Abstract
Planar cell polarity (PCP) signaling has been shown in different studies to either promote or inhibit the malignancy of breast cancer. Using the 21T cell lines, which were derived from an individual patient and represent distinct stages of progression, we show that the prototypical PCP ligand, WNT5A, is expressed highest in 21MT-1 cells (invasive mammary carcinoma) and lowest in 21PT (atypical ductal hyperplasia) and 21NT (ductal carcinoma in situ) cells. Overexpression of WNT5A decreased spherical colony formation and increased invasion and in vivo extravasation only in 21NT cells; whereas overexpression increased migration of both 21PT and 21NT cells. WNT5A overexpression also increased RHOA expression of both cell lines and subsequent RHOA knockdown blocked WNT5A-induced migration, but only partially blocked WNT5A-induced invasion of 21NT cells. PCP can signal through VANGL1 to modulate AP-1 target genes (e.g. MMP3) and induce invasion. VANGL1 knockdown inhibited WNT5A-induced invasion of 21NT cells, but had no effect on WNT5A-induced migration of either 21PT or 21NT cells. WNT5A-induced MMP3 expression was seen only in 21NT cells, an effect that was VANGL1 dependent, but independent of AP-1. We thus provide evidence that PCP signaling can act in a context dependent manner to promote breast cancer progression.
Histological and molecular evidence has led to a model of breast cancer progression in which cells from the terminal duct lobular unit give rise to atypical ductal hyperplasia (ADH), which can progress to ductal carcinoma in situ (DCIS), and eventually to invasive mammary carcinoma (IMC)1,2,3,4,5,6. This transition, from a pre-invasive in situ lesion to an invasive lesion, is a critical step in breast cancer progression. These histological patterns observed during breast cancer progression are likely rough phenotypic indications of underlying molecular changes. Hence, there is interest in identifying the cellular and molecular regulators involved in breast cancer progression especially during earlier non-invasive stages.
Using microarray analysis, we have previously identified WNT5A, the prototypical non-canonical Wnt/planar cell polarity (PCP) ligand, to be differentially expressed in 21T series cells, all derived from the same patient, which have been shown to represent distinct stages of breast cancer7. The non-canonical PCP pathway exerts an important role in cell differentiation by regulating key components of the cytoskeleton that lead to cell shape and cell motility changes. Different PCP components have been shown to be involved in modulating cancer progression due to their role in cell motility. For example, WNT5A itself has been shown to promote metastasis of breast cancer by activating Rac and JNK8. WNT5A has also been implicated in metastasis of melanoma and gastric cancer9,10. Wnt5a acts via binding to Fzd family receptors and co-receptors (ROR-2, Ryk)11, which in the PCP pathway have been shown to signal through JNK and Rho11,12,13. Activation of Fzd7 in particular has been shown to promote invasion of colon carcinoma14,15 and migration of hepatocellular carcinoma cells16. Despite the large body of evidence that implicates PCP signaling in promoting invasion and metastasis, it is unclear if key components of PCP signaling are drivers of breast cancer progression alone or if they work in combination with other pathways. Conversely, it is also possible that PCP signaling may in some instances/cellular contexts inhibit cancer progression, as occurs due to antagonism between the different Wnt pathways, or when β-catenin signaling is upregulated during tumor development17. Importantly, what also remains unclear is the role of certain accessory molecules involved in PCP pathway signaling, such as VANGL1, in these different cellular contexts. We have previously shown that expression of VANGL1 is increased with malignancy of the 21T series cells7, although a potential functional role for VANGL1/PCP pathway in this system has yet to be explored. According to recent reports, downregulation of VANGL1 expression inhibits progression of hepatocellular carcinoma cells18, and this has been shown to be associated with decreased expression of AP-1 target genes such as COX-2 and MMP319. VANGL1 has also been shown to bind to the metastasis suppressor KAI1/CD82 in the mouse colon cancer cell line CT-26, increasing invasiveness and adhesion to fibronectin in vitro and increasing tumorigenicity and metastasis in vivo20. VANGL1 overexpression also increases invasion and migration of squamous carcinoma cells in vitro and promotes metastasis in a mouse squamous tumor model in vivo21. Additionally, suppression of VANGL1 via small interfering RNA (siRNA) has been shown to decrease colon cancer metastasis in mice, thereby supporting VANGL1's role as a metastasis promoter, likely through PCP signaling and increased cell motility and/or invasiveness22. In certain cases, this can occur by promotion of migration and invasion through the interaction of VANGL1 in a functional complex with Dvl and PKCδ23.
The dichotomy of effects observed in PCP signaling is also observed with its ligands, whereby WNT5A acts as both an oncogene or tumor suppressor depending on the context. A number of studies suggest that WNT5A can act as a tumor suppressor by antagonizing the Wnt/β-catenin pathway. For example, disruption of WNT5A expression with a WNT5A antisense expression vector has been found to mimic transformation caused by Wnt1 in the mouse mammary epithelial cell line C57MG, suggesting that WNT5A is an important regulator of cell growth and differentiation of these cells24. Additionally, there is evidence that in hematopoietic malignancies, thyroid carcinoma, colorectal cancer, hepatocellular carcinoma, and breast cancer, WNT5A is downregulated and in some cases there is an inverse correlation between WNT5A and β-catenin expression25,26,27,28,29,30. However, in more advanced stages of cancer progression, WNT5A has been shown to be upregulated and to promote invasion and metastasis in melanoma, gastric cancer, and breast cancer8,9,10. The PCP pathway may thus have a dual role in breast cancer progression, by acting as an inhibitor of progression during earlier stages and as a promoter of cancer progression during later stages. It is therefore important to further characterize the influence of the PCP pathway, to better understand the specific context that allows WNT5A to act as either a tumor suppressor or an oncogene.
Here, we show that WNT5A can promote progression to an invasive phenotype in a cell line representing DCIS (21NT), but not in a cell line representing ADH (21PT). We show that WNT5A promotes cell migration in a RHOA dependent manner and induces COX-2 expression in either cell type (21PT or 21NT), but that it only promotes invasiveness in 21NT (DCIS-like) cells, in a VANGL1 dependent, AP-1 independent manner, that involves increased expression of MMP3. This study thus provides evidence that the ability of WNT5A to promote cancer progression is dependent on the cellular context.
Results
WNT5A was differentially expressed among the 21T cell lines and was overexpressed in WNT5A transfectants vs. empty vector controls
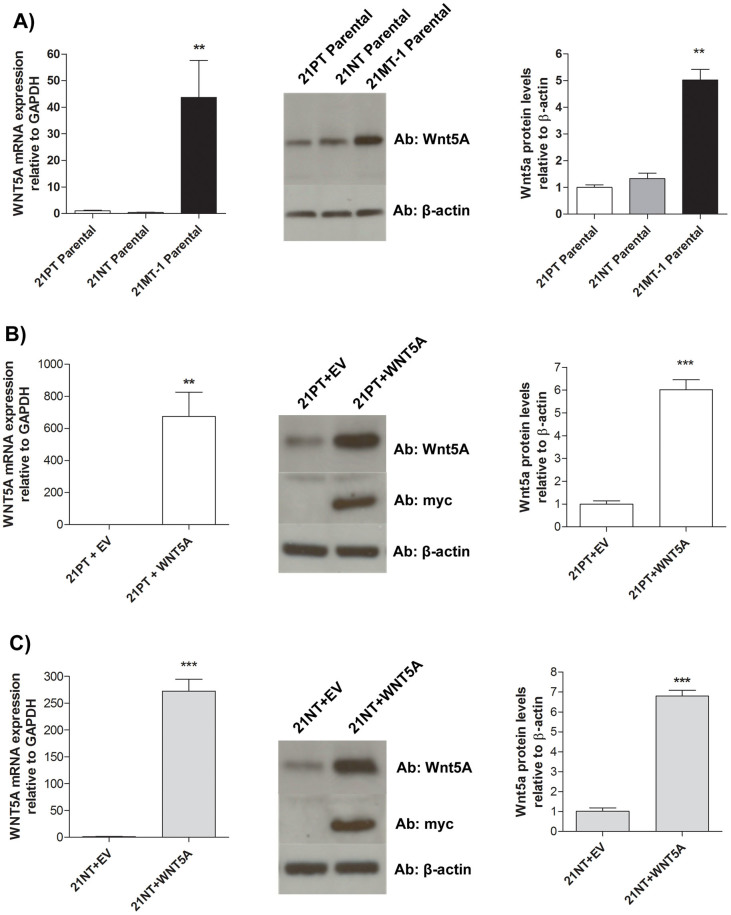
We have previously reported that WNT5A is present in the 21T cell lines and that its expression is relatively low in 21PT and 21NT cells and increased in 21MT-1 cells7. This finding is consistent with the view that PCP signaling potentially inhibits progression during earlier stages and promotes cancer progression during later stages8,9,10. To first confirm the relative levels of WNT5A in the 21T cell lines, WNT5A expression was examined at the mRNA and protein levels. Total RNA was extracted from the 21PT, 21NT and 21MT-1 cell lines and reverse transcribed into cDNA. Primers specific to WNT5A were used in qRT-PCR to examine relative levels of WNT5A among the three cell lines. WNT5A mRNA expression was higher in 21MT-1 cells compared to both 21PT and 21NT cells by 44-fold (P < 0.01) and 109-fold (P < 0.01) respectively; however, WNT5A mRNA levels were not significantly different between the 21PT and 21NT parental cell lines (Figure 1A left panel). Protein was extracted from the 21T parental cell lines and analyzed by western blot, with an antibody specific to Wnt5a protein (Figure 1A middle panel). (Full length blot is shown in Supplementary Figure S1). Wnt5a expression was higher in 21MT-1 cells compared to 21PT and 21NT cells by 5-fold (P < 0.01) and 4-fold (P < 0.01) respectively; however, levels of expression of Wnt5a were not significantly different between 21PT and 21NT (Figure 1A right panel).
Figure 1. WNT5A is differentially expressed among the 21T parental cell lines and expression is increased in WNT5A transfectants.
(A) left panel WNT5A mRNA expression is higher in parental 21MT-1 cells compared to both parental 21PT and 21NT cells. WNT5A expression was similar in both parental 21PT and 21NTcells. mRNA expression levels were calculated as a ratio to the housekeeping gene GAPDH. middle panel and right panel Wnt5a protein expression was higher in parental 21MT-1 cells compared to parental 21PT and 21NT cells. Wnt5a protein levels were similar in both parental 21PT and 21NT cells. (B) left panel WNT5A mRNA expression is increased in 21PT + WNT5A cells compared to 21PT + EV. middle panel Western blotting for Wnt5a and myc showed a band at the predicated molecular weight of a Wnt5a-myc fusion protein in the 21PT + WNT5A and 21PT + EV cells. right panel Densitometric quantification of Wnt5a protein blot showed increased expression in 21PT + WNT5A cells compared to 21PT + EV. (C) left panel WNT5A mRNA expression is increased in 21NT + WNT5A cells compared to 21NT + EV. middle panel Western blotting for Wnt5a and myc showed a band at the predicated molecular weight of a Wnt5a-myc fusion protein in the 21NT + WNT5A and 21NT + EV cells. right panel Densitometric quantification of Wnt5a protein blot showed increased expression in 21NT + WNT5A cells compared to 21NT + EV. Full-length blots are presented in Supplementary Figure 1S and 2S. For ease of comparison, the expression levels for the empty vector controls were set to 1. mRNA and protein expression was calculated as a ratio to GAPDH and β-actin respectively. To analyze mRNA and protein levels a Student's t-test (2 groups) or a one-way ANOVA (three groups) was performed and P < 0.05 was considered statistically significant. **P < 0.01, ***P < 0.001.
To examine whether WNT5A plays a functional role in the transition between stages of breast cancer progression, WNT5A was overexpressed in 21PT and 21NT cells. A WNT5A expression vector containing a myc-His epitope was constructed and transfected into 21PT cells (21PT + WNT5A) and 21NT cells (21NT + WNT5A). The expression vector alone (empty vector [EV], without WNT5A) was also transfected into 21PT cells (21PT + EV) and 21NT cells (21NT + EV). Both qRT-PCR and Western blot were used to determine levels of WNT5A expression in 21PT + WNT5A cells and 21NT + WNT5A cells compared to empty vector controls.
WNT5A mRNA was increased in both the 21PT + WNT5A cells and 21NT + WNT5A cells compared to their respective empty vector controls (675-fold for 21PT + WNT5A vs. 21PT + EV (P < 0.001); 272-fold for 21NT + WNT5A vs. 21NT + EV (P < 0.001)) (Figure 1B and C left panels). Similarly, Wnt5a protein was increased in both the 21PT + WNT5A cells and 21NT + WNT5A cells compared to their respective empty vector controls (6-fold for 21PT + WNT5A vs. 21PT + EV (P < 0.001); (7-fold for 21NT + WNT5A vs. 21NT + EV (P < 0.001)) (Figure 1B and C middle and right panels). (Full length blots are shown in Supplementary Figure S2). In order to show that the increase in Wnt5a was due to transfection of a WNT5A-myc construct, an anti-myc antibody was used to detect myc epitope expression. Western blotting of protein from 21PT + WNT5A and 21NT + WNT5A cells showed a band at the predicted molecular weight of Wnt5a-myc (Figure 1B and C middle panel).
WNT5A induced altered colony morphology of 21NT, but not 21PT cells in 3D Matrigel
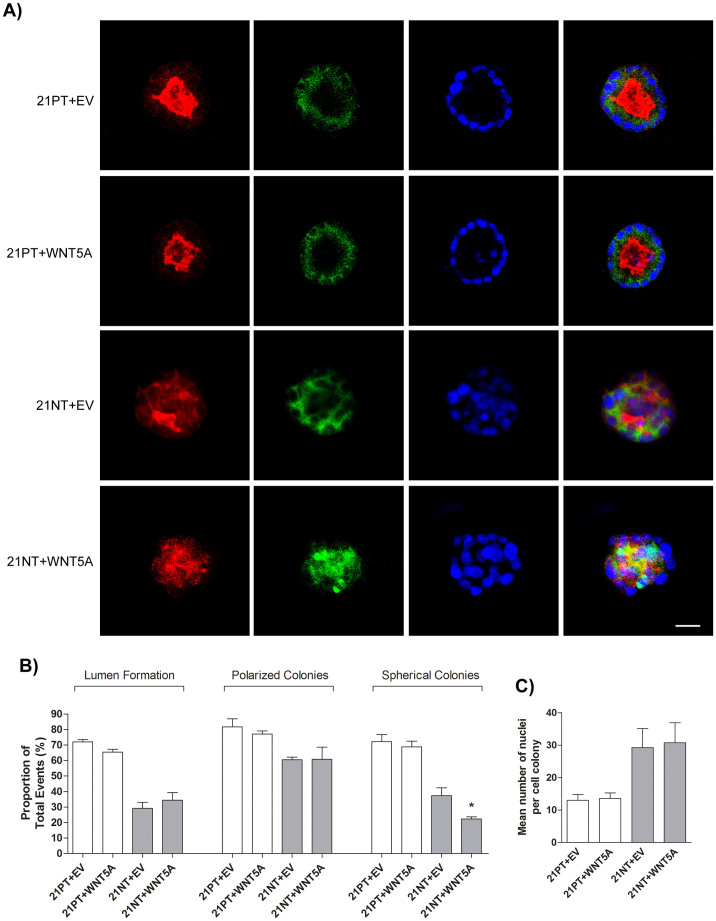
To first assess the role of WNT5A in the progression of early breast cancer, 21PT and 21NT transfectants were assessed using a well-established 3D morphology assay31,32. The 21PT + EV, 21PT + WNT5A, 21NT + EV, and 21NT + WNT5A cells were grown on top of 100% Matrigel in a αHE media containing 2% Matrigel and 2% FBS for 12 days (Figure 2A). WNT5A overexpression had no effect on 3D morphology with respect to lumen formation and polarized colonies in either the 21PT or 21NT cell lines. However, WNT5A overexpression decreased the percentage of colonies that were spherical (i.e. more irregular/dispersed colonies) in 21NT (not 21PT) cells. The percentage of spherical colonies decreased from 37.4% (SE 5.00) in 21NT + EV cells to 22.3% (SE 1.46) in 21NT + WNT5A cells (P < 0.05) (Figure 2B). There was no effect on the number of nuclei per cell colony after WNT5A overexpression (Figure 2C) indicating no change in overall growth characteristics in the 3D Matrigel system.
Figure 2. WNT5A induces altered colony morphology of 21NT, but not 21PT cells in 3D Matrigel.
(A) 21T transfectants were grown using a 3D Matrigel system for 12 days. Cell colonies were fixed, permeabilized, stained, and imaged using confocal microscopy. Representative cell colonies are shown. Actin is shown in red, E-cadherin is shown in green, and the nuclear stain Hoechst is shown in blue. An overlay of all three channels is shown in the final set of panels. (B) There was a decrease in the proportion of spherical colonies formed by 21NT cells when overexpressing WNT5A, but no change in the proportion of lumen formation or % polarized colonies. (C) WNT5A overexpression had no effect on the number of nuclei per cell colony. Scale bar represents 30 μm. To analyze groups a one-way ANOVA was performed with Tukey's post hoc test and P < 0.05 was considered statistically significant. *P < 0.05.
WNT5A promoted migration of 21PT and 21NT, but increased invasion only of 21NT cells
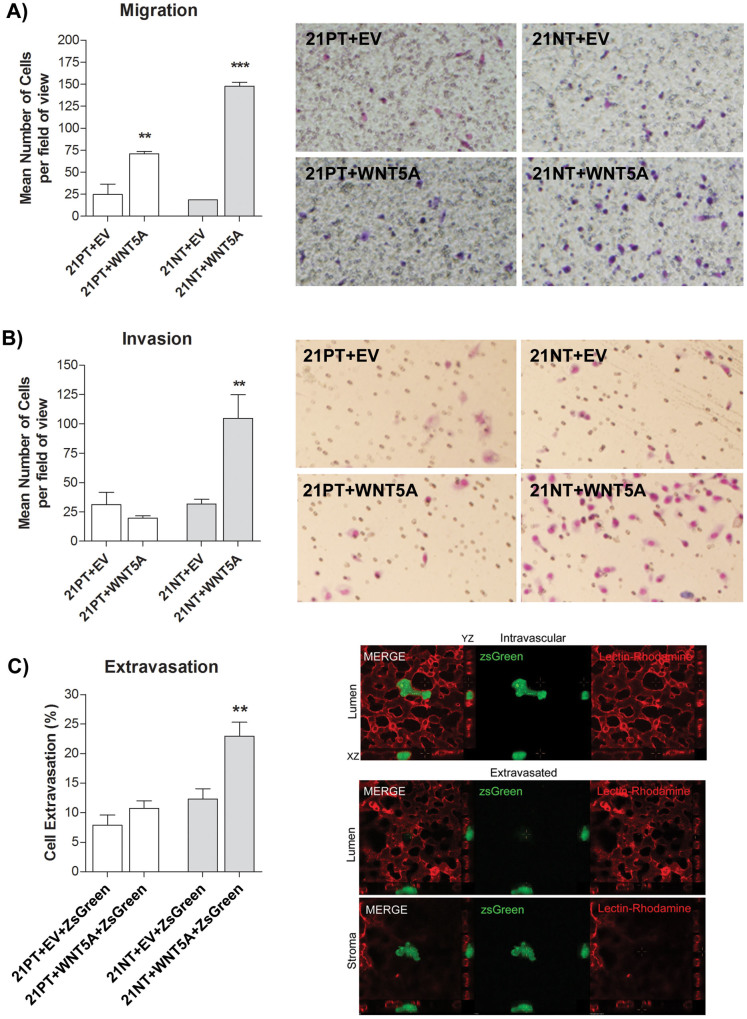
The migratory ability of WNT5A transfectants was assessed using transwell migration assay. 21PT + EV, 21PT + WNT5A, 21NT + EV or 21NT + WNT5A cells were placed in the upper chamber of a transwell migration system (membranes with 8.0 μm pores coated with 6 μg of gelatin) and allowed to migrate through the pores, with FBS as the chemoattractant in the lower chamber. WNT5A overexpression increased migration of both 21PT and 21NT cell lines. WNT5A overexpression increased 21PT cell migration from 24.7 (SE 11.8) cells per field of view (21PT + EV) to 71.0 (SE 2.66) cells per field of view (21PT + WNT5A) (P < 0.01) (Figure 3A). WNT5A overexpression increased 21NT cell migration from 18.5 (SE 0.276) cells per field of view (21NT + EV) to 148 (SE 4.40) cells per field of view (21NT + WNT5A) (P < 0.001) (Figure 3A).
Figure 3. WNT5A promotes migration of 21PT and 21NT, but increases invasion and extravasation of 21NT cells only.
(A) Transwell migration assay (transwells coated with gelatin) using 21T transfectants towards 10% FBS for 5 hours. Both 21PT + WNT5A cells and 21NT + WNT5A cells showed an increase in the number of cells that migrated to the underside of transwell membranes, compared to their respective empty vector control. Representative transwell images are shown (right panel). (B) Transwell invasion assay (transwells coated with Matrigel) using 21T transfectants towards 10% FBS for 20 hours. 21NT + WNT5A cells showed an increase in the number of cells that invaded to the underside of the transwell membrane compared to the empty vector control. However, WNT5A overexpression did not increase invasion of 21PT cells. Representative transwell images are shown (right panel). (C) In vivo (chick embryo) cancer cell extravasation assay using 21T transfectants labeled by transduction with ZsGreen. WNT5A overexpression significantly increased the ability of 21NT, but not 21PT cells to extravasate. Representative (21NT transfected) cells are shown in the intravascular (luminal) and extravascular (stromal) state (right panel). To analyze groups a one-way ANOVA was performed with Tukey's post hoc test and P < 0.05 was considered statistically significant. **P < 0.01; ***P < 0.001.
To directly assess the ability of WNT5A overexpression to influence invasiveness of the 21T cell lines, the WNT5A transfected cells were tested in a transwell invasion assay. 21PT + EV, 21PT + WNT5A, 21NT + EV and 21NT + WNT5A cells were placed in the upper chamber of a transwell system (membranes with 8.0 μm pores precoated with Matrigel) and allowed to invade through the pores, with FBS as the chemoattractant in the lower chamber. WNT5A overexpression increased invasion of 21NT cells only, from 31.7 (SE 4.04) cells per field of view for 21NT + EV cells, to 105 (SE 20.1) cells per field of view for 21NT + WNT5A (P < 0.01) cells (Figure 3B).
In order to assess cell invasiveness in vivo, we used a chick embryo chorioallantoic membrane vein injection assay for cell extravasation33,34,35,36. In this assay, tumor cells are injected intravenously into ex ovo chicken embryos. The extent of tumor cell extravasation is quantified using intravital imaging. After 24 hours, we confirmed that the majority of cells that were present in the CAM had extravasated and that 21NT cells that overexpressed WNT5A (21NT + WNT5A + ZsGreen vs. 21NT + EV + ZsGreen) exhibited significantly higher rates of cancer cell extravasation (Figure 3C). In contrast, 21PT cells that overexpressed WNT5A (21PT + WNT5A + ZsGreen vs. 21PT + EV + ZsGreen) did not exhibit any significant change in extravasation rates compared to the empty vector control (Figure 3C).
WNT5A overexpression upregulated RHOA mRNA expression, which mediated migration and invasion
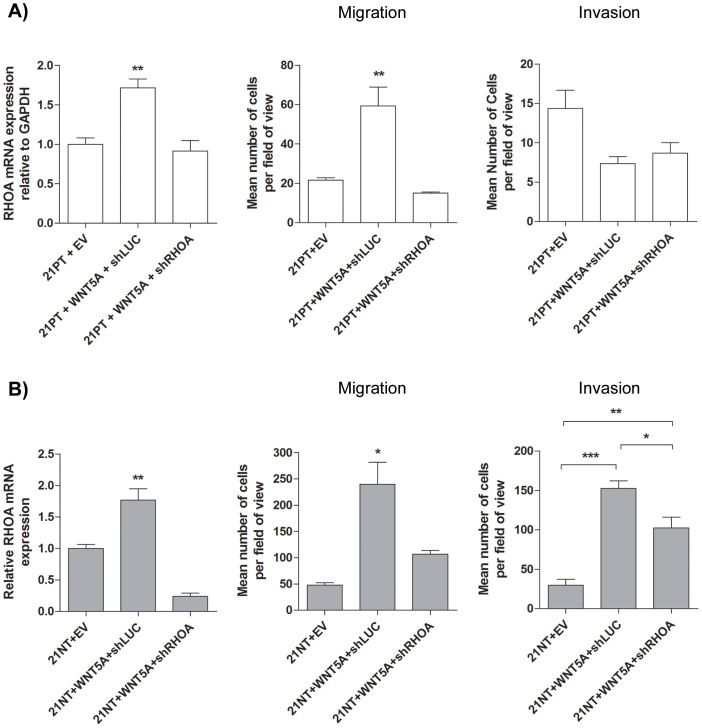
To further investigate the functional response to WNT5A overexpression in 21PT and 21NT cells, the downstream effector RHOA was examined. Overexpression of WNT5A in 21PT or 21NT cells increased the expression of RHOA at the mRNA level. RHOA expression was increased by 1.75-fold in WNT5A transfected 21PT cells (P < 0.01) (Figure 4A left panel), and by 1.77-fold in WNT5A transfected 21NT cells (P < 0.01) (Figure 4B left panel). To determine if WNT5A-induced RHOA expression plays a functional role in 21PT or 21NT cell migration or invasion, RHOA expression was knocked down in the WNT5A transfectants, using an shRNA specific for RHOA. In RHOA knockdowns of both WNT5A transfected 21PT and 21NT cells, RHOA expression was decreased to a level significantly less that of the off target (luciferase) knockdown control, to a level similar to, or lower than that of non-WNT5A transfected (empty vector control) cells (Figure 4A and B left panels).
Figure 4. Involvement of RHOA in WNT5A-induced migration and invasion.
(A) left panel WNT5A overexpression in 21PT cells increased the expression of total RHOA at the mRNA level. RHOA expression was knocked down to a level similar to 21PT, compared to an off target knockdown control. middle panel Transwell migration assay using 21PT derived cells. Knockdown of RHOA in 21PT + WNT5A cells decreased migration to levels similar to 21PT + EV. Such an effect was not seen using an off target knockdown. right panel Transwell invasion assay using 21PT derived cells. Knockdown of RHOA in 21PT + WNT5A cells had no significant effect on invasion. (B) left panel WNT5A overexpression in 21NT cells increased the expression of total RHOA at the mRNA level. RHOA expression was knocked down to a level lower than 21NT, compared to an off target knockdown control. middle panel Transwell migration assay using 21NT derived cells. Knockdown of RHOA in 21NT + WNT5A cells decreased migration to levels similar to 21NT + EV. right panel Transwell invasion assay using 21NT derived cells. Knockdown of RHOA in 21NT + WNT5A cells decreased WNT5A-induced invasion compared to an off target knockdown control. The reduction in invasiveness was incomplete in that it did not reach levels of the 21NT + EV cells. Representative transwell images are shown in Supplementary Figure S3. mRNA expression levels were calculated as a ratio to the housekeeping gene GAPDH. For ease of comparison, the expression levels for empty vector control cells were set to 1. To analyze groups a one-way ANOVA was performed with Tukey's post hoc test and P < 0.05 was considered statistically significant. *P < 0.05, **P < 0.01, ***P < 0.001.
To assess the change in migration due to RHOA knockdown in 21PT + WNT5A and 21NT + WNT5A cells, transwell migration assays were used. Cells were placed in the upper chamber of a transwell migration system and allowed to migrate through the pores in response to FBS as the chemoattractant in the lower chamber. RHOA knockdown decreased migration of 21PT + WNT5A cells from 59.3 (SE 9.53) cells per field of view to 15.0 (SE 0.580) cells per field of view (P < 0.01), a level comparable to that of 21PT + EV (Figure 4A middle panel and Supplementary Figure S3). Similarly, RHOA knockdown decreased the migration of 21NT + WNT5A cells from 240 (SE 41.6) cells per field of view to 107 (SE 6.88) cells per field of view (P < 0.05), a level comparable to that of 21NT + EV (Figure 4B middle panel and Supplementary Figure S3). To assess the change in invasion due to RHOA knockdown in 21PT + WNT5A and 21NT + WNT5A cells, transwell invasion assays were again used. Cells were placed in the upper chamber of the transwell invasion system (membranes precoated with Matrigel) and allowed to invade through the pores in order to reach FBS (chemoattractant) that was present in the lower chamber. Interestingly, although there was no difference in invasion of the 21PT + WNT5A cells with RHOA knockdown (Figure 4A right panel and Supplementary Figure S3), there was a significant reduction in invasion of 21NT + WNT5A, from 153 (SE 9.49) cells per field of view to 102 (SE 13.7) cells per field of view in the shRHOA knockdowns (P < 0.05), The degree of reduction in invasion of 21NT + WNT5A + shRHOA cells was incomplete, however, in that it did not reach the baseline levels of invasion exhibited by the non-WNT5A transfected vector control 21NT cells (21NT + EV) (P < 0.01) (Figure 4B right panel and Supplementary Figure S3).
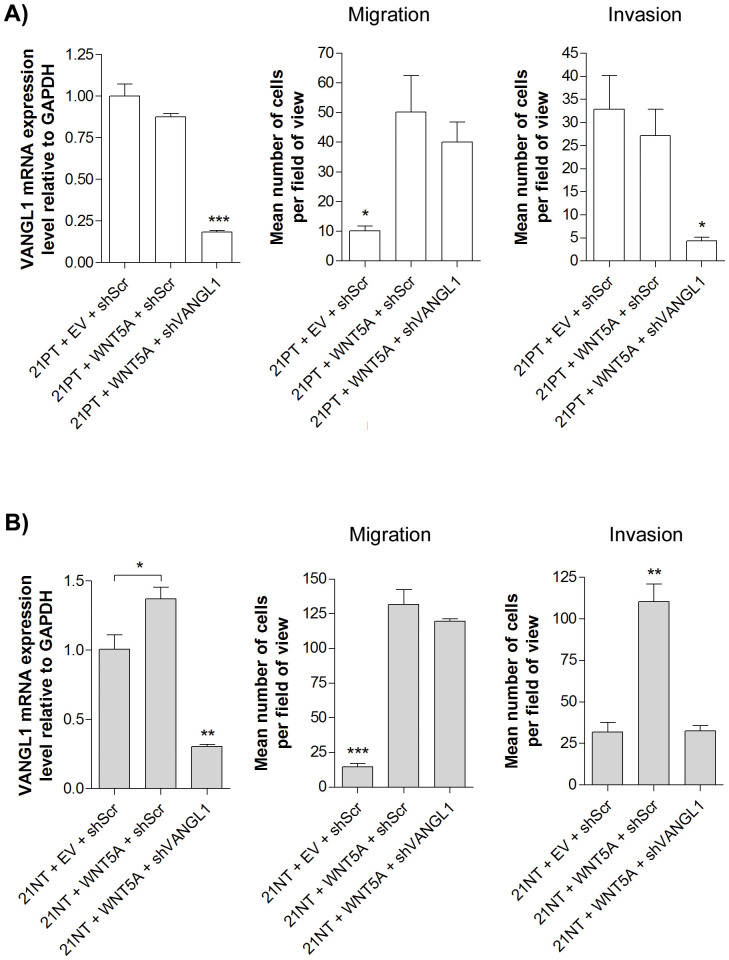
VANGL1 knockdown blocked WNT5A induced invasion of 21NT cells
To determine if VANGL1 is required for PCP-based migration or invasion of the 21T cell lines, VANGL1 was knocked down using an shRNA approach, with shRNA specific for VANGL1 (shVANGL1) vs. scrambled shRNA control (shScr). In 21PT cells, VANGL1 mRNA levels remained unchanged after WNT5A overexpression (21PT + WNT5A + shScr vs. 21PT + EV + shScr), but were significantly reduced by 5.50-fold and 4.81-fold (P < 0.001) after VANGL1 knockdown (21PT + WNT5A + shVANGL1 vs. 21PT + EV + shScr and 21PT + WNT5A + shScr cells respectively (Figure 5A left panel)). In 21NT cells, VANGL1 mRNA levels were increased by 1.37-fold (P < 0.05) after WNT5A overexpression (21NT + WNT5A + shScr vs. 21NT + EV + shScr), and were significantly reduced by 3.26-fold and 4.47-fold (P < 0.01) after VANGL1 knockdown (21NT + WNT5A + shVANGL1 vs. 21NT + EV + shScr and 21NT + WNT5A + shScr cells respectively (Figure 5B left panel)).
Figure 5. Effect of VANGL1 knockdown on migration and invasion of 21PT and 21NT cells in the presence of WNT5A overexpression.
(A) left panel Level of VANGL1 expression in 21PT cells is not altered by WNT5A overexpression (21PT + WNT5A + shScr vs. 21PT + EV + shScr), but is significantly down-regulated by VANGL1 shRNA, compared to a scrambled shRNA control (21PT + WNT5A + shVANGL1 vs. 21PT + WNT5A + shScr). middle panel Transwell migration assay using 21PT derived cells. Migration of 21PT cells is significantly induced by WNT5A (21PT + WNT5A + shScr vs. 21PT + EV + shScr), but knockdown of VANGL1 in 21PT + WNT5A has no significant effect on migration compared to a scrambled shRNA control (21PT + WNT5A + shVANGL1 vs. 21PT + WNT5A + shScr). right panel Transwell invasion assay using 21PT derived cells. WNT5A overexpression does not alter invasiveness of 21PT (21PT + WNT5A + shScr vs. 21PT + EV + shScr), while down-regulation of VANGL1 significantly decreases invasiveness (21PT + WNT5A + shVANGL1 vs. 21PT + WNT5A + shScr). (B) left panel WNT5A overexpression 21NT cells increases the expression of VANGL1. VANGL1 expression of 21NT cells is significantly increased by overexpression of WNT5A (21NT + WNT5A + shScr vs. 21NT + EV + shScr), but is significantly down-regulated by VANGL1 shRNA compared to a scrambled shRNA control (21NT + WNT5A + shVANGL1 vs. 21NT + WNT5A + shScr). middle panel Transwell migration assay using 21NT derived cells. Migration of 21NT cells is significantly induced by WNT5A (21NT + WNT5A + shScr vs. 21NT + EV + shScr), but knockdown of VANGL1 in 21NT + WNT5A cells has no significant effect on migration compared to a scrambled shRNA control (21NT + WNT5A + shVANGL1 vs. 21NT + WNT5A + shScr). right panel Transwell invasion assay using 21NT derived cells. WNT5A overexpression significantly increases invasiveness of 21NT cells (21NT + WNT5A + shScr vs. 21NT + EV + shScr), while knockdown of VANGL1 in 21NT + WNT5A cells reverses this effect (21NT + WNT5A + shVANGL1 vs. 21NT + WNT5A + shScr). Representative transwell images are shown in Supplementary Figure S4. mRNA expression levels were calculated as a ratio to GAPDH. For ease of comparison, the expression levels for control cells were set to 1. To analyze groups a one-way ANOVA was performed with Tukey's post hoc test and P < 0.05 was considered statistically significant. *P < 0.05, **P < 0.01, ***P < 0.001.
Transwell assays, performed as described above, were used to assess the change in migration and invasion due to VANGL1 knockdown in 21PT + WNT5A and 21NT + WNT5A cells. VANGL1 knockdown did not significantly alter the migratory ability of either 21PT + WNT5A or 21NT + WNT5A cells (Figure 5A and B middle panels and Supplementary Figure S4), but did result in decreased cell invasion of both cell lines (Figure 5A and B right panels and Supplementary Figure S4). This effect was particularly marked for the 21NT cells, in which invasion had been significantly enhanced by WNT5A transfection (21NT + WNT5A + shScr vs. 21NT + EV + shScr), and in which invasiveness was knocked down to baseline (21NT + EV + shScr) levels from 110 to 32.5 cells per field of view (P < 0.01) by down-regulation of VANGL1 (21NT + WNT5A + shScr vs. 21NT + WNT5A + shVANGL1 (Figure 5A and B right panels) and Supplementary Figure S4).
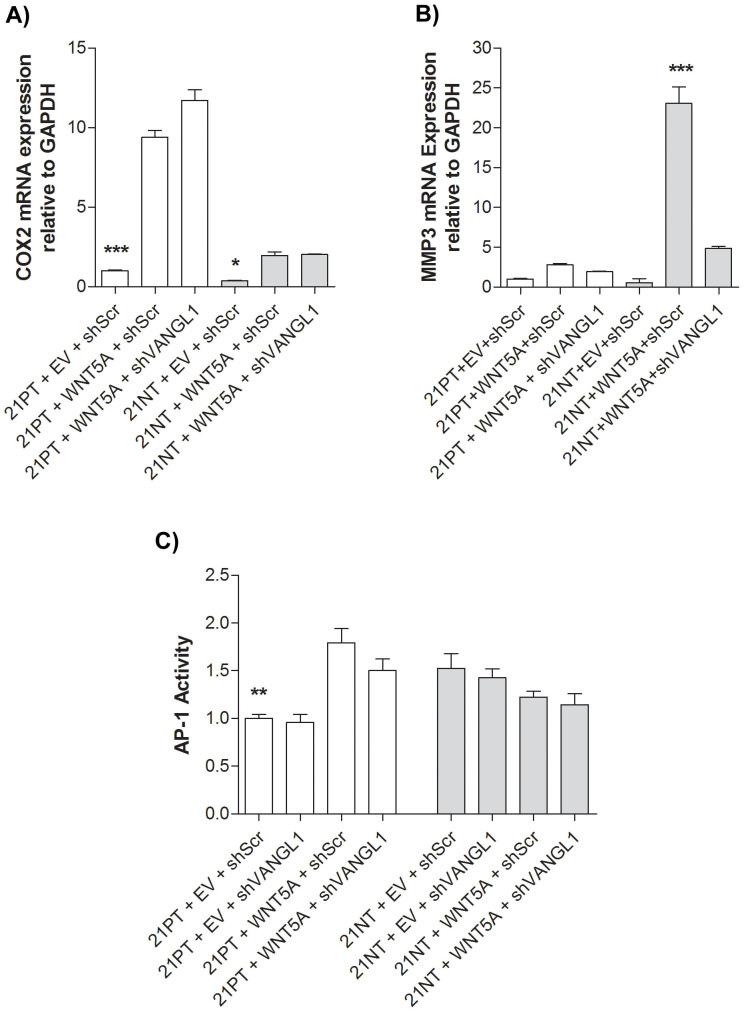
WNT5A overexpression increased COX-2 expression in both 21PT and 21NT cells, in a VANGL1 independent manner, and MMP3 in 21NT cells only, in a VANGL1 dependent manner
Signaling through VANGL1, a core PCP component, has previously been reported to increase invasion of colorectal, gastric, and liver cancer cells19,23,37,38 through AP-1 target genes. In addition, in hepatocellular and colorectal carcinoma cells, expression of AP-1 regulated target genes COX-2 and MMP3 was associated with VANGL1 expression and was decreased upon down-regulation of VANGL119,37, suggesting their role in VANGL1 dependent signaling. These genes were thus here examined to determine their potential involvement in WNT5A-induced invasion, particularly in the 21NT cells. Using real-time PCR and primers specific to COX-2, and MMP3, we found that WNT5A overexpression resulted in a 9.40-fold and 5.38-fold increase in COX-2 expression in 21PT and 21NT cells respectively (21PT + WNT5A + shScr vs. 21PT + EV + shScr and 21NT + WNT5A + shScr vs. 21NT + EV + shScr) (Figure 6A); however, VANGL1 knockdown did not alter the elevated levels of COX-2 present in either cell line overexpressing WNT5A (21PT + WNT5A + shVANGL1 vs. 21PT + WNT5A + shScr and 21NT + WNT5A + shVANGL1 vs. 21NT + WNT5A + shScr) (Figure 6A). In contrast, WNT5A overexpression resulted in a 40.3-fold increase in MMP3 expression only in 21NT cells (21NT + WNT5A + shScr vs. 21NT + EV + shScr), while VANGL1 knockdown resulted in a 4.74-fold decrease in this WNT5A induced MMP3 expression (21NT + WNT5A + shVANGL1 vs. 21NT + WNT5A + shScr) (Figure 6B). Neither WNT5A overexpression (21PT + WNT5A + shScr vs. 21PT + EV + shScr) nor VANGL1 knockdown (21PT + WNT5A + shVANGL1 vs. 21PT + WNT5A + shScr) resulted in a change in MMP3 expression in the 21PT cells (Figure 6B). In work not shown here, we found no change in MMP1 expression in either the 21PT or 21NT cells in response to WNT5A overexpression or with knockdown of VANGL1.
Figure 6. WNT5A overexpression increases COX2 in both 21PT and 21NT cells and MMP3 only in 21NT cells, in a VANGL1 dependent manner.
(A) WNT5A overexpression significantly increases COX2 mRNA expression in both the 21PT and 21NT cells (21PT + WNT5A + shScr vs. 21PT + EV + shScr and 21NT + WNT5A + shScr vs. 21NT + EV + shScr). VANGL1 knockdown does not significantly change the expression levels of COX2 in either the WNT5A overexpressing 21PT or 21NT cells (21PT + WNT5A + shVANGL1 vs. 21PT + WNT5A + shScr and 21NT + WNT5A + shVANGL1 vs. 21NT + WNT5A + shScr). (B) WNT5A overexpression significantly increases MMP3 mRNA expression in 21NT cells (21NT + WNT5A + shScr vs. 21NT + EV + shScr), but not 21PT cells (21PT + WNT5A + shScr vs. 21PT + EV + shScr). VANGL1 knockdown significantly reduces the WNT5A-induced MMP3 expression of 21NT cells (21NT + WNT5A + shVANGL1 vs. 21NT + WNT5A + shScr), but has no significant effect on MMP3 expression of 21PT cells (21PT + WNT5A + shVANGL1 vs. 21PT + WNT5A + shScr or 21PT + EV + shScr). (C) AP-1 activity was measured in the 21T transfectants using a Luciferase reporter. WNT5A significantly increased overall AP-1 activity in the 21PT cells (21PT + WNT5A + shScr vs. 21PT + EV + shScr), but this was not significantly reduced by VANGL1 knockdown (21PT + WNT5A + shVANGL1 vs. 21PT + WNT5A + shScr). In 21NT cells, AP-1 activity was not significantly affected by WNT5A overexpression (21NT + WNT5A + shScr vs. 21NT + EV + shScr) or VANGL1 knockdown (21NT + WNT5A + shVANGL1 vs. 21NT + WNT5A + shScr or 21NT + EV + shVANGL1 vs. 21NT + EV + shScr). mRNA expression levels were calculated as a ratio to GAPDH. For ease of comparison, mRNA expression and AP-1 activity levels in the 21PT control cells were set to 1. To analyze groups a one-way ANOVA was performed with Tukey's post hoc test and P < 0.05 was considered statistically significant. *P < 0.05, **P < 0.01, ***P < 0.001.
To examine whether changes in COX-2 and MMP3 expression in response to WNT5A were AP-1 mediated, we looked at AP-1 activity in the WNT5A transfectants of both 21PT and 21NT cells. Luciferase assays revealed increased AP-1 activity in WNT5A overexpressing 21PT cells (21PT + WNT5A + shScr vs. 21PT + EV + shScr), that was not altered by VANGL1 knockdown (21PT + WNT5A + shVANGL1 vs. 21PT + WNT5A + shScr) (Figure 6C). In the 21NT cells, AP-1 activity was endogenously higher than in 21PT cells (21NT + EV + shScr vs. 21PT + EV + shScr), but did not change when WNT5A was increased (21NT + WNT5A + shScr vs. 21NT + EV + shScr) or when VANGL1 was knocked down (21NT + WNT5A + shVANGL1 vs. 21NT + WNT5A + shScr) (Figure 6C), thus indicating that the significant increases in MMP3 activity in 21NT + WNT5A cells occur independently of the AP-1 transcription factor.
Discussion
WNT5A, the prototypical Wnt ligand which activates non-canonical Wnt signaling, has been shown to act as either an oncogene or a tumor suppressor in various studies8,9,10,24,25,26,27,28,29,30. This dichotomy of WNT5A effects may be related to other PCP signaling components such as VANGL1 and the cellular context in which the signaling occurs. Given the differential levels of WNT5A and VANGL1 expression we have previously observed in the 21T cell lines (both higher in invasive, metastatic 21MT-1 cells)7, we here undertook to modulate these factors and determine their effects on migration and invasion in the different cellular contexts, in order to better understand the significance of the PCP pathway in the early, pre-malignant stages of breast cancer progression.
Here we have used the 21T series cells to show that cells representing different stages of early breast cancer progression respond differently to activation of the Wnt/PCP pathway by WNT5A. Although both ADH-like 21PT cells and DCIS-like 21NT cells respond to WNT5A induction by increased migration (which we show to be a RHOA-dependent phenomenon associated with induction of COX-2 expression in both cell lines), only 21NT cells exhibit an increased ability to invade (a phenomenon associated with increased MMP3 expression and dependent upon VANGL1). Although VANGL1 knockdown in WNT5A overexpressing 21PT or 21NT did not significantly affect cell migration, cell invasiveness was markedly inhibited, suggesting a role for VANGL1 in the signaling involved in cell invasiveness. These findings are consistent with previous reports that VANGL1 activation is associated with invasiveness of multiple different tumor types18,19,20,21,22,23,37,38, and that increased invasiveness requires both migratory ability and the ability to elaborate or induce matrix-degrading proteases (such as MMP3)39. In 21NT cells, this VANGL1-dependent increase in MMP3 expression and cellular invasiveness appeared to be independent of AP-1 induction. Interestingly, only 21NT cells showed an increase in VANGL1 expression in response to WNT5A, perhaps reflecting a different state of responsiveness than in 21PT cells. One may speculate that the two cell types are primed differently, such that cell migration induced in 21PT cells by WNT5A is more associated with morphologic organization (they tend to form more extracellular lumina and more compact, spherical colonies than 21NT), whereas in 21NT, there is an associated activation of VANGL1 and MMP3 expression, increased cellular invasiveness and decreased propensity to form organized structure. The 21T series cells have previously been shown to express very low levels of estrogen receptor protein and to overexpress HER2/neu, a phenotype that is shared by both 21PT and 21NT cells7,43, such that the hormone receptor and HER2/neu status alone could not account for the differential responsiveness of these two cell lines. The cellular context thus determines the ultimate nature of the response to a WNT/PCP pathway signal, although the specifics of the different cellular contexts have yet to be determined.
The association of increased COX-2 expression with induction of cell migration by WNT5A in both 21PT and 21NT is also of interest. In 21PT, this was associated with a significant increase in AP-1 activity, whereas AP-1 activity, already higher in 21NT cells, showed no change in response to WNT5A. Increased COX-2 expression has been previously associated with increased propensity of DCIS (or ADH) to progress to invasion, particularly when seen in association with increased proliferation (Ki-67 index), and in the case of DCIS, with high p1640,41,42. This has been suggested to be a reflection of an altered stress response in cells more likely to progress. In many instances, it thus appears that increased COX-2 expression may be a common, but insufficient alteration associated with increased propensity for progression to invasive malignancy. In the instance of the 21T series cells, this does appear to be the case, in that 21PT cells, which show increased COX-2 in response to WNT5A do not progress to invasion (in vitro or in vivo in extravasation assay), whereas 21NT cells do. The cellular background of 21NT cells allows for the additional activation of VANGL1 and MMP3 (and likely other invasiveness-associated gene) expression by WNT5A, which is associated with further progression to an actively invasive phenotype.
Although we have not in this study examined the specific receptor mechanism(s) involved in transducing the WNT5A signal, there is recent evidence in the literature to implicate possible roles of Ror2 and Ryk in tumor cell migration and invasion43,44,45. For example, WNT5A and Ror2 are induced during progression to an invasive phenotype in human osteosarcoma and epidermoid carcinoma cells, with corresponding changes in MMP2 and MMP1343. Moreover, migration is mediated by Ror2 overexpression and WNT5A treatment in murine B16 melanoma cells44. Ryk has also been observed to be important during WNT5A-induced invasion in human glioma cells through regulation of MMP245. Interestingly, knockdown of Ryk but not Ror2 reduced WNT5A-induced invasion and MMP2 activity of the glioma cells45. Hence, the involvement of either Ryk and/or Ror2 in WNT5A signaling may be dependent on the tumor cell type and stage of progression. Further studies examining the role of WNT5A's receptors and co-receptors in cell lines such as the 21T series are required for a complete understanding of WNT5A-induced progression of breast cancer cells.
This work thus provides evidence that the WNT/PCP pathway can promote early breast cancer progression, both by a RHOA-dependent increase in cell migration, associated with increased COX-2 expression, and a further VANGL1-dependent increase in invasiveness, associated with increased MMP3 expression, the latter VANGL1-dependent effect only seen in susceptible cells of a later stage of progression (21NT). The nature of these differences in cellular context of a PCP pathway signal in breast cancer precursor cells will be important to delineate, as it may reveal key, targetable factors involved in control of progression to invasion. Understanding the cellular context in which Wnt/PCP pathway signaling leads to invasiveness is also of potential importance in identifying precursor (ADH, DCIS) lesions which are more likely to progress and thus in need of more aggressive follow-up and management.
Methods
Cell lines and culture conditions
The 21T parental cell lines (21PT, 21NT, and 21MT-1), all derived from the same breast cancer patient, were a kind gift from Dr. Vimla Band (Dana Farber Cancer Institute, Boston, MA)46 and were cultured in alpha modification of Eagle's medium (α-MEM) supplemented with 2 mM L-glutamine, insulin (1 mg/mL), epidermal growth factor (12.5 ng/mL), hydrocortisone (2.8 mM), 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 1 mM sodium pyruvate, 0.1 mM non-essential amino acids, and 50 mg/mL gentamycin reagent (now called αHE; all supplements from Wisent Bioproducts, Montreal, QC) as previously described46. For regular culture conditions, the αHE media was further supplemented with 10% fetal bovine serum (FBS; Sigma) and named αHE10F. The 21T stable clones were cultured in the above supplemented growth media with the addition of 500 μg/mL G418 (Wisent Bioproducts) or 0.8 μg/mL puromycin (Sigma-Aldrich, St. Louis, MO) as selection markers.
Generation of WNT5A expression vector
An expression vector containing WNT5A was constructed in order to overexpress Wnt5a protein in 21PT and 21NT cells. To construct the expression vector, WNT5A cDNA was PCR-amplified using a pCMV-Sport expression vector containing WNT5A (Genbank accession number: NM_003392), provided by the Centre for Applied Genomics at The Hospital for Sick Children (University of Toronto, Toronto, ON) as the cDNA template. PCR amplification was performed using Phusion High-Fidelity DNA Taq Polymerase (New England BioLabs, Pickering, ON) at the following conditions: 98°C for 30 seconds, followed by 40 cycles of 98°C for 7 seconds, 62°C for 15 seconds and 72°C for 45 seconds and finally a 72°C hold for 8 minutes. The primers for PCR amplification of WNT5A were: forward: 5′- GCC ACC ATG AAG AAG TCC ATT GGA ATA TTA A-3′ and reverse: 5′- GGA GAT CGT GGA CCA GTT TGT GTG CAA G-3′. Five separate PCR reactions were completed and run on a 1% agarose gel. Bands at the appropriate size (~1500 kb) were gel extracted, using an EZ-10 Spin Column DNA Gel Extraction Kit (Bio Basic, Markham, ON) and pooled. Next, the WNT5A PCR product was incubated with T4 polynucleotide kinase (T4 PNK; New England BioLabs) to phosphorylate the insert. Finally, the WNT5A PCR product was purified using a QIAquick PCR purification kit (Qiagen, Mississauga, ON), following the manufacturer's protocol and the concentration was determined by spectrophotometry, using a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Mississauga, ON).
WNT5A was inserted into a pcDNA3.1/myc-His B plasmid (Invitrogen Life Technologies, Mississauga, ON), such that the myc-His tag would be fused to the C-terminus of WNT5A. The plasmid was digested with EcoRV (New England BioLabs) so that the WNT5A sequence would be inserted downstream of the pCMV promoter into the multiple cloning site. The plasmid was subsequently incubated with calf intestinal phosphatase (CIP; New England BioLabs) to dephosphorylate the plasmid. The plasmid was then electrophoresed on a 1% agarose gel and extracted using an EZ-10 Spin Column DNA Gel Extraction Kit (Bio Basic).
The prepared plasmid and WNT5A PCR product were incubated overnight with ATP (adenosine triphosphate) and T4 DNA ligase (both from New England BioLabs) at 16°C to complete the ligation. Competent bacteria (DH5alpha, made in-house) were then transformed with the ligated plasmid and streaked on Luria Bertani (LB) agar (Invitrogen Life Technologies) plates with 50 μg/mL ampicillin (Gibco Life Technologies, Grand Island, NY). Twenty bacteria colonies were picked and expanded in LB + ampicillin media, and DNA was isolated using an EZ-10 Spin Column Plasmid DNA Miniprep Kit (Bio Basic). The DNA was digested with Xhol and BsrGl (both from New England BioLabs) and run on a 1% agarose gel to see which ligation resulted in correct insertion. Four of the 20 clones were sent to the DNA Sequencing Facility at Robarts Research Institute (London, ON) for sequencing. Clones with the proper sequence were used for stable transfections.
Stable transfections
Stably transfected cell lines were generated to overexpress WNT5A in the 21PT and 21NT cells using PolyJet DNA In Vitro Transfection Reagent (SignaGen Laboratories, Rockville, MD). Plasmids lacking the gene of interest were also transfected into each cell line as an empty vector control. The day before transfection, 21PT or 21NT cells were seeded in 10cm dishes at a density that would result in 70–80% confluency the next day. Cells were transfected with 5 μg of the appropriate expression vector following the manufacturer's protocol. PolyJet/DNA complex-containing medium was replaced 18 hours post-transfection with αHE10F. Forty-eight hours post-transfection, the media was replaced with αHE10F containing 500 μg/mL G418. Approximately two weeks post-transfection, G418 resistant clones were pooled, expanded, and frozen down in vials for later use.
Generation of lentivirus particles and transduction
To generate shRNA containing lentivirus particles against RHOA, VANGL1, and Luciferase, HEK293T cells were plated in 2 mL of high glucose Dulbecco's Modified Eagle Media (DMEM) + 10% FBS in 6 well plates and incubated for 24 hours prior to transfection. Co-transfection of a three plasmid system was carried out using hairpin-pLKO.1 vector (1 ug), envelope plasmid (VSVG/pMD2.G; 100 ng) and packaging plasmid (pCMV-R8.74psPAX2; 900 ng) as previously described47. SuperFect Transfection Reagent (Qiagen) was used according to the manufacturer's protocol. Cells were incubated for 18 hours post transfection and the media containing the transfection reagent was replaced with 2 mL high glucose DMEM + 10% FBS. Approximately 48 hours post-transfection, the viral supernatant (2 mL) was collected, passed through a 0.2 μm filter, and stored at 4°C. Media (2 mL) was replenished, and viral harvesting was repeated a second time 24 hours later, pooled with previous harvest, aliquoted, and stored at -80°C until further use. The shRNA target sequence for RHOA was GAA AGC AGG TAG AGT TGG CTT; the target sequence for VANGL1 was CTC GTA GTC AAT GTG AAG AAA; and the shRNA target sequence for Luciferase was ACG CTG AGT ACT TCG AAA TGT. Viral supernatant for the scrambled shRNA control was purchased from Santa Cruz Biotechnology. HEK293T cells, pLKO.1-shRNA clones (RHOA, VANGL1 and Luciferase), pMD2.G, pCMV-R8.74psPAX2 plasmids, viral supernatant with viral particles containing pLVX-ZsGreen1 were all gifts from Dr. Jason Moffat (University of Toronto).
To knockdown expression of RHOA and VANGL1 and to express ZsGreen for extravasation assay, cells were seeded into 6-well plates at a density of 5 × 104 cells per well into 2 mL of αHE10F. 500 μL of viral supernatant was added to each well (for a total of 2.5 mL of media per well) for the shRNAs against RHOA, VANGL1 and Luciferase (which was used as an off-target knockdown control) and for the ZsGreen1 expression vector. 20 uL of the viral supernatant was added to each well for the scramble control shRNA, as per the manufacture's protocol. Seventy-two hours post viral transduction, cells were expanded in 10 cm dishes with 0.8 ug/mL puromycin. Approximately one week post-transduction, puromycin-resistant clones were pooled, expanded, and frozen down in vials for later use.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from 3 biological replicates of each cell line grown in 10 cm dishes using TRIZOL (Invitrogen Life Technologies) following the manufacturer's protocol. RNA concentration was determined using a NanoDrop spectrophotometer (Thermo Scientific) and 5 ug of RNA for each sample was DNase I (Invitrogen Life Technologies) treated prior to reverse transcription to cDNA using SuperScript III Reverse Transcriptase with random hexamers (both from Invitrogen Life Technologies). qRT-PCR was performed on cDNA samples and water-primer set only controls using RT2 SYBR-Green qPCR Master Mix (SABiosciences, Frederick, MD). Reactions were carried out in a 96-well plate and were spun at 2500 RPM for 30 seconds prior to placing the plate in the Thermo Cycler. Samples amplified with WNT5A, VANGL1, RHOA, COX-2, or MMP3 specific primers were reported relative to GAPDH as fold expression normalized to control cell lines. Primers used for RHOA were: forward: 5′- GGG ACA CAG CTG GGC AGG AGG A -3′ and reverse: 5′- GGT TTC ACC GGC TCC TGC TTC A -3′. Primers used for GAPDH were: forward: 5′- AGG CTG GGG CTC ATT TGC AG-3′ and reverse: 5′- CCA TCC ACA GTC TTC TGG GTG-3′. All custom primers used for qRT-PCR were purchased from Sigma-Aldrich. Primers for WNT5A, VANGL1, COX-2, MMP3, and MMP1 were purchased from SABiosciences.
Preparation of 21T cell lysates
The 21T cells were grown on 10 cm dishes. Subconfluent monolayers were washed once with ice cold PBS. RIPA buffer (10 nM Tris-HCl pH 7.5, 1 mM EDTA, 0.5 mM EGTA, 150 nM NaCl, 1% Triton-X 100, 0.5% DOC, and 1% SDS) containing one protease inhibitor tablet per 10 mL (cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail, Roche; Mannheim, Germany) was applied directly to the plate to lyse the cells (400 uL per 10 cm dish). The cells were removed with a cell scraper, collected in a clean microcentrifuge tube, and placed on a rotator for 20 minutes at 4°C. Tubes were spun at 13000 RPM for 10 minutes at 4°C and the resulting supernatant was collected and transferred to a clean microcentrifuge tube. The concentrations of the protein samples were determined using a BioRad Bradford Assay (Mississauga, ON) by comparing to BSA standards.
Electrophoresis and western blotting
Protein was separated via SDS-PAGE on 8% polyacrylamide gels. Separated proteins were transferred onto PVDF membrane and probed for proteins of interest by western blot. Membranes were blocked with 5% skim-milk in TBST (Tris-buffered saline with 0.1% Tween-20) overnight at 4°C. After washing membranes once with TBST, they were incubated with required primary antibodies for 1.5 hours at room temperature. Rat anti-Wnt5a antibody was purchased from R&D Systems (Burlington, ON, catalogue no. MAB645 – diluted to 1:100 in 5% skim milk) and the mouse anti-myc antibody was purchased from Abcam (Toronto, ON, catalogue no. ab32 – diluted to 1:1000 in 5% skim milk). Membranes were washed with TBST 3 times for 5 minutes each and incubated with horseradish peroxidase (HRP)-conjugated goat anti-rat secondary antibody (R&D Systems catalogue no. HAF005 1:1000 in 5% skim milk TBST) or HRP-conjugated goat anti-mouse secondary antibody (Amersham GE Healthcare, Piscataway NJ, catalogue no. NXA931 – diluted 1:10000 in 5% skim milk TBST) for 1 hour at room temperature. Membranes were washed 5 times for 3 minutes each. Detection was performed using the ECL Plus Western Blotting Detection Reagents (Amersham GE Healthcare) and then exposed to film in a dark room. Densitometric quantification was performed using ImageJ (Open source software, National Institutes of Health, USA).
Three-dimensional Matrigel culture and immunofluorescence
Subconfluent monolayers were trypsinized, washed, resuspended in αHE media containing 2% growth factor reduced Matrigel (BD Biosciences, Mississauga, ON) and 2% FBS, and plated as single cell suspensions on 100% Matrigel in 8-well chambers slides as previously described31,48. αHE media containing 2% Matrigel and 2% FBS was changed every 3 days. Following the indicated time points, the cell colonies were fixed with 10% (w/v) formalin in phosphate buffered saline (PBS) for 20 minutes at room temperature. Cells were then permeabilized with 0.5% Triton X-100 in PBS for 20 minutes at 4°C and blocked with 10% normal goat serum (Invitrogen Life Technologies) for 45 minutes at room temperature. Cell colonies were probed using a purified mouse anti-E-cadherin primary antibody (BD Biosciences, catalogue no. 61081) for 1.5 hours in 10% normal goat serum at room temperature. Following primary antibody incubation, the cell colonies were washed 3 times with PBS for 5 minutes per wash. Cell colonies were then probed with Alexa Fluor 488 Goat anti-mouse secondary antibody (Invitrogen Life Technologies) and Alexa Fluor 546 conjugated phalloidin (Invitrogen Life Technologies) to stain for F-actin in 10% normal goat serum for 1 hour at room temperature. Following secondary antibody and phalloidin incubation, the cell colonies were washed 5 times with PBS for 3 minutes each. Finally, cell colonies were incubated for 15 minutes at room temperature with 3 μg/mL Hoechst 33342, trihydrochloride, trihydrate (Invitrogen Life Technologies) to stain for the nuclei. Chamber slide gaskets were removed prior to mounting with Vectashield mounting medium (Vector Laboratories, Burlington, ON). Images were captured with an Olympus Confocal Imaging System (FluoView FV1000 coupled to the IX81 Motorized Inverted System Microscope) at the Victoria Research Laboratory Confocal Microscope Core Facility (London, ON) and processed using Volocity software (PerkinElmer, Woodbridge, ON).
Transwell migration assay
The migration of the 21PT and 21NT cells overexpressing WNT5A was tested using a transwell chamber system (Corning Life Sciences; Lowell, MA). The day before the assay, 6.5 mm transwell inserts (8.0 μm pores) were each coated with 6 μg of gelatin (diluted in 60 μL of sterile Milli-Q water). Plates were allowed to dry overnight at room temperature. On the day of the assay, the gelatin coated transwell inserts were reconstituted by adding 0.1 mL of αHE media supplemented with 0.1% BSA to the upper chamber for 90 minutes at room temperature. Subconfluent cells were washed twice with PBS, and resuspended in serum free media containing 0.1% BSA at a concentration of 5 × 105 cells/mL. Media containing chemoattractant (0.8 mL of αHE10F) was added to the bottom chambers and 0.1 mL of the cell suspension (5.0 × 104 cells per well) was immediately added to the upper chambers in triplicate. The transwell chambers were incubated at 37°C and 5% CO2 for 5 hours. After the assay was complete, the inner chamber inserts were removed and fixed/stained/washed using the Diff-Quick staining system (modified Wright Giemsa stain, Sigma-Aldrich) according to manufacturer's protocol. The cells that did not migrate to the underside of the membrane were removed using a cotton swab. The insert wells were allowed to dry overnight and images of 5 non-overlapping fields of view were acquired using Image-Pro Analysis Software (Media Cybernetics, Acton, MA) coupled to an inverted microscope. Images were captured at 10× objective. Cells were counted from these pictures using ImageJ (Open source software).
Transwell invasion assay
The invasion of the 21PT and 21NT cells overexpressing WNT5A was tested using a transwell chamber system precoated with Matrigel (reconstituted basement membrane proteins - BD Biosciences). Two hours prior to plating, the Matrigel precoated transwells (6.5 mm inserts and 8.0 μm pores) were reconstituted by adding 0.5 mL of αHE media supplemented with 0.1% BSA to the upper and lower chambers at 37°C and 5% CO2. Cells were washed twice with PBS, and resuspended in serum free media containing 0.1% BSA at a concentration of 1 × 105 cells/mL. The media used for reconstitution of the Matrigel in the lower and upper chambers was removed. Media containing chemoattractant (0.75 mL of αHE10F) was added to the bottom chambers and 0.5 mL of the cell suspension (5 × 104 cells) was immediately added to the upper chambers in triplicate. The transwell chambers were incubated at 37°C and 5% CO2 for 22 hours. After the assay was complete, the inner chamber inserts were removed and fixed/stained/washed using the Diff-Quick staining system (Sigma-Aldrich) according to manufacturer's protocol. The cells that did not invade to the underside of the membrane were removed using a cotton swab. The inserts were allowed to dry overnight and images of 5 non-overlapping fields of view were acquired using Image-Pro Analysis Software (Media Cybernetics) coupled to an inverted microscope. Images were captured at 10× objective. Cells were counted from these pictures using ImageJ (Open source software).
Cancer cell extravasation assay
The chorioallantoic membrane (CAM) of the chicken embryo is a well-established model for quantifying extravasation rates by IV-injected tumor cells33,34,35,36. Immediately after IV-injection, ZsGreen positive 21T cells arrest within the intravascular portion of the CAM as identified by lectin-rhodamine injected IV. Using fluorescence confocal microscopy, the number of cells that extravasate into the underlying stromal tissue of the CAM in a specific field of view over 24 hours may be quantified. Extravasated cells are those that appear to be fully present within the stroma and do not have any cell processes left within the CAM lumen. Cells that are imaged midway are considered to be intravascular. Using day 12 embryos, 50,000 ZsGreen positive cells (50 μL of 1 × 106 cells/mL) were IV-injected and after 10 minutes, lectin-rhodamine (75 μL of 0.1 mg/mL, Vector Laboratories Inc.) was intravenously injected to label the luminal surface of the endothelial cells in the CAM. Cells were washed once with PBS prior to IV injection. Immediately after IV injection of lectin-rhodamine, a 1 cm × 1 cm field of the CAM marked by foil markers at the corners was made for each animal and the number of cells that were intravascular, in the process of extravasation and extravasated into the CAM stroma were counted at T = 0 hours and T = 24 hours. A resonance scanner confocal microscope equipped with three lasers and various filter sets were used to determine each cell's location within the CAM in respect to lectin-rhodamine signal in real time.
AP-1 luciferase assay
To measure AP-1 activity using the 21T transfectants, commercially available AP-1 dual reporter constructs were used (Qiagen). Cells were plated into 96-well plates at a density of 1 × 104 cells per well and transfected with dual reporter construct, negative control construct (no AP-1 inducible promoter), and positive control (constitutively active promoter) using Attractene (Qiagen). Twenty-four hours post transfection media was changed with complete αHE10F media. Forty-eight hours post-transfection, Dual Luciferase Assay was performed (Promega, Madison, MI), using the manufacturer's protocol.
Statistical analysis
Statistical analyses were done using GraphPad Prism 5.0 software (La Jolla, CA). Colony morphology experiments, migration and invasion assays, and mRNA and protein levels were analyzed using ANOVA followed by Tukey's post hoc test (for comparison between more than two groups) or Student's t-test (for comparison between two groups). P-values less than 0.05 were considered statistically significant.
Author Contributions
C.D.M., H.S.L., A.F.C., J.D.L. and A.B.T. conceived and designed the experiments. C.D.M., H.S.L., D.W.D. and A.E.R. performed the experiments. C.D.M., H.S.L., A.F.C. and A.B.T. interpreted results. C.D.M., H.S.L., A.F.C. and A.B.T. wrote the manuscript. All authors have read and have approved the final manuscript.
Supplementary Material
Supplementary Figures
Acknowledgments
This work was supported by a Catalyst/Small Grants award from the London Regional Cancer Program (to ABT and AFC). CDM was a recipient of an NSERC Alexander Graham Bell Canada Graduate Scholarship, an Ontario Graduate Scholarship and a Translational Breast Cancer Traineeship sponsored in part by the Breast Cancer Society of Canada. HSL was a recipient of a Canadian Breast Cancer Foundation Post-Doctoral Fellowship. AFC is a Canada Research Chair in Oncology supported by the Canada Research Chairs Program.
References
- Page D. L., Dupont W. D., Rogers L. W. & Rados M. S. Atypical hyperplastic lesions of the female breast. A long-term follow-up study. Cancer 55, 2698–2708, 10.1097/00006534-198604000-00059 (1985). [DOI] [PubMed] [Google Scholar]
- Page D. L. & Dupont W. D. Anatomic indicators (histologic and cytologic) of increased breast cancer risk. Breast Cancer Res Treat 28, 157–166, 10.1007/BF00666428 (1993). [DOI] [PubMed] [Google Scholar]
- Lakhani S. R., Collins N., Stratton M. R. & Sloane J. P. Atypical ductal hyperplasia of the breast: clonal proliferation with loss of heterozygosity on chromosomes 16q and 17p. J Clin Pathol 48, 611–615, 10.1136/jcp.48.7.611 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allred D. C., Mohsin S. K. & Fuqua S. A. Histological and biological evolution of human premalignant breast disease. Endocr Relat Cancer 8, 47–61, 10.1677/erc.0.0080047 (2001). [DOI] [PubMed] [Google Scholar]
- Allred D. C. et al. Ductal carcinoma in situ and the emergence of diversity during breast cancer evolution. Clin Cancer Res 14, 370–378, 10.1158/1078-0432.CCR-07-1127 (2008). [DOI] [PubMed] [Google Scholar]
- Arpino G., Laucirica R. & Elledge R. M. Premalignant and in situ breast disease: biology and clinical implications. Ann Intern Med 143, 446–457 (2005). [DOI] [PubMed] [Google Scholar]
- Souter L. H. et al. Human 21T breast epithelial cell lines mimic breast cancer progression in vivo and in vitro and show stage-specific gene expression patterns. Lab Invest 90, 1247–1258, 10.1038/labinvest.2010.97 (2010). [DOI] [PubMed] [Google Scholar]
- Pukrop T. et al. Wnt 5a signaling is critical for macrophage-induced invasion of breast cancer cell lines. Proc Natl Acad Sci U S A 103, 5454–5459, 10.1073/pnas.0509703103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeraratna A. T. et al. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell 1, 279–288, 10.1016/S1535-6108(02)00045-4 (2002). [DOI] [PubMed] [Google Scholar]
- Kurayoshi M. et al. Expression of Wnt-5a is correlated with aggressiveness of gastric cancer by stimulating cell migration and invasion. Cancer Res 66, 10439–10448, 10.1158/0008-5472.CAN-06-2359 (2006). [DOI] [PubMed] [Google Scholar]
- Medina A., Reintsch W. & Steinbeisser H. Xenopus frizzled 7 can act in canonical and non-canonical Wnt signaling pathways: implications on early patterning and morphogenesis. Mech Dev 92, 227–237 (2000). [DOI] [PubMed] [Google Scholar]
- Kinoshita N., Iioka H., Miyakoshi A. & Ueno N. PKC delta is essential for Dishevelled function in a noncanonical Wnt pathway that regulates Xenopus convergent extension movements. Genes Dev 17, 1663–1676, 10.1101/gad.1101303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanegashima K., Zhao H. & Dawid I. B. WGEF activates Rho in the Wnt-PCP pathway and controls convergent extension in Xenopus gastrulation. EMBO J 27, 606–617, 10.1038/emboj.2008.9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincan E., Swain R. K., Brabletz T. & Steinbeisser H. Frizzled7 dictates embryonic morphogenesis: implications for colorectal cancer progression. Front Biosci 12, 4558–4567 (2007). [DOI] [PubMed] [Google Scholar]
- Ueno K. et al. Frizzled-7 as a potential therapeutic target in colorectal cancer. Neoplasia 10, 697–705 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merle P. et al. Functional consequences of frizzled-7 receptor overexpression in human hepatocellular carcinoma. Gastroenterology 127, 1110–1122 (2004). [DOI] [PubMed] [Google Scholar]
- Veeman M. T., Axelrod J. D. & Moon R. T. A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev Cell 5, 367–377, 10.1016/S1534-5807(03)00266-1 (2003). [DOI] [PubMed] [Google Scholar]
- Yagyu R. et al. Isolation and characterization of a novel human gene, VANGL1, as a therapeutic target for hepatocellular carcinoma. Int J Oncol 20, 1173–1178 (2002). [PubMed] [Google Scholar]
- Cho S. B. et al. KITENIN is associated with activation of AP-1 target genes via MAPK cascades signaling in human hepatocellular carcinoma progression. Oncology research 19, 115–123, 10.3727/096504011X12935427587722 (2011). [DOI] [PubMed] [Google Scholar]
- Lee J. H. et al. KAI1 COOH-terminal interacting tetraspanin (KITENIN), a member of the tetraspanin family, interacts with KAI1, a tumor metastasis suppressor, and enhances metastasis of cancer. Cancer Res 64, 4235–4243, 10.1158/0008-5472.CAN-04-0275 (2004). [DOI] [PubMed] [Google Scholar]
- Lee J. K. et al. KITENIN increases invasion and migration of mouse squamous cancer cells and promotes pulmonary metastasis in a mouse squamous tumor model. FEBS Lett 583, 711–717, 10.1016/j.febslet.2009.01.014 (2009). [DOI] [PubMed] [Google Scholar]
- Lee J. H. et al. Suppression of progression and metastasis of established colon tumors in mice by intravenous delivery of short interfering RNA targeting KITENIN, a metastasis-enhancing protein. Cancer Res 65, 8993–9003, 10.1158/0008-5472.CAN-05-0590 (2005). [DOI] [PubMed] [Google Scholar]
- Kho D. H. et al. KITENIN recruits Dishevelled/PKC delta to form a functional complex and controls the migration and invasiveness of colorectal cancer cells. Gut 58, 509–519, 10.1136/gut.2008.150938 (2009). [DOI] [PubMed] [Google Scholar]
- Olson D. J. & Gibo D. M. Antisense wnt-5a mimics wnt-1-mediated C57MG mammary epithelial cell transformation. Exp Cell Res 241, 134–141, 10.1006/excr.1998.4030 (1998). [DOI] [PubMed] [Google Scholar]
- Liang H. et al. Wnt5a inhibits B cell proliferation and functions as a tumor suppressor in hematopoietic tissue. Cancer Cell 4, 349–360, 10.1016/S1535-6108(03)00268-X (2003). [DOI] [PubMed] [Google Scholar]
- Kremenevskaja N. et al. Wnt-5a has tumor suppressor activity in thyroid carcinoma. Oncogene 24, 2144–2154, 10.1038/sj.onc.1208370 (2005). [DOI] [PubMed] [Google Scholar]
- Leris A. C., Roberts T. R., Jiang W. G., Newbold R. F. & Mokbel K. WNT5A expression in human breast cancer. Anticancer research 25, 731–734 (2005). [PubMed] [Google Scholar]
- Ying J. et al. WNT5A is epigenetically silenced in hematologic malignancies and inhibits leukemia cell growth as a tumor suppressor. Blood 110, 4130–4132, 10.1182/blood-2007-06-094870 (2007). [DOI] [PubMed] [Google Scholar]
- Ying J. et al. WNT5A exhibits tumor-suppressive activity through antagonizing the Wnt/beta-catenin signaling, and is frequently methylated in colorectal cancer. Clin Cancer Res 14, 55–61, 10.1158/1078-0432.CCR-07-1644 (2008). [DOI] [PubMed] [Google Scholar]
- Liu X. H. et al. Expression of Wnt-5a and its clinicopathological significance in hepatocellular carcinoma. Dig Liver Dis 40, 560–567, 10.1016/j.dld.2007.12.011 (2008). [DOI] [PubMed] [Google Scholar]
- Lee G. Y., Kenny P. A., Lee E. H. & Bissell M. J. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods 4, 359–365, 10.1038/nmeth1015 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebner C., Weaver V. M. & Debnath J. Modeling morphogenesis and oncogenesis in three-dimensional breast epithelial cultures. Annu Rev Pathol 3, 313–339, 10.1146/annurev.pathmechdis.3.121806.151526 (2008). [DOI] [PubMed] [Google Scholar]
- Arpaia E. et al. The interaction between caveolin-1 and Rho-GTPases promotes metastasis by controlling the expression of alpha5-integrin and the activation of Src, Ras and Erk. Oncogene 31, 884–896, 10.1038/onc.2011.288 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkovic D. et al. KISS1R induces invasiveness of estrogen receptor-negative human mammary epithelial and breast cancer cells. Endocrinology 154, 1999–2014, 10.1210/en.2012-2164 (2013). [DOI] [PubMed] [Google Scholar]
- Leong H. S., Chambers A. F. & Lewis J. D. Assessing cancer cell migration and metastatic growth in vivo in the chick embryo using fluorescence intravital imaging. Methods Mol Biol 872, 1–14, 10.1007/978-1-61779-797-2_1 (2012). [DOI] [PubMed] [Google Scholar]
- Leong H. S. et al. Intravital imaging of embryonic and tumor neovasculature using viral nanoparticles. Nature protocols 5, 1406–1417, 10.1038/nprot.2010.103 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. et al. Expression of KITENIN in human colorectal cancer and its relation to tumor behavior and progression. Pathology international 61, 210–220, 10.1111/j.1440-1827.2011.02646.x (2011). [DOI] [PubMed] [Google Scholar]
- Ryu H. S. et al. KITENIN is associated with tumor progression in human gastric cancer. Anticancer research 30, 3479–3486 (2010). [PubMed] [Google Scholar]
- Chang C. & Werb Z. The many faces of metalloproteases: cell growth, invasion, angiogenesis and metastasis. Trends in cell biology 11, S37–43, 10.1016/S0962-8924(01)02122-5 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier M. L. et al. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell 12, 479–491, 10.1016/j.ccr.2007.10.017 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerlikowske K. et al. Biomarker expression and risk of subsequent tumors after initial ductal carcinoma in situ diagnosis. J Natl Cancer Inst 102, 627–637, 10.1093/jnci/djq101 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisky D. C. et al. p16(INK4a) expression and breast cancer risk in women with atypical hyperplasia. Cancer Prev Res (Phila) 4, 1953–1960, 10.1158/1940-6207.CAPR-11-0282 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Minami Y. & Nishita M. Critical role of Wnt5a-Ror2 signaling in motility and invasiveness of carcinoma cells following Snail-mediated epithelial-mesenchymal transition. Genes Cells 16, 304–315, 10.1111/j.1365-2443.2011.01487.x (2011). [DOI] [PubMed] [Google Scholar]
- Lai S. S. et al. Ror2-Src signaling in metastasis of mouse melanoma cells is inhibited by NRAGE. Cancer Genet 205, 552–562, 10.1016/j.cancergen.2012.09.002 (2012). [DOI] [PubMed] [Google Scholar]
- Habu M. et al. Ryk is essential for Wnt-5a-dependent invasiveness in human glioma. J Biochem 156, 29–38, 10.1093/jb/mvu015 (2014). [DOI] [PubMed] [Google Scholar]
- Band V. et al. Tumor progression in four mammary epithelial cell lines derived from the same patient. Cancer Res 50, 7351–7357 (1990). [PubMed] [Google Scholar]
- Moffat J. et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298, 10.1016/j.cell.2006.01.040 (2006). [DOI] [PubMed] [Google Scholar]
- Debnath J., Muthuswamy S. K. & Brugge J. S. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268, 10.1016/S1046-2023(03)00032-X (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures