Abstract
Each year about 1.4 million people die from lung cancer worldwide. Despite efforts in prevention, diagnosis and treatment, survival rate remains poor for this disease. This unfortunate situation is largely due to the fact that a high proportion of cases are diagnosed at advanced stages, highlighting the great need for identifying new biomarkers in order to improve early diagnosis and treatment. Recent studies on microRNAs have not only shed light on their involvement in tumor development and progression, but also suggested their potential utility as biomarkers for subtype diagnostics, staging and prediction of treatment response. This review article summarizes the impact of microRNAs on lung cancer biology, and highlights their role in the detection and classification of lung cancer as well as direct targets for drug development.
Role Of MicroRNAs In Lung Cancer
Lung cancer is the leading cause of cancer-related death worldwide [1]. Tobacco smoke is unquestionably the main etiological agent, although ~25% of cases occur in never smokers, and are mostly associated with environmental carcinogens [2]. Lung cancer patients are often diagnosed at advanced stages and despite efforts made, the mortality remain high, with the five-year survival rate around 15% [3]. However, when detected at early stages, patient outcomes are substantially improved [4]. In this context, search for new biomarkers is necessary in order to improve early diagnosis.
Based on histopathologic features, lung cancer is classified as small cell lung cancer (SCLC) or non-small cell lung cancer (NSCLC), with lung squamous cell carcinomas (SqCC) and adenocarcinomas (AdC) being the major histological subtypes of NSCLC [5–9]. Both clinical and molecular characteristics of these subtypes have been associated with different patterns of genetic and epigenetic aberrations, including changes in microRNA (miRNA) expression profiles [10, 11].
MicroRNAs are small non-coding RNAs (~21–25 nucleotides in length) that interact with homologous mRNAs and regulate gene expression at the post-transcriptional level [12]. In animals, miRNAs act mainly through inhibition of the translation process, while in plants the literature indicates the major mechanism of action is enzymatic cleavage of target mRNAs [13, 14]. MiRNA genes are transcribed as long precursors called “pri-miRNA”. These precursors are cleaved in the nucleus by a complex called “microprocessor”, which contains the enzymes RNASEN (Drosha) and DiGeorge critical region 8 (DGCR8). The processed intermediate “pre-miRNA” is about 70 nucleotides in length, and adopts a hairpin-containing secondary structure through imperfect complementarities of bases between the two halves of its sequence. This pre-miRNA is transported to the cytosol and cleaved by a DICER family enzyme to release a small double stranded “miRNA”. Subsequently, this miRNA interacts with a protein of the Argonaute family (AGO1 or AGO2) to form the RNA-induced Silencing Complex (RISC) containing only the active single strand miRNA. The target mRNA is then loaded into RISC where it is silenced by either AGO2-mediated degradation or AGO1-mediated translational repression [13–15].
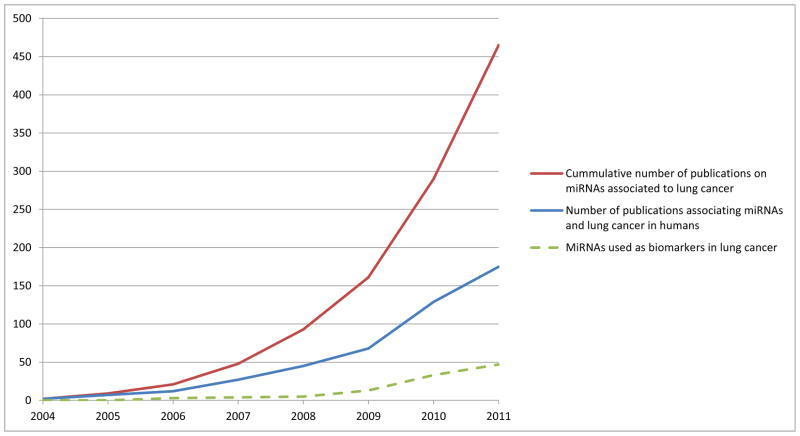
Alterations in miRNA expression have been shown to play major roles in cancer (reviewed in [16]). As miRNAs play important roles in development, cell proliferation and apoptosis, their deregulation has been implicated in cancer initiation and progression, indicating that miRNAs may function as tumor suppressor genes or oncogenes in different cancer types, including lung [13, 17]. Although their potential as biomarkers has been intensively explored, the contribution of miRNAs to lung cancer biology is not completely understood. Moreover, the study of microRNAs in lung cancer is an emerging field. This is evident from the increasing trend of scientific publications on this topic, including their use as biomarkers (Figure 1). In this context, the focus of this review will be the impact of miRNAs on lung cancer biology, and the pros and cons of their role as biomarkers for detection and classification of lung cancer.
Figure 1. Number of publications annotated on PubMed database from the US National Library of Medicine.
Search was performed using the following terms: “miRNA” OR “microRNA” AND “cancer” AND “lung”. Red line represents the cumulative number of entries (between 2004 and 2011). Blue line indicates number of entries per year. Green dotted line indicates the results of the same search with “biomarker” as an additional search term.
Biological Relevance Of MicroRNAs In Lung Cancer
A variety of miRNAs have been shown to play a role in lung cancer initiation and progression (Table 1 and Supplementary Table 1). Furthermore, disruption of the machinery of miRNA biosynthesis has also been documented in lung cancer. For example, reduction in DICER expression has been reported in NSCLC and has been significantly associated with poor prognosis [18]. In a mouse K-Ras-induced lung cancer model, conditional deletion of Dicer enhances lung tumor development [19]. Moreover, miRNAs might also play an important role in early events in lung tumorigenesis, since changes in miRNA expression patterns have been described in premalignant lesions of the bronchial epithelium [20]. Early involvement of miRNAs is also evident in carcinogen-induced lung cancer animal models. Treatment with tobacco carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) in rats resulted in deregulation of several miRNAs, including loss of miR-126* expression. Interestingly, cytochrome P450 2A3 (CYP2A3), which activates NNK carcinogenicity, is a target of miR-126* [21]. Changes in miR-101, miR-126* and miR-199 observed in early tumorigenesis in rats have also been reported to be underexpressed in human NSCLC [22–25]. Next we will discuss the relevance of miRNAs in deregulation of key cellular functions in lung cancer. Some of those major miRNA-regulated pathways are summarized in Figure 2.
Table 1.
Frequently deregulated miRNAs in lung cancer
| miRNA | Expression
|
Confirmed targets | Associated with | Reference | ||||
|---|---|---|---|---|---|---|---|---|
| NSCLC | SCLC | |||||||
|
|
|
|||||||
| Tumors | Lines | Tumors | Lines | Diagnosis | Survival1 | |||
| let-7 | Dn | Dn | UC | HMGA2, RAS, MYC, E2Fs, CDKs | SCC | NSCLC | [24, 25, 54, 65, 77, 78, 80, 81, 105] | |
| miR-7 | Up | Up | ETS2, EGFR, Raf1 | [24, 54] | ||||
| miR-9 | Dn/Up | Up | TGFBI, TRIM2, SIRT1, BTBD3 | SCLC | [24, 25, 54, 106] | |||
| miR-15 | Up | NC | Up | Dn/Up | Bcl-2, CCND1, CCNE | NSCLC | [10, 54, 77–79] | |
| miR-16 | Dn | NC | Dn/Up | Bcl-2, CCND1, CCNE | NSCLC | [10, 24, 54, 65, 79] | ||
| miR-17-3p,5p | Up | Up | Up | p21, E2F, Bim, CCND1, PTEN, Rb | NSCLC | [25, 40, 54, 77, 78, 107] | ||
| miR-17-92 cluster | Up | Up | Up | Up | p21, E2F, CCND1, PTEN, Rb, HIF-1α | SCLC | SCLC | [40, 54] |
| miR-20 | Up | Up | p21, E2F, Bim, Cyclin D1, PTEN, Rb | NSCLC | [40, 54, 77, 78] | |||
| miR-21 | Up | Up | UC | Dn | Apaf-1, PTEN, PDCD4, Faslg, Spry1-2, Btg2 | NSCLC; ADC | NSCLC | [10, 23–25, 43, 44, 54, 64, 65, 77–79, 95, 107, 108] |
| miR-22 | UC | Dn | PTEN, ESR1, TIAM1 | [10, 109, 110] | ||||
| miR-23a,b | Up | NC | Dn | Has2, LaminB1, Brn-3b | NSCLC | [10, 54, 79] | ||
| miR-24 | Up | NC | Dn | E2F2, Myc, Net1A | NSCLC | [10, 25, 79] | ||
| miR-25 | Up | Up | Up | Bim, TRAIL | NSCLC | NSCLC | [54, 60, 78, 111] | |
| miR-27b | Dn | UC | UC | ST14, PPARα | NSCLC | [25, 54, 79] | ||
| miR-29a,b,c | Dn | NC | Dn | Dn | DNMT3 (A&B) | [10, 23, 25, 54] | ||
| miR-30 | Dn | Dn | p53, B-Myb, CTGF | [22–25, 54, 65] | ||||
| miR-30c, d | Dn | Up | Up | Serpine1 | NSCLC | [10, 65, 79, 82, 112] | ||
| miR-31 | Up | Dn | Dn | LATS2, PPP2R2A | ADC | [10, 23, 46, 64, 65] | ||
| miR-32 | Dn | Dn | Bim | [25, 54] | ||||
| miR-33 | Dn | Dn | ABCA1, p53 | [25, 54, 113, 114] | ||||
| miR-34a | Dn/Up | Dn | DLL1, CDK4, Myc, CCND1 | NSCLC | [23, 54, 78, 115, 116] | |||
| miR-92 | UC | Up | Bim | [10, 54, 117] | ||||
| miR-93 | UC | Up | p21, FUS1 | SCLC | [10, 54, 111, 118] | |||
| miR-95 | Dn | Up | Up | SNX1 | SCLC | [25, 54, 119] | ||
| miR-96 | Up | Up | Up | Up | GPC3, FOXO1 | NSCLC | [10, 54, 120] | |
| miR-98 | UC | Up | Up | FUS1 | SCLC | [10, 54, 93, 118] | ||
| miR-99 | Dn | Dn | SMARCA5, SMARCD1, mTOR | [10, 23, 65, 121] | ||||
| miR-100 | Dn | Dn | ATM, PLK1, MTOR, RPTOR, IGF1R, Gli1 | SCC | [23, 54, 77, 78] | |||
| miR-101 | Dn | Up | Up | Mcl-1 | [10, 22–25, 47–49] | |||
| miR-106a | Up | Up | Up | E2F1 | ADC | OS-NSCLC | [10, 54, 77, 78, 111] | |
| miR-106b | Up | Up | p21, E2F, Bim, Cyclin D1, PTEN, Rb | [10, 54, 77, 78, 111] | ||||
| miR-125 | Dn | Dn | Dn | Smo, invasion and migration factors | NSCLC | [23, 25, 54, 65, 77, 122] | ||
| miR-126 | Dn | Dn | Dn | Spred-1 | SCC, ADC | SCC | [22–25, 64, 65, 77, 78] | |
| miR-126* | Dn | ERBB2IP | NSCLC | [23–25, 95] | ||||
| miR-128 | Dn | Up | EGFR | SCLC | EGFR-TKi2 | [45, 54] | ||
| miR-130a | Dn | Dn | MET | NSCLC | [23, 24, 54, 65, 79] | |||
| miR-130b | Up | Up | Up | CSF-1, TP53INP1, RUNX3 | SCC | [54, 64, 65] | ||
| miR-133b | Dn | Dn | Mcl-1, BCLw | [24, 54] | ||||
| miR-139 | Dn | ROCK2, FOXO1, CXCR4 | SCC | [24, 64, 65, 123–125] | ||||
| miR-140 | Dn | Sp1, BMP2 | ADC | [25, 64] | ||||
| miR-142-3p,5p | Up | Dn/Up | ADAM9, GRα, PKA | [10, 54] | ||||
| miR-143 | Dn | Dn | ERK5, KRAS, Bcl-2, DNMT3A, ELK1 | ADC | [23–25, 54, 64] | |||
| miR-145 | Dn | Dn | c-MYC, CDK4, eIF4E4 | NSCLC; ADC | [23, 25, 54, 64, 95] | |||
| miR-146b | Up | Dn | MMP16, TRAF6, IRAK1, | ADC | SCC | [25, 54, 77, 78, 107] | ||
| miR-149 | Up | Up | Up | E-cadherin | SCC | [10, 55, 65] | ||
| miR-150 | Up/Dn | Dn | Dn | MUC4, P2RX7 | ADC | [25, 54, 64] | ||
| miR-155 | Up | Dn | TAB2, FADD, RIP-1, IKKε, ET -1 | ADC | NSCLC | [25, 54, 61, 77, 78, 107] | ||
| miR-181a,b | Dn | Dn | Dn | Mcl-1, Bcl-2 | [23, 54, 65] | |||
| miR-181c | Dn | NC | Up | TGFBI, TRIM2, SIRT1, BTBD3 | [10, 65, 106] | |||
| miR-182 | Up | Up | Up | FOXO1 | NSCLC, ADC | NSCLC | [24, 54, 61, 64, 65, 77, 78, 80, 95, 120] | |
| miR-183 | Up | Up | Up | FOXO3, FOXO1, EGR1, PTEN | NSCLC | NSCLC | [24, 54, 65, 77, 95, 120] | |
| miR-185 | Dn | Dn | Dn | c-Myc, DNMT1, SMG6, CDK6, AKT1 | [54, 126] | |||
| miR-191 | Up | Dn | CDK6, TIMP3, NDST1 | ADC | SCC | [25, 54, 77, 78, 107] | ||
| miR-192 | Up | ERCC3, ERCC4, Rb1 | [25, 107] | |||||
| miR-195 | Dn | NC | Dn/Up | CCNE, BCLw | [10, 24, 54, 65] | |||
| miR-197 | Up | Up | Up | FUS1 | [10, 25, 61] | |||
| miR-199 | Dn | Dn | Dn | Dn | HES, ET-1, HIF-1α | [10, 21, 25, 54] | ||
| miR-200a,b | Dn/Up | Up | Up | C-ets-1, Zeb1, ECM proteins, p38α | ADC | NSCLC | [54, 64, 79, 127] | |
| miR-200c | Up | Dn | Up | Up | C-ets-1, Zeb1, ECM proteins | SCC | NSCLC | [54, 65, 77, 128] |
| miR-203 | Up | . | DeltaNp63, Smo | NSCLC, ADC | [25, 77, 107] | |||
| miR-205 | Up | Dn | Dn | E2F1, Zeb1, Sip1 | SCC; ADC | [10, 25, 65, 74, 75, 78, 93, 107] | ||
| miR-210 | Up | Up | Up | HOXA1, FGFRL1, HOXA9, NDUFA4, EFNA3 | NSCLC, SCC, ADC | [24, 25, 54, 57, 64, 65, 77, 95, 107, 108, 129] | ||
| miR-214 | Up | Dn | Dn | EZH2, Brn-3b | ADC | [25, 54, 107] | ||
| miR-218 | Dn | RICTOR | ADC | [24, 25, 64] | ||||
| miR-221 | Up/UC | Dn | p27, PTEN, TIMP3 | NSCLC | [10, 50, 80] | |||
| miR-222 | Up/UC | Dn | p27, PTEN, TIMP3 | [10, 50, 54] | ||||
| miR-223 | Dn/Up | Dn | Dn | LMO2-L/-S, CEBP-β | NSCLC | [54, 60, 65] | ||
| miR-224 | Dn/Up | Dn | DIO1 | [77] | ||||
| miR-324-5p | Up | Up | Up | Up/Dn | Smo, Gli1 | SCC | [10, 54, 65, 130] | |
| miR-326 | Up | Up | MRP-1/ABCC1, Ets-1 | [10, 131, 132] | ||||
| miR-335 | Dn | Up | Up | SOD2, Txnrd2, BCLw, SP1, ERα, IGF1R | [24, 54, 133–135] | |||
| miR-338 | Dn | Up | Up/Dn | Runx2, Smo | [10, 24, 54, 130, 136] | |||
| miR-375 | Up | Dn | Up | Up | IGF1R, SP1, LDHB, YAP1 | ADC | SCLC | [54, 64, 137–140] |
| miR-423 | Up | p21 | SCC | [24, 65, 141] | ||||
| miR-429 | Dn | Up | ALDH1L2, PSAT1, ZEB1 | SCC | [54, 65, 142] | |||
| miR-451 | Dn | Dn | RAB14, apoptosis | [23, 24, 54] | ||||
| miR-453 | Dn | ESR1 | SCC | [77, 78, 82] | ||||
| miR-486 | Dn | CD40 | ADC | NSCLC | [64, 65, 108] | |||
| miR-494 | Dn | Up | Nucleolin, Bmal1 | SCC | [54, 77, 78, 143, 144] | |||
| miR-497 | Dn | Dn | BCL2, CCND2 | [24, 54, 145]{Xing, 2010 #85 | ||||
| miR-504 | Up | p53, DRD1 | [23, 146, 147] | |||||
| miR-511 | Up | Dn | TLR4, CD80 | SCC | [54, 77, 78, 148] | |||
| miR-512 | Dn | c-FLIP, Mcl-1 | [54, 149, 150] | |||||
Expression trend can be either positively or negatively correlated to overall survival
Associated with drug response and not to overall survival
Abbreviations: Lines = cell lines, SCC = lung squamous cell carcinoma, ADC = lung adenocarcinoma, Up = increased expression, Dn = decreased expression, UC = no change
Figure 2. Main cancer-related cell functions targeted by miRNA in lung cancer.
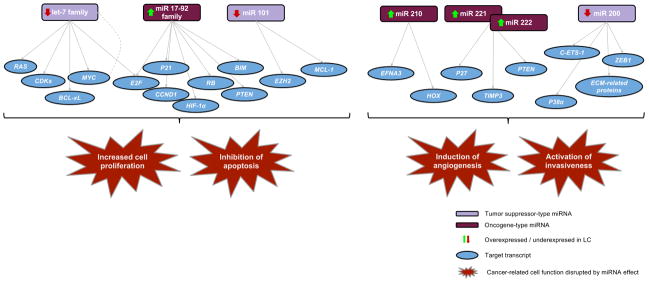
Multiple targets of lung cancer related key miRNAs are evidenced, as well as the variety of downstream effects. Members of the Let-7 and miR17-92 families are involved in disruption of cell cycle and apoptotic processes in lung cancer. The let-7 cluster negatively regulates multiple oncogenes, including Ras and Myc. Myc is thought to negatively regulate let-7 members by binding directly to their promoters (grey dotted line). Over-expression of miR-17-92 family and miR-101 contribute to cell survival by reducing apoptosis. On the other hand, miRNAs are involved in lung cancer progression (mainly through induction of angiogenesis and activation of invasiveness). This is the case of miR-221 and miR-222 that target PTEN and TIMP3 tumor suppressors and enhance cellular migration through activation of the Akt pathway and metallopeptidases; and miR- 210, which is involved in angiogenesis by targeting EFNA3 and the HOX family of transcription factors. The miR-200 cluster restricts metastasis and EMT by targeting E-cadherin transcriptional regulators such as ZEB proteins.
MicroRNAs involved in cell cycle and apoptosis in lung cancer
The let-7 family
The tumor suppressive let-7 family comprises 13 members in the human genome that are dispersed over chromosomes 3, 9, 10, 11, 12, 19, 21, 22, and X [26]. Members are highly expressed in normal lung tissue and negatively regulate multiple oncogenes, including RAS and MYC [27, 28]. MYC is thought to negatively regulate let-7 members by binding directly to their promoters (specifically through the E-box 3 domain), representing a negative-feedback loop between these two elements [26, 29]. Expression of let-7 members also affects cell-cycle regulators, such as cyclins, CDKs and E2F transcription factors, and anti-apoptotic factors, like Bcl-xL [28, 30]. In NSCLC cell lines, inhibition of let-7 leads to increased cell division, while overexpression results on cell growth inhibition [28]. Similar effects have been observed in xenografts and transgenic mouse models [31].
Importantly, pathogenic mechanisms of let-7 disruption in lung cancer are not limited to alterations in expression levels. A single nucleotide polymorphism in let-7 complementary site 6 of the KRAS mRNA 3′-UTR is associated with an increased risk of NSCLC among moderate smokers. The frequency of this variant is ~ 20% in NSCLC patients compared to only 6% in the general population [32]. These findings illustrate that even at biologically normal levels, let-7 function can be impaired by variations in its target mRNAs.
The miR-17-92 family
The oncogenic function of the miR-17-92 cluster on chromosome 13q31.3, also known as oncomir-1, was first demonstrated in a mouse B-cell lymphoma model requiring over-expression of c-MYC [33]. Subsequent studies have further demonstrated MYC-induced expression of miR-17-92, and interestingly, induction of its paralog cluster, miR-106b-25, located on chromosome 7 [34, 35]. In addition to MYC, miR-17-92 expression can be induced by E2F proteins, which in turn are targeted by miR-17-92 members miR-17 and miR-20a, and miR-106b-25 member miR-106b. Through MYC and E2F proteins, the two clusters regulate cell cycle and apoptosis via feedback mechanisms as well as through targeting of a common set of downstream cell cycle regulators and apoptotic factors including BIM, PTEN, RB1, p21, and cyclin-dependent kinases [34–38].
In NSCLC, loss of let-7 and up-regulation of MYC are frequent events, and together can lead to an increase in miR-17-92 levels and excessive activation of E2F proteins. Excessive levels of E2F1 can induce cell cycle inhibition and apoptosis [39], yet in cancer, the net effect of this signalling network is the overall enhanced activity of E2F transcription factor activity and subsequent cell proliferation [36]. Clearly, the miR-17-92 cluster plays a key role in the cancer promoting function of E2F upregulation to promote proliferation and evade apoptosis.
Both SCLC and NSCLC exhibit over-expression of the miR-17-92 cluster, although the effects of this overexpression are perhaps more pronounced in SCLC [40, 41]. In SCLC cell lines, over-expression of miR-17-92 was shown to influence genomic instability upon RB inactivation. It was observed that RB inactivation induced γ-H2AX foci formation, a marker of DNA damage, as well as growth inhibition and generation of reactive oxygen species, effects that were offset by miR-17-92 overexpression [40]. It is possible that in an RB inactivated model of SCLC, miR-17-92 allows cell tolerance of DNA damage and increased genomic instability.
Other miRNAs
Other miRNAs have also been observed to be involved in cell cycle and apoptosis regulation. This is the case for miR-15 and miR-16, which are transcriptional targets of E2F1 and limit E2F-induced excessive proliferation by targeting CYCLIN E [42]. Another example is miR-21, which is one of the most frequently over-expressed miRNAs in NSCLC. This miRNA is up-regulated by activation of EGFR and RAS pathways, and it is implicated in lung tumorigenesis in never smokers with KRAS and EGFR activating mutations and amplifications [43, 44]. Over-expression of miR-21 enhances tumorigenesis by targeting SPRY1, SPRY2, BTG2 and PDCD4, which act as negative regulators of the RAS/MEK/ERK pathway, and APAF-1, FASLG, PDCD4 and RHOB, which promote apoptosis [43]. Conversely, the anti-tumoral miR-128b, which directly targets EGFR, is frequently under-expressed in NSCLC patients [45].
MiR-31 has been shown to be up-regulated in murine lung cancers relative to adjacent normal lung tissues and in primary SqCC [23]. It may function as an oncomir by directly repressing the tumor suppressors LATS2 and PPP2R2, thereby leading to increased cell division [46]. Conversely, miR-101 is down-regulated in NSCLC [22–25], leading to enhanced expression of its MCL-1 target gene in NSCLC, thus favoring tumor progression via inhibition of apoptosis [47]. Additionally, miR-101 is affected by DNA copy number losses, which is associated with reduced expression in NSCLC. This alteration can lead to overexpression of its target gene, EZH2 (a mammalian histone methyltransferase), and consequently induce disruption of cell proliferation-related processes, among other effects [48, 49].
MicroRNAs involved in invasiveness and angiogenesis
Besides being implicated in the regulation of early steps of lung tumorigenesis, some miRNAs are involved in lung cancer progression. Examples include miR-221 and miR-222, which are over-expressed in NSCLC via C-JUN, and target PTEN and TIMP3 tumor suppressors [50]. These miRNAs induce TRAIL resistance and enhance cellular migration through activation of the AKT pathway and metallopeptidases. Similarly, miR-191, a miRNA up-regulated in NSCLC [25], targets TIMP3 and has been shown to promote epithelial-to-mesenchymal transition (EMT) in hepatocellular carcinoma [51]. This process can be hampered by over-expression of miR-130a, which targets MET, an upstream activator of C-JUN, and induces down-regulation of miR-221 and miR-222 while increasing TRAIL-sensitivity in NSCLC cell lines [52]. In fact, miR-130a down-regulation is observed in NSCLC [23].
The miR-200 cluster consists of another tumor suppressive family that restricts metastasis and EMT in lung adenocarcinoma by targeting E-CADHERIN transcriptional regulators such as ZEB proteins [53]. This family appears to undergo reciprocal regulation between lung cancer subtypes, as it has been reported to be under-expressed in NSCLC while over-expressed in SCLC [54]. The observed up-regulation in SCLC is particularly intriguing knowing the high metastatic propensity of these tumors. Additionally, miR-149 is known to directly inhibit E-CADHERIN expression [55] and has been shown to be over-expressed in both SCLC and NSCLC cell lines [10].
Some miRNAs highlighted in lung cancer may also play major roles in angiogenesis. A recent study observed that miR-21, miR-106a, miR-126, miR-155, miR-182, miR-210 and miR-424 were associated with angiogenesis in NSCLC [56]. In fact, hypoxia inducible factors enhance the expression of miR-210, which promotes angiogenesis by targeting EFNA3 [57].
Sequence variations in miRNAs and their targets
The change in expression levels is not the only mechanism by which miRNAs can disrupt biological functions. Like protein-coding genes, miRNA genes exhibit sequence variations, such as single-nucleotide polymorphisms (SNPs). Tian and colleagues have shown that the homozygous C/C rs11614913 variant of miR-196a2 was associated with a significantly elevated risk of lung cancer compared with the wild-type T/T or T/C counterparts [58]. It was suggested that the rs11614913 variant can influence miR-196a expression, which regulates metastasis by targeting HOX genes. In another example, a SNP in the 3′UTR of KRAS was associated with increased risk for NSCLC presumably through disruption of let-7 binding affinity, which in turns affects KRAS regulation [32].
Clinical Relevance Of MiRNAs In Lung Cancer
Different studies associating roles of miRNAs with deregulation of key cellular functions in lung cancer have lead to the idea that miRNAs could be used as biomarkers for this disease, especially for subtype classification, prognosis as well as directing or design of new targeted therapies. Moreover, the stability and availability of these molecules in clinical sample materials such as FFPE and serum reinforces their potential [59]. Here we present correlations established between miRNAs and clinical characteristics of lung cancer, and their potential role as biomarkers as well as therapeutic targets.
MicroRNA as biomarkers for lung cancer early diagnosis
The implication of some miRNAs in the development of lung cancer has led to a cottage industry exploring the utility of miRNAs as biomarkers for early stage diagnosis. Moreover, some studies demonstrated that the levels of miRNAs in serum are stable, consistent among individuals, and reproducible enough to be taken into account in clinical routines. For example, miR-223 and miR-25 are over-represented in serum of lung cancer patients [60]. In fact, this study revealed a 76-miRNA lung cancer signature detected only in patient serum (relative to blood cells) when compared to healthy subjects. MiRNAs detected at elevated levels in the plasma of stage I lung cancer patients include miR-155, miR-197, and miR-182 [61]. MiRNA signatures in plasma [62] as well as in serum [63] can discriminate between screening detected early-stage (I and II) NSCLC and non-cancer controls, with a high degree of diagnostic accuracy. It has been suggested that serum signatures could even be used to detect lung cancer among asymptomatic high-risk individuals [62, 63]. MiRNAs profiles are also discernible in sputum of early-stage lung cancer patients. Indeed, studies have been able to identify specific miRNA signatures with high sensitivity and specificity in sputum of early stage AdC (81% and 92% of sensitivity and specificity, respectively) and SqCC patients (73% of sensitivity and 96% of specificity) [64, 65]. These observations have led to the consideration that analysis of miRNA in sputum could also be explored for identifying biomarkers in healthy smokers that would have the ability to distinguish individuals who will develop lung cancer in the future [66].
Despite these promising findings, it is important to note that several confounding issues may contribute to circulating miRNA variability. Disturbances in the immune system, such as inflammation, which can be independent of the tumor status, may considerably influence miRNA profiles observed in serum and plasma [67–69]. MiRNA profiles from sputum have also been observed to be modified by the inflammation status of smokers and the presence or absense of inflammatory diseases such as chronic obstructive pulmonary disease [70]. Additionally, other confounding issues are related to sample collection and processing conditions [71, 72].
Biomarkers for histopathological classification
Relevance of miRNA profiling for subclassification of solid tumors has been confirmed in several studies including lung cancer subtypes [25, 73]. Nevertheless, their accuracy compared to histopathology remains uncertain. For instance, miR-205 has been previously identified as a highly accurate marker for lung SqCC with sensitivity of 96% and specificity of 90% [74], and showed 100% concordance between the diagnoses established by histopathologic and qRT-PCR based diagnostic assay [75]. However, more recent work has indicated that the use of miR-205 alone led to the misclassification of certain cases [76]. Another subtype-specific miRNA is miR-155, which is under-expressed in SCLC [54] and over-expressed in NSCLC [25, 77]. This example illustrates the complexity of miRNA-related functions, since the same miRNA can display both oncogenic and tumor suppressive behavior depending on the cellular context.
Another strategy to distinguish lung cancer subtypes has been the application of miRNA-specific signatures. A recent study identified a set of 34 miRNAs that are differentially expressed between AdC and SqCC, while another study identified 15 miRNAs that can differentiate SqCC from normal lung [77, 78]. However, only five miRNAs of the 15 miRNA SqCC set (miR-17, miR-20a, let-7e, miR-106a, miR-106b) overlap with the subtype specific panel of 34 miRNAs. Let-7e is the only miRNA downregulated in SqCC compared to AdC [78] as well as compared to normal lung [77], while miR-17, miR-20a, miR-106a, miR-106b are upregulated in SqCC, but even more in AdC [77, 78]. Sputum is an easily accessible source of material to derive panels of subtype specific miRNAs. Early stage SqCC patients were distinguished from normal subjects by detection of over-expression of miR-205, miR-210, miR-21 and miR-708, and under-expression of miR-429, miR-139, miR-30a and miR-126 in sputum, with 73% sensitivity and 96% specificity [65]. A similar study in AdC revealed over-expression of miR-21, miR-182, miR-200b, miR-375, and under-expression of miR-486, miR-126 and miR-145 in sputum of AdC patients [64]. The use of miR-21, miR-200b, miR-375 and miR-486 alone was able to distinguish AdC patients with a sensitivity and specificity of 81% and 92%, respectively. Taken together, these studies identified several miRNAs (miR-486, miR-126, miR-139, miR-130b or miR-210) with similar expression in the two major subtypes of NSCLC.
In summary, although a new classification method based on miRNA expression should be considered in the future, current literature lacks strong evidence to support unequivocal subtype-specific miRNAs. The most recurrent subtype-specific miRNAs are listed in Table 2.
Table 2.
miRNAs differentially expressed in histological subtypes of lung cancer
| miRNA | Expression in subtype | Confirmed targets | References | ||
|---|---|---|---|---|---|
|
| |||||
| SqCC | AdC | SCLC | |||
|
|
|||||
| let-7 | Dn | HMGA2, RAS, MYC, E2Fs, CDKs | [24, 25, 54, 65, 77, 78, 80, 81, 105] | ||
| miR-9 | Up | TGFBI, TRIM2, SIRT1, BTBD3 | [24, 25, 54, 106] | ||
| miR-17-92 cluster | Up | p21, E2F, CCND1, PTEN, Rb, HIF-1α | [40, 54] | ||
| miR-31 | Up | LATS2, PPP2R2A | [10, 23, 46, 64, 65] | ||
| miR-93 | Up | p21, FUS1 | [10, 54, 111, 118] | ||
| miR-95 | Up | SNX1 | [25, 54, 119] | ||
| miR-98 | Up | FUS1 | [10, 54, 93, 118] | ||
| miR-128 | Up | EGFR | [45, 54] | ||
| miR-130b | Up | CSF-1, TP53INP1, RUNX3 | [54, 64, 65] | ||
| miR-139 | Dn | ROCK2, FOXO1, CXCR4 | [24, 64, 65, 123–125] | ||
| miR-140 | Dn | Sp1, BMP2 | [25, 64] | ||
| miR-143 | Dn | ERK5, KRAS, Bcl-2, DNMT3A, ELK1 | [23–25, 54, 64] | ||
| miR-146b | Up | MMP16, TRAF6, IRAK1, | [25, 54, 77, 78, 107] | ||
| miR-149 | Up | E-cadherin | [10, 55, 65] | ||
| miR-150 | Dn | MUC4, P2RX7 | [25, 54, 64] | ||
| miR-155 | Up | TAB2, FADD, RIP-1, IKKε, ET-1 | [25, 54, 61, 77, 78, 107] | ||
| miR-182 | Up | FOXO1 | [24, 54, 61, 64, 65, 77, 78, 80, 95, 120] | ||
| miR-191 | Up | CDK6, TIMP3, NDST1 | [25, 54, 77, 78, 107] | ||
| miR-192 | Up | ERCC3, ERCC4, Rb1 | [25, 107] | ||
| miR-200a,b | Up | C-ets-1, Zeb1, ECM proteins, p38α | [54, 64, 79, 127] | ||
| miR-200c | Up | C-ets-1, Zeb1, ECM proteins | [54, 65, 77, 128] | ||
| miR-203 | Up | DeltaNp63, Smo | [25, 77, 107] | ||
| miR-205 | Up | E2F1, Zeb1, Sip1 | [10, 25, 65, 74, 75, 78, 93, 107] | ||
| miR-218 | Dn | RICTOR | [24, 25, 64] | ||
| miR-324-5p | Up | Smo, Gli1 | [10, 54, 65, 130] | ||
| miR-375 | Up | IGF1R, SP1, LDHB, YAP1 | [54, 64, 137–140] | ||
| miR-423 | Up | p21 | [24, 65, 141] | ||
| miR-429 | Dn | ALDH1L2, PSAT1, ZEB1 | [54, 65, 142] | ||
| miR-486 | Dn | CD40 | [64, 65, 108] | ||
Note: Targets cited are confirmed by experiments
Biomarkers for prognostics and treatment prediction
MiRNAs have been associated with patient prognosis and response to chemo- and radiotherapy in lung cancer. Over- and under-expression of miRNAs has been associated with recurrence of stage I disease and disease-free survival in NSCLC patients [79, 80]. In terms of survival, higher expression levels of some miRNAs (such hsa-miR-221 and hsa-let-7a) seem to have protective functions, while others (hsa-miR-137, hsa-miR-372, and hsa-miR-182*) are associated with decreased survival [80]. Additionally, poor survival of NSCLC patients has been linked to low expression of the let-7 family, and miRNA processing enzymes DICER and DROSHA [18, 81]. Some studies have tested the utility of miRNA-signatures as non-invasive serum biomarkers for overall survival in NSCLC patients who received surgical treatment and adjuvant chemotherapy. A 4-miRNA signature (miR-486, miR-30d, miR-1 and miR-499) was identified as a predictor of overall survival [82]. Moreover, expression of let-7 is related to survival time after curative resection [81].
Alterations in miRNA levels are also correlated with prognosis in a subtype-specific manner. Poor survival in AdC has been correlated with low expression of let-7a-2 and high expression of miR-155, while changes in miR-21 and other miRNA levels have been recently associated with shortened survival time in patients with SqCC [25, 77, 78]. Additionally, elevated expression of miR-31 inversely correlates with the survival of Chinese SqCC patients [83].
MiRNAs have been shown to modulate chemo- and radiosensitivity of lung tumor cells [84, 85]. The let-7 family of miRNAs modulates response to cytotoxic anticancer therapy and also affects response to radiation of lung cancer cells through a RAS dependent mechanism [85]. Loss of heterozygosity of miR-128b, which directly targets EGFR, correlates significantly with positive NSCLC patient response and survival with EGFR-TKI treatment [45]. Another study found that low plasma levels of miR-21 correlated with sensitivity to platinum-based chemotherapy [86]. In lung cancer cell models, miR-181a and miR-630 conferred sensitization to cisplatin [87], while downregulation of miR-200 was associated with chemoresistance to cisplatin and cetuximab [88].
MicroRNAs as therapeutic agents and targets in lung cancer
The multiple molecular pathways targeted by miRNAs make them candidates for developing novel molecular therapeutics agents [89]. Antagonizing a single miRNA could affect a number of vital signalling pathways in the tumor cell simultaneously; however, the same characteristic could have undesirable side effects on other pathways. The potential broad biological impact of microRNA perturbation needs to be considered when considering miRNAs as therapeutic targets.
Currently, direct strategies to target miRNAs in cancer involve the use of oligonucleotides or virus-based constructs to either suppress the expression of oncogenic miRNAs or to replace the loss of expression of tumor suppressor miRNAs. Indirect strategies involve the use of drugs to regulate miRNA expression by targeting their transcription and processing [90]. In this context, let-7 has been intensively studied. In mouse models, delivery of let-7 miRNA via adenoviral infection induces regression of lung tumors [31], and intranasal let-7 administration reduces lung tumor formation in KRAS mutant models [91]. Additionally, ectopic let-7g miRNA expression in xenografts and in a mouse lung tumor model was found to reduce tumor burden [92]. Even if under-expression of the let-7 family is stronger in SqCC [93], let-7 is associated with survival in both subtypes and could still be a useful target in both subtypes [25].
The therapeutic potential of other miRNAs has been explored in lung cancer. In vivo knockdown of oncogenic miR-31 has been shown to repress lung cancer in FVB syngeneic mice [46]. Likewise, administration of miR-34a (either locally or systemically) inhibited NSCLC tumor growth in mouse models by blocking CDK4, c-Met and Bcl-2 expression [94]. Promising results on cell growth inhibition have also been obtained by administering miR-145 to EGFR mutant human AdC cell lines [95] and efficacy of miR-133b delivery in lung tissue has been proved in mice [96]. The potential use of the miR-17-92 cluster as a therapeutic target was also recently demonstrated in lung cancer cell models. Antagonization of miR-17 and miR-20a was found to induce apoptosis in vitro [97].
To date, most of the investigations into the therapeutic use of miRNAs in lung cancer have been performed in cell lines or animal models. Results suggest a reasonable degree of toxicity of these molecules; however, a final decision for human use must be based on results derived from clinical trials. Clinical trials have already been initiated to investigate the safety and efficacy of miRNA treatment in human patients, such as the NCT00979927 protocol (http://clinicaltrials.gov). This study aims to assess the potential of the SPC3649 oligonucleotide as an antagonist of miR-122, a liver specific miRNA, in HCV therapy. The potential success of this clinical trial represents a proof of concept for the field of miRNA therapeutics, although lung cancer clinical trials have yet to begin.
Conclusions and Perspectives
There is a great need for the identification of non-invasive biomarkers for early detection, subtype classification and drug response prediction in lung cancer. The presence and stability of miRNAs in body fluids, such as in peripheral blood or sputum, and in FFPE samples makes them attractive candidates for this purpose. Furthermore, new techniques such as nanopore-based detection of miRNA in plasma samples of lung cancer patients reinforces their potential [98].
Despite all the advantages, several concerns need to be addressed before these molecules can be reliably translated as cancer biomarkers into clinical settings. First, research in this field is still in its infancy, with pilot studies showing promising results; however, most of them still involve a small number of individuals which could lead to the omission of confounding issues (e.g. effects derived from other diseases and lifestyle) that can alter miRNA expression. Another concern is related to the variability of miRNA profiles in different body fluids, as it is the case between profiles from serum and plasma, as well as for profiles between body fluid and tumor samples [99]. It is noteworthy to consider that miRNA profiles may change due to reasons not directly related to the tumor itself. For example, miRNA signatures can be influenced by miRNA expression from immune system and inflammation status [67–70]. On the other hand, miRNA expression levels from the peripheral immune system are dependent on the tumor status and can revert after tumor resection [100].
Subtype specificity of miRNAs should also be carefully considered, as some miRNAs (such as miR-486, miR-126, miR-139, miR-130b, and miR-210) have been identified as markers for AdC as well as for SqCC. Nevertheless, a few miRNAs such as the let-7 cluster in SqCC, miR-155/miR-191 in AdC, and miR-375/miR-98 in SCLC, show consistent results between studies and have a great potential to be used as reliable biomarkers for subtype identification.
A major concern related to the development of miRNA-based therapies is the complexity and widespread effects exerted by miRNAs. Since a single miRNA can regulate various genes, the potential off-target effects of miRNA therapeutics can lead to unpredictable side effects. Moreover, miRNAs can function in a cell type-specific manner, as some miRNAs can act as oncogenes in some subtypes of lung cancer and as tumor suppressors in others. It is also important to note that little information is available concerning epigenetic modulation of miRNAs [101–104] as well as SNPs or nucleotide variations in miRNAs and their target binding sites [32, 58].
Recent research has only begun to unveil the utility of some miRNAs as biomarkers and therapeutic targets. Future studies should focus on a better understanding of miRNA functions, regulation, and on maximizing the advantages related to target diversity. Certainly, the next decade will reveal new important features of these players in lung tumorigenesis, which will help with their translation from the bench to the clinical practice.
Supplementary Material
Acknowledgments
This work was supported by funds from the Canadian Institutes for Health Research (MOP 86731, MOP 94867, MOP-110949), Canadian Cancer Society (CCS20485), NIH/NCI 1R01CA164783-01, U.S. Department of Defense (CDMRP W81XWH-10-1-0634), NCI Early Detection Research Network and the Canary Foundation.
References
- 1.Ferlay J, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International journal of cancer Journal international du cancer. 2010;127(12):2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers--a different disease. Nature reviews Cancer. 2007;7(10):778–90. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, et al. Cancer statistics, 2009. CA: a cancer journal for clinicians. 2009;59(4):225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 4.Lam S, et al. EarlyCDT-Lung: an immunobiomarker test as an aid to early detection of lung cancer. Cancer Prev Res (Phila) 2011;4(7):1126–34. doi: 10.1158/1940-6207.CAPR-10-0328. [DOI] [PubMed] [Google Scholar]
- 5.Travis WD, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6(2):244–85. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gazdar AF. Should we continue to use the term non-small-cell lung cancer? Ann Oncol. 2010;21(Suppl 7):vii225–9. doi: 10.1093/annonc/mdq372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato M, et al. A translational view of the molecular pathogenesis of lung cancer. J Thorac Oncol. 2007;2(4):327–43. doi: 10.1097/01.JTO.0000263718.69320.4c. [DOI] [PubMed] [Google Scholar]
- 8.Minna JD, Roth JA, Gazdar AF. Focus on lung cancer. Cancer Cell. 2002;1(1):49–52. doi: 10.1016/s1535-6108(02)00027-2. [DOI] [PubMed] [Google Scholar]
- 9.Zochbauer-Muller S, Gazdar AF, Minna JD. Molecular pathogenesis of lung cancer. Annu Rev Physiol. 2002;64:681–708. doi: 10.1146/annurev.physiol.64.081501.155828. [DOI] [PubMed] [Google Scholar]
- 10.Du L, et al. MicroRNA expression distinguishes SCLC from NSCLC lung tumor cells and suggests a possible pathological relationship between SCLCs and NSCLCs. J Exp Clin Cancer Res. 2010;29:75. doi: 10.1186/1756-9966-29-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salgia R, Skarin AT. Molecular abnormalities in lung cancer. J Clin Oncol. 1998;16(3):1207–17. doi: 10.1200/JCO.1998.16.3.1207. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Lynam-Lennon N, Maher SG, Reynolds JV. The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos Soc. 2009;84(1):55–71. doi: 10.1111/j.1469-185X.2008.00061.x. [DOI] [PubMed] [Google Scholar]
- 14.Hartmann C, et al. MicroRNAs: a new class of gene expression regulators. Med Sci (Paris) 2004;20(10):894–8. doi: 10.1051/medsci/20042010894. [DOI] [PubMed] [Google Scholar]
- 15.Winter J, et al. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 16.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–79. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 17.Lin PY, Yu SL, Yang PC. MicroRNA in lung cancer. Br J Cancer. 2010;103(8):1144–8. doi: 10.1038/sj.bjc.6605901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karube Y, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96(2):111–5. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar MS, et al. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39(5):673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 20.Mascaux C, et al. Evolution of microRNA expression during human bronchial squamous carcinogenesis. Eur Respir J. 2009;33(2):352–9. doi: 10.1183/09031936.00084108. [DOI] [PubMed] [Google Scholar]
- 21.Kalscheuer S, et al. Differential expression of microRNAs in early-stage neoplastic transformation in the lungs of F344 rats chronically treated with the tobacco carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis. 2008;29(12):2394–9. doi: 10.1093/carcin/bgn209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, et al. The role of microRNA in human lung squamous cell carcinoma. Cancer Genet Cytogenet. 2010;200(2):127–33. doi: 10.1016/j.cancergencyto.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 23.Gao W, et al. MiR-21 overexpression in human primary squamous cell lung carcinoma is associated with poor patient prognosis. J Cancer Res Clin Oncol. 2011;137(4):557–66. doi: 10.1007/s00432-010-0918-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crawford M, et al. MicroRNA 133B targets pro-survival molecules MCL-1 and BCL2L2 in lung cancer. Biochem Biophys Res Commun. 2009;388(3):483–9. doi: 10.1016/j.bbrc.2009.07.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, et al. MYC protein inhibits transcription of the microRNA cluster MC-let-7a-1~let-7d via noncanonical E-box. J Biol Chem. 2011;286(46):39703–14. doi: 10.1074/jbc.M111.293126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He XY, et al. The let-7a microRNA protects from growth of lung carcinoma by suppression of k-Ras and c-Myc in nude mice. J Cancer Res Clin Oncol. 2010;136(7):1023–8. doi: 10.1007/s00432-009-0747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson CD, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67(16):7713–22. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 29.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–16. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 30.Shimizu S, et al. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J Hepatol. 2010;52(5):698–704. doi: 10.1016/j.jhep.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 31.Trang P, et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29(11):1580–7. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chin LJ, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68(20):8535–40. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68(20):8191–4. doi: 10.1158/0008-5472.CAN-08-1768. [DOI] [PubMed] [Google Scholar]
- 35.O’Donnell KA, et al. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 36.Trompeter HI, et al. MicroRNAs MiR-17, MiR-20a, and MiR-106b act in concert to modulate E2F activity on cell cycle arrest during neuronal lineage differentiation of USSC. PLoS One. 2011;6(1):e16138. doi: 10.1371/journal.pone.0016138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grillari J, Hackl M, Grillari-Voglauer R. miR-17-92 cluster: ups and downs in cancer and aging. Biogerontology. 2010;11(4):501–6. doi: 10.1007/s10522-010-9272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol. 2010;42(8):1348–54. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Putzer BM. E2F1 death pathways as targets for cancer therapy. J Cell Mol Med. 2007;11(2):239–51. doi: 10.1111/j.1582-4934.2007.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ebi H, et al. Counterbalance between RB inactivation and miR-17-92 overexpression in reactive oxygen species and DNA damage induction in lung cancers. Oncogene. 2009;28(38):3371–9. doi: 10.1038/onc.2009.201. [DOI] [PubMed] [Google Scholar]
- 41.Hayashita Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65(21):9628–32. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 42.Ofir M, Hacohen D, Ginsberg D. MiR-15 and miR-16 are direct transcriptional targets of E2F1 that limit E2F-induced proliferation by targeting cyclin E. Mol Cancer Res. 2011;9(4):440–7. doi: 10.1158/1541-7786.MCR-10-0344. [DOI] [PubMed] [Google Scholar]
- 43.Hatley ME, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18(3):282–93. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seike M, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(29):12085–90. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss GJ, et al. EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann Oncol. 2008;19(6):1053–9. doi: 10.1093/annonc/mdn006. [DOI] [PubMed] [Google Scholar]
- 46.Liu X, et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest. 2010;120(4):1298–309. doi: 10.1172/JCI39566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo L, et al. MiR-101 and Mcl-1 in non-small-cell lung cancer: expression profile and clinical significance. Med Oncol. 2011 doi: 10.1007/s12032-011-0085-8. [DOI] [PubMed] [Google Scholar]
- 48.Thu KL, et al. miR-101 DNA copy loss is a prominent subtype specific event in lung cancer. J Thorac Oncol. 2011;6(9):1594–8. doi: 10.1097/JTO.0b013e3182217d81. [DOI] [PubMed] [Google Scholar]
- 49.Varambally S, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322(5908):1695–9. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garofalo M, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16(6):498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.He Y, et al. Hypomethylation of the hsa-miR-191 locus causes high expression of hsa-mir-191 and promotes the epithelial-to-mesenchymal transition in hepatocellular carcinoma. Neoplasia. 2011;13(9):841–53. doi: 10.1593/neo.11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Acunzo M, et al. miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene. 2012;31(5):634–42. doi: 10.1038/onc.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schliekelman MJ, et al. Targets of the tumor suppressor miR-200 in regulation of the epithelial-mesenchymal transition in cancer. Cancer Res. 2011;71(24):7670–82. doi: 10.1158/0008-5472.CAN-11-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miko E, et al. Differentially expressed microRNAs in small cell lung cancer. Exp Lung Res. 2009;35(8):646–64. doi: 10.3109/01902140902822312. [DOI] [PubMed] [Google Scholar]
- 55.Luo Z, et al. miR-149 promotes epithelial-mesenchymal transition and invasion in nasopharyngeal carcinoma cells. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2011;36(7):604–9. doi: 10.3969/j.issn.1672-7347.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 56.Donnem T, et al. MicroRNA Signatures in Tumor Tissue Related to Angiogenesis in Non-Small Cell Lung Cancer. PLoS One. 2012;7(1):e29671. doi: 10.1371/journal.pone.0029671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang X, Le QT, Giaccia AJ. MiR-210--micromanager of the hypoxia pathway. Trends Mol Med. 2010;16(5):230–7. doi: 10.1016/j.molmed.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tian T, et al. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev. 2009;18(4):1183–7. doi: 10.1158/1055-9965.EPI-08-0814. [DOI] [PubMed] [Google Scholar]
- 59.Xi Y, et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13(10):1668–74. doi: 10.1261/rna.642907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 61.Zheng D, et al. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int J Clin Exp Pathol. 2011;4(6):575–86. [PMC free article] [PubMed] [Google Scholar]
- 62.Bianchi F, et al. A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol Med. 2011;3(8):495–503. doi: 10.1002/emmm.201100154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Keller A, et al. Stable serum miRNA profiles as potential tool for non-invasive lung cancer diagnosis. RNA Biol. 2011;8(3):506–16. doi: 10.4161/rna.8.3.14994. [DOI] [PubMed] [Google Scholar]
- 64.Yu L, et al. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. International journal of cancer Journal international du cancer. 2010;127(12):2870–8. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xing L, et al. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod Pathol. 2010;23(8):1157–64. doi: 10.1038/modpathol.2010.111. [DOI] [PubMed] [Google Scholar]
- 66.Malerba M, Montuschi P. Non-invasive biomarkers of lung inflammation in smoking subjects. Curr Med Chem. 2012;19(2):187–96. doi: 10.2174/092986712803414204. [DOI] [PubMed] [Google Scholar]
- 67.Zernecke A. MicroRNAs in the regulation of immune cell functions - implications for atherosclerotic vascular disease. Thromb Haemost. 2012;107(4) doi: 10.1160/TH11-08-0603. [DOI] [PubMed] [Google Scholar]
- 68.Cermelli S, et al. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6(8):e23937. doi: 10.1371/journal.pone.0023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yao R, et al. The altered expression of inflammation-related microRNAs with microRNA-155 expression correlates with Th17 differentiation in patients with acute coronary syndrome. Cell Mol Immunol. 2011;8(6):486–95. doi: 10.1038/cmi.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pottelberge GR, et al. MicroRNA expression in induced sputum of smokers and patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2011;183(7):898–906. doi: 10.1164/rccm.201002-0304OC. [DOI] [PubMed] [Google Scholar]
- 71.Duttagupta R, et al. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS One. 2011;6(6):e20769. doi: 10.1371/journal.pone.0020769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McDonald JS, et al. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem. 2011;57(6):833–40. doi: 10.1373/clinchem.2010.157198. [DOI] [PubMed] [Google Scholar]
- 73.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(7):2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lebanony D, et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol. 2009;27(12):2030–7. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- 75.Bishop JA, et al. Accurate classification of non-small cell lung carcinoma using a novel microRNA-based approach. Clin Cancer Res. 2010;16(2):610–9. doi: 10.1158/1078-0432.CCR-09-2638. [DOI] [PubMed] [Google Scholar]
- 76.Del Vescovo V, et al. miR-205 Expression levels in nonsmall cell lung cancer do not always distinguish adenocarcinomas from squamous cell carcinomas. Am J Surg Pathol. 2011;35(2):268–75. doi: 10.1097/PAS.0b013e3182068171. [DOI] [PubMed] [Google Scholar]
- 77.Raponi M, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69(14):5776–83. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 78.Landi MT, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16(2):430–41. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Patnaik SK, et al. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res. 2010;70(1):36–45. doi: 10.1158/0008-5472.CAN-09-3153. [DOI] [PubMed] [Google Scholar]
- 80.Yu SL, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13(1):48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 81.Takamizawa J, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64(11):3753–6. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 82.Hu Z, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28(10):1721–6. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 83.Tan X, et al. A 5-microRNA signature for lung squamous cell carcinoma diagnosis and hsa-miR-31 for prognosis. Clin Cancer Res. 2011;17(21):6802–11. doi: 10.1158/1078-0432.CCR-11-0419. [DOI] [PubMed] [Google Scholar]
- 84.Blower PE, et al. MicroRNAs modulate the chemosensitivity of tumor cells. Mol Cancer Ther. 2008;7(1):1–9. doi: 10.1158/1535-7163.MCT-07-0573. [DOI] [PubMed] [Google Scholar]
- 85.Weidhaas JB, et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67(23):11111–6. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wei J, et al. Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non-small cell lung cancer. Chin J Cancer. 2011;30(6):407–14. doi: 10.5732/cjc.010.10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Galluzzi L, et al. miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res. 2010;70(5):1793–803. doi: 10.1158/0008-5472.CAN-09-3112. [DOI] [PubMed] [Google Scholar]
- 88.Ceppi P, et al. Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Mol Cancer Res. 2010;8(9):1207–16. doi: 10.1158/1541-7786.MCR-10-0052. [DOI] [PubMed] [Google Scholar]
- 89.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nature reviews Cancer. 2006;6(4):259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 90.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9(10):775–89. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Esquela-Kerscher A, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7(6):759–64. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 92.Kumar MS, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(10):3903–8. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fassina A, Cappellesso R, Fassan M. Classification of non-small cell lung carcinoma in transthoracic needle specimens using microRNA expression profiling. Chest. 2011;140(5):1305–11. doi: 10.1378/chest.11-0708. [DOI] [PubMed] [Google Scholar]
- 94.Wiggins JF, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70(14):5923–30. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cho WC, Chow AS, Au JS. Restoration of tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. Eur J Cancer. 2009;45(12):2197–206. doi: 10.1016/j.ejca.2009.04.039. [DOI] [PubMed] [Google Scholar]
- 96.Wu Y, et al. MicroRNA delivery by cationic lipoplexes for lung cancer therapy. Mol Pharm. 2011;8(4):1381–9. doi: 10.1021/mp2002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Matsubara H, et al. Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene. 2007;26(41):6099–105. doi: 10.1038/sj.onc.1210425. [DOI] [PubMed] [Google Scholar]
- 98.Wang Y, et al. Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat Nanotechnol. 2011;6(10):668–74. doi: 10.1038/nnano.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Heegaard NH, et al. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. International journal of cancer Journal international du cancer. 2012;130(6):1378–86. doi: 10.1002/ijc.26153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kossenkov AV, et al. Resection of non-small cell lung cancers reverses tumor-induced gene expression changes in the peripheral immune system. Clin Cancer Res. 2011;17(18):5867–77. doi: 10.1158/1078-0432.CCR-11-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tanaka N, et al. Frequent methylation and oncogenic role of microRNA-34b/c in small-cell lung cancer. Lung Cancer. 2011 doi: 10.1016/j.lungcan.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 102.Kitano K, et al. CpG island methylation of microRNAs is associated with tumor size and recurrence of non-small-cell lung cancer. Cancer Sci. 2011;102(12):2126–31. doi: 10.1111/j.1349-7006.2011.02101.x. [DOI] [PubMed] [Google Scholar]
- 103.Watanabe K, et al. Genome structure-based screening identified epigenetically silenced microRNA associated with invasiveness in non-small-cell lung cancer. International journal of cancer. Journal international du cancer. 2011 doi: 10.1002/ijc.26254. [DOI] [PubMed] [Google Scholar]
- 104.Wang Z, et al. DNA hypermethylation of microRNA-34b/c has prognostic value for stage non-small cell lung cancer. Cancer Biol Ther. 2011;11(5):490–6. doi: 10.4161/cbt.11.5.14550. [DOI] [PubMed] [Google Scholar]
- 105.Landi MT, et al. MicroRNA Expression Differentiates Histology and Predicts Survival of Lung Cancer. Clinical Cancer Research. 2010;16(2):430–441. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schonrock N, et al. Target Gene Repression Mediated by miRNAs miR-181c and miR-9 Both of Which Are Down-regulated by Amyloid-beta. J Mol Neurosci. 2012;46(2):324–35. doi: 10.1007/s12031-011-9587-2. [DOI] [PubMed] [Google Scholar]
- 107.Rabinowits G, et al. Exosomal MicroRNA: A Diagnostic Marker for Lung Cancer. Clinical Lung Cancer. 2009;10(1):42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 108.Shen J, et al. Diagnosis of lung cancer in individuals with solitary pulmonary nodules by plasma microRNA biomarkers. BMC Cancer. 2011;11:374. doi: 10.1186/1471-2407-11-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bar N, Dikstein R. miR-22 Forms a Regulatory Loop in PTEN/AKT Pathway and Modulates Signaling Kinetics. PLoS One. 2010;5(5):e10859. doi: 10.1371/journal.pone.0010859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li J, et al. An inhibitory effect of miR-22 on cell migration and invasion in ovarian cancer. Gynecol Oncol. 2010;119(3):543–8. doi: 10.1016/j.ygyno.2010.08.034. [DOI] [PubMed] [Google Scholar]
- 111.Kan T, et al. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology. 2009;136(5):1689–700. doi: 10.1053/j.gastro.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Patel N, et al. Involvement of miR-30c and miR-301a in immediate induction of plasminogen activator inhibitor-1 by placental growth factor in human pulmonary endothelial cells. Biochem J. 2011;434(3):473–82. doi: 10.1042/BJ20101585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rayner KJ, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478(7369):404–7. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Herrera-Merchan A, et al. miR-33-mediated downregulation of p53 controls hematopoietic stem cell self-renewal. Cell Cycle. 2010;9(16):3277–3285. doi: 10.4161/cc.9.16.12598. [DOI] [PubMed] [Google Scholar]
- 115.de Antonellis P, et al. MiR-34a Targeting of Notch Ligand Delta-Like 1 Impairs CD15+/CD133+ Tumor-Propagating Cells and Supports Neural Differentiation in Medulloblastoma. PLoS One. 2011;6(9):e24584. doi: 10.1371/journal.pone.0024584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gallardo E, et al. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009;30(11):1903–1909. doi: 10.1093/carcin/bgp219. [DOI] [PubMed] [Google Scholar]
- 117.Tsuchida A, et al. miR-92 is a key oncogenic component of the miR-17-92 cluster in colon cancer. Cancer Sci. 2011;102(12):2264–71. doi: 10.1111/j.1349-7006.2011.02081.x. [DOI] [PubMed] [Google Scholar]
- 118.Du L, et al. miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol Cancer Res. 2009;7(8):1234–43. doi: 10.1158/1541-7786.MCR-08-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Huang Z, et al. MicroRNA-95 promotes cell proliferation and targets sorting Nexin 1 in human colorectal carcinoma. Cancer Res. 2011;71(7):2582–9. doi: 10.1158/0008-5472.CAN-10-3032. [DOI] [PubMed] [Google Scholar]
- 120.Zhu W, et al. Overexpression of members of the microRNA-183 family is a risk factor for lung cancer: a case control study. BMC Cancer. 2011;11:393. doi: 10.1186/1471-2407-11-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sun D, et al. miR-99 family of MicroRNAs suppresses the expression of prostate-specific antigen and prostate cancer cell proliferation. Cancer Res. 2011;71(4):1313–24. doi: 10.1158/0008-5472.CAN-10-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jiang L, et al. Hsa-miR-125a-3p and hsa-miR-125a-5p are downregulated in non-small cell lung cancer and have inverse effects on invasion and migration of lung cancer cells. BMC Cancer. 2010;10:318. doi: 10.1186/1471-2407-10-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wong CC, et al. The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating Rho-kinase 2. Gastroenterology. 2011;140(1):322–31. doi: 10.1053/j.gastro.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 124.Hasseine LK, et al. miR-139 impacts FoxO1 action by decreasing FoxO1 protein in mouse hepatocytes. Biochem Biophys Res Commun. 2009;390(4):1278–82. doi: 10.1016/j.bbrc.2009.10.135. [DOI] [PubMed] [Google Scholar]
- 125.Bao W, et al. HER2 interacts with CD44 to up-regulate CXCR4 via epigenetic silencing of microRNA-139 in gastric cancer cells. Gastroenterology. 2011;141(6):2076–2087. e6. doi: 10.1053/j.gastro.2011.08.050. [DOI] [PubMed] [Google Scholar]
- 126.Takahashi Y, et al. MiR-107 and MiR-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. PLoS One. 2009;4(8):e6677. doi: 10.1371/journal.pone.0006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gibbons DL, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23(18):2140–51. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hurteau GJ, et al. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007;67(17):7972–6. doi: 10.1158/0008-5472.CAN-07-1058. [DOI] [PubMed] [Google Scholar]
- 129.Chan SY, Loscalzo J. MicroRNA-210: a unique and pleiotropic hypoxamir. Cell Cycle. 2010;9(6):1072–83. doi: 10.4161/cc.9.6.11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ferretti E, et al. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 2008;27(19):2616–2627. doi: 10.1038/emboj.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liang Z, et al. Involvement of miR-326 in chemotherapy resistance of breast cancer through modulating expression of multidrug resistance-associated protein 1. Biochem Pharmacol. 2010;79(6):817–24. doi: 10.1016/j.bcp.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 132.Du C, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10(12):1252–9. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 133.Bai XY, et al. miR-335 and miR-34a Promote renal senescence by suppressing mitochondrial antioxidative enzymes. J Am Soc Nephrol. 2011;22(7):1252–61. doi: 10.1681/ASN.2010040367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Xu Y, et al. MicroRNA-335 acts as a metastasis suppressor in gastric cancer by targeting Bcl-w and specificity protein 1. Oncogene. 2011 doi: 10.1038/onc.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Heyn H, et al. MicroRNA miR-335 is crucial for the BRCA1 regulatory cascade in breast cancer development. International journal of cancer Journal international du cancer. 2011;129(12):2797–806. doi: 10.1002/ijc.25962. [DOI] [PubMed] [Google Scholar]
- 136.Zhang Y, et al. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proceedings of the National Academy of Sciences. 2011;108(24):9863–9868. doi: 10.1073/pnas.1018493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kong KL, et al. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut. 2012;61(1):33–42. doi: 10.1136/gutjnl-2011-300178. [DOI] [PubMed] [Google Scholar]
- 138.Wang F, et al. miR-375 is down-regulated in squamous cervical cancer and inhibits cell migration and invasion via targeting transcription factor SP1. Am J Pathol. 2011;179(5):2580–8. doi: 10.1016/j.ajpath.2011.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kinoshita T, et al. Tumor suppressive microRNA-375 regulates lactate dehydrogenase B in. Int J Oncol. 2012;40(1):185–93. doi: 10.3892/ijo.2011.1196. [DOI] [PubMed] [Google Scholar]
- 140.Nishikawa E, et al. miR-375 is activated by ASH1 and inhibits YAP1 in a lineage-dependent manner in lung cancer. Cancer Res. 2011;71(19):6165–73. doi: 10.1158/0008-5472.CAN-11-1020. [DOI] [PubMed] [Google Scholar]
- 141.Lin J, et al. MicroRNA-423 promotes cell growth and regulates G(1)/S transition by targeting p21Cip1/Waf1 in hepatocellular carcinoma. Carcinogenesis. 2011;32(11):1641–7. doi: 10.1093/carcin/bgr199. [DOI] [PubMed] [Google Scholar]
- 142.Chen J, et al. Overexpression of miR-429 induces mesenchymal-to-epithelial transition (MET) in metastatic ovarian cancer cells. Gynecol Oncol. 2011;121(1):200–5. doi: 10.1016/j.ygyno.2010.12.339. [DOI] [PubMed] [Google Scholar]
- 143.Shende VR, et al. Expression and rhythmic modulation of circulating microRNAs targeting the clock gene Bmal1 in mice. PLoS One. 2011;6(7):e22586. doi: 10.1371/journal.pone.0022586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tominaga K, et al. Competitive regulation of nucleolin expression by HuR and miR-494. Mol Cell Biol. 2011;31(20):4219–31. doi: 10.1128/MCB.05955-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yadav S, et al. miR-497 and miR-302b regulate ethanol-induced neuronal cell death through BCL2 protein and cyclin D2. J Biol Chem. 2011;286(43):37347–57. doi: 10.1074/jbc.M111.235531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Huang W, Li MD. Differential allelic expression of dopamine D1 receptor gene (DRD1) is modulated by microRNA miR-504. Biol Psychiatry. 2009;65(8):702–5. doi: 10.1016/j.biopsych.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hu W, et al. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol Cell. 2010;38(5):689–99. doi: 10.1016/j.molcel.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tserel L, et al. MicroRNA expression profiles of human blood monocyte-derived dendritic cells and macrophages reveal miR-511 as putative positive regulator of Toll-like receptor 4. J Biol Chem. 2011;286(30):26487–95. doi: 10.1074/jbc.M110.213561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Saito Y, et al. Chromatin remodeling at Alu repeats by epigenetic treatment activates silenced microRNA-512-5p with downregulation of Mcl-1 in human gastric cancer cells. Oncogene. 2009;28(30):2738–44. doi: 10.1038/onc.2009.140. [DOI] [PubMed] [Google Scholar]
- 150.Chen F, et al. Inhibition of c-FLIP expression by miR-512-3p contributes to taxol-induced apoptosis in hepatocellular carcinoma cells. Oncol Rep. 2010;23(5):1457–62. doi: 10.3892/or_00000784. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.