SUMMARY
The inflammatory microenvironment promotes skin tumorigenesis. However, the mechanisms of how cells protect themselves from inflammatory signals have yet to be revealed. Downregulation of IKKα promotes skin tumor progression from papillomas to squamous cell carcinomas, which is frequently accompanied by genomic instability, including aneuploid chromosomes and extra centrosomes. In this study, we found that IKKα promoted oligomerization of nucleophosmin (NPM), a negative centrosome duplication regulator, which further enhanced NPM and centrosome association, inhibited centrosome amplification, and maintained genome integrity. Levels of NPM hexamers and IKKα were conversely associated with skin tumor progression. Importantly, pro-inflammatory cytokine-induced IKKα activation promoted the formation of NPM oligomers and reduced centrosome numbers in mouse and human cells, whereas kinase-dead IKKα blocked this connection. Therefore, our findings suggest a previously unknown mechanism in which an IKKα-NPM axis may use the inflammatory signal to suppress centrosome amplification, promote genomic integrity, and prevent tumor progression.
INTRODUCTION
IKKα (also called Chuk) is a serine/threonine protein kinase that can be activated by various cytokines through different receptors in a kinase-dependent manner (Ghosh and Karin, 2002). However, IKKα regulates keratinocyte differentiation and proliferation in the skin independently of its kinase activity (Hu et al., 2001). IKKα downregulation promotes skin carcinogenesis and its deletion induces spontaneous squamous cell carcinomas (SCCs) of the skin, lungs, and forestomach in mice (Liu et al., 2008; Park et al., 2007; Xiao et al., 2013). In humans, IKKα downregulation has been reported in SCCs of the skin, lungs, esophagus, and head and neck (Marinari et al., 2008). A single mutation-generated IKKα deletion has been identified in human lethal syndrome (cocoon syndrome), in which multiple organs are malformed (Lahtela et al., 2010). These findings highlight the importance of IKKα in the pathogenesis of these human diseases. Recently, we have demonstrated that inflammation promotes IKKα reduction-initiated SCC development in mice (Xiao et al., 2013). Similar inflammatory phenotypes have been observed in human SCCs. This evidence prompted us to investigate whether pro-inflammatory cytokine-induced IKKα activation may antagonize skin tumorigenesis.
The centrosome, a small organelle composed of pericentriolar material and tubulin proteins, generates polar spindles that segregate chromosomes into two daughter cells during mitosis (Nigg, 2002). Normal cells contain one or two centrosomes. The centrosome duplication cycle is incorporated into the cell cycle. Many cell cycle regulators, tumor suppressors, and oncogenic proteins regulate centrosome duplication. The presence of extra centrosomes is frequently associated with cancer (D’Assoro et al., 2002; Nigg, 2002). The extra centrosomes have been demonstrated to generate multipolar spindles that directly mediate merotelic attachments, a type of exacerbating erroneous attachment of spindle microtubules to chromosomes during chromosome segregation, thereby inducing chromosome missegregation and promoting chromosomal instability and aneuploidy (Ganem et al., 2009). Centrosome amplification can initiate tumorigenesis in flies (Basto et al., 2008). Therefore, proper centrosome replication may be pivotal for maintaining genomic stability and preventing tumor development.
NPM regulates centrosome duplication and binds to centrosomes at the M phase (Okuda et al., 2000). After Cdk2 and cyclin E phosphorylate Thr199 in NPM, NPM dissociates from centrosomes, which in turn triggers centrosome duplication at the late G1 phase. Blocking NPM dissociation from centrosomes inhibits centrosome duplication. Conversely, NPM loss leads to unrestricted centrosome duplication and genomic instability in mouse embryonic fibroblasts (MEFs) (Grisendi et al., 2005). Npm−/− mice exhibit embryonic lethality, and NPM reduction promotes c-Myc-induced leukemia development in mice. The N-terminal region of the Npm gene can be fused with the anaplastic lymphoma kinase, retinoic acid receptor α, or myeloid leukemia factor 1 gene through genomic DNA breaks in several types of human leukemia (Grisendi and Pandolfi, 2005). NPM has multiple functions and partners. On the other hand, NPM has been proposed to have an oncogenic activity (Yung, 2007). The controversial functions of NPM may be partially due to its various partners in particular cellular events. We have observed increased NPM levels in benign skin papillomas and reduced NPM levels in malignant skin carcinomas (Zhu et al., 2009), indicating that NPM may regulate skin tumorigenesis.
Here, we show that IKKα enhances the association between NPM and centrosomes through phosphorylating NPM, which prevents centrosome amplification and genomic instability. Unexpectedly, this IKKα-NPM axis responds to inflammatory signals. Since, IKKα expression is downregulated in SCCs, decreased IKKα cannot provide a connection between the inflammatory microenvironment and control of centrosome duplication, thereby promoting centrosome amplification and genomic instability. Our finding reveals a mechanism for how IKKα may prevent skin tumor progression in the inflammatory microenvironment.
RESULTS
IKKα Interacts with NPM and Phosphorylates S125 of NPM
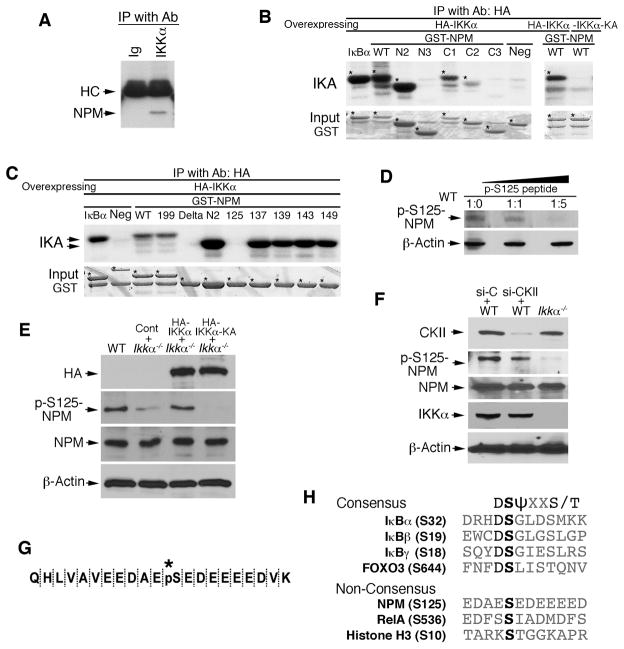
To study the relationship between IKKα and NPM in keratinocytes, we first compared the status of NPM using two-dimensional gel electrophoresis and found that its isoelectric status was different in primary cultured Ikkα−/− keratinocytes compared to wild-type (WT) keratinocytes (Figure S1A). Using immunoprecipitation (IP), an interaction between NPM and IKKα was detected, but not between NPM and IKKβ in keratinocytes (Figures 1A and S1B). We identified the interaction regions of IKKα and NPM by co-expressing Flag-tagged NPM with HA-tagged WT IKKα and four other mutant forms or co-expressing WT HA-IKKα with WT Flag-NPM and six other deletion forms, followed by IP and western blotting (Figures S1C–S1E). The results showed that WT IKKα interacted with WT NPM, although IKKα bound weakly to an N-terminal-truncated NPM, suggesting that the interaction between IKKα and NPM requires both proteins in their entirety.
Figure 1. NPM Interacts with IKKα and Phosphorylates S125 of NPM.
(A) Interaction of IKKα and NPM in primary cultured keratinocytes detected by immunoprecipitation (IP) with anti-IKKα antibody (Ab) and western blot with anti-NPM antibody. Immunoglobulin (Ig), control for IP; HC, antibody heavy chain.
(B–C) Phosphorylation of NPM by IKKα was analyzed with an immunocomplex kinase assay (IKA). HA-IKKα was precipitated with anti-HA antibody using lysates of HEK293 cells overexpressing HA-IKKα or HA-IKKα-KA. GST-NPM proteins were used as kinase substrates. Five phosphorylation sites within aa 119–195 of NPM were examined using IKA. GST IκBα, positive control for IKK; Neg, GST-14-3-3σ protein as a kinase-negative control; *, bands indicating labeled proteins.
(D) The specificity of p-S125-NPM from WT MEF lysates was analyzed by western blot with anti-p-S125-NPM antibody that was incubated with a specific p-S125-NPM peptide for 2 hr at room temperature. β-Actin, protein-loading control. Ratio, antibody:peptide.
(E) Western blot shows HA-IKKα (HA), p-S125-NPM and NPM levels in WT and Ikkα−/− MEFs transfected with HA-IKKα, HA-IKKα-KA, or control vector (Cont).
(F) Western blot shows CKII, p-S125NPM, NPM, and IKKα levels in Ikkα−/− and WT MEFs treated with control siRNA (si-C) or CKII siRNA (si-CKII).
(G) Phosphorylated S125-NPM sequence (*, pS) was analyzed using trypsin digestion and mass spectrometry.
(H) The consensus sequences containing serine sites that can be phosphorylated by IKKα and IKKβ. S, serine; D, aspartic acid; T, threonine; ψ, hydrophobic amino acid; X, any amino acid. See also Figure S1.
To determine whether IKKα phosphorylates NPM, we used different NPM deletion and mutation forms as kinase substrates and found that IKKα phosphorylated the amino acid (aa) 119–195 region, which contains five serine sites, but a kinase-dead IKKα mutant (IKKα-KA) did not phosphorylate NPM (Figures 1B and 1C; Figure S1E). We then replaced each serine (S) with alanine (A) to generate five mutated NPM proteins (S125A, S137A, S139A, S143A, and S149A). S125A and a delta form lacking the aa 119–195 region completely abolished NPM phosphorylation by IKKα (Figure 1C), suggesting that IKKα phosphorylates S125 of NPM. IKKα efficiently phosphorylated NPMThr199A (Thr199 was replaced by alanine [Okuda et al., 2000]) (Figure 1C). Although IKKβ also phosphorylated the aa 119–195 region of NPM, it did not show specificity for one of these serine sites (Figure S1F). Because IKKα is specifically associated with NPM, we focused on understanding the relationship between IKKα and NPM.
We generated an anti-phospho-(p)-S125-NPM antibody and the specific p-S125-NPM peptide blocked the p-S125 antibody activity in a dose-dependent manner in WT cells (Figure 1D). Re-expressing WT HA-IKKα restored S125-NPM phosphorylation in Ikkα−/− cells, but re-expressing HA-IKKα-KA did not (Figure 1E). Previous reports suggest that CKII kinase phosphorylates S125-NPM (Chan et al., 1986; Szebeni et al., 2003). To determine whether IKKα requires CKII for phosphorylating S125-NPM, we knocked down CKII expression in WT MEFs using small interfering RNA (siRNA), which did not affect NPM levels. Although CKII siRNA slightly reduced p-S125-NPM level in WT MEFs, the p-S125-NPM level in Ikkα−/− cells was lower than that in WT cells lacking CKII (Figure 1F), suggesting that IKKα induced the phosphorylation of S125-NPM independently of CKII.
Mass spectrometry analysis further confirmed that p-S125 was associated with IKKα and not IKKα-KA (Figure 1G; Figures S3C–S3E). Together, these results indicate that IKKα specifically phosphorylates S125-NPM. The sequence surrounding S125 of NPM is not a consensus phosphorylation site like the consensus S32 of IκBα (Figures 1G and 1H). A comparison of the similarities and differences between the phosphorylation sequences of several proteins and the activities of IKKα and IKKβ on these proteins suggests that IKKβ likely shows stronger kinase activity for those proteins containing consensus sequences than IKKα does (Hu et al., 2004; Mercurio et al., 1997), whereas IKKα preferentially phosphorylates proteins containing non-consensus sequences (Anest et al., 2003; Hoberg et al., 2006) (Figure 1H). Thus, IKKα and IKKβ kinase activities are not identical.
Immunofluorescent (IF) staining with the anti-p-S125-NPM antibody revealed substantially reduced staining in the skin of kinase-dead Ikkα (IkkαKA/KA) mice compared to WT mice (Figure S1G). Western blot consistently showed reduced p-S125-NPM levels in the skin and heart of Ikkα−/− newborn mice compared to WT (Figure S1H). These results suggest a physiological relationship between IKKα and S125-NPM.
S125 Phosphorylation Promotes NPM-NPM Interaction and NPM Hexamer Formation
To determine the functional relationship between IKKα and S125-NPM, we examined NPM levels in primary cultured Ikkα+/+, Ikkα+/−, and Ikkα−/− keratinocytes and found that, although the NPM levels at the expected molecular size (37 kD) remained similar, increased levels of NPM hexamers with a molecular size of approximately 230 kD were correlated with increased IKKα levels in a dose-dependent manner. Conversely, increased levels of small NPM oligomers were associated with decreased IKKα levels in the keratinocytes (Figure 2A, left and right panels). IKKα siRNA treatment reduced NPM hexamer levels in a dose-dependent manner (Figure 2B). Previous studies showed the presence of NPM monomers and hexamers in cells by using either native or denaturing protein gel electrophoresis, and the NPM hexamer levels were lower in denaturing gels than in native gels (Herrera et al., 1996; Yung and Chan, 1987). We detected NPM hexamer levels using the two different gel methods (data not shown). As the denaturing gel produced the most consistent results, we used it in this study. NPM hexamer formation can be altered by many factors, such as cell cycle regulation, small molecule inhibitors, salts, and several NPM mutations (Chou and Yung, 1995; Herrera et al., 1996; Liu and Chan, 1991; Zirwes et al., 1997). Here, our results showed that IKKα regulated NPM hexamer formation.
Figure 2. IKKα Regulates NPM Oligomerization via S125-NPM.
(A) Western blot shows NPM monomer and hexamer, IKKα, and γ-tubulin (γ-tub) levels in primary cultured Ikkα−/−, Ikkα+/−, and Ikkα+/+ keratinocytes.
(B) The effect of IKKα siRNA on NPM oligomerization was detected with western blot. Mock, no siRNA; Cont, nonspecific siRNA (400 nM).
(C) Binding of Flag-NPM, Flag-NPMS125A (S125A), and Flag-NPMThr199A (T199A) in HEK293 cells was compared using IP with anti-Flag antibody and western blot (WB) with anti-NPM antibody at short (left) and long (right) exposures.
(D) Binding abilities of Flag-NPM, Flag-NPMS125A, and Flag-NPMThr199A from HEK293 cells with GST-NPM protein were compared using GST pull-down assay and western blot with anti-Flag antibody. GST-NPM, loaded GST-NPM protein.
(E) Western blot shows NPM, NPM hexamer, and IKKα levels in normal skin (Sk), papillomas (Pa) and carcinomas (Ca). Regress, tumor regress; Progress, tumor progression; *, fine band. βActin, protein loading control.
(F) Western blot shows IKKα, p-IKKα, p-IKKβ, p-S125-NPM, NPM, and NPM hexamer levels in HEK293 cells following TNFα (10 ng/ml) stimulation.
(G) Western blot shows NPM hexamer, NPM, IKKα, IKKβ, p-S125-NPM, and p-IKKα levels in WT and IkkαKA/KA MEFs following anti-LtβR antibody (500 ng/ml) treatment.
(H) qPCR shows relative expression levels of Ltα in normal skin and skin tumors induced by chemical carcinogens. p<0.05 examined by Student’s t-test. The results are presented as mean ± SD of four samples. See also Figure S2.
We then investigated whether S125 regulates NPM oligomerization by introducing Flag-tagged NPM, NPMS125A and NPMThr199A into cells, followed by IP with an anti-Flag antibody and western blott with an anti-NPM antibody. Because NPM interacts with NPM, the anti-Flag antibody should pull down both Flag-tagged NPM and endogenous NPM. Flag-NPMS125A showed a much weaker ability to pull down Flag-NPMS125A and endogenous NPM, and formed fewer hexamers than Flag-NPM, whereas Flag-NPMThr199A showed a stronger ability to perform these activities than Flag-NPM and Flag-NPMS125A (Figure 2C, left and right panels). A GST-NPM protein consistently pulled down substantially more Flag-NPMThr199A and Flag-NPM, including NPM monomers and hexamers than Flag-NPMS125A (Figure 2D). We also found reduced NPM hexamer and p-S125-NPM levels in IkkαKA/KA cells and that reintroducing WT IKKα increased NPM hexamer and p-S125-NPM levels (Figure S2A). These results indicate that S125 may stabilize NPM hexamers by strengthening NPM oligomerization.
We then examined the correlation between NPM hexamer levels, IKKα and skin tumors, including papillomas and carcinomas derived from BL6 mice, induced by the chemical carcinogens 7,12-dimethylbenz[a]-anthracene (DMBA) and 12-O-tetradecanoylphorbol-13-acetate (TPA) (Park et al., 2007). In this setting, approximately 95% of the papillomas eventually repress and only 5% progress to malignant carcinomas. TPA treatment increases IKKα levels in the skin (Gu et al., 2004). Western blot showed significantly increased NPM hexamer and IKKα levels in papillomas compared to untreated normal skin (Figure 2E). Conversely, the NPM hexamer and IKKα levels in carcinomas were markedly decreased compared to normal skin and papillomas (Figure 2E). These results indicate a good correlation between reduced NPM hexamer and IKKα levels and tumor progression. NPM loss increased centrosome numbers (Grisendi et al., 2005). Consistently, we observed increased centrosome numbers in carcinomas compared to papillomas and normal skin (Figure S2B). We also detected increased centrosome numbers in skin SCCs derived from IkkαKA/KA mice (Figure S2C). Previously, we reported spontaneous skin SCCs in a proportion of IkkαKA/KA mice, associated with IKKα reduction and marked inflammation (Xiao et al., 2013). Increased TNFα, a major pro-inflammatory cytokine, has been observed in the inflammatory microenvironment during skin and lung SCC development and increased IKKα has been reported to inhibit skin tumor progression (Liu et al., 2006; Park et al., 2007; Xiao et al., 2013). We found that TNFα treatment induced IKKα phosphorylation and increased NPM hexamers, which were correlated with increased NPM phosphorylation (Figure 2F). Because TNFα also activated IKKβ (Figure 2F), we next treated WT and IkkαKA/KA MEFs with an anti-lymphotoxin beta receptor (LtβR) antibody (agonist) (Dejardin et al., 2002) to specifically activate IKKα. This agonist markedly elevated NPM hexamers in WT cells, but not in IkkαKA/KA cells (Figure 2G), and the increased hexamers correlated with the increased IKKα phosphorylation in WT cells, indicating that IKKα kinase is required for hexamerization. Previously, we reported reduced IKKα in IkkαKA/KA cells (Xiao et al., 2013). Although increased NPM hexamer levels were associated with IKKα phosphorylation in both the HEK293 cells and WT MEFs, the NPM hexamer peak then decreased quickly, suggesting that a mechanism that downregulates NPM hexamer formation is present. We will investigate this in the future.
Lymphotoxin alpha (Ltα) is a ligand of LtβR. We found increased Ltα and TNFα expression in mouse skin tumors induced by DMBA and TPA (Park et al., 2007) compared to normal skin (Figure 2H; Figure S2D). Together, these results suggest that this IKKα-NPM axis may use the pro-inflammatory cytokine-induced signal to promote NPM hexamer formation in vivo.
IKKα Co-localizes with NPM in Centrosomes, and NPM Hexamers Stabilize the Association of NPM and Centrosomes
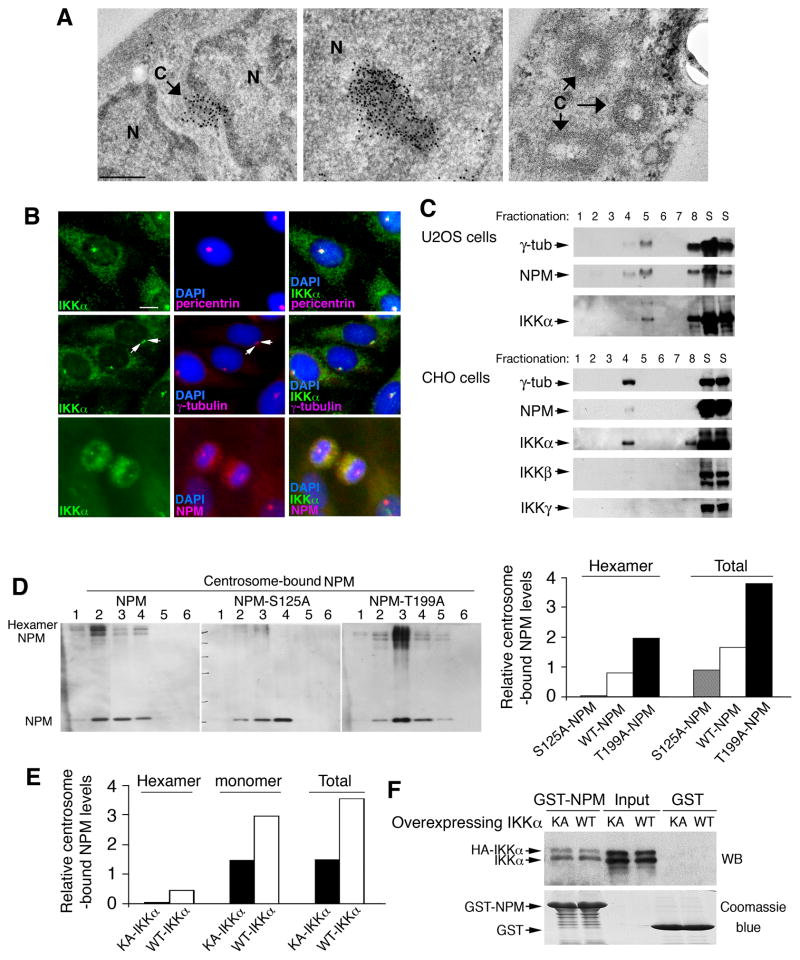
The function of NPM hexamers in centrosomes was previously unknown. An immunoelectron microscopy approach confirmed that immunostained NPM polymers were associated with centrosomes (Figure 3A, left). Immunostained NPM polymers associated with nucleoli were used as a positive control (Bertwistle et al., 2004) (Figure 3A, middle). Chinese hamster ovary (CHO) and U2OS human osteosarcoma cells have been widely used to study centrosome duplication (Meraldi et al., 1999). IF staining showed that IKKα co-localized with one or two centrosomes and that IKKα co-localized with NPM in centrosomes at the M phase (Figure 3B), suggesting that IKKα associates with centrosomes in both the M phase and interphase in CHO cells. NPM was also found to co-localize with centrosomes at the M phase in primary cultured keratinocytes (Figure S3A). We confirmed that the antibody against γ-tubulin or centrin (centrosome components) stained the same centrosomes in cells (Figure S3B). Also, purified centrosomes (γ-tubulin containing fractions) from U2OS and CHO cells contained IKKα and NPM, but not IKKβ and IKKγ (Figure 3C).
Figure 3. S125 of NPM Regulates Centrosome-Bound NPM Levels.
(A) Left: Immunoelectron microscopy shows the association of NPM and centrosomes. N, nucleus; C, cen trosome; dark spots, immunostained NPM. Scale bar, 250 nm. Middle: Immunostained NPM associates with a nucleolus as positive-staining control. Right: unstained centrosomes as negative-staining control.
(B) Co-localization of IKKα with pericentrin, γ-tubulin, and NPM in CHO cells was detected with IF staining. Each arrow indicates a centrosome. Blue, DAPI staining. Scale bar, 10 μm.
(C) Fractions of purified centrosomes obtained from U2OS and CHO cells using small-scale sucrose-gradient ultracentrifugation, followed by western blot. Numbers indicate fractions. S, supernatants of cells; γ-tub, γ-tubulin.
(D) Left: Levels of centrosome-bound Flag-NPM, Flag-NPMS125A (S125A), and Flag-NPMThr199A (T199A) hexamers and monomers, detected by western blot with anti-Flag antibody. Number, centrosome fractions. Right: Relative density levels of centrosome-bound NPM were measured using densitometry (Image Station 440; Eastman Kodak, Rochester, NY) and analyzed using an image analysis software program (ImageQuant TL v2003.01; Amersham Biosciences Corp.). Each relative value was determined from six fractions and then divided by the density levels of γ-tubulin and overexpressed Flag-NPM.
(E) Centrosome-bound NPM from CHO cells overexpressing IKKα or IKKα-KA was detected using western blot with anti-NPM antibody. Signals from the western blot were converted to relative density levels using densitometry (Image Station 440) and analyzed using the ImageQuant TL software program (version v2003.01). Each relative value was collected from six fractions.
(F) Interaction of GST-NPM with IKKα (WT) or IKKα-KA (KA) in CHO cells was analyzed by GST pull-down followed by western blot (WB) with anti-IKKα antibody. GST protein loading was stained with Coomassie blue. Input, HA-IKKα and HA-IKKα-KA levels in total cell lysates. See also Figure S3.
To examine whether NPM hexamers regulate the association of NPM and centrosomes through S125-NPM, we isolated centrosomes from CHO cells transfected with Flag-NPM, Flag-NPMS125A, and Flag-NPMThr199A and examined centrosome-bound NPM levels. The total levels of centrosome-bound Flag-NPMS125A (NPM monomers and hexamers) were markedly lower than the total levels of centrosome-bound Flag-NPM (Figure 3D, left and right panels). In contrast, the total levels of centrosome-bound Flag-NPMThr199A (monomers and hexamers) were markedly increased compared to Flag-NPM. Moreover, overexpressed IKKα-KA reduced centrosome-bound NPM hexamer levels in cells compared to overexpressed WT IKKα (Figure 3E). Both WT IKKα and IKKα-KA were found to interact with NPM (Figure 3F). To determine whether IKKα-KA can compete with WT IKKα for interaction with NPM, we co-expressed Flag-NPM/HA-IKKα or Flag-NPM/HA-IKKα-KA in HEK293 cells and immunoprecipitated Flag-NPM with an anti-Flag antibody. The isolated Flag-NPM protein was digested with trypsin and was analyzed using mass spectrometry (Figures S3C–S3E). The S125 site is located in a 31-aa peptide. We found more peptides containing unphosphorylated S125 in cells co-expressing Flag-NPM/HA-IKKα-KA than in cells co-expressing Flag-NPM/HA-IKKα. These results suggest that IKKα-KA may block S125-NPM phosphorylation, reducing centrosome-bound NPM hexamers and NPM. Collectively, the S125-NPM phosphorylation enhanced NPM hexamer levels to stabilize the association of NPM with centrosomes.
IKKα Loss Induces Centrosome Amplification and Increases Chromosome Numbers
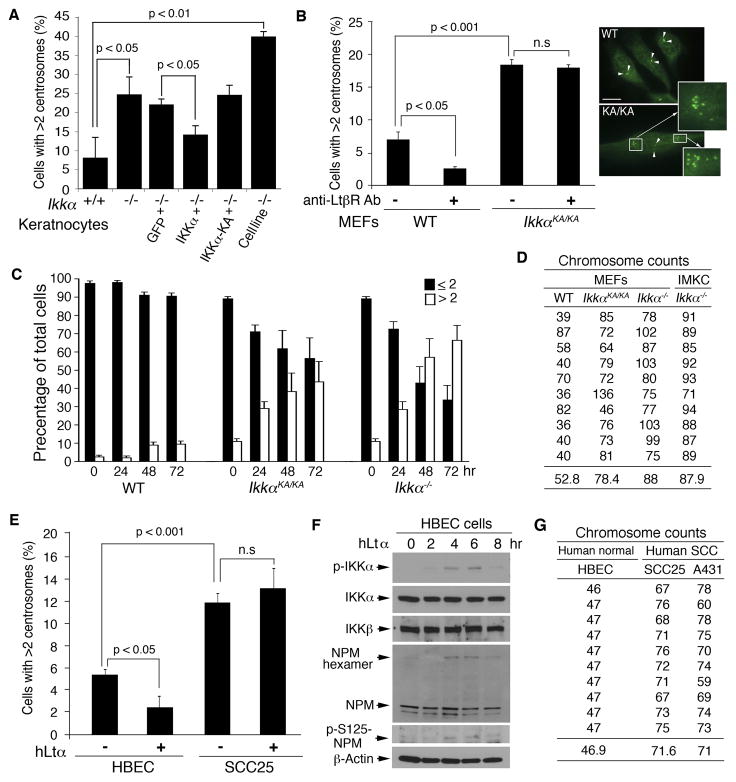
We further examined the effect of IKKα loss on centrosome numbers. IF staining showed increased centrosome numbers in primary Ikkα−/− keratinocytes and Ikkα−/− immortal keratinocytes (Liu et al., 2009) compared to WT keratinocytes (Figure 4A). Reintroducing WT IKKα, but not IKKα-KA, reduced centrosome numbers in Ikkα−/− keratinocytes. Consistently, centrosome numbers were increased in IkkαKA/KA MEFs compared to WT MEFs (Figure 4B, left and right). The anti-LtβR antibody treatment reduced centrosome numbers in WT MEFs, but not in IkkαKA/KA MEFs (Figure 4B, left), suggesting that IKKα kinase activation specifically represses centrosome amplification.
Figure 4. IKKα Deficiency Is Associated with Centrosome Amplification and Increased Chromosome Numbers.
(A) Centrosome numbers in primary cultured WT (+/+) and Ikkα−/− (−/−) keratinocytes and Ikkα−/− immortalized keratinocytes (Cell line) detected by IF staining. p value was determined by Student’s t-test. The results are presented as mean ± SD from 3 experiments.
(B) Left: Comparison of untreated and anti-LtβR antibody (500 ng/ml) treated WT and IkkαKA/KA (KA/KA) MEFs containing more than two centrosomes detected by IF staining. The statistical result (p) was analyzed by Student’s t-test. n.s, no significant difference. The results are presented as mean ± SD from 3 experiments. Right: Stained centrosomes (green) indicated by arrows. Multiple centrosomes are enlarged from small boxes to large boxes (arrows). Scale bars, 10 μm.
(C) Analysis for centrosome duplication in WT, IkkαKA/KA and Ikkα−/− MEFs treated with 2 mM hydroxyurea. Centrosome numbers were analyzed by staining with anti-γ-tubulin antibody. Black column, cells containing one or two centrosomes; White column, cells containing more than two centrosomes. The results represent as average ± SD from 3 experiments.
(D) Comparison of chromosome numbers from 10 cells in each type of cells: WT, Ikkα−/−, and IkkαKA/KA MEFs, as detected by SKY.
(E) Centrosome numbers in HBEC and SCC25 cells treated with hLtα (50 ng/ml) detected by IF staining. n.s, no significant difference. The results are presented as mean ± SD from 3 experiments.
(F) Western blot shows the indicated protein levels in HBEC cells treated with hLtα (50 ng/ml). β-Actin, protein loading control.
(G) Comparison of chromosome numbers from 10 cells in each type of cells: HBEC, A431, and SCC25 cells as detected by SKY. See also Figure S4.
We further examined whether centrosome duplication is deregulated in Ikkα−/− and IkkαKA/KA cells using a standard centrosome duplication assay (Meraldi et al., 1999; Nigg, 2002). We treated WT, IkkαKA/KA, and Ikkα−/− MEFs with hydroxyurea and examined centrosome numbers at 24, 48, and 72 hr. The numbers of IkkαKA/KA and Ikkα−/− cells with amplified centrosomes increased progressively; however, the number of WT cells with amplified centrosomes did not increase (Figure 4C). The progression of centrosome amplification was more severe in Ikkα−/− than in IkkαKA/KA cells. These results suggest that IKKα loss or inactivation is associated with centrosome amplification. Using spectral karyotyping (SKY) analysis, we assayed chromosome instability and found dramatically increased chromosome numbers in Ikkα−/− and IkkαKA/KA MEFs and immortal Ikkα−/− immortal keratinocytes compared to WT cells (Figure 4D; Figure S4A). These results suggest that IKKα deficiency induces centrosome amplification, which may be associated with chromosome instability, although the detailed mechanism remains to be investigated. To determine the relevance of this study to human tumor development, we compared numbers of centrosomes in a normal human bronchial epithelial cell line (HBEC) (Xiao et al., 2013) and a human tongue SCC line (SCC25), and found markedly increased centrosomes in SCC25 cells compared to HBEC cells (Figure 4E). Human Ltα (hLtα) treatment reduced centrosome numbers in HBEC cells, but not in SCC25 cells (Figure 4F). Furthermore, hLtα treatment increased levels of IKKα and NPM phosphorylation and NPM hexamers in human HBEC cells, indicating that human and mouse cells share a common mechanism for the control of centrosome duplication. Chromosome numbers were also increased in SCC25 and the A431 line, derived from a human epidermal tumor, compared to HEBC cells (Figure 4G; Figure S4B). Western blot showed reduced levels of IKKα and increased levels of Aurora A, a kinase that promotes cell mitosis (Prajapati et al., 2006) and phosphorylated Aurora A in A431 and SCC25 cells compared to HBEC cells; while reintroduced IKKα reduced Aurora A and phosphorylated Aurora A levels in A431 and SCC25 cells (Figure S4C), suggesting that IKKα downregulates Aurora A expression, which is different from the result reported by Prajapati et al (2006). We will further investigate whether IKKα acts differently in different types of cells and how IKKα regulates Aurora A expression.
p53 has been shown to regulate centrosome duplication (D’Assoro et al., 2002; Fukasawa et al., 1996). Therefore, we examined whether IKKα-null cells have a defect in p53 expression. Following treatment with ultraviolet B (UVB) or doxorubicin, p53 and p21 were appropriately induced in both primary WT and Ikkα−/− keratinocytes (Figure S4D), suggesting that IKKα can regulate centrosomes independent of p53 in primary keratinocytes.
Mutant S125A-NPM Induces Centrosome Amplification
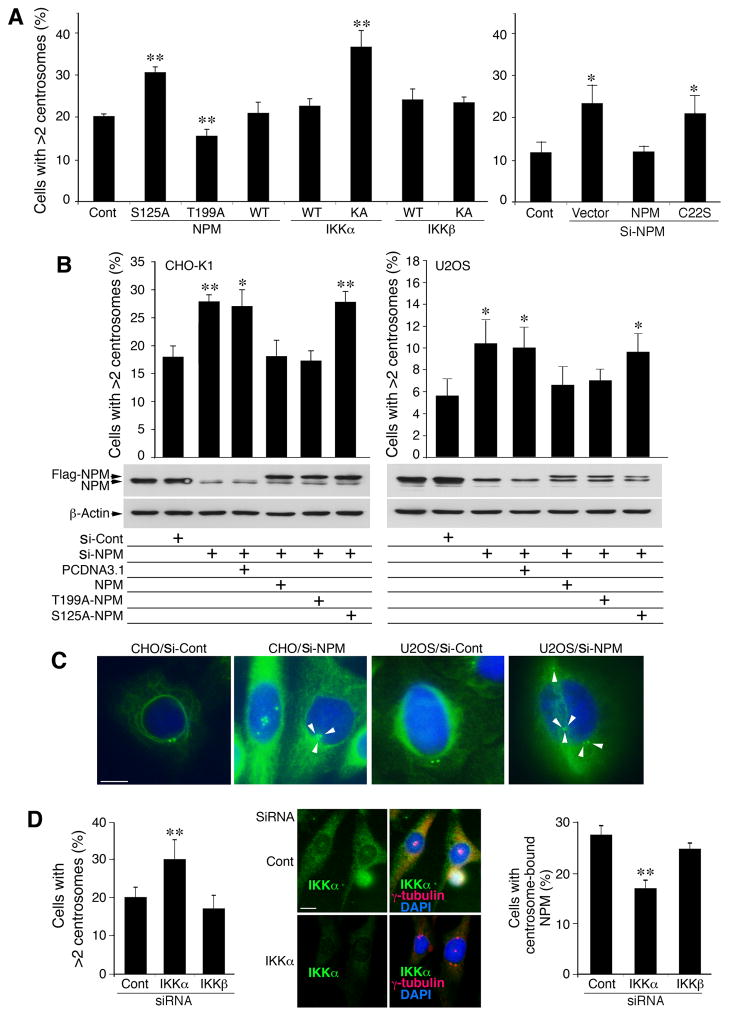
We further hypothesized that the S125A-NPM mutant or other NPM mutations that reduce NPM hexamers may increase centrosome amplification. We then introduced NPM and NPMS125A into CHO cells. NPMThr199A was used as a functional control because it inhibits centrosome duplication (Tokuyama et al., 2001); IKKα, IKKα-KA, IKKβ and IKKβ-KA were used as controls. NPMS125A or IKKα-KA induced significantly more centrosomes than WT NPM, WT IKKα, IKKβ, or IKKβ-KA, whereas NPMThr199A reduced centrosomes significantly (Figure 5A). To determine whether NPM oligomerization regulates centrosomes as a common mechanism, we generated three NPM mutants (Figure S5A). A mimic NPMS125D slightly increased NPM hexamer levels and did not cause centrosome amplification (Figure 5A; Figures S5A–S5C). NPMC22S and NPMM579L, previously reported to reduce NPM oligomerization (Liu and Chan, 1991; Zirwes et al., 1997), were found to reduce NPM hexamers and increase centrosome amplification (Figures 5A; Figures S5A–S5C). These results support the notion that NPM oligomerization is a mechanism involved in preventing centrosome amplification.
Figure 5. S125 of NPM Suppresses Centrosome Amplification.
(A) Effects of NPM, NPMS125A (S125A), NPMThr199A (T199A), IKKα, IKKα-KA, IKKβ, IKKβ-KA, and NPMC22S (C22S; see Figure S6A) on centrosome amplification in CHO cells. Each bar is compared to the control (Cont, untreated cells). Si-, siRNA; Cont, untreated cells. **, p < 0.01; *, p < 0.05 examined by Student’s t-test. Bar graphs are presented as mean ± SD from 3 experiments.
(B) Top: Effects of WT NPM, NPMS125A (S125A) and NPMThr199A (T199A) on centrosome amplification in CHO and U2OS cells lacking endogenous NPM. Each column is compared to the control (Cont). Si-, siRNA. **, p < 0.01; *, p < 0.05 examined by Student’s t-test. Bar graphs are presented as mean ± SD. Middle and bottom: western blot shows NPM and Flag-NPM levels in cells treated with NPM siRNA (si-NPM), control siRNA (si-Cont) or overexpression of Flag-NPM, T199A-NPM or S125A-NPM. PCDNA3.1, vector control.
(C) Centrosomes in CHO and U2OS cells treated with NPM siRNA or control siRNA were stained with IF with anti-γ-tubulin antibody (green). Blue, DAPI for nuclear staining. Scale bar, 10 μm.
(D) Left: Effects of reduced IKKα on centrosome amplification in CHO cells treated with IKKα siRNA, IKKβ siRNA or control siRNA (Cont). Middle: Reduced endogenous IKKα expression and increased centrosome numbers in CHO cells treated with IKKα siRNA (siRNA) or control siRNA (Cont), detected by IF staining. Scale bar, 10 μm. Right: Effects of IKKα siRNA and IKKβ siRNA on centrosome-bound NPM levels in CHO cells, detected by IF staining with antibodies against NPM and γ-tubulin. NPM colocalized with γ-tubulin was considered centrosome-bound NPM. **, p < 0.01 examined by Student’s t-test. Bar graphs are presented as mean ± SD from 3 experiments. See also Figure S5.
We depleted endogenous NPM from CHO and U2OS cells by NPM siRNA. NPM knockdown dramatically increased centrosome numbers in these cells (Figures 5B and 5C). In cells treated with NPM siRNA, re-expressing NPM and NPMThr199A reconstituted endogenous NPM function, but NPMS125A failed to do so (Figure 5B). The result further demonstrated that S125-NPM prevented centrosome amplification.
We next tested whether the kinase activity of IKKα plays a role in preventing centrosome amplification by co-expressing NPM/IKKα, NPM/IKKα-EE (a constitutively active form of IKKα [Mercurio et al., 1997]), or NPM/IKKα-KA in CHO cells, after deleting the endogenous NPM with siRNA. NPM/IKKα and NPM/IKKα-EE decreased centrosome numbers, while NPM/IKKα-KA increased centrosome numbers (Figure S5D). Moreover, re-expressed IKKα or IKKα-EE repressed centrosome amplification in KA/KA cells, but IKKα-KA did not (Figure S5D). Furthermore, we depleted endogenous IKKα in CHO cells using siRNA and found that IKKα siRNA increased centrosome numbers, but IKKβ siRNA and the control siRNA did not (Figure 5D, left and middle panels). We used IF staining to localize NPM in the centrosomes of those cells treated with control siRNA, IKKα siRNA, or IKKβ siRNA (Tokuyama et al., 2001). The results showed that the percentage of cells containing centrosome-bound NPM was lower among the cells treated with IKKα siRNA than those treated with control siRNA and IKKβ siRNA (Figure 5D, right panel), suggesting that IKKα enhances the association of NPM and centrosomes. Thus, both S125-NPM and IKKα kinase are required for preventing centrosome amplification.
To verify the effect of the presence or absence of IKKα kinase on S125-NPM phosphorylation associated with centrosomes in cells, we performed IF staining and found that the anti-p-S125-NPM antibody stained centrosomes in cells containing one centrosome (G1 phase) and in dividing cells (M phase; Figure S5E), but did not stain centrosomes well in cells containing two centrosomes (S or G2 phase; Figure S5E). These results are consistent with the relationship between NPM and the centrosome cycle during the cell cycle (Okuda et al., 2000). However, the anti-p-S125-NPM antibody did not stain centrosomes in dividing IkkαKA/KA cells (Figure S5E), suggesting a relationship between p-S125-NPM and IKKα kinase activity in centrosomes during the cell cycle regulation.
NPM Is a Potential Target in Human Skin SCCs
Moreover, we examined both NPM and IKKα levels in human skin samples in tissue arrays containing human skin SCCs, basal cell carcinomas (BCCs), melanomas, condyloma acuminata of the cunnus (keratinocytic tumor-like lesions), hyperplastic epidermis, epidermal tissues adjacent to skin tumors, and normal skin. The immunohistochemical staining showed that NPM was well expressed in normal human epidermal and epithelial tissues, in the hyperproliferating epidermis with inflammation, and in tumor-adjacent tissues (Figures 6A and 6B, left). This NPM staining pattern was consistent with the pattern in the NPM-stained human lung epithelial cells reported previously (Mascaux et al., 2008). Approximately 45% of skin SCCs and a proportion of BCCs and melanomas expressed reduced NPM (Figure 6A, left). Interestingly, approximately half of the human condylomas expressed little or markedly reduced NPM. IKKα expression was reduced in 72% of human SCCs and in 90% of human BCCs (Figure 6A, right). Further analysis of the relationship between IKKα and NPM levels showed that 47.8% of human skin SCCs and 26.7% of BCCs with downregulated NPM expressed reduced IKKα. However, we did not observe a correlation between the expression of IKKα and NPM in condylomas. Decreased NPM levels also impair NPM activity, suggesting that reduced NPM, IKKα, or NPM/IKKα may contribute to human SCC development via the mechanism identified in this study (Figure 7).
Figure 6. NPM Is a Potential Target for the Development of Human Skin Cancers.
(A) The graph shows NPM and IKKα levels in human tissue arrays containing skin SCCs, adjacent tissues surrounding SCCs, and hyperplasia of squamous epithelium, detected using immunohistochemical staining with anti-NPM (left) or anti-IKKα antibody (right). Staining was classified as +++, ++, + and −, according to staining intensity.
(B) Human tissue sections from arrays that were immunohistochemically stained with anti-NPM or anti-IKKα antibody. Brown, positive for NPM or IKKα; blue, nuclear counterstaining; epidermis stained as +++; skin SCC stained as −; condyloma stained as −; PC, breast tissue stained with anti-NPM antibody as positive control; NC, breast tissue stained with isotype control antibody as negative control within black lines. Scale bars, 50 μm.
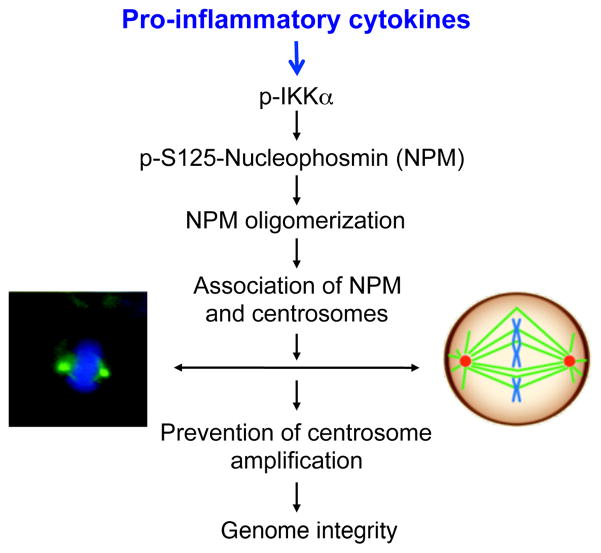
Figure 7. A Working Model for the IKKα-NPM Axis.
Left photo shows two centrosomes stained by γ-tubulin (green) during cell mitosis. Blue, nucleus stained with DAPI. Right picture indicates two centrosomes (red) that link to chromosomes (blue) through spindles (green) during mitosis. The IKKα and NPM axis contributes to maintaining normal centrosome numbers through enhancing the association of NPM hexamers and centrosomes. P-, phosphorylation.
DISCUSSION
Appropriate molecular surveillants at various cellular events protect cells, maintaining organ homeostasis. Defects in these crucial molecules may be involved in the pathogenesis of the diseases and cancer. Chronic inflammation surrounding tumor loci is a common phenomenon and promotes tumor initiation and progression (Xiao et al., 2013). Whether the specific molecules can use the harmful inflammatory signaling to protect cells during tumorigenesis remains to be revealed. Here, we identified a new function of IKKα in suppressing centrosome amplification. IKKα interacted with and phosphorylated NPM, which further prevented centrosome amplification and genomic instability and inflammatory signaling activated IKKα to regulate centrosome duplication. The tumor progression requires multi-molecular alterations. IKKα downregulation impairs this protective connection in the inflammatory microenvironment, which may cooperate with other non-immune mechanisms to promote tumor progression.
NPM monomers can form hexamers (Chou and Yung, 1995). In this study, we showed that IKKα regulated the formation of NPM hexamers in a dose-dependent manner in primary cultured keratinocytes and in other types of cells. Importantly, in BL6 mice, increased NPM hexamer and IKKα levels were associated with papillomas induced by DMBA and TPA (Park et al., 2007), most of which eventually regressed. Conversely, reduced NPM hexamer and IKKα levels correlated with skin tumor progression. These good correlations suggest that IKKα and NPM hexamers may play a preventive role during skin carcinogenesis.
Our study further demonstrated that IKKα interacted with NPM and phosphorylated S125-NPM, which promoted NPM NPM interaction and NPM hexamer formation. The increased NPM hexamer levels promoted the association of NPM and centrosomes, which prevented centrosome amplification. IKKα loss, kinase-dead IKKα, NPM loss, or NPMS125A decreased levels of NPM hexamers and the association of NPM with centrosomes, thereby promoting centrosome amplification. Furthermore, the NPMThr199A mutant increased NPM hexamer levels and repressed centrosome numbers, and the NPMC22S and NPMM579L mutants decreased hexamer levels and enhanced centrosome amplification (Liu and Chan, 1991; Zirwes et al., 1997). These results suggest that NPM hexamerization may serve as a common mechanism for regulating the association of NPM and centrosomes. On the other hand, overexpressed IKKα or kinase-active IKKα repressed centrosome amplification, which may partially explain the result where overexpressed IKKα in keratinocytes repressed skin tumor progression in Lori. IKKα mice with FVB background treated with chemical carcinogens (Liu et al., 2006) and in K5. IKKα mice treated with UVB radiation (Xia et al., 2013). The increased inflammation is required to cooperate with UVB or DMBA/TPA carcinogen in skin carcinogenesis.
IKKα and IKKβ, two highly conserved protein kinases, display distinct kinase activities for some proteins, which may also contribute to their physiological functions (Mercurio et al., 1997). Interestingly, IKKα has been reported to phosphorylate serine 10 of histone H3, but not IKKβ (Anest et al., 2003; Yamamoto et al., 2003), however, we did not detect this H3 phosphorylation in keratinocytes (Zhu et al., 2007), suggesting that IKKα kinase activity has a cell-type specificity, which may partially explain why the combined molecular alterations, including IKKα downregulation, kinase-dead IKKα, and inflammation, can induce spontaneous skin, lung, and forestomach SCCs in IkkαKA/KA mice, although kinase-independent IKKα activities also contribute to the tumorigenesis (Liu et al., 2008; Xiao et al., 2013).
The association of extra (supernumerary) centrosomes with malignant tumors has been known for more than 100 years (Godinho et al., 2009). Only recently have studies provided direct evidence demonstrating how extra centrosomes cause chromosome instability (Ganem et al., 2009). In addition, although extra centrosomes may cause cell cycle arrest or cell death in normal cells, tumor and immortalized cells possessing supernumerary centrosomes usually divide successfully (Godinho et al., 2009). Human SCCs show increased chromosome numbers (Weaver and Cleveland, 2006). In the current study, we also detected increased centrosomes and chromosomes in human SCC cells compared to normal human epithelial cells. Consistently, chromosome numbers were abnormally increased in Ikkα−/− and IkkαKA/KA mouse cells compared to WT cells. Together, because centrosome abnormalities are one of the mechanisms for causing aneuploidy and our results demonstrated a good correlation between centrosome amplification/increased chromosomes and mutated IKKα/NPM, lack of IKKα and NPM function may increase centrosome amplification, which further cooperates with other molecular events to facilitate chromosome and genome changes; thus, the benign tumor cells may be prone to malignant transformation in the inflammatory microenvironment. In the future, we will investigate underlying mechanisms for increased chromosome numbers in these mouse and human cells since additional molecules may be required for maintaining the proper chromosome numbers. Reduced NPM was detected in human and mouse skin SCCs, but how NPM is downregulated in malignant skin tumors still needs to be addressed in the future. Notably, the percentage of human skin SCCs expressing reduced IKKα was higher than the skin SCCs expressing reduced NPM. Since IKKα deletion is able to induce spontaneous skin carcinomas (Liu et al., 2008), downregulated IKKα may play a predominant role in driving skin tumor development, while NPM is one of IKKα’s targets. In addition, Npm mutations have been reported in human leukemia (Grisendi and Pandolfi, 2005), and Ikkα mutations have been detected in human SCCs and other types of cancers (Greenman et al., 2007; Liu et al., 2006). Here, we showed that mutations and deletions in IKKα and/or NPM abolished the interaction between IKKα and NPM and attenuated their function in repressing centrosome amplification.
Chronic inflammation is frequently found surrounding tumors. Here, we demonstrated that inflammatory cytokines Ltα and TNFα, and an anti-LtβR antibody (agonist) specifically activated IKKα to induce NPM hexamer formation and repress centrosome amplification in mouse and human cells. These results suggest that the presence of IKKα may suppress tumor progression through maintaining proper centrosome duplication and genomic integrity in the inflammatory microenvironment.
EXPERIMENTAL PROCEDURES
Cell Culture, Antibodies and Reagents
HEK293, CHO and U2OS cells were purchased from the American Type Culture Collection (Manassas) and were cultured in Dulbecco’s Modified Eagle’s medium or F12K medium supplemented with 10% serum. Cultured cells were transfected with different expression vectors using a transfection reagent (Lipofectamine with PLUS Reagent; Invitrogen), according to the manufacturer’s recommendations. Primary keratinocytes were isolated from mice and cultured as previously described (Hu et al., 2001). A431 (human epidermoid carcinoma cells) and SCC25 (human tongue SCC) cell lines were purchased from ATCC. Purified human Ltα (BMS302) was from eBioscience. Human tissue arrays (SK2081 and CR602) were obtained from US Biomax, Inc. Antibodies used in this study included those against γ-tubulin (T3559, Sigma-Aldrich; ab11316, Abcam, Inc.), centrin 1 (ab11257, Abcam, Inc.), α-tubulin (E-19, Santa Cruz Biotechnology), β-tubulin (D-10, Santa Cruz Biotechnology), CKII (N18, Santa Cruz Biotechnology), EGFR (sc-03, Santa Cruz Biotechnology), p-EGFR (sc-12351, Santa Cruz Biotechnology), HA (H-9658, Sigma-Aldrich), Flag (F-3165, Sigma-Aldrich), NPM (3542, Cell Signaling Technology; and 32–5200, Zymed-Invitrogen), IKKα (IMG-136, Imgenex), IKKβ (05–535, Upstate Cell Signaling Technology), IKKγ (559675, BD Pharmingen), lymphotoxin beta (LtβR, 16-5671-82, eBioscience), and rabbit IgG(H+L) (Invitrogen). An antibody against phosphorylated Ser125 in NPM was generated using the peptide EDAE{SER}EDEDEEDVKC (GenScript USA, Inc.), and the peptide was used to block anti-p-S125-NPM antibody activity. Plasmid DNA containing different Flag-NPM deletions and GST-NPM WT and Thr199A mutant plasmids were provided by Dr. Charles J. Sherr (St. Jude Children’s Research Hospital) and Dr. Kenji Fukasawa (University of Cincinnati).
Mice
All of the mice used in this study were cared for in accordance with the guidelines of the Animal Care and Use Committee (animal protocols 08–075 and 08–074) at the National Cancer Institute.
Centrosome Isolation
Centrosome isolation using sucrose gradient centrifugation was performed as previously described (Hsu and White, 1998), with minor modifications. Briefly, exponentially growing CHO or U2OS cells were incubated in a medium containing 1 μg/ml of cytochalasin D and 0.2 μM of nocodazole for 1 hr at 37°C to depolymerize the actin and microtubule filaments. Cells were then harvested and lysed. Swollen nuclei and chromatin aggregates were removed using centrifugation at 2500 × g for 10 min, and the supernatant was filtered through a 40-μm nylon mesh (BD Biosciences). The concentration of HEPES was adjusted to 10 mM, 2 units/ml DNase I (Ambion, Inc.). The mixture was incubated for 30 min on ice. The lysate was then underlaid with sucrose solutions and centrifuged at 10,000 × g for 30 min. This crude centrosome preparation was further purified using discontinuous sucrose gradient centrifugation at 30,000 × g for 1 hr, and the fractions were collected from the bottom of each tube. Each fraction was diluted in 1 ml of 10 mM PIPES buffer (pH 7.2) and centrifuged at 15,000 rpm for 15 min in a microcentrifuge to recover the centrosomes. The centrosomes were then dissolved in a loading buffer.
Mass Spectrometry (MS) Analysis
Coomassie blue-stained SDS-PAGE gel bands corresponding to NPM were reduced, alkylated, and digested with an in-gel tryptic digestion to extract the peptides (Simkus et al., 2009). Each sample was loaded on an Agilent 1100 nano-capillary high-performance liquid chromatography system (Agilent Technologies) with 10-cm integrated μRPLC-electrospray ionization emitter columns (made in-house), coupled online with an LTQ XP mass spectrometer (Thermo Fisher Scientific) for μRPLC-MS2-MS3 analysis. Peptides were eluted using a linear gradient of 2% mobile phase B (acetonitrile with 0.1% formic acid) to 42% mobile phase B within 40 min at a constant flow rate of 0.25 μl/min. The five most intense molecular ions in the MS scan were sequentially selected for MS2 and MS3 (if phosphate neutral loss was observed in MS2 spectra) by CID using 35% normalized collision energy. The mass spectra were acquired at the mass range of m/z 1000–1300. The ion-source capillary voltage and temperature were set at 1.7 kV and 200°C, respectively. MS2 and MS3 data were searched against the NPM protein sequence using BioWorks-interfaced SEQUEST (Thermo Fisher Scientific). Up to 2 missed cleavage sites were allowed during the database search. The cut-offs for legitimate identifications were: charge state-dependent cross correlation (Xcorr) ≥ 2.0 for [M+H]1+, ≥ 2.5 for [M+2H]2+, and ≥ 3.0 for [M+3H]3+.
Supplementary Material
Highlight.
Inflammation-led IKKα activation promotes genomic integrity in tumor microenvironment
IKKα prevents centrosome amplification through phosphorylating Ser125-NPM
IKKα phosphoryaltes NPM to enhance the association of NPM and centrosomes
Downregulation of IKKα and NPM is found in human squamous cell carcinomas
Acknowledgments
We thank D. Butcher, I. Gimenez-Conti, and N. W. Otto for the immunohistochemical staining. This work was supported by funding from the National Cancer Institute (CA-102510, ZIA BC011212, and ZIA BC 011391) to Y.H.
Footnotes
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anest V, Hanson JL, Cogswell PC, Steinbrecher KA, Strahl BD, Baldwin AS. A nucleosomal function for IkappaB kinase-alpha in NF-kappaB-dependent gene expression. Nature. 2003;423:659–663. doi: 10.1038/nature01648. [DOI] [PubMed] [Google Scholar]
- Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, Raff JW. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertwistle D, Sugimoto M, Sherr CJ. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol Cell Biol. 2004;24:985–996. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PK, Aldrich M, Cook RG, Busch H. Amino acid sequence of protein B23 phosphorylation site. J Biol Chem. 1986;261:1868–1872. [PubMed] [Google Scholar]
- Chou YH, Yung BY. Cell cycle phase-dependent changes of localization and oligomerization states of nucleophosmin / B23. Biochem Biophys Res Commun. 1995;217:313–325. doi: 10.1006/bbrc.1995.2779. [DOI] [PubMed] [Google Scholar]
- D’Assoro AB, Lingle WL, Salisbury JL. Centrosome amplification and the development of cancer. Oncogene. 2002;21:6146–6153. doi: 10.1038/sj.onc.1205772. [DOI] [PubMed] [Google Scholar]
- Dejardin E, Droin NM, Delhase M, Haas E, Cao Y, Makris C, Li ZW, Karin M, Ware CF, Green DR. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity. 2002;17:525–535. doi: 10.1016/s1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude GF. Abnormal centrosome amplification in the absence of p53. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81–96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- Godinho SA, Kwon M, Pellman D. Centrosomes and cancer: how cancer cells divide with too many centrosomes. Cancer Metastasis Rev. 2009;28:85–98. doi: 10.1007/s10555-008-9163-6. [DOI] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisendi S, Bernardi R, Rossi M, Cheng K, Khandker L, Manova K, Pandolfi PP. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437:147–153. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- Grisendi S, Pandolfi PP. NPM mutations in acute myelogenous leukemia. N Engl J Med. 2005;352:291–292. doi: 10.1056/NEJMe048337. [DOI] [PubMed] [Google Scholar]
- Gu L, Zhu N, Findley HW, Woods WG, Zhou M. Identification and characterization of the IKKalpha promoter: positive and negative regulation by ETS-1 and p53, respectively. J Biol Chem. 2004;279:52141–52149. doi: 10.1074/jbc.M407915200. [DOI] [PubMed] [Google Scholar]
- Herrera JE, Correia JJ, Jones AE, Olson MO. Sedimentation analyses of the salt- and divalent metal ion-induced oligomerization of nucleolar protein B23. Biochemistry. 1996;35:2668–2673. doi: 10.1021/bi9523320. [DOI] [PubMed] [Google Scholar]
- Hoberg JE, Popko AE, Ramsey CS, Mayo MW. IkappaB kinase alpha-mediated derepression of SMRT potentiates acetylation of RelA/p65 by p300. Mol Cell Biol. 2006;26:457–471. doi: 10.1128/MCB.26.2.457-471.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu LC, White RL. BRCA1 is associated with the centrosome during mitosis. Proc Natl Acad Sci USA. 1998;95:12983–12988. doi: 10.1073/pnas.95.22.12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, Zou Y, Bao S, Hanada N, Saso H, et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- Hu Y, Baud V, Oga T, Kim KI, Yoshida K, Karin M. IKKa controls formation of the epidermis independently of NF-kB. Nature. 2001;410:710–714. doi: 10.1038/35070605. [DOI] [PubMed] [Google Scholar]
- Lahtela J, Nousiainen HO, Stefanovic V, Tallila J, Viskari H, Karikoski R, Gentile M, Saloranta C, Varilo T, Salonen R, et al. Mutant CHUK and severe fetal encasement malformation. N Engl J Med. 2010;363:1631–1637. doi: 10.1056/NEJMoa0911698. [DOI] [PubMed] [Google Scholar]
- Liu B, Park E, Zhu F, Bustos T, Liu J, Shen J, Fischer SM, Hu Y. A critical role for I{kappa}B kinase {alpha} in the development of human and mouse squamous cell carcinomas. Proc Natl Acad Sci USA. 2006;103:17202–17207. doi: 10.1073/pnas.0604481103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Xia X, Zhu F, Park E, Carbajal S, Kiguchi K, DiGiovanni J, Fischer SM, Hu Y. IKKalpha is required to maintain skin homeostasis and prevent skin cancer. Cancer Cell. 2008;14:212–225. doi: 10.1016/j.ccr.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zhu F, Xia X, Park E, Hu Y. A tale of terminal differentiation: IKKalpha, the master keratinocyte regulator. Cell Cycle. 2009;8:527–531. doi: 10.4161/cc.8.4.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QR, Chan PK. Formation of nucleophosmin/B23 oligomers requires both the amino- and the carboxyl-terminal domains of the protein. Eur J Biochem. 1991;200:715–721. doi: 10.1111/j.1432-1033.1991.tb16236.x. [DOI] [PubMed] [Google Scholar]
- Marinari B, Moretti F, Botti E, Giustizieri ML, Descargues P, Giunta A, Stolfi C, Ballaro C, Papoutsaki M, Alema S, et al. The tumor suppressor activity of IKKalpha in stratified epithelia is exerted in part via the TGF-beta antiproliferative pathway. Proc Natl Acad Sci USA. 2008;105:17091–17096. doi: 10.1073/pnas.0809288105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascaux C, Bex F, Martin B, Burny A, Haller A, Paesmans M, Willard-Gallo K, Ninane V, Sculier JP. The role of NPM, p14arf and MDM2 in precursors of bronchial squamous cell carcinoma. Eur Respir J. 2008;32:678–686. doi: 10.1183/09031936.00008408. [DOI] [PubMed] [Google Scholar]
- Meraldi P, Lukas J, Fry AM, Bartek J, Nigg EA. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat Cell Biol. 1999;1:88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A, et al. IKK-1 and IKK-2: cytokine-activated IkB kinases essential for NF-kB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- Okuda M, Horn HF, Tarapore P, Tokuyama Y, Smulian AG, Chan PK, Knudsen ES, Hofmann IA, Snyder JD, Bove KE, et al. 2000 doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 103:127–140. [Google Scholar]
- Park E, Zhu F, Liu B, Xia X, Shen J, Bustos T, Fischer SM, Hu Y. Reduction in IkappaB kinase alpha expression promotes the development of skin papillomas and carcinomas. Cancer Res. 2007;67:9158–9168. doi: 10.1158/0008-5472.CAN-07-0590. [DOI] [PubMed] [Google Scholar]
- Prajapati S, Tu Z, Yamamoto Y, Gaynor RB. IKKalpha regulates the mitotic phase of the cell cycle by modulating Aurora A phosphorylation. Cell Cycle. 2006;5:2371–2380. doi: 10.4161/cc.5.20.3359. [DOI] [PubMed] [Google Scholar]
- Simkus C, Bhattacharyya A, Zhou M, Veenstra TD, Jones JM. Correlation between recombinase activating gene 1 ubiquitin ligase activity and V(D)J recombination. Immunology. 2009;128:206–217. doi: 10.1111/j.1365-2567.2009.03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebeni A, Hingorani K, Negi S, Olson MO. Role of protein kinase CK2 phosphorylation in the molecular chaperone activity of nucleolar protein b23. J Biol Chem. 2003;278:9107–9115. doi: 10.1074/jbc.M204411200. [DOI] [PubMed] [Google Scholar]
- Tokuyama Y, Horn HF, Kawamura K, Tarapore P, Fukasawa K. Specific phosphorylation of nucleophosmin on Thr(199) by cyclin-dependent kinase 2-cyclin E and its role in centrosome duplication. J Biol Chem. 2001;276:21529–21537. doi: 10.1074/jbc.M100014200. [DOI] [PubMed] [Google Scholar]
- Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18:658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Xia X, Park E, Fischer SM, Hu Y. Mouse genetic models reveal surprising function of Ikappa B kinase alpha in skin development and skin carcinogenesis. Cancers. 2013;5:170–183. doi: 10.3390/cancers5010170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Jiang Q, Willette-Brown J, Xi S, Zhu F, Burkett S, Back T, Song NY, Datla M, Sun Z, et al. The Pivotal Role of IKKalpha in the Development of Spontaneous Lung Squamous Cell Carcinomas. Cancer Cell. 2013;23:527–540. doi: 10.1016/j.ccr.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Verma UN, Prajapati S, Kwak YT, Gaynor RB. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature. 2003;423:655–659. doi: 10.1038/nature01576. [DOI] [PubMed] [Google Scholar]
- Yung BY. Oncogenic role of nucleophosmin/B23. Chang Gung Med J. 2007;30:285–293. [PubMed] [Google Scholar]
- Yung BY, Chan PK. Identification and characterization of a hexameric form of nucleolar phosphoprotein B23. Biochim Biophys Acta. 1987;925:74–82. doi: 10.1016/0304-4165(87)90149-8. [DOI] [PubMed] [Google Scholar]
- Zhu F, Park E, Liu B, Xia X, Fischer SM, Hu Y. Critical role of IkB kinase alpha in embryonic devleopment and skin carcinogenesis. Histol Histopathol. 2009;24:265–271. doi: 10.14670/HH-24.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Xia X, Liu B, Shen J, Hu Y, Person M, Hu Y. IKKalpha Shields 14-3-3sigma, a G(2)/M Cell Cycle Checkpoint Gene, from Hypermethylation, Preventing Its Silencing. Mol Cell. 2007;27:214–227. doi: 10.1016/j.molcel.2007.05.042. [DOI] [PubMed] [Google Scholar]
- Zirwes RF, Kouzmenko AP, Peters JM, Franke WW, Schmidt-Zachmann MS. Topogenesis of a nucleolar protein: determination of molecular segments directing nucleolar association. Mol Biol Cell. 1997;8:231–248. doi: 10.1091/mbc.8.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.