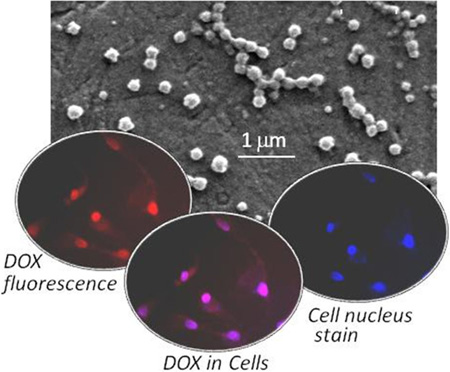
Abstract
Esterification of microbial poly(malic acid) is performed with either ethanol or 1-butanol to obtain polymalate conjugates capable to form nanoparticles (100–350 nm). Degradation under physiological conditions takes place with release of malic acid and the corresponding alcohol as unique degradation products. The anticancer drugs Temozolomide and Doxorubicin are encapsulated in nanoparticles with efficiency of 17 and 37%, respectively. In vitro drug release assays show that Temozolomide is almost completely discharged in a few hours whereas Doxorubicin is steadily released along several days. Drug-loaded nano-particles show remarkable effectiveness against cancer cells. Partially ethylated poly(malic acid) nano-particles are those showing the highest cellular uptake.
Keywords: iodegradable drug delivery systems, poly(malic acid), poly(malic acid) nanoparticles
1. Introduction
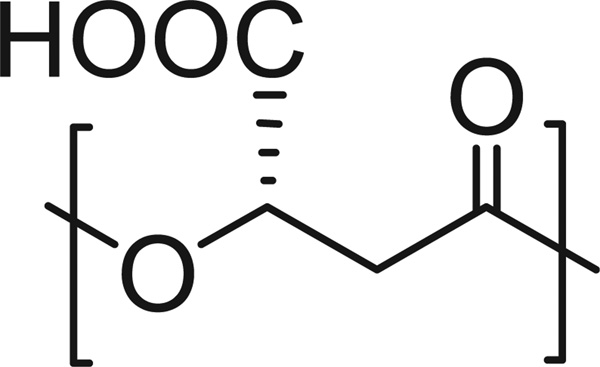
Nowadays, biodegradation and bioassimilation are indispensable qualities of any polymer intended for temporal applications in human therapy.[1] Accordingly biodegradable and safe polymers are the preferred materials for the manufacture of many devices that are today used in medicine and pharmacology.[2] Such requirements have stimulated efforts towards both the modification of naturally occurring biopolymers and the synthesis of new polymers with biodegradable and biocompatible properties.[3,4] In this regard, poly(β,l-malic acid) (PMLA) and its derivatives constitute a family of promising candidates. PMLA is a poly(β-hydroxy propionate) derivative with a carboxylic group stereoregularly attached to the β-carbon of the repeating unit (Scheme 1). The polymer is water-soluble, nontoxic, biodegradable, bioresorbable and nonimmunogenic.[5,6] PMLA can be produced by either chemical synthesis [4,7] or by biosynthesis.[8] Degradation of PMLA produces easily metabolizable l-malic acid.[9] In contrast to most common biodegradable polyesters such as polylactides, polyalkanoates or polycaprolactone, PMLA is a functional polymer with properties adjustable trough chemical modification of the pendant carboxylic side group;[10] not only the overall hydrophobicity of the polymer may be controlled by partial esterification but also bioactive ligands may be incorporated by reaction with appropriate agents.[11]
Scheme 1.
Chemical formula of poly(β,l-malic acid).
The application of drug delivery systems (DDS) in chemotherapy has spread out rapidly in the last years since their use reduces systemic toxicity and enables to increase drug concentration at the specific site. Although cancer cells are more vulnerable than normal cells to the effect of chemotherapy agents, drugs are non-selective and can affect normal tissues. The use of polymeric DDS in cancer therapy is increasing in popularity because they are less immunogenic than protein-based vectors, and they allow repetitive administration without acute or chronic host immune response.[12] Furthermore, DDS based on polymer particles, either nanoparticles or microparticles, are clearly advantageous for several reasons: a) particle size and surface can be engineered for passive or active drug targeting, b) drugs can be incorporated without chemical reaction, c) drug activity is optimally preserved during transportation to the site of action, and d) different routes of administration are optional for drug delivery.[13–17]
PMLA and its derivatives have been used either as platform in the synthesis of nanoparticles for drug delivery[1,3,18–21] or as backbone in macromolecular conjugates bearing several functionalities to treat human brain and breast tumors in mouse models.[11,22–24] It was concluded from these investigations that PMLA is a very suitable polymer for building efficient DDS. Recently we reported on methylated PMLA nanoparticles and showed that the cell toxicity of this system increased significantly after long time periods of incubation due to the noxious effect of the released methanol.[20] In this work we report on other esters of PMLA, which are expected to display less cytotoxicity. These new esters are obtained by partial or total esterification of PMLA with ethanol or 1-butanol, and the nanoparticles made from them are explored for their suitability as DDS for the anticancer drugs Temozolomide (TMZ) and Doxorubicin (DOX).
2. Experimental Section
2.1. Materials
PMLA used in this work was biotechnologically produced by aerobic cultivation of Physarum polycephalum and isolated and purified as described elsewhere.[8] The purity of the sample was ascertained by 300 MHz 1H NMR, and it had a weight-averaged molecular weight of 30 000 g mol−1 with a dispersity of 1.2 as determined by Gel Permeation Chromatography (GPC). TMZ (3,4-dihydro-3-methyl-4-oxoimidazo[5,1-d]-as-tetrazine-8-carboxamide) and DOX ((7S,9S)-7-[(2R,4S,5S,6S)-4-amino-5-hydroxy-6-methyloxan-2-yl]oxy-6,9,11-trihydroxy-9-(2-hydroxyacetyl)-4-methoxy-8,10-dihydro-7H-tetracene-5,12-dione) hydrochloride were obtained from AKSci (Union City, CA, USA). All organic solvents were either analytical or high performance liquid chromatography (HPLC) grade and they were used without further purification.
2.2. PMLA Esterification
Esterification of PMLA with either ethanol or 1-butanol was performed at room temperature with the polyacid dissolved in the corresponding alcohol and using dicyclohexylcarbodiimide (DCC) for activation of the carboxylic side group. Briefly, to 1 mmol of PMLA in 3 mL of either ethanol or 1-butanol, 0.5 or 1.0 mmol of DCC dissolved in 2 mL of the same alcohol, according to the desired esterification degree, were added drop-wise under stirring, and the reaction was left to proceed for 2 h. Exhaustive removal of dicyclohexylurea (DCU) was achieved by successive dialysis of the reaction solution against methanol for 24 h and water for 6 h using a cellulose membrane of 8 kDa cut-off. The dialyzed solution was roto-evaporated to remove the methanol and then lyophilized. The conversion degree and purity of the resulted polyesters was ascertained by 1H NMR.
2.3. Hydrolytic Degradation
Hydrolytic degradation of PMLA derivatives was evaluated by following the change in molecular weight with time of samples incubated in aqueous buffers at 37 °C. About 2 mg of nanoparticles, prepared as it will be described below, were immersed in citrate buffer pH 5.0 or phosphate buffer pH 7.4 and aliquots were collected at scheduled times. After centrifugation, the degraded particles sediments were recovered and subjected to GPC analysis. For the assessment of the water degradation mechanism, 10 mg of polymer were placed in NMR tubes containing 1 mL of deuterated water at 60°C and analyzed by 1H NMR at scheduled times. Degradation products released to the incubation medium were identified and changes in their relative amounts were monitored with time.
2.4. Nanoparticle Preparation and Drug Encapsulation
Two methods were employed for the formation of nanoparticles depending on the esterification degree of the polymer. For 100% modified polymers, which are the most hydrophobic samples, the emulsion-solvent evaporation method was applied. Briefly, 10 mg of polymer were dissolved in 0.5 mL of dichloromethane (DCM), added to 5 mL of 1% poly(vinyl acetate) (PVA) (M̅w ≈ 2000) aqueous solution and emulsified by sonication for 45 s with a tip probe apparatus (Bandelin, Berlin, Germany, Sonoplus, 200W) operating at 50% of amplitude. DCM was evaporated under reduced pressure and the nanoparticles were collected from the aqueous suspension by centrifugation, washed 3 times with distilled water to eliminate the emulsifier excess, and freeze-dried for storage.
The precipitation-dialysis method was applied for nanoparticle formation when partially esterified PMLA was used. In this case, to a solution of 10 mg mL−1 of the copolymalate in dimethylsulfoxide (DMSO), 1 mL of water was added drop-wise under magnetic stirring. The mixture was dialyzed against distilled water for 24 h using a cellulose membrane with a molecular weight cut-off of 8 kDa. Nanoparticles formed inside the bag were recovered by centrifugation and then freeze-dried. Particle morphology was monitored by Scanning Electron Microscopy (SEM) and their average hydrodynamic diameters were determined by dynamic light scattering.
For TMZ and DOX loading, 10% (w/w) of drug to polymer was added to the initial organic solution used either in the emulsion solvent-evaporation or in the precipitation-dialysis method. Drug contents in the nanoparticles were determined by dissolving 5 mg of drug-loaded nanoparticles in DMSO and quantifying the drug concentration by UV–vis spectrophotometry at 330 and 480 nm for TMZ and DOX, respectively. Encapsulation efficiency (EE) was calculated according to the well-known expression % EE = 100 × ([Drug]final/[Drug]initial).
2.5. In Vitro Drug Release Assays
In vitro TMZ and DOX release was evaluated by the dialysis method. Briefly, 10 mg of freeze-dried drug-loaded nanoparticles were re-suspended in 1 mL of phosphate buffer at pH 7.4, and the solution transferred into a dialysis tube with 8 kDa molecular weight cut-off. The tube was then immersed into 10 mL of buffer and left at 37 °C under slight stirring. Aliquots (0.5 mL) of the releasing medium were taken at scheduled times and the drawn volume was replaced by fresh buffer every time. Drug concentration was determined by HPLC at 330 and 480 nm for TMZ and DOX, respectively, using known amounts of free drugs as standards. Since TMZ is hydrolytically labile, its degradation product 5-aminoimidazole-4-carbox-amide (AIC) absorbing at 254 nm was also accounted for determination.
2.6. Cell Viability Assays and Drug Cell Uptake
Primary glioma cell line U87MG and invasive breast carcinoma cell line MDA-MB468 were obtained from American Type Culture Collection (ATCC, USA). U87MG cells were cultured in minimum essential media (MEM) supplemented with 10% fetal bovine serum, 1% MEM NEAA, 1 × 10−3 m sodium pyruvate and 2 × 10−3 m l-glutamine. For MDA-MB468, Lebovitz’s L-15 medium with 10% fetal bovine serum was used. Cells were seeded at 103 per well (0.1 mL) in 96-well flat-bottom plate and incubated overnight at 37 °C in a humid atmosphere for MDA-MB468 and in an atmosphere containing 5% of CO2 for U87MG. Exposure time to loaded and unloaded nanoparticles was 24 h. Cell viability was measured on day 2 for DOX containing nanoparticles and day 7 for TMZ containing nanoparticles, using the CellTiter 96 Aqueous One Solution Cell Proliferation Assay kit (Promega, USA). Yellow [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] (MTS) is bioreduced by cells into formazan that is soluble in the culture medium. Absorbance at 490 nm is directly proportional to cell viability.[25] Viability of untreated cells was taken as 100%.
DOX uptake was measured by fluorescent microscopy with cells prepared and treated as described above. After 2 h of incubation with DOX loaded nanoparticles, cells were fixed with 4% paraformaldehyde at room temperature for 10 min, and the nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI). The fluorescence exhibited by DOX was observed using the free drug and unloaded drug nanoparticles for positive and negative control, respectively. For all experiments the constant concentration of DOX was 3 × 10−6 m.
2.7. Measurements
SEM images were taken with a field-emission JEOL JSM-7001F instrument (JEOL, Japan) from uncoated samples. Particle size measurements, based on dynamic light scattering, were performed with a ZetaSizer NS, (Malvern Instruments, UK) with particles suspended in deionized water. 1H NMR spectra were recorded at 25 °C on a Bruker AMX-300 instrument from samples immersed in D2O or dissolved in DMSO-d6. Spectra were registered with the equipment operating at 300.1 MHz and acquisition of 128 scans with 32000 data points and relaxations of 2 s. Differencial Scanning Calorimetry (DSC) profiles were recorded using a Perkin-Elmer Pyris-1 under a nitrogen flow. Thermograms were obtained from 2–4 mg samples at heating and cooling rates of 20 K min−1 to determine the glass transition temperature (Tg) of polymers. Indium and zinc were used as standards for calibration. Cell fluorescence was accounted by using an inverted fluorescence microscope (Leica) at 40× magnification, exposure time of 25 ms and provided with an appropriate filter set.
For GPC a Waters 515 HPLC pump equipped with a Waters 410IR detector and a Waters Styragel HR 5E column (7.8 × 300 mm) (Waters, Massachusetts, USA) was used. Solvent consisting of 0.05 m sodium trifluoroacetate in hexafluoro-2-propanol at 30°C and 0.5 mL min−1 flow rate was applied. Chromatograms were calibrated against poly(methyl methacrylate) standards (Varian, USA). HPLC was carried out with a Waters 600 system consisting of Waters 996 photodiode array detector and a GL Sciences Inertsil ODS-3V column (5 µm, 4.6 × 250 mm) (California, USA) at an elution rate of 1 mL min−1. Mobile phase for TMZ quantification was a mixture of methanol and 0.5% aqueous acetic acid (10:90), and for DOX, a mixture (60:40) of 0.02 m sodium hydrogen phosphate and acetonitrile.
3. Results and Discussion
3.1. Synthesis and Characterization
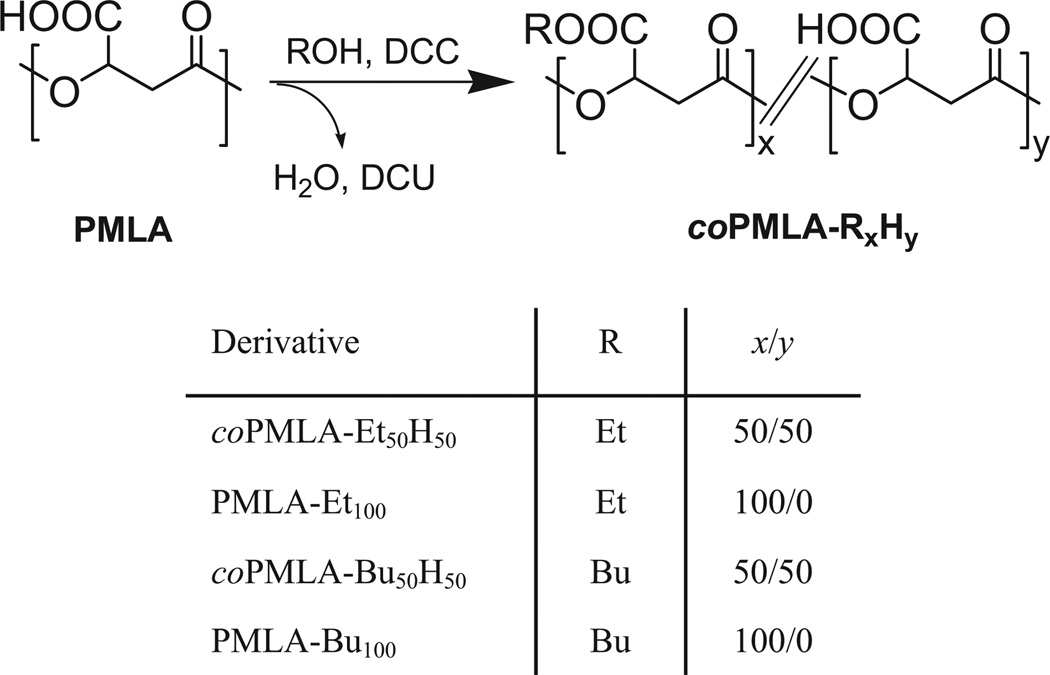
Ethyl and butyl PMLA esters with esterification degrees of approximately 50% and fully esterified were obtained by reaction of PMLA with ethanol and 1-butanol, respectively, using DCC as activator (Scheme 2). Esterification results are summarized in Table 1.
Scheme 2.
Esterification of PMLA with ethanol and 1-butanol.
Table 1.
Results for PMLA esterification.
| Solubility |
||||||||
|---|---|---|---|---|---|---|---|---|
| Conversion [%] |
Yield [%] |
Mwa [g mol−1] |
Ða) | Tg [°C] |
H2O | DMSO | CHCl3 | |
| PMLA | – | – | 30000 | 1.2 | 110 | + | + | − |
| coPMLA-Et50H50 | 56 | 62 | 33 000 (33 600) | 2.4 | 31 | − | + | − |
| PMLA-Et100 | 100 | 72 | 36 000 (37 200) | 1.8 | 19 | − | + | + |
| coPMLA-Bu50H50 | 53 | 57 | 34 000 (37 200) | 2.6 | 13 | − | + | − |
| PMLA-Bu100 | 100 | 53 | 41 000 (44 500) | 2.3 | −12 | − | + | + |
Weight-average molecular weights and dispersities determined by GPC. In parenthesis, values estimated on the basis of the attained esterification degree.
The esterification degree was controlled by adjusting the added amount of DCC so that conversions close to the used DCC/PMLA molar ratios were obtained in both cases. Reaction yields were around 50–70% with higher values attained in the esterification with ethanol. Product losses during polymer isolation and purification are the most probable reasons accounting for such relatively low yields. Average molecular weights of esterified products were found to be higher than that of PMLA and they show a coherent increasing correlation with the average molecular weight values that should be expected from ethyl and butyl grafting for the respective attained conversions. However, these experimental values are slightly lower than the theoretically calculated ones and dispersity was noticeably increased. Such results may be taken as an indication that some degradation could take place during the esterification reaction, an event that was more significant when the butyl group was introduced. However, it cannot be discarded that the differences observed in GPC results are merely due to the application of the same standard calibration to two different compounds with different hydrodynamic volumes. Nevertheless, the esterified polymers were spectroscopically pure giving 1H NMR spectra in full agreement with the expected constitution and without showing any sign of chain end groups (see Supporting Information, Figure S1–S4); apparently low molecular weight species eventually generated by degradation were removed along the treatments applied for isolation and purification with the subsequent lowering of yields, as observed.
The reason for determining Tg values will be explained later. It can be observed that the insertion of ethyl and butyl groups has a significant effect in lowering Tg, showing a bigger effect with increasing number and length of the alkyl grafted chains, so 100% esterified PMLA presented lower Tg than their respective copolymers (Table 1).
3.2. Hydrolytic Degradation
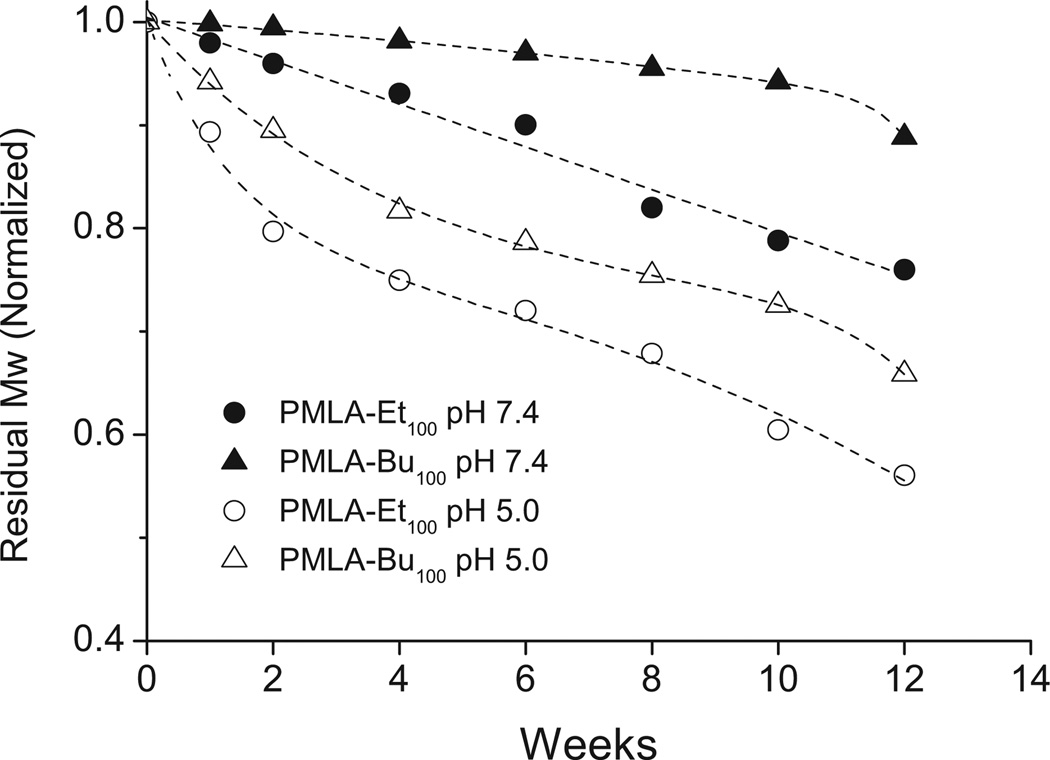
Hydrolytic degradation assays were carried under physiological conditions (pH 7.4, 37 °C), and also in a slightly acidic medium (pH 5.0, 37 °C) intended to simulate the environment occurring inside of mature lysosomes. Results obtained by GPC for the incubated PMLA-Et100 and PMLA-Bu100 samples are compared in Figure 1. These polyesters, after degradation, produced monomodal GPC chromatograms with single peaks and values almost steadily decreasing with time, following a steeper slope for the ethyl than for the butyl derivative, as well as for the acidic than for the neutral solutions. Maximum degradability differences were found between PMLA-Bu100 incubated at pH 7.4, and PMLA-Et100 incubated at pH 5.0 which showed a decreasing in their original M̅w around 10% and 50%, respectively. The differences observed in the degradation rate are attributed, in addition to the pH effect, to differences in the length of the alkyl side groups of the polyesters. Larger alkyl groups increase the hydrophobic character and hinder the water attack on both main chain and side chain ester groups.
Figure 1.
Evolution of the molecular weight of PMLA-Et100 and PMLA-Bu100 incubated in aqueous buffer at pH 7.4 and 5.0 at 37 °C
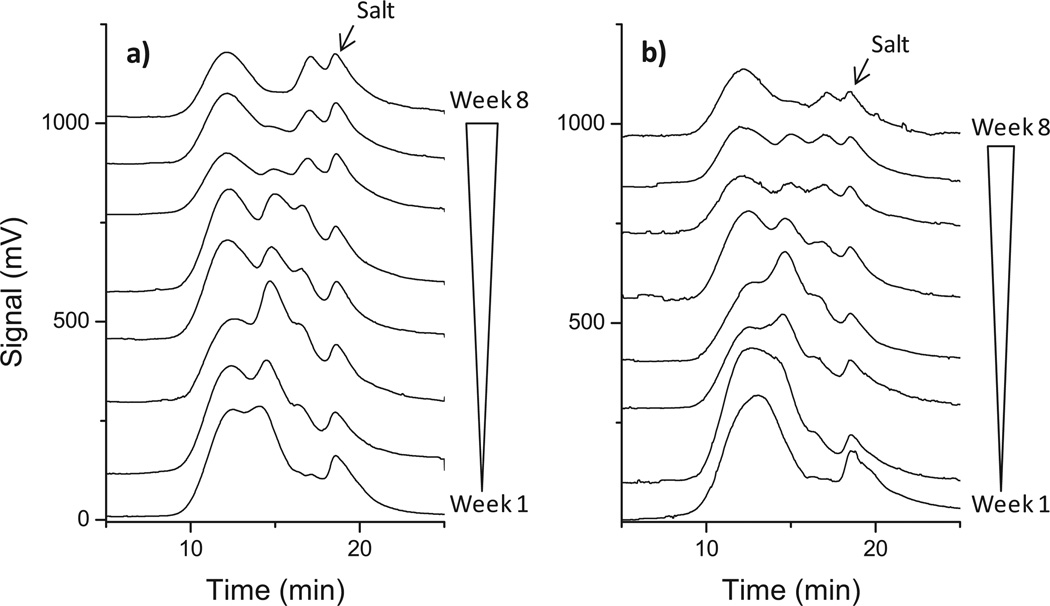
Degradation of coPMLA-Et50H50 and coPMLA-Bu50H50 followed a more complex pattern. Bimodal or trimodal GPC chromatograms, indicative of the occurrence of populations with different molecular weights, were invariably registered from the residues left by these copolyesters upon incubation (Figure 2). To understand these results it is necessary to make clear that chains with low esterification degree are water soluble and therefore escape the GPC analysis because they either remained in the discarded aqueous incubation solution or they were not carried by the HFIP used as mobile phase. Chromatograms were deconvoluted using Peakfit software to monitor the two main peak distributions with time (Figure S5, Supporting Information). Taking into account the evolution of the two deconvoluted peaks allows inferring that the one appearing at shorter retention times must represent the original polymer chains, while the second one with a much lower molecular weight must correspond to oligomeric products originating from the parent chain with a relatively high degree of esterification. The change in signal intensity of the low M̅w chains between the third and fourth week of incubation suggests the occurrence firstly of significant cleavage of the initial polymer to generate oligomers and then the degradation of these oligomers taking place at higher rate than that of the remaining higher molecular weight chains.
Figure 2.
GPC chromatograms of: a) coPMLA-Et50H50 and b) coPMLA-Bu50H50, after incubation in aqueous buffer at pH 7.4 and 37°C for the indicated times.
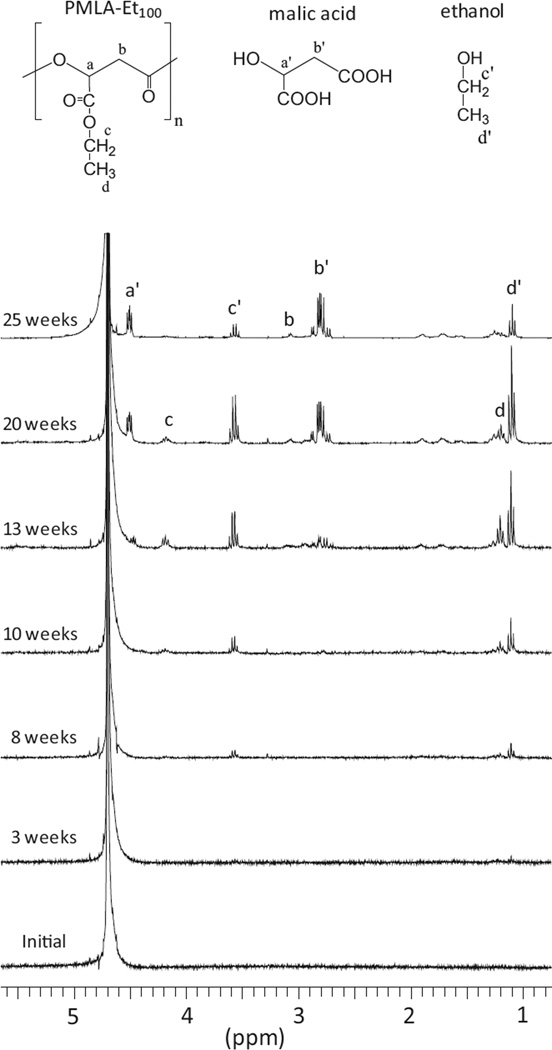
1H NMR was employed for getting insight into the degradation mechanism by identifying and monitoring the soluble products that are generated upon incubation of the polymers. Degradation spectra of PMLA-Et100 are presented in Figure 3. Results obtained with PMLA-Bu100 (Figure S6, Supporting Information) were almost identical, indicating that the same degradation mechanism must operate in both systems. The first detectable NMR signals appeared after three weeks of incubation and they corresponded to the alcohol (ethanol or 1-butanol) released from the hydrolysis of the ester side group. Differences between the two polymers started to be apparent after 8th week. At this time, the spectrum recorded from the PMLA-Et100 degradation medium showed the signals characteristic of the ethyloxycarbonyl group, while those of the butyloxycarbonyl group did not appear in the incubation medium of PMLA-Bu100 until 13th week. These results are a clear indication of the faster degradation and/or easier solubilization that takes place in PMLA-Et100 compared to PMLA-Bu100. Signals corresponding to PMLA oligomers, partially esterified polymer and free malic acid are present in both samples after 13th week, but displayed higher intensity in the case of the ethyl derivative. After twenty weeks of incubation of PMLA-Et100, ethanol and free malic acid were the only products detected in the incubation medium of PMLA-Et100. In the degradation of PMLA-Bu100, signals corresponding to the butyloxycarbonyl group were still observable at 25th week according to the higher reluctance of this group to be hydrolyzed.
Figure 3.
Evolution of 1H NMR spectra of the incubation medium of PMLA-Et100 degraded at pH 7.4 and 37 °C.
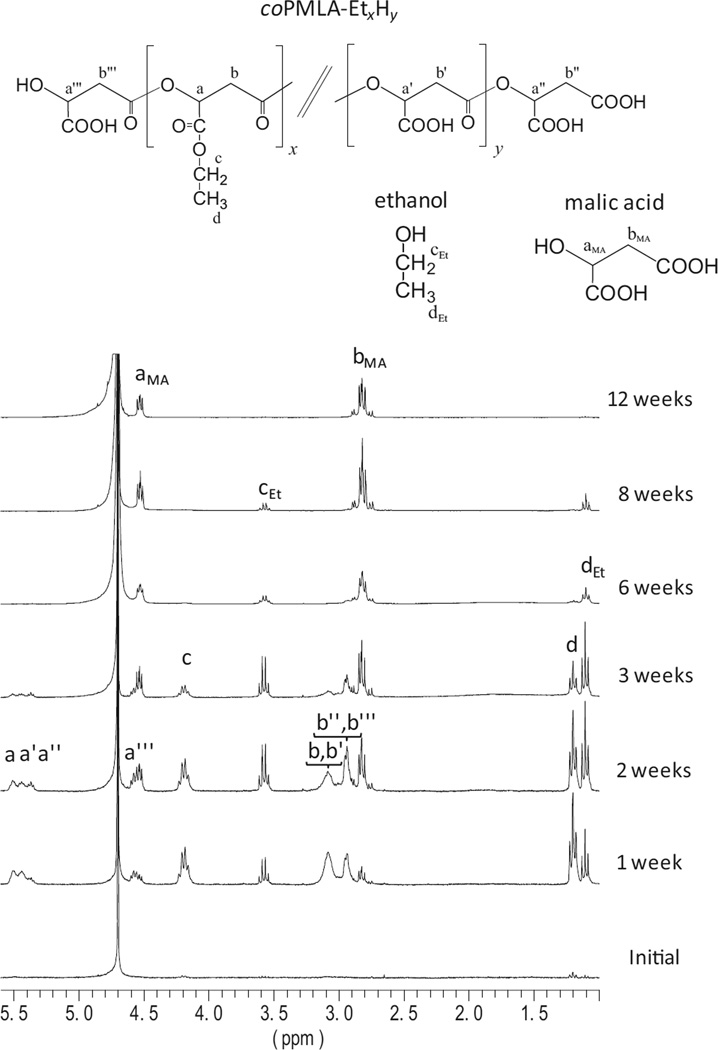
Degradation of copolymers proceeded following a pattern similar to homopolymers but at higher rates since their unmodified carboxylic units confer them a marked hydrophilicity. 1H NMR spectra of coPMLA-Et50H50 incubated at pH 7.4 and 37°C for three months are presented in Figure 4. Degradation started after only a week of incubation as it is revealed by the spectrum recorded at that time from the incubation medium. Signals indicative of the presence of ethanol, malic acid and oligomeric compounds are detected in the spectrum recorded from the supernatant after just a week of incubation. After two weeks, signals arising from terminal groups increased whereas those arising from the main chain diminished. This is taken as an unequivocal indication of the occurrence of main chain breaking with generation of oligomers. These oligomers still contain unhydrolysed malate units as it is revealed by the presence of signals arising from ethyl and butyl side groups. At the third week signals from free alcohol and malic acid increased in intensity, while those from the alkyl esters and oligomers became weaker as it is expected to result from the progressive hydrolysis of the lateral and main chain ester bonds. After 6th week, the only observable signals are those arising from malic acid and the alcohol, which are obviously the final products of degradation. Note that the weakening observed in the alcohols signals over time is due to partial evaporation of these volatile compounds. A similar mechanism was concluded that must be operating in the degradation of coPMLA-Bu50H50 (Figure S7, Supporting Information).
Figure 4.
1H NMR spectra registered from the supernatant of coPMLA-Et50H50 immersed in aqueous buffer at pH 7.4 and 37 °C for the indicated incubation times.
3.3. Nanoparticle Formation
Given the potential application intended for these polyesters as DDS, their capability to form nanoparticles was assessed. Due to differences in polarity between fully and partially esterified PMLAs, two different methods were employed for nanoparticle formation. The emulsion solvent-evaporation method was preferred for the most hydrophobic homopolymers (PMLA-Et100 and PMLA-Bu100), whereas for the copolymers coPMLA-Et50H50 and coPMLA-Bu50H50, which are amphiphilic polymers, the precipitation-dialysis method was employed instead. The chemical microstructure of the PMLA copolyesters prepared in this work is not easily accessible by NMR and it has not been determined. A random microstructure should be assumed however for these copolyesters, according to the method that has been used for their synthesis. Nevertheless, the occurrence of short homogeneous blocks of both esterified and non-esterified units of varying length are compatible with such microstructure. In consequence, and in agreement with what is largely experienced in the self-assembly of amphiphilic polymers,[26] a heterogeneous distribution of hydrophobic and hydrophilic domains preferentially located in the inner and outer regions of the nanoparticle respectively, should be expected for the topology of polymer.
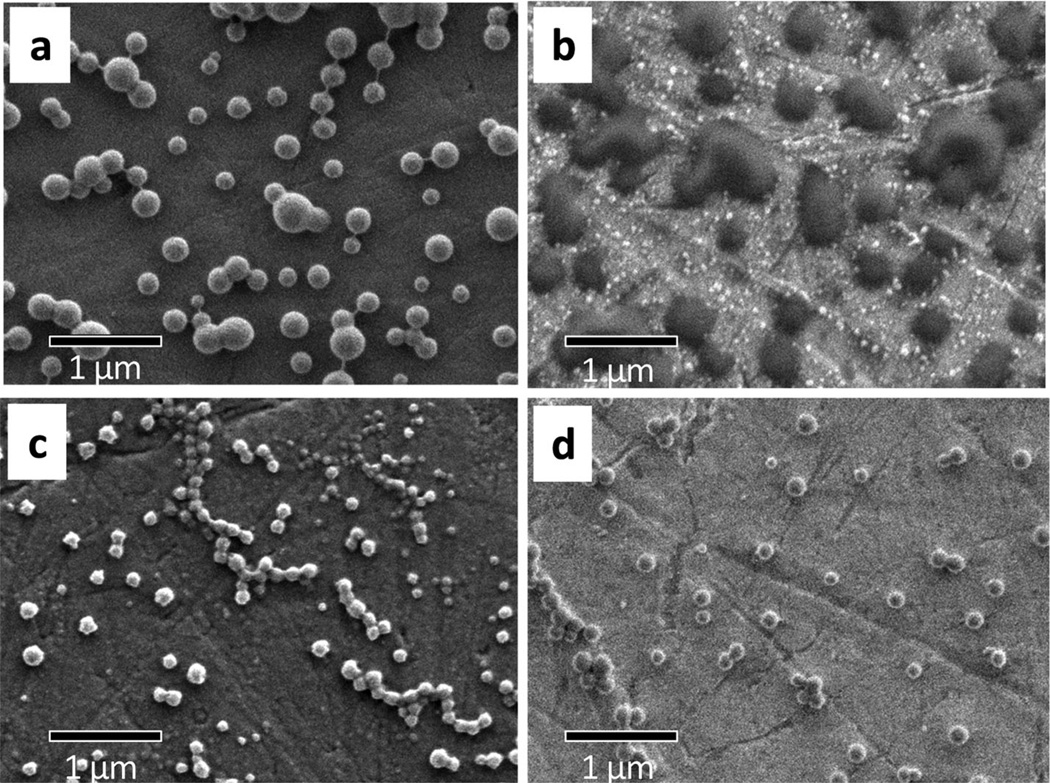
Light scattering (LS) measurements (see size distribution profiles in Figure S8, Supporting Information) revealed that particles with average hydrodynamic diameters in the 250– 350 nm range were obtained for the homopolymers by using the emulsion-evaporation method (Table 2). Much smaller particles with average diameters between 100 and 200 nm were obtained from the copolymers by precipitation-dialysis method. Moreover, SEM observations revealed significant morphological differences among them. As shown in Figure 5, nanoparticles with pretty defined spherical shape were observed for all the polymers except for PMLA-Bu100 (Figure 5b). In this case the particles displayed a much larger size than that determined by LS and their shapes were not well outlined. This is interpreted as the result of the coalescence probably occurring upon deposition of the particles on the support used for sample preparation. Such particular behavior displayed by PMLA-Bu100 is in agreement with its relative low Tg (Table 1) and its homogeneous constitution. The combination of these two factors could render sticky particles prone to coalesce when in contact with each other. Some signs of a similar behavior are also detected for PMLA-Et100 in Figure 5a, although in this case the particles still retain their spherical shape. Given the unsatisfactory behavior of PMLA-Bu100 particles, they were discarded in subsequent drug encapsulation and release assays.
Table 2.
Particle size and polydispersity of nanospheres made of different polymers.
| TMZ |
DOX |
||||||
|---|---|---|---|---|---|---|---|
| Average diameter (nm) |
Dispersity | Preparation Method |
Cont.a) [%] |
EEb) [%] |
Cont.a) [%] |
EEb) [%] |
|
| PMLA-Et100 | 279 ± 27 | 0.114 ± 0.044 | emulsion-evaporation | 3.75 ± 0.14 | 37.5 | 3.70± 0.21 | 37.0 |
| PMLA-Bu100 | 345 ± 33 | 0.059 ± 0.010 | emulsion-evaporation | – | – | – | – |
| coPMLA-Et50H50 | 136 ± 25 | 0.221 ± 0.038 | precipitation-dialysis | 1.36 ± 0.09 | 13.6 | 1.71 ± 0.07 | 17.1 |
| coPMLA-Bu50H50 | 163 ± 28 | 0.116 ± 0.084 | precipitation-dialysis | 2.03 ± 0.02 | 20.3 | 3.27 ± 0.13 | 32.7 |
Drug content (%-w/w).
Encapsulation efficiency.
Figure 5.
SEM images of polymer nanoparticles: a) PMLA-Et100; b) PMLA-Bu100; c) coPMLA-Et50H50; d) coPMLA-Bu50H50.
3.4. Drug Encapsulation and in Vitro Release
For encapsulation of TMZ and DOX in the nanoparticles, the same procedure as for nanoparticle formation was applied but with the drug added to the initial polymer solution. Results obtained for esterified PMLA excluding PMLA-Bu100, are presented in Table 2 for the two drugs. Encapsulation efficiencies (EE) in the 13–38% and 17–37% ranges were achieved for TMZ and DOX, respectively. It is remarkable that EE values are very similar for both drugs encapsulated in PMLA-Et100, and that higher values were attained when the emulsion method was used. Low EEs and drug contents obtained for copolymers could be explained by drug loss taking place during solvent removal by dialysis during nanoparticle formation. Nevertheless, it is remarkable that the amount of encapsulated drug can be correlated with the nanoparticle diameter (Table 2), which suggests a dependence of the encapsulation efficiency on particle size. The higher content obtained for DOX compared to TMZ in the partially esterified PMLA particles is probably due to the capability of this cationic drug to form ionic complexes with the carboxylic groups remaining present in the copolymer, in a similar manner as it has been reported to occur in other polyelectrolyte systems.[27] Limited drug loading and burst drug release are features usually associated to encapsulation and delivery in nanoparticles due to their small available volume and large surface area.[14]
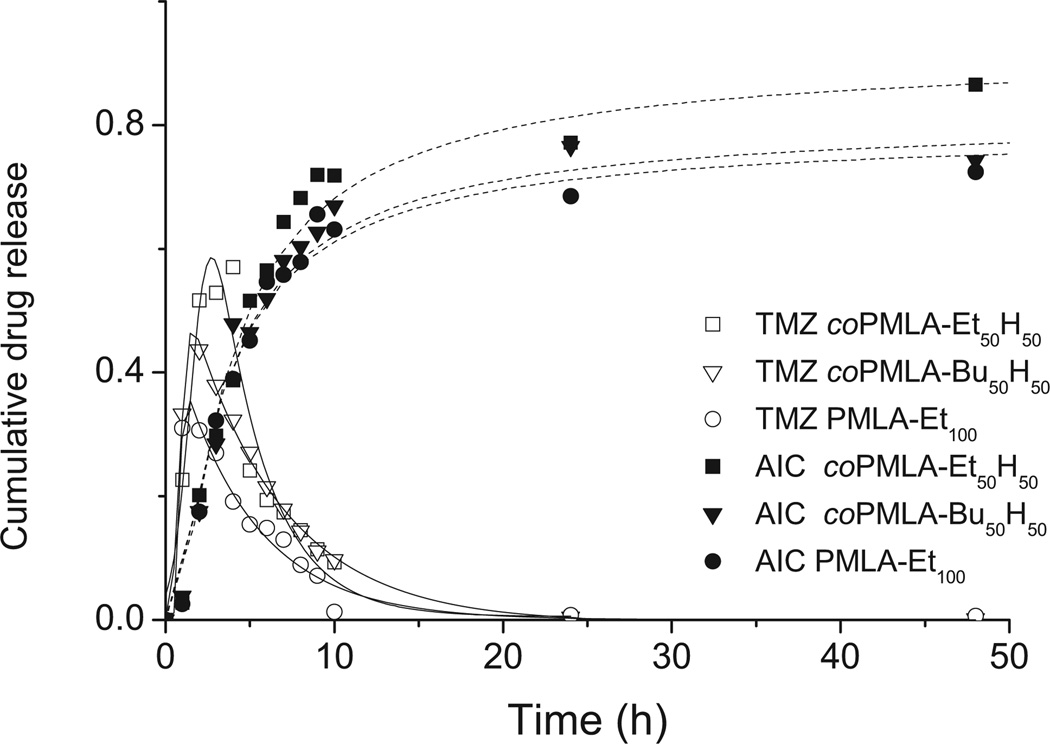
Drug release was measured under conditions close to physiological ones, that is, at pH 7.4 and 37 °C. It should be taken in to account that TMZ is susceptible to degradation in water at pH 7 with generation of AIC; the half-life time of TMZ under such conditions is 2 h.[28] Both compounds must be monitored therefore in order to evaluate the actual release of TMZ. In Figure 6 it is shown that the maximum TMZ release took place between 2 and 4 h of incubation with a cumulative release of 60% of the content in the case of coPMLA-Et50H50. Later the TMZ concentration peak decreased drastically due to its decomposition, whereas the AIC concentration increased to reach a constant level after 24 h. It is worthy to note that polymer nanoparticles still released TMZ after 24 h of incubation. Comparison of the releasing profiles for the three polymers revealed that most of the drug was delivered within the first few hours of incubation, and that the release was faster as the hydrophilicity of the polymer increased. In fact, traces of TMZ releasing from PMLA-Et100 nanoparticles were detected even after 48 h of incubation (Figure 6). The long retention time of TMZ in these particles correlates with the relative great resistance against hydrolysis of the polymer and is therefore probably due to the high hydrophobic character of the fully ethylated polymalate.
Figure 6.
TMZ release from polymer nanoparticles at pH 7.4 and 37 °C.
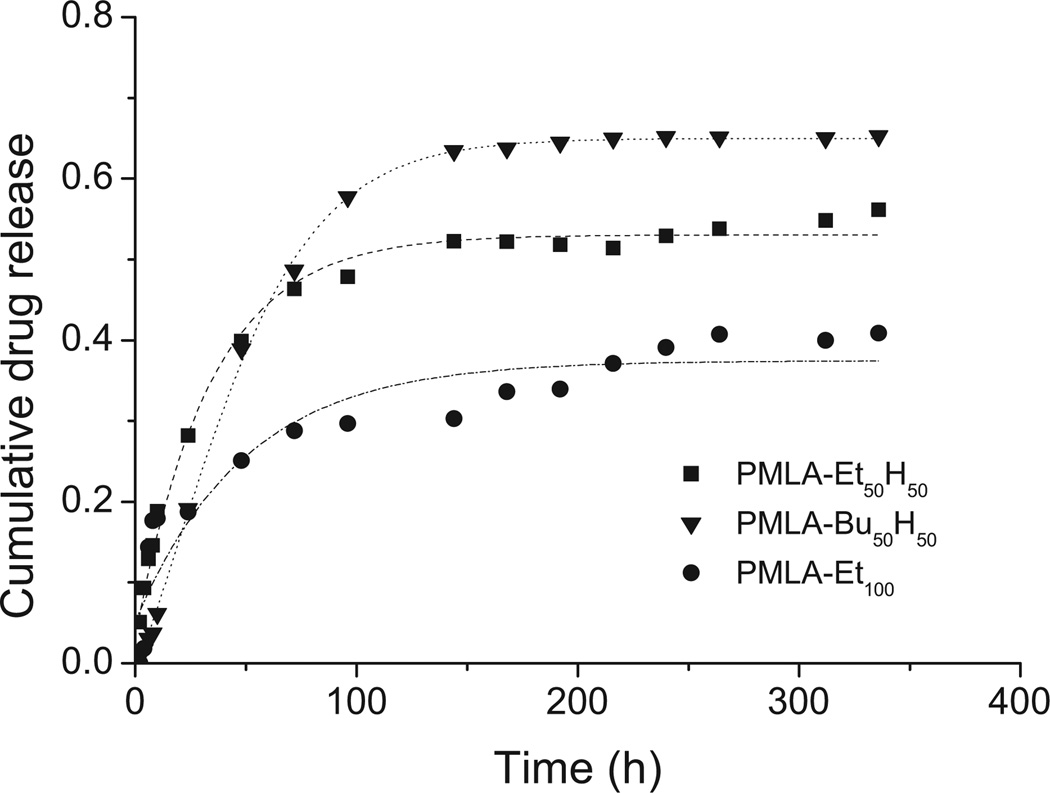
DOX was released much more slowly that TMZ(Figure 7), following an ascendant cumulative drug-time profile that after several days reached a constant value. The slower release observed for DOX compared to TMZ might be attributed to both its higher hydrophobicity and the probable interactions taking place between this drug and the unmodified carboxylic residues of the partially esterified PMLA. The fastest drug release was observed for coPMLA-Bu50H50 particles whereas those made of PMLA-Et100 showed the slowest one. It is also remarkable that differences in release rate among the polymers were larger for DOX than for TMZ.
Figure 7.
DOX release from polymer nanoparticles at pH 7.4 and 37 °C.
3.5. Cell Viability and Nanoparticles Cellular Uptake
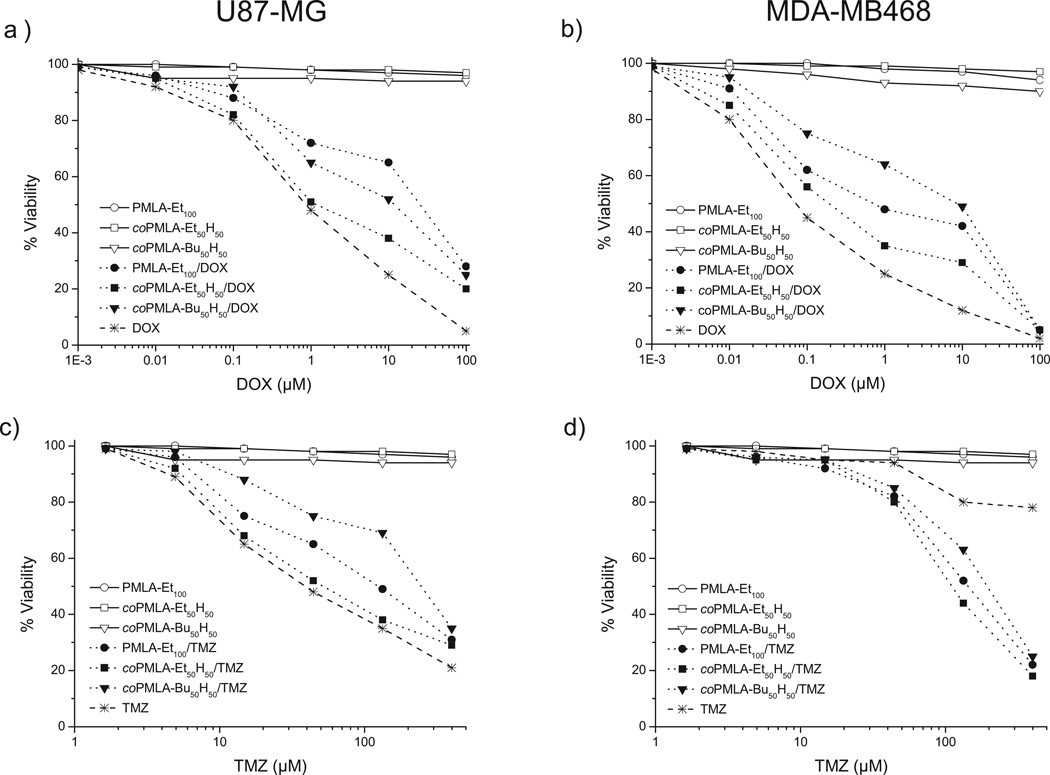
Cytotoxic tests were performed with unloaded and loaded drug nanoparticles on cell lines U87-MG and MDA-MB468; U87-MG cells are used as an in vitro model of human glioblastoma, while MDA-MB468 is a cell line for breast carcinoma cells. Both are extensively used to investigate the cytotoxic effect of chemotherapeutic drugs on cancer cells. Cytotoxicities of unloaded nanoparticles were practically negligible, since at the tested concentrations cell viability remained above 94%, except for PMLA-H50Bu50 which caused decays to 90% viability for MDA-MB468 cell line at higher concentrations (Figure 8). The cytotoxicity of these derivatives (ethyl and butyl polymalates) was found to be significantly lower than that observed for methyl polymalate in which the cell viability decayed to less than 80% after 24 h of exposure of the cells to the nanoparticles. The higher toxicity displayed by the methyl derivative was attributed to the action of methanol released during polymer degradation.[29]
Figure 8.
Cytotoxicity test of unloaded and drug-loaded nanoparticles and free drugs on U-87-MG and MDA-MB468 cell lines. a, b) DOX treatment; c, d) TMZ treatment.
In toxicology, half maximal effective concentration (EC50) refers to the concentration of a drug for which either 50% of its maximal effect is observed or 50% of the cell population exhibits a response after a specified exposure time. In our case EC50 has been taken for 50% of cell viability at the 2nd or 7th day after the 24 h-exposure drug treatment was applied, depending on the drug used. Cytotoxicity response of cell lines is plotted in Figure 8, as a function of drug concentration, and the obtained EC50 values are summarized in Table 3. In general, drugs loaded in nanoparticles needed higher concentrations than the free drugs to exert the same effect. coPMLA-Et50H50/drug showed the closest behavior to free drugs, followed by PMLA-Et100/drug, with the exception of the response of U87-MG cell line to coPMLA-Bu50H50/DOX. The most remarkable case was observed for MDA-MB468 cell line treated with TMZ and TMZ-NPs, in which free TMZ was shown to be ineffective at all concentrations while TMZ loaded nanoparticles reached EC50 at concentrations between 1–2 × 10−4 m. Two reasons can be invoked to explain such differences. A more sustained presence of encapsulated TMZ in the media (free TMZ has a half-life of only 2 h in aqueous media), and/or the nanoparticel internalization by cells that allows TMZ to be released directly in the cytosol.
Table 3.
Cytotoxicity EC50 values for free and encapsulated drugs in polymer nanoparticles.
| TMZ |
DOX |
|||
|---|---|---|---|---|
| U87-MG [µM] |
MDA MB468 [µM] |
U87-MG [µM] |
MDA MB468 [µM] |
|
| Free drug | 37 | >400 | 1 | 0.08 |
| PMLA-Et100/drug | 133 | 130 | 30 | 0.8 |
| PMLA-Et50H50/drug | 50 | 105 | 1 | 0.2 |
| PMLA-Bu50H50/drug | 260 | 200 | 10 | 10 |
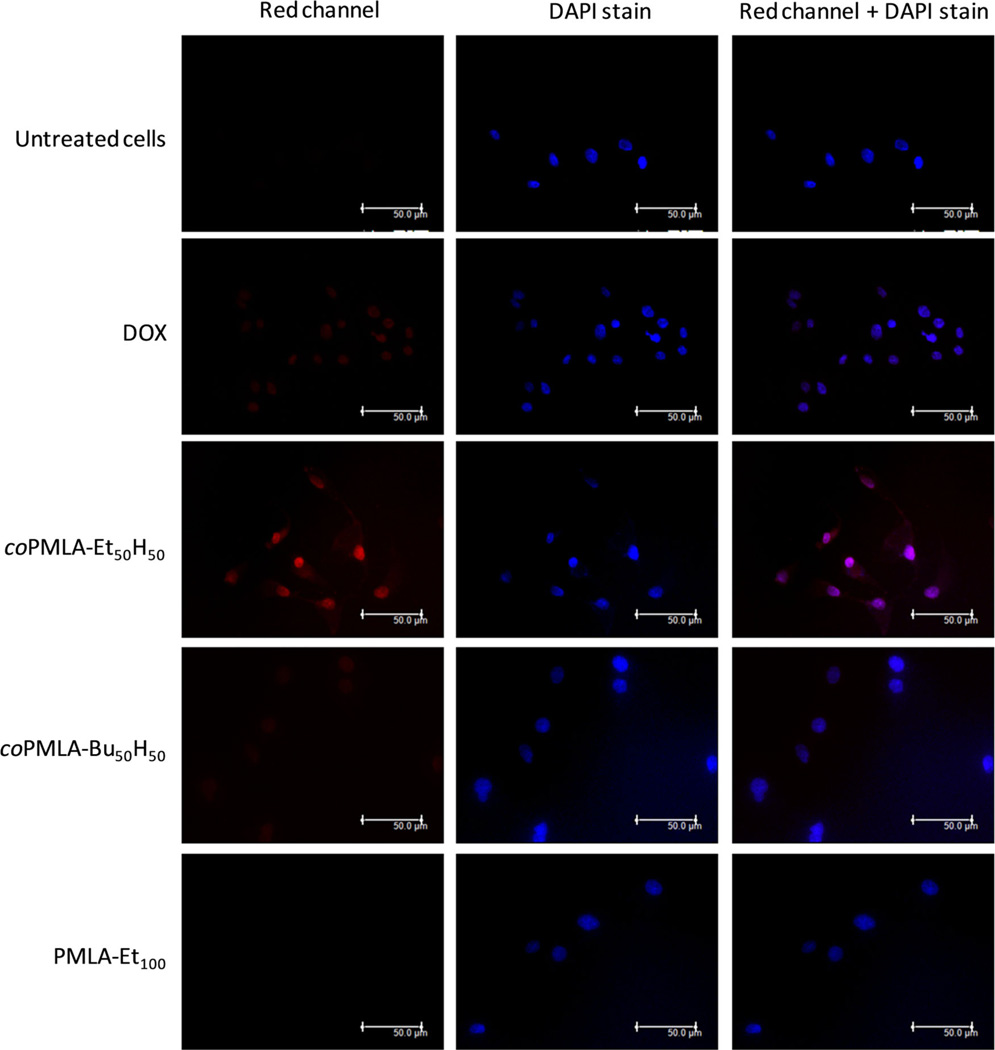
To evaluate the cellular uptake and trafficking of DOX-loaded nanoparticles in U87-MG cells, we performed microscopic studies based on red auto-fluorescence of DOX (Figure 9). DOX-loaded coPMLA-Et50H50 nanoparticles showed the most intense auto-fluorescence compared to the other samples; this auto-fluorescence is localized in the cytoplasm and cell nucleus, while cells treated with free DOX demonstrated significantly less auto-fluorescence and only inside the nucleus. Loaded nanoparticles from coPMLA-Bu50H50 and PMLA-Et100 showed a limited internalization which is consistent with cytotoxicity results. The effective protection of TMZ against degradation, the slow release of DOX, the low cytotoxicity and the effective internalization mainly of coPMLA-Et50H50 nanoparticles, make this derivative a potential material for the encapsulation and delivery of drugs for cancer treatment.
Figure 9.
Fluorescence microscopy of U87-MG cells incubated with free DOX and DOX-loaded nanoparticles. DOX auto-fluorescence in the red channel.
4. Conclusion
Esterification of microbial poly(malic acid) with ethanol or 1-butanol rendered easily hydrolysable polyesters and copolyesters, suitable for building nanoparticles useful for drug encapsulation and controlled delivery. The final degradation products of these polymers are the innocuous corresponding alcohol and easily metabolyzable malic acid. Nanoparticles diameters oscillate from 100 to 350 nm, depending on the polymer and on the methodology used for particle formation and encapsulation. DOX and TMZ can be encapsulated in these nanoparticles and released upon incubation under physiological conditions. Most of the TMZ was released within a few hours with subsequent hydrolytic degradation into AIC, while DOX was steadily released in a time scale of days. Furthermore, TMZ encapsulation afforded protection for the drug against hydrolytic decomposition. Drug-unloaded nanoparticles were not cytotoxic for the tested cell lines, whereas drug-loaded nanoparticles were cytotoxic for cancer cell lines. In the case of MDA MB468 cells, drug-loaded particles were highly efficient, while free TMZ did not show a measurable effect. The most efficient polymer nanoparticles were coPMLA-Et50H50 which showed better internalization of DOX by cells than the free drug.
Supplementary Material
Acknowledgements
This work received financial support from MCINN of Spain with Grant MAT2009-14053-C02-01 and MAT2012-38044-C03-03, AGAUR with grant 2009SGR1469 and from National Institute of Health, USA, Grant U01 CA151815. Thanks also to AGAUR (Generalitat de Catalunya) for the Ph.D. grant awarded to Alberto Lanz.
Footnotes
Supporting Information is available at Wiley Online Library or from the author.
Contributor Information
Alberto Lanz-Landázuri, Departament d’Enginyeria Química, Universitat Politècnica deCatalunya, ETSEIB, Diagonal 647, 08028 Barcelona, Spain.
José Portilla-Arias, Nanomedicine Research Center, Department of Neurosurgery, Cedars-Sinai Medical Center, 127 S. San Vicente Blvd. AHSP-A8220, Los Angeles, CA 90048, USA.
Antxon Martínez de Ilarduya, Departament d’Enginyeria Química, Universitat Politècnica deCatalunya, ETSEIB, Diagonal 647, 08028 Barcelona, Spain.
Montserrat García-Alvarez, Departament d’Enginyeria Química, Universitat Politècnica deCatalunya, ETSEIB, Diagonal 647, 08028 Barcelona, Spain.
Eggehard Holler, Nanomedicine Research Center, Department of Neurosurgery, Cedars-Sinai Medical Center, 127 S. San Vicente Blvd. AHSP-A8220, Los Angeles, CA 90048, USA.
Julia Ljubimova, Nanomedicine Research Center, Department of Neurosurgery, Cedars-Sinai Medical Center, 127 S. San Vicente Blvd. AHSP-A8220, Los Angeles, CA 90048, USA.
Sebastián Muñoz-Guerra, Email: sebastian.munoz@upc.edu, Departament d’Enginyeria Química, Universitat Politècnica deCatalunya, ETSEIB, Diagonal 647, 08028 Barcelona, Spain.
References
- 1.Abdellaoui K, Boustta M, Vert M, Morjani H, Manfait M. Eur. J. Pharm. Sci. 1998;6:61. doi: 10.1016/s0928-0987(97)00069-9. [DOI] [PubMed] [Google Scholar]
- 2.Zambaux MF, Bonneaux F, Gref R, Maincent P, Dellacherie E, Alonso MJ, Labrude P, Vigneron C. J. Controlled Release. 1998;50:31. doi: 10.1016/s0168-3659(97)00106-5. [DOI] [PubMed] [Google Scholar]
- 3.Martinez-Barbosa ME, Cammas S, Appel M, Ponchel G. Biomacromolecules. 2004;5:137. doi: 10.1021/bm0300608. [DOI] [PubMed] [Google Scholar]
- 4.Lenz RW, Vert M. Polym. Prepr. 1979;20:608. [Google Scholar]
- 5.Lee BS, Vert M, Holler E. Water-soluble aliphatic polyesters: Poly(malic acid)s. In: Doi Y, Steinbuechel A, editors. Biopolymers, Volume 3a: Polyesters I. Germany: Wiley-VCH, Weinheim; 2002. pp. 75–103. [Google Scholar]
- 6.Ljubimova JY, Portilla-Arias J, Patil R, Ding H, Inoue S, Markman JL, Rekechenetskiy A, Konda B, Gangalum P, Chesnokova A, Ljubimov AV, Black KL, Holler E. J. Drug Target. 2013;21:956. doi: 10.3109/1061186X.2013.837470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cammas S, Renard I, Langlois V, Guèrin Ph. Polymer. 1996;37:4215. [Google Scholar]
- 8.Holler E. In: Handbook of Engineering Polymeric Materials. Cheremisinoff NP, editor. New York: Marcel Dekker; 1997. pp. 93–103. [Google Scholar]
- 9.Holler E, Lee BS. Recent Res. Dev. Anal Chem. 2002;2:177. [Google Scholar]
- 10.Fernández CE, Mancera M, Holler E, Galbis JA, Muñoz-Guerra S. Polymer. 2006;47:6501. [Google Scholar]
- 11.Lee BS, Fujita M, Khazenzon NM, Wawrowsky KA, Wachsmann-Hogiu S, Farkas DL, Black KL, Ljubimova JY, Holler E. Bioconjugate Chem. 2006;17:317. doi: 10.1021/bc0502457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patil R, Portilla-Arias J, Ding H, Inoue S, Konda B, Hu J, Wawrowsky KA, Shin PK, Black KL, Holler E, Ljubimova J. Pharmaceut. Res. 2010;27:2317. doi: 10.1007/s11095-010-0091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soppimath KS, Aminabhavi TM, Kulkarni AR, Rudzinski WE. J. Controlled Release. 2001;70:1. doi: 10.1016/s0168-3659(00)00339-4. [DOI] [PubMed] [Google Scholar]
- 14.Mohanraj VJ, Chen Y, Pharm Trop. J. Res. 2006;5:561. [Google Scholar]
- 15.Manocha B, Margaritis A. Crit. Rev. Biotechnol. 2008;28:83. doi: 10.1080/07388550802107483. [DOI] [PubMed] [Google Scholar]
- 16.Hoffman AS. J. Controlled Release. 2008;132:153. doi: 10.1016/j.jconrel.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 17.Malam Y, Loizidou M, Seifalian AM. Trends Pharmacol Sci. 2009;30:592. doi: 10.1016/j.tips.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Osanai S, Nakamura K. Biomaterials. 2000;21:867. doi: 10.1016/s0142-9612(99)00210-0. [DOI] [PubMed] [Google Scholar]
- 19.Cammas S, Bèar MM, Harada A, Guèrin Ph, Kataoka K. Macromol. Chem. Phys. 2000;201:355. [Google Scholar]
- 20.Portilla-Arias JA, García-Alvarez M, Galbis JA, Muñoz-Guerra S. Macromol. Biosci. 2008;8:551. doi: 10.1002/mabi.200700249. [DOI] [PubMed] [Google Scholar]
- 21.Lanz-Landázuri A, García-Alvarez M, Portilla-Arias J, Martınez de Ilarduya A, Holler E, Ljubimova JY, Muñoz-Guerra S. Macromol. Chem. Phys. 2012;213:1623. doi: 10.1002/macp.201200134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ljubimova JY, Fujita M, Khazenzon NM, Lee BS, Wachsmann-Hogiu S, Farkas DL, Black KL, Holler E. Chem. Biol. Interact. 2008;171:195. doi: 10.1016/j.cbi.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding H, Inoue S, Ljubimov LV, Patil R, Portilla-Arias JA, Hu J, Konda B, Wawrowsky KA, Fujita M, Karabalin N, Sasaki T, Black KL, Holler E, Ljubimova JY. Proc. Natl. Acad Sci. USA. 2010;107:18143. doi: 10.1073/pnas.1003919107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding H, Helguera G, Rodríguez JA, Markman J, Luria-Pèrez R, Gangalum P, Portilla-Arias J, Inoue S, Daniels-Wells TR, Black K, Holler E, Penichet ML, Ljubimova JY. J. Controlled Release. 2013;10:322. doi: 10.1016/j.jconrel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mosmann TJ. Immunol. Methods. 1983;65:55. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 26.Akagi T, Higashi M, Kaneko T, Kida T, Akashi M. Biomacromolecules. 2006;7:297. doi: 10.1021/bm050657i. [DOI] [PubMed] [Google Scholar]
- 27.Manocha B, Margaritis A. J. Nanomater. 2010 [Google Scholar]
- 28.Baker SD, Wirth M, Statkevich P, Reidenberg P, Alton K, Sartorius SE, Dugan M, Cutler D, Batra V, Grochow LB, Donehower RC, Rowinsky EK. Clin. Cancer Res. 1999;5:309. [PubMed] [Google Scholar]
- 29.Lanz-Landazuri A, García-Alvarez M, Portilla-Arias J, Martínez de Ilarduya A, Patil R, Holler E, Ljubimova JY, Muñoz-Guerra S. Macromol. Biosci. 2011;10:1370. doi: 10.1002/mabi.201100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.