Abstract
Mass spectrometric imaging (MSI) is a powerful tool that grants the ability to investigate a broad mass range of molecules, from small molecules to large proteins, by creating detailed distribution maps of selected compounds. To date, MSI has demonstrated its versatility in the study of neurotransmitters and neuropeptides of different classes toward investigation of neurobiological functions and diseases. These studies have provided significant insight in neurobiology over the years and current technical advances are facilitating further improvements in this field. neurotransmitters, focusing specifically on the challenges and recent Herein, we advances of MSI of neurotransmitters.
Background
Mass spectrometric imaging (MSI) shows great promise for biological analyses because it allows for molecular analysis of tissue while retaining information about the spatial distribution of different analytes, including proteins, peptides, lipids and small molecules [1]. During an MSI experiment, a multitude of mass spectra are collected from a tissue slice in a predefined raster, resulting in a 2D distribution map for each mass measured. One of the advantages of MSI is that it allows for the analysis of thousands of analytes at once, without the need of labels or prior knowledge of the analytes, and provides spatial information along with the mass analysis. MSI is often used in tandem with other techniques to obtain more molecular information, while using MSI to visualize the results [2–7]. Many excellent reviews have already been published on the subject of MSI [8–17]. Herein, we review current publications that highlight the critical role that MSI plays in the study of neurotransmitters and neuropeptides, focusing on the challenges and recent advances in the field.
Ionization techniques: advantages & disadvantages
There are three main ionization methods used for MSI: MALDI [18,19], secondary ion MS (SIMS) [20–22] and DESI [23,24].
MALDI-MSI has proven to be a valuable technology with numerous applications for analyzing proteins [25,26], lipids [27], neuropeptides [28,29] and small molecules [2] at both organ and cellular levels. One advantage of using MALDI for MSI on biological samples is that it allows for the generation of larger ions, such as peptides and proteins, which is one of the reasons why MALDI is the most widely used method for MSI [1,30–32]. Sample preparation is extremely important for this ionization technique. Immediately after the sacrifice of an animal specimen, rapid molecular degradation occurs. To limit the degradation of analytes, the tissue can be embedded in supporting media such as gelatin [28,29] or sucrose [33], and snap-frozen in dry ice or liquid nitrogen. Polymer-containing material, such as optimal cutting temperature compound, Tissue-Tek® and carboxymethylcellulose, should be avoided as they often introduce polymer interferences into the mass spectrometer [34]. The embedded and frozen tissue should be stored at −80°C until use [35]. Prior to long-term storage, the tissue sample can be stabilized using various methods, such as microwave irradiation [36] or heat denaturation by Denator Stabilizor® T1 (Gothenburg, Sweden), to deactivate proteolytic enzymes, preventing postmortem degradation of proteins or peptides of interest [37–39].
Next, the frozen tissue can be cut into thin sections with a cryostat. MSI experiments typically require 10–20 μm thick tissue sections [40]. The previous step of embedding tissue in supporting media allows for precise sectioning of tissue samples. Once sliced, tissue sections are then transferred and mounted onto a target plate or glass slide [41,42] by thaw-mounting [34]. When working with higher mass analytes, such as peptides and proteins, washing tissue sections with organic solvents is a recommended step to fix tissues, and remove ion-suppressing salts and lipids, thus increasing the analyte signal [43–45].
The next step of the MALDI-MSI sample-preparation process involves coating the tissue with a thin layer of matrix and irradiating the sample with a laser beam. The matrix is designed to absorb much of the incident laser, providing ‘soft’ ionization for analyte compounds, which allows for the ionization of larger molecules (m/z over 100 kDa) [46]. Matrix selection and application is a critical step for MALDI-MSI, greatly impacting the sensitivity, spatial resolution and selective analyte ionization of the experiment [43]. Conventional matrices include α-cyano-4-hydroxycinnamic acid (CHCA) and 2,5-dihydroxy benzoic acid (DHB) [28,29,34]. One disadvantage of using these conventional matrices is that they produce ions themselves, which can interfere or mask analyte ions when looking at small molecules. High-resolution instrumentation may negate some of the interference; however, less conventional matrices, such as ionic matrices, made by mixing conventional matrices with organic bases [47–49], TiO2 nanoparticles [50], 1,5-diaminonapthalene [51], 2,3,4,5-Tetrakis(3′,4′-dihydroxylphenyl)thiophene [52], 1,8-bis(dimethyl-amino) naphthalene [53,54] and ferulic acid [55] are being used more frequently, and are reported to improve spectral quality, crystallization and vacuum stability. The matrix application technique plays a crucial role in the quality of mass-spectral images, especially when obtaining high spatial resolution images [10,51,56].
Spatial resolution and reproducibility of results are limited by the matrix crystal size and application consistency, among other instrumental parameters such as raster step size and laser beam diameter [10]. Matrix application methods can be divided into two categories: solvent-based and solvent-free. Solvent-based methods apply matrix dissolved in solution and are typically performed by manual airbrush or automated systems [56]. For solvent-free methods, dry matrix can be sublimated onto tissue under low pressure [57], filtered onto tissue through a small (~20 μm) sieve [58] or ground through fine mesh (~10–1 μm) above a tissue slice using a ball-mill [59]. These solvent-free methods were developed to limit spatial delocalization of soluble analytes that often occurs from excessive amounts of solvent during matrix application from solvent-based methods [60]. In comparison with the solvent-based matrix application methods, solvent-free methods yield very fine crystals amenable to high spatial-resolution MSI, but may also suffer from relatively low sensitivities due to the limited analyte–matrix interactions [51]. Several researchers have modified these dry-matrix application techniques by rehydrating the sections following dry-coating to improve the detection sensitivity of analytes [25,61].
SIMS is a long-established technique in which a sample is put under high vacuum and bombarded with high energy primary ions, which facilitates the ionization of analytes [62,63]. The ionized analytes are referred to as secondary ions and are sputtered from the sample surface and then drawn into the mass analyzer for analysis. The highly focused ion beam in SIMS provides excellent spatial resolution; however, the primary ion source is fairly limited to small molecules, under 1000 Da [2], due to its high energy that is prone to fragment large molecules in the ablation/ionization process. Recent advances in SIMS, such as using beams of gold trimers (Au3+) [64] or buckminsterfullerenes (C60) [65], can provide significantly greater yields of higher mass ions. SIMS imaging does not require special preparation after sectioning and mounting the tissue, but there are some optional methods that improve the imaging results. The tissue can be coated with a thin layer of gold, silver or matrix (e.g., DHB, α-CHCA or sinapinic acid) to improve the ionization and reduce the fragmentation of larger molecules [66–68].
In contrast to traditional MALDI and SIMS, which require a vacuum (although atmospheric pressure-MALDI has been developed), DESI is a simple, ambient ionization technique [69–72] that channels charged solvent droplets and ions from an electrospray source onto the sample surface [73]. The surface is impacted by the charged particles, yielding gaseous analyte ions. MSI had been mainly viewed as an invasive process until the development of ambient ionization techniques, such as DESI. DESI’s ambient nature and softness allow for the examination of various natural surfaces with no need for matrices [74]. An advantage of DESI, is its capability for high-throughput analysis with minimal tissue adulteration, again due to its ambient nature [13]. Since a vacuum is not required for DESI, it has a greater advantage of being used for clinical or field application, as the removal of the vacuum makes it more portable [75]. The ability to perform in situ analysis and the convenience of portable mass spectrometers suggests the potential role of DESI-MSI and other ambient ionization techniques lie in histology guided diagnoses and therapies in the clinical setting [75]. A disadvantage of DESI when compared with vacuum MS methods, SIMS and MALDI, is that it has lower spatial resolution of approximately 180–200 μm [74]. Recently, the nano-DESI probe was developed and reported an increase in the spatial resolution of approximately 12 μm. With the nano-DESI probe, controlled desorption of analytes present in a restricted region of specimen is achieved by using a minute amount of solvent between two capillaries [71]. Table 1 summarizes the optimal analytes, mass range, and spatial resolutions of MALDI, SIMS and DESI.
Table 1.
Comparison of MS imaging approaches detailing ionization methods, mass analyzers, optimal analytes, mass range and spatial resolution.
| Type of MSI | Ionization source | Common mass analyzer | Optimal analytes | Mass range (Da) | Spatial resolution (μm) |
|---|---|---|---|---|---|
| MALDI | UV/IR laser Soft ionization |
TOF | Lipids, peptides, proteins, small molecules | 0–50,000 | 30–50 |
| SIMS | Ion gun Hard ionization |
TOF Magnetic sector Orbitrap |
Lipids, small peptides, small molecules | 0–2000 | 0.5–1 |
| DESI | Solvent spray Soft ionization |
Orbitrap | Lipids, peptides, small molecules | 0–2000 | 100 |
MSI: MS imaging; SIMS: Secondary ion MS.
Reprinted with permission from [16] © Elsevier (2013).
A fourth ionization technique worth mentioning is nanostructure-initiator MS (NIMS) [76]. NIMS is a matrix-free ionization technique that shows promise for the analysis of small molecules. NIMS uses a liquid initiator to facilitate desorption instead of a crystalline MALDI-matrix. NIMS initiators are different from MALDI matrices in that the initiators do not absorb UV energy and most do not ionize, thus, do not produce interfering background ions. The advantages of the NIMS initiator are that the NIMS surface is stable in ambient air, has improved reproducibility, enables direct biofluid analysis and tissue imaging, and allows for a significantly expanded mass range [77]. In NIMS desorption/ionization, the analyte is adsorbed on top of the NIMS surface and laser irradiation results in rapid surface heating, causing the NIMS initiator to vaporize, triggering the desorption of adsorbed materials [76]. Several other MSI techniques have been developed more recently, including IR matrix-assisted laser DESI [78–80], liquid extraction surface analysis [81] and laser ablation ESI [82–86], but are beyond the scope of this review.
Neurotransmitters
Neurotransmitters are endogenous substances released by synaptic terminals upon depolarization, which transmit signals between nerve cells [87,88]. More than 100 substances have been identified as neurotransmitters since 1914, when the first neurotransmitter, acetylcholine, was discovered [89]. In the nervous system, neurotransmitters are the most common chemical messengers [90]. Neurotransmitters can be divided into six groups: acetylcholine, purines, amino acids, biogenic amines, peptides and unconventional neurotransmitters. Except for peptides, all other neurotransmitters are considered to be small-molecule neurotransmitters due to their lower molecular weights (<1000 Da). Most small-molecule neurotransmitters modulate activities requiring fast responses. Therefore, they are rapidly inactivated by highly specific transporter systems or enzymes in order to prevent continuous activation of the postsynaptic nerve cells [90]. Peptides and some small neurotransmitters tend to modulate slower and ongoing actions, and these neurotransmitters have slower turnover rates [87]. Due to differences in modes of action, neuropeptides are sometimes considered to be neuromodulators or neurohormones. Neuropeptides with neurotransmitter functions are defined as putative neurotransmitters or co-transmitters [90].
Neurotransmitter MSI
Recently, many studies have successfully mapped the distributions of neurotransmitters using MSI by developing new sample preparation techniques, matrix application methods, ionization methods and mass analyzers, which are summarized in Table 2. MSI studies of different classes of neurotransmitters are reviewed in the following sections.
Table 2.
Summary of neurotransmitter groups, sample preparation, suitable matrices, MSI acquisition methods and related studies.
| Neurotransmitter groups | Examples | Sample preparation | Matrices | MSI methods | Related studies |
|---|---|---|---|---|---|
| Small molecule neurotransmitters | |||||
| Acetylcholine | Acetylcholine | In situ freezing | Deuterated matrix (D4-CHCA) | MALDI tandem mass MSI; high-resolution and mass-accuracy MSI | [92–94] |
| Purines | ATP, AMP, adenosine | In situ freezing | 9-AA | MALDI, high-resolution and mass-accuracy MSI | [7,93,95,96] |
| Amino acids | Glutamate, γ-aminobutyric acid, glycine | NA | DHB | MALDI, high-resolution and mass-accuracy MSI | [93,103–105] |
| Biogenic amines | Dopamine, epinephrine, serotonin | DPP derivatization | CHCA | DESI MALDI | [105,109] |
| Peptide neurotransmitters | |||||
| Neuropeptides | Substance P, somatostatin | NA | CHCA, DHB and so on | MALDI, SIMS, NIMS | [26,114,128,129,132] |
9-AA: 9-aminoacridine; CHCA: α-cyano-4-hydroxycinnamic acid; D4-CHCA: Deuterated α-cyano-4-hydroxycinnamic acid; DHB: 2,5-dihydroxy benzoic acid; DPP: 2,4-diphenyl pyrylium; MSI: MS imaging; NIMS: Nanostructure-initiator MS; SIMS: Secondary ion MS.
MSI of small-molecule neurotransmitters
MSI of small-molecule neurotransmitters is much more challenging than MSI of other bio-molecules due to the following factors: conventional matrix compounds generate interference in the mass range of small molecules; small molecule neurotransmitters have rapid turnover rates, which introduces difficulties for MSI sample preparations; and small-molecule neurotransmitters have low in vivo concentrations, which requires a sensitive detection method. Despite the difficulties, several MSI studies have analyzed the distribution of small molecule neurotransmitters.
MSI of acetylcholine
Acetylcholine acts as an excitatory transmitter to modulate motor systems in the CNS and PNS, except in the heart, where it acts as an inhibitory transmitter [91]. Both DHB and CHCA, two common MALDI matrices used in small molecule profiling and imaging, produce strong interference for acetylcholine. Therefore, innovative MALDI matrices, high-mass accuracy and high-resolution mass analyzers, or alternative data acquisition methods are needed to solve the matrix interference problem. Additionally, due to the rapid turnover rate of acetylcholine, a well-developed sample preparation procedure is needed to minimize postmortem degradation. Several recent studies have successfully detected endogenous acetylcholine in brain tissues [92–94].
Shariatgorji et al. developed a deuterated matrix, D4-α-CHCA (D4-CHCA), to solve the signal interference problem associated with small molecule MSI [94]. D4-CHCA has mass shifts of +4, +8 and +12 Da. As shown in Figure 1, by switching between D4-CHCA and CHCA, matrix peaks can be differentiated from acetylcholine as well as other small molecules. The distribution of acetylcholine was also confirmed by MS/MS with a characteristic fragmentation peak at m/z 87.0 [94]. Sugiura et al. developed a MSI method combining both tandem mass imaging and in situ freezing techniques to visualize acetylcholine distribution in mouse brain [92]. In this study, acetylcholine transition signatures m/z 146→87 and 146→92 were monitored to avoid matrix interference at the parent ion mass. The observed acetylcholine signal distribution at m/z 87 overlapped with the acetylcholinesterase in situ hybridization, proving the validity of this method. Furthermore, using in situ freezing techniques, which freeze anesthetized mouse brain in liquid nitrogen during dissection, postmortem changes were minimized, and dynamic range and sensitivity were improved [92].
Figure 1. D4-α-cyano-4-hydroxycinnamic acid has been synthesized for use as a matrix for MALDI-MS and MALDI-MS imaging of small-molecule drugs and endogenous compounds.
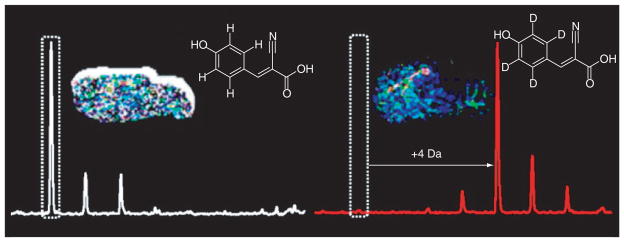
MALDI-MS analysis of small molecules has historically been hindered by interference from matrix ion clusters and fragment peaks that mask signals of low molecular weight compounds of interest. By using D4-α-cyano-4-hydroxycinnamic acid, the cluster and fragment peaks of α-cyano-4-hydroxycinnamic acid, the most common matrix for analysis of small molecules, are shifted by +4, +8 and +12 Da, which expose signals across areas of the previously concealed low mass range. Here, obscured MALDI-MS signals of a synthetic small molecule pharmaceutical, a naturally occurring isoquinoline alkaloid, and endogenous compounds including the neurotransmitter acetylcholine have been unmasked and imaged directly from biological tissue sections. Reprinted with permission from [94] © American Chemical Society (2012).
The problem of matrix peaks interfering with the acetylcholine signal in MSI can also be solved by using a high-resolution and high-accuracy mass analyzer. Ye et al. successfully mapped acetylcholine in rat brain tissue with standard DHB matrix with an LTQ Orbitrap™ mass analyzer (Thermo Fisher Scientific, MA, USA) [93]. The high resolving power of the LTQ Orbitrap made it possible to separate the acetylcholine peaks from the matrix peaks. Ye et al. have also mapped several other amino acid and biogenic amine neurotransmitters, such as histidine and AMP in rat and crab brain tissue using this method [93]. Figure 2 summarizes the metabolites and neurotransmitters detected in rat brain tissue using MSI [93]. The successful detection of acetylcholine by MSI is very useful for biologists and neuroscientists to rapidly monitor the distribution and relative quantitation changes during various physiological states.
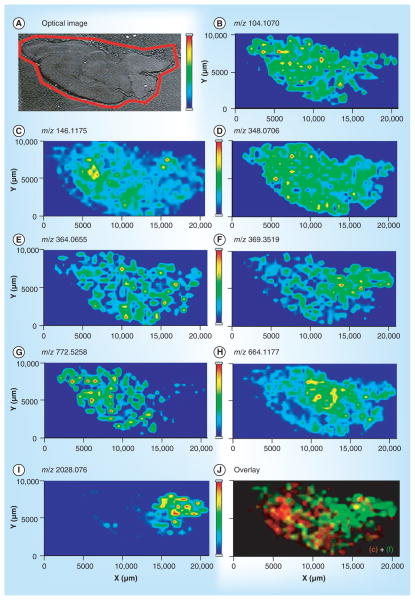
Figure 2. MS images of metabolites and neurotransmitters identified from rat CNS.
(A) Optical image of a rat brain section subjected to MS imaging sample preparation. MS images of metabolites and neurotransmitters, including: (B) choline at m/z 104.1070; (C) acetylcholine at m/z 146.1175; (D) AMP at m/z 348.0706; (E) GMP at m/z 364.0655; (F) cholesterol with neutral loss at m/z 369.3519; (G) potassiated phosphatidylcholine (32:0) at m/z 772.5258; and (H) nicotinamide adenine dinucleotide at m/z 644.1177. Other than low MW molecules, (I) MS image of the acetylated peptide ASQKRPSQRHGSKYLATA at m/z 2028.076 was also shown. (J) Overlaid image of a neurotransmitter, acetylcholine, as in (C) and cholesterol-derived ion as in (F). Adapted with permission from [93] © American Chemical Society (2013).
MSI of purines
Purine neurotransmitters, including ATP, AMP, adenosine and other purines are small signaling molecules that contain purine rings. Most purines have similar molecular structures, similar molecular weights of 300–1000 Da and similar in situ concentrations on the micromolar order of magnitude [95]. ATP was first thought to be a co-transmitter with an energy supply, but was later found to have signal transmitting capability. It acts on spinal cord motor neurons and sensory and autonomic ganglia as an excitatory neurotransmitter. Adenosine, which is derived from ATP by enzymatic activities, is also widespread in the CNS as an excitatory neurotransmitter [87].
Similar to acetylcholine, MSI of purine neurotransmitters also faces the challenges of rapid turnover rates, low concentrations and matrix interference. Nevertheless, several MSI studies have successfully mapped the distributions of purines, mainly ATP, ADP and AMP, in brain tissues [7,93,95,96].
Benabdellah et al. first reported the detection and identification of 13 primary metabolites, including ADP, AMP, GTP and UDP, in rat brain sections using MALDI-TOF/TOF-MSI [95]. Nine common MALDI matrices were tested on tissue surfaces in either positive or negative ion mode. Only 9-aminoacridine did not generate interfering matrix peaks on the target mass range (m/z 300→1000) under negative ion mode. A robotic sprayer was also used for matrix application in order to generate a homogenous and thin layer of matrix, thus improving reproducibility. However, the distribution of ATP was not detected in the tissue due to low abundance of ATP [95]. Hattori et al. developed a quantitative MSI technique for purines by combining in situ freezing, MALDI-MS and quantitative CE-ESI-MS on ischemic penumbra mouse brain samples [96]. In this study MSI results of samples prepared by in situ freezing and conventional postmortem freezing were compared. Both samples were coated with 9-aminoacridine and analyzed in negative ion mode. ATP could only be detected on in situ frozen samples as postmortem freezing reduced ATP autolysis. Insitu metabolite concentrations were quantified in this study by parallel CE-ESI-MS experiments [96]. Sugiura et al. also utilized a similar quantitative MSI technique by combining MALDI-MSI with quantitative CE-ESI-MS for purines for monitoring spatiotemporal energy dynamics of hippocampal neurons in a kainite-induced seizure mouse model during the seizure process [7]. They reported the ATP depletion and recovery process, along with the changes in other metabolites in the tricarboxylic acid cycle during the seizure, using quantitative MSI. The successful detection and quantitation of purine neurotransmitters, especially ATP, not only allows researchers to monitor the changes of neurotransmitters, but also allows the observation of the energy consumption at different histological regions under different conditions.
MSI of amino acid neurotransmitters
Amino acid neurotransmitters are individual amino acids that act as signaling molecules between neurons, including glutamate, γ-aminobutyric acid (GABA), glycine, D-serine and D-aspartate. Glutamate is the most common neurotransmitter in the CNS with an excitatory postsynaptic effect [97]. GABA is a common inhibitory neurotransmitter in the nervous system [98], although a recent study reported the existence of GABA excitatory synapses in the cerebellar interneuron network [99]. Glycine is also a common, but more localized, inhibitory neurotransmitter in the CNS, especially in the spinal cord. These amino acid neurotransmitters can be removed from the synaptic cleft via reuptake under the actions of various transporters [87]. For instance, GABA can be removed by the highly specific GABA transporters [90].
MSI of amino acid neurotransmitters also faces similar challenges to other small-molecule neurotransmitters. Even though several studies have analyzed amino acid neurotransmitters in various nervous systems using GC [100], LC [101] or microfluidic devices [102] coupled with ESI-type mass spectrometers, few MSI studies have successfully visualized and quantified the distribution of amino acid neurotransmitters [93,103–105].
Goto-Inoue et al. detected GABA (m/z 104.07) in eggplant by MALDI-MSI [103]. DHB was chosen as matrix and produced no interference at the target mass. No specific sample-handling procedures or detection methods were used due to the higher concentration of GABA in eggplant when compared with the nervous system. Toue et al. visualized several amino acids in human colon cancer xenograft tissues using quantitative MSI, combining MALDI-MS with CE-ESI-MS [104]. Due to the low concentration of endogenous amino acids, p-N,N,N-trimethylammonioanilyl N′-hydroxysuccinimidyl carbamate iodide was applied on the surface of the tissue as a derivatization reagent to enhance ionization efficiency. Tandem MSI was used to solve the matrix interference problem. The rest of the tissues were homogenized for quantitative CE-ESI-MS analysis. Several amino acids, including glutamate and glycine, were detected on the tissue surface by MALDI-MS and their concentrations were reported by CE-ESI-MS. Shariatgorji et al. has successfully quantified GABA brain concentrations by MALDI-MSI using their in-house imaging software [105]. In this study, a standard curve was established by measuring GABA standard intensities with various concentrations on tissue surface; the GABA concentration on brain tissue could then be calculated from the standard curve. This method was also successfully applied to many other drugs and neurotransmitters, such as acetylcholine and ATP.
MSI of biogenic amine neurotransmitters
Biogenic amine neurotransmitters are small signaling molecules derived from amino acids [98]. Three catecholamine neurotransmitters: dopamine, epinephrine and norepinephrine, are derived from tyrosine; histamine is derived from histidine, and serotonin and 5-hydroxytryptamine are derived from tryptophan [87]. Dopamine plays an important role in modulation of arterial blood-flow, cognition, motor control and anxiety-related behavior [106]. Parkinson’s disease is caused by a lack of dopamine in the brain [107]. Norepinephrine regulates sleep, attention, and feeding in both the CNS and the PNS, while epinephrine is mostly restricted to the PNS [108]. Epinephrine and norepinephrine also act as hormones, regulating heart rate, respiratory rate, muscle contraction and other actions. Histamine regulates attention and arousal, and is also related to allergic reactions. Serotonin works on CNS neurons that control circadian rhythm, emotions and motor behaviors [87].
Similar to MSI of amino acid neurotransmitters, few studies have successfully performed MSI of biogenic amine neurotransmitters due to the previously mentioned challenges. Wu et al. detected epinephrine and norepinephrine in the adrenal gland of adult porcine by DESI-MSI, which is a matrix free method that avoids the matrix interference problem [109]. Recently, Shariatgorji et al. reported that dopamine was visualized and quantified by MALDI-MSI [105]. 2,4-diphenyl pyrylium was used to derivatize dopamine, which could then be detected by high sensitivity MALDI-MS using CHCA as the matrix. The derivatized dopamine concentration on tissue could be determined by the in-house imaging software mentioned above.
MSI of neuropeptides
Peptide neurotransmitters, or neuropeptides, such as substance P, endorphin and opioid peptides, are larger signaling molecules that are typically composed of three to 70 amino acids. They are derived from polypeptides, prepropeptides and propeptides [87]. Substance P, the first discovered neuropeptide [58], is an excitatory neurotransmitter in pain perception. In contrast, endorphin is an inhibitory neurotransmitter in pain reduction and euphoria production [98]. Other neuropeptides are reported to be involved in anxiety, autism and panic attacks [110–113]. Since the development of MSI for biological systems, MSI analysis of neuropeptides has been an intensive research topic and widely applied to various biological systems. MSI has been used to map the distribution of endogenous neuropeptides within the nervous systems of various organisms, including the medicinal leech [114], terrestrial and fresh water snails [115,116], the sea slug Aplysia californica [117,118], numerous species of crustaceans [29,119–121] and rodents [122]. These MSI studies provide valuable information on the neuropeptide distributions and relative quantitation within various nervous systems, thus providing a new approach for neuroscience research in monitoring neuropeptidomic changes under different physiological conditions. Various ionization techniques have been used in neuropeptide MSI, such as MALDI, SIMS [122] and NIMS [123]; several advanced techniques have been developed for MSI, such as 3D MSI [124,125] and quantitative MSI [126]. Additionally, MSI has been used for biomarker discovery studies in several disease models, including Parkinson’s disease [127], Alzheimer’s disease [128] and cortical spreading depression [125]. Numerous excellent reviews and book chapters have summarized the MSI workflow, techniques and applications to neuropeptides [47,129–132]. Ye et al. used a high-mass-resolution and high-mass-accuracy MALDI-Fourier transform ion cyclotron resonance instrument to map neuropeptide distribution in the crustacean stomatogastric nervous system at cellular resolution [121]. Hanrieder et al. used MSI to study L-DOPA-induced dyskinesia in rat brain [127]. They noted elevated levels of neuropeptides, including substance P and dynorphin B, in the dorsolateral striatum of high-dyskinesia rats compared with control rats.
Challenges of neurotransmitter MSI
Matrix interference
Choosing an appropriate matrix and its application method is one of the main challenges in MALDI-MSI of small molecules, such as neurotransmitters, and there have been extensive method developments in this area. When the sample is ablated conventional matrices produce matrix ions that potentially mask analyte ions in the low mass region (<500 Da). Some matrix application methods, such as manual airbrush, produce matrix crystals of varying sizes, which leads to nonreproducible results. The use of solvent-based matrix application methods can also cause analyte diffusion when working with small molecules. As mentioned previously, Shariatgorji et al. developed a different strategy for reducing matrix interference by using a deuterated matrix [94]. The research group employed the traditional matrix, CHCA, but also synthesized a deuterated version, D4-CHCA, giving a 4, 8, or 12 Da mass shift of interfering matrix peaks between the two matrices. When these two matrices are used in conjunction, the mass shift characteristic of matrix interference allows analyte peaks in the lower mass range to be un-masked. In this work, the neurotransmitter, acetylcholine, was detected when employing D4-CHCA, but was masked by matrix cluster peaks when employing traditional CHCA [94]; thus, researchers were able to eliminate matrix interferences while still keeping the advantages of CHCA ionization efficiency. Other strategies for eliminating matrix interference have been developed for small-molecule MSI and could be applied to MSI of neurotransmitters in the future. For targeted analysis, Porta et al. demonstrated the use of MALDI-SRM/MS [133]. Using this technique, the analyte was fragmented and several of the unique fragment ions were monitored; thus, the matrix ions were no longer being detected and therefore did not interfere with the analyte signal. Recently, Tang et al. used solvent-free argon ion sputtering to apply gold nanoparticles to mouse brain tissue and were able to directly detect neurotransmitters and other small molecules via MALDI-MSI [134]. Argon ion sputtering is a solvent-free matrix application method that has been reported to produce a thin, even layer of matrix coating onto the sample surface [134]. The chemical interference from the gold nanoparticles was very minimal, thus alleviating the common problem of matrix ions masking analyte ions in the lower molecular weight region. Using this technique, the researchers were able to differentiate five unique regions of mouse tumor tissue based on the distributions of characteristic ions known to be metabolic markers of tumors. Water ice has been used as a matrix in IR-MALDI-MS, but would be difficult to use on MS systems requiring high vacuum pressure, such as TOF instruments, which are most commonly used for MALDI-MSI. Pirkl et al. employed a cooling stage to generate ions at high vacuum conditions using water ice as a matrix [135]. Ice is an ideal matrix for water-rich biological tissue because it produces matrix-ion-free spectra while still facilitating analyte ionization. Thus far, ice has been demonstrated as a suitable matrix for the detection of peptides, such as substance P [135], from tissue sections with IR-MALDI-MS profiling. MSI has yet to be performed on tissue samples utilizing ice as the matrix. Another matrix-free imaging strategy was developed by Qian et al. [136]. In this research, a graphene paper was engineered using a pulse-laser process to form densely packed nanospheres. This graphene paper was used as a substrate for matrix-free laser desorption/ionization-MSI. This method is reported to enhance the detection of small molecules because the method used for engineering this material creates a highly homogenous surface, eliminating the variability that is often seen when matrix is applied to the sample, producing more reliable and reproducible results.
Reproducibility in sample preparation
Aside from matrix selection and application, there are several other challenges of MALDI-MSI sample preparation that are being improved upon for enhanced MSI of small molecules (neurotransmitters) and neuropeptides. Consistency and reproducibility of techniques during sample preparation can be a challenge. Chang et al. developed a technique to obtain more reproducible sample preparations for SIMS-MSI of small molecules [137]. In this method, powerful magnets were used to fracture cells of interest. Typically cells are fractured via cryomicrotoming and freeze–fracture techniques that damage the cells and complicate SIMS-MSI results. By using powerful magnets to fracture the cells, the surface environment can be more controlled, which could provide more reproducible and reliable sample preparations. A comparison between the standard freeze–fracture technique and the powerful magnet method showing the differences in sample surface uniformity is shown in Figure 3. Another challenge in MSI of neurotransmitters is the low signal intensity caused by low abundance of analytes and/or signal suppression from other endogenous compounds in the tissue. Typically sample washing causes delocalization of small molecules and is therefore not part of the MSI workflow for neurotransmitters and small molecules. Shariatgorji et al. demonstrated a washing protocol with a pH-controlled buffer system for the enhanced detection of small molecules [138]. This approach can only be used for targeted analyses because the protocol employs an aqueous, buffered wash solution with the pH adjusted to the point where the target analyte is insoluble. This washing step removes soluble, endogenous salts and other compounds that suppress signal, thus increasing the detection of small molecules. Goodwin et al. developed a method for stabilizing delicate tissue sections for MALDI-MSI [139]. Double-sided conductive carbon tape was used to collect, stabilize and mount delicate, heat-treated rat brain tissue. Neuropeptides are rapidly modified postmortem; heat stabilization, as mentioned previously, can halt the postmortem degradation process, but can also result in more fragile tissues. Sectioning thin or delicate tissues using a cryostat can be challenging and may often result in tearing or folding of the tissue. The carbon tape did not interfere with the detection of small molecules or neuropeptides and could be used to reduce the variability in sample quality when attempting to section delicate tissues. The researchers used this method to detect and identify many endogenous compounds and show their unique distributions in a variety of rat tissues.
Figure 3. TOF-secondary ion MS.
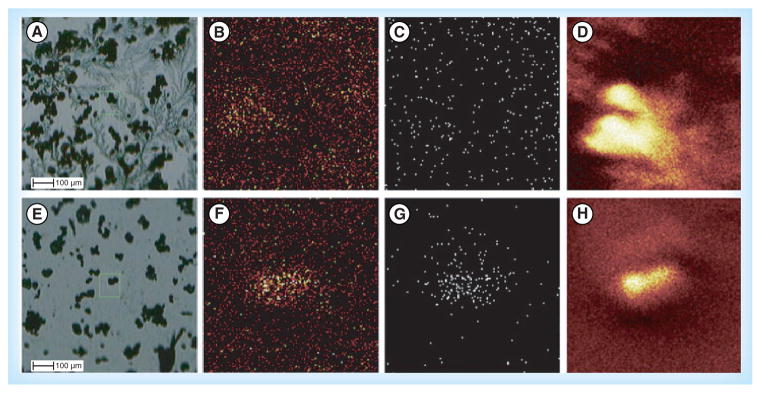
(A–D) Using the freeze–fracture method; (E–H) with the powerful magnet method. (A) and (E) are bright fields; (B) and (F) are Ca, m/z 40; (C) and (G) are Cd, m/z 48; (D) and (H) are total ions.
Reprinted with permission from [137] © John Wiley & Sons, Ltd (2012).
Quantitative MSI
The intensity of an MS signal is related to the analyte’s ionization efficiency as well as its concentration. With MALDI-MSI the smallest difference in matrix inhomogeneity can cause signal suppression and inaccurate quantitation. One technique that is being used for quantitative imaging is multi-isotope imaging MS (MIMS), which has been reviewed previously [140]. Steinhauser et al. demonstrate this technique for the quantification of stem cell division and metabolism on the sub-micrometer scale [141]. MIMS is based on SIMS technology. The instrument can simultaneously measure data from multiple isotopes in the same region of the tissue. MIMS uses isotopic tags to localize and measure their incorporation into intracellular compartments and quantitative data is extracted based on the isotope ratios [142]. The research team used MIMS to test the ‘immortal strand hypothesis’, which predicts that during stem cell division, chromosomes containing older template DNA are segregated to the daughter cell destined to remain a stem cell. When the isotopic tag is introduced, if the hypothesis held true, there would be one completely unlabeled daughter cell after the first cell division. The results showed no unlabeled daughter cells and, thus, do not support the ‘immortal strand hypothesis’. Reviews specifically focused on quantitative MSI have recently been published [13,15,140]. Another, more typical, approach to quantitative MSI employs an IS for quantification, which involves generating a standard curve by measuring standards spotted on the tissue surface and calculating the concentration of the analyte in the tissue based on this standard curve [105]. Pirman et al. quantified cocaine from brain tissue using MALDI-MSI by employing deuterated cocaine as the IS [143]. In order to limit the background interference during the experiment, MS/MS MSI was performed with a wide isolation window to incorporate the analyte and the deuterated IS. A calibration curve was generated using control tissue and used to determine concentrations of the analyte. The quantitative MSI data were compared with quantitative LC–MS/MS and the results were comparable. Furthermore, several semi-quantitative studies have been successfully conducted [127,144–151]. Recently, new software tools have been developed to aid in quantitative MSI and will be discussed below.
Low mass resolution & spatial resolution of MSI instruments
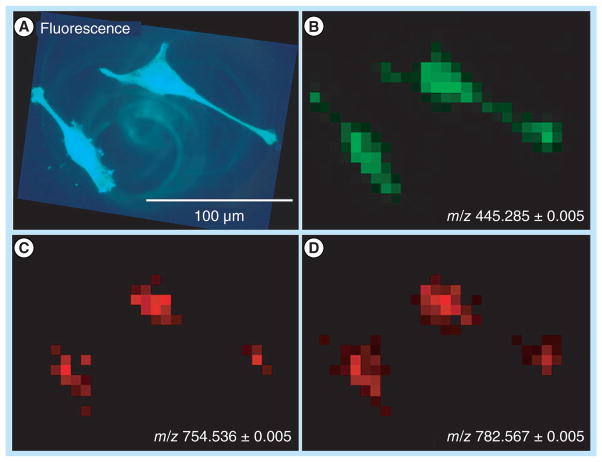
The main, on-going challenges for MSI instrumentation are obtaining faster acquisition speeds, and higher spatial and chemical resolution. Spraggins and Caprioli report high-speed MALDI-MSI using a prototype TOF mass spectrometer [152]. This work incorporates a neodymium-doped yttrium lithium fluoride solid state laser capable of repetition rates up to 5 kHz, which reportedly produce more than a tenfold increase in sample throughput when compared with commercially available instruments. This technique was applied to MSI of lipids, but should translate to MSI of other chemical species, including other small molecules or peptides. As mentioned previously, Ye et al. focused on increased mass resolution of MSI experiments [93]. HRMS and high-accuracy MS is able to distinguish analytes from interfering matrix ions. In this study, Ye et al. performed MALDI-MSI of neurotransmitters in rodent and crustacean nervous systems and were able to resolve peaks that were merged in lower resolution TOF/TOF instruments. Schober et al. tackled the problem of high-spatial resolution imaging, while still maintaining high-mass-accuracy and high-mass-resolution [153]. In this work, HeLa cells were imaged with optical fluorescence imaging and with a high-resolution atmospheric-pressure-MALDI source attached to an Exactive™ Orbitrap mass spectrometer. A spatial resolution of 7 μm was achieved in this work as shown in Figure 4.
Figure 4. HeLa cells on indium tin oxide slide.
(A) Optical fluorescence of DIOC6(3) stained HeLa cells, λ = 501 nm. (B–D) MALDI imaging (28 × 21 pixels, pixel size 7 μm). (B) Selected ion image of staining agent DIOC6(3) ([M+]). (C) Selected ion image of PC(32:1) ([M + Na]+). (D) Selected ion image of PC(34:1) ([M + Na]+).
Reprinted with permission from [153] © American Chemical Society (2012).
Data processing
MSI data analysis software, such as BioMap [201] and proprietary programs for MSI systems (e.g., FlexImaging™ from Bruker Daltonics [MA, USA], ImageQuest™ from Thermo Fisher Scientific and TissueView™ from Applied Biosystems/MDS SCIEX [CA, USA]), are primarily used to produce distribution maps for selected analytes. These software packages have different features but mostly allow the user to adjust color scales, overlay ion density maps and integrate MS images with acquired histological pictures. MSI data typically requires manual extraction of ion images, which can be extremely time-consuming for large-scale analyses, such as untargeted studies. Paschke et al. have developed a free software package, called Mirion™, for automatic processing of MS images [154]. With Mirion, the user selects an m/z range of interest and the software creates the ion images. Manual image generation is also available with Mirion, since no software algorithm is as good as the eye of the MS expert. The capabilities of this software were demonstrated using peptide standards and tissue from mice. Another open-source MSI data processing software has been developed, called MSiReader [155]. This software allows the user to select a region of interest and a reference region. The software can generate a mass list of compounds found in the region of interest, excluding peaks in the reference region. The created mass list or a manually made mass list, can then be imported and an ion imaged will be extracted for every ion in the list. Software such as Mirion and MSiReader significantly improve MSI data processing in comparison to manual processing by drastically decreasing the amount of time it takes to work through a data set. Recently, software tools have been developed for quantitation of analytes from MSI data, such as Quantinetix™ from ImaBiotech (Loos, France). Källback et al. have developed a novel MSI software for labeled quantitation and normalization of endogenous neuropeptides [126]. Shariatgorji et al. have developed in-house software for MSI quantitation based on signal intensities. This software can be used to define region of interest, process spectra, normalize spectra and quantify analytes based on pre-established calibration curves. Several neurotransmitters, drugs and neuropeptides have been successfully quantified using this software [105].
Conclusion
Over the past decade, MSI has obtained increasing attention from biologists and become more routinely employed to map various classes of bio-molecules from biological specimens. Its novel applications to the study of neurotransmitters and neuropeptides have provided valuable knowledge regarding disease mechanisms and related reparative processes. These studies offer significant insights into neurobiology and hold promise for our continuous search of effective treatment for diseases. We anticipate MSI for neurotransmitter mapping will lead to a better understanding of the biology of neurological diseases and improvements in clinical diagnostics.
Future perspective
Exciting technical advances are further improving the capabilities of MSI for biological applications from various aspects, such as sample preparation and instrumentation. The development and use of novel matrices and matrix-free imaging techniques, along with improvements in sample preparation are generating more reproducible, clean and reliable results with improved analyte detection. The advancements in instrumentation for shorter acquisition times are increasing the throughput of MSI techniques, making MSI more applicable to large-scale study of complex biological systems. The improved mass accuracy, mass resolution and spatial resolution are exciting developments that could greatly enhance the sensitivity of MSI and make this technique capable of studying biological problems on a cellular scale. Data processing and analysis is the current bottleneck of MSI, but progress is certainly being made in this area. Again, once software becomes commercially available and used enough to inspire confident, reliable results, MSI has the potential to be a powerful analytical tool, expanding into many new areas of research and industries.
Executive summary.
Small molecule neurotransmitter mass spectrometric imaging
Mass spectrometric imaging (MSI) of small molecules is challenging due to matrix interference, rapid turnover rate and low in situ concentration. Innovative MSI techniques, such as in situ freezing, novel matrices and matrix free techniques, high-resolution and high-mass-accuracy mass analyzer have been used to overcome these difficulties.
Neuropeptide MSI
MSI of neuropeptides is well developed and widely applied to many species and disease states. Advanced MSI techniques such as 3D imaging and quantitative imaging have been developed to further improve neuropeptide detection and visualization.
Novel matrices
Advancements in the use of novel matrices and matrix-free methods enhance signal detection and produce cleaner spectra in the low mass region, which will allow for improved metabolomics and small molecule studies.
Sample preparation
Challenges in sample preparation are being solved in novel ways, leading to more reproducible results that could even turn MSI into a quantitative technique.
Data processing
There have been several recent advances for improved mass resolution, spatial resolution, acquisition time and data processing. Making these advancements commercially available could produce significant progress in many different fields of study.
Key Terms
- Mass spectrometric imaging,
Analytical technique that allows for molecular analysis of tissue while retaining information about the spatial distribution of different analytes by collecting mass spectra in a predefined raster across a tissue sample, resulting in a 2D distribution map for each mass measured
- Neurotransmitters
Endogenous substances released by synaptic terminals to transmit signals between nerve cells
- Neuropeptides
Endogenous peptides synthesized in nerve cells or peripheral organs that are involved in cell–cell signaling. Often considered as peptide neurotransmitters and peptide hormones with sizes ranging from three to 70 amino acids
- Matrix
Typically a small organic compound that is used to coat MALDI samples and assist in the ionization of analytes
Footnotes
For reprint orders, please contact reprints@future-science.com
No writing assistance was utilized in the production of this manuscript.
Financial & competing interests disclosure
Preparation of this manuscript was supported in part by National Science Foundation (NSF CHE-0957784) and National Institutes of Health through grant 1R01DK071801. Gemperline acknowledges an NSF Graduate Research Fellowship (DGE-1256259). L Li acknowledges an H. I. Romnes Faculty Fellowship. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
- 1.Balluff B, Schone C, Hofler H, Walch A. MALDI imaging mass spectrometry for direct tissue analysis: technological advancements and recent applications. Histochem Cell Biol. 2011;136(3):227–244. doi: 10.1007/s00418-011-0843-x. [DOI] [PubMed] [Google Scholar]
- 2.Ye H, Gemperline E, Venkateshwaran M, et al. MALDI mass spectrometry-assisted molecular imaging of metabolites during nitrogen fixation in the Medicago truncatula– Sinorhizobium meliloti symbiosis. Plant J. 2013;75(1):130–145. doi: 10.1111/tpj.12191. [DOI] [PubMed] [Google Scholar]
- 3.Burnum KE, Cornett DS, Puolitaival SM, et al. Spatial and temporal alterations of phospholipids determined by mass spectrometry during mouse embryo implantation. J Lipid Res. 2009;50(11):2290–2298. doi: 10.1194/jlr.M900100-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yanagisawa K, Shyr Y, Xu BGJ, et al. Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet. 2003;362(9382):433–439. doi: 10.1016/S0140-6736(03)14068-8. [DOI] [PubMed] [Google Scholar]
- 5.Skold K, Svensson M, Nilsson A, et al. Decreased striatal levels of PEP-19 following MPTP lesion in the mouse. J Proteome Res. 2006;5(2):262–269. doi: 10.1021/pr050281f. [DOI] [PubMed] [Google Scholar]
- 6.Stauber J, Lemaire R, Franck J, et al. MALDI imaging of formalin-fixed paraffin-embedded tissues: application to model animals of Parkinson disease for biomarker hunting. J Proteome Res. 2008;7(3):969–978. doi: 10.1021/pr070464x. [DOI] [PubMed] [Google Scholar]
- 7.Sugiura Y, Taguchi R, Setou M. Visualization of spatiotemporal energy dynamics of hippocampal neurons by mass spectrometry during a kainate-induced seizure. PLoS ONE. 2011;6(3):e17952. doi: 10.1371/journal.pone.0017952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterji B, Pich A. MALDI imaging mass spectrometry and analysis of endogenous peptides. Expert Rev Proteomics. 2013;10(4):381–388. doi: 10.1586/14789450.2013.814939. [DOI] [PubMed] [Google Scholar]
- 9.Hanrieder J, Phan NTN, Kurczy ME, Ewing AG. Imaging mass spectrometry in neuroscience. ACS Chem Neurosci. 2013;4(5):666–679. doi: 10.1021/cn400053c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodwin RJA. Sample preparation for mass spectrometry imaging: small mistakes can lead to big consequences. J Proteomics. 2012;75(16):4893–4911. doi: 10.1016/j.jprot.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 11.Jones EA, Deininger SO, Hogendoorn PCW, Deelder AM, McDonnell LA. Imaging mass spectrometry statistical analysis. J Proteomics. 2012;75(16):4962–4989. doi: 10.1016/j.jprot.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 12.Gilmore IS. SIMS of organics: advances in 2D and 3D imaging and future outlook. J Vac Sci Technol A. 2013;31(5):050819. [Google Scholar]
- 13.Lietz CB, Gemperline E, Li L. Qualitative and quantitative mass spectrometry imaging of drugs and metabolites. Adv Drug Deliv Rev. 2013;65(8):1074–1085. doi: 10.1016/j.addr.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nimesh S, Mohottalage S, Vincent R, Kumarathasan P. Current status and future perspectives of mass spectrometry imaging. Int J Mol Sci. 2013;14(6):11277–11301. doi: 10.3390/ijms140611277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun N, Walch A. Qualitative and quantitative mass spectrometry imaging of drugs and metabolites in tissue at therapeutic levels. Histochem Cell Biol. 2013;140(2):93–104. doi: 10.1007/s00418-013-1127-4. [DOI] [PubMed] [Google Scholar]
- 16.Weaver EM, Hummon AB. Imaging mass spectrometry: from tissue sections to cell cultures. Adv Drug Deliv Rev. 2013;65(8):1039–1055. doi: 10.1016/j.addr.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Norris JL, Caprioli RM. Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem Rev. 2013;113(4):2309–2342. doi: 10.1021/cr3004295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10000 Daltons. Anal Chem. 1988;60(20):2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 19.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69(23):4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 20.Liebl H. Ion microprobe mass analyzer. J Appl Phys. 1967;38(13):5277–5283. [Google Scholar]
- 21.Brown A, Vickerman JC. Static SIMS, FABMS and SIMS imaging in applied surface-analysis. Analyst. 1984;109(7):851–857. [Google Scholar]
- 22.Cliff B, Lockyer NP, Corlett C, Vickerman JC. Development of instrumentation for routine ToF-SIMS imaging analysis of biological material. Appl Surf Sci. 2003;203:730–733. [Google Scholar]
- 23.Wiseman JM, Ifa DR, Song QY, Cooks RG. Tissue imaging at atmospheric pressure using desorption electrospray ionization (DESI) mass spectrometry. Angew Chem Int Ed Engl. 2006;45(43):7188–7192. doi: 10.1002/anie.200602449. [DOI] [PubMed] [Google Scholar]
- 24.Takats Z, Wiseman JM, Cooks RG. Ambient mass spectrometry using desorption electrospray ionization (DESI): instrumentation, mechanisms and applications in forensics, chemistry, and biology. J Mass Spectrom. 2005;40(10):1261–1275. doi: 10.1002/jms.922. [DOI] [PubMed] [Google Scholar]
- 25.Cazares LH, Troyer D, Mendrinos S, et al. Imaging mass spectrometry of a specific fragment of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase 2 discriminates cancer from uninvolved prostate tissue. Clin Cancer Res. 2009;15(17):5541–5551. doi: 10.1158/1078-0432.CCR-08-2892. [DOI] [PubMed] [Google Scholar]
- 26.Deininger SO, Ebert MP, Futterer A, Gerhard M, Rocken C. MALDI imaging combined with hierarchical clustering as a new tool for the interpretation of complex human cancers. J Proteome Res. 2008;7(12):5230–5236. doi: 10.1021/pr8005777. [DOI] [PubMed] [Google Scholar]
- 27.Murphy RC, Hankin JA, Barkley RM. Imaging of lipid species by MALDI mass spectrometry. J Lipid Res. 2009;50:S317–S322. doi: 10.1194/jlr.R800051-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen R, Hui L, Sturm RM, Li L. Three dimensional mapping of neuropeptides and lipids in crustacean brain by mass spectral imaging. J Am Soc Mass Spectrom. 2009;20(6):1068–1077. doi: 10.1016/j.jasms.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeKeyser SS, Kutz-Naber KK, Schmidt JJ, Barrett-Wilt GA, Li L. Mass spectral imaging of neuropeptides in decapod crustacean neronal tissues. J Proteome Res. 2007;6(5):1782–1791. doi: 10.1021/pr060603v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang WC, Huang LCL, Wang YS, et al. Matrix-assisted laser desorption/ionization (MALDI) mechanism revisited. Anal Chim Acta. 2007;582(1):1–9. doi: 10.1016/j.aca.2006.08.062. [DOI] [PubMed] [Google Scholar]
- 31.Karas M, Kruger R. Ion formation in MALDI: the cluster ionization mechanism. Chem Rev. 2003;103(2):427–439. doi: 10.1021/cr010376a. [DOI] [PubMed] [Google Scholar]
- 32.Knochenmuss R. A quantitative model of ultraviolet matrix-assisted laser desorption/ionization including analyte ion generation. Anal Chem. 2003;75(10):2199–2207. doi: 10.1021/ac034032r. [DOI] [PubMed] [Google Scholar]
- 33.Verhaert PDEM, Pinkse MWH, Strupat K, Conaway MCP. Imaging of similar mass neuropeptides in neuronal tissue by enhanced resolution MALDI MS with an ion trap-Orbitrap™ hybrid instrument. Methods Mol Biol. 2010;656:433–449. doi: 10.1007/978-1-60761-746-4_25. [DOI] [PubMed] [Google Scholar]
- 34.Schwartz SA, Reyzer ML, Caprioli RM. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J Mass Spectrom. 2003;38(7):699–708. doi: 10.1002/jms.505. [DOI] [PubMed] [Google Scholar]
- 35.Jehl B, Bauer R, Dorge A, Rick R. The use of propane-isopentane mixtures for rapid freezing of biological specimens. J Microsc. 1981;123(Pt 3):307–309. doi: 10.1111/j.1365-2818.1981.tb02475.x. [DOI] [PubMed] [Google Scholar]
- 36.Che FY, Lim J, Pan H, Biswas R, Fricker LD. Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Mol Cell Proteomics. 2005;4(9):1391–1405. doi: 10.1074/mcp.T500010-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Svensson M, Boren M, Skold K, et al. Heat stabilization of the tissue proteome: a new technology for improved proteomics. J Proteome Res. 2009;8(2):974–981. doi: 10.1021/pr8006446. [DOI] [PubMed] [Google Scholar]
- 38.Rountree CB, Van Kirk CA, You HN, et al. Clinical application for the preservation of phosphoproteins through in situ tissue stabilization. Proteome Sci. 2010;8(61) doi: 10.1186/1477-5956-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sturm RM, Greer T, Woodards N, Gemperline E, Li LJ. Mass spectrometric evaluation of neuropeptidomic profiles upon heat stabilization treatment of neuroendocrine tissues in crustaceans. J Proteome Res. 2013;12(2):743–752. doi: 10.1021/pr300805f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crossman L, McHugh NA, Hsieh YS, Korfmacher WA, Chen JW. Investigation of the profiling depth in matrix-assisted laser desorption/ionization imaging mass spectrometry. Rapid Commun Mass Spectrom. 2006;20(2):284–290. doi: 10.1002/rcm.2259. [DOI] [PubMed] [Google Scholar]
- 41.Chaurand P, Fouchecourt S, DaGue BB, et al. Profiling and imaging proteins in the mouse epididymis by imaging mass spectrometry. Proteomics. 2003;3(11):2221–2239. doi: 10.1002/pmic.200300474. [DOI] [PubMed] [Google Scholar]
- 42.Chaurand P, Schriver KE, Caprioli RM. Instrument design and characterization for high resolution MALDI-MS imaging of tissue sections. J Mass Spectrom. 2007;42(4):476–489. doi: 10.1002/jms.1180. [DOI] [PubMed] [Google Scholar]
- 43.Kaletas BK, van der Wiel IM, Stauber J, et al. Sample preparation issues for tissue imaging by imaging MS. Proteomics. 2009;9(10):2622–2633. doi: 10.1002/pmic.200800364. [DOI] [PubMed] [Google Scholar]
- 44.Lemaire R, Wisztorski M, Desmons A, et al. MALDI-MS direct tissue analysis of proteins: improving signal sensitivity using organic treatments. Anal Chem. 2006;78(20):7145–7153. doi: 10.1021/ac060565z. [DOI] [PubMed] [Google Scholar]
- 45.Seeley EH, Oppenheimer SR, Mi D, Chaurand P, Caprioli RM. Enhancement of protein sensitivity for MALDI imaging mass spectrometry after chemical treatment of tissue sections. J Am Soc Mass Spectrom. 2008;19(8):1069–1077. doi: 10.1016/j.jasms.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka K, Waki H, Ido Y, et al. Protein and polymer analyses up to m/z 100000 by laser ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 1988;2(8):151–153. [Google Scholar]
- 47.Ye H, Gemperline E, Li L. A vision for better health: mass spectrometry imaging for clinical diagnostics. Clin Chim Acta. 2013;420:11–22. doi: 10.1016/j.cca.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fitzgerald JJD, Kunnath P, Walker AV. Matrix-enhanced secondary ion mass spectrometry (ME-SIMS) using room temperature ionic liquid matrices. Anal Chem. 2010;82(11):4413–4419. doi: 10.1021/ac100133c. [DOI] [PubMed] [Google Scholar]
- 49.Lemaire R, Tabet JC, Ducoroy P, Hendra JB, Salzet M, Fournier I. Solid ionic matrixes for direct tissue analysis and MALDI imaging. Anal Chem. 2006;78(3):809–819. doi: 10.1021/ac0514669. [DOI] [PubMed] [Google Scholar]
- 50.Shrivas K, Hayasaka T, Sugiura Y, Setou M. Method for simultaneous imaging of endogenous low molecular weight metabolites in mouse brain using TiO2 nanoparticles in nanoparticle-assisted laser desorption/ionization-imaging mass spectrometry. Anal Chem. 2011;83(19):7283–7289. doi: 10.1021/ac201602s. [DOI] [PubMed] [Google Scholar]
- 51.Thomas A, Charbonneau JL, Fournaise E, Chaurand P. Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: enhanced information in both positive and negative polarities after 1,5-diaminonapthalene deposition. Anal Chem. 2012;84(4):2048–2054. doi: 10.1021/ac2033547. [DOI] [PubMed] [Google Scholar]
- 52.Chen S, Chen L, Wang J, et al. 2,3,4,5-Tetrakis(3′,4′-dihydroxylphenyl) thiophene: a new matrix for the selective analysis of low molecular weight amines and direct determination of creatinine in urine by MALDI-TOF MS. Anal Chem. 2012;84(23):10291–10297. doi: 10.1021/ac3021278. [DOI] [PubMed] [Google Scholar]
- 53.Shroff R, Rulisek L, Doubsky J, Svatos A. Acid-base-driven matrix-assisted mass spectrometry for targeted metabolomics. Proc Natl Acad Sci USA. 2009;106(25):10092–10096. doi: 10.1073/pnas.0900914106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shroff R, Svatos A. Proton sponge: a novel and versatile MALDI matrix for the analysis of metabolites using mass spectrometry. Anal Chem. 2009;81(19):7954–7959. doi: 10.1021/ac901048z. [DOI] [PubMed] [Google Scholar]
- 55.Mainini V, Bovo G, Chinello C, et al. Detection of high molecular weight proteins by MALDI imaging mass spectrometry. Mol Biosyst. 2013;9(6):1101–1107. doi: 10.1039/c2mb25296a. [DOI] [PubMed] [Google Scholar]
- 56.Baluya DL, Garrett TJ, Yost RA. Automated MALDI matrix deposition method with inkjet printing for imaging mass spectrometry. Anal Chem. 2007;79(17):6862–6867. doi: 10.1021/ac070958d. [DOI] [PubMed] [Google Scholar]
- 57.Hankin JA, Barkley RM, Murphy RC. Sublimation as a method of matrix application for mass spectrometric imaging. J Am Soc Mass Spectrom. 2007;18(9):1646–1652. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Puolitaival SM, Burnum KE, Cornett DS, Caprioli RM. Solvent-free matrix dry-coating for MALDI imaging of phospholipids. J Am Soc Mass Spectrom. 2008;19(6):882–886. doi: 10.1016/j.jasms.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trimpin S, Herath TN, Inutan ED, et al. Automated solvent-free matrix deposition for tissue imaging by mass spectrometry. Anal Chem. 2010;82(1):359–367. doi: 10.1021/ac902065u. [DOI] [PubMed] [Google Scholar]
- 60.Troendle FJ, Reddick CD, Yost RA. Detection of pharmaceutical compounds in tissue by matrix-assisted laser desorption/ionization and laser desorption/chemical ionization tandem mass spectrometry with a quadrupole ion trap. J Am Soc Mass Spectrom. 1999;10(12):1315–1321. [Google Scholar]
- 61.Deutskens F, Yang JH, Caprioli RM. High spatial resolution imaging mass spectrometry and classical histology on a single tissue section. J Mass Spectrom. 2011;46(6):568–571. doi: 10.1002/jms.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fletcher JS, Lockyer NP, Vickerman JC. Developments in molecular SIMS depth profiling and 3D imaging of biological systems using polyatomic primary ions. Mass Spectrom Rev. 2011;30(1):142–174. doi: 10.1002/mas.20275. [DOI] [PubMed] [Google Scholar]
- 63.Liebl H. SIMS instrumentation and imaging techniques. Scanning. 1980;3(2):79–89. [Google Scholar]
- 64.Touboul D, Halgand F, Brunelle A, et al. Tissue molecular ion imaging by gold cluster ion bombardment. Anal Chem. 2004;76(6):1550–1559. doi: 10.1021/ac035243z. [DOI] [PubMed] [Google Scholar]
- 65.Jones EA, Lockyer NP, Vickerman JC. Mass spectral analysis and imaging of tissue by ToF-SIMS - the role of buckminsterfullerene, C-60(+), primary ions. Int J Mass Spectrom. 2007;260(2–3):146–157. [Google Scholar]
- 66.Keune K, Boon JJ. Enhancement of the static SIMS secondary ion yields of lipid moieties by ultrathin gold coating of aged oil paint surfaces. Surf Interface Anal. 2004;36(13):1620–1628. [Google Scholar]
- 67.Nygren H, Malmberg P, Kriegeskotte C, Arlinghaus HF. Bioimaging TOF-SIMS: localization of cholesterol in rat kidney sections. FEBS Lett. 2004;566(1–3):291–293. doi: 10.1016/j.febslet.2004.04.052. [DOI] [PubMed] [Google Scholar]
- 68.Adriaensen L, Vangaever F, Lenaerts J, Gijbels R. Matrix-enhanced secondary ion mass spectrometry: the influence of MALDI matrices on molecular ion yields of thin organic films. Rapid Commun Mass Spectrom. 2005;19(8):1017–1024. [Google Scholar]
- 69.Dill AL, Eberlin LS, Costa AB, et al. Multivariate statistical identification of human bladder carcinomas using ambient ionization imaging mass spectrometry. Chemistry. 2011;17(10):2897–2902. doi: 10.1002/chem.201001692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao YH, Yang CH, Liu X, Ma RJ, Kong DL, Shi LQ. A multifunctional nanocarrier based on nanogated mesoporous silica for enhanced tumor-specific uptake and intracellular delivery. Macromol Biosci. 2012;12(2):251–259. doi: 10.1002/mabi.201100208. [DOI] [PubMed] [Google Scholar]
- 71.Laskin J, Heath BS, Roach PJ, Cazares L, Semmes OJ. Tissue imaging using nanospray desorption electrospray ionization mass spectrometry. Anal Chem. 2012;84(1):141–148. doi: 10.1021/ac2021322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vismeh R, Waldon DJ, Teffera Y, Zhao ZY. Localization and quantification of drugs in animal tissues by use of desorption electrospray ionization mass spectrometry imaging. Anal Chem. 2012;84(12):5439–5445. doi: 10.1021/ac3011654. [DOI] [PubMed] [Google Scholar]
- 73.Takats Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306(5695):471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 74.Ifa DR, Wu CP, Ouyang Z, Cooks RG. Desorption electrospray ionization and other ambient ionization methods: current progress and preview. Analyst. 2010;135(4):669–681. doi: 10.1039/b925257f. [DOI] [PubMed] [Google Scholar]
- 75.Cooks RG, Manicke NE, Dill AL, et al. New ionization methods and miniature mass spectrometers for biomedicine: DESI imaging for cancer diagnostics and paper spray ionization for therapeutic drug monitoring. Faraday Discuss. 2011;149:247–267. doi: 10.1039/c005327a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Northen TR, Yanes O, Northen MT, et al. Clathrate nanostructures for mass spectrometry. Nature. 2007;449(7165):1033–1036. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- 77.Greving MP, Patti GJ, Siuzdak G. Nanostructure-initiator mass spectrometry metabolite analysis and imaging. Anal Chem. 2011;83(1):2–7. doi: 10.1021/ac101565f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Robichaud G, Barry JA, Garrard KP, Muddiman DC. Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) imaging source coupled to a FT-ICR mass spectrometer. J Am Soc Mass Spectrom. 2013;24(1):92–100. doi: 10.1007/s13361-012-0505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sampson JS, Hawkridge AM, Muddiman DC. Generation and detection of multiply-charged peptides and proteins by matrix-assisted laser desorption electrospray ionization (MALDESI) Fourier transform ion cyclotron resonance mass spectrometry. J Am Soc Mass Spectrom. 2006;17(12):1712–1716. doi: 10.1016/j.jasms.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 80.Sampson JS, Hawkridge AM, Muddiman DC. Construction of a versatile high precision ambient ionization source for direct analysis and imaging. J Am Soc Mass Spectrom. 2008;19(10):1527–1534. doi: 10.1016/j.jasms.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Eikel D, Vavrek M, Smith S, et al. Liquid extraction surface analysis mass spectrometry (LESA-MS) as a novel profiling tool for drug distribution and metabolism analysis: the terfenadine example. Rapid Commun Mass Spectrom. 2011;25(23):3587–3596. doi: 10.1002/rcm.5274. [DOI] [PubMed] [Google Scholar]
- 82.Nemes P, Barton AA, Li Y, Vertes A. Ambient molecular imaging and depth profiling of live tissue by infrared laser ablation electrospray ionization mass spectrometry. Anal Chem. 2008;80(12):4575–4582. doi: 10.1021/ac8004082. [DOI] [PubMed] [Google Scholar]
- 83.Nemes P, Barton AA, Vertes A. Three-dimensional imaging of metabolites in tissues under ambient conditions by laser ablation electrospray ionization mass spectrometry. Anal Chem. 2009;81(16):6668–6675. doi: 10.1021/ac900745e. [DOI] [PubMed] [Google Scholar]
- 84.Nemes P, Vertes A. Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal Chem. 2007;79(21):8098–8106. doi: 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- 85.Nemes P, Vertes A. Laser ablation electrospray ionization for atmospheric pressure molecular imaging mass spectrometry. Methods Mol Biol. 2010;656:159–171. doi: 10.1007/978-1-60761-746-4_9. [DOI] [PubMed] [Google Scholar]
- 86.Nemes P, Woods AS, Vertes A. Simultaneous imaging of small metabolites and lipids in rat brain tissues at atmospheric pressure by laser ablation electrospray ionization mass spectrometry. Anal Chem. 2010;82(3):982–988. doi: 10.1021/ac902245p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Purves D, Augustine GJ, Fitzpatrick D, Hall WC, LaMantia A-S, White LE. Neuroscience. Sinauer Associates, Inc; MA, USA: 2012. [Google Scholar]
- 88.Werman R. Criteria for identification of a central nervous system transmitter. Comp Biochem Physiol. 1966;18(4):745. doi: 10.1016/0010-406x(66)90209-x. [DOI] [PubMed] [Google Scholar]
- 89.Fishman MC. Sir Henry Hallett Dale and acetylcholine story. Yale J Biol Med. 1972;45(2):104–118. [PMC free article] [PubMed] [Google Scholar]
- 90.von Bohlen und Halboch O, Dermietzel R. Neurotransmitters and Neuromodulators: Handbook of Receptors and Biological Effects. Wiley-Blackwell; NJ, USA: 2006. [Google Scholar]
- 91.Dale HH, Feldberg W, Vogt M. Release of acetylcholine at voluntary motor nerve endings. J Physiol. 1936;86(4):353–380. doi: 10.1113/jphysiol.1936.sp003371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sugiura Y, Zaima N, Setou M, Ito S, Yao I. Visualization of acetylcholine distribution in central nervous system tissue sections by tandem imaging mass spectrometry. Anal Bioanal Chem. 2012;403(7):1851–1861. doi: 10.1007/s00216-012-5988-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ye H, Wang J, Greer T, Strupat K, Li L. visualizing neurotransmitters and metabolites in the central nervous system by high resolution and high accuracy mass spectrometric imaging. ACS Chem Neurosci. 2013;4(7):1049–1056. doi: 10.1021/cn400065k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shariatgorji M, Nilsson A, Goodwin RJ, et al. Deuterated matrix-assisted laser desorption ionization matrix uncovers masked mass spectrometry imaging signals of small molecules. Anal Chem. 2012;84(16):7152–7157. doi: 10.1021/ac301498m. [DOI] [PubMed] [Google Scholar]
- 95.Benabdellah F, Touboul D, Brunelle A, Laprevote O. In situ primary metabolites localization on a rat brain section by chemical mass spectrometry imaging. Anal Chem. 2009;81(13):5557–5560. doi: 10.1021/ac9005364. [DOI] [PubMed] [Google Scholar]
- 96.Hattori K, Kajimura M, Hishiki T, et al. Paradoxical ATP elevation in ischemic penumbra revealed by quantitative imaging mass spectrometry. Antioxid Redox Signal. 2010;13(8):1157–1167. doi: 10.1089/ars.2010.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Curtis DR, Phillis JW, Watkins JC. Chemical excitation of spinal neurones. Nature. 1959;183(4661):611–612. doi: 10.1038/183611a0. [DOI] [PubMed] [Google Scholar]
- 98.Reece JB, Campbell NA. Campbell Biology. Benjamin Cummings; CA, USA: 2011. [Google Scholar]
- 99.Chavas J, Marty A. Coexistence of excitatory and inhibitory GABA synapses in the cerebellar interneuron network. J Neurosci. 2003;23(6):2019–2031. doi: 10.1523/JNEUROSCI.23-06-02019.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kondrat RW, Kanamori K, Ross BD. In vivo microdialysis and gas-chromatography/mass-spectrometry for 13C-enrichment measurement of extracellular glutamate in rat brain. J Neurosci Methods. 2002;120(2):179–192. doi: 10.1016/s0165-0270(02)00201-7. [DOI] [PubMed] [Google Scholar]
- 101.Buck K, Voehringer P, Ferger B. Rapid analysis of GABA and glutamate in microdialysis samples using high performance liquid chromatography and tandem mass spectrometry. J Neurosci Methods. 2009;182(1):78–84. doi: 10.1016/j.jneumeth.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 102.Wei H, Li H, Gao D, Lin JM. Multi-channel microfluidic devices combined with electrospray ionization quadrupole time-of-flight mass spectrometry applied to the monitoring of glutamate release from neuronal cells. Analyst. 2010;135(8):2043–2050. doi: 10.1039/c0an00162g. [DOI] [PubMed] [Google Scholar]
- 103.Goto-Inoue N, Setou M, Zaima N. Visualization of spatial distribution of gamma-aminobutyric acid in eggplant (Solanum melongena) by matrix-assisted laser desorption/ionization imaging mass spectrometry. Anal Sci. 2010;26(7):821–825. doi: 10.2116/analsci.26.821. [DOI] [PubMed] [Google Scholar]
- 104.Toue S, Sugiura Y, Kubo A, et al. Microscopic imaging mass spectrometry assisted by on-tissue chemical derivatization for visualizing multiple amino acids in human colon cancer xenografts. Proteomics. 2013 doi: 10.1002/pmic.201300041. [DOI] [PubMed] [Google Scholar]
- 105.Shariatgorji M, Nilsson A, Goodwin R, et al. MALDI-MS imaging and quantitation of primary amine neurotransmitters dopamine, GABA and glutamate directly in brain tissue sections. Proceedings for the 61st American Society for Mass Spectrometry Annual Conference.; Minneapolis, MN. 9–13 June 2013. [Google Scholar]
- 106.Bannon MJ. The dopamine transporter: role in neurotoxicity and human disease. Toxicol Appl Pharmacol. 2005;204(3):355–360. doi: 10.1016/j.taap.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 107.Greer M, Williams CM. Dopamine metabolism in Parkinson’s disease. Neurology. 1963;13:73–76. doi: 10.1212/wnl.13.1.73. [DOI] [PubMed] [Google Scholar]
- 108.Berecek KH, Brody MJ. Evidence for a neurotransmitter role for epinephrine derived from the adrenal medulla. Am J Physiol. 1982;242(4):H593–H601. doi: 10.1152/ajpheart.1982.242.4.H593. [DOI] [PubMed] [Google Scholar]
- 109.Wu C, Ifa DR, Manicke NE, Cooks RG. Molecular imaging of adrenal gland by desorption electrospray ionization mass spectrometry. Analyst. 2010;135(1):28–32. doi: 10.1039/b919816d. [DOI] [PubMed] [Google Scholar]
- 110.Mallimo EM, Kusnecov AW. The role of orphanin FQ/nociceptin in neuroplasticity: relationship to stress, anxiety and neuroinflammation. Front Cell Neurosci. 2013;7 doi: 10.3389/fncel.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bowers ME, Choi DC, Ressler KJ. Neuropeptide regulation of fear and anxiety: Implications of cholecystokinin, endogenous opioids, and neuropeptide Y. Physiol Behav. 2012;107(5):699–710. doi: 10.1016/j.physbeh.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Teng BL, Nonneman RJ, Agster KL, et al. Prosocial effects of oxytocin in two mouse models of autism spectrum disorders. Neuropharmacology. 2013;72:187–196. doi: 10.1016/j.neuropharm.2013.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zwanzger P, Domschke K, Bradwejn J. Neuronal network of panic disorder: the role of the neuropeptide cholecystokinin. Depress Anxiety. 2012;29(9):762–774. doi: 10.1002/da.21919. [DOI] [PubMed] [Google Scholar]
- 114.Meriaux C, Arafah K, Tasiemski A, et al. Multiple changes in peptide and lipid expression associated with regeneration in the nervous system of the medicinal leech. PLoS ONE. 2011;6(4):e18359. doi: 10.1371/journal.pone.0018359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mark L, Maasz G, Pirger Z. High resolution spatial distribution of neuropeptides by maldi imaging mass spectrometry in the terrestrial snail, Helix pomatia. Acta Biol Hung. 2012;63(Suppl 2):113–122. doi: 10.1556/ABiol.63.2012.Suppl.2.15. [DOI] [PubMed] [Google Scholar]
- 116.Altelaar AF, van Minnen J, Jimenez CR, Heeren RM, Piersma SR. Direct molecular imaging of Lymnaea stagnalis nervous tissue at subcellular spatial resolution by mass spectrometry. Anal Chem. 2005;77(3):735–741. doi: 10.1021/ac048329g. [DOI] [PubMed] [Google Scholar]
- 117.Zimmerman TA, Rubakhin SS, Romanova EV, Tucker KR, Sweedler JV. MALDI mass spectrometric imaging using the stretched sample method to reveal neuropeptide distributions in aplysia nervous tissue. Anal Chem. 2009;81(22):9402–9409. doi: 10.1021/ac901820v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rubakhin SS, Greenough WT, Sweedler JV. Spatial profiling with MALDI MS: distribution of neuropeptides within single neurons. Anal Chem. 2003;75(20):5374–5380. doi: 10.1021/ac034498+. [DOI] [PubMed] [Google Scholar]
- 119.Chansela P, Goto-Inoue N, Zaima N, Sroyraya M, Sobhon P, Setou M. Visualization of neuropeptides in paraffin-embedded tissue sections of the central nervous system in the decapod crustacean, Penaeus monodon, by imaging mass spectrometry. Peptides. 2012;34(1):10–18. doi: 10.1016/j.peptides.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 120.Chen R, Hui L, Cape SS, Wang J, Li L. Comparative neuropeptidomic analysis of food intake via a multi-faceted mass spectrometric approach. ACS Chem Neurosci. 2010;1(3):204–214. doi: 10.1021/cn900028s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ye H, Hui L, Kellersberger K, Li L. Mapping of neuropeptides in the crustacean stomatogastric nervous system by imaging mass spectrometry. J Am Soc Mass Spectrom. 2013;24(1):134–147. doi: 10.1007/s13361-012-0502-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Monroe EB, Annangudi SP, Hatcher NG, Gutstein HB, Rubakhin SS, Sweedler JV. SIMS and MALDI MS imaging of the spinal cord. Proteomics. 2008;8(18):3746–3754. doi: 10.1002/pmic.200800127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sturm RM, Greer T, Chen R, Hensen B, Li L. Comparison of NIMS and MALDI platforms for neuropeptide and lipid mass spectrometric imaging in brain tissue. Anal Methods. 2013;5(6):1623–1628. doi: 10.1039/C3AY26067D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Andersson M, Groseclose MR, Deutch AY, Caprioli RM. Imaging mass spectrometry of proteins and peptides: 3D volume reconstruction. Nat Methods. 2008;5(1):101–108. doi: 10.1038/nmeth1145. [DOI] [PubMed] [Google Scholar]
- 125.Jones EA, Shyti R, van Zeijl RJ, et al. Imaging mass spectrometry to visualize biomolecule distributions in mouse brain tissue following hemispheric cortical spreading depression. J Proteomics. 2012;75(16):5027–5035. doi: 10.1016/j.jprot.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 126.Källback P, Shariatgorji M, Nilsson A, Andren PE. Novel mass spectrometry imaging software assisting labeled normalization and quantitation of drugs and neuropeptides directly in tissue sections. J Proteomics. 2012;75(16):4941–4951. doi: 10.1016/j.jprot.2012.07.034. [DOI] [PubMed] [Google Scholar]
- 127.Hanrieder J, Ljungdahl A, Falth M, Mammo SE, Bergquist J, Andersson M. L-DOPA-induced dyskinesia is associated with regional increase of striatal dynorphin peptides as elucidated by imaging mass spectrometry. Mol Cell Proteomics. 2011;10(10):M111.009308. doi: 10.1074/mcp.M111.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Stoeckli M, Knochenmuss R, McCombie G, et al. MALDI MS imaging of amyloid. Methods Enzymol. 2006;412:94–106. doi: 10.1016/S0076-6879(06)12007-8. [DOI] [PubMed] [Google Scholar]
- 129.Ljungdahl A, Hanrieder J, Bergquist J, Andersson M. Analysis of neuropeptides by MALDI imaging mass spectrometry. Methods Mol Biol. 2013;1023:121–136. doi: 10.1007/978-1-4614-7209-4_7. [DOI] [PubMed] [Google Scholar]
- 130.Andersson M, Andren P, Caprioli RM. MALDI imaging and profiling mass spectrometry in neuroproteomics. In: Alzate O, editor. Neuroproteomics. CRC Press; FL, USA: 2010. [PubMed] [Google Scholar]
- 131.Ye H, Greer T, Li L. Probing neuropeptide signaling at the organ and cellular domains via imaging mass spectrometry. J Proteomics. 2012;75(16):5014–5026. doi: 10.1016/j.jprot.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ye H, Greer T, Li L. From pixel to voxel: a deeper view of biological tissue by 3D mass spectral imaging. Bioanalysis. 2011;3(3):313–332. doi: 10.4155/bio.10.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Porta T, Grivet C, Kraemer T, Varesio E, Hopfgartner G. Single hair cocaine consumption monitoring by mass spectrometric imaging. Anal Chem. 2011;83(11):4266–4272. doi: 10.1021/ac200610c. [DOI] [PubMed] [Google Scholar]
- 134.Tang HW, Wong MYM, Lam W, Cheng YC, Che CM, Ng KM. Molecular histology analysis by matrix-assisted laser desorption/ionization imaging mass spectrometry using gold nanoparticles as matrix. Rapid Commun Mass Spectrom. 2011;25(24):3690–3696. doi: 10.1002/rcm.5281. [DOI] [PubMed] [Google Scholar]
- 135.Pirkl A, Soltwisch J, Draude F, Dreisewerd K. Infrared matrix-assisted laser desorption/ionization orthogonal-time-of-flight mass spectrometry employing a cooling stage and water ice as a matrix. Anal Chem. 2012;84(13):5669–5676. doi: 10.1021/ac300840b. [DOI] [PubMed] [Google Scholar]
- 136.Qian K, Zhou L, Liu J, et al. Laser engineered graphene paper for mass spectrometry imaging. Sci Rep. 2013;3:1415. doi: 10.1038/srep01415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chang CC, Chen CN, Lin WJ, Lo TY, Lei SL, Mai FD. Novel method to prepare biological samples using powerful magnets on TOF-SIMS analysis. Surf Interface Anal. 2013;45(1):248–250. [Google Scholar]
- 138.Shariatgorji M, Kallback P, Gustavsson L, et al. Controlled-pH tissue cleanup protocol for signal enhancement of small molecule drugs analyzed by MALDI-MS imaging. Anal Chem. 2012;84(10):4603–4607. doi: 10.1021/ac203322q. [DOI] [PubMed] [Google Scholar]
- 139.Goodwin RJA, Nilsson A, Borg D, et al. Conductive carbon tape used for support and mounting of both whole animal and fragile heat-treated tissue sections for MALDI MS imaging and quantitation. J Proteomics. 2012;75(16):4912–4920. doi: 10.1016/j.jprot.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 140.Steinhauser ML, Lechene CP. Quantitative imaging of subcellular metabolism with stable isotopes and multi-isotope imaging mass spectrometry. Semin Cell Dev Biol. 2013;24(8–9):661–667. doi: 10.1016/j.semcdb.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Steinhauser ML, Bailey AP, Senyo SE, et al. Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature. 2012;481(7382):516–519. doi: 10.1038/nature10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.McMahon G, Glassner BJ, Lechene CP. Quantitative imaging of cells with multi-isotope imaging mass spectrometry (MIMS)-nanoautography with stable isotope tracers. Appl Surf Sci. 2006;252(19):6895–6906. [Google Scholar]
- 143.Pirman DA, Reich RF, Kiss A, Heeren RMA, Yost RA. Quantitative MALDI tandem mass spectrometric imaging of cocaine from brain tissue with a deuterated internal standard. Anal Chem. 2013;85(2):1081–1089. doi: 10.1021/ac302960j. [DOI] [PubMed] [Google Scholar]
- 144.Jones EA, van Remoortere A, van Zeijl RJM, et al. Multiple statistical analysis techniques corroborate intratumor heterogeneity in imaging mass spectrometry datasets of myxofibrosarcoma. PLoS ONE. 2011;6(9):e24913. doi: 10.1371/journal.pone.0024913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kriegsmann M, Seeley EH, Schwarting A, et al. MALDI MS imaging as a powerful tool for investigating synovial tissue. Scand J Rheumatol. 2012;41(4):305–309. doi: 10.3109/03009742.2011.647925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Karlsson O, Roman E, Berg AL, Brittebo EB. Early hippocampal cell death, and late learning and memory deficits in rats exposed to the environmental toxin BMAA (beta-N-methylamino-L-alanine) during the neonatal period. Behav Brain Res. 2011;219(2):310–320. doi: 10.1016/j.bbr.2011.01.056. [DOI] [PubMed] [Google Scholar]
- 147.Ljungdahl A, Hanrieder J, Falth M, Bergquist J, Andersson M. Imaging mass spectrometry reveals elevated nigral levels of dynorphin neuropeptides in L-DOPA-induced dyskinesia in rat model of Parkinson’s disease. PLoS ONE. 2011;6(9):e25653. doi: 10.1371/journal.pone.0025653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Meistermann H, Norris JL, Aerni HR, et al. Biomarker discovery by imaging mass spectrometry: transthyretin is a biomarker for gentamicin-induced nephrotoxicity in rat. Mol Cell Proteomics. 2006;5(10):1876–1886. doi: 10.1074/mcp.M500399-MCP200. [DOI] [PubMed] [Google Scholar]
- 149.Onishi S, Tatsumi Y, Wada K, et al. Sulfatide accumulation in the dystrophic terminals of gracile axonal dystrophy mice: lipid analysis using matrix-assisted laser desorption/ionization imaging mass spectrometry. Med Mol Morphol. 2013;46(3):160–165. doi: 10.1007/s00795-013-0019-y. [DOI] [PubMed] [Google Scholar]
- 150.Oppenheimer SR, Mi DM, Sanders ME, Caprioli RM. molecular analysis of tumor margins by MALDI mass spectrometry in renal carcinoma. J Proteome Res. 2010;9(5):2182–2190. doi: 10.1021/pr900936z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Willems SM, van Remoortere A, van Zeijl R, Deelder AM, McDonnell LA, Hogendoorn PCW. Imaging mass spectrometry of myxoid sarcomas identifies proteins and lipids specific to tumour type and grade, and reveals biochemical intratumour heterogeneity. J Pathol. 2010;222(4):400–409. doi: 10.1002/path.2771. [DOI] [PubMed] [Google Scholar]
- 152.Spraggins JM, Caprioli R. High-speed MALDI-TOF imaging mass spectrometry: rapid ion image acquisition and considerations for next generation instrumentation. J Am Soc Mass Spectrom. 2011;22(6):1022–1031. doi: 10.1007/s13361-011-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Schober Y, Guenther S, Spengler B, Rompp A. Single cell matrix-assisted laser desorption/ionization mass spectrometry imaging. Anal Chem. 2012;84(15):6293–6297. doi: 10.1021/ac301337h. [DOI] [PubMed] [Google Scholar]
- 154.Paschke C, Leisner A, Hester A, et al. Mirion-A software package for automatic processing of mass spectrometric images. J Am Soc Mass Spectrom. 2013;24(8):1296–1306. doi: 10.1007/s13361-013-0667-0. [DOI] [PubMed] [Google Scholar]
- 155.Robichaud G, Garrard KP, Barry JA, Muddiman DC. MSiReader: an open-source interface to view and analyze high resolving power MS imaging files on Matlab platform. J Am Soc Mass Spectrom. 2013;24(5):718–721. doi: 10.1007/s13361-013-0607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Website
- 201.msimaging. www.maldi-msi.org