Abstract
Lysosomal proteases perform critical functions in protein turnover and are essential for normal growth and development. Cathepsin P is a member of a newly discovered family of lysosomal cysteine proteases uniquely expressed in rodent placenta (PECs), and is closely related to human cathepsin L. Using the rat choriocarcinoma cell line model, Rcho-1, mRNA for the PECs cathepsins P, M, Q, R, 1, 2 was found to increase in expression during differentiation into a trophoblast giant cell phenotype. By contrast, expression of cathepsin L was not regulated. A specific enzyme assay was developed to show that activity of cathepsin P mirrored mRNA expression during differentiation. Cathepsin P protein co-localizes with cathepsin B, indicating that the enzyme probably functions in the endosomal-lysosomal compartment. This study demonstrates that the PEC genes produce functional proteases that can perform specific placental roles that are probably performed by broader specificity proteases in human placenta.
INTRODUCTION
Lysosomal proteases play important roles in placental function. Recent clinical studies have implicated cathepsins in pre-eclampsia and recurrent miscarriage [1, 2], so it is important to understand the roles of cathepsins in placental function. Leupeptin, which inhibits a range of proteases including the lysosomal enzymes cathepsins B and L, has a teratogenic effect when administered to pregnant rats at gestational day 8.5 [3]. E-64 (trans-epoxysuccinyl-L-leucylaminido-(4-guanidino)-butane), a commonly used cysteine proteases inhibitor, caused failure of implantation when administered to pregnant mice at day 4.5 and caused teratogenic effects and growth retardation when administered later in gestation [4]. The more specific cathepsin inhibitor, Cbz-Phe-Ala-CHN2 (Cbz = carboxybenzyl), also caused malformation of mouse embryonic tissues when administered between days 10 and 12 of pregnancy [5]. Leupeptin also causes failure of blastocyst hatching in hamsters [6]. Genetic deletion of cathepsin L, B, or both has no obvious impact in mouse development until birth [7]. It seems that these two genes are dispensable for rodent placental function. A series of gene duplications has resulted in the generation of a sub-family of proteases that are uniquely expressed in rodent placenta (PECs, for placentally expressed cathepsins), and the protease inhibitors may be targeting specific placental functions that are performed by these genes [8–10]. Most of the PEC genes are expressed later in gestation and have been shown to be located in the labyrinthine layer and spongiotrophoblasts of mature placenta [8, 11, 12]. The expression of the placental cathepsins early in gestation is less clear. Cathepsins 1 and 2 were originally identified as being expressed in early placenta in a differential expression study that compared genes in early and late placenta, but subsequent studies have also shown high levels of expression later in gestation [10, 11, 13]. Cathepsin P mRNA expression was detected in ectoplacental cone from the day 7.5 mouse conceptus, but expression at any earlier times has not been reported. Recombinant cathepsin P has been prepared and shown to yield an active enzyme but nothing is known about the activity for any of these proteases in vivo.
This study is designed to examine expression of PECs in cellular systems. A well-established cell model of trophoblast cell differentiation is Rcho-1, a rat choriocarcinoma cell line that can be manipulated in culture to differentiate into cells that resemble trophoblast giant cells (TGs) [14–16]. The cells become polyploid due to multiple rounds of DNA synthesis without mitosis (endoreduplication). Like true TGs, the cells become highly invasive and produce placental lactogen type 1 (PL-1). TGs penetrate maternal endometrial epithelia cells and remodel the endometrial stroma to connect the maternal blood to placenta during implantation. Inhibition of proteases in TGs would likely cause implantation failure and possibly impairs placental function, resulting in embryonic malformations and growth retardation. In this study we examine expression of cathepsins in both placental tissues and this Rcho-1 cellular model of trophoblast cell differentiation.
MATERIALS AND METHODS
Materials
Fluorogenic substrate MOCAc-Pro-Leu-Gly-Leu-A2pr(Dnp)-Ala-Arg-NH2 (where MOCAc = (7-Methoxycoumarin-4-acyl) and A2pr(Dnp) = N-(2,4-Dinitrophenyl)-L-2,3-Diaminopropionyl) was purchased from Peptides International, (Louisville, KY, USA). Cbz-Phe-Arg-N-methylcoumarin, and E-64 were from Sigma-Aldrich (St Louis, MO, USA). Primers for PCR and real time-PCR were from Integrated DNA Technologies (Coralville, AL, USA). NCTC-135 medium was purchased from Sigma Aldrich, (St Louis, MO, USA). FBS (fetal bovine serum), HS (horse serum), trypsin and HBSS were purchased from Mediatech Inc, (Herndon, VA, USA). TRIzol, avian myeloblastosis reverse transcriptase kits, and PCR kits were from Invitrogen, (Carlsburg, CA, USA). Rcho-1 cells were a kind gift from Michael J. Soares (Kansas Medical Center, Kansas City, KS, USA). PC-12 and JEG-3 cell lines were from ATCC (Rockville, MD, USA). iQ-SYBER Green Supermix kit was from Biorad Laboratories, (Hercules, CA, USA). 4-chambered glass slides were from NalgeNun International (Rochester, NY, USA). Texas red-labeled mouse anti-rabbit IgG antibody and FITC-labeled donkey anti-sheep IgG antibody were from Jackson Immuno Research Inc (West Grove, PA, USA). Sheep anti-human cathepsin B antibody [17] was a gift from David Buttle (Sheffield, U.K.). Other reagents used in this study were analytical grade and were obtained from Sigma Aldrich, (St Louis, MO, USA).
Animal tissues
FVB/N mice (Taconic farms Inc, Hudson, NY, USA) were kept in standard controlled conditions at 22–24°C and a 12 h light-dark cycle. Animals were fed standard laboratory chow diet and had free access to water. Females were mated with males from the same strain (FVB/N) overnight during pro-estrus period. The morning of finding vaginal plug was designated as day 0.5. At day 18.5, females (9) were euthanized and placenta, embryo, umbilical cord, and maternal liver and brain harvested for enzymatic activity analysis. The fetal portion of the placentae was separated from maternal decidua and other embryonic tissues. Tissues were homogenized directly in ice-cold cathepsin P activity buffer (400 mM sodium phosphate, 4 mM EDTA, pH 6.5) or cathepsin B activity buffer (400 mM sodium acetate, 4 mM EDTA, pH 5.5). Homogenates were centrifuged at 15000 g for 5 min. The supernatants were collected and assayed for protein and activity as described below. Sprague Dawley rats (Taconic farms Inc, Hudson, NY, USA) were kept under similar conditions and tissues obtained and processed from pregnant animals (day 15.5).
Cell culture
Rcho-1 cells were maintained in a proliferative state by culturing in hormone and growth factor-rich medium (NCTC-135 containing 20% FBS, 50 µM β-mercaptoethanol and 1% pyruvic acid). Cells were split every 2–3 days as described [15, 16]. To induce differentiation into trophoblast giant cells, medium from confluent Rcho-1 cells was replaced with growth factor-deficient medium (NCTC-135 containing 10% HS, 50 µM β-mercaptoethanol, 1% pyruvic acid). Rat pheochromocytoma PC12 cells were cultured in DMEM supplemented with 10% HS, and 5% FBS. JEG-3 cells were cultured in DMEM with 10% FBS as recommended by ATCC.
RNA isolation and PCR analysis
Primers were designed using MacVector (version 7.2.3 from Accelyrs, San Diego, CA, USA) and Primer-3 software (http://workbench.sdsc.edu from University of California, San Diego, USA). Primer pairs and amplicon sizes are shown in table 1. Total RNA was isolated from Rcho-1 cells using TRIzol. To synthesize cDNA, 500 ng total RNA was incubated with avian myeloblastosis reverse transcriptase for 30 min at 50°C. Reverse transcriptase was replaced with water as a negative control (−RT). For RT/PCR analysis, cDNA was amplified by PCR using specific primers in the presence of Taq-DNA polymerase. Typical conditions were 30 s at 94°C, 1 min at 57°C and 1 min at 72°C for 35 cycles with a final extension of 7 min at 72°C. The RT-PCR products were separated in 2% agarose gels and visualized with ethidium bromide using UV light. Rat β-actin was used as a control for mRNA loading. Quantitative-PCR was performed using cathepsin P and L and β-actin specific primers designed with Primer-3 software. Real time PCR was performed according to the protocols recommended by iQ SYBR green super mix and results were analyzed using a MyiQ optical system software analysis package (Biorad, Hercules, CA, USA). cDNA was subjected to PCR with each primer pair (95°C for 3 min, 55°C for 45 sec, 40 cycles). Melt curves performed at the end of cycling were used to confirm specific amplification. Threshold cycles (Ct) of cathepsins P and L were normalized to the corresponding value of β-actin and relative abundance is compared to expression levels in undifferentiated cells.
Table 1.
PCR primer list.
| Target | Primers, forward and reverse | Amplicon (bp) |
|---|---|---|
| cathepsin P (qPCR) | TGCAGTTGGTGCGATAGAAG | 110 |
| GGCAGCCATTATTTCCTTCA | ||
| cathepsin L (qPCR) | CAAAGACCGGAACAACCACT | 103 |
| CCTTCGGATGTAGTGTCCGT | ||
| β-actin (qPCR) | AGCCATGTACGTAGCCATCC | 115 |
| ACCCTCATAGATGGGCACAG | ||
| cathepsin P | TGAAGAGTTGAGTCTGTGGAGGACC | 526 |
| GTTTTTGAGAACATCTGACC | ||
| cathepsin L | TGTGGTTCTTGCTGGGCTTTTAG | 397 |
| AACTGGAGAGACGGATGGCTTG | ||
| cathepsin 1 | GGAGTGGAAGAGGAACAATGCG | 592 |
| TGGGACAACAAAAAAGCGGG | ||
| cathepsin 2 | ATGAAGGTCAATGCCTTTGG | 376 |
| TTGACTCAGCCTCCAGACCT | ||
| cathepsin M | CCTGGGCTGTTATTTAGGCA | 603 |
| ACAACTGGCCTTCCAATGAC | ||
| cathepsin R | CAGGACCTTGGGACATGACT | 603 |
| TGCACACTCAGAGGGATGAG | ||
| placental lactogen | CTCTGAAACACTTGGTCGTCTGC | 247 |
| CGGCACAGGTTACAAATGGC | ||
| β-actin | ACCCTCATAGATGGGCACAG | 600 |
| AGCCATGTACGTAGCCATCC | ||
| Mash2 | CTCGAGGACTTGTCGAGGAC | 528 |
| GCAGCGTCTCCACCTTACTC | ||
| Hand1 | CCTCAAGTCCGCAGGTC | 252 |
| ACCAAAGAGGAAGGGTTCGT | ||
| Stra13 | AACTTACAAATTGCCGCACC | 560 |
| ACTGGCACACAGTTTTTCCC |
Immunostaining
Immunofluorescent staining to detect cathepsin P expression in fixed Rcho-1 cells was used to determine the subcellular distribution of the enzyme before and after differentiation using techniques described previously [18]. Briefly, cells were grown to confluence in 4-chambered glass slides. Media was removed and cells were washed thoroughly with PBS prior to fixing in methanol: acetone (1:1 ratio) at −20°C for 5 min. Fixed cells were permeabilized by adding 0.25% Triton 100-X for 15 min and then washing with PBS to eliminate any trace of detergent. After permeabilization, non-specific binding of IgG was blocked by incubating the slides in PBS containing 5% horse serum and 5% donkey serum (blocking solution) for 1 h at 25°C. Rabbit anti-mouse cathepsin P antibody (1:300 in blocking solution) [19] was then added to the slides and incubated at 4°C for 24 h. Texas red-labeled mouse anti-rabbit IgG antibody (1:500 dilution) was used to visualize the primary antibody. Sheep anti-human cathepsin B antibody [17](1:200 dilution) followed by FITC-labeled donkey anti-sheep IgG antibody (1:500 dilution) was used to identify lysosomal/endosomal compartments in the cells. DAPI stain was used as a nuclear marker. PC12 and JEG3 cells were treated similarly and used as negative controls for cathepsin P. Separate slides from all three cell lines tested were incubated without primary antibodies or with pre-immune serum to control for nonspecific signals. Immunofluorescence was analyzed by standard fluorescent microscopy techniques with a Leica DM RXA2 microscope, and images were analyzed using Openlab 3.13 software (www.improvision.com).
Cathepsin activity assays
Cells were homogenized in 100 mM sodium phosphate, 1% Triton X-100, 1 mM EDTA, and 1 mM DTT, pH 6.5. Homogenates were then centrifuged at 15000 g for 5 min and supernatants harvested for protein determination and enzyme assay. Protein was determined using a Biorad protein assay. Aliquots of extracted proteins were pre-incubated for 20 min at 37°C in the presence or absence of protease inhibitors; 1 µM E-64 to inactivate cathepsins B and L, or 1 µM CtP-I (cathepsin P specific inhibitor) to inactivate cathepsin P (M. Hassanein, C.T. Seto, and R.W. Mason unpublished data). MOCAc-Pro-Leu-Gly-Leu-Dap(Dnp)-Ala-Arg-NH2 (1 µM) was then added and incubation continued for 30 min [19]. Appearance of fluorescent products was detected continuously (excitationmax = 325 nm and emissionmax = 400 nm) in a Perkin Elmer Wallac 1420 plate reader. Proteins extracted from adult and embryonic tissues were assayed for cathepsin P activity similarly. Cathepsin B and L activities were assayed in 100 mM sodium acetate, 1% Triton X-100, 1 mM EDTA, and 1 mM DTT, pH 5.5 using Cbz-Phe-Arg-N-methylcoumarin (5 µM) as described previously [20].
RESULTS
Cathepsin P expression is induced during differentiation of Rcho-1 cells into a trophoblast giant cell phenotype
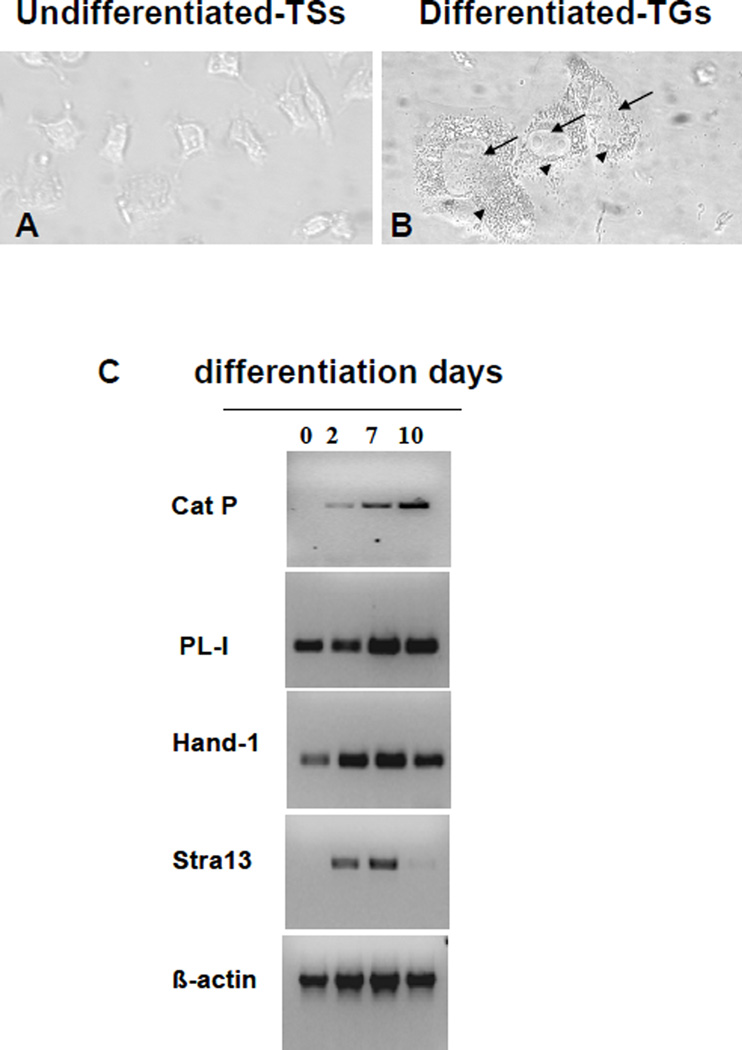
Undifferentiated Rcho-1 cells share characteristics of proliferative trophoblast stem cells from ectoplacental cone [21]. These cells were induced to differentiate to resemble trophoblast giant cells [15, 22–24]. The morphological differences between undifferentiated and differentiated cells are clearly shown in fig 1. While the stem cells appear as angular shaped cells (fig 1A), the differentiated cells exhibit large nuclei with multiple nucleoli and a highly vesicularized cytoplasm (fig 1B). Numbers of cells with this phenotype increased during culture in HS and by 10 days most of the remaining cells showed this appearance. Reverse transcription polymerase chain reaction analyses were performed to determine the relationship between expression of PECs and markers of trophoblast giant cells during differentiation of these cells. PL-1, a glycopeptide hormone that is expressed by trophoblast giant cells, was elevated during differentiation, consistent with previous studies with these cells (fig1C) [25, 26]. The basic helix-loop-helix transcription factors Hand1 and Stra13 showed a similar pattern of expression. After 10 days in differentiation media, cells begin to die, showing reduced levels of Hand1 and Stra13 as well as control β-actin (fig 1C). These results are consistent with previous studies that have shown induction of these transcription factors during differentiation of mouse trophoblast stem cells into trophoblast giant cells [14, 27–29].
Figure 1. Cathepsin P expression is elevated in cells with a trophoblast giant cell phenotype.
Phase contrast images of undifferentiated cells (panel A) and differentiated Rcho-1 cells (10 days, panel B) show morphological differences between the cells. In panel B, arrows point to the enlarged nucleus and arrow heads point to the highly vesicularized cytoplasm of the differentiated cells. Total RNA was isolated from Rcho-1 cells before (0 days) and after differentiation (days 2, 7 and 10) into giant cells. RT/PCR was performed to evaluate expression of cathepsin P and genes characteristic of TGs (PL-1, Hand1 and Stra13). β-Actin expression was examined as a loading control (panel C).
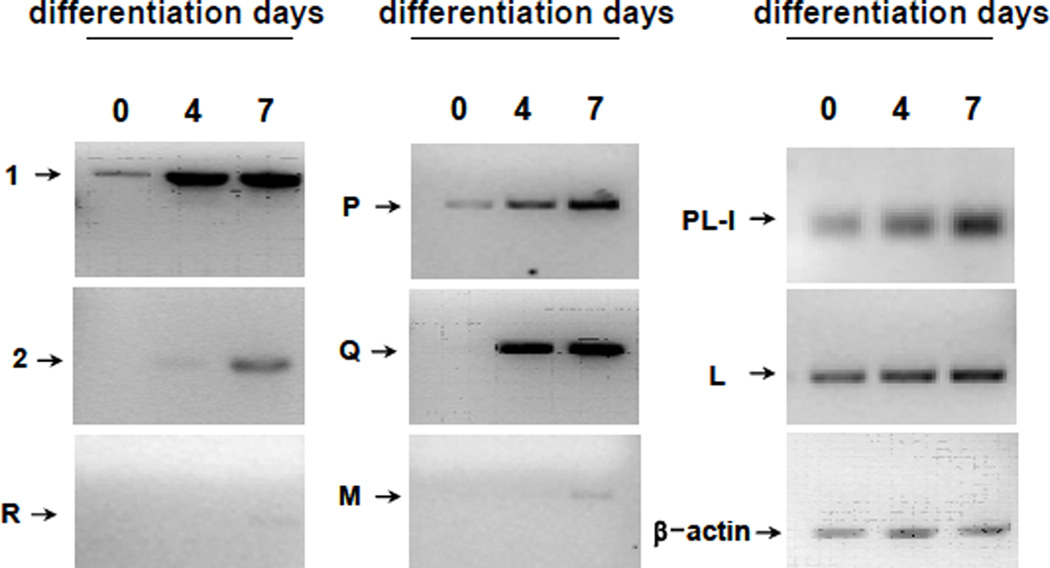
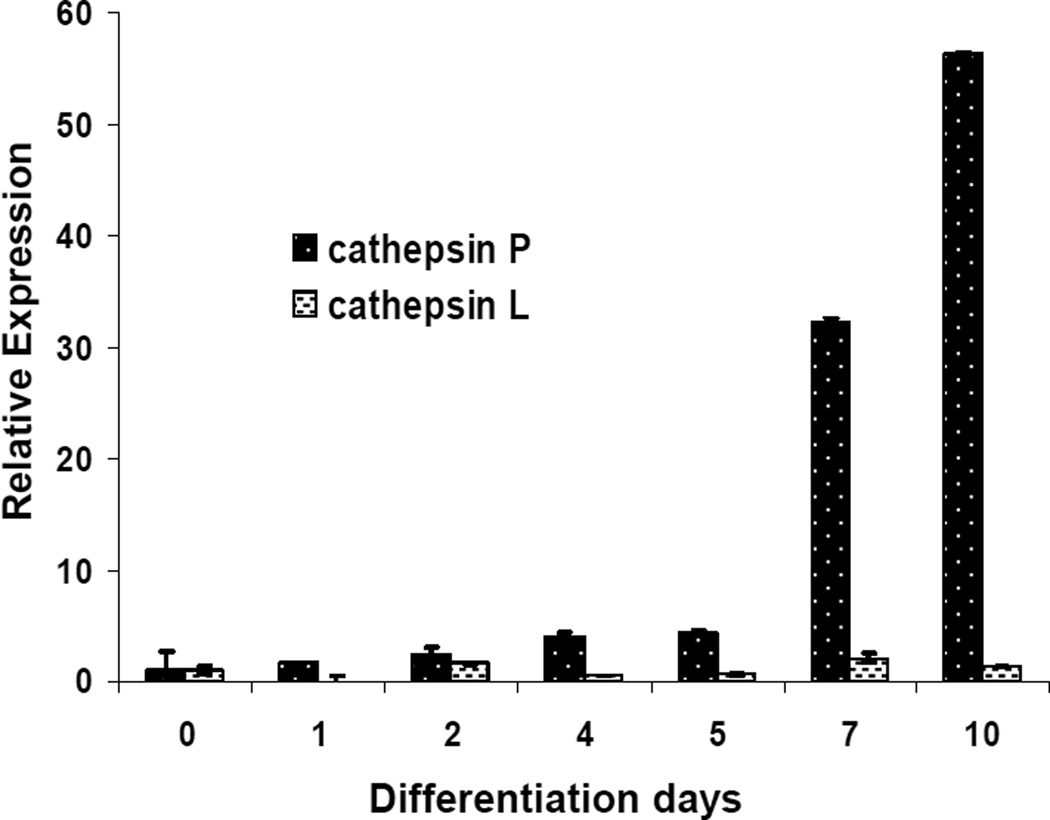
Only low levels of cathepsin P mRNA were detected in undifferentiated cells but levels of expression increased during differentiation (fig 1C). Cathepsins M, Q, R, 1, and 2 were also shown to be up-regulated to varying degrees during differentiation of Rcho-1 cells towards a giant cell phenotype (Fig 2). Up-regulation of cathepsins P, Q and 1 were most pronounced while only low levels of cathepsins 2, M, and R could be detected. By contrast, levels of cathepsin L were regulated to a lesser extent (fig 2). Quantitative analysis using real time-PCR demonstrated that cathepsin P message levels increased approximately 50 fold after 10 days of differentiation whereas there was little induction of expression of cathepsin L (fig 3). These results point to potentially important roles of the PECs in trophoblast giant cells.
Figure 2. Expression of several PECs increase during Rcho-1 differentiation.
Total RNA was isolated from Rcho-1 cells before (0 days) and after differentiation (days 4 and 7) into giant cells. RT/PCR was performed to evaluate expression of cathepsins L, M, P, Q, R, 1 and 2. Placental lactogen-I was analyzed as a marker of differentiation and β-Actin expression was examined as a loading control.
Figure 3. Real Time PCR quantitation of expression of cathepsins P and L during differentiation of trophoblast cells.
Total RNA was extracted from undifferentiated cells and differentiated cells (days 1, 2, 4, 7 and 10). Ct values were obtained for cathepsins P and L and β-actin. Changes in expression of cathepsins P and L relative to β-actin are shown, normalized to expression levels in undifferentiated cells. Error bars indicate standard deviations (3 samples each) of a representative experiment that was repeated three times.
Cathepsin P activity in Rcho-1 cells
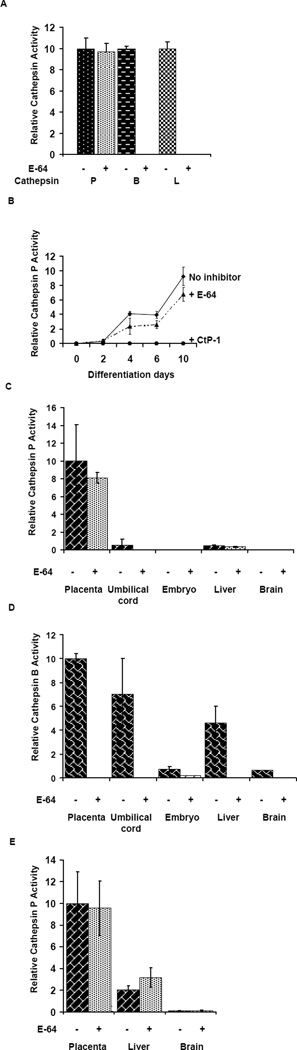
To measure cathepsin P activity in crude tissue extracts, we have developed a specific assay for this enzyme. Recombinant cathepsin P hydrolyzes the fluorogenic substrate, MOCAc-Pro-Leu-Gly-Leu-A2pr(Dnp)-Ala-Arg-NH2 at pH 6.5 [19] but activity was not inhibited by pre-incubation with the general cysteine protease inhibitor, E-64 (1 µM, fig4A). This inhibitor was previously shown to be a very slow binding inhibitor of cathepsin P [19]. By contrast, cathepsin B, and L exhibited little activity against this cathepsin P substrate and activities against Cbz-Phe-Arg-N-methylcoumarin were completely inhibited by E-64 (fig 4A). These results demonstrate that MOCAc-Pro-Leu-Gly-Leu-A2pr(Dnp)-Ala-Arg-NH2 may be used to monitor activity of cathepsin P in tissue extracts. Cathepsin P activity was assayed in crude protein extracts of Rcho-1 trophoblast stem cells (undifferentiated cells) and Rcho-1 cells induced to differentiate for up to 10 days. Specific activity of extracts increased during differentiation of Rcho-1 cells, consistent with the increased expression of mRNA for this gene (fig 4B). Activity against this substrate in Rcho-1 extracts was not blocked by E-64, but was blocked by CtP-I, a cathepsin P specific inhibitor (Hassanein, M Christopher T. Seto and R W. Mason unpublished data).
Figure 4. Activity of cathepsin P in cells and tissues.
Recombinant cathepsin P was assayed using MOCAc-Pro-Leu-Gly-Leu-Dap(Dnp)-Ala-Arg-NH2 in the presence or absence of E-64 (panel A). For comparison, cathepsin B and L activity were assayed using Cbz-Phe-Arg-N-methylcoumarin in the presence and absence of E-64 using pure enzymes (panel A). Activities are normalized to activity in the absence of inhibitor. Rcho-1 cells at different times after differentiation were homogenized in activity buffer and cathepsin P activity was determined in the absence (♦) and presence of E-64 (▲) and CtP-I (●) (panel B). Cathepsin P and B activities were measured in tissues from pregnant mice (panels C and D) and pregnant rats (panel E). In panels B, C, D and E, activities were calculated per mg protein and are normalized to tissues that exhibited maximal activity. Error bars indicate standard deviations and are derived from at least 3 separate samples.
Cathepsin P activity in tissues
MOCAc-Pro-Leu-Gly-Leu-A2pr(Dnp)-Ala-Arg-NH2 was also used to measure cathepsin P activity in mouse tissue extracts (fig 4C). The highest specific activity was found in placenta, consistent with the known tissue distribution of cathepsin P mRNA. By contrast, cathepsin B activity was found in all tissues, although placenta is shown to be the richest source of cathepsin B activity (fig 4D). The cathepsin B activity was inhibited completely by E-64. Cathepsin P activity could also be measured in rat placenta using the same assay conditions (fig 4E). Some activity against this substrate was found in liver from both rodents and this activity is probably due to another protease that is not cathepsins P, B or L.
Cathepsin P is a lysosomal/endosomal enzyme
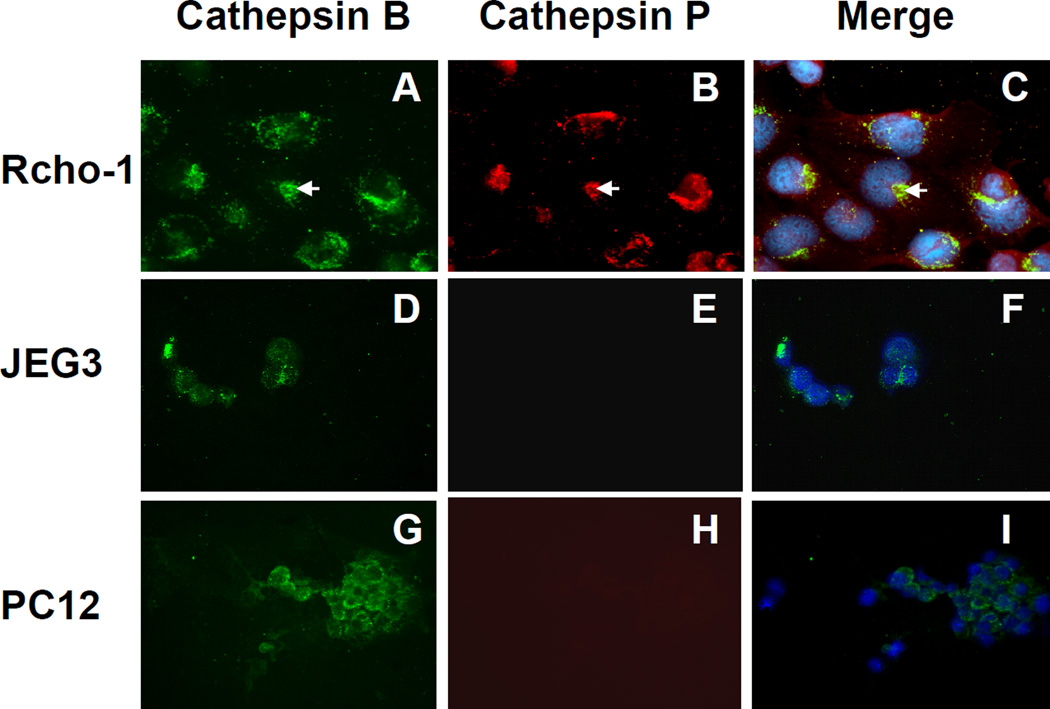
The sequence homology of cathepsin P to other well-characterized cathepsins indicates that it is likely to be a lysosomal enzyme. To determine the subcellular distribution of cathepsin P in Rcho-1 trophoblasts we compared its localization with the well known lysosomal/endosomal enzyme, cathepsin B. Cathepsin P was shown to be expressed in Rcho-1 trophoblast cells (fig 5, panel B) but not in PC12 (rat pheochromocytoma) (fig 5, panel E) or JEG3 (human trophoblasts) (fig 5, panel H). PC12 and JEG3 cells serve as valuable controls to show specificity of the antibody for proteins expressed in rodent trophoblasts. The sheep anti-human cathepsin B antibody showed peri-nuclear expression of cathepsin B in all three cell types, consistent with its known lysosomal location (fig 5, panels A, D, and G). The merged images show widespread co-localization of cathepsin P with cathepsin B (fig 5, panels C, F and I). Some non-overlapping regions indicating possible additional cellular locations of cathepsin P in the Rcho-1 cells are also apparent (red in fig 5, panel C).
Figure 5. Cellular localization of cathepsins B and P in Rcho-1 cells.
Rcho-1 (A–C), JEG3 (D–F), and PC12 (G–I) cells were fixed, permeabilized, and incubated with rabbit anti-cathepsin P and sheep anti-cathepsin B specific antibodies. Species-specific fluorescent antibodies (Texas red mouse anti-rabbit IgG antibody and FITC donkey anti-sheep IgG antibody) were used to identify the primary IgGs. Cathepsin B is shown as green (A, D, and G) and cathepsin P as red (B, E, and H). Merged images are shown in C, F, and I, with nuclei stained blue with DAPI.
DISCUSSION
Rcho-1 cells have become one of the most common culture models for studying placental development at the cellular level in rodents [16]. Many studies have validated this cell model for studying aspects of trophoblast biology including cell cycle regulation, cell differentiation regulation, trophoblast phenotypic characterization, cell invasion, and trophoblast tumor development [14, 16, 24, 30–35]. Differentiation of Rcho-1 cells to yield cells with a trophoblast giant cell phenotype helps overcome the inherent difficulties in obtaining purified populations of these cells from tissues [16]. Although the ubiquitously expressed cysteine protease, cathepsin L, has been proposed to play an important role in giant cell mediated implantation and invasion [4], this enzyme was not significantly up regulated on differentiation of Rcho-1 cells. In this study we clearly show that expression of cathepsin P correlates with the differentiation of Rcho-1 cells into a cell that resembles a trophoblast giant cell. Immunofluorescent detection of cathepsin P in Rcho-1 cells but not in the non-placental rat cell line, PC12, or the human trophoblast cell line, JEG3, confirms the tissue and species-specific expression of this enzyme.
The majority of studies on mRNA expression of the PECS have focused on elevated expression in term placenta [10–13, 36] with little information of expression in TGs. Re-examination of earlier in situ hybridization data does reveal the location of cathepsin P mRNA in a sub-population of cells on the apical surface of mouse ectoplacental cone, even though expression is higher later in gestation in the labyrinthine layer of the mature placenta [11, 12]. RT/PCR has also shown expression of cathepsin P mRNA in mouse ectoplacental cone, and low levels of cathepsin P mRNA have been detected in the visceral yolk sac [8]. The apical portion of ectoplacental cone comprises a population of trophoblast stem cells that give a rise to secondary trophoblast giant cells that are responsible for bringing the maternal blood to the implantation site by invading through the decidual portion of uterus until reaching the spiral arties [37–41]. Results from the present study indicate that the PECs may have evolved to perform important functions in trophoblast cells at the fetal- maternal interface.
The cathepsin P-specific activity assay was developed from studies with the recombinant mouse enzyme [19]. In the present study we used this assay to provide the first evidence that a PEC gene can produce a functional protease in rodent cells and tissues. Cathepsin P activity showed a direct correlation with the increase in cathepsin P mRNA expression during differentiation of Rcho-1 cells. This cell line provides a valuable source of trophoblast giant cells that are not readily obtainable from primary tissues in sufficient quantities to determine specific enzyme activities. Cathepsin P activity was also detected in protein extracts from mature placenta, consistent with the high expression of mRNA for this protein at later stages of gestation [8]. The assay has now been shown to be able to measure cathepsin P activity in mouse and rat placental tissues, a rat placental cell line and recombinant mouse enzyme.
The similarities between human and rodent placenta and the feasibility of genetic manipulation of rodents have resulted in rats and mice to be widely used to dissect molecular pathways that control placental development and function. Evolutionarily, the placenta is a young organ and must adopt enzymes that originally evolved to perform functions in other organs. These enzymes may not be optimal for functions that are unique to placenta. The duplications that gave rise to the PEC genes may have enabled them to acquire new functions that are unique to rodents, allowing adaptation to environmental challenges. It is proposed that the evolution of a family of prolactin genes may have enabled rodents to adapt to oxidative stress [42].
Alternatively, gene duplications may have allowed the PECs to evolve more specific functions that are performed by a related enzyme such as cathepsin L in other mammals. Elevated levels of expression of cathepsin L may fulfill functions of the PECs in human placenta [43]. A similar situation may have evolved in a primate-specific duplication of cathepsin L that gave rise to the thymus-specific expression of human cathepsin V [44]. The thymus is the richest source of mouse cathepsin L. While evolutionary pressure to maintain both placental and adult functions of cathepsin L in humans would prevent this enzyme from evolving into a more specific enzyme, the gene duplications that resulted in tissue-specific enzymes would relieve these constraints and allow the PECs to evolve more specific functions. The PECs may prove to be valuable tools to define placental functions of human cathepsin L.
ACKNOWLEDGMENTS
We would like to thank J.Q. Zheng for help in setting up the immunohistochemistry and our colleagues for many valuable discussions. This work was supported by Nemours Research Programs and the Center for Pediatric Research (P20RR20173).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nakanishi T, Ozaki Y, Blomgren K, Tateyama H, Sugiura-Ogasawara M, Suzumori K. Role of cathepsins and cystatins in patients with recurrent miscarriage. Mol. Hum. Reprod. 2005;11:351–355. doi: 10.1093/molehr/gah172. [DOI] [PubMed] [Google Scholar]
- 2.Varanou A, Withington SL, Lakasing L, Williamson C, Burton GJ, Hemberger M. The importance of cysteine cathepsin proteases for placental development. J Mol Med. 2006;84:305–317. doi: 10.1007/s00109-005-0032-2. [DOI] [PubMed] [Google Scholar]
- 3.Freeman SJ, Lloyd JB. Inhibition of proteolysis in rat yolk sac as a cause of teratogenesis. Effects of leupeptin in vitro and in vivo. J.Embryol.Exp.Morph. 1983;78:183–193. [PubMed] [Google Scholar]
- 4.Afonso S, Romagnano L, Babiarz B. The expression and function of cystatin C and cathepsin B and cathepsin L during mouse embryo implantation and placentation. Development. 1997;124:3415–3425. doi: 10.1242/dev.124.17.3415. [DOI] [PubMed] [Google Scholar]
- 5.Ambroso JL, Harris C. In vitro embryotoxicity of the cysteine proteinase inhibitors benzyloxycarbonyl-phenylalanine-alanine-diazomethane(Z-Phe-Ala-CHN2) and benzyloxycarbonyl-phenylalanine-phenylalanine-diazomethane (Z-Phe-Phe-CHN2) Teratology. 1994;50:214–228. doi: 10.1002/tera.1420500307. [DOI] [PubMed] [Google Scholar]
- 6.Mishra A, Seshagiri PB. Evidence for the involvement of a species-specific embryonic protease in zona escape of hamster blastocysts. Mol.Hum.Reprod. 2000;6:1005–1012. doi: 10.1093/molehr/6.11.1005. [DOI] [PubMed] [Google Scholar]
- 7.Felbor U, Kessler B, Mothes W, Goebel HH, Ploegh HL, Bronson RT, Olsen BR. Neuronal loss and brain atrophy in mice lacking cathepsins B and L. Proc.Natl.Acad.Sci.U.S.A. 2002;99:7883–7888. doi: 10.1073/pnas.112632299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sol-Church K, Frenck J, Troeber D, Mason RW, Cathepsin P. a novel protease in mouse placenta. Biochem.J. 1999;343:307–309. [PMC free article] [PubMed] [Google Scholar]
- 9.Sol-Church K, Picerno GN, Stabley DL, Frenck J, Xing S, Bertenshaw GP, Mason RW. Evolution of placentally expressed cathepsins. Biochem.Biophys.Res.Commun. 2002;293:23–29. doi: 10.1016/S0006-291X(02)00167-5. [DOI] [PubMed] [Google Scholar]
- 10.Deussing J, Kouadio M, Rehman S, Werber I, Schwinde A, Peters C. Identification and Characterization of a Dense Cluster of Placenta- Specific Cysteine Peptidase Genes and Related Genes on Mouse Chromosome 13. Genomics. 2002;79:225–240. doi: 10.1006/geno.2002.6696. [DOI] [PubMed] [Google Scholar]
- 11.Hemberger M, Himmelbauer H, Ruschmann J, Zeitz C, Fundele R. cDNA subtraction cloning reveals novel genes whose temporal and spatial expression indicates association with trophoblast invasion. Dev.Biol. 2000;222:158–169. doi: 10.1006/dbio.2000.9705. [DOI] [PubMed] [Google Scholar]
- 12.Nakajima A, Kataoka K, Takata Y, Huh NH. Cathepsin-6, a novel cysteine proteinase showing homology with and co- localized expression with cathepsin J/P in the labyrinthine layer of mouse placenta. Biochem.J. 2000;349:689–692. doi: 10.1042/bj3490689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemberger M, Cross JC, Ropers HH, Lehrach H, Fundele R, Himmelbauer H. UniGene cDNA array-based monitoring of transcriptome changes during mouse placental development. Proc.Natl.Acad.Sci.U.S.A. 2001;98:13126–13131. doi: 10.1073/pnas.231396598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grummer R, Hellmann P, Traub O, Soares MJ, el-Sabban ME, Winterhager E. Regulation of connexin31 gene expression upon retinoic acid treatment in rat choriocarcinoma cells. Exp Cell Res. 1996;227:23–32. doi: 10.1006/excr.1996.0245. [DOI] [PubMed] [Google Scholar]
- 15.Faria TN, Soares MJ. Trophoblast cell differentiation: establishment, characterization, and modulation of a rat trophoblast cell line expressing members of the placental prolactin family. Endocrinology. 1991;129:2895–2906. doi: 10.1210/endo-129-6-2895. [DOI] [PubMed] [Google Scholar]
- 16.Sahgal N, Canham LN, Canham B, Soares MJ. Rcho-1 trophoblast stem cells: a model system for studying trophoblast cell differentiation. Methods Mol Med. 2006;121:159–178. [PubMed] [Google Scholar]
- 17.Buttle DJ, Bonner BC, Burnett D, Barrett AJ. A catalytically active high molecular weight form of human cathepsin B from sputum. Biochem.J. 1988;254:693–699. doi: 10.1042/bj2540693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng JQ, Kelly TK, Chang B, Ryazantsev S, Rajasekaran AK, Martin KC, Twiss JL. A functional role for intra-axonal protein synthesis during axonal regeneration from adult sensory neurons. J Neurosci. 2001;21:9291–9303. doi: 10.1523/JNEUROSCI.21-23-09291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mason RW, Bergman CA, Lu G, Frenck Holbrook J, Sol-Church K. Expression and characterization of cathepsin P. Biochem. J. 2004;378:657–663. doi: 10.1042/BJ20031548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mason RW, Barrett AJ. Properties of human liver cathepsin L. Progress.in Clinical.& Biological.Research. 1985;180:217–219. [PubMed] [Google Scholar]
- 21.Verstuyf A, Fonteyn E, Sobis H, Vandeputte M. A rat cytotrophoblast antigen defined by a monoclonal antibody. Am J Reprod Immunol. 1992;28:6–11. doi: 10.1111/j.1600-0897.1992.tb00749.x. [DOI] [PubMed] [Google Scholar]
- 22.Lu XJ, Deb S, Soares MJ. Spontaneous differentiation of trophoblast cells along the spongiotrophoblast cell pathway: expression of members of the placental prolactin gene family and modulation by retinoic acid. Dev. Biol. 1994;163:86–97. doi: 10.1006/dbio.1994.1125. [DOI] [PubMed] [Google Scholar]
- 23.Peters TJ, Chapman BM, Soares MJ. Trophoblast differentiation. An in vitro model for trophoblast giant cell development. Methods Mol.Biol. 2000;137:301–311. doi: 10.1385/1-59259-066-7:301. [DOI] [PubMed] [Google Scholar]
- 24.Peters TJ, Albieri A, Bevilacqua E, Chapman BM, Crane LH, Hamlin GP, Seiki M, Soares MJ. Differentiation-dependent expression of gelatinase B/matrix metalloproteinase-9 in trophoblast cells. Cell Tissue Res. 1999;295:287–296. doi: 10.1007/s004410051235. [DOI] [PubMed] [Google Scholar]
- 25.Shida MM, Jackson-Grusby LL, Ross SR, Linzer DI. Placental-specific expression from the mouse placental lactogen II gene promoter. Proc.Natl.Acad.Sci.U.S.A. 1992;89:3864–3868. doi: 10.1073/pnas.89.9.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamlin GP, Lu XJ, Roby KF, Soares MJ. Recapitulation of the pathway for trophoblast giant cell differentiation in vitro: stage-specific expression of members of the prolactin gene family. Endocrinology. 1994;134:2390–2396. doi: 10.1210/endo.134.6.8194465. [DOI] [PubMed] [Google Scholar]
- 27.Cross JC, Anson-Cartwright L, Scott IC. Transcription factors underlying the development and endocrine functions of the placenta. Recent Prog. Horm. Res. 2002;57:221–234. doi: 10.1210/rp.57.1.221. [DOI] [PubMed] [Google Scholar]
- 28.Loregger T, Pollheimer J, Knofler M. Regulatory transcription factors controlling function and differentiation of human trophoblast--a review. Placenta. 2003;24(Suppl A):S104–S110. doi: 10.1053/plac.2002.0929. [DOI] [PubMed] [Google Scholar]
- 29.Cross JC, Baczyk D, Dobric N, Hemberger M, Hughes M, Simmons DG, Yamamoto H, Kingdom JC. Genes, development and evolution of the placenta. Placenta. 2003;24:123–130. doi: 10.1053/plac.2002.0887. [DOI] [PubMed] [Google Scholar]
- 30.Verstuyf A, Sobis H, Goebels J, Fonteyn E, Cassiman JJ, Vandeputte M. Establishment and characterization of a continuous in vitro line from a rat choriocarcinoma. Int J Cancer. 1990;45:752–756. doi: 10.1002/ijc.2910450430. [DOI] [PubMed] [Google Scholar]
- 31.Verstuyf A, Goebels J, Sobis H, Vandeputte M. Influence of different growth factors on a rat choriocarcinoma cell line. Tumour Biol. 1993;14:46–54. doi: 10.1159/000217824. [DOI] [PubMed] [Google Scholar]
- 32.Verstuyf A, Sobis H, Vandeputte M. Morphological and immunological characteristics of a rat choriocarcinoma. Int J Cancer. 1989;44:879–884. doi: 10.1002/ijc.2910440522. [DOI] [PubMed] [Google Scholar]
- 33.Basyuk E, Cross JC, Corbin J, Nakayama H, Hunter P, Nait-Oumesmar B, Lazzarini RA. Murine Gcm1 gene is expressed in a subset of placental trophoblast cells. Dev.Dyn. 1999;214:303–311. doi: 10.1002/(SICI)1097-0177(199904)214:4<303::AID-AJA3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 34.Yamaguchi M, Murakami T, Yasui Y, Otani S, Kawai M, Kishi K, Kurachi H, Shima K, Aono T, Murata Y. Mouse placental cells secrete soluble leptin receptor (sOB-R): cAMP inhibits sOB-R production. Biochem.Biophys.Res.Commun. 1998;252:363–367. doi: 10.1006/bbrc.1998.9636. [DOI] [PubMed] [Google Scholar]
- 35.Yamaguchi M, Ogren L, Endo H, Thordarson G, Bigsby RM, Talamantes F. Production of mouse placental lactogen-I and placental lactogen-II by the same giant cell. Endocrinology. 1992;131:1595–1602. doi: 10.1210/endo.131.4.1396305. [DOI] [PubMed] [Google Scholar]
- 36.Sol-Church K, Shipley J, Beckman DA, Mason RW. Expression of cysteine proteases in extraembryonic tissues during mouse embryogenesis. Arch.Biochem.Biophys. 1999;372:375–381. doi: 10.1006/abbi.1999.1520. [DOI] [PubMed] [Google Scholar]
- 37.Cross JC, Werb Z, Fisher SJ. Implantation and the placenta: key pieces of the development puzzle. Science. 1994;266:1508–1518. doi: 10.1126/science.7985020. [DOI] [PubMed] [Google Scholar]
- 38.Cross JC. Genetic insights into trophoblast differentiation and placental morphogenesis. Semin Cell Dev Biol. 2000;11:105–113. doi: 10.1006/scdb.2000.0156. [DOI] [PubMed] [Google Scholar]
- 39.Carney EW, Prideaux V, Lye SJ, Rossant J. Progressive expression of trophoblast-specific genes during formation of mouse trophoblast giant cells in vitro. Mol Reprod Dev. 1993;34:357–368. doi: 10.1002/mrd.1080340403. [DOI] [PubMed] [Google Scholar]
- 40.Zybina TG, Zybina EV. Genome multiplication in the tertiary giant trophoblast cells in the course of their endovascular and interstitial invasion into the rat placenta decidua basalis. Early Pregnancy. 2000;4:99–109. [PubMed] [Google Scholar]
- 41.Zybina EV, Zybina TG, Stein GI. Trophoblast cell invasiveness and capability for the cell and genome reproduction in rat placenta. Early Pregnancy. 2000;4:39–57. [PubMed] [Google Scholar]
- 42.Ain R, Dai G, Dunmore JH, Godwin AR, Soares MJ. A prolactin family paralog regulates reproductive adaptations to a physiological stressor. Proc Natl Acad Sci U S A. 2004;101:16543–16548. doi: 10.1073/pnas.0406185101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bromme D, Li ZQ, Barnes M, Mehler E. Human cathepsin V functional expression, tissue distribution, electrostatic surface potential, enzymatic characterization, and chromosomal localization. Biochemistry. 1999;38:2377–2385. doi: 10.1021/bi982175f. [DOI] [PubMed] [Google Scholar]