Abstract
Denitrifying phosphorus removal is an attractive wastewater treatment process due to its reduced carbon source demand and sludge minimization potential. Two lab-scale sequencing batch reactors (SBRs) were operated in alternating anaerobic-anoxic (A-A) or anaerobic-oxic (A-O) conditions to achieve denitrifying enhanced biological phosphate removal (EBPR) and traditional EBPR. No significant differences were observed in phosphorus removal efficiencies between A-A SBR and A-O SBR, with phosphorus removal rates being 87.9% and 89.0% respectively. The community structures in denitrifying and traditional EBPR processes were evaluated by high-throughput sequencing of the PCR-amplified partial 16S rRNA genes from each sludge. The results obtained showed that the bacterial community was more diverse in A-O sludge than in A-A sludge. Taxonomy and β-diversity analyses indicated that a significant shift occurred in the dominant microbial community in A-A sludge compared with the seed sludge during the whole acclimation phase, while a slight fluctuation was observed in the abundance of the major taxonomies in A-O sludge. One Dechloromonas-related OTU outside the 4 known Candidatus “Accumulibacter” clades was detected as the main OTU in A-A sludge at the stationary operation, while Candidatus “Accumulibacter” dominated in A-O sludge.
Keywords: EBPR, denitrifying phosphorous removal, microbial community, pyrosequencing
Enhanced biological phosphorus removal (EBPR) is considered to be the most cost-effective and environmentally friendly technology to meet the increasingly stringent discharge standard of wastewater treatment plants (2). In the EBPR process, polyphosphate-accumulating organisms (PAO) are favored and enriched for phosphorus removal through alternating anaerobic-oxic (A-O) conditions. Under the anaerobic condition, PAO are capable of storing organic substances (particularly volatile fatty acids (VFAs)) as intracellular poly-β-hydroxyalkanoates (PHA) through the release of orthophosphate and degradation of glycogen. Under the subsequent oxic condition, the stored PHA is oxidized to produce energy for the uptake of excessive amounts of orthophosphate, biomass growth, and regeneration of glycogen. By discharging polyphosphate-enriched excess sludge, phosphorus is ultimately removed from the wastewater.
In addition to the alternating anaerobic-aerobic condition, phosphorus can also be removed under anaerobic-anoxic (A-A) cycling with nitrate or nitrite as an electron acceptor (1, 20, 25, 34). The organisms responsible for anaerobic-anoxic phosphorus removal are referred as denitrifying PAO (DPAO). The internal carbon source (typically PHA) is utilized for both phosphorus removal and denitrification in the same process; therefore, the demand for an often-limiting carbon source can be reduced, with approximately 50% of the carbon source being saved. Moreover, the denitrifying phosphorus removal process can save aeration by approximately 30% and minimize excess sludge (21, 30).
The activities of DPAO have been widely studied and reported since the discovery of denitrifying phosphorus removal (4, 10, 32). However, it remains controversial whether the same organisms are responsible for phosphorus removal under oxic and anoxic conditions. DPAO were hypothesized to be the same organisms as PAO, and the different activities observed under different environments were only attributed to the inducing conditions (35, 39). Unlike PAO, which can use only oxygen as an electron acceptor, DPAO can utilize oxygen or nitrate for phosphorus removal (6, 23, 38). Previous studies also demonstrated that DPAO were a fraction of PAO (1, 20, 41).
Recently developed culture-independent approaches have identified the Rhodocyclus-related organism, Candidatus “Accumulibacter” as one of the most important PAO in EBPR systems (13, 18, 26). Based on an elaborate online survey and subsequent phylogenetic analysis of reported 16S rRNA gene sequences from full scale wastewater treatment plants (WWTPs) and bench scale reactors (22, 27), four clades (namely Acc-SG1, Acc-SG2, Acc-SG3 and Acc-SG4) were assigned by Kim et al. (13). Candidatus “Accumulibacter” may have also adapted to different ecological environments and exhibits various physiological properties similar to DPAO (5, 11, 12, 15). In addition to the clades of Candidatus “Accumulibacter”, Dechloromonas-related bacteria had been detected in several EBPR processes operated under partially anoxic conditions (15, 17, 19). However, whether they function as PAO remains controversial. According to Kong’s study (19), a Dechloromonas-like organism (Bet135 FISH probe positive) was found to have a polyphosphate-accumulating phenotype by culture-independent methods. Although another Dechloromonas-like organism (Dech453 FISH probe positive) was abundant in A-A-O SBR, it was predicted to only reduce nitrate to nitrite in the reactor, and Candidatus “Accumulibacter” acted as the real PAO in this system (15). Due to their wide distribution in wastewater treatment-related environments and disputed functions, the phosphorous removal activity of Dechloromonas-related bacteria needs to be elucidated in more detail.
In the present study, two sequencing batch reactors (SBRs) were operated under alternating anaerobic-anoxic (A-A) or anaerobic-oxic (A-O) conditions with nitrate or oxygen as the electron acceptor in order to establish strict denitrifying phosphorous removal and conventional EBPR environments, respectively. A comparative study of the phosphorus removal performance of the two SBRs was firstly reported. The whole community structure, dominant taxonomy, and community dynamics during acclimation were systematically compared with the aid of high-throughput sequencing to detect DPAO. The further characterization of the denitrifying phosphorous removal sludge community and identification of DPAO in this study has provided an insight into denitrifying phosphorus removal and process efficiency.
Materials and Methods
Reactor operation
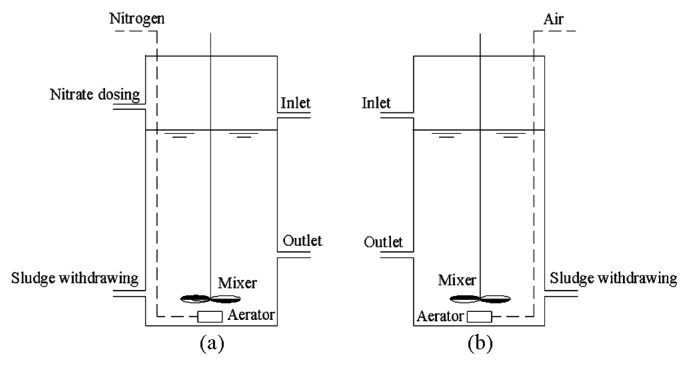
Two parallel sequencing batch reactors with an effective working volume of 10.9 L were configured in the present study (Fig. 1) for continuous operation in order to conduct phosphorus removal studies. The A-A SBR was operated under an alternating anaerobic-anoxic condition for the acclimation of denitrifying phosphate removal sludge, while the A-O SBR was operated under an anaerobic- oxic condition for the acclimation of traditional phosphate removal sludge as a control reactor for the A-A SBR.
Fig. 1.
Configuration of the two lab-scale phosphorous removal SBRs.
Both SBRs were inoculated with the sludge from a full-scale sewage WWTP with an A2/O (anaerobic-anoxic-aerobic) process and operated at room temperature with an operation cycle of 8 h. Each of the above cycles consisted of five stages: filling (0.25 h), anaerobic (2.5 h), anoxic or oxic (4.0 h), settling (1.0 h), and withdrawing (0.25 h). Mixers were operated during the anaerobic and anoxic (or oxic) stages for better mixing at a rate of 40 r min−1. In the A-A reactor, nitrate was dosed continuously for the initial 1.0 h of the anoxic stage with a peristaltic pump (Lange, BT-100). The initial nitrate concentration was designed at 20 mg L−1 based on the influent COD concentration (the C: N ratio was approximately 7.5:1), and the dosing flow of the nitrate solution was timely adjusted (±5%–10%) to achieve an effluent nitrate concentration of 2–7 mg L−1 during the whole operation. These parameters were used to satisfy complete anoxic phosphorus uptake and avoid excessive nitrate at the beginning of the anaerobic phase. In the A-O reactor, air was aerated at flow rate of 0.3 m−3 h−1 during the oxic stage with an aerator to supply oxygen for aerobic phosphorus removal sludge. All of the above operations were automatically controlled with time controllers. Moreover, pH was uncontrolled except that the initial pH was adjusted to approximately 7.0 at the beginning of the anaerobic stage. The SRT of the sludge in the A-A and A-O SBRs were 15–20 d and 10–15 d, respectively.
At the end of each cycle, 7.2 L of supernatant was exchanged with an equal volume of synthetic wastewater. Each liter of the wastewater contained 200 mg CH3COONa, 14 mg NH4Cl, 20 mg NaH2PO4, and 0.3 mL trace element solution, which consisted (1 L) of 10 g EDTA, 0.12 g ZnSO4·7H2O, 0.12 g MnSO4·7H2O, 1.0 g FeCl3, 0.03 g CuSO4·5H2O, 0.15 g CoCl2·6H2O, 0.06 g (NH4)2Mo4O13·2H2O, 0.15 g H3BO3, and 0.18 g KI. Real primary settled sewage wastewater was also mixed with the synthetic wastewater at a ratio of 1:10 by volume to maintain microbial diversity.
Chemical analysis
Sampling was conducted regularly at the end of the anaerobic and anoxic (or oxic) stages as well as the synthetic influent for the concentration analysis of COD, NH4+-N, NO3−-N, NO2−N, and PO43−-P. COD was measured using the potassium dichromate method and PO43−-P was quantified by the molybdate colorimetric method using a spectrophotometer (Shimadzu, UVmini-1240). The N-component including NH4+-N, NO3−N, and NO2−-N were measured automatically by colorimetric methods with a Cleverchem200 (DeChem-Tech.Gmbh, Germany).
DNA extraction, PCR amplification, and High-throughput Sequencing
Total DNA were extracted from sludge samples using the FastDNA® SPIN Kit for Soil (MP Biomedicals, Illkirch, France). The PCR primer set (33) of 967F (CAACGCGAAGAACCTTACC) and 1046R (CGACAGCCATGCANCACCT) was chosen to target the V6 hypervariable region of the bacterial 16S rRNA gene. Each 30 μL PCR reaction system contained 0.75 μL of MightyAmp® DNA Polymerase (Takara, Dalian, China), 15 μL of 2×Buffer, 1.5 μL of each primer, and 20–50 ng of genomic DNA. The amplification was conducted in an i-Cycler (BioRad Laboratories, CA, USA) under the following thermo steps: initial denaturation at 98°C for 2 min, followed by 28 cycles at 98°C for 20 s, 55°C for 20 s, 68°C for 1 min, and a final extension step at 68°C for 5 min. In order to minimize the impact of potential early-round PCR errors, three parallel amplifications of each DNA were conducted simultaneously and then mixed together for further analysis. The quality of the amplification products was examined by agarose (2.0%) gel electrophoresis. Lastly, PCR amplicons were purified with the quick Midi Purification Kit (Qiagen) and quantified using spectrometry (NanoDrop-1000).
Barcodes that allowed sample multiplexing during sequencing were incorporated into both the forward and reverse primers for V6 amplicon sequencing (31). The amplicons from different samples were then mixed together by ensuring equal mass concentrations in the final mixture, which was sent out for library construction and sequencing on the Illumina-HiSeq 2000 at the Beijing Genomics Institute (BGI) of Shenzhen with the strategy of paired-end sequencing (2×100 bp).
Independent metagenomic-based community profiling of one sludge DNA sample from the A-A SBR on day 274 (March 22, 2013) was carried out in parallel with V6 amplicon-based quantification to validate the accuracy of community analysis based on 16S rRNA gene partial amplicons. Briefly, DNA was mechanically fragmented to an enrichment size of –170 bp. The DNA fragments were then gel purified and quality checked. Recycled DNA was used for shotgun library construction, which was finally sequenced on an Illumina HiSeq 2000 platform using the Paired End 100 bp sequencing strategy at the BGI of Shenzhen.
Bioinformatic analysis
Routine amplicon sequence processing, including primer removal, low quality sequence screening, and sample sorting, were firstly carried out using the Pyrosequencing Pipeline of the Ribosomal Database Project (RDP). The qualified sequences of each sample were then denoised to remove sequencing-induced errors by the shhh.seqs command in Mothur. Removal of the archaeal sequence was based on the RDP classifier (Version 2.5) and carried out by a self-written C++ program. OTUs were obtained by processing the incorporated cluster file through another two self-written C++ program, and OTU-based principle coordinate analysis (PCoA) and cluster analysis (CA) were performed using PAST software, as described by Zhang et al. (40). The microbial diversity indices in terms of Chao 1, Chao 2, and ACE (Abundance-based Coverage Estimator) as well as the Good’s coverage were calculated using the EstmateS tool. The comprehensive taxonomy information at different levels was confirmed via Global Alignment for Sequence Taxonomy (GAST).
In the metagenomic-based analysis, low quality reads screening was performed by PRINSEQ (http://sourceforge.net/), then pairedend (PE) reads were pair-aligned by FLASH (http://www.cbcb.umd.edu/software/flash) to screen 10–50 bp overlapping PE reads and subsequently assemble them into 150–190 bp iTags. Finally, iTags were processed with MetaPhlAn (http://huttenhower.sph.harvard.edu/metaphlan/) for comprehensive population profiling of the sludge microbial community.
Accession number
The V6 amplicon sequences of 16S rRNA reported in this study have been deposited into the short reads archive (SRA) database of NCBI (http://www.ncbi.nlm.nih.gov/) with the accession number of SRR768434. The metagenomic data sets of this study have been deposited in the MG-RAST server (http://metagenomics.anl.gov/) and the accession number was 4524971.3.
Results and Discussion
Performance of the two SBRs
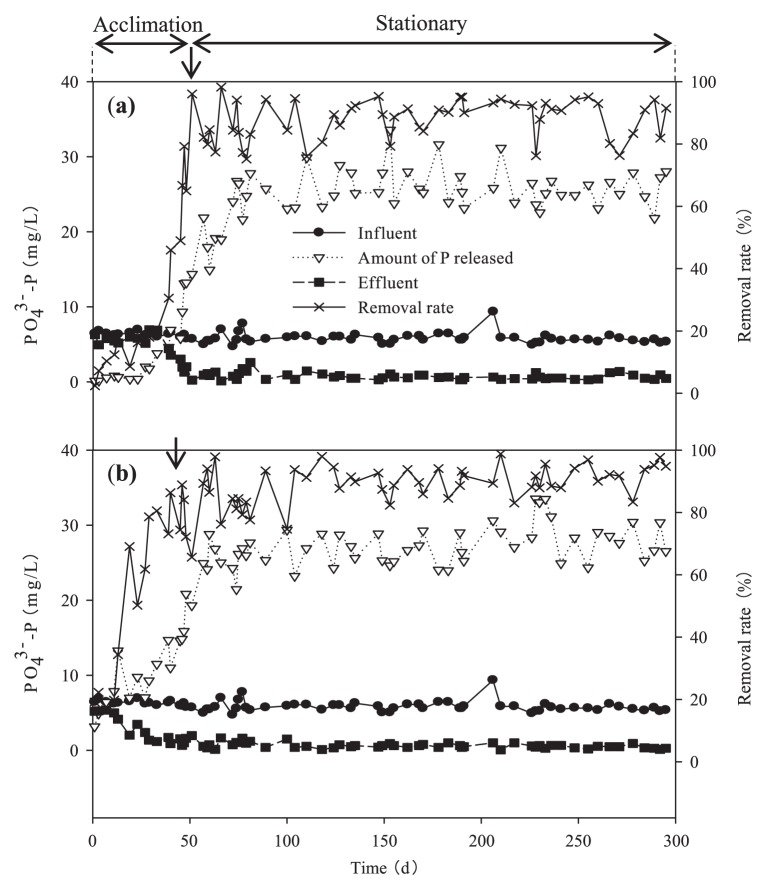
The whole profiles of the PO43−-P concentration and phosphorus removal rate from inoculation until stationary operation were shown in Fig. 2. The concentration of PO43−-P in the influent was controlled at approximately 6.0 mg L−1 during the entire study, and it took 5 L and 40 d for the A-A and A-O SBRs to achieve stationary operation, respectively. In the A-A SBR, the phosphorus removal rate increased gradually and stabilized at approximately 87.9%. The acclimation period in the A-O SBR was shorter, which was attributed to more PAO in the seed sludge being inherited due to the similar redox conditions between the A-O SBR and full-scale A2/O WWTP environment, and the phosphorus removal rate in the A-O SBR stabilized at approximately 89.0%. Thus, after acclimation, the two SBRs were continuously operated until day 295 to evaluate the operation stability of denitrifying phosphorus removal. During the entire stationary operation, the PO43−-P concentration in the effluent was 0.225–1.76 mg L−1 (averaged at 0.728 mg L−1) for the A-A SBR and 0.072–1.61 mg L−1 (averaged at 0.664 mg L−1) for A-O SBR, respectively. These results indicated the stability of denitrifying phosphorus removal process. No significant differences were observed in the phosphorus removal efficiencies between the two SBRs, which was consistent with the findings by Kapagiannidis et al. (12).
Fig. 2.
Overall phosphorus removal during a 295-day operation.
Sample selection for microbial community structure analysis
The sludge samples used in the microbial community analysis were summarized in Table 1. Firstly, the seed sludge from WWTP (noted as D0) was collected to obtain initial microbial information on the two SBRs before acclimation. Sludge samples were collected from the two SBRs at both the acclimation and stationary phases, according to the phosphorus removal efficiency and amount of anaerobic phosphorus released, to demonstrate the succession of the microbial community during the acclimation phase. As shown in Table 1, sludge samples taken on days 9 and 35 were selected to represent the acclimation phase and samples on days 55 and 76 were selected to represent the stationary phase.
Table 1.
Sludge samples selected for microbial community analysis
| Origin | Seed | A-A SBR | A-O SBR | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||
| Phase | Inoculation | Acclimation phase | Stationary phase | Acclimation phase | Stationary phase | ||||
| Code | SEED-D0 | AA-D9 | AA-D35 | AA-D55 | AA-D76 | AO-D9 | AO-D35 | AO-D55 | AO-D76 |
| Sampling time | Jun.20 | Jun.28 | Jul.24 | Aug.15 | Sep.05 | Jun.28 | Jul.24 | Aug.15 | Sep.05 |
| PO43−-P release (mg L−1) | / | 0.67 | 4.63 | 23.9 | 25.4 | 7.14 | 10.6 | 24.9 | 26.9 |
| PO43−-P removal (%) | / | 11.3 | 25.6 | 82.1 | 84.7 | 18.3 | 73.2 | 89.3 | 84.5 |
Microbial diversity of different EBPR sludges
As shown in Table 2, a total of 19,331–28,419 raw sequences were generated for the nine samples through high-throughput sequencing. After filtering out low quality and archaeal sequences and denoising, 8,373–11,572 sequences remained and were considered to be the effective sequences for different samples. In order to achieve a fair comparison at the same depth, the sequences of all nine samples were normalized to 8,373, which was the smallest of these samples, for downstream analysis.
Table 2.
Microbial diversity of the 9 sludge samples
| Code | Sequence | 3% cut-off | 6% cut-off | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||||||||
| Raw | Effective | NO after normalization | OTUs | Chao1 mean | Chao2 mean | ACE mean | Bootstrap mean | GOOD’s coverage | OTUs | Chao1 mean | Chao2 mean | ACE mean | Bootstrap mean | GOOD’s coverage | |
| SEED-D0 | 20290 | 8979 | 8373 | 1784 | 3672.8 | 1701 | 3493.4 | 1701 | 88.21 | 1468 | 2392.9 | 1384 | 2338.7 | 1384 | 91.76 |
| AA-D9 | 20967 | 9113 | 8373 | 579 | 1103.8 | 525 | 1005.2 | 525 | 95.98 | 416 | 680.8 | 354 | 638.5 | 354 | 97.40 |
| AO-D9 | 26104 | 10031 | 8373 | 1100 | 2298.1 | 1096 | 2378.8 | 1096 | 92.76 | 846 | 1282.6 | 777 | 1264.2 | 777 | 95.45 |
| AA-D35 | 24325 | 9416 | 8373 | 868 | 1898.0 | 773 | 1797.0 | 773 | 93.49 | 623 | 1063.8 | 537 | 1023.3 | 537 | 95.95 |
| AO-D35 | 9501 | 8373 | 8373 | 1012 | 1624.5 | 907 | 1776.9 | 907 | 93.26 | 764 | 1047.8 | 666 | 1089 | 666 | 95.90 |
| AA-D55 | 28232 | 10861 | 8373 | 626 | 1220.9 | 555 | 1147.6 | 555 | 95.52 | 401 | 652.4 | 346 | 585.4 | 346 | 97.65 |
| AO-D55 | 25193 | 10792 | 8373 | 1023 | 1780.9 | 925 | 1851.9 | 925 | 92.99 | 727 | 927.0 | 648 | 958.1 | 648 | 96.38 |
| AA-D76 | 24447 | 10022 | 8373 | 747 | 1348.6 | 680 | 1476.1 | 680 | 94.83 | 519 | 765.2 | 450 | 798.9 | 450 | 97.00 |
| AO-D76 | 28278 | 11572 | 8373 | 1003 | 1952.6 | 930 | 2089.6 | 930 | 92.99 | 744 | 1119.6 | 668 | 1120.4 | 668 | 95.71 |
The diversity indices, namely, OTUs, Chao1, Chao2, ACE, Bootstrap, and GOOD’s coverage based on the above-described normalized sequences at both the 3% and 6% cut-off levels, were summarized in Table 2. On the basis of OTUs number, the microbial diversity of the seed sludge was the highest because it was taken from the full scale WWTP, which dealt with more complex pollutants in municipal wastewater than the synthetic influent. Moreover, sludge in the A-O SBR was more diverse than that in the A-A SBR, and this may have been because of the anaerobic-anoxic condition of the A-A SBR laying additional burden on the microbial community, thereby resulting in a less diverse community after the acclimation phase. Similar results to the OTUs analysis were observed with Chao1, Chao2, ACE, and Bootstrap as well as the rarefaction curve (Fig. S1).
β-diversity
Two independent methods, Cluster analysis (CA) and Principal coordinate analysis (PCoA) were adopted to compare the similarity of the sludge microbial community among the two SBRs and seed sludge. Based on the abundances of the OTUs at the 3% cut-off and phylum levels, Cluster analysis (Fig. S2) and Principal coordinate analysis (Fig. S3) showed that sludge samples from the A-A SBR were clustered together as one group and samples from the A-O SBR formed another group, which indicated the diverse microbial community in denitrifying phosphorus removal sludge and conventional phosphorus removal sludge. Moreover, after acclimation under different operation modes, the community structure in the A-O SBR was more similar to the seed sludge (A2/O process) than the denitrifying A-A SBR sludge. This may have been because of the similar redox conditions between the A-O SBR and real A2/O process.
The identity of denitrifying phosphorus removal bacteria remains controversial. Based on the results of the above β-diversity analysis, the introduction of the A-A condition may have induced a shift in the bacterial community structure in EBPR, which has been reported in a previous FISH study (15).
Dominant taxonomy
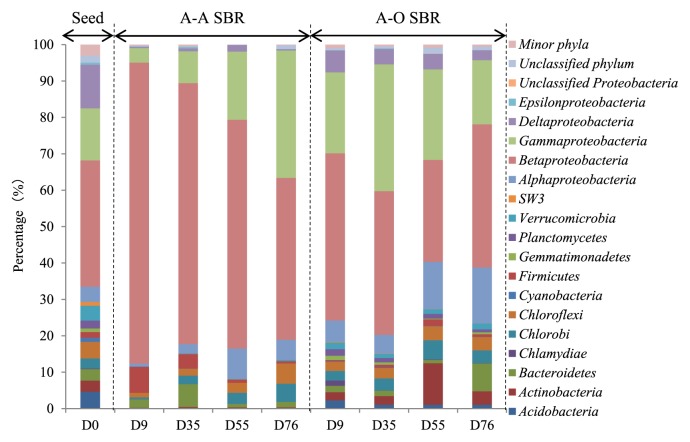
The 8,373 selected effective bacterial sequences in each sample were assigned to different taxa (from phylum to genus) via Global Alignment for Sequence Taxonomy (GAST), as shown in Fig. 3.
Fig. 3.
Abundances of different phyla and classes in Proteobacteria in the 9 sludge samples.
Similar to the community structure of globally distributed WWTP (28, 36, 40), Proteobacteria were the most abundant phylum in all samples, accounting for 65.8%–91.8% of the total bacterial sequences. Other dominant phyla were Chloroflexi (1.27–5.54%, averaged at 3.20%), Chlorobi (0.47–5.27%, averaged at 3.16%), Bacteroidetes (0.97– 7.64%, averaged at 2.89%), and Actinobacteria (0.16–11.4%, averaged at 2.64%), followed by several other major (average abundance>1%) phyla including Firmicutes (1.97%), Acidobacteria (1.16%) and Verrucomicrobia (1.15%). In addition, Acidobacteria, Actinobacteria, and Verrucomicrobia were the major phyla in the seed and A-O sludges, with abundances in the A-A sludge being markedly low (averaging at 0.09%, 0.24%, and 0.12% respectively).
Within Proteobacteria, Betaproteobacteria was the most dominant class in all nine samples, followed by Gammaproteobacteria, Alphaproteobacteria, and Deltaproteobacteria in the eight samples from the two SBRs, while the abundant classes in the seed sludge were Gammaproteobacteria, Deltaproteobacteria, and Alphaproteobacteria. Moreover, the abundances of Epsilonproteobacteria were only 0–0.59% (average of 0.16%) in the nine sludge samples. In addition to the four classes of Proteobacteria, there were some other dominant shared classes (abundance>1% occurred in at least 50% of samples) including Chlorobia, Actinobacteria, Anaerolineae, Sphingobacteria, and Flavobacteria.
The abundance of Betaproteobacteria decreased from 82.7% to 44.6% in sludge samples collected from the A-A SBR during the 76-day operation, while the abundance of Gammaproteobacteria increased by nearly 9-fold from 3.99% to 34.9%. The abundances of Alphaproteobacteria and Deltaproteobacteria also slightly increased after the acclimation phase. As for the A-O SBR, the abundances of Betaproteobacteria and Gammaproteobacteria fluctuated within a small range during acclimation, while that of Alphaproteobacteria slightly increased and Deltaproteobacteria decreased.
Apart from the important phosphorous removal relevant functional groups, which have been discussed below, the abundances of filamentous organisms that cause sludge bulking and foaming were also compared. The abundances of filamentous genera including Caldilinea, Gordonia, Microthrix, Nostocoida, Trichococcus, and Zoogloea (10) in denitrifying and aerobic phosphorus removal sludges were 1.76% and 0.681%, respectively, in samples collected on day 76. Their abundances were slightly higher in denitrifying phosphorus removal sludge, which may have been the reason for its slightly worse sludge-setting ability than that of aerobic phosphorus removal sludge.
These results indicated that microbial communities changed during the acclimation phase, especially in the A-A SBR, which led to a significant shift in the microbial community as a result of the A-A operation.
Validation of V6 amplicon-based microbial population quantification by independent metagenomic analysis
In the present study, taxonomic profiling of sludge microbial communities mainly relied on the sequencing and classification of the PCR-amplified 16S ribosomal RNA gene V6 region. Owing to its universality in prokaryotes and the availability of comprehensive reference databases, the 16S rRNA gene is regarded as a powerful phylogenetic marker; however, it has been criticized for biases introduced by copy-number variations, inconsistent amplification efficiencies, and heterogeneity when targeting different 16S rRNA regions (http://www.nature.com/nmeth/journal/vaop/ncurrent/full/nmeth.2693.html#ref4).
Due to the possible bias in PCR-based quantification, an independent metagenomic-based analysis MetaPhlAn was applied to evaluate the microbial community analysis of sludge samples (March 22, 2013) collected from the A-A SBR based on V6 amplicon. MetaPhlAn is a newly developed analysis platform for profiling microbial communities that uses unique clade-specific marker genes as a BLAST database selected from more than 3,000 reference microbial genomes (29). Due to its current cost disadvantage over 16S rRNA gene amplicon-based methods, MetaPhlAn is rarely utilized to routinely profile community shift dynamics; however, its strength in comprehensive and accurate taxonomic profiling have been demonstrated for representative samples analysis ranging from the human microbiome (24) to wastewater treatment plant (3).
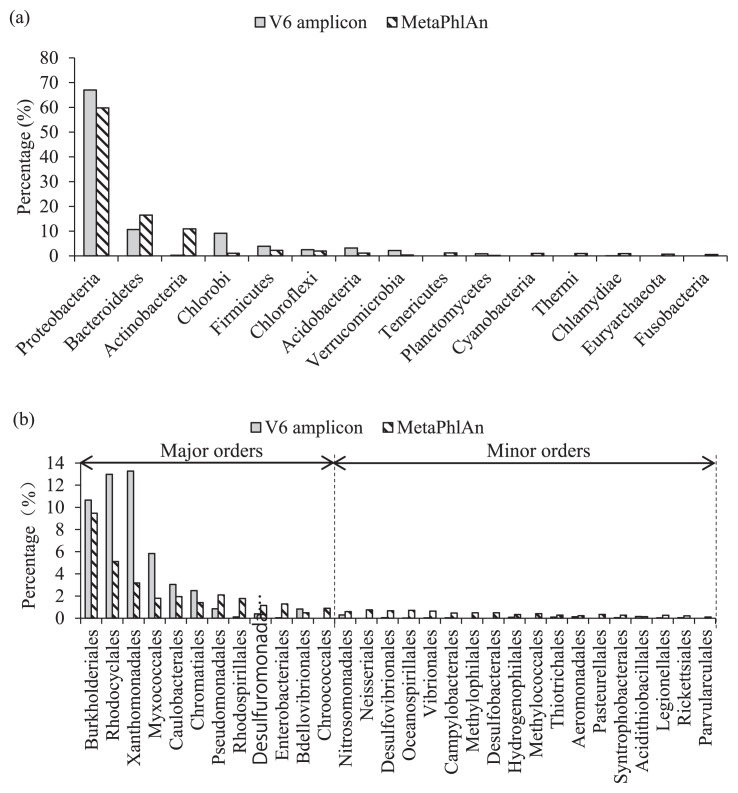
Fig. 4(a) shows the distribution pattern of both methods at the phylum level. Seven major phyla (including Proteobacteria and Bacteroidetes) were ranked as the top phyla by both methods. These top phyla accounted for 97.7% and 92.9% of the total community, as revealed by V6 amplicon and MetaPhlAn respectively. The minor difference observed in the phylum level distribution may have been due to the lack of clade-specific marker genes in the environmental metagenome (soil, sediment, or sludge).
Fig. 4.
Abundances of phyla (a) and orders within Proteobacteria (b) of the A-A SBR sludge (day 274) by V6 amplicon and MetaPhlAn methods.
As was demonstrated in the present and previous studies (28, 36, 40), Proteobacteria dominated in all sludge samples; therefore, the abundances of different orders within Proteobacteria were comprehensively compared. Fig. 4(b) showed the abundances of covered orders within Proteobacteria (8) by the two methods. In spite of certain abundance fluctuations, both methods revealed similar distribution characteristics of the major and minor orders.
These results supported the effectiveness of the current V6 amplicon-based quantitative analysis of microbial communities.
DPAO in a strict A-A system
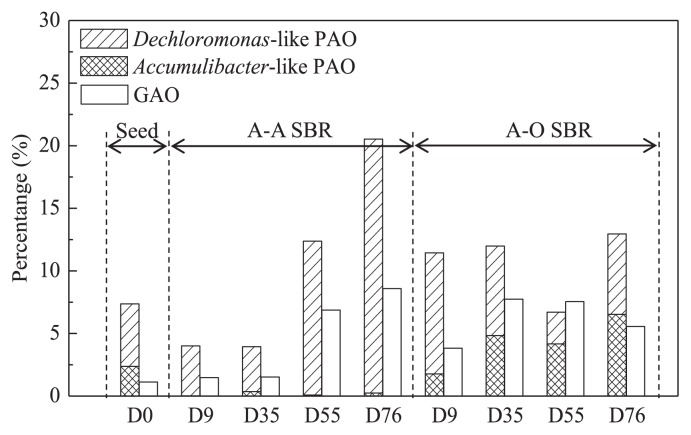
An important task was to identify the PAO (or DPAO) responsible for phosphorus removal in the current A-A and A-O SBRs. The traditional strategy for this relied on sequence similarity comparisons against already identified PAO. As previously described, Candidatus “Accumulibacter” was the most frequently reported and solely well accepted PAO in the activated sludge system. Thus, local BLAST (Basic Local Alignment Search Tool) was firstly attempted using a self-made local database containing all 41 typical Candidatus “Accumulibacter” sequences that had summarized in a phylogenetic tree by Kim et al. (13). The results shown in Fig. 5 indicated that 2.36% of the sequences in the seed sludge could be assigned to Candidatus “Accumulibacter”- related sequences (OTU196). Regarding the A-O sludge, the abundances of Candidatus “Accumulibacter”-related sequences were 1.77–4.83% and 4.16–6.53% for the acclimation and stationary phases. These two ratios were slightly below the average reported PAO percentages of 6–22% (7, 43), but were still acceptable. However, only 0.02–0.36% (0.16% on average) of sequences in the A-A sludge were significantly similar to the known Candidatus “Accumulibacter”, which was in contrast to the similar phosphorus removal efficiencies between A-A and A-O SBR. If we accepted that the nearly negligible 0.16% Candidatus “Accumulibacter”- related population could not have performed the observed luxury phosphorus uptake flux, the only explanation was that a novel DPAO must exist in the current A-A system. Therefore, to identify this novel DPAO, another strategy emphasizing the isolation source of the related NCBI Genbank sequences was attempted, as illustrated in Fig. S4 and Table S1. After screening, OTU-1 came to the fore based on its significant abundance, as shown in Fig. 5, and frequent detection records in very similar phosphorous-removing SBR systems (9, 42) and full-scale EBPR (19, 37). Based on all these findings, OTU-1 was proposed as the main DPAO responsible for phosphorus removal in the current strict A-A system.
Fig. 5.
Dynamics of the Accumulibacter-like PAO (OTU-196), Dechloromonas-like PAO (OTU-1), and GAO (OTUs-87 and 312) populations in A-A and A-O systems.
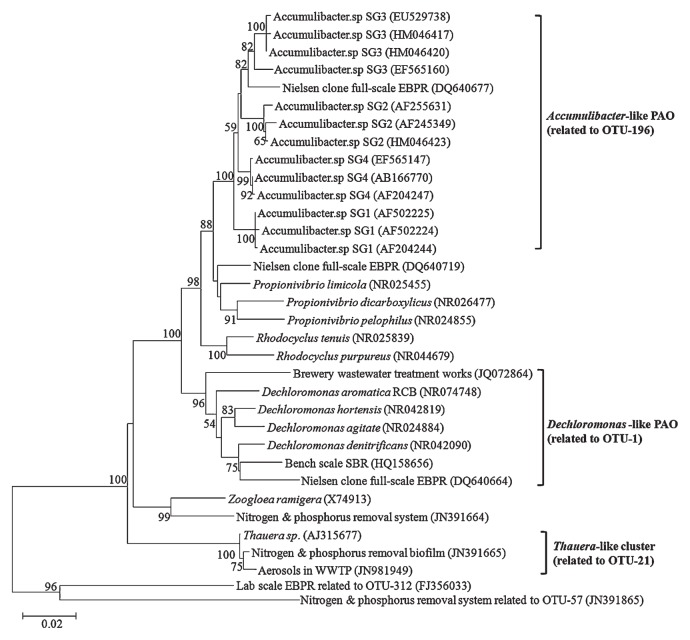
The phylogenetic relationship between OTU-1, the Rhodocyclus-related 4 clades (13) of Candidatus “Accumulibacter”, and the other important species within the family Rhodocyclaceae was shown in Fig. 6. All selected environmental sequences related to OTU-1 fell in a separate cluster that was related more to Dechloromonas denitrificans. OTU-1 shared similar sequences in the V6 region to Dechloromonas-related organisms from a full-scale EBPR clone (DQ640664) identified by Kong et al. (19). In their study, this group of organisms, which matched probe Bet135, was shown to be positive for both polyphosphates and PHA staining. Thus, the Bet135-defined organism was proposed to be putative DPAO and was suspected to behave similarly to Candidatus “Accumulibacter” in terms of the uptake of substrates and storage of PHA and polyphosphate (19). This group of PAO may play an important role in WWTP because many identical sequences that originated in the phosphorus removal sludge, e.g. HQ158656 and JQ072864, have been deposited to the NCBI GenBank. Unfortunately, their potential phosphorus removal function has not been properly annotated. In the present study, the Dechloromonas-related organism was further detected as putative DPAO in an independent environment, and was able to tolerate long-term strict A-A operation (Fig. 5), which was attributed to its competitive advantage over Candidatus “Accumulibacter”. Thus, our results supported and also extended previous findings (15, 19).
Fig. 6.
Phylogenetic tree.
One clear advantage of our identification strategy was its independence on known PAO sequence information. The ability of organisms to cycle phosphorus for energy during EBPR was exhibited by organisms from a range of different phylogenies inclusive of, but not restricted to, members of Gammaproteobacteria, Betaproteobacteria, and Actinobacteria (26). In our approach, each abundant OTU was checked by the isolation source of their close relatives in the NCBI. This was the reason why one Dechloromonas-related organism outside of the four known clades could be successfully targeted. In the current case, the uptake and storage behavior of phosphorous by the organism had already been comprehensively characterized by Kong’s pioneering work. Otherwise, the candidates identified by our approach should be strictly examined using in situ techniques.
Dynamics of PAO and the glycogen-accumulating organisms (GAO) population in A-A and A-O systems
The dynamics of PAO, including the Accumulibacter-like PAO and Dechloromonas-like PAO population, during the whole acclimation phase was shown in Fig. 5. Only minor abundance variations were observed in PAO in the A-O SBR (6.06%–12.0%), and this was attributed to a similar redox condition to that of A2O/WWTP. Regarding the A-A SBR, PAO abundances decreased in the initial acclimation phase (3.98% on average) due to the pressure associated with the anaerobic-anoxic condition. After acclimation for approximately two months, the PAO percentage gradually increased to 16.9% on average at the stable phase, which was consistent with the demonstrated phosphorus removal performance.
The presence of GAO is known to potentially compete with PAO due to its uptake of VFA under anaerobic conditions, but not the accumulation of polyphosphate under aerobic conditions (14, 16). GAO was quantified by Local BLAST of each sample with a local database containing 48 known GAO sequences (Table S2). As shown in Fig. 5, the abundance of GAO in the seed sludge was low (0.93%) as a result of operation control in WWTP. In the acclimation phase, the percentage of GAO in A-O SBR was markedly higher than that in A-A SBR, with an average percentage of 4.65% and 1.22%, respectively. In the stable phase, the abundances of GAO in the A-O and A-A SBRs averaged 6.19% and 5.26%, respectively, which suggested that the abundance of GAO was higher under the comparatively singular conditions of anaerobic-anoxic or anaerobic-oxic than full-scale WWTP.
Conclusion
The quantification of PAO and GAO in the present study provided a simple and convenient approach for monitoring the important functional microbial population in wastewater treatment systems.
Supplementary Information
Acknowledgements
We gratefully acknowledge the financial support of the China Major Science and Technology Program for Water Pollutant Control and Treatment (2012ZX07313001), National Natural Science Foundation of China (31200104), Shenzhen Basic Research Project (JC201105160582A), Guangdong Natural Science Foundation Funding (S2012040006350), and China Postdoctoral Science Foundation Funded Project (2012M510975).
References
- 1.Barker PS, Dold PL. Denitrification behaviour in biological excess phosphorus removal in activated sludge systems. Water Res. 1996;30:769–780. [Google Scholar]
- 2.Broughton B, Pratt S, Shilton A. Enhanced biological phosphorus removal for high-strength wastewater with a low rbCOD: P ratio. Bioresource Technol. 2008;99:1236–1241. doi: 10.1016/j.biortech.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Cai L, Zhang T. Detecting human bacterial pathogens in wastewater treatment plants by a high-throughput shotgun sequencing technique. Environ Sci Technol. 2013;47:5433–5441. doi: 10.1021/es400275r. [DOI] [PubMed] [Google Scholar]
- 4.Carvalho G, Lemos PC, Oehmen A, Reis MAM. Denitrifying phosphorus removal: linking the process performance with the microbial community structure. Water Res. 2007;41:4383–4396. doi: 10.1016/j.watres.2007.06.065. [DOI] [PubMed] [Google Scholar]
- 5.Flowers JJ, He S, Yilmaz S, Noguera DR, McMahon KD. Denitrification capabilities of two biological phosphorus removal sludges dominated by different “Candidatus Accumulibacter” clades. Env Microbiol Rep. 2009;1:583–588. doi: 10.1111/j.1758-2229.2009.00090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freitas F, Temudo M, Reis MAM. Microbial population response to changes of the operating conditions in a dynamic nutrient-removal sequencing batch reactor. Bioproc Biosyst Eng. 2005;28:199–209. doi: 10.1007/s00449-005-0029-9. [DOI] [PubMed] [Google Scholar]
- 7.Fukushima T, Uda N, Okamoto M, Onuki M, Satoh H, Mino T. Abundance of Candidatus‘Accumulibacter phosphatis’ in enhanced biological phosphorus removal activated sludge acclimatized with different carbon sources. Microbes Environ. 2007;22:346–354. [Google Scholar]
- 8.Garrity GM, Bell JA, Lilburn TG. Bergey’s Manual of Systematic Bacteriology. 2nd ed. Springer New York Inc; New York: 2004. Taxonomic Outline of the Prokaryotes. [Google Scholar]
- 9.Gonzalez-Gil G, Holliger C. Dynamics of microbial community structure of and enhanced biological phosphorus removal by aerobic granules cultivated on propionate or acetate. Appl Environ Microb. 2011;77:8041–8051. doi: 10.1128/AEM.05738-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo F, Zhang T. Profiling bulking and foaming bacteria in activated sludge by high throughput sequencing. Water Res. 2012;46:2772–2782. doi: 10.1016/j.watres.2012.02.039. [DOI] [PubMed] [Google Scholar]
- 11.He S, Gall DL, McMahon KD. ‘Candidatus Accumulibacter’ population structure in enhanced biological phosphorus removal sludges as revealed by polyphosphate kinase genes. Appl Environ Microb. 2007;73:5865–5874. doi: 10.1128/AEM.01207-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapagiannidis AG, Zafiriadis I, Aivasidis A. Comparison between UCT type and DPAO biomass phosphorus removal efficiency under aerobic and anoxic conditions. Water Sci Technol. 2009;60:2695–2703. doi: 10.2166/wst.2009.703. [DOI] [PubMed] [Google Scholar]
- 13.Kim JM, Lee HJ, Kim SY, Song JJ, Park W, Jeon CO. Analysis of the fine-scale population structure of “Candidatus Accumulibacter phosphatis” in enhanced biological phosphorus removal sludge, using fluorescence in situ hybridization and flow cytometric. Appl Environ Microb. 2010;76:3825–3835. doi: 10.1128/AEM.00260-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim JM, Lee HJ, Lee DS, Lee K, Jeon CO. Identification of a novel subgroup of uncultured gammaproteobacterial glycogen-accumulating organisms in enhanced biological phosphorus removal sludge. Microbiol-SGM. 2011;157:2287–2296. doi: 10.1099/mic.0.047779-0. [DOI] [PubMed] [Google Scholar]
- 15.Kim JM, Lee HJ, Lee DS, Jeon CO. Characterization of the denitrification-associated phosphorus uptake properties of “Candidatus Accumulibacter phosphatis” clades in sludge subjected to enhanced biological phosphorus removal. Appl Environ Microb. 2013;79:1969–1979. doi: 10.1128/AEM.03464-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo T, Ebie Y, Tsuneda S, Inamori Y. Detection of Defluvicoccus-related glycogen-accumulating organisms in enhanced biological phosphorus removal processes. Microbes Environ. 2007;22:190–195. [Google Scholar]
- 17.Kondo T, Tsuneda S, Ebie Y, Inamori Y, Xu KQ. Characterization of the microbial community in the anaerobic/oxic/anoxic process combined with sludge ozonation and phosphorus adsorption. J Water Environ Technol. 2009;7:155–162. [Google Scholar]
- 18.Kong YH, Nielsen JL, Nielsen PH. Micro-autoradiographic study of Rhodocyclus-related polyphosphate-accumulating bacteria in full-scale enhanced biological phosphorus removal plants. Appl Environ Microb. 2004;70:5383–5390. doi: 10.1128/AEM.70.9.5383-5390.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong YH, Xia Y, Nielsen JL, Nielsen PH. Structure and function of the microbial community in a full-scale enhanced biological phosphorus removal plant. Microbiol-SGM. 2007;153:4061– 4073. doi: 10.1099/mic.0.2007/007245-0. [DOI] [PubMed] [Google Scholar]
- 20.Kuba T, Smolders G, van Loosdrecht MCM, Heijnen JJ. Biological phosphorus removal from wastewater by anaerobic-anoxic sequencing batch reactor. Water Sci Technol. 1993;27:241–252. [Google Scholar]
- 21.Kuba T, van Loosdrecht MCM, Heijnen JJ. Phosphorus and nitrogen removal with minimal cod requirement by integration of denitrifying dephosphatation and nitrification in a two-sludge system. Water Res. 1996;30:1702–1710. [Google Scholar]
- 22.Lee N, Nielsen PH, Aspegren H, Henze M, Schleifer KH, Jansen JL. Long-term population dynamics and in situ physiology in activated sludge systems with enhanced biological phosphorus removal operated with and without nitrogen removal. Syst Appl Microbiol. 2003;26:211–227. doi: 10.1078/072320203322346065. [DOI] [PubMed] [Google Scholar]
- 23.Meinhold J, Filipe CDM, Daigger GT, Isaacs S. Characterization of the denitrifying fraction of phosphate accumulating organisms in biological phosphate removal. Water Sci Technol. 1999;39:31–42. [Google Scholar]
- 24.Neville BA, Sheridan PO, Harris HMB, et al. Proinflammatory flagellin proteins of prevalent motile commensal bacteria are variably abundant in the intestinal microbiome of elderly humans. PloS One. 2013;8:e68919. doi: 10.1371/journal.pone.0068919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng WJ, Ong SL, Hu JY. Denitrifying phosphorus removal by anaerobic/anoxic sequencing batch reactor. Water Sci Technol. 2001;43:139–146. [PubMed] [Google Scholar]
- 26.Okunuki S, Nakamura K, Kawaharasaki M, Tanaka H, Uchiyama H, Noda N. Quantification of Rhodocyclus-related and actinobacterial polyphosphate-accumulating organisms in an enhanced biological phosphorus removal process using quenching probe PCR. Microbes Environ. 2007;22:106–115. [Google Scholar]
- 27.Peterson SB, Warnecke F, Madejska J, McMahon KD, Hugenholtz P. Environmental distribution and population biology of Candidatus Accumulibacter, a primary agent of biological phosphorus removal. Environ Microbiol. 2008;10:2692–2703. doi: 10.1111/j.1462-2920.2008.01690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satoh H, Oshima K, Suda W, Ranasinghe P, Li N, Gunawardana EGW, Hattori M, Mino T. Bacterial population dynamics in a laboratory activated sludge reactor monitored by pyrosequencing of 16S rRNA. Microbes Environ. 2013;28:65–70. doi: 10.1264/jsme2.ME12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva AF, Carvalho G, Oehmen A, Lousada-Ferreira M, van Nieuwenhuijzen A, Reis MAM, Crespo MTB. Microbial population analysis of nutrient removal-related organisms in membrane bioreactors. Appl Microbiol Biotechnol. 2012;93:2171–2180. doi: 10.1007/s00253-011-3499-5. [DOI] [PubMed] [Google Scholar]
- 31.Sul WJ, Cole JR, Jesus ED, Wang Q, Farris RJ, Fish JA, Tiedje JM. Bacterial community comparisons by taxonomy-supervised analysis independent of sequence alignment and clustering. Proc Natl Acad Sci USA. 2011;108:14637–14642. doi: 10.1073/pnas.1111435108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsuneda S, Ohno T, Soejima K, Hirata A. Simultaneous nitrogen and phosphorus removal using denitrifying phosphate-accumulating organisms in a sequencing batch reactor. Biochem Eng J. 2006;27:191–196. [Google Scholar]
- 33.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–487. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vargas M, Guisasol A, Artigues A, Casas C, Baeza JA. Comparison of a nitrite-based anaerobic-anoxic EBPR system with propionate or acetate as electron donors. Process Biochem. 2011;46:714–720. [Google Scholar]
- 35.Wachtmeister A, Kuba T, van Loosdrecht MCM, Heijnen JJ. A sludge characterization assay for aerobic and denitrifying phosphate removing sludge. Water Res. 1997;31:471–478. [Google Scholar]
- 36.Wang XH, Wen XH, Yan HJ, Ding K, Zhao F, Hu M. Bacterial community dynamics in a functionally stable pilot-scale wastewater treatment plant. Bioresource Technol. 2011;102:2352–2357. doi: 10.1016/j.biortech.2010.10.095. [DOI] [PubMed] [Google Scholar]
- 37.Yang C, Zhang W, Liu R, Li Q, Li B, Wang S, Song C, Qiao C, Mulchandani A. Phylogenetic diversity and metabolic potential of activated sludge microbial communities in full-scale wastewater treatment plants. Environ Sci Technol. 2011;45:7408–7415. doi: 10.1021/es2010545. [DOI] [PubMed] [Google Scholar]
- 38.You SJ, Shen YJ, Ouyang CF, Hsu CL. The effect of residual chemical oxygen demand on anoxic and aerobic phosphate uptake and release with various intracellular polymer levels. Water Environ Res. 2004;76:149–154. doi: 10.2175/106143004x141672. [DOI] [PubMed] [Google Scholar]
- 39.Zeng RJ, Saunders AM, Yuan ZG, Blackall LL, Keller J. Identification and comparison of aerobic and denitrifying polyphosphate- accumulating organisms. Biotechnol Bioeng. 2003;83:140–148. doi: 10.1002/bit.10652. [DOI] [PubMed] [Google Scholar]
- 40.Zhang T, Shao MF, Ye L. 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J. 2011;6:1137–1147. doi: 10.1038/ismej.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou SQ, Zhang XJ, Feng LY. Effect of different types of electron acceptors on the anoxic phosphorus uptake activity of denitrifying phosphorus removing bacteria. Bioresource Technol. 2010;101:1603–1610. doi: 10.1016/j.biortech.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 42.Zhu X, Chen Y. Reduction of N2O and NO generation in anaerobic-aerobic (low dissolved oxygen) biological wastewater treatment process by using sludge alkaline fermentation liquid. Environ Sci Technol. 2011;45:2137–2143. doi: 10.1021/es102900h. [DOI] [PubMed] [Google Scholar]
- 43.Zilles JL, Peccia J, Kim MW, Hung CH, Noguera DR. Involvement of rhodocyclus-related organisms in phosphorus removal in full-scale wastewater treatment plants. Appl Environ Microb. 2002;68:2763–2769. doi: 10.1128/AEM.68.6.2763-2769.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.