback inhibited by intermediates of the pathway of porphyrin biosynthesis as well as competitively inhibited by malonate in the reverse direction but is activated by 2OG and inhibited by both citrate and isocitrate and all downstream intermediates of the TCA cycle when assayed in the forward direction (331).
Succinate dehydrogenase (SDH), also commonly referred to as complex II, plays a dual role in mitochondrial metabolism both as a member of the electron transport chain and TCA cycle (9, 148). The regulation of the enzyme was investigated in coupled mitochondria by simultaneously measuring oxygen uptake rates and ubiquinone reduction levels (2). This study revealed that the activation state level of the enzyme is unambiguously reflected in the kinetic dependence of the succinate oxidation rate on the ubiquinone redox poise. Kinetic results indicated that it is additionally activated by both ATP and ADP (2).
Allosteric properties of fumarase from pea (Pisum sativum L.) revealed inhibition of this enzyme by physiological concentrations of pyruvate, 2-OG and the adenine nucleotides ATP, ADP, and AMP (33). Accordingly, downregulation of this enzyme in tomato resulted in a relatively large reduction in the rate of respiration in comparison to the majority of other enzymes of the cycle (254). The cycle is completed by NAD-dependent MDH, which catalyzes the reversible oxidation of malate to produce OAA (255, 352). While the equilibrium position favors malate and NAD+ production, the in vivo removal of OAA by CS, coupled with the removal of NADH by the respiratory chain, causes the reaction to function in the direction of malate oxidation in most tissues (253). Therefore, it is again likely that accumulation of NADH would lead to an inhibition of the mitochondrial MDH activity, although as shown by the modeling study of (126) mentioned earlier, the additional presence of malic enzyme in plant mitochondria can generate the pyruvate required to operate in the direction of malate production.
In addition to these allosteric changes, some of the enzymes of the TCA cycle are regulated at the level of protein abundance—for example, in response to oxidative or flooding stress or during the diel cycle (179, 193, 341, 342). Similarly, they are potentially regulated by a wide number of post-translational modifications such as phosphorylation, Trx-mediated redox status regulation, and lysine acetylation (21, 89, 134). Indeed, interrogation of compiled metabolomics responses to abiotic stress reveals that the cellular levels of TCA cycle intermediates respond dramatically to multiple cues, including pharmacological manipulation of redox status, tissue oxygenation, and circadian clock-mediated processes (40, 99, 177, 257, 294). Returning to the sum effect of the TCA cycle dehydrogenases, it is clear that the redox balance of the NADH/NAD+ pool is set by the balance of electron influx and efflux (see Fig. 5)—an equation complicated considerably in plants by the action of alternative NADH dehydrogenases which are upregulated under conditions of stress (279, 309) and, to a lesser extent, by the operation of the NAD+ transporter, NDT2 (263). Furthermore, as stated earlier, the finding that the free NADH concentration is kept constant in plant mitochondria needs to be taken into account when analyzing the regulation of the TCA cycle. Indeed, the identification that many of the enzymes of the cycle bind to Trxs (21, 389) suggests that redox status regulation is more likely mediated by the NTR system than by direct allosteric considerations.
Mitochondrial contribution to energy metabolism—(photo)respiration, amino acid, lipid, and vitamin synthesis
In addition to the TCA cycle, other major redox-related pathways in mitochondria include the photorespiratory amino-acid metabolism mentioned earlier as well as cysteine, proline, and branched chain amino-acid and lipid and vitamin metabolism (334). In photosynthetically active tissues, the photorespiratory enzymes, glycine decarboxylase and serine hydroxymethyltransferase, are among the most prominent proteins of the mitochondrial matrix (31). As mentioned earlier, it is well established that glycine produced by photorespiration is taken up by mitochondria and oxidized by these two enzymes (85). Oxidation of NADH produced on glycine oxidation occurs preferentially over that from other substrates such as malate or succinate (37, 73). These findings have led to suggestions that protein complexes located in the vicinity of the respiratory chain may give preferential access of specific reducing equivalents to the respiratory chain (185); however, direct evidence for this is currently lacking. Irrespective of how glycine oxidation is prioritized, it is clear that the NADH requirement of peroxisomal hydroxypyruvate reductase is stoichiometrically equivalent to the NADH production by glycine oxidation in the mitochondrial matrix, leading to the proposal that the NADH produced in the mitochondria is utilized in the peroxisome (162). However, given that only 25% to 50% of the redox equivalents produced in the mitochondrial matrix are exported, another source of NADH is clearly required to cover this shortfall (304). Experimental evidence suggests that the activity of the malate-OAA shuttle in the chloroplastic envelope is sufficient to meet this demand (132). However, it remains likely that both chloroplasts and mitochondria simultaneously allocate some NADH to the peroxisomes although the exact contribution of the two sources remains to be quantified (186). Recently, a mechanism for facilitating the oxidation of photorespiratory NADH in the mitochondrion has been revealed. Biochemical and physiological analyses of a T-DNA insertional mutant of Arabidopsis deficient in the expression of the uncoupling protein AtUCP1 revealed a specific inhibition of photorespiration (337). Uncoupling proteins (UCPs) are integral to the inner mitochondrial membrane, where they catalyze proton conductance across this membrane, dissipating the mitochondrial proton gradient as heat (184, 366). It has been postulated that this is important when the demand for oxidation of NADH is high and may facilitate high TCA cycle flux (317). Consistent with this suggestion, the ucp1 mutants displayed a reduced photosynthetic carbon assimilation rate that was linked to a reduced rate of oxidation of photorespiratory glycine (337). The role of UCPs in other aspects of redox biology is discussed in detail next. As for the TCA cycle, operation of the mitochondrial steps of this pathway also requires the recycling of NAD+ in the mitochondrial matrix, a function that could be performed by the internal NADH dehydrogenase. Circumstantial support for this comes from the observation that the expression of a gene encoding an internal NADH dehydrogenase is strictly light dependent (332). Intriguingly, the abundance of this protein highly correlates with that of the alternative oxidase (AOX), raising the possibility of the operation of a truncated and entirely non-phosphorylating electron transport chain (85, 221).
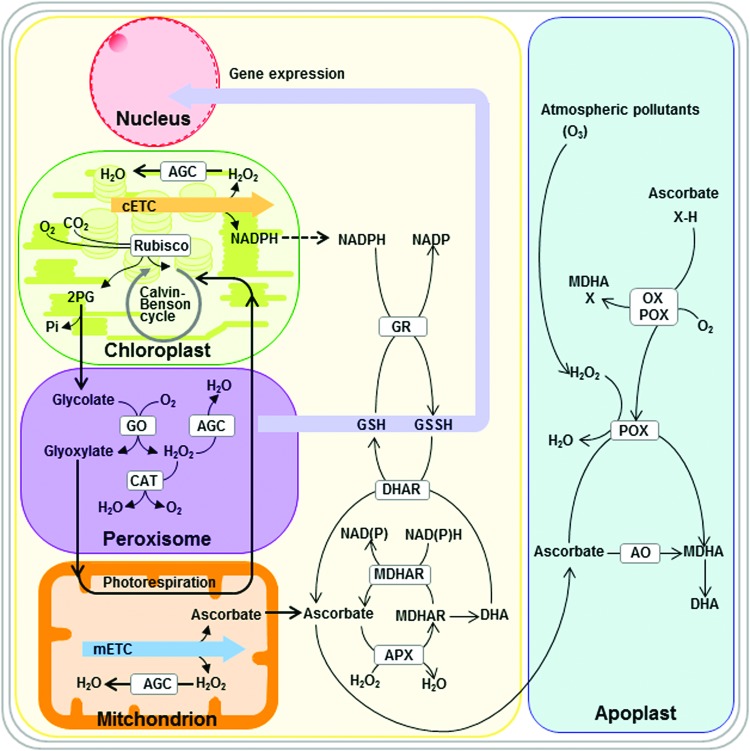
Also partially localized in the mitochondria are the enzymes of cysteine synthesis, although a full complement of these enzymes is also duplicated in the plastid and the cytosol (137). That said, the mitochondria is clearly highly important for the production of O-acetyl serine and, as such, is likely of high importance for the overall cellular redox balance (123, 380). Similarly, redox reactions play an important role in proline degradation with mutants of the delta1-pyrroline-5-carboxylate dehydrogenase, catalyzing the second step in this pathway, being hypersensitive to proline and leading to programmed cell death, callose deposition, ROS production, and DNA laddering via a salicylic acid-mediated signal transduction pathway (69). Catabolism of proline has been demonstrated to be highly important in plants, not only in programmed cell death (138), but also in protection of catalase, peroxidase, and complex II activities (127, 262), as well as potentially having an important role in stimulating seed germination (130). Beyond these metabolites and the alternative substrates of the mETC (discussed in the section below), the mitochondria are also the site of metabolism for, among others, fatty acids, vitamins, and haem (334) as well as have their own ascorbate-glutathione cycle (AGC) (57, 157). Due to space constraints, we cannot detail redox status constraints of the constituent reactions in the former processes nor the subsequent roles in redox regulation of their products. However, readers are referred to earlier articles for details thereof (157, 334) as well as to the next section on the cytosol in the case of the AGC.
The mETC
As already implied in the earlier sections, the mETC is intimately linked to the redox status of the mitochondria. As with other eukaryotes, the mETC of plants comprises four large protein complexes (Fig. 5). In contrast, to other eukaryotes, however, the plant mETC is highly branched (6, 277) and features several alternative pathways catalyzed by NAD(P)H dehydrogenases and by the AOX with the former bypassing complex I and the latter complexes III and IV (230, 233, 363). Both of these proteins decrease respiratory ATP output by 30%–60%, while the UCP discussed earlier enables proton flow that bypasses ATP synthesis, conferring additional flexibility to plant metabolism (277). That said, although the role for UCP is at least partially elucidated in plants, that for AOX remains elusive despite many hypotheses, including roles in minimizing ROS production and as a defence against metabolic oscillations (122, 215, 276, 291).
Recently, several additional electron inputs into the mETC have been characterized in plants; for example, the electron transfer flavoprotein: quinone oxidoreductase (ETFQO), L-galactono-1, 4-lactone dehydrogenase, and glycerol 3phosphate dehydrogenase (28, 155, 255, 310) and, indeed, many more redox centers are involved. Depending on their respective contributions, these alternative inputs could potentially have a major effect on the redox status of the mitochondria and beyond. However, to get a better understanding, this requires that we initially consider the conventional cytochrome pathway. In all mitochondria, the principal respiratory transfer is through four membrane-bound proton complexes that catalyze electron transfer from NADH and FADH2 to oxygen. The two-electron reduction of ½ O2 by NADH involves a reduction potential difference of 1.14 V, which translates into 219.2 kJ of free energy released for every mole of NADH oxidized. The mETC, thus, facilitates stepwise modestly exergonic redox reactions rather than a single explosive one. As mentioned earlier, the plant mETC is augmented by a wide variety of novel or newly uncovered pathways for the oxidation of NAD(P)H and the reduction of oxygen. In brief, the cytochrome pathway links four multiple subunit protein complexes. Complex I is an NADH dehydrogenase that oxidizes the NADH generated in the mitochondrial matrix by the operation of the TCA cycle. Complex II, which includes one of the constituent enzymes of the TCA cycle, namely SDH, oxidizes succinate to fumarate. Similar to Complex I, Complex II transfers electrons to ubiquinone. Ubiquinone can carry one or two electrons, and both fully oxidized and fully reduced molecules are freely diffusible within the inner membrane, enabling it to shuttle electrons from Complexes I and II to Complex III. Complex III, subsequently, transfers electrons from ubiquinone to cytochrome c, a small peripheral membrane protein that carries one electron at a time from Complex III to Complex IV (also known as cytochrome c oxidase), the terminal electron carrier in the chain. For every four molecules of cytochrome c oxidized, one molecule of oxygen is reduced to two molecules of water.
While of critical importance to plants, the cytochrome pathway is augmented by multiple other elements in plants; arguably, the best researched are the alternative dehydrogenases and the AOX. The type II NAD(P)H dehydrogenases are located on the internal and external sides of the inner mitochondrial membrane and oxidize NADH or NADPH from the mitochondrial matrix or the cytosol (206, 276, 277). These reactions bypass the first energy conservation step maintained by the proton pumping complex I, which oxidizes matrix NADH. Among the type II NAD(P)H dehydrogenases, NDB proteins of potato (Solanum tuberosum) and Arabidopsis (Arabidopsis thaliana) are present on the external surface of the inner mitochondrial membrane and can be subject to calcium-mediated regulation (80, 107, 276). NDA and NDC proteins are, by contrast, matrix directed (276).
The function of these internal dehydrogenase is by no means fully elucidated; however, the fact that the Km for NADH of the internal alternative NADH dehydrogenase(s) is 10-fold higher than that of complex I (235, 278) renders this enzyme likely to oxidize NADH only under high matrix NADH concentrations, for example those attained during photorespiration (74, 150, 151). Circumstantial support for such a role is provided by the characterized light- and photoreceptor-dependent upregulation of nda1 gene expression in A. thaliana (81, 275). However, further study is required to ascertain how the interplay between complex I and these dehydrogenases is regulated.
The role of the NDB, however, is much better understood in plants. Nicotiana sylvestris overexpressing StNDB1 displayed a decrease in the NADPH/NADP+ ratio independent of changes in the NADH/NAD+ ratio in illuminated leaves. This, thus, demonstrated that the NDB1 enzyme was active and able to specifically modify the cellular NADP(H) pools, although given that no visual phenotype was observed it was difficult to conclude the physiological importance of mitochondrial NADPH oxidation (205). However, in a subsequent study, it was demonstrated that plants overexpressing StNDB1 exhibit an earlier transition from rosette stage to bolting, whereas a line suppressing the expression of both StNDB1 and NsNDB1 is delayed in this parameter. The phenotype was strongly correlated with stem-specific changes in NADPH reduction levels (206), supporting an important role of this enzyme in mediation of stem physiology. It remains highly likely, however, that the NDBs as well as NDAs and NDC have further functions which remain to be elucidated.
Another protein proving rather difficult to pin down a physiological role on is the AOX. As mentioned earlier, this protein, similar to the UCP, provides a means to relax the coupling between respiration and ATP production and it appears to be particularly important under stress (359). A small number of thermogenic plants use AOX in order to generate heat from respiration (228, 370, 377); however, given the limited range of plants possessing this capacity, the function of AOX should differ from this in most species. Specific AOX gene family members are strongly induced at the transcript and protein level by complex III or complex IV dysfunction (167, 361), suggesting that AOX expression is highly responsive to insufficient cytochrome pathway capacity downstream of the ubiquinone pool. However, AOX is also commonly induced by complex I dysfunction and by other disruptions in respiratory metabolism such as the inhibition of ATP synthase, uncoupling of the mETC, and inhibition of the TCA cycle (164, 362). However, since some other studies reported no change in AOX levels in response to dramatic changes in the ETC (319), it would seem reasonable to conclude that the expression level of AOX is governed by multiple and complex signals from the mETC (359). Isotope discrimination studies have revealed that AOX activity does not correlate directly with protein abundance (369) and that it is subject to a wide range of post-translational modulations. First, the plant AOX is embedded within the inner mitochondrial membrane as a homodimer that is either non-covalently linked (the reduced, active form) or covalently linked by a regulatory disulfide bond between the two monomers (the oxidized, inactive form) (354). The responsible residue, a conserved cysteine toward the N-terminus and exposed within the matrix, is responsible for the formation of this disulfide bond (364). Reduction of the disulfide bond is facilitated by the oxidation of specific TCA cycle substrates and, based on the substrate specificity, it is hypothesized that specifically NADPH provides the reducing power for this regulatory reduction (360). Furthermore, a mitochondrially localized Trx has been demonstrated to be able to reduce this disulfide bond (108). Once reduced, AOX is sensitive to activation by specific organic acids, most notably pyruvate (229, 260, 360). Recent studies indicate that this effect is due to the enzyme's apparent Vmax resulting from the ability of pyruvate to stabilize the active AOX (54).
L-galactono-1, 4-lactone dehydrogenase, the terminal enzyme of ascorbate biosynthesis is associated to the cytochrome pathway although its precise localization was unclear (28, 227). A recent study showed that it is a component of Complex I (305), while reverse genetics and inhibitor studies have indicated the responsiveness of ascorbate biosynthesis to inhibition of respiration (227, 255). This link is rather intriguing, as it may represent a powerful manner by which to integrate organellar energy production; however, recent observations summarized earlier suggest that it is far from fully understood.
In addition, an input from a mitochondrial ubiquinone-reducing glycerol-3-phosphate dehydrogenase has been characterized in plants (310, 311). Thus, as in animals, it seems likely that a glycerol-3-phosphate shuttle exists in which an NADH-dependent glycerol-3-phosphate dehydrogenase produces glycerol-3-phosphate which permeates the outer mitochondrial membrane and gets oxidized by the ubiquinone reducing glycerol-3-phosphate dehydrogenase is present on the outer surface of the inner mitochondrial membrane with the dihydroacetone phosphate formed being recycled back to the cytosol (46, 129). However, despite the presence of the required enzymatic complement in plants, as yet no transporter of glycerol 3 phosphate has been identified. Furthermore, transcript levels of components of the putative glycerol 3 phosphate shuttle neither correlate with each other nor correlate with the NADH dehydrogenases (277); however, as the authors of this analysis suggest, it is conceivable that their co-regulation occurs at the enzyme level.
A third recently uncovered route of electron donation is that afforded by the ETF complex which was first discovered in plants after transcript profiling of senescent plants (47) and subsequently directly demonstrated to be functionally linked to branched chain amino-acid and lysine degradation by the isovaleryl and hydroxyglutarate dehydrogenases, respectively (6, 155). In utilizing this system, the plant cell considerably increases the energy efficiency of protein degradation by both providing substrates for the TCA cycle and directly donating electrons to the mETC (10).
Since genotypes of Arabidopsis deficient in the expression of at least some of the proteins involved in these alternate electron donor systems exist (6, 113, 155, 205, 310, 311, 330), future studies should be able to assess the hierarchy of their contribution to respiration across a range of environmental conditions.
Redox status signaling and other mitochondrially emitted signals
Given the extensive discussion of the metabolic control of the major redox systems and the fact that mitochondrial redox signaling has been recently comprehensively and expertly reviewed (309), we will only highlight a few brief aspects of this vast topic here and refer the reviewer to this and other previous reviews (93, 334) for details. Similar to the plastid, the mitochondrion originated from a bacterial symbiosis and still harbors its own genome. As such, redox status signaling from the mitochondria alongside other has wide implications. Although not traditionally regarded as major sources of ROS in leaves, it is well known that reactions associated with Complex I and Complex III produce superoxide (233) and as mentioned earlier, many mitochondrial enzymes are highly susceptible to oxidative damage. Even though on a cellular basis mitochondrial ROS production in the illuminated leaf is relatively modest, this does not preclude a role in setting the cellular redox status, particularly given that the ROS detoxification capacity is relatively small in comparison to that housed by the plastid or peroxisome (93). However, similar to the other organelles, mitochondria house both enzymic and non-enzymic antioxidants (157, 278), including a Trx system. While retrograde signaling from mitochondria is far less well characterized than its plastidial counterpart with no single pathway being established thus far (309), several candidate redox-related signals have been proposed, including superoxide, NO, H2O2, ascorbate, glutathione, lipid peroxide, peroxynitrite, lipoic acid, cysteine, NAD+/NADH, and oxidized proteins [for a detailed discussion, see Ref. (309)] as well as TCA cycle organic acids (88). The superoxide radical anion is not membrane permeable and has a very short half life in the mitochondrion. Hence, a potential superoxide sensor would need to reside in close proximity to the location of its generation, particularly given the high activity of Mn-superoxide dismutase. However, several mitochondrial proteins, including aconitase, are highly susceptible to oxidation by this radical (240); therefore, a possible signaling role for it should not be dismissed too quickly. The possibility that mitochondrial NO signaling plays an important role in cellular co-ordination is reasonably supported by experimental evidence, despite the fact that a classical NO synthase has as yet not been identified in plants (122). NO inhibits aconitase and via nitrosylation glycine decarboxylase and constituents of the mETC (122, 263). It furthermore reacts with superoxide to form peroxynitrite, which can influence tyrosine-dependent kinase signaling pathways (357). Little is currently known with regard to mitochondrially initiated H2O2 signaling. As mentioned earlier, ascorbate is produced in the mitochondria and has strong effects on the transcription of a wide range of nuclear and photosynthetic genes (174, 316, 355), and recent studies imply important roles of ABI4 and the cullin protein degradation network in setting or mediating this signaling system (172, 374). While synthesized in the plastid rather than in the mitochondria, the mitochondrial pool of glutathione is particularly large, stable, and independent from variations in cellular levels (390). This pool is, however, sensitive (in terms of oxidation) to changes envoked by mutations in either the mETC or other elements of the mitochondrial antioxidant system (75, 87, 240, 396), suggesting that it may work as a good signaling molecule most likely exerting its effects by oxidation of nearby protein thiols. Lipoic acid and free cysteine are proposed to signal in an analogous manner (309). Protein oxidation could be mediated by a broad number of different systems, including peroxidases, Trxs, and glutaredoxins, with the evidence for the importance of each of these being critically discussed by (309). Finally, returning to molecules intimately related to the TCA cycle, both NAD+/NADH and the TCA themselves need to be considered in the context of mitochondrial signaling. We have discussed the regulation of mitochondrial NAD+ levels in detail earlier. Further to this, it is intriguing that the tobacco cytoplasmic male-sterile (CMS) II mutant revealed a massively altered cellular redox status homeostasis as a consequence of an impaired function of Complex I (75). In mammals, sirtuin type protein deacetylases act as metabolite sensors for NAD+ (367). The study of sirtuins in plants, alongside that of mutants of the recently identified mitochondrial NAD+ transporter (264) will be instrumental in furthering our understanding as to the extent of NAD+ signaling in plants. Similarly, poorly characterized as yet in plants are carboxylic acid signaling pathways, although first works explicitly addressing this are now beginning to be published (88). Earlier studies focused on using the AOX transcript as a marker for such retrograde signaling due to the large increases in the expression of this gene after incubation with exogenous carboxylic acids (120, 260). More recent studies demonstrated that a similar response can be achieved by a non-metabolizable analog of citrate, suggesting specificity and that this response was not abolished in known hormone signaling mutants (88). However, as for all of the candidate signals, far more work will be needed to firmly establish the underlying reception and relay pathways. To summarize, signaling from the mitochondria is likely to be a highly complex network of overlapping responses. Once one of these pathways is fully elucidated, it would seem likely that we will be able to get a better handle on the others and finally an understanding of the hierarchy of their control and influence.
Peroxisomal Redox Biology
Although emerging roles for peroxisomes include participation in the biosynthesis of the plant hormone auxin and the signaling molecule jasmonic acid (JA) and role in sulfur and nitrogen metabolism (147, 182, 251, 284, 329, 394), the defining role of this organelle, at least in photosynthetically active tissues, is participation in photorespiration and concomitant production of H2O2 (Fig. 6). Indeed, a large proportion of proteins identified in peroxisomal proteomic studies are identified as reductases or dehydrogenases [see Ref. (147) and references therein], reflecting the importance of this organelle in cellular redox balance. The β-oxidation of fatty acids, as well as the role of peroxisomes in JA and auxin metabolism, is dependent on the redox status aspect of the peroxisome; while its role within photorespiration is also intimately related to the redox balance (31, 95, 147). For this reason, we will briefly describe the operation of the steps of each of these metabolic pathways, which is confined within the peroxisome before detailing their metabolic interaction with other compartments and signaling aspects of peroxisomally generated redox species.
FIG. 6.
Cellular and apoplastic reactions involved in hydrogen peroxide (H2O2) metabolism, its generation by photorespiration (operating in the chloroplast, peroxisome, mitochondria, and even in the cytosol; not shown), and signaling through the ascorbate/glutathione cycle as well as the production of ascorbate, of which the terminal reaction catalyzed by galacto-lactone dehydrogenase is coupled to the mETC from where it is transported via an as yet unknown mechanism to the apoplast. AGC, ascorbate-glutathione cycle; AO, ascorbate oxidase; CAT, catalase; cETC, chloroplast electron transport chain; DHA, dehydroascorbate; DHAR, DHA reductase; GO, glycine oxidase; GR, glutathione reductase; MDHA, monodehydroascorbate; MDHAR, MDHA reductase; OX, oxidase; POX, peroxidase. Modified from Munne-Bosch et al. (242). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
β-oxidation of fatty acids
The β-oxidation of fatty acids is an essential process in the mobilization of the oil reserves that seeds lay down during development, predominantly as triacylglycerol, in order to support post-germinative growth during seedling establishment (76, 118, 171). Fatty acids are transported into peroxisomes by a peroxisomal ABC transporter protein variously known as CTS, PXA1, or PED3 (91, 133, 208, 394). Subsequent β-oxidation is particularly important for initial stages of germination (16) as well as in reserve mobilization during developmental or dark induced senescence (72, 188, 315). The first step after import into the peroxisome of straight chain saturated fatty acids is oxidation catalyzed by a family of acyl-CoA oxidases (173) that are linked to flavin adenine dinucleotide and pass electrons to molecular oxygen to produce H2O2. Next, multifunctional proteins containing both hydratase and dehydrogenase domains act on the resultant 2-trans-enoyl CoA (12). Finally, the product of these reactions, 3-ketoacyl Co A is cleaved by thiolase to produce acetyl-CoA and acyl-CoA (109, 296). The β-oxidation of unsaturated fatty acids is more complicated and is reliant on the activity of accessory enzymes to convert these metabolites into suitable substrates for the pathway described earlier (116). However, essentially it operates in a very similar manner. The acyl-CoA oxidase reactions described earlier generate considerable amounts of H2O2, the majority of which is metabolized by catalase; however, when production is excessively high, for example during the mobilization of TAG that occurs during seedling establishment, membrane-bound ascorbate peroxidase and monodehydroascorbate reductase also operate to prevent leakage of H2O2 into the cytosol (77). In addition, an active ascorbate/glutathione system has been reported to help maintain redox balance in the peroxisome of pea leaves (157, 220). The acetyl-CoA produced during β-oxidation is respired by either mitochondria (189) or feeds into the glyoxylate cycle in which it is converted to succinate and malate and used for gluconeogenesis (274). During β-oxidation, the operation of the hydroxyacyl-CoA dehydrogenase activity produces NADH. The reoxidation of the co-factor, and by implication continued activity of the process, therefore, depends on a malate-OAA shuttle involving peroxisomal and cytosolic isoforms of MDH and operating in a similar manner to that defined earlier in the mitochondria. Intriguingly, mutants deficient in both peroxisomal isoforms of MDH germinate but are dependent on sucrose for establishment and mobilize TAGs at a considerably slower rate (273).
Hormone biosynthesis
Recently, redox status links to hormone biosynthesis and modification have been uncovered largely due to the clarification of the structure and localization of these pathways. The JA hormone family comprises JA itself, derivatives of JA, and its precursor 12-oxo-phytodienoic acid. The latter is produced in the chloroplast from which it is released by an as yet undefined mechanism (1). After the uptake of 12-oxo-phytodienoic acid into the peroxisome, it is reduced to 3-oxo-2-(29-[Z]-penenyl)cytopentane-1-octanoic acid (62, 300, 325). This product subsequently undergoes three rounds of β-oxidation to form JA (65, 175, 182). Importantly, 12-oxo-phytodienoic acid regulates seed germination with severe mutants in core β-oxidation function only being able to germinate if the testa is manually ruptured (90, 271). Evaluation of a range of peroxisomal uptake and activity mutants has recently indicated that the peroxisomal import and metabolism of 12-oxo-phytodienoic acid are important for germination (68). Similarly, the conversion of IBA to IAA occurs in the peroxisome. IBA is structurally similar to IAA, the principal form of auxin, and is known for efficacy in root induction (387). However, it is actually a protoauxin that is transferred or stored without auxin activity (314, 327). The predicted pathway for the metabolism of IBA also parallels β-oxidation of fatty acids, but its exact constitution has not yet been clarified [see Refs. (147, 328, 395) for details].
Photorespiration
Whilst the pathways described earlier clearly play highly important roles in seedling establishment as mentioned earlier, the four compartment-spanning pathway of photorespiration is the major role of the peroxisomes in photosynthetically active tissues (32, 147). Peroxisomally localized enzymes include glycolate oxidase (124), the catalase (220) serine-glycolate and glutamate-glycolate aminotransferases mentioned earlier (152, 202), the major isoform of hydroxypyruvate reductase (351), and a considerable MDH activity (64), rendering the peroxisome a major site of photorespiration (32). Glycolate is formed in the plastid via the dephosphorylation of 2PG (32), the product of the oxygenase reaction of Rubisco. It is exported to the cytosol by the glycolate/glycerate transporter (269) and is considered, by many, to simply diffuse into the matrix of peroxisomes (147). There, glycolate is oxidized, yielding H2O2 and glyoxylate as products (124, 295). As detailed earlier, the mitochondrial glycine decarboxylase decomposes glycine to CO2, NH3, and NADH transferring a C group to 5, 10-methyl tetrahydrofolate in the process (60); while SHMT attaches this methylene unit to a second glycine molecule to produce Ser, which diffuses back to the peroxisome and is transaminated by the serine-glycolate transferase to yield hydroxypyruvate. This intermediate is reduced by hydroxypyruvate reductase and NADH, which is provided by the operation of the peroxisomal MDH to form glycerate (64, 351). Given the importance of the hydroxypyruvate reductase reaction, a minor isoform of this enzyme is also localized to the cytosol (351). The exact reason for this dual localization is, at present, unknown; however, it has been speculated that it enables greater flexibility under a wide range of subcellular redox conditions (335). Whichever the route of hydroxypyruvate reduction, the glycerate produced is converted via glycerate kinase to the Calvin–Benson cycle intermediate 3PGA (43, 214). Intriguingly, by contrast to its C3 counterparts, the maize glycerate kinase enzyme is redox regulated via an additional C-terminal autoinhibitory domain, which forms a disulfide bridge in the dark, rendering the oxidized enzyme inactive (29). Attempts to optimize the potential of C3 plants by manipulating the photorespiratory pathway have been undertaken. Although promising, these have exposed unanticipated problems in that they exhibit many additional consequences likely due to the fact that the photorespiratory pathway is strongly embedded in both plant primary metabolism and subcellular redox balances (83, 169, 219, 267).
Peroxisomal transporters
Having described the major metabolic routes operating in the peroxisome, an obvious question is how they are sustained with regard to energy, reducing equivalents, and other co-factors. Great strides have been taken toward addressing these questions by the use of proteomics and reverse genetic studies in recent years. Arabidopsis PNC1 and PNC2 proteins are members of the mitochondrial carrier family (263), but they function as adenine nucleotide carriers. Both proteins import cytosolic ATP to support energy-consuming processes such as those regulated by the many kinases identified in the peroxisomal proteome (284, 289), as well as, perhaps more significantly, the pathway of β-oxidation described earlier. Interestingly, simultaneous repression of both genes after an RNAi strategy severely restricted this pathway, indicating the absence of a major, peroxisome-autonomous, ATP-generating system such as substrate level phosphorylation (5, 203). Similarly, the peroxisomal NAD+ transporter, PXN, is a highly abundant peroxisomal membrane protein (82, 98) that was additionally isolated in a screen for mutants of abnormal peroxisomal morphology (211). Despite having very high similarity to the PNCs, transport studies (39) revealed that it is functionally more closely related to the recently characterized mitochondrial and plastidic NAD+ transporters (264). Given that NAD+ is known to be synthesized exclusively in the cytosol (131, 249), it seems reasonable to assume, on the basis of the kinetic characterization of the recombinant protein (39), that PXN mediates an NAD+import/AMPexport antiport (147). By contrast to the specific transport of adenylates and NAD+, diffusion of carboxylic acids appears to be facilitated by a peroxisomal pore-forming channel (147, 286). Those from spinach leaves and castor beans are anion selective and enable diffusion of small carboxylic acids such as intermediates of photorespiration, β-oxidation, and the glyoxylate cycle (285–288). It is likely that further transport functions will be uncovered given that our understanding of peroxisomal metabolism has had to be expanded to include enzymes of the OPP (218), purine catabolism (93, 191), and polyamine metabolism (320).
The transporters described earlier are likely to be vital in co-ordinating not only cellular redox metabolism, but also transmission of redox-based signals. Peroxisomes are known to release signals that regulate nuclear gene expression, photomorphogenesis, plant development, light signaling, and stress responses as well as peroxisomal biogenesis itself (93, 100, 358, 379). The nature of the signals responsible are not fully defined; however, three candidates that are regularly discussed are H2O2, NO, and redox hormone interactions (93). In the case of H2O2, insufficient activity of catalase in C3 plants has been documented, during high rates of photorespiration, and results in a marked accumulation of oxidized glutathione (379). Furthermore, even with a complete complement of catalase, photorespiration-linked alteration of the redox states of ascorbate and glutathione can occur transiently (250). It is possible that the glycollate oxidase reaction acts as an important mediator of H2O2-derived signals (295) as could the superoxide produced on the conversion of xanthine to urate (220) and even the acyl-CoA oxidase (220). It is still unclear how the signal is relayed within the cell with both direct signaling (232) and signaling by oxidized peptide intermediaries (236) being debated. The strongest arguments for the latter theory would be their specificity as well as the fact that a combination of cytoplasmic streaming and degradation of H2O2 would likely render direct signaling unlikely. Indeed, elegant mathematical modeling provides strong support for this theory in that while it demonstrated that signaling over the distance equivalent to that typical between the plasma membrane and the nucleus would theoretically be sustainable tellingly, this requires a much faster enzymatic degradation and a much lower background concentration of H2O2 than has been observed experimentally (234, 368). Another possible signaling candidate is the well characterized, yet somewhat controversial NO (93, 121). This intermediate is produced in peroxisomes by the reduction of nitrite as catalyzed by the peroxisomal enzyme xanthine oxidoreductase—the redox regulation of which can reversibly convert the enzyme function from a xanthine dehydrogenase into a xanthine oxidase. A role for this enzyme in signaling has been postulated in pea (318) as well as on phosphate deficiency in cluster roots of white lupin (Lupinus albus) (373), and there is a large body of evidence that NO has a signaling function in plants (121). The third possibility is that the redox status of the peroxisome interacts with hormone signaling particulary with regard to those phytohormones whose synthesis are closely linked to β-oxidation-like processes in the mitochondria. This possibility has recently been expertly critically discussed in the context of the overlap between peroxisomal function and plant defence (320). In addition to these direct signals, the impact of peroxisomal activity on cytosolic metabolite pool sizes and redox status should also be considered as potential indirect signaling mechanisms by which the peroxisome can influence both nuclear gene expression and, either subsequently or in parallel, cellular phenotypes. As recently discussed (52), the application of integrated functional genomics approaches and modeling will likely enable greater resolution of these aspects.
Cytosolic Redox Biology
ROS have received great attention due to both their inherent reactivity and their potential as signaling molecules (4, 66, 231, 242). That said, despite intense research activity, the exact mechanisms underlying this signaling remain controversial and it is still uncertain whether superoxide, hydrogen peroxide, or singlet oxygen are themselves signaling molecules or whether they rather merely provide an environment that is a suitable conduit for signaling by other pathways (242). The primary targets of ROS signals appear to be amino acids such as Cys and small molecule thiols such as glutathione (66), although their abundance is additionally closely linked to that of the signal molecule nitric oxide (245), rendering the unraveling of the underlying mechanisms even more problematic. The halftimes of ROS turnover are determined by the antioxidant environment, which includes two factors. First, antioxidant enzymes such as catalase in the peroxisome, ascorbate peroxidase in the apoplast, and enzymes of the ascorbate/glutathione cycle in the cytosol (Fig. 6), and second, small-molecule antioxidants such as ascorbate and glutathione as well as the plastidial carotenoids and tocopherols (242). Alongside peroxiredoxins localized in the plastid, cytosol, and mitochondria, these small-molecule antioxidants play a major role in the detoxification of ROS (97). Returning specifically to the cytosol while both ascorbate and glutathione pools have been clearly demonstrated to act as redox buffers, our current understanding suggests that these pools, nevertheless, appear to have quite distinctive functions within ROS signaling. The principal effect of ascorbate has been suggested to be in setting the thresholds for cytoplasmic and apoplastic signaling (242). However, several recent studies provide evidence which would appear to suggest that its role transcends such limitations, suggesting roles in plant defence and maintenance of optimal photosynthesis (172, 255, 374). In contrast, glutathione appears to exert a greater influence via the hydrogen peroxide signaling pathways and is widely regarded as a common arbiter of the intracellular redox potential (94, 242). However, the roles of both cytosolic ascorbate and glutathione are reviewed in detail in several excellent recent articles (94, 96, 242), so we will not dwell on them here.
Relatively little is known about the functional role of cytosolic Trxs, comprising mainly Trx h isoforms (h1-h6 in Arabidopsis), which are reduced by NTRA and NTRB using NADPH as a redox donor (282). The redox status of the NADP(H) system in the cytosol is most likely determined by (i) the activities of G6PDH and 6PGDH regulating NADPH provision by the OPP and (ii) NADP-ICDH catalyzing NADPH metabolism as a part of the amino-acid synthesis pathway. In Arabidopsis, there are six G6PDH isoforms with two (G6PD5 and G6PD6) located in the cytosol, with G6PD5 being insensitive to redox changes, while G6PD6 is subject to inactivation by oxidation (372). The latter has been found to be regulated by protein phosphorylation with cytosolic glycogen synthase kinase 3 leading to its activation (67). Glycogen synthase kinase 3 might play a crucial role in counterbalancing oxidative inhibition of G6PD6 by phosphorylating and thereby enhancing cytosolic G6PD6 activity. Intriguingly, Arabidopsis plants overexpressing glycogen synthase kinase 3 have increased G6PDH activity and lower levels of ROS in response to stress and are more tolerant to salt stress (67). In confirmation to this, changes in G6PDH expression in the cytosol of transgenic tobacco plants provide evidence for a role of cytosolic G6PDH in biotic and abiotic stress tolerance (301).
While the redox status of the cytosol seems to be well buffered under normal conditions, large changes in the cytosolic redox balance can occur in response to environmental stress conditions, such as hypoxia. Oxygen deficiency inhibits respiration and leads to a subsequent increase in the NADH/NAD+ ratio in the cytosol. The resulting decrease in NAD+ recycling is critical for the operation of glycolysis, as it will limit NAD-GAPDH activity. Plants, therefore, respond to low oxygen concentrations by inducing hypoxic genes involved in fermentative pathways in the cytosol, which convert pyruvate to lactate, ethanol, or other products to recycle NAD+ from NADH to enable glycolysis and its attendant ATP production to proceed (102). There has been rapid progress in our understanding of the underlying low oxygen sensing and signaling pathways in the last years. Recent reports indicate group VII ethylene response factor (ERF VII) transcription factors such as HRE1, HRE2 (200), RAP2.2, and RAP2.12 (142) as important regulators of hypoxic gene expression and survival in Arabidopsis plants. Intriguingly, it was shown that group-VII ERFs are stabilized under hypoxia and degraded on re-oxygenation via the N-end rule pathway of targeted proteolysis, functioning as an oxygen-sensing mechanism in Arabidopsis plants (110, 199). In animals and plants, N-end rule-regulated proteolysis involves spontaneous or enzymatic oxidation of an exposed Cys to Cys-sulfinate or further to Cys-sulfonate in an oxygen-, ROS-, or NO-dependent manner (365). The role of the redox status of the tissue in modulating oxidation of the penultimate Cys of group-VII ERFs in response to changes in oxygen concentrations has not yet been investigated. Moreover, it will be interesting to study the impact of this oxygen-sensing pathway on adaptive responses in metabolism to balance the redox and energy status of the tissue in response to changes in oxygen concentrations.
Integration at the Cellular Level
Plastid signals play important roles in various cellular processes that are vital to the plant by influencing nuclear gene expression during different stress conditions (24). This includes different signal components related to photosynthetic electron transport, changes of the chloroplast redox state, accumulation of ROS, and metabolic intermediates (i.e., protoporphyrin IX or sugars). These aspects of plastid retrograde signaling to the nucleus have been presented by many excellent recent reviews and will not be covered here (24, 58, 195, 268, 340, 344, 386). However, there are also signals transferred between plastids and mitochondria to coordinate their metabolic activities. In the next section, we will discuss the role of redox status signals involved in the communication between these two organelles.
Turning attention to cellular processes that are regulated by metabolic redox signals emanating from the mitochondria, recent reverse genetic and functional genomic studies have consolidated the role of the mitochondria in optimizing photosynthesis (252) and elucidated roles for the mitochondria in the regulation of normal fruit ripening (55). Considerable cumulative evidence has accrued suggesting a vital role for mitochondrial function during the photosynthetic process (256). Depending on a plant`s developmental stage and/or environmental considerations, reducing equivalents generated by the photochemical reactions accumulate in the chloroplast stroma, causing over-reduction of the photosynthetic electron transport chain and the generation of ROS, leading to photoinhibition (92). It is generally accepted that this excess of reducing equivalents can be dissipated by their export from the chloroplast to the mitochondria via the malate valve (186, 302); once within the mitochondria, these reducing equivalents are oxidized by the mitochondrial respiratory chain (248), thus enabling continued high rates of photosynthesis. However, while the reduced rates of photosynthesis in UCP mutants provided some weak circumstantial evidence for the importance of the operation of the malate valve (337), mutants in the plastidial NADP-dependent MDH essential for its operation revealed that the plant harbors additional mechanisms to protect against photoinhibition (135). A second link between these organelles, one working in the opposite direction, was recently uncovered after the downregulation of the mitochondrial isoform of MDH in tomato (255). These studies demonstrated that plants deficient in this TCA cycle enzyme were able to effectively utilize L-galactono-1,4—lactone as an alternative electron donor to the mETC, resulting in a considerable increase in ascorbate content. Intriguingly clear and direct evidence for such an interaction between respiration and ascorbate biosynthesis was also provided by Bartoli et al. (28). Another consequence of the inhibition of the mitochondrial MDH is that the transgenic plants have enhanced rates of photosynthesis and growth. However, the exact mechanism by which this is achieved is not known. It is speculated that this is a direct result of the upregulated ascorbate biosynthesis (255) likely via one of the myriad of processes by which ascorbate can exert an effect on photosynthesis (316). Perhaps unsurprisingly, this effect seems to be highly environmentally dependent, neither being observed in short day conditions in tomato (256) nor in corresponding mutants in Arabidopsis (352). The role of photorespiration in linking chloroplastic and mitochondrial metabolism and function has also been the subject of multiple studies (32, 84, 349). While we have mainly dealt with it in the preceding sections, one recent study which is particularly pertinent to this discussion is the recent finding that overexpression of the H-protein of glycine decarboxylase considerably enhanced net photosynthesis and growth of A. thaliana (350). At the molecular level, lower glycine levels confirmed elevated glycine decarboxylase activity in vivo, and lower levels of the CO2 acceptor RuBP indicated higher drain from CO2 fixation. Thus, the photorespiratory enzyme glycine decarboxylase in mitochondria appears to constitute an important feedback mechanism that contributes to the control of the Calvin–Benson cycle in the plastid and, hence, carbon flow through both photosynthesis and photorespiration (350).
The second example is likely also to be mediated by the malate valve described earlier but this time in reverse direction that is, the redox status of the mitochondria is transmitted to the plastid and influences plastidial metabolism and function with subsequent whole cell and, subsequently, whole organ consequences (Fig. 7). It was again, at least partially, uncovered by the suppression of the expression of mitochondrial MDH alongside independent suppression of fumarase in tomato, but this time in a fruit-specific manner (55). Detailed characterization suggested that, although the rate of ripening was essentially unaltered in these lines, there were minor changes in the accumulation of pigments which are modulated in a redox-mediated manner (154, 243, 382). Furthermore, lines containing higher levels of malate were characterized by lower levels of transitory starch and lower soluble sugar content at harvest, whereas those with lower malate contained higher levels of these carbohydrates. Analysis of the redox-activation state of AGPase revealed that it correlated with the accumulation of transitory starch (55). Intriguingly, most likely as a consequence of the altered sugar content, these lines were characterized as having elevated or reduced shelf life and inversely reduced or elevated susceptibility to post-harvest bacterial infection. More recent studies have revealed that the operation of this reverse malate shunt is context dependent and does not work in fully heterotrophic tissues such as the potato tuber (339), but that similar effects can be brought about by altering the malate-related redox status balance in other compartments of the tomato fruit (261).
FIG. 7.
Model of the influence of mitochondrially derived malate on tomato fruit starch, soluble sugar content, post-harvest shelf life, and bacterial infection. Data are presented on the basis of analysis of transgenics lines described in (55). Mitochondrial malate dehydrogenase (MDH) lines [(A); increased malate]; fumarase lines [(B); decreased malate]. The same ripening and postharvest sequence is presented for both transgenic sets. (i) Alterations in mitochondrial redox status are transmitted, either within the same cell type or from adjacent tissues, to the plastid via the malate valve as described by (302). (ii) Altered plastidial redox status results in a decreased (MDH lines) or enhanced (fumarase lines) redox-activation state of AGPase and concomitant starch synthesis (as well as similar changes in the activation state of the plastidial MDH); whether this is mediated by the Trx or the NTRC pathway (Fig. 4) is currently unknown. (iii) This leads to redox-mediated alterations in pigment biosynthesis during ripening. (iv) Starch is rapidly broken down, leading to a decreased soluble solid content in red-ripe fruit in the MDH lines and an increased soluble solid content in the fumarase lines. (v) Potentially as a result of differences in cellular osmolarity, the transgenic sets oppositely display an increased water loss and wrinkling (MDH lines) or a decreased water loss and wrinkling (fumarase lines) that appears to be cell wall independent. (vi) These changes in water loss and wrinkling correlate positively to the rate of opportunistic pathogen infection in the transgenic sets, while the MDH lines are increasingly susceptible to Botrytis cincerea infection. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
In this review, we have discussed recent developments with regard to the redox status control of metabolism and the metabolic control of redox status, at both the organellar and cellular levels. It is becoming clear that redox status plays a central role in sensing physiological and environmental alterations within chloroplasts and mitochondria. These organelles respond actively to these inputs by regulating their own central metabolic processes via internal redox systems and by emitting interorganellar redox signals to maintain the homeostasis at a cellular level. Further work is required to resolve the network of intra- and interorganellar redox signals. This will require genetic and metabolomics approaches (52, 346) as well as the application of techniques to analyze redox states at the subcellular level (187, 309). Such methodological advances will be imperative to get a comprehensive understanding of how compartmental issues affect both the metabolic control of redox and its corollary, the redox control of metabolism.
Abbreviations Used
- 2OG
2-oxoglutarate
- 2PG
2-phosphoglycolate
- 3PGA
glycerate-3-phosphate
- 6PGDH
6-phosphogluconate dehydrogenase
- ACCase
acetyl-CoA carboxylase
- ADPGlc
ADP-glucose
- AGC
ascorbate-glutathione cycle
- AGPase
ADP-glucose pyrophosphorylase
- ALA
5-aminolevulinic acid
- AO
ascorbate oxidase
- AOX
alternative oxidase
- APS1
small subunit of AGPase
- CAT
catalase
- cETC
chloroplast electron transport chain
- Chl
chlorophyll
- CHLM
Mg protoporphyrin IX methyltransferase
- CS
citrate synthase
- Cyt b6f
cytochrome b6f complex
- DHA
dehydroascorbate
- DHAR
dehydroascorbate reductase
- ERF VII
group VII ethylene response factor
- ETFQO
electron transfer flavoprotein: quinone oxidoreductase
- FAS
fatty acid synthase
- FBPase
fructose-1,6-bisphosphatase
- Fdx
ferredoxin
- FNR
Fdx-NADP-reductase
- Fru6P
fructose-6-phosphate
- FTR
Fdx-Trx-reductase
- G6PDH
glucose-6-phosphate dehydrogenase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GAT
glycerol-3-phosphate acyltransferase
- GDC
glycine decarboxylase
- Glc1P
glucose-1-phosphate
- Gln
glutamine
- Glu
glutamate
- GluTR
glutamyl-transfer RNA reductase
- GO
glycine oxidase
- GOGAT
glutamine:oxoglutarate amino transferase
- GR
glutathione reductase
- GS
glutamine synthetase
- GWD
glucan water dikinase
- ICDH
isocitrate dehydrogenase
- ISA
isoamylase
- JA
jasmonic acid
- MDH
malate dehydrogenase
- MDHA
monodehydroascorbate
- MDHAR
monodehydroascorbate reductase
- mETC
mitochondrial electron transport chain
- MGD
monogalactosyldiacylglycerol synthase
- NDB
external mitochondrial NADPH dehydrogenase
- NDT
NAD+ transporter
- NiR
nitrite reductase
- NR
nitrate reductase
- NTR
NADPH-dependent thioredoxin reductase
- NTRC
NADPH-dependent thioredoxin reductase C
- OAA
oxaloacetate
- OGDHC
2OG dehydrogenase complex
- OPP
oxidative pentose phosphate pathway
- PC
plastocyanin
- PDC
pyruvate decarboxylase
- PDH
pyruvate dehydrogenase
- PGI
phosphoglucose isomerase
- PGK
phosphoglycerate kinase
- PGM
phosphoglucomutase
- PGRL1
proton-gradient-regulation-like protein 1
- Pi
inorganic phosphate
- PK
pyruvate kinase
- POR
NADPH:protochlorophyllide oxido-reductase
- POX
peroxidase
- PPi
inorganic pyrophosphate
- PQ
plastoquinone
- PRK
phosphoribulokinase
- Prx
peroxyredoxin
- PS
photosystem
- ROS
reactive oxygen species
- Rubisco
ribulose-1,5-bisphosphate carboxylase/oxygenase
- RuBP
ribulose-1,5-bisphosphate
- SBE
starch branching enzyme
- SBPase
sedoheptulose bisphosphatase
- ScoAL
succinyl CoA ligase
- SDH
succinate dehydrogenase
- SEX4
starch-excess-4 type glucan phosphatase
- SnRK1
SNF1-related protein kinase
- SS
starch synthase
- TCA
tricarboxylic acid
- TK
transketolase
- TR-BAMY
thioredoxin-dependent beta-amylase
- Tre6P
trehalose-6-phosphate
- TROL
thylakoid rhodanese-like protein
- Trx
thioredoxin
- UCP
uncoupling protein
Acknowledgments
This work on redox has been supported by the Deutsche Forschungsgemeinschaft (grants Ge 878/5-1 & 8-1 to P.G. and the Research Unit Promics FOR 1186 to A.R.F.).
References
- 1.Acosta IF. and Farmer EE. Jasmonates. Arabidopsis Book 8: e0129, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Affourtit C, Krab K, Leach GR, Whitehouse DG, and Moore AL. New insights into the regulation of plant succinate dehydrogenase—on the role of the protonmotive force. J Biol Chem 276: 32567–32574, 2001 [DOI] [PubMed] [Google Scholar]
- 3.Andriotis VME, Kruger NJ, Pike MJ, and Smith AM. Plastidial glycolysis in developing Arabidopsis embryos. New Phytol 185: 649–662, 2010 [DOI] [PubMed] [Google Scholar]
- 4.Apel K. and Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Arai Y, Hayashi M, and Nishimura M. Proteomic Identification and Characterization of a novel peroxisomal adenine nucleotide transporter supplying ATP for fatty acid beta-oxidation in soybean and Arabidopsis. Plant Cell 20: 3227–3240, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Araujo WL, Ishizaki K, Nunes-Nesi A, Larson TR, Tohge T, Krahnert I, Witt S, Obata T, Schauer N, Graham IA, Leaver CJ, and Fernie AR. Identification of the 2-Hydroxyglutarate and Isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Araujo WL, Nunes-Nesi A, and Fernie AR. Fumarate: multiple functions of a simple metabolite. Phytochemistry 72: 838–843, 2011 [DOI] [PubMed] [Google Scholar]
- 8.Araujo WL, Nunes-Nesi A, Nikoloski Z, Sweetlove LJ, and Fernie AR. Metabolic control and regulation of the tricarboxylic acid cycle in photosynthetic and heterotrophic plant tissues. Plant Cell Environ 35: 1–21, 2012 [DOI] [PubMed] [Google Scholar]
- 9.Araujo WL, Nunes-Nesi A, Osorio S, Usadel B, Fuentes D, Nagy R, Balbo I, Lehmann M, Studart-Witkowski C, Tohge T, Martinoia E, Jordana X, DaMatta FM, and Fernie AR. Antisense inhibition of the iron-sulphur subunit of succinate dehydrogenase enhances photosynthesis and growth in tomato via an organic acid-mediated effect on stomatal aperture. Plant Cell 23: 600–627, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Araujo WL, Tohge T, Ishizaki K, Leaver CJ, and Fernie AR. Protein degradation—an alternative respiratory substrate for stressed plants. Trends Plant Sci 16: 489–498, 2011 [DOI] [PubMed] [Google Scholar]
- 11.Araujo WL, Tohge T, Osorio S, Lohse M, Balbo I, Krahnert I, Sienkiewicz-Porzucek A, Usadel B, Nunes-Nesi A, and Fernie AR. Antisense inhibition of the 2-oxoglutarate dehydrogenase complex in tomato demonstrates its importance for plant respiration and during leaf senescence and fruit maturation. Plant Cell 24: 2328–2351, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arent S, Christensen CE, Pye VE, Norgaard A, and Henriksen A. The multifunctional protein in peroxisomal beta-oxidation structure and substrate specificity of the Arabidopsis thaliana protein MFP2. J Biol Chem 285: 24066–24077, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnaud N, Ravet K, Borlotti A, Touraine B, Boucherez J, Fizames C, Briat JF, Cellier F, and Gaymard F. The iron-responsive element (IRE)/iron-regulatory protein 1 (IRP1)-cytosolic aconitase iron-regulatory switch does not operate in plants. Biochem J 405: 523–531, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arsova B, Hoja U, Wimmelbacher M, Greiner E, Ustun S, Melzer M, Petersen K, Lein W, and Bornke F. Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 22: 1498–1515, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asada K. The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol 50: 601–639, 1999 [DOI] [PubMed] [Google Scholar]
- 16.Baker A, Graham IA, Holdsworth M, Smith SM, and Theodoulou FL. Chewing the fat: beta-oxidation in signalling and development. Trends Plant Sci 11: 124–132, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Ballicora MA, Frueauf JB, Fu YB, Schurmann P, and Preiss J. Activation of the potato tuber ADP-glucose pyrophosphorylase by thioredoxin. J Biol Chem 275: 1315–1320, 2000 [DOI] [PubMed] [Google Scholar]
- 18.Ballicora MA, Iglesias AA, and Preiss J. ADP-glucose pyrophosphorylase: a regulatory enzyme for plant starch synthesis. Photosynth Res 79: 1–24, 2004 [DOI] [PubMed] [Google Scholar]
- 19.Balmer Y, Koller A, del Val G, Manieri W, Schurmann P, and Buchanan BB. Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc Natl Acad Sci U S A 100: 370–375, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balmer Y, Vensel WH, Cai N, Manieri W, Schurmann P, Hurkman WJ, and Buchanan BB. A complete ferredoxin/thioredoxin system regulates fundamental processes in amyloplasts. Proc Natl Acad Sci U S A 103: 2988–2993, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balmer Y, Vensel WH, Tanaka CK, Hurkman WJ, Gelhaye E, Rouhier N, Jacquot JP, Manieri W, Schuurmann P, Droux M, and Buchanan BB. Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc Natl Acad Sci U S A 101: 2642–2647, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balsera M, Goetze TA, Kovacs-Bogdan E, Schurmann P, Wagner R, Buchanan BB, Soll J, and Bolter B. Characterization of Tic110, a channel-forming protein at the inner envelope membrane of chloroplasts, unveils a response to Ca2+ and a stromal regulatory disulfide bridge. J Biol Chem 284: 2603–2616, 2009 [DOI] [PubMed] [Google Scholar]
- 23.Bao XM, Focke M, Pollard M, and Ohlrogge J. Understanding in vivo carbon precursor supply for fatty acid synthesis in leaf tissue. Plant J 22: 39–50, 2000 [DOI] [PubMed] [Google Scholar]
- 24.Barajas-Lopez JD, Blanco NE, and Strand A. Plastid-to-nucleus communication, signals controlling the running of the plant cell. Biochim Biophys Acta 1833: 425–437, 2013 [DOI] [PubMed] [Google Scholar]
- 25.Barajas-Lopez JD, Tezycka J, Travaglia CN, Serrato AJ, Chueca A, Thormahlen I, Geigenberger P, and Sahrawy M. Expression of the chloroplast thioredoxins f and m is linked to short-term changes in the sugar and thiol status in leaves of Pisum sativum. J Exp Bot 63: 4887–4900, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barajas-Lopez JDD, Serrato AJ, Cazalis R, Meyer Y, Chueca A, Reichheld JP, and Sahrawy M. Circadian regulation of chloroplastic f and m thioredoxins through control of the CCA1 transcription factor. J Exp Bot 62: 2039–2051, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barkan A. Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol 155: 1520–1532, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartoli CG, Pastori GM, and Foyer CH. Ascorbate biosynthesis in mitochondria is linked to the electron transport chain between complexes III and IV. Plant Physiol 123: 335–343, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartsch O, Mikkat S, Hagemann M, and Bauwe H. An autoinhibitory domain confers redox regulation to maize glycerate kinase. Plant Physiol 153: 832–840, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartsch S, Monnet J, Selbach K, Quigley F, Gray J, von Wettstein D, Reinbothe S, and Reinbothe C. Three thioredoxin targets in the inner envelope membrane of chloroplasts function in protein import and chlorophyll metabolism. Proc Natl Acad Sci U S A 105: 4933–4938, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bauwe H, Hagemann M, and Fernie AR. Photorespiration: players, partners and origin. Trends Plant Sci 15: 330–336, 2010 [DOI] [PubMed] [Google Scholar]
- 32.Bauwe H, Hagemann M, Kern R, and Timm S. Photorespiration has a dual origin and manifold links to central metabolism. Curr Opin Plant Biol 15: 269–275, 2012 [DOI] [PubMed] [Google Scholar]
- 33.Behal RH. and Oliver DJ. Biochemical and molecular characterization of fumarase from plants: purification and characterization of the enzyme—Cloning, sequencing, and expression of the gene. Arch Biochem Biophys 348: 65–74, 1997 [DOI] [PubMed] [Google Scholar]
- 34.Benitez-Alfonso Y, Cilia M, Roman AS, Thomas C, Maule A, Hearn S, and Jackson D. Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc Natl Acad Sci U S A 106: 3615–3620, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benson A. and Calvin M. The dark reductions of photosynthesis. Science 105: 648–649, 1947 [DOI] [PubMed] [Google Scholar]
- 36.Benz JP, Stengel A, Lintala M, Lee YH, Weber A, Philippar K, Guegel IL, Kaieda S, Ikegami T, Mulo P, Soll J, and Boelter B. Arabidopsis Tic62 and ferredoxin-NADP(H) oxidoreductase form light-regulated complexes that are integrated into the chloroplast redox poise. Plant Cell 21: 3965–3983, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergman A. and Ericson I. Effects of PH, NADH, succinate and malate on the oxidation of glycine in spinach leaf mitochondria. Physiol Plant 59: 421–427, 1983 [Google Scholar]
- 38.Bernal-Bayard P, Hervas M, Cejudo FJ, and Navarro JA. Electron transfer pathways and dynamics of chloroplast NADPH-dependent thioredoxin reductase C (NTRC). J Biol Chem 287: 33865–33872, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernhardt K, Wilkinson S, Weber APM, and Linka N. A peroxisomal carrier delivers NAD plus and contributes to optimal fatty acid degradation during storage oil mobilization. Plant J 69: 1–13, 2012 [DOI] [PubMed] [Google Scholar]
- 40.Bocobza SE, Malitsky S, Araujo WL, Nunes-Nesi A, Meir S, Shapira M, Fernie AR, and Aharoni A. Orchestration of thiamin biosynthesis and central metabolism by combined action of the thiamin pyrophosphate riboswitch and the circadian clock in Arabidopsis. Plant Cell 25: 288–307, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bohne AV, Schwarz C, Schottkowski M, Lidschreiber M, Piotrowski M, Zerges W, and Nickelsen J. Reciprocal regulation of protein synthesis and carbon metabolism for thylakoid membrane biogenesis. PLoS Biol 11: e1001482, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bohrer AS, Massot V, Innocenti G, Reichheld JP, Issakidis-Bourguet E, and Vanacker H. New insights into the reduction systems of plastidial thioredoxins point out the unique properties of thioredoxin z from Arabidopsis. J Exp Bot 63: 6315–6323, 2012 [DOI] [PubMed] [Google Scholar]
- 43.Boldt R, Edner C, Kolukisaoglu U, Hagemann M, Weckwerth W, Wienkoop S, Morgenthal K, and Bauwe H. D-GLYCERATE 3-KINASE, the last unknown enzyme in the photorespiratory cycle in Arabidopsis, belongs to a novel kinase family. Plant Cell 17: 2413–2420, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brautigam K, Dietzel L, Kleine T, Stroher E, Wormuth D, Dietz KJ, Radke D, Wirtz M, Hell R, Dormann P, Nunes-Nesi A, Schauer N, Fernie AR, Oliver SN, Geigenberger P, Leister D, and Pfannschmidt T. Dynamic plastid redox signals integrate gene expression and metabolism to induce distinct metabolic states in photosynthetic acclimation in Arabidopsis. Plant Cell 21: 2715–2732, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Breazeale VD, Buchanan BB, and Wolosiuk RA. Chloroplast sedoheptulose 1,7-bisphosphatase—evidence for regulation by ferredoxin-thioredoxin system. Z Naturforsch [C] 33: 521–528, 1978 [Google Scholar]
- 46.Brown LJ, Macdonald MJ, Lehn DA, and Moran SM. Sequence of rat mitochondrial glycerol-3-phosphate dehydrogenase cDNA—evidence for ef-hand calcium-binding domains. J Biol Chem 269: 14363–14366, 1994 [PubMed] [Google Scholar]
- 47.Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, and Leaver CJ. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585, 2005 [DOI] [PubMed] [Google Scholar]
- 48.Buchanan BB. Role of light in the regulation of chloroplast enzymes. Annu Rev Plant Physiol Plant Mol Biol 31: 341–374, 1980 [Google Scholar]
- 49.Buchanan BB. and Balmer Y. Redox regulation: a broadening horizon. Annu Rev Plant Biol 56: 187–220, 2005 [DOI] [PubMed] [Google Scholar]
- 50.Buchanan BB. and Wong JH. A conversation with Andrew Benson: reflections on the discovery of the Calvin-Benson cycle. Photosynth Res 114: 207–214, 2013 [DOI] [PubMed] [Google Scholar]
- 51.Buchanan BB, Gruissem W, and Jones RL. Biochemistry & Molecular Biology of Plants. Rockville, MD: American Society of Plant Physiologists, 2000 [Google Scholar]
- 52.Caldana C, Fernie AR, Willmitzer L, and Steinhauser D. Unraveling retrograde signaling pathways: finding candidate signaling molecules via metabolomics and systems biology driven approaches. Front Plant Sci 3: 267, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carrari F, Nunes-Nesi A, Gibon Y, Lytovchenko A, Loureiro ME, and Fernie AR. Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol 133: 1322–1335, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carre JE, Affourtit C, and Moore AL. Interaction of purified alternative oxidase from thermogenic Arum maculatum with pyruvate. FEBS Lett 585: 397–401, 2011 [DOI] [PubMed] [Google Scholar]
- 55.Centeno DC, Osorio S, Nunes-Nesi A, Bertolo ALF, Carneiro RT, Araujo WL, Steinhauser M-C, Michalska J, Rohrmann J, Geigenberger P, Oliver SN, Stitt M, Carrari F, Rose JKC, and Fernie AR. Malate plays a crucial role in starch metabolism, ripening, and soluble solid content of tomato fruit and affects postharvest softening. Plant Cell 23: 162–184, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.This reference has been deleted.
- 57.Chew O, Whelan J, and Millar AH. Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J Biol Chem 278: 46869–46877, 2003 [DOI] [PubMed] [Google Scholar]
- 58.Chi W, Sun X, and Zhang L. Intracellular signaling from plastid to nucleus. Annu Rev Plant Biol 64: 559–582, 2013 [DOI] [PubMed] [Google Scholar]
- 59.Choi YA, Kim SG, and Kwon YM. The plastidic glutamine synthetase activity is directly modulated by means of redox change at two unique cysteine residues. Plant Sci 149: 175–182, 1999 [Google Scholar]
- 60.Collakova E, Goyer A, Naponelli V, Krassovskaya I, Gregory JF, III, Hanson AD, and Shachar-Hill Y. Arabidopsis 10-formyl tetrahydrofolate deformylases are essential for photorespiration. Plant Cell 20: 1818–1832, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Collin V, Issakidis-Bourguet E, Marchand C, Hirasawa M, Lancelin JM, Knaff DB, and Miginiac-Maslow M. The Arabidopsis plastidial thioredoxins—new functions and new insights into specificity. J Biol Chem 278: 23747–23752, 2003 [DOI] [PubMed] [Google Scholar]
- 62.Costa CL, Arruda P, and Benedetti CE. An Arabidopsis gene induced by wounding functionally homologous to flavoprotein oxidoreductases. Plant Mol Biol 44: 61–71, 2000 [DOI] [PubMed] [Google Scholar]
- 63.Courteille A, Vesa S, Sanz-Barrio R, Cazale AC, Becuwe-Linka N, Farran I, Havaux M, Rey P, and Rumeau D. Thioredoxin m4 controls photosynthetic alternative electron pathways in Arabidopsis. Plant Physiol 161: 508–520, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cousins AB, Pracharoenwattana I, Zhou W, Smith SM, and Badger MR. Peroxisomal malate dehydrogenase is not essential for photorespiration in Arabidopsis but its absence causes an increase in the stoichiometry of photorespiratory CO2 release. Plant Physiol 148: 786–795, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cruz Castillo M, Martinez C, Buchala A, Metraux J-P, and Leon J. Gene-specific involvement of beta-oxidation in wound-activated responses in Arabidopsis. Plant Physiol 135: 85–94, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.D'Autreaux B. and Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol 8: 813–824, 2007 [DOI] [PubMed] [Google Scholar]
- 67.Dal Santo S, Stampfl H, Krasensky J, Kempa S, Gibon Y, Petutschnig E, Rozhon W, Heuck A, Clausen T, and Jonak C. Stress-induced GSK3 regulates the redox stress response by phosphorylating glucose-6-phosphate dehydrogenase in Arabidopsis. Plant Cell 24: 3380–3392, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dave A. and Graham IA. Oxylipin signaling: a distinct role for the jasmonic acid precursor cis-(+)-12-oxo-phytodienoic acid (cis-OPDA). Front Plant Sci 3: 42, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deuschle K, Funck D, Forlani G, Stransky H, Biehl A, Leister D, van der Graaff E, Kunzee R, and Frommer WB. The role of Delta(1)-Pyrroline-5-carboxylate dehydrogenase in proline degradation. Plant Cell 16: 3413–3425, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dietz KJ. and Pfannschmidt T. Novel regulators in photosynthetic redox control of plant metabolism and gene expression. Plant Physiol 155: 1477–1485, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dixon DP, Skipsey M, Grundy NM, and Edwards R. Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiol 138: 2233–2244, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dong C-H, Zolman BK, Bartel B, Lee B-H, Stevenson B, Agarwal M, and Zhu J-K. Disruption of Arabidopsis CHY1 reveals an important role of metabolic status in plant cold stress signaling. Mol Plant 2: 59–72, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dry IB, Day DA, and Wiskich JT. Preferential oxidation of glycine by the respiratory-chain of pea leaf mitochondria. FEBS Lett 158: 154–158, 1983 [Google Scholar]
- 74.Dry IB. and Wiskich JT. Characteristics of glycine and malate oxidation by pea leaf mitochondria—evidence of differential access to nad and respiratory chains. Aust J Plant Physiol 12: 329–339, 1985 [Google Scholar]
- 75.Dutilleul C, Garmier M, Noctor G, Mathieu C, Chetrit P, Foyer CH, and de Paepe R. Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 15: 1212–1226, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eastmond PJ. SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18: 665–675, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eastmond PJ. MONODEHYROASCORBATE REDUCTASE4 is required for seed storage oil hydrolysis and postgerminative growth in Arabidopsis. Plant Cell 19: 1376–1387, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eberhard S, Drapier D, and Wollman F-A. Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J 31: 149–160, 2002 [DOI] [PubMed] [Google Scholar]
- 79.Ekkehard Neuhaus H. and Stitt M. Control analysis of photosynthate partitioning. Planta 182: 445–454, 1990 [DOI] [PubMed] [Google Scholar]
- 80.Elhafez D, Murcha MW, Clifton R, Soole KL, Day DA, and Whelan J. Characterization of mitochondrial alternative NAD(P)H dehydrogenases in Arabidopsis: intraorganelle location and expression. Plant Cell Physiol 47: 43–54, 2006 [DOI] [PubMed] [Google Scholar]
- 81.Escobar MA, Franklin KA, Svensson AS, Salter MG, Whitelam GC, and Rasmusson AG. Light regulation of the Arabidopsis respiratory chain. Multiple discrete phatoreceptor responses contribute to induction of type IINAD(P)H dehydrogenase genes. Plant Physiol 136: 2710–2721, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eubel H, Meyer EH, Taylor NL, Bussell JD, O'Toole N, Heazlewood JL, Castleden I, Small ID, Smith SM, and Millar AH. Novel proteins, putative membrane transporters, and an integrated metabolic network are revealed by quantitative proteomic analysis of Arabidopsis cell culture peroxisomes. Plant Physiol 148: 1809–1829, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fahnenstich H, Scarpeci TE, Valle EM, Flugge UI, and Maurino VG. Generation of hydrogen peroxide in chloroplasts of Arabidopsis overexpressing glycolate oxidase as an inducible system to study oxidative stress. Plant Physiol 148: 719–729, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fernie AR, Bauwe H, Eisenhut M, Florian A, Hanson DT, Hagemann M, Keech O, Mielewczik M, Nikoloski Z, Peterhänsel C, Roje S, Sage R, Timm S, von Cammerer S, Weber AP, and Westhoff P. Perspectives on plant photorespiratory metabolism. Plant Biol 15: 748–753, 2013 [DOI] [PubMed] [Google Scholar]
- 85.Fernie AR, Carrari F, and Sweetlove LJ. Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opin Plant Biol 7: 254–261, 2004 [DOI] [PubMed] [Google Scholar]
- 86.Fernie AR. and Martinoia E. Malate. Jack of all trades or master of a few? Phytochemistry 70: 828–832, 2009 [DOI] [PubMed] [Google Scholar]
- 87.Finkemeier I, Goodman M, Lamkemeyer P, Kandlbinder A, Sweetlove LJ, and Dietz KJ. The mitochondrial type II peroxiredoxin F is essential for redox homeostasis and root growth of Arabidopsis thaliana under stress. J Biol Chem 280: 12168–12180, 2005 [DOI] [PubMed] [Google Scholar]
- 88.Finkemeier I, König A-C, Heard W, Nunes-Nesi A, Pham PA, Leister D, Fernie AR, and Sweetlove LJ. Transcriptomic analysis of the role of carboxylic acids in metabolite signaling in Arabidopsis leaves. Plant Physiol 162: 239–253, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Finkemeier I, Laxa M, Miguet L, Howden AJM, and Sweetlove LJ. Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis. Plant Physiol 155: 1779–1790, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Footitt S, Marquez J, Schmuths H, Baker A, Theodoulou FL, and Holdsworth M. Analysis of the role of COMATOSE and peroxisomal beta-oxidation in the determination of germination potential in Arabidopsis. J Exp Bot 57: 2805–2814, 2006 [DOI] [PubMed] [Google Scholar]
- 91.Footitt S, Slocombe SP, Larner V, Kurup S, Wu YS, Larson T, Graham I, Baker A, and Holdsworth M. Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J 21: 2912–2922, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Foyer CH. and Noctor G. Oxygen processing in photosynthesis: regulation and signalling. New Phytol 146: 359–388, 2000 [Google Scholar]
- 93.Foyer CH. and Noctor G. Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol Plant 119: 355–364, 2003 [Google Scholar]
- 94.Foyer CH. and Noctor G. Oxidant and antioxidant signalling in plants: a re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ 28: 1056–1071, 2005 [Google Scholar]
- 95.Foyer CH. and Noctor G. Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid Redox Signal 11: 861–905, 2009 [DOI] [PubMed] [Google Scholar]
- 96.Foyer CH. and Noctor G. Ascorbate and glutathione: the heart of the redox hub. Plant Physiol 155: 2–18, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Foyer CH. and Shigeoka S. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol 155: 93–100, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fukao Y, Hayashi M, and Nishimura M. Proteomic analysis of leaf peroxisomal proteins in greening cotyledons of Arabidopsis thaliana. Plant Cell Physiol 43: 689–696, 2002 [DOI] [PubMed] [Google Scholar]
- 99.Fukushima A, Kusano M, Nakamichi N, Kobayashi M, Hayashi N, Sakakibara H, Mizuno T, and Saito K. Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc Natl Acad Sci U S A 106: 7251–7256, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inze D, Mittler R, and Van Breusegem F. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141: 436–445, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gardemann A, Stitt M, and Heldt HW. Control of CO2 fixation. Regulation of spinach ribulose-5-phosphate kinase by stromal metabolite levels. Biochim Biophys Acta 722: 51–60, 1983 [Google Scholar]
- 102.Geigenberger P. Response of plant metabolism to too little oxygen. Curr Opin Plant Biol 6: 247–256, 2003 [DOI] [PubMed] [Google Scholar]
- 103.Geigenberger P. Regulation of starch biosynthesis in response to a fluctuating environment. Plant Physiol 155: 1566–1577, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Geigenberger P, Kolbe A, and Tiessen A. Redox regulation of carbon storage and partitioning in response to light and sugars. J Exp Bot 56: 1469–1479, 2005 [DOI] [PubMed] [Google Scholar]
- 105.Geigenberger P. and Stitt M. Diurnal changes in sucrose, nucleotides, starch synthesis and AGPS transcript in growing potato tubers that are suppressed by decreased expression of sucrose phosphate synthase. Plant J 23: 795–806, 2000 [DOI] [PubMed] [Google Scholar]
- 106.Geiger DR. and Servaites JC. Diurnal regulation of photosynthetic carbon metabolism in C-3 plants. Annu Rev Plant Physiol Plant Mol Biol 45: 235–256, 1994 [Google Scholar]
- 107.Geisler DA, Broselid C, Hederstedt L, and Rasmusson AG. Ca2+-binding and Ca2+-independent respiratory NADH and NADPH dehydrogenases of Arabidopsis thaliana. J Biol Chem 282: 28455–28464, 2007 [DOI] [PubMed] [Google Scholar]
- 108.Gelhaye E, Rouhier N, Gerard J, Jolivet Y, Gualberto J, Navrot N, Ohlsson PI, Wingsle G, Hirasawa M, Knaff DB, Wang HM, Dizengremel P, Meyer Y, and Jacquot JP. A specific form of thioredoxin h occurs in plant mitochondria and regulates the alternative oxidase. Proc Natl Acad Sci U S A 101: 14545–14550, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Germain V, Rylott EL, Larson TR, Sherson SM, Bechtold N, Carde JP, Bryce JH, Graham IA, and Smith SM. Requirement for 3-ketoacyl-CoA thiolase-2 in peroxisome development, fatty acid beta-oxidation and breakdown of triacylglycerol in lipid bodies of Arabidopsis seedlings. Plant J 28: 1–12, 2001 [DOI] [PubMed] [Google Scholar]
- 110.Gibbs DJ, Lee SC, Isa NM, Gramuglia S, Fukao T, Bassel GW, Correia CS, Corbineau F, Theodoulou FL, Bailey-Serres J, and Holdsworth MJ. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479: 415–418, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gibon Y, Blasing OE, Palacios-Rojas N, Pankovic D, Hendriks JHM, Fisahn J, Hohne M, Gunther M, and Stitt M. Adjustment of diurnal starch turnover to short days: depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J 39: 847–862, 2004 [DOI] [PubMed] [Google Scholar]
- 112.Giege P, Heazlewood JL, Roessner-Tunali U, Millar AH, Fernie AR, Leaver CJ, and Sweetlove LJ. Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell 15: 2140–2151, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Giraud E, Ho LHM, Clifton R, Carroll A, Estavillo G, Tan Y-F, Howell KA, Ivanova A, Pogson BJ, Millar AH, and Whelan J. The absence of alternative oxidase1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol 147: 595–610, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Giraud E, Van Aken O, Uggalla V, and Whelan J. REDOX regulation of mitochondrial function in plants. Plant Cell Environ 35: 271–280, 2012 [DOI] [PubMed] [Google Scholar]
- 115.Glaring MA, Skryhan K, Kotting O, Zeeman SC, and Blennow A. Comprehensive survey of redox sensitive starch metabolising enzymes in Arabidopsis thaliana. Plant Physiol Biochem 58: 89–97, 2012 [DOI] [PubMed] [Google Scholar]
- 116.Goepfert S, Hiltunen JK, and Poirier Y. Identification and functional characterization of a monofunctional peroxisomal enoyl-CoA hydratase 2 that participates in the degradation of even cis-unsaturated fatty acids in Arabidopsis thaliana. J Biol Chem 281: 35894–35903, 2006 [DOI] [PubMed] [Google Scholar]
- 117.Graf A. and Smith AM. Starch and the clock: the dark side of plant productivity. Trends Plant Sci 16: 169–175, 2011 [DOI] [PubMed] [Google Scholar]
- 118.Graham IA. Seed storage oil mobilization. Annu Rev Plant Biol 59: 115–142, 2008 [DOI] [PubMed] [Google Scholar]
- 119.Graham JWA, Williams TCR, Morgan M, Fernie AR, Ratcliffe RG, and Sweetlove LJ. Glycolytic enzymes associate dynamically with mitochondria in response to respiratory demand and support substrate channeling. Plant Cell 19: 3723–3738, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gray GR, Maxwell DP, Villarimo AR, and McIntosh L. Mitochondria/nuclear signaling of alternative oxidase gene expression occurs through distinct pathways involving organic acids and reactive oxygen species. Plant Cell Rep 23: 497–503, 2004 [DOI] [PubMed] [Google Scholar]
- 121.Gupta KJ, Hebelstrup KH, Mur LAJ, and Igamberdiev AU. Plant hemoglobins: important players at the crossroads between oxygen and nitric oxide. FEBS Lett 585: 3843–3849, 2011 [DOI] [PubMed] [Google Scholar]
- 122.Gupta KJ, Zabalza A, and van Dongen JT. Regulation of respiration when the oxygen availability changes. Physiol Plant 137: 383–391, 2009 [DOI] [PubMed] [Google Scholar]
- 123.Haas FH, Heeg C, Queiroz R, Bauer A, Wirtz M, and Hell R. Mitochondrial serine acetyltransferase functions as a pacemaker of cysteine synthesis in plant cells. Plant Physiol 148: 1055–1067, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hackenberg C, Kern R, Huege J, Stal LJ, Tsuji Y, Kopka J, Shiraiwa Y, Bauwe H, and Hagemann M. Cyanobacterial lactate oxidases serve as essential partners in N-2 fixation and evolved into photorespiratory glycolate oxidases in plants. Plant Cell 23: 2978–2990, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Haedrich N, Hendriks JHM, Koetting O, Arrivault S, Feil R, Zeeman SC, Gibon Y, Schulze WX, Stitt M, and Lunn JE. Mutagenesis of cysteine 81 prevents dimerization of the APS1 subunit of ADP-glucose pyrophosphorylase and alters diurnal starch turnover in Arabidopsis thaliana leaves. Plant J 70: 231–242, 2012 [DOI] [PubMed] [Google Scholar]
- 126.Hagedorn PH, Flyvbjerg H, and Møller IM. Modelling NADH turnover in plant mitochondria. Physiol Plant 120: 370–385, 2004 [DOI] [PubMed] [Google Scholar]
- 127.Hamilton EW. and Heckathorn SA. Mitochondrial adaptations to NaCl. Complex I is protected by anti-oxidants and small heat shock proteins, whereas complex II is protected by proline and betaine. Plant Physiol 126: 1266–1274, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hanke G. and Mulo P. Plant type ferredoxins and ferredoxin-dependent metabolism. Plant Cell Environ 36: 1071–1084, 2013 [DOI] [PubMed] [Google Scholar]
- 129.Hansford RG. and Chappell JB. Effect of Ca2+ on oxidation of glycerol phosphosphate by blowfly flight-muscle mitochondria. Biochem Biophys Res Commun 27: 686–692, 1967 [DOI] [PubMed] [Google Scholar]
- 130.Hare PD, Cress WA, and van Staden J. A regulatory role for proline metabolism in stimulating Arabidopsis thaliana seed germination. Plant Growth Regul 39: 41–50, 2003 [Google Scholar]
- 131.Hashida S-N, Takahashi H, and Uchimiya H. The role of NAD biosynthesis in plant development and stress responses. Ann Bot 103: 819–824, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hatch MD, Droscher L, Flugge UI, and Heldt HW. A specific translocator for oxaloacetate transport in chloroplasts. FEBS Lett 178: 15–19, 1984 [Google Scholar]
- 133.Hayashi M, Nito K, Takei-Hoshi R, Yagi M, Kondo M, Suenaga A, Yamaya T, and Nishimura M. Ped3p is a peroxisomal ATP-binding cassette transporter that might supply substrates for fatty acid beta-oxidation. Plant Cell Physiol 43: 1–11, 2002 [DOI] [PubMed] [Google Scholar]
- 134.Heazlewood JL, Durek P, Hummel J, Selbig J, Weckwerth W, Walther D, and Schulze WX. PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res 36: D1015–D1021, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hebbelmann I, Selinski J, Wehmeyer C, Goss T, Voss I, Mulo P, Kangasjarvi S, Aro EM, Oelze ML, Dietz KJ, Nunes-Nesi A, Do PT, Fernie AR, Talla SK, Raghavendra AS, Linke V, and Scheibe R. Multiple strategies to prevent oxidative stress in Arabidopsis plants lacking the malate valve enzyme NADP-malate dehydrogenase. J Exp Bot 63: 1445–1459, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Heber UW. and Santariu KA. Compartmentation and reduction of pyridine nucleotides in relation to photosynthesis. Biochim Biophys Acta 109: 390–408, 1965 [DOI] [PubMed] [Google Scholar]
- 137.Heeg C, Kruse C, Jost R, Gutensohn M, Ruppert T, Wirtz M, and Hell R. Analysis of the Arabidopsis O-acetylserine(thiol)lyase gene family demonstrates compartment-specific differences in the regulation of cysteine synthesis. Plant Cell 20: 168–185, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hellmann H, Funck D, Rentsch D, and Frommer WB. Hypersensitivity of an Arabidopsis sugar signaling mutant toward exogenous proline application. Plant Physiol 123: 779–790, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hendriks JHM, Kolbe A, Gibon Y, Stitt M, and Geigenberger P. ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol 133: 838–849, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hennen-Bierwagen TA, Lin Q, Grimaud F, Planchot V, Keeling PL, James MG, and Myers AM. Proteins from multiple metabolic pathways associate with starch biosynthetic enzymes in high molecular weight complexes: a model for regulation of carbon allocation in maize amyloplasts. Plant Physiol 149: 1541–1559, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hertle AP, Blunder T, Wunder T, Pesaresi P, Pribil M, Armbruster U, and Leister D. PGRL1 is the elusive ferredoxin-plastoquinone reductase in photosynthetic cyclic electron flow. Mol Cell 49: 511–523, 2013 [DOI] [PubMed] [Google Scholar]
- 142.Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, Sauter M, and Dolferus R. Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol 153: 757–772, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hodges M, Flesch V, Gálvez S, and Bismuth E. Higher plant NADP+-dependent isocitrate dehydrogenases, ammonium assimilation and NADPH production. Plant Physiol Biochem 41: 577–585, 2003 [Google Scholar]
- 144.Hoff T, Truong HN, and Caboche M. The use of mutants and transgenic plants to study nitrate assimilation. Plant Cell Environ 17: 489–506, 1994 [Google Scholar]
- 145.Holmgren A. Thioredoxin. Annu Rev Biochem 54: 237–271, 1985 [DOI] [PubMed] [Google Scholar]
- 146.Howard TP, Metodiev M, Lloyd JC, and Raines CA. Thioredoxin-mediated reversible dissociation of a stromal multiprotein complex in response to changes in light availability. Proc Natl Acad Sci U S A 105: 4056–4061, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hu J, Baker A, Bartel B, Linka N, Mullen RT, Reumann S, and Zolman BK. Plant peroxisomes: biogenesis and function. Plant Cell 24: 2279–2303, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Huang S. and Millar AH. inate dehydrogenase: the complex roles of a simple enzyme. Curr Opin Plant Biol 16: 344–349, 2013 [DOI] [PubMed] [Google Scholar]
- 149.Hunter SC. and Ohlrogge JB. Regulation of spinach chloroplast acetyl-CoA carboxylase. Arch Biochem Biophys 359: 170–178, 1998 [DOI] [PubMed] [Google Scholar]
- 150.Igamberdiev AU, Bykova NV, and Gardestrom P. Involvement of cyanide-resistant and rotenone-insensitive pathways of mitochondrial electron transport during oxidation of glycine in higher plants. FEBS Lett 412: 265–269, 1997 [DOI] [PubMed] [Google Scholar]
- 151.Igamberdiev AU. and Gardestrom P. Regulation of NAD- and NADP-dependent isocitrate dehydrogenases by reduction levels of pyridine nucleotides in mitochondria and cytosol of pea leaves. Biochim Biophys Acta 1606: 117–125, 2003 [DOI] [PubMed] [Google Scholar]
- 152.Igarashi D, Tsuchida H, Miyao M, and Ohsumi C. Glutamate: glyoxylate aminotransferase modulates amino acid content during photorespiration. Plant Physiol 142: 901–910, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ikegami A, Yoshimura N, Motohashi K, Takahashi S, Romano PGN, Hisabori T, Takamiya K-I, and Masuda T. The CHLI1 subunit of Arabidopsis thaliana magnesium chelatase is a target protein of the chloroplast thioredoxin. J Biol Chem 282: 19282–19291, 2007 [DOI] [PubMed] [Google Scholar]
- 154.Isaacson T, Ohad I, Beyer P, and Hirschberg J. Analysis in vitro of the enzyme CRTISO establishes a poly-cis-carotenoid biosynthesis pathway in plants. Plant Physiol 136: 4246–4255, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ishizaki K, Larson TR, Schauer N, Fernie AR, Graham IA, and Leaver CJ. The critical role of Arabidopsis electron-transfer flavoprotein: ubiquinone oxidoreductase during dark-induced starvation. Plant Cell 17: 2587–2600, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jacoby RP, Li L, Huang S, Pong Lee C, Millar AH, and Taylor NL. Mitochondrial composition, function and stress response in plants. J Integr Plant Biol 54: 887–906, 2012 [DOI] [PubMed] [Google Scholar]
- 157.Jimenez A, Mateos L, Pedrajas JR, Miranda-Vizuete A, and Revuelta JL. The txl1(+) gene from Schizosaccharomyces pombe encodes a new thioredoxin-like 1 protein that participates in the antioxidant defence against tert-butyl hydroperoxide. Yeast 24: 481–490, 2007 [DOI] [PubMed] [Google Scholar]
- 158.Johnson GN. Physiology of PSI cyclic electron transport in higher plants. Biochim Biophys Acta 1807: 384–389, 2011 [DOI] [PubMed] [Google Scholar]
- 159.Johnson HS. NADP-malate dehydrogenase: photoactivation in leaves of plants with Calvin cycle photosynthesis. Biochem Biophys Res Commun 43: 703–709, 1971 [DOI] [PubMed] [Google Scholar]
- 160.Joliot P. and Johnson GN. Regulation of cyclic and linear electron flow in higher plants. Proc Natl Acad Sci U S A 108: 13317–13322, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jossier M, Bouly JP, Meimoun P, Arjmand A, Lessard P, Hawley S, Grahame Hardie D, and Thomas M. SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. Plant J 59: 316–328, 2009 [DOI] [PubMed] [Google Scholar]
- 162.Journet EP, Neuburger M, and Douce R. Role of glutamate-oxaloacetate transaminase and malate-dehydrogenase in the regeneration of NAD+ for glycine oxidation by spinach leaf mitochondria. Plant Physiol 67: 467–469, 1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Juric S, Hazler-Pilepic K, Tomasic A, Lepedus H, Jelicic B, Puthiyaveetil S, Bionda T, Vojta L, Allen JF, Schleiff E, and Fulgosi H. Tethering of ferredoxin:NADP plus oxidoreductase to thylakoid membranes is mediated by novel chloroplast protein TROL. Plant J 60: 783–794, 2009 [DOI] [PubMed] [Google Scholar]
- 164.Juszczuk IM, Szal B, and Rychter AM. Oxidation-reduction and reactive oxygen species homeostasis in mutant plants with respiratory chain complex I dysfunction. Plant Cell Environ 35: 296–307, 2012 [DOI] [PubMed] [Google Scholar]
- 165.Kaplan F. and Guy CL. RNA interference of Arabidopsis beta-amylase8 prevents maltose accumulation upon cold shock and increases sensitivity of PSII photochemical efficiency to freezing stress. Plant J 44: 730–743, 2005 [DOI] [PubMed] [Google Scholar]
- 166.Kaplan F, Sung DY, and Guy CL. Roles of β-amylase and starch breakdown during temperatures stress. Physiol Plant 126: 120–128, 2006 [Google Scholar]
- 167.Karpova OV, Kuzmin EV, Elthon TE, and Newton KJ. Differential expression of alternative oxidase genes in maize mitochondrial mutants. Plant Cell 14: 3271–3284, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kasimova MR, Grigiene J, Krab K, Hagedorn PH, Flyvbjerg H, Andersen PE, and Moller IM. The free NADH concentration is kept constant in plant mitochondria under different metabolic conditions. Plant Cell 18: 688–698, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kebeish R, Niessen M, Thiruveedhi K, Bari R, Hirsch HJ, Rosenkranz R, Stabler N, Schonfeld B, Kreuzaler F, and Peterhansel C. Chloroplastic photorespiratory bypass increases photosynthesis and biomass production in Arabidopsis thaliana. Nat Biotechnol 25: 593–599, 2007 [DOI] [PubMed] [Google Scholar]
- 170.Kelly AA. and Dormann P. Green light for galactolipid trafficking. Curr Opin Plant Biol 7: 262–269, 2004 [DOI] [PubMed] [Google Scholar]
- 171.Kelly AA, Quettier A-L, Shaw E, and Eastmond PJ. Seed storage oil mobilization is important but not essential for germination or seedling establishment in Arabidopsis. Plant Physiol 157: 866–875, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kerchev PI, Pellny TK, Vivancos PD, Kiddle G, Hedden P, Driscoll S, Vanacker H, Verrier P, Hancock RD, and Foyer CH. The transcription factor ABI4 is required for the ascorbic acid-dependent regulation of growth and regulation of jasmonate-dependent defense signaling pathways in Arabidopsis. Plant Cell 23: 3319–3334, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Khan BR, Adham AR, and Zolman BK. Peroxisomal Acyl-CoA oxidase 4 activity differs between Arabidopsis accessions. Plant Mol Biol 78: 45–58, 2012 [DOI] [PubMed] [Google Scholar]
- 174.Kiddle G, Pastori GM, Bernard S, Pignocchi C, Antoniw J, Verrier PJ, and Foyer CH. Effects of leaf ascorbate content on defense and photosynthesis gene expression in Arabidopsis thaliana. Antioxid Redox Signal 5: 23–32, 2003 [DOI] [PubMed] [Google Scholar]
- 175.Kienow L, Schneider K, Bartsch M, Stuible H-P, Weng H, Miersch O, Wasternack C, and Kombrink E. Jasmonates meet fatty acids: functional analysis of a new acyl-coenzyme A synthetase family from Arabidopsis thaliana. J Exp Bot 59: 403–419, 2008 [DOI] [PubMed] [Google Scholar]
- 176.Kirchsteiger K, Ferrandez J, Pascual MB, Gonzalez M, and Cejudo FJ. NADPH thioredoxin reductase C is localized in plastids of photosynthetic and nonphotosynthetic tissues and is involved in lateral root formation in Arabidopsis. Plant Cell 24: 1534–1548, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kolbe A, Oliver SN, Fernie AR, Stitt M, van Dongen JT, and Geigenberger P. Combined transcript and metabolite profiling of Arabidopsis leaves reveals fundamental effects of the thiol-disulfide status on plant metabolism. Plant Physiol 141: 412–422, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kolbe A, Tiessen A, Schluepmann H, Paul M, Ulrich S, and Geigenberger P. Trehalose 6-phosphate regulates starch synthesis via posttranslational redox activation of ADP-glucose pyrophosphorylase. Proc Natl Acad Sci U S A 102: 11118–11123, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Komatsu S, Yamamoto A, Nakamura T, Nouri MZ, Nanjo Y, Nishizawa K, and Furukawa K. Comprehensive analysis of mitochondria in roots and hypocotyls of soybean under flooding stress using proteomics and metabolomics techniques. J Proteome Res 10: 3993–4004, 2011 [DOI] [PubMed] [Google Scholar]
- 180.Konig J, Baier M, Horling F, Kahmann U, Harris G, Schurmann P, and Dietz KJ. The plant-specific function of 2-Cys peroxiredoxin-mediated detoxification of peroxides in the redox-hierarchy of photosynthetic electron flux. Proc Natl Acad Sci U S A 99: 5738–5743, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Konno H, Nakane T, Yoshida M, Ueoka-Nakanishi H, Hara S, and Hisabori T. Thiol modulation of the chloroplast ATP synthase is dependent on the energization of thylakoid membranes. Plant Cell Physiol 53: 626–634, 2012 [DOI] [PubMed] [Google Scholar]
- 182.Koo AJK, Chung HS, Kobayashi Y, and Howe GA. Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. J Biol Chem 281: 33511–33520, 2006 [DOI] [PubMed] [Google Scholar]
- 183.Kozaki AK, Mayumi K, and Sasaki Y. Thiol-disulfide exchange between nuclear-encoded and chloroplast-encoded subunits of pea acetyl-CoA carboxylase. J Biol Chem 276: 39919–39925, 2001 [DOI] [PubMed] [Google Scholar]
- 184.Krauss S, Zhang CY, and Lowell BB. The mitochondrial uncoupling-protein homologues. Nat Rev Mol Cell Biol 6: 248–261, 2005 [DOI] [PubMed] [Google Scholar]
- 185.Kromer S. Respiration during photosynthesis. Annu Rev Plant Physiol Plant Mol Biol 46: 45–70, 1995 [Google Scholar]
- 186.Kromer S. and Scheibe R. Function of the chloroplastic malate valve for respiration during photosynthesis. Biochem Soc Trans 24: 761–766, 1996 [DOI] [PubMed] [Google Scholar]
- 187.Kueger S, Steinhauser D, Willmitzer L, and Giavalisco P. High-resolution plant metabolomics: from mass spectral features to metabolites and from whole-cell analysis to subcellular metabolite distributions. Plant J 70: 39–50, 2012 [DOI] [PubMed] [Google Scholar]
- 188.Kunz H-H, Scharnewski M, Feussner K, Feussner I, Fluegge U-I, Fulda A, and Gierth M. The ABC transporter PXA1 and peroxisomal beta-oxidation are vital for metabolism in mature leaves of Arabidopsis during extended darkness. Plant Cell 21: 2733–2749, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kunze M, Pracharoenwattana I, Smith SM, and Hartig A. A central role for the peroxisomal membrane in glyoxylate cycle function. Biochim Biophys Acta 1763: 1441–1452, 2006 [DOI] [PubMed] [Google Scholar]
- 190.Lam HM, Coschigano KT, Oliveira IC, MeloOliveira R, and Coruzzi GM. The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu Rev Plant Physiol Plant Mol Biol 47: 569–593, 1996 [DOI] [PubMed] [Google Scholar]
- 191.Lamberto I, Percudani R, Gatti R, Folli C, and Petrucco S. Conserved Alternative splicing of Arabidopsis transthyretin-like determines protein localization and S-allantoin synthesis in peroxisomes. Plant Cell 22: 1564–1574, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lancien M, Gadal P, and Hodges M. Molecular characterization of higher plant NAD-dependent isocitrate dehydrogenase: evidence for a heteromeric structure by the complementation of yeast mutants. Plant J 16: 325–333, 1998 [DOI] [PubMed] [Google Scholar]
- 193.Lee CP, Eubel H, and Millar AH. Diurnal changes in mitochondrial function reveal daily optimization of light and dark respiratory metabolism in Arabidopsis. Mol Cell Proteomics 9: 2125–2139, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lehmann M, Schwarzlander M, Obata T, Sirikantaramas S, Burow M, Olsen CE, Tohge T, Fricker MD, Moller BL, Fernie AR, Sweetlove LJ, and Laxa M. The metabolic response of Arabidopsis roots to oxidative stress is distinct from that of heterotrophic cells in culture and highlights a complex relationship between the levels of transcripts, metabolites, and flux. Mol Plant 2: 390–406, 2009 [DOI] [PubMed] [Google Scholar]
- 195.Leister D. Retrograde signaling in plants: from simple to complex scenarios. Front Plant Sci 3: 135, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lepisto A. NADPH-dependent Thioredoxin System in Regulation of Chloroplast Functions. PHD thesis, University of Turku, Finland, 2011 [Google Scholar]
- 197.Lepisto A, Pakula E, Toivola J, Krieger-Liszkay A, Vignols F, and Rintamaki E. Deletion of chloroplast NADPH-dependent thioredoxin reductase results in inability to regulate starch synthesis and causes stunted growth under short-day photoperiods. J Exp Bot 64: 3843–3854, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Li J, Ezquer I, Bahaji A, Montero M, Ovecka M, Baroja-Fernández E, Muñoz FJ, Mérida Á, Almagro G, Hidalgo M, Sesma MT, and Pozueta-Romero J. Microbial volatile-induced accumulation of exceptionally high levels of starch in Arabidopsis leaves is a process involving NTRC and starch synthase classes III and IV. Mol Plant Microbe Interact 24: 1165–1178, 2011 [DOI] [PubMed] [Google Scholar]
- 199.Licausi F, Kosmacz M, Weits DA, Giuntoli B, Giorgi FM, Voesenek LACJ, Perata P, and van Dongen JT. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479: 419–422, 2011 [DOI] [PubMed] [Google Scholar]
- 200.Licausi F, van Dongen JT, Giuntoli B, Novi G, Santaniello A, Geigenberger P, and Perata P. HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J 62: 302–315, 2010 [DOI] [PubMed] [Google Scholar]
- 201.Lichter A. and Haberlein I. A light-dependent redox signal participates in the regulation of ammonia fixation in chloroplasts of higher plants—ferredoxin: glutamate synthase is a thioredoxin-dependent enzyme. J Plant Physiol 153: 83–90, 1998 [Google Scholar]
- 202.Liepman AH. and Olsen LJ. Peroxisomal alanine: glyoxylate aminotransferase (AGT1) is a photorespiratory enzyme with multiple substrates in Arabidopsis thaliana. Plant J 25: 487–498, 2001 [DOI] [PubMed] [Google Scholar]
- 203.Linka N, Theodoulou FL, Haslam RP, Linka M, Napier JA, Neuhaus HE, and Weber APM. Peroxisomal ATP import is essential for seedling development in Arabidopsis thaliana. Plant Cell 20: 3241–3257, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lintala M, Schuck N, Thormahlen I, Jungfer A, Weber KL, Weber AP, Geigenberger P, Soll J, Bolter B, and Mulo P. Arabidopsis tic62 trol mutant lacking thylakoid-bound ferredoxin-NADP+ oxidoreductase shows distinct metabolic phenotype. Mol Plant 7: 45–57, 2014 [DOI] [PubMed] [Google Scholar]
- 205.Liu Y-J, Norberg FEB, Szilagyi A, De Paepe R, Akerlund H-E, and Rasmusson AG. The mitochondrial external NADPH dehydrogenase modulates the leaf NADPH/NADP(+) ratio in transgenic Nicotiana sylvestris. Plant Cell Physiol 49: 251–263, 2008 [DOI] [PubMed] [Google Scholar]
- 206.Liu Y-J, Nunes-Nesi A, Wallstrom SV, Lager I, Michalecka AM, Norberg FEB, Widell S, Fredlund KM, Fernie AR, and Rasmusson AG. A redox-mediated modulation of stem bolting in transgenic Nicotiana sylvestris differentially expressing the external mitochondrial NADPH dehydrogenase. Plant Physiol 150: 1248–1259, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.This reference has been deleted.
- 208.Lousa CD, van Roermund CWT, Postis VLG, Dietrich D, Kerr ID, Wanders RJA, Baldwin SA, Baker A, and Theodoulou FL. Intrinsic acyl-CoA thioesterase activity of a peroxisomal ATP binding cassette transporter is required for transport and metabolism of fatty acids. Proc Natl Acad Sci U S A 110: 1279–1284, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lunn JE, Feil R, Hendriks JHM, Gibon Y, Morcuende R, Osuna D, Scheible WR, Carillo P, Hajirezaei MR, and Stitt M. Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem J 397: 139–148, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Luo T, Fan T, Liu Y, Rothbart M, Yu J, Zhou S, Grimm B, and Luo M. Thioredoxin redox regulates ATPase activity of magnesium chelatase CHLI subunit and modulates redox-mediated signaling in tetrapyrrole biosynthesis and homeostasis of reactive oxygen species in pea plants. Plant Physiol 159: 118–130, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Mano S, Nakamori C, Fukao Y, Araki M, Matsuda A, Kondo M, and Nishimura M. A defect of peroxisomal membrane protein 38 causes enlargement of peroxisomes. Plant Cell Physiol 52: 2157–2172, 2011 [DOI] [PubMed] [Google Scholar]
- 212.Marchand C, Le Marechal P, Meyer Y, Miginiac-Maslow M, Issakidis-Bourguet E, and Decottignies P. New targets of Arabidopsis thioredoxins revealed by proteomic analysis. Proteomics 4: 2696–2706, 2004 [DOI] [PubMed] [Google Scholar]
- 213.Marri L, Zaffagnini M, Collin V, Issakidis-Bourguet E, Lemaire SD, Pupillo P, Sparla F, Miginiac-Maslow M, and Trost P. Prompt and easy activation by specific thioredoxins of calvin cycle enzymes of Arabidopsis thaliana associated in the GAPDH/CP12/PRK supramolecular complex. Mol Plant 2: 259–269, 2009 [DOI] [PubMed] [Google Scholar]
- 214.Maurino VG. and Peterhansel C. Photorespiration: current status and approaches for metabolic engineering. Curr Opin Plant Biol 13: 249–256, 2010 [DOI] [PubMed] [Google Scholar]
- 215.Maxwell DP, Wang Y, and McIntosh L. The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc Natl Acad Sci U S A 96: 8271–8276, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.McKibbin RS, Muttucumaru N, Paul MJ, Powers SJ, Burrell MM, Coates S, Purcell PC, Tiessen A, Geigenberger P, and Halford NG. Production of high-starch, low-glucose potatoes through over-expression of the metabolic regulator SnRK1. Plant Biotechnol J 4: 409–418, 2006 [DOI] [PubMed] [Google Scholar]
- 217.McKinney DW, Buchanan BB, and Wolosiuk RA. Activation of chloroplast atpase by reduced thioredoxin. Phytochemistry 17: 794–795, 1978 [Google Scholar]
- 218.Meyer T, Hoelscher C, Schwoeppe C, and von Schaewen A. Alternative targeting of Arabidopsis plastidic glucose-6-phosphate dehydrogenase G6PD1 involves cysteine-dependent interaction with G6PD4 in the cytosol. Plant J 66: 745–758, 2011 [DOI] [PubMed] [Google Scholar]
- 219.Meyer Y, Belin C, Delorme-Hinoux V, Reichheld JP, and Riondet C. Thioredoxin and glutaredoxin systems in plants: molecular mechanisms, crosstalks, and functional significance. Antioxid Redox Signal 17: 1124–1160, 2012 [DOI] [PubMed] [Google Scholar]
- 220.Mhamdi A, Noctor G, and Baker A. Plant catalases: peroxisomal redox guardians. Arch Biochem Biophys 525: 181–194, 2012 [DOI] [PubMed] [Google Scholar]
- 221.Michalecka AM, Svensson AS, Johansson FI, Agius SC, Johanson U, Brennicke A, Binder S, and Rasmusson AG. Arabidopsis genes encoding mitochondrial type IINAD(P)H dehydrogenases have different evolutionary orgin and show distinct responses to light. Plant Physiol 133: 642–652, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Michalska J, Zauber H, Buchanan BB, Cejudo FJ, and Geigenberger P. NTRC links built-in thioredoxin to light and sucrose in regulating starch synthesis in chloroplasts and amyloplasts. Proc Natl Acad Sci U S A 106: 9908–9913, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Michelet L, Zaffagnini M, Morisse S, Sparla F, Perez-Perez ME, Francia F, Danon A, Marchand CH, Fermani S, Trost P, and Lemaire SD. Redox regulation of the Calvin-Benson cycle: something old, something new. Front Plant Sci 4: 470, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Michelet L, Zaffagnini M, Vanacker H, Le Marechal P, Marchand C, Schroda M, Lemaire SD, and Decottignies P. In vivo targets of S-thiolation in Chlamydomonas reinhardtii. J Biol Chem 283: 21571–21578, 2008 [DOI] [PubMed] [Google Scholar]
- 225.Miginiac-Maslow M. and Lancelin JM. Intrasteric inhibition in redox signalling: light activation of NADP-malate dehydrogenase. Photosynth Res 72: 1–12, 2002 [DOI] [PubMed] [Google Scholar]
- 226.Mikkelsen R, Mutenda KE, Mant A, Schürmann P, and Blennow A. α-Glucan, water dikinase (GWD): a plastidic enzyme with redox-regulated and coordinated catalytic activity and binding affinity. Proc Natl Acad Sci U S A 102: 1785–1790, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Millar AH, Mittova V, Kiddle G, Heazlewood JL, Bartoli CG, Theodoulou FL, and Foyer CH. Control of ascorbate synthesis by respiration and its implications for stress responses. Plant Physiol 133: 443–447, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Millar AH, Whelan J, Soole KL, and Day DA. Organization and regulation of mitochondrial respiration in plants. Annu Rev Plant Biol 62: 79–104, 2011 [DOI] [PubMed] [Google Scholar]
- 229.Millar AH, Wiskich JT, Whelan J, and Day DA. Organic-acid activation of the alternative oxidase of plant-mitochondria. FEBS Lett 329: 259–262, 1993 [DOI] [PubMed] [Google Scholar]
- 230.Millenaar FF. and Lambers H. The alternative oxidase: in vivo regulation and function. Plant Biol 5: 2–15, 2003 [Google Scholar]
- 231.Miller G, Shulaev V, and Mittler R. Reactive oxygen signaling and abiotic stress. Physiol Plant 133: 481–489, 2008 [DOI] [PubMed] [Google Scholar]
- 232.Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, and Van Breusegem F. ROS signaling: the new wave? Trends Plant Sci 16: 300–309, 2011 [DOI] [PubMed] [Google Scholar]
- 233.Moller IM. Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol 52: 561–591, 2001 [DOI] [PubMed] [Google Scholar]
- 234.Moller IM, Jensen PE, and Hansson A. Oxidative modifications to cellular components in plants. Annu Rev Plant Biol 58: 459–481, 2007 [DOI] [PubMed] [Google Scholar]
- 235.Moller IM. and Palmer JM. Direct evidence for the presence of a rotenone-resistant NADH dehydrogenase on the inner surface of the inner membrane of plant-mitochondria. Physiol Plant 54: 267–274, 1982 [Google Scholar]
- 236.Moller IM. and Sweetlove LJ. ROS signalling—specificity is required. Trends Plant Sci 15: 370–374, 2010 [DOI] [PubMed] [Google Scholar]
- 237.Montrichard F, Alkhalfioui F, Yano H, Vensel WH, Hurkman WJ, and Buchanan BB. Thioredoxin targets in plants: the first 30 years. J Proteomics 72: 452–474, 2009 [DOI] [PubMed] [Google Scholar]
- 238.Mooney BP, Miernyk JA, and Randall DD. The complex fate of α-ketoacids. Annu Rev Plant Biol 53: 357–375, 2002 [DOI] [PubMed] [Google Scholar]
- 239.Mora-Garcia S, Rodriguez-Suarez R, and Wolosiuk RA. Role of electrostatic interactions on the affinity of thioredoxin for target proteins. Recognition of chloroplast fructose-1, 6-bisphosphatase by mutant Escherichia coli thioredoxins. J Biol Chem 273: 16273–16280, 1998 [DOI] [PubMed] [Google Scholar]
- 240.Morgan MJ, Lehmann M, Schwarzlander M, Baxter CJ, Sienkiewicz-Porzucek A, Williams TCR, Schauer N, Fernie AR, Fricker MD, Ratcliffe RG, Sweetlove LJ, and Finkemeier I. Decrease in manganese superoxide dismutase leads to reduced root growth and affects tricarboxylic acid cycle flux and mitochondrial redox homeostasis. Plant Physiol 147: 101–114, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Motohashi K, Kondoh A, Stumpp MT, and Hisabori T. Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc Natl Acad Sci U S A 98: 11224–11229, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Munne-Bosch S, Queval G, and Foyer CH. The impact of global change factors on redox signaling underpinning stress tolerance. Plant Physiol 161: 5–19, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Nashilevitz S, Melamed-Bessudo C, Izkovich Y, Rogachev I, Osorio S, Itkin M, Adato A, Pankratov I, Hirschberg J, Fernie AR, Wolf S, Usadel B, Levy AA, Rumeau D, and Aharoni A. An orange ripening mutant links plastid NAD(P)H dehydrogenase complex activity to central and specialized metabolism during tomato fruit maturation. Plant Cell 22: 1977–1997, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Nee G, Zaffagnini M, Trost P, and Issakidis-Bourguet E. Redox regulation of chloroplastic glucose-6-phosphate dehydrogenase: a new role for f-type thioredoxin. FEBS Lett 583: 2827–2832, 2009 [DOI] [PubMed] [Google Scholar]
- 245.Neill S, Desikan R, and Hancock J. Hydrogen peroxide signalling. Curr Opin Plant Biol 5: 388–395, 2002 [DOI] [PubMed] [Google Scholar]
- 246.Nickelsen K. The path of carbon in photosynthesis: how to discover a biochemical pathway. Ambix 59: 266–293, 2012 [Google Scholar]
- 247.Nishizawa AN. and Buchanan BB. Enzyme regulation in C4 photosynthesis. Purification and properties of thioredoxin-linked fructose bisphosphatase and sedoheptulose bisphosphatase from corn leaves. J Biol Chem 256: 6119–6126, 1981 [PubMed] [Google Scholar]
- 248.Noctor G, De Paepe R, and Foyer CH. Mitochondrial redox biology and homeostasis in plants. Trends Plant Sci 12: 125–134, 2007 [DOI] [PubMed] [Google Scholar]
- 249.Noctor G, Queval G, and Gakiere B. NAD(P) synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J Exp Bot 57: 1603–1620, 2006 [DOI] [PubMed] [Google Scholar]
- 250.Noctor G, Veljovic-Jovanovic S, Driscoll S, Novitskaya L, and Foyer CH. Drought and oxidative load in the leaves of C-3 plants: a predominant role for photorespiration? Ann Bot 89: 841–850, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Nowak K, Luniak N, Witt C, Wustefeld Y, Wachter A, Mendel RR, and Hansch R. Peroxisomal localization of sulfite oxidase separates it from chloroplast-based sulfur assimilation. Plant Cell Physiol 45: 1889–1894, 2004 [DOI] [PubMed] [Google Scholar]
- 252.Nunes-Nesi A, Araujo WL, and Fernie AR. Targeting mitochondrial metabolism and machinery as a means to enhance photosynthesis. Plant Physiol 155: 101–107, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Nunes-Nesi A, Araujo WL, Obata T, and Fernie AR. Regulation of the mitochondrial tricarboxylic acid cycle. Curr Opin Plant Biol 16: 335–343, 2013 [DOI] [PubMed] [Google Scholar]
- 254.Nunes-Nesi A, Carrari F, Gibon Y, Sulpice R, Lytovchenko A, Fisahn J, Graham J, Ratcliffe RG, Sweetlove LJ, and Fernie AR. Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J 50: 1093–1106, 2007 [DOI] [PubMed] [Google Scholar]
- 255.Nunes-Nesi A, Carrari F, Lytovchenko A, Smith AMO, Loureiro ME, Ratcliffe RG, Sweetlove LJ, and Fernie AR. Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol 137: 611–622, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Nunes-Nesi A, Sulpice R, Gibon Y, and Fernie AR. The enigmatic contribution of mitochondrial function in photosynthesis. J Exp Bot 59: 1675–1684, 2008 [DOI] [PubMed] [Google Scholar]
- 257.Obata T. and Fernie AR. The use of metabolomics to dissect plant responses to abiotic stresses. Cell Mol Life Sci 69: 3225–3243, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Obata T, Matthes A, Koszior S, Lehmann M, Araujo WL, Bock R, Sweetlove LJ, and Fernie AR. Alteration of mitochondrial protein complexes in relation to metabolic regulation under short-term oxidative stress in Arabidopsis seedlings. Phytochemistry 72: 1081–1091, 2011 [DOI] [PubMed] [Google Scholar]
- 259.Ohlrogge J. and Browse J. Lipid biosynthesis. Plant Cell 7: 957–970, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Oliver SN, Lunn JE, Urbanczyk-Wochniak E, Lytovchenko A, van Dongen JT, Faix B, Schmaelzlin E, Fernie AR, and Geigenberger P. Decreased expression of cytosolic pyruvate kinase in potato tubers leads to a decline in pyruvate resulting in an in vivo repression of the alternative oxidase. Plant Physiol 148: 1640–1654, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Osorio S, Vallarino JG, Szecowka M, Ufaz S, Tzin V, Angelovici R, Galili G, and Fernie AR. Alteration of the interconversion of pyruvate and malate in the plastid or cytosol of ripening tomato fruit invokes diverse consequences on sugar but similar effects on cellular organic acid, metabolism, and transitory starch accumulation. Plant Physiol 161: 628–643, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Ozturk L. and Demir Y. In vivo and in vitro protective role of proline. Plant Growth Regul 38: 259–264, 2002 [Google Scholar]
- 263.Palmieri F, Pierri CL, De Grassi A, Nunes-Nesi A, and Fernie AR. Evolution, structure and function of mitochondrial carriers: a review with new insights. Plant J 66: 161–181, 2011 [DOI] [PubMed] [Google Scholar]
- 264.Palmieri F, Rieder B, Ventrella A, Blanco E, Do PT, Nunes-Nesi A, Trauth AU, Fiermonte G, Tjaden J, Agrimi G, Kirchberger S, Paradies E, Fernie AR, and Neuhaus HE. Molecular identification and functional characterization of Arabidopsis thaliana mitochondrial and chloroplastic NAD(+) carrier proteins. J Biol Chem 284: 31249–31259, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Pastori GM. and Foyer CH. Common components, networks, and pathways of cross-tolerance to stress. The central role of “redox” and abscisic acid-mediated controls. Plant Physiol 129: 460–468, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Perez-Ruiz JM, Spinola MC, Kirchsteiger K, Moreno J, Sahrawy M, and Cejudo FJ. Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell 18: 2356–2368, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Peterhansel C, Krause K, Braun HP, Espie GS, Fernie AR, Hanson DT, Keech O, Maurino VG, Mielewczik M, and Sage RF. Engineering photorespiration: current state and future possibilities. Plant Biol 15: 754–758, 2013 [DOI] [PubMed] [Google Scholar]
- 268.Pfannschmidt T. and Yang C. The hidden function of photosynthesis: a sensing system for environmental conditions that regulates plant acclimation responses. Protoplasma 249Suppl 2: S125–S136, 2012 [DOI] [PubMed] [Google Scholar]
- 269.Pick TR, Brautigam A, Schulz MA, Obata T, Fernie AR, and Weber APM. PLGG1, a plastidic glycolate glycerate transporter, is required for photorespiration and defines a unique class of metabolite transporters. Proc Natl Acad Sci U S A 110: 3185–3190, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Pignocchi C. and Foyer CH. Apoplastic ascorbate metabolism and its role in the regulation of cell signalling. Curr Opin Plant Biol 6: 379–389, 2003 [DOI] [PubMed] [Google Scholar]
- 271.Pinfield-Wells H, Rylott EL, Gilday AD, Graham S, Job K, Larson TR, and Graham IA. Sucrose rescues seedling establishment but not germination of Arabidopsis mutants disrupted in peroxisomal fatty acid catabolism. Plant J 43: 861–872, 2005 [DOI] [PubMed] [Google Scholar]
- 272.Plaxton WC. and Podesta FE. The functional organization and control of plant respiration. Crit Rev Plant Sci 25: 159–198, 2006 [Google Scholar]
- 273.Pracharoenwattana I, Cornah JE, and Smith SM. Arabidopsis peroxisomal malate dehydrogenase functions in beta-oxidation but not in the glyoxylate cycle. Plant J 50: 381–390, 2007 [DOI] [PubMed] [Google Scholar]
- 274.Pracharoenwattana I. and Smith SM. When is a peroxisome not a peroxisome? Trends Plant Sci 13: 522–525, 2008 [DOI] [PubMed] [Google Scholar]
- 275.Rasmusson AG. and Escobar MA. Light and diurnal regulation of plant respiratory gene expression. Physiol Plant 129: 57–67, 2007 [Google Scholar]
- 276.Rasmusson AG, Fernie AR, and van Dongen JT. Alternative oxidase: a defence against metabolic fluctuations? Physiol Plant 137: 371–382, 2009 [DOI] [PubMed] [Google Scholar]
- 277.Rasmusson AG, Geisler DA, and Moller IM. The multiplicity of dehydrogenases in the electron transport chain of plant mitochondria. Mitochondrion 8: 47–60, 2008 [DOI] [PubMed] [Google Scholar]
- 278.Rasmusson AG. and Moller IM. Effect of calcium-ions and inhibitors on internal NAD(P)H dehydrogenases in plant-mitochondria. Eur J Biochem 202: 617–623, 1991 [DOI] [PubMed] [Google Scholar]
- 279.Rasmusson AG. and Wallstrom SV. Involvement of mitochondria in the control of plant cell NAD(P)H reduction levels. Biochem Soc Trans 38: 661–666, 2010 [DOI] [PubMed] [Google Scholar]
- 280.Reichard P. The Biosynthesis of deoxyribonucleotides. Eur J Biochem 3: 259–266, 1968 [DOI] [PubMed] [Google Scholar]
- 281.Reichert A, Baalmann E, Vetter S, Backhausen JE, and Scheibe R. Activation properties of the redox-modulated chloroplast enzymes glyceraldehyde 3-phosphate dehydrogenase and fructose-1,6-bisphosphatase. Physiol Plant 110: 330–341, 2000 [Google Scholar]
- 282.Reichheld JP, Khafif M, Riondet C, Droux M, Bonnard G, and Meyer Y. Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in Arabidopsis development. Plant Cell 19: 1851–1865, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Reinbothe C, Bartsch S, Eggink LL, Hoober JK, Brusslan J, Andrade-Paz R, Monnet J, and Reinbothe S. A role for chlorophyllide a oxygenase in the regulated import and stabilization of light-harvesting chlorophyll a/b proteins. Proc Natl Acad Sci U S A 103: 4777–4782, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, Antonicelli GE, Rasche N, Lueder F, Weckwerth W, and Jahn O. Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19: 3170–3193, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Reumann S, Bettermann M, Bent R, and Heldt HW. Evidence for the presence of a porin in the membrane of glyoxysomes of castor bean. Plant Physiol 115: 891–899, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Reumann S, Maier E, Benz R, and Heldt HW. The membrane of leaf peroxisomes contains a porin-like channel. J Biol Chem 270: 17559–17565, 1995 [DOI] [PubMed] [Google Scholar]
- 287.Reumann S, Maier E, Benz R, and Heldt HW. A specific porin is involved in the malate shuttle of leaf peroxisomes. Biochem Soc Trans 24: 754–757, 1996 [DOI] [PubMed] [Google Scholar]
- 288.Reumann S, Maier E, Heldt HW, and Benz R. Permeability properties of the porin of spinach leaf peroxisomes. Eur J Biochem 251: 359–366, 1998 [DOI] [PubMed] [Google Scholar]
- 289.Reumann S, Quan S, Aung K, Yang P, Manandhar-Shrestha K, Holbrook D, Linka N, Switzenberg R, Wilkerson CG, Weber APM, Olsen LJ, and Hu J. In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol 150: 125–143, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Rey P, Sanz-Barrio R, Innocenti G, Ksas B, Courteille A, Rumeau D, Issakidis-Bourguet E, and Farran I. Overexpression of plastidial thioredoxins f and m differentially alters photosynthetic activity and response to oxidative stress in tobacco plants. Front Plant Sci 4: 390, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Rhoads DM. and Subbaiah CC. Mitochondrial retrograde regulation in plants. Mitochondrion 7: 177–194, 2007 [DOI] [PubMed] [Google Scholar]
- 292.Richter AS, Peter E, Rothbart M, Schlicke H, Toivola J, Rintamaki E, and Grimm B. Posttranslational influence of NADPH-dependent thioredoxin reductase C on enzymes in tetrapyrrole synthesis. Plant Physiol 162: 63–73, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Riesmeier JW, Flugge UI, Schulz B, Heineke D, Heldt HW, Willmitzer L, and Frommer WB. Antisense repression of the chloroplast triose phosphate translocator affects carbon partitioning in transgenic potato plants. Proc Natl Acad Sci U S A 90: 6160–6164, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Rocha M, Licausi F, Araujo WL, Nunes-Nesi A, Sodek L, Fernie AR, and van Dongen JT. Glycolysis and the tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus. Plant Physiol 152: 1501–1513, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Rojas CM, Senthil-Kumar M, Wang K, Ryu C-M, Kaundal A, and Mysore KS. Glycolate oxidase modulates reactive oxygen species-mediated signal transduction during nonhost resistance in Nicotiana benthamiana and Arabidopsis. Plant Cell 24: 336–352, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Rylott EL, Eastmond PJ, Gilday AD, Slocombe SP, Larson TR, Baker A, and Graham IA. The Arabidopsis thaliana multifunctional protein gene (MFP2) of peroxisomal beta-oxidation is essential for seedling establishment. Plant J 45: 930–941, 2006 [DOI] [PubMed] [Google Scholar]
- 297.Sakakibara Y, Kimura H, Iwamura A, Saitoh T, Ikegami T, Kurisu G, and Hase T. A new structural insight into differential interaction of cyanobacterial and plant ferredoxins with nitrite reductase as revealed by NMR and X-ray crystallographic studies. J Biochem 151: 483–492, 2012 [DOI] [PubMed] [Google Scholar]
- 298.Sanz-Barrio R, Corral-Martinez P, Ancin M, Segui-Simarro JM, and Farran I. Overexpression of plastidial thioredoxin f leads to enhanced starch accumulation in tobacco leaves. Plant Biotechnol J 11: 618–627, 2013 [DOI] [PubMed] [Google Scholar]
- 299.Sasaki Y, Kozaki A, and Hatano M. Link between light and fatty acid synthesis: thioredoxin-linked reductive activation of plastidic acetyl-CoA carboxylase. Proc Natl Acad Sci U S A 94: 11096–11101, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Schaller A. and Stintzi A. Enzymes in jasmonate biosynthesis—structure, function, regulation. Phytochemistry 70: 1532–1538, 2009 [DOI] [PubMed] [Google Scholar]
- 301.Scharte J, Schoen H, Tjaden Z, Weis E, and von Schaewen A. Isoenzyme replacement of glucose-6-phosphate dehydrogenase in the cytosol improves stress tolerance in plants. Proc Natl Acad Sci U S A 106: 8061–8066, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Scheibe R. Malate valves to balance cellular energy supply. Physiol Plant 120: 21–26, 2004 [DOI] [PubMed] [Google Scholar]
- 303.Scheibe R. and Anderson LE. Dark modulation of NADP-dependent malate dehydrogenase and glucose-6-phosphate dehydrogenase in the chloroplast. Biochim Biophys Acta 636: 58–64, 1981 [DOI] [PubMed] [Google Scholar]
- 304.Scheibe R. and Jacquot JP. NADP regulates the light activation of NADP-dependent malate-dehydrogenase. Planta 157: 548–553, 1983 [DOI] [PubMed] [Google Scholar]
- 305.Schertl P, Sunderhaus S, Klodmann J, Gergoff Grozeff GE, Bartoli CG, and Braun H-P. L-Galactono-1,4-lactone dehydrogenase (GLDH) forms part of three subcomplexes of mitochondrial complex I in Arabidopsis thaliana. J Biol Chem 287: 14412–14419, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Schindler I, Renz A, Schmid FX, and Beck E. Activation of spinach pullulanase by reduction results in a decrease in the number of isomeric forms. Biochim Biophys Acta 1548: 175–186, 2001 [DOI] [PubMed] [Google Scholar]
- 307.Schurmann P. and Buchanan BB. The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid Redox Signal 10: 1235–1273, 2008 [DOI] [PubMed] [Google Scholar]
- 308.Schwarz C, Bohne AV, Wang F, Cejudo FJ, and Nickelsen J. An intermolecular disulfide-based light switch for chloroplast psbD gene expression in Chlamydomonas reinhardtii. Plant J 72: 378–389, 2012 [DOI] [PubMed] [Google Scholar]
- 309.Schwarzlaender M. and Finkemeier I. Mitochondrial energy and redox signaling in plants. Antioxid Redox Signal 18: 2122–2144, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Shen WY, Wei YD, Dauk M, Tan YF, Taylor DC, Selvaraj G, and Zou JT. Involvement of a glycerol-3-phosphate dehydrogenase in modulating the NADH/NAD(+) ratio provides evidence of a mitochondrial glycerol-3-phosphate shuttle in Arabidopsis. Plant Cell 18: 422–441, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Shen WY, Wei YD, Dauk M, Zheng ZF, and Zou JT. Identification of a mitochondrial glycerol-3-phosphate dehydrogenase from Arabidopsis thaliana: evidence for a mitochondrial glycerol-3-phosphate shuttle in plants. FEBS Lett 536: 92–96, 2003 [DOI] [PubMed] [Google Scholar]
- 312.Shlizerman L, Marsh K, Blumwald E, and Sadka A. Iron-shortage-induced increase in citric acid content and reduction of cytosolic aconitase activity in Citrus fruit vesicles and calli. Physiol Plant 131: 72–79, 2007 [DOI] [PubMed] [Google Scholar]
- 313.Silver DM, Silva LP, Issakidis-Bourguet E, Glaring MA, Schriemer DC, and Moorhead GBG. Insight into the redox regulation of the phosphoglucan phosphatase SEX4 involved in starch degradation. FEBS J 280: 538–548, 2013 [DOI] [PubMed] [Google Scholar]
- 314.Simon S. and Petrasek J. Why plants need more than one type of auxin. Plant Sci 180: 454–460, 2011 [DOI] [PubMed] [Google Scholar]
- 315.Slocombe SP, Cornah J, Pinfield-Wells H, Soady K, Zhang Q, Gilday A, Dyer JM, and Graham IA. Oil accumulation in leaves directed by modification of fatty acid breakdown and lipid synthesis pathways. Plant Biotechnol J 7: 694–703, 2009 [DOI] [PubMed] [Google Scholar]
- 316.Smirnoff N. Ascorbic acid: metabolism and functions of a multi-facetted molecule. Curr Opin Plant Biol 3: 229–235, 2000 [PubMed] [Google Scholar]
- 317.Smith AMO, Ratcliffe RG, and Sweetlove LJ. Activation and function of mitochondrial uncoupling protein in plants. J Biol Chem 279: 51944–51952, 2004 [DOI] [PubMed] [Google Scholar]
- 318.Smith PMC. and Atkins CA. Purine biosynthesis. Big in cell division, even bigger in nitrogen assimilation. Plant Physiol 128: 793–802, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Solheim C, Li L, Hatzopoulos P, and Millar AH. Loss of Lon1 in Arabidopsis changes the mitochondrial proteome leading to altered metabolite profiles and growth retardation without an accumulation of oxidative damage. Plant Physiol 160: 1187–1203, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Sørhagen K, Laxa M, Peterhänsel C, and Reumann S. The emerging role of photorespiration and non-photorespiratory peroxisomal metabolism in pathogen defence. Plant Biol 15: 723–736, 2013 [DOI] [PubMed] [Google Scholar]
- 321.Sparla F, Costa A, Lo Schiavo F, Pupillo P, and Trost P. Redox regulation of a novel plastid-targeted β-amylase of Arabidopsis. Plant Physiol 141: 840–850, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Spielbauer G, Li L, Romisch-Margl L, Do PT, Fouquet R, Fernie AR, Eisenreich W, Gierl A, and Settles AM. Chloroplast-localized 6-phosphogluconate dehydrogenase is critical for maize endosperm starch accumulation. J Exp Bot 64: 2231–2242, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Stenbaek A, Hansson A, Wulff RP, Hansson M, Dietz KJ, and Jensen PE. NADPH-dependent thioredoxin reductase and 2-Cys peroxiredoxins are needed for the protection of Mg-protoporphyrin monomethyl ester cyclase. FEBS Lett 582: 2773–2778, 2008 [DOI] [PubMed] [Google Scholar]
- 324.Stenbaek A. and Jensen PE. Redox regulation of chlorophyll biosynthesis. Phytochemistry 71: 853–859, 2010 [DOI] [PubMed] [Google Scholar]
- 325.Stintzi A. and Browse J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci U S A 97: 10625–10630, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Stitt M. Progress in understanding and engineering primary plant metabolism. Curr Opin Biotechnol 24: 229–238, 2013 [DOI] [PubMed] [Google Scholar]
- 327.Strader LC. and Bartel B. Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol Plant 4: 477–486, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Strader LC, Wheeler DL, Christensen SE, Berens JC, Cohen JD, Rampey RA, and Bartel B. Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin. Plant Cell 23: 984–999, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Strassner J, Schaller F, Frick UB, Howe GA, Weiler EW, Amrhein N, Macheroux P, and Schaller A. Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. Plant J 32: 585–601, 2002 [DOI] [PubMed] [Google Scholar]
- 330.Strodtkoetter I, Padmasree K, Dinakar C, Speth B, Niazi PS, Wojtera J, Voss I, Do PT, Nunes-Nesi A, Fernie AR, Linke V, Raghavendra AS, and Scheibe R. Induction of the AOX1D isoform of alternative oxidase in A-thaliana T-DNA insertion lines lacking isoform AOX1A is insufficient to optimize photosynthesis when treated with antimycin A. Mol Plant 2: 284–297, 2009 [DOI] [PubMed] [Google Scholar]
- 331.Studart-Guimaraes C, Gibon Y, Frankel N, Wood CC, Zanor MI, Fernie AR, and Carrari F. Identification and characterisation of the alpha and beta subunits of succinyl CoA ligase of tomato. Plant Mol Biol 59: 781–791, 2005 [DOI] [PubMed] [Google Scholar]
- 332.Svensson AS. and Rasmusson AG. Light-dependent gene expression for proteins in the respiratory chain of potato leaves. Plant J 28: 73–82, 2001 [DOI] [PubMed] [Google Scholar]
- 333.Sweetlove LJ, Beard KFM, Nunes-Nesi A, Fernie AR, and Ratcliffe RG. Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci 15: 462–470, 2010 [DOI] [PubMed] [Google Scholar]
- 334.Sweetlove LJ, Fait A, Nunes-Nesi A, Williams T, and Fernie AR. The mitochondrion: an integration point of cellular metabolism and signalling. Crit Rev Plant Sci 26: 17–43, 2007 [Google Scholar]
- 335.Sweetlove LJ. and Fernie AR. The spatial organisation of metabolism within the plant cell. Annu Rev Plant Biol 64: 723–746, 2013 [DOI] [PubMed] [Google Scholar]
- 336.Sweetlove LJ, Heazlewood JL, Herald V, Holtzapffel R, Day DA, Leaver CJ, and Millar AH. The impact of oxidative stress on Arabidopsis mitochondria. Plant J 32: 891–904, 2002 [DOI] [PubMed] [Google Scholar]
- 337.Sweetlove LJ, Lytovchenko A, Morgan M, Nunes-Nesi A, Taylor NL, Baxter CJ, Eickmeier I, and Fernie AR. Mitochondrial uncoupling protein is required for efficient photosynthesis. Proc Natl Acad Sci U S A 103: 19587–19592, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Szecowka M, Heise R, Tohge T, Nunes-Nesi A, Vosloh D, Huege J, Feil R, Lunn J, Nikoloski Z, Stitt M, Fernie AR, and Arrivault S. Metabolic fluxes in an illuminated Arabidopsis rosette. Plant Cell 25: 694–714, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Szecowka M, Osorio S, Obata T, Araujo WL, Rohrmann J, Nunes-Nesi A, and Fernie AR. Decreasing the mitochondrial synthesis of malate in potato tubers does not affect plastidial starch synthesis, suggesting that the physiological regulation of ADPglucose pyrophosphorylase is context dependent. Plant Physiol 160: 2227–2238, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Tadini L, Romani I, Pribil M, Jahns P, Leister D, and Pesaresi P. Thylakoid redox signals are integrated into organellar-gene-expression-dependent retrograde signaling in the prors1-1 mutant. Front Plant Sci 3: 282, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Taylor NL, Heazlewood JL, Day DA, and Millar AH. Differential impact of environmental stresses on the pea mitochondrial proteome. Mol Cell Proteomics 4: 1122–1133, 2005 [DOI] [PubMed] [Google Scholar]
- 342.Taylor NL, Tan YF, Jacoby RP, and Millar AH. Abiotic environmental stress induced changes in the Arabidopsis thaliana chloroplast, mitochondria and peroxisome proteomes. J Proteomics 72: 367–378, 2009 [DOI] [PubMed] [Google Scholar]
- 343.Tcherkez G, Cornic G, Bligny R, Gout E, and Ghashghaie J. In vivo respiratory metabolism of illuminated leaves. Plant Physiol 138: 1596–1606, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Terry MJ. and Smith AG. A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front Plant Sci 4: 14, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Tetlow IJ, Wait R, Lu Z, Akkasaeng R, Bowsher CG, Esposito S, Kosar-Hashemi B, Morell MK, and Emes MJ. Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein–protein interactions. Plant Cell 16: 694–708, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Thormahlen I, Ruber J, Von Roepenack-Lahaye E, Ehrlich SM, Massot V, Hummer C, Tezycka J, Issakidis-Bourguet E, and Geigenberger P. Inactivation of thioredoxin f1 leads to decreased light activation of ADP-glucose pyrophosphorylase and altered diurnal starch turnover in leaves of Arabidopsis plants. Plant Cell Environ 36: 16–29, 2013 [DOI] [PubMed] [Google Scholar]
- 347.Tiessen A, Hendriks JHM, Stitt M, Branscheid A, Gibon Y, Farre EM, and Geigenberger P. Starch synthesis in potato tubers is regulated by post-translational redox modification of ADP-glucose pyrophosphorylase: a novel regulatory mechanism linking starch synthesis to the sucrose supply. Plant Cell 14: 2191–2213, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Tiessen A, Prescha K, Branscheid A, Palacios N, McKibbin R, Halford NG, and Geigenberger P. Evidence that SNF1-related kinase and hexokinase are involved in separate sugar-signalling pathways modulating post-translational redox activation of ADP-glucose pyrophosphorylase in potato tubers. Plant J 35: 490–500, 2003 [DOI] [PubMed] [Google Scholar]
- 349.Timm S. and Bauwe H. The variety of photorespiratory phenotypes—employing the current status for future research directions on photorespiration. Plant Biol 15: 737–747, 2013 [DOI] [PubMed] [Google Scholar]
- 350.Timm S, Mielewczik M, Florian A, Frankenbach S, Dreissen A, Hocken N, Fernie AR, Walter A, and Bauwe H. High-to-low CO2 acclimation reveals plasticity of the photorespiratory pathway and indicates regulatory links to cellular metabolism of Arabidopsis. PLoS One 7: e42809, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Timm S, Nunes-Nesi A, Pamik T, Morgenthal K, Wienkoop S, Keerberg O, Weckwerth W, Kleczkowski LA, Fernie AR, and Bauwe H. A cytosolic pathway for the conversion of hydroxypyruvate to glycerate during photorespiration in Arabidopsis. Plant Cell 20: 2848–2859, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Tomaz T, Bagard M, Pracharoenwattana I, Linden P, Lee CP, Carroll AJ, Stroher E, Smith SM, Gardestrom P, and Millar AH. Mitochondrial malate dehydrogenase lowers leaf respiration and alters photorespiration and plant growth in Arabidopsis. Plant Physiol 154: 1143–1157, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Tovar-Mendez A, Miernyk JA, and Randall DD. Regulation of pyruvate dehydrogenase complex activity in plant cells. Eur J Biochem 270: 1043–1049, 2003 [DOI] [PubMed] [Google Scholar]
- 354.Umbach AL. and Siedow JN. Covalent and noncovalent dimers of the cyanide-resistant alternative oxidase protein in higher-plant mitochondria and their relationship to enzyme-activity. Plant Physiol 103: 845–854, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Urbanczyk-Wochniak E, Usadel B, Thimm O, Nunes-Nesi A, Carrari F, Davy M, Blasing O, Kowalczyk M, Weicht D, Polinceusz A, Meyer S, Stitt M, and Fernie AR. Conversion of MapMan to allow the analysis of transcript data from Solanaceous species: effects of genetic and environmental alterations in energy metabolism in the leaf. Plant Mol Biol 60: 773–792, 2006 [DOI] [PubMed] [Google Scholar]
- 356.Valerio C, Costa A, Marri L, Issakidis-Bourguet E, Pupillo P, Trost P, and Sparla F. Thioredoxin-regulated β-amylase (BAM1) triggers diurnal starch degradation in guard cells, and in mesophyll cells under osmotic stress. J Exp Bot 62: 545–555, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Vandelle E. and Delledonne M. Peroxynitrite formation and function in plants. Plant Sci 181: 534–539, 2011 [DOI] [PubMed] [Google Scholar]
- 358.Vandenabeele S, Vanderauwera S, Vuylsteke M, Rombauts S, Langebartels C, Seidlitz HK, Zabeau M, Van Montagu M, Inze D, and Van Breusegem F. Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J 39: 45–58, 2004 [DOI] [PubMed] [Google Scholar]
- 359.Vanlerberghe GC. Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int J Mol Sci 14: 6805–6847, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Vanlerberghe GC, Day DA, Wiskich JT, Vanlerberghe AE, and McIntosh L. Alternative oxidase activity in tobacco leaf mitochondria—dependence on tricarboxylic-acid cycle-mediated redox regulation and pyruvate activation. Plant Physiol 109: 353–361, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Vanlerberghe GC. and McIntosh L. Mitochondrial electron-transport regulation of nuclear gene-expression—studies with the alternative oxidase gene of tobacco. Plant Physiol 105: 867–874, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Vanlerberghe GC. and McIntosh L. Signals regulating the expression of the nuclear gene encoding alternative oxidase of plant mitochondria. Plant Physiol 111: 589–595, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Vanlerberghe GC. and McIntosh L. Identification of the cysteine residue responsible for covalent redox regulation of alternative oxidase in tobacco mitochondria. Plant Physiol 114: 1025, 1997 [Google Scholar]
- 364.Vanlerberghe GC, McIntosh L, and Yip JYH. Molecular localization of a redox-modulated process regulating plant mitochondrial electron transport. Plant Cell 10: 1551–1560, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci 20: 1298–1345, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Vercesi AE, Borecky J, Godoy Maia ID, Arruda P, Cuccovia IM, and Chaimovich H. Plant uncoupling mitochondrial proteins. Annu Rev Plant Biol 57: 383–404, 2006 [DOI] [PubMed] [Google Scholar]
- 367.Verdin E, Hirschey MD, Finley LWS, and Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci 35: 669–675, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Vestergaard CL, Flyvbjerg H, and Moller IM. Intracellular signaling by diffusion: can waves of hydrogen peroxide transmit intracellular information in plant cells? Front Plant Sci 3: 295, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Vidal G, Ribas-Carbo M, Garmier M, Dubertret G, Rasmusson AG, Mathieu C, Foyer CH, and De Paepe R. Lack of respiratory chain complex I impairs alternative oxidase engagement and modulates redox signaling during elicitor-induced cell death in tobacco. Plant Cell 19: 640–655, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Wagner AM, Krab K, Wagner MJ, and Moore AL. Regulation of thermogenesis in flowering Araceae: the role of the alternative oxidase. Biochim Biophys Acta 1777: 993–1000, 2008 [DOI] [PubMed] [Google Scholar]
- 371.Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, and Schmid M. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339: 704–707, 2013 [DOI] [PubMed] [Google Scholar]
- 372.Wakao S. and Benning C. Genome-wide analysis of glucose-6-phosphate dehydrogenases in Arabidopsis. Plant J 41: 243–256, 2005 [DOI] [PubMed] [Google Scholar]
- 373.Wang BL, Tang XY, Cheng LY, Zhang AZ, Zhang WH, Zhang FS, Liu JQ, Cao Y, Allan DL, Vance CP, and Shen JB. Nitric oxide is involved in phosphorus deficiency-induced cluster-root development and citrate exudation in white lupin. New Phytol 187: 1112–1123, 2010 [DOI] [PubMed] [Google Scholar]
- 374.Wang J, Yu Y, Zhang Z, Quan R, Zhang H, Ma L, Deng XW, and Huang R. Arabidopsis CSN5B interacts with VTC1 and modulates ascorbic acid synthesis. Plant Cell 25: 625–636, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Wang P, Liu J, Liu B, Feng D, Da Q, Wang P, Shu S, Su J, Zhang Y, Wang J, and Wang HB. Evidence for a role of chloroplastic m-Type thioredoxins in the biogenesis of photosystem II in Arabidopsis. Plant Physiol 163: 1710–1728, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Wang Y, Yu B, Zhao J, Guo J, Li Y, Han S, Huang L, Du Y, Hong Y, Tang D, and Liu Y. Autophagy contributes to leaf starch degradation. Plant Cell 25: 1383–1399, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Watling JR, Robinson SA, and Seymour RS. Contribution of the alternative pathway to respiration during thermogenesis in flowers of the sacred lotus. Plant Physiol 140: 1367–1373, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Wendt UK, Wenderoth I, Tegeler A, and von Schaewen A. Molecular characterization of a novel glucose-6-phosphate dehydrogenase from potato (Solanum tuberosum L.). Plant J 23: 723–733, 2000 [DOI] [PubMed] [Google Scholar]
- 379.Willekens H, Chamnongpol S, Davey M, Schraudner M, Langebartels C, VanMontagu M, Inze D, and VanCamp W. Catalase is a sink for H2O2 and is indispensable for stress defence in C-3 plants. EMBO J 16: 4806–4816, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Wirtz M, Berkowitz O, Droux M, and Hell R. The cysteine synthase complex from plants—mitochondrial serine acetyltransferase from Arabidopsis thaliana carries a bifunctional domain for catalysis and protein-protein interaction. Eur J Biochem 268: 686–693, 2001 [DOI] [PubMed] [Google Scholar]
- 381.Witt W. and Sauter JJ. Purification and characterization of α-amylase from poplar leaves. Phytochemistry 41: 365–372, 1996 [Google Scholar]
- 382.Woitsch S. and Romer S. Expression of xanthophyll biosynthetic genes during light-dependent chloroplast differentiation. Plant Physiol 132: 1508–1517, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Wolosiuk RA. and Buchanan BB. Thioredoxin and glutathione regulate photosynthesis in chloroplasts. Nature 266: 565–567, 1977 [Google Scholar]
- 384.Wolosiuk RA. and Buchanan BB. Activation of chloroplast NADP-linked glyceraldehyde-3-phosphate dehydrogenase by ferredoxin-thioredoxin system. Plant Physiol 61: 669–671, 1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Wolosiuk RA. and Buchanan BB. Regulation of chloroplast phosphoribulokinase by ferredoxin-thioredoxin system. Arch Biochem Biophys 189: 97–101, 1978 [DOI] [PubMed] [Google Scholar]
- 386.Woodson JD. and Chory J. Organelle signaling: how stressed chloroplasts communicate with the nucleus. Curr Biol 22: R690–R692, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Woodward AW. and Bartel B. A receptor for auxin. Plant Cell 17: 2425–2429, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Yamaryo Y, Motohashi K, Takamiya K-I, Hisabori T, and Ohta H. In vitro reconstitution of monogalactosyldiacylglycerol (MGDG) synthase regulation by thioredoxin. FEBS Lett 580: 4086–4090, 2006 [DOI] [PubMed] [Google Scholar]
- 389.Yoshida K, Noguchi K, Motohashi K, and Hisabori T. Systematic exploration of thioredoxin target proteins in mitochondria. Plant Cell Physiol 54: 875–892, 2013 [DOI] [PubMed] [Google Scholar]
- 390.Zechmann B, Mauch F, Sticher L, and Muller M. Subcellular immunocytochemical analysis detects the highest concentrations of glutathione in mitochondria and not in plastids. J Exp Bot 59: 4017–4027, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Zeeman SC, Kossmann J, and Smith AM. Starch: its metabolism, evolution, and biotechnological modification in plants. In: Annual Review of Plant Biology, Vol. 61, edited by Merchant S, Briggs WR, and Ort D. Palo Alto, CA: Annual Reviews, 2010, pp. 209–234 [DOI] [PubMed] [Google Scholar]
- 392.Zhang N. and Portis AR. Mechanism of light regulation of Rubisco: a specific role for the larger Rubisco activase isoform involving reductive activation by thioredoxin-f. Proc Natl Acad Sci U S A 96: 9438–9443, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Zhang N, Schurmann P, and Portis AR., Jr.Characterization of the regulatory function of the 46-kDa isoform of Rubisco activase from Arabidopsis. Photosynth Res 68: 29–37, 2001 [DOI] [PubMed] [Google Scholar]
- 394.Zolman BK, Silva ID, and Bartel B. The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiol 127: 1266–1278, 2001 [PMC free article] [PubMed] [Google Scholar]
- 395.Zolman BR, Martinez N, Millius A, Adham AR, and Bartel B. Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics 180: 237–251, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Zsigmond L, Tomasskovics B, Deak V, Rigo G, Szabados L, Banhegyi G, and Szarka A. Enhanced activity of galactono-1,4-lactone dehydrogenase and ascorbate-glutathione cycle in mitochondria from complex III deficient Arabidopsis. Plant Physiol Biochem 49: 809–815, 2011 [DOI] [PubMed] [Google Scholar]