Abstract
The nutrient sensor, O-linked N-acetylglucosamine (O-GlcNAc), cycles on and off nuclear and cytosolic proteins to regulate many cellular processes, including transcription and signaling. Dysregulated O-GlcNAcylation and its interplay with phosphorylation contribute to the etiology of diabetes, cancer and neurodegeneration. Herein, we review recent findings about O-GlcNAc's regulation of cell physiology.
Introduction
Complex molecular mechanisms ensure cellular homeostasis between nutrient sensing and energy metabolism. Strong evidence from many laboratories support a role for the hexosamine biosynthetic pathway (HBP), and its end product UDP-GlcNAc, as important nutrient sensors (Bond and Hanover, 2013; Hart et al., 2011; Vaidyanathan et al., 2014). The biosynthesis of UDP-GlcNAc is regulated by nearly every major metabolic pathway (Figure 1a). Production of fructose-6-phosphate by the glycolytic pathway is the first committed step of the HBP. Depending upon the cell type and metabolic state, about 2 to 5% of glucose that enters the cell is directed to the HBP to make UDP-GlcNAc. Amino acid metabolism is linked to flux through the HBP at several steps: via glucose through gluconeogenesis, via glutamine serving as the nitrogen donor to form glucosamine-6-phosphate, and via Acetyl-CoA serving as the donor for acetylation of glucosamine-6-phosphate through ketogenesis. Fatty acid metabolism is linked to the HBP via acetyl-CoA as well. The latter is not only essential to fatty acid metabolism but also serves to balance flux between carbohydrates and fatty acids. UTP, which serves as the donor to form UDP-GlcNAc, links flux through the HBP to both energy and nucleotide metabolism. Thus, UDP-GlcNAc sits at major nexus of cellular metabolic pathways and is responsive to flux through them.
Figure 1. O-GlcNAcylation is a key link between nutrient sensing and signaling.
a) The biosynthesis of UDP-GlcNAc by HBP integrates the flux through many metabolic pathways linked to nutrient intake. Variation in part or all of these pathways impacts directly the flux through HBP, the production of UDP-GlcNAc and the rate of O-GlcNAcylation. b) Schematic depicting the relationship of O-GlcNAcylation with phosphorylation for a single protein. c) Depending upon UDP-GlcNAc concentration, OGT modifies specific kinases. Regulation of substrate specificity and/or activity of upstream kinases by O-GlcNAcylation have profound effects on signaling and cellular physiology.
Although UDP-GlcNAc is used for the synthesis of secreted or membrane glycoproteins, where its concentration appears to control branching of complex glycans, the nucleotide sugar also serves as the donor substrate for O-GlcNAc transferase (OGT) catalyzed O-GlcNAcylation of myriad nuclear, cytoplasmic and mitochondrial proteins. Although discovered in the early 1980's, O-GlcNAcylation's key roles in cellular regulation are only more recently being widely understood. Unlike phosphorylation that requires over 600 kinases and phosphatases in humans, only two genes encode the enzymes that catalyze O-GlcNAc cycling. OGT catalyzes the transfer of a single O-GlcNAc moiety from UDP-GlcNAc to a serine or threonine residue in proteins, whereas O-GlcNAcase (OGA) removes the O-GlcNAc. OGT enzymatic activity is not only highly responsive to UDP-GlcNAc across a very broad range of concentrations but also its substrate specificity is dependent upon the concentration of the nucleotide sugar (Shen et al., 2012). UDP-GlcNAc concentrations vary dramatically between tissues, during differentiation, during inflammation, and in response to stimuli and nutrient availability. OGT is therefore a “control center” that integrates metabolic flux to send signals to effectors via O-GlcNAcylation.
This short review cannot summarize all the extensive work about O-GlcNAcylation of proteins and their regulation (we refer interested readers to recent comprehensive reviews: Bond and Hanover, 2013; Hart et al., 2011; Ma and Hart, 2013; Ruan et al., 2013; Vaidyanathan et al., 2014). Here we discuss only very recent findings that illustrate how OGT and O-GlcNAcylation regulate key downstream effectors to modulate cell physiology in response to nutrients.
Nutrient Regulation of Signaling via O-GlcNAcylation
Crosstalk between protein phosphorylation and O-GlcNAcylation at the same or proximal sites on proteins or by one modification regulating the other's cycling enzymes is extensive and well documented (Figure 1b). Competition for the same or nearby site has been previously described for c-Myc, estrogen receptor β (ERβ), C-terminal domain (CTD) of the RNA polymerase II, p53, forkhead box O1 (FoxO1), calcium/calmodulin-dependent protein kinase IV (CamKIV), casein kinase 2 (CK2), period 2 (PER2) and many other proteins. The interplay between O-GlcNAcylation and phosphorylation can also occur at more distant sites on proteins, which has been previously shown for cytokeratins. Both O-GlcNAc and O-phosphate may also exist simultaneously on the same protein at different sites, as seen on insulin receptor substrate (IRS), cardiac myosin light chain or AMP-activated protein kinase (AMPK). Finally, phosphorylation can enhance O-GlcNAcylation, such as it has been shown for cAMP response element-binding protein (CREB) (Bullen et al., 2014; Hart et al., 2011; Kaasik et al., 2013; Tarrant et al., 2012).
Recent glycomic and phosphoproteomic analyses indicate that the interplay between O-GlcNAcylation and phosphorylation are more extensive and complex than previously anticipated. Both OGT and OGA have been found in complexes with kinases or phosphatases, suggesting that they are acting within molecular complexes able to both phosphorylate and/or O-GlcNAcylate. Integration of cell stimuli and signals, as well as nutrient availability, determines which post-translational modifications (PTMs) are added to polypeptides (Figure 1b).
Numerous kinases belonging to various signaling pathways are O-GlcNAcylated (Han et al., unpublished data), and those studied so far are regulated by O-GlcNAcylation. Modulation of upstream kinases by O-GlcNAcylation may drastically change the activation or inhibition of downstream signaling pathways (Figure 1c). O-GlcNAcylation of CamKIV at Ser189 inhibits its phosphorylation at Thr200 and blocks its ability to bind ATP and to catalyze phosphorylation (Hart et al., 2011). Since the Km for UDP-GlcNAc of OGT with CamKIV is over 20μM, it was proposed that in vivo O-GlcNAcylation of CamKIV could vary significantly in response to the rate of HBP flux (Shen et al., 2012). Calcium/Calmodulin-dependant protein Kinase II (CaMKII) is a multifunctional serine/threonine kinase important for a variety of cellular functions, including cell division, differentiation, cardiac contraction, and synaptic plasticity. During hyperglycemia, as occurs in diabetes, O-GlcNAcylation of CaMKII is increased in the heart and brain of rats and humans. Its O-GlcNAcylation at Ser279 leads to an autonomous constitutive activation even after Ca2+ concentration declines (Erickson et al., 2013), contributing to diabetic cardiomyopathy. Recently, it has been shown that CaMKII is activated by glucagon and fasting in vivo in a calcium- and IP3R-dependent manner. Activated CaMKII stimulates hepatic glucose production through FoxO1 phosphorylation. In obese mice, hepatic CaMKII remains activated and mediates hepatic glucose production (Ozcan et al., 2012). Owing to the tight relationship between obesity, type II diabetes and O-GlcNAcylation, it is possible that CaMKII O-GlcNAcylation might also contribute to the abnormal enhanced hepatic glucose production associated with diabetes and obesity, as it has already been shown for O-GlcNAcylation of PPAR-gamma coactivator-1-alpha (PGC-1α), FoxO1 and CREB-regulated transcription coactivator 2 (CRTC2) transcription factors. O-GlcNAcylation also controls the substrate specificity of certain kinases. CK2 is a serine/threonine kinase that regulates cell physiology via the phosphorylation of hundreds of proteins involved in several major cellular processes such as cell cycle, apoptosis or cell differentiation. Although its regulation remains poorly understood, it has been shown that the CK2α subunit is O-GlcNAcylated at Ser347. This modification inhibits its phophorylation at Thr344 by Cdk1/Cyclin B and impairs its stabilization. In vitro assays using protein microarrays found the surprising result that although O-GlcNAcylation of CK2α doesn't affect its catalytic activity, it does regulate its substrate specificity (Tarrant et al., 2012). In vitro analyses of the OGT affinity toward CK2 suggest that its substrate selectivity may be nutrient sensitive depending on its O-GlcNAcylation state (Lubas and Hanover, 2000).
In turn, there is evidence showing that enzymes that govern O-GlcNAc cycling are substrate for kinases. Although, OGT and OGA are both phosphorylated, the role of the phosphorylation of OGA remains unknown. Phosphorylation by insulin receptor, CaMKIV, glycogen synthase kinase 3 beta (GSK3β), and AMPK activates OGT or modulates its interactions with its binding partners and substrates (Bullen et al., 2014; Hart et al., 2011; Kaasik et al., 2013). Phosphorylation and O-GlcNAcylation are in competition for the modification of the residues S3 or S4 of OGT and it has been shown that phosphorylation by GSK3β at these residues enhance OGT activity (Kaasik et al., 2013). AMPK is an energy sensor protein kinase activated in response to reduction of intracellular ATP levels. AMPK regulates hundreds of substrates involved in energy metabolism by phosphorylation. It was recently shown that T444 of OGT is phosphorylated by AMPK and it appears that this modification regulates OGT substrate selectivity (Bullen et al., 2014). Considering that the activity of AMPK is positively regulated by O-GlcNAcylation (Xu et al., 2014), these results suggest a regulatory feed back control of AMPK through O-GlcNAcylation, giving a key role to OGT/AMPK in the regulation of energy metabolism.
While there is still much to learn, it is already clear that nutrient regulation of signaling pathways via O-GlcNAcylation impacts nearly all regulatory mechanisms and cellular physiology.
Fundamental Importance of O-GlcNAcylation to Transcription
While regulation of gene expression by hormones, growth factors and tissue-specific factors is fairly well established, the regulation of gene expression by nutrients is not well understood. The first evidence linking O-GlcNAcylation and chromatin regulation emerged in the late 1980s. OGT and O-GlcNAcylation were shown to be highly abundant on chromatin, associated with transcriptionally active loci on polytene chromosomes in Drosophilia, and the CTD of a subset of RNA polymerase is extensively O-GlcNAcylated. More recently, OGT was shown to be a polycomb gene. The polycomb group proteins are master regulators, repressing the expression of large sets of genes that control tissue patterning during development. In 2011, Myers et al. confirmed the link between OGT and polycomb group proteins in mammals by showing that the polycomb repressive complex 2 is required to maintain normal levels of OGT and for correct cellular distribution of O-GlcNAcylation (Myers et al., 2011). Deep sequencing technology established that OGT, OGA and O-GlcNAcylation are enriched at the active site of transcription in thousands of genes in both human and in C. Elegans (Deplus et al., 2013; Love et al., 2010). O-GlcNAcylation regulates many RNA polymerase II transcription factors. Depending upon the transcription factor, the sugar modification regulates their DNA binding activity (c-Rel, p53, pancreatic-duodenal homeobox factor1…), transcriptional activity (c-Myc, FoxO1, CREB…) and/or stability (mER-β, p53, delta-lactoferrin, beta-catenin…) (Bond and Hanover, 2013; Hardiville et al., 2010; Olivier-Van Stichelen et al., 2014).
O-GlcNAcylation also regulates the basal transcriptional machinery. Nearly all of the components of the basal transcription machinery in mammals are modified by O-GlcNAc. The CTD of RNA polymerase II is O-GlcNAcylated at the pre-initiation step of the transcription cycle, and the sugar modification is reciprocal to its phosphorylation, which is required for elongation (Figure 2). O-GlcNAcylation occurs during the preinitiation complex assembly and may regulate the initiation of transcription on a multitude of genes in higher eukaryotes (Bond and Hanover, 2013).
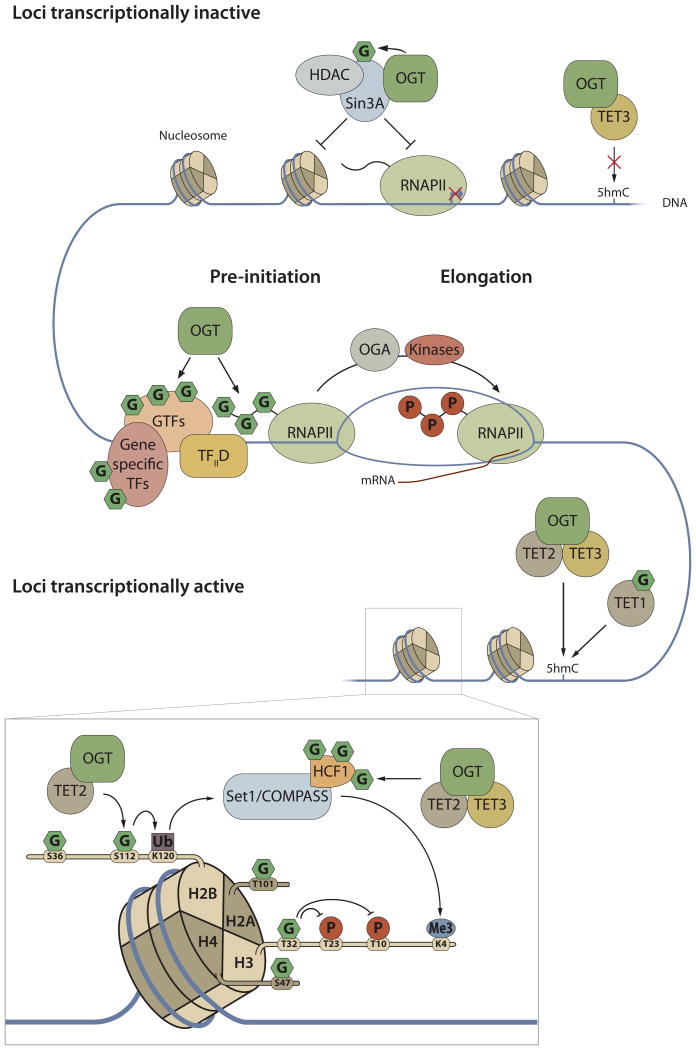
Figure 2. Regulation of gene transcription and epigenetics by O-GlcNAcylation.
O-GlcNAc cycling on gene specific transcription factors and components of the basal transcriptional machinery regulates gene activation. Sp1 is one of the most extensively modified transcription factors. The CTD of the RNA polymerase II (RNAP II) is itself O-GlcNAcylated at the transcription initiation step. The switch from transcription initiation to elongation requires OGA activity to remove the sugar. O-GlcNAcylation or OGT interaction with master regulators of epigenetic marks also switch on and off gene expression. The complex comprised of Sin3A, HDAC and OGT has been proposed to inhibit transcription. OGT O-GlcNAcylates and/or forms complexes with TETs that drive DNA methylation. H2B O-GlcNAcylation and ubiquitination are associated with transcriptionally active loci. Insert emphasizes the role of O-GlcNAcylation in the regulation of the histone code.
Epigenetic modifications are heritable changes in chromatin structure that govern temporal and spatial gene expression. O-GlcNAcylation is not only part of the histone code but also influences other epigenetic modifications, such as DNA or histone methylation. All four subunits of the core histones are O-GlcNAcylated. Some sites of modification are known: H2A at T101, H2B at S36 and S112, H3 at T32 and H4 at S47 (Vaidyanathan et al., 2014). Histone O-GlcNAcylation is increased during the G1 phase of the cell cycle, declines during S phase and increases through G2 and mitosis phases to drop at M/G1. Although the role of sugar on histone subunits remains poorly understood some of the O-GlcNAcylation sites directly interact with DNA. O-GlcNAcylation of S112 of histone H2B serves as the docking site for the ubiquitin ligase that modifies H2B K120, which is a transcriptionally active gene mark (Vaidyanathan et al., 2014). H2B ubiquitination and its ubiquitination complex stimulate H3K4me3 through the SET1/COMPASS complex. Host Cell Factor 1 (HCF-1), is an extensively O-GlcNAcylated (19 known attachment sites) component of SET1/COMPASS. OGT not only O-GlcNAcylates HCF-1, but also acts as a unique protease to cleave the protein into its functional fragments by a totally novel protease mechanism involving UDP-GlcNAc in the reaction mechanism (Lazarus et al., 2013).
Methylation of cytosine is another key epigenetic event critical for various cellular processes and is often associated with transcription repression. Ten-eleven translocation (Tet) family proteins are evolutionarily conserved dioxygenases that convert 5-methylcytosine (5mC) to 5-Hydroxylmethylcytosine (5hmC). Hydroxylmethylation of CpG-rich DNA promotes DNA demethylation. Tet-1, -2, and -3 interact strongly with OGT and undergo O-GlcNAcylation (Chen et al., 2013; Deplus et al., 2013; Shi et al., 2013a; Zhang et al., 2014). Mutation of Tet1 O-GlcNAc sites stabilizes Tet1 and the knock down of OGT by siRNA reduces the amounts of Tet1 and 5hmC on Tet1 target genes (Shi et al., 2013a). Interaction between Tet3 and OGT drives Tet3 out of the nucleus, concomitant with decreased 5hmC on DNA. Under pathologic glucose concentrations (25mM), Tet3 exhibits a cytoplasmic localization and the HBP inhibitor deoxynorleucine (DON) increases DNA 5hmC content (Zhang et al., 2014). Conversely, Tet2 and Tet3 co-localize with OGT at promoters, clustered around the transcription start sites and CpG islands, and they overlap at promoters with H3K4me3 (Chen et al., 2013; Deplus et al., 2013). Depletion of Tet2 reduces O-GlcNAcylation of H2B at Ser112 suggesting that Tet2 targets OGT to the nucleosome for H2B O-GlcNAcylation (Chen et al., 2013) supporting the link between DNA methylation and the histone code through OGT and O-GlcNAcylation. Taken together, these studies reveal a central role of OGT and O-GlcNAcylation in basal and gene specific transcription, as well as in the epigenetic modifications on DNA and histones (Figure 2).
O-GlcNAcylation and Cell Physiology
To maintain proper homeostasis, cell physiology has to adapt to fluctuations in nutrient intake. Cell proliferation, for instance, must be regulated in response to nutrient levels to ensure that sufficient materials are synthesized to generate viable daughter cells. O-GlcNAcylation is intimately linked to the cell cycle since it varies on many proteins all along the cell cycle, and its dysregulation leads to impaired cell cycle progression at major checkpoints (Hart et al., 2011).
Circadian rhythms are biological oscillations set up by clock transcription factors for a period of about 24h. At the cellular level, rhythms in nutrient intake must coincide with fluctuations in gene expression for proper homeostasis. Not surprisingly, nutrient regulation of circadian rhythms is linked to O-GlcNAcylation. Protein O-GlcNAcylation and OGT protein levels peak in the middle of the dark phase in mouse heart. OGT mRNA levels increased 2 hours before dark, suggesting a regulation of OGT expression by the circadian transcription factors (Durgan et al., 2011). In the primary feedback loop, the positive elements CLOCK and aryl hydrocarbon receptor nuclear translocator-like (BMAL1) initiate transcription of targeted genes, including PER and Cryptochrome (CRY) genes. In return, negative feedback is achieved by PER and CRY repressing transcriptional activity of CLOCK/BMAL1. BMAL1, CLOCK, PER1 and PER2 are all O-GlcNAcylated (Durgan et al., 2011; Kaasik et al., 2013). BMAL1 and PER1 appeared to be O-GlcNAcylated with a peak in the middle of the dark phase in mouse heart (Durgan et al., 2011). O-GlcNAcylation stabilizes BMAL1 and CLOCK by inhibiting their ubiquitination, which promotes expression of their target genes (Li et al., 2013). PER2 can be O-GlcNAcylated at 10 different residues, and phosphorylation at S662 inhibits its O-GlcNAcylation. The reciprocal modification of PER2 S662 was shown to be at the core of regulation of PER2 transcriptional repressor activity with a stronger repressor activity for the O-GlcNAcylated isoform. The OGT inhibitor (Alloxan) or OGT knock down (siRNA and OGT conditional KO mice) shortened the circadian rhythm, while the OGA inhibitor (PUGNAC) lengthened the circadian rhythmicity of PER2-luc oscillation (Durgan et al., 2011; Kaasik et al., 2013), suggesting that the circadian rhythm is adjusted by O-GlcNAcylation through PER2. Elevated glucose significantly prevents phosphorylation of PER2, even under OGA overexpression (Kaasik et al., 2013) and knockdown of glutamine-fructose-6-phosphate aminotransferase 1 or OGT reduce expression of BMAL1 and CRY1 (Li et al., 2013). These studies suggest that nutrient sensing by the HBP entrains circadian rhythm via O-GlcNAcylation of the core circadian clock transcription factors. Likewise, circadian oscillation is involved in the control of nutrient homeostasis. A recent study showed that knockout of Bmal1, which disrupts the clock rhythmicity, leads to profound insulin resistance, hyperglycemia and fat accumulation in mice (Shi et al., 2013b). Since OGT modulates the diurnal rhythm of glucose homeostasis (Li et al., 2013), it is possible that O-GlcNAcylation could be the molecular link between the circadian cycle, glucose homeostasis and insulin-resistance.
O-GlcNAcylation regulates the insulin-signaling pathway at several steps, and has been shown to be involved in insulin resistance. Downstream of the insulin-signaling cascade, hyperglycemia targets OGT to O-GlcNAcylate the transcription factor CRTC2, increasing transcription of PGC-1α, which in turn interacts with OGT to target the transferase to FoxO1, resulting in its O-GlcNAcylation and enhanced transcriptional activity. This cascade of events leads to inappropriate gluconeogenesis associated with diabetes. Upstream of the insulin-signaling pathway, OGT is activated by the insulin receptor and translocated to the plasma membrane and bound to phosphoinositides. The O-GlcNAcylation of IRS1 inhibits phosphorylation required for its interaction with the p85 regulatory subunit of phosphatidylinositol-3-kinase. The phospho-activation of protein kinase B (AKT) at T308 is inhibited by its O-GlcNAcylation, leading to down-regulation of AKT function (Ma and Hart, 2013). GSK3β activity is inhibited by AKT. Phosphorylation by GSK3β promotes IRS1 degradation. GSK3β is O-GlcNAcylated and regulated by the sugar modification (Lubas and Hanover, 2000). Interestingly, the inhibitory phosphorylation at S9 of GSK3β shows a robust circadian oscillation (Kaasik et al., 2013), and the stress-induced S9 phosphorylation is delayed in an OGT null mouse cell line, suggesting that O-GlcNAcylation may link circadian cycle, synthesis of glycogen and the insulin pathway through GSK3β. Since OGT is activated by GSK3β phosphorylation, it is possible that there may be a synergistic effect between O-GlcNAcylation and GSK3β phosphorylation leading to insulin resistance.
O-GlcNAcylation also appears to regulate glucose metabolism. All enzymes in the glycolytic pathway are substrates for OGT. Yi et al., have shown that OGT overexpression or OGA inhibition lowered the rate of glucose metabolism by decreasing PFK1 activity. The authors mapped the O-GlcNAc site as Ser529, a highly conserved residue known to be important for allosteric regulation of PFK1 by fructose-2,6-bisphosphate. Inhibition of PFK1 by O-GlcNAcylation redirects glucose flux from the glycolytic pathway to the oxidative pentose phosphate pathway, which has been proposed to benefit cancer cell growth (Yi et al., 2012). Yi et al. also observed that increase O-GlcNAcylation reduced cellular ATP concentrations, suggesting a relationship between the sugar modification and energy production. This effect might in part be due to decreased glycolytic flux but not entirely since members of mitochondrial respiratory chain complexes are also O-GlcNAcylated, such as the subunit NDUFA9 of complex I, core 1 and 2 of complex III and subunit COX I of complex IV. Hyperglycemia increases mitochondrial O-GlcNAcylation and leads to impaired activity of complex I, III and IV, which results to lower cellular ATP content (Ma and Hart, 2013).
Reduced glucose metabolism contributes to the etiology of neurodegeneration. Several studies have shown that reduced O-GlcNAcylation of neural proteins, such as the tau protein, neurofilaments and synaptic vesicle proteins, and their concomitant increased phosphorylation, contributes directly to the etiology of Alzhemier's Disease (AD). Inhibitors of OGA, which help maintain or increase O-GlcNAcylation in the brain, may prove useful for the treatment of AD (Hart et al., 2011). Clearly, understanding more fully the ties that exist between nutrient sensing by O-GlcNAcylation, phosphorylation, circadian rhythm regulation, cell cycle and metabolism will provide novel approaches regarding treatment of obesity, diabetes, cancer or AD.
Conclusions
It is clear that HBP, O-GlcNAcylation and OGT act as key molecular components to sense nutrient availability. In normal physiological conditions, OGT integrates those changes and transmits the information to many effector proteins involved in myriad cellular processes, such as transcription or signaling. OGT and O-GlcNAcylation regulate cellular processes to maintain homeostasis at different states of nutrient availability. A growing body of evidence links changes in nutritional status, cellular homeostasis and the establishment of chronic diseases beyond those typically referred to as metabolic related pathologies. Improper nutrient homeostasis or nutrient excess leads to abnormal O-GlcNAcylation of proteins with corresponding effects on cell signaling and transcription. Prolonged nutrient excess leads to abnormally high protein O-GlcNAcylation. Such hyper-O-GlcNAcylation not only contributes to abnormal regulation of phosphorylation signaling cascades but also leads to incorrect control of gene expression and chromatin architecture, all of which contribute to nutrient toxicity, as is seen in diabetes. Likewise, defective glucose utilization, which results in abnormally low O-GlcNAcylation, contributes directly to the onset of neurodegeneration. Faulty epigenetic information induced by deregulation of O-GlcNAcylation could also be transmitted to the daughter cells. The latter is particularly important in the light of cancer etiology and the higher risk of cancer in individuals with diabetes and obesity. Future research will need to elucidate how the crosstalk between different PTMs on proteins work in concert to modulate polypeptide functions in response to metabolism and environmental clues. Clearly, understanding how the nutrient sensor, O-GlcNAcylation, controls signaling pathways and cellular processes, and how it cross talks with other PTMs, will be essential not only in understand cellular physiology but also in the development new therapeutics.
Highlights.
OGT & O-GlcNAc act as a nutrient sensors to regulate signaling
Kinases in various signaling pathways are regulated by O-GlcNAcylation
O-GlcNAcylation regulates gene expression in response to nutrient availability
Misregulation of O-GlcNAcylation is associated with chronic disease.
Acknowledgments
Supported by NIH R01CA42486, R01DK61671 and P01HL107153, N01-HV-00240. Dr. Hart receives a share of royalty received by the university on sales of the CTD 110.6 antibody, which are managed by JHU. We would like to thank Dr. SB Peterson and Dr. PS Banerjee for critical reading of the manuscript. We apologize to our colleagues whose original work could not be cited due to space constraints.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Stéphan Hardivillé, Email: shardiv1@jhmi.edu.
Gerald W. Hart, Email: gwhart@jhmi.edu.
References
- Bond MR, Hanover JA. O-GlcNAc cycling: a link between metabolism and chronic disease. Annu Rev Nutr. 2013;33:205–229. doi: 10.1146/annurev-nutr-071812-161240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen JW, Balsbaugh JL, Chanda D, Shabanowitz J, Hunt DF, Neumann D, Hart GW. Crosstalk between two essential nutrient-sensitive enzymes: O-GlcNAc transferase (OGT) and AMP-activated protein kinase (AMPK) J Biol Chem. 2014 doi: 10.1074/jbc.M113.523068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durgan DJ, Pat BM, Laczy B, Bradley JA, Tsai JY, Grenett MH, Ratcliffe WF, Brewer RA, Nagendran J, Villegas-Montoya C, et al. O-GlcNAcylation, novel post-translational modification linking myocardial metabolism and cardiomyocyte circadian clock. J Biol Chem. 2011;286:44606–44619. doi: 10.1074/jbc.M111.278903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson JR, Pereira L, Wang L, Han G, Ferguson A, Dao K, Copeland RJ, Despa F, Hart GW, Ripplinger CM, et al. Diabetic hyperglycaemia activates CaMKII and arrhythmias by O-linked glycosylation. Nature. 2013;502:372–376. doi: 10.1038/nature12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardiville S, Hoedt E, Mariller C, Benaissa M, Pierce A. O-GlcNAcylation/phosphorylation cycling at Ser10 controls both transcriptional activity and stability of delta-lactoferrin. J Biol Chem. 2010;285:19205–19218. doi: 10.1074/jbc.M109.080572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaasik K, Kivimae S, Allen JJ, Chalkley RJ, Huang Y, Baer K, Kissel H, Burlingame AL, Shokat KM, Ptacek LJ, et al. Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 2013;17:291–302. doi: 10.1016/j.cmet.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus MB, Jiang J, Kapuria V, Bhuiyan T, Janetzko J, Zandberg WF, Vocadlo DJ, Herr W, Walker S. HCF-1 is cleaved in the active site of O-GlcNAc transferase. Science. 2013;342:1235–1239. doi: 10.1126/science.1243990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Ruan HB, Hughes ME, Lee JS, Singh JP, Jones SP, Nitabach MN, Yang X. O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab. 2013;17:303–310. doi: 10.1016/j.cmet.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love DC, Ghosh S, Mondoux MA, Fukushige T, Wang P, Wilson MA, Iser WB, Wolkow CA, Krause MW, Hanover JA. Dynamic O-GlcNAc cycling at promoters of Caenorhabditis elegans genes regulating longevity, stress, and immunity. Proc Natl Acad Sci U S A. 2010;107:7413–7418. doi: 10.1073/pnas.0911857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas WA, Hanover JA. Functional expression of O-linked GlcNAc transferase. Domain structure and substrate specificity. J Biol Chem. 2000;275:10983–10988. doi: 10.1074/jbc.275.15.10983. [DOI] [PubMed] [Google Scholar]
- Ma J, Hart GW. Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev Proteomics. 2013;10:365–380. doi: 10.1586/14789450.2013.820536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SA, Panning B, Burlingame AL. Polycomb repressive complex 2 is necessary for the normal site-specific O-GlcNAc distribution in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2011;108:9490–9495. doi: 10.1073/pnas.1019289108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier-Van Stichelen S, Dehennaut V, Buzy A, Zachayus JL, Guinez C, Mir AM, El Yazidi-Belkoura I, Copin MC, Boureme D, Loyaux D, et al. O-GlcNAcylation stabilizes beta-catenin through direct competition with phosphorylation at threonine 41. FASEB J. 2014 doi: 10.1096/fj.13-243535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan L, Wong CC, Li G, Xu T, Pajvani U, Park SK, Wronska A, Chen BX, Marks AR, Fukamizu A, et al. Calcium signaling through CaMKII regulates hepatic glucose production in fasting and obesity. Cell Metab. 2012;15:739–751. doi: 10.1016/j.cmet.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen DL, Gloster TM, Yuzwa SA, Vocadlo DJ. Insights into O-linked N-acetylglucosamine ([0-9]O-GlcNAc) processing and dynamics through kinetic analysis of O-GlcNAc transferase and O-GlcNAcase activity on protein substrates. J Biol Chem. 2012;287:15395–15408. doi: 10.1074/jbc.M111.310664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi FT, Kim H, Lu W, He Q, Liu D, Goodell MA, Wan M, Songyang Z. Ten-eleven translocation 1 (Tet1) is regulated by O-linked N-acetylglucosamine transferase (Ogt) for target gene repression in mouse embryonic stem cells. J Biol Chem. 2013a;288:20776–20784. doi: 10.1074/jbc.M113.460386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi SQ, Ansari TS, McGuinness OP, Wasserman DH, Johnson CH. Circadian disruption leads to insulin resistance and obesity. Curr Biol. 2013b;23:372–381. doi: 10.1016/j.cub.2013.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrant MK, Rho HS, Xie Z, Jiang YL, Gross C, Culhane JC, Yan G, Qian J, Ichikawa Y, Matsuoka T, et al. Regulation of CK2 by phosphorylation and O-GlcNAcylation revealed by semisynthesis. Nat Chem Biol. 2012;8:262–269. doi: 10.1038/nchembio.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan K, Durning S, Wells L. Functional O-GlcNAc modifications: implications in molecular regulation and pathophysiology. Crit Rev Biochem Mol Biol. 2014;49:140–163. doi: 10.3109/10409238.2014.884535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Yang C, Du Y, Chen Y, Liu H, Deng M, Zhang H, Zhang L, Liu T, Liu Q, et al. AMPK regulates histone H2B O-GlcNAcylation. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA, 3rd, Peters EC, Driggers EM, Hsieh-Wilson LC. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science. 2012;337:975–980. doi: 10.1126/science.1222278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Liu X, Gao W, Li P, Hou J, Li J, Wong J. Differential Regulation of Ten-Eleven Translocation Family of Dioxygenases by O-Linked beta-N-Acetylglucosamine Transferase OGT. J Biol Chem. 2014 doi: 10.1074/jbc.M113.524140. [DOI] [PMC free article] [PubMed] [Google Scholar]