Abstract
Liver X receptors (LXRs) attenuate inflammation by modulating the expression of key inflammatory genes, making LXRs and their ligands particularly attractive candidates for therapeutic intervention in cardiovascular, metabolic, and/or inflammatory diseases. In this study, we demonstrate enhanced proresolving activity of nanoparticles (NPs) containing the synthetic LXR agonist GW3965 (LXR-NPs), developed from a combinatorial library of more than 70 formulations with variations in critical physicochemical parameters. The library of LXR-NPs was engineered via self-assembly of biodegradable poly(lactide)- and poly(lactide-co-glycolide)-based polymers. In vitro studies on peritoneal macrophages confirmed that LXR-NPs were significantly more effective than the free agonist (GW3965) at downregulating pro-inflammatory mediators (MCP-1 and TNFα), as well as inducing the expression of LXR target genes (ABCA1 and SREBP1c). Through a zymosan-induced acute peritonitis in vivo model, LXR-NPs were found to be more efficient than free LXR agonist at limiting the recruitment of polymononuclear neutrophils (50% vs. 17%) and at decreasing the resolution interval up to 4 h. In addition, treatment with LXR-NPs significantly suppressed gene expression and secretion of pro-inflammatory factors MCP-1 and TNFα in peritoneal macrophages when compared to LXR agonist alone. Furthermore, the LXR-NPs suppressed secretion of these pro-inflammatory factors by monocytes and macrophages more efficiently than the commercial drug dexamethasone. Overall, these findings demonstrate that LXR-NPs are capable of promoting resolution of inflammation and highlight the prospect of LXR-based nanotherapeutics for inflammatory diseases.
Keywords: nanomedicine, drug delivery systems, polymeric nanoparticles, liver X receptor agonist, Inflammation
1. Introduction
Liver X receptors (LXRs) play a crucial role in regulating several metabolic pathways including lipid, carbohydrate, and bile acid metabolism.[1] LXRs are thought to have atherosclerosis protective properties that include their contribution to the reverse cholesterol transport process (by stimulating cholesterol efflux from macrophages) and their attenuation of inflammation in macrophages by trans-repressing Nf-kB.[2] For example, Tontonoz et al. recently reported that LXR ligands inhibit the expression of inflammatory mediators in peritoneal macrophages in response to bacterial infection or lipopolysaccharide (LPS) stimulation.[3] Several in vivo studies have highlighted the profound impact of LXR agonists on controlling inflammation in mouse models of several acute and chronic diseases such as contact dermatitis,[4] experimental autoimmune encephalomyelitis,[5] Alzheimer's disease,[6] lupus-like autoimmunity, experimental stroke, infection with Mycobacterium tuberculosis,[7] and atherosclerosis.[3] In addition, activation of LXR plays an important role in mitigating inflammation by modulating inflammatory gene expression in many cell types, such as macrophages, CD-positive lymphocytes, microglia, astrocytes, and dendritic cells.[8] In this context, synthetic LXR agonists (T0901317 and GW3965)[9] have been shown to promote cholesterol efflux and inhibit inflammation, and are promising candidates in the development of therapeutics for atherosclerosis and inflammatory diseases.
Nanotechnology is one of the fastest-growing areas in medicine, and nanotherapeutics are poised to influence the landscape of both pharmaceutical and biotechnology industries.[10] The many advantages of therapeutic NPs include improving the pharmaceutical and pharmacological properties of existing drugs, enhancing therapeutic efficacy, and delivering drugs across a range of biological barriers including epithelial and endothelial layers.[11] Over the past decade, nanomedicines have made significant progress towards human use, with exploration of their application beyond oncology, e.g., in infectious diseases and cardiovascular diseases.[12] For example, in a recent study, humanized NPs termed nano-proresolving medicines, have been shown to limit acute inflammation, enhance resolution, and reduce joint damage in a peritonitis model.[13] In addition, we previously demonstrated that proresolving NPs containing the annexin A1/lipocortin A1 mimetic peptide; Ac2-26, were capable of enhancing inflammation resolution in vivo and blunting excessive inflammation in a hind-limb ischemia-reperfusion model.[14]
In the present study, we investigated the anti-inflammatory effects of polymeric NPs containing the encapsulated and/or covalently conjugated LXR agonist GW3965. The LXR agonist–containing NPs (LXR-NPs) were synthesized via self-assembly of biodegradable PLGA, PLA-based polymeric derivatives. Lactide- and glycolide-based polymers and polymeric derivatives are versatile building blocks with distinctive properties, making them important ingredients of various nanotechnology tools developed for drug delivery applications.[15] In combination with polyethylene glycol (PEG), they have long been used safely in several pharmaceutical products and medical devices.[16] These derivatives also have unique biocompatibility and degradation properties, and their physicochemical properties can easily be manipulated to customize and control release of therapeutics.[17] To address the multifaceted challenges of optimizing NPs, high-throughput technologies and combinatorial biomaterial libraries with a wide diversity of physicochemical properties have been utilized for in vitro screening to identify NPs suitable for in vivo testing.[18] Recently, a library of NPs targeted to Prostate Specific Membrane Antigen (PSMA) was developed by introducing physicochemical diversity into the NP design while restricting the particle makeup to a clinically validated set of biomaterials.[19]
Here we present a platform to design and develop a LXR-NP library (Figure 1) with variations in critical parameters including size, surface charge, drug loading, and drug release, all of which can affect the pharmacokinetics and biodistribution of the LXR agonist GW3965. Our LXR-NP library contained more than 70 distinct NP formulations, with the LXR-NPs with desirable physicochemical properties (< 120 nm in size and high drug loading) being further evaluated for their anti-inflammatory effects in vitro, by inhibiting LPS-induced inflammatory response in peritoneal macrophages. The selected LXR-NP from in vitro evaluation was shown to be more potent than LXR agonist alone or dexamethasone, a potent anti-inflammatory and immunosuppressant drug,[20] at blocking the recruitment of zymosan-stimulated polymorphonuclear neutrophils (PMNs) in an acute peritonitis model.[14] Additionally, these NPs were shown to significantly decrease levels of inflammatory cytokines (MCP-1, monocyte chemotactic protein-1; and Tumor necrosis factor TNF-α) in zymosan-induced inflammation in vivo. The anti-inflammatory LXR-NPs developed in this study may prove to be beneficial for other chronic diseases involving excessive inflammation.
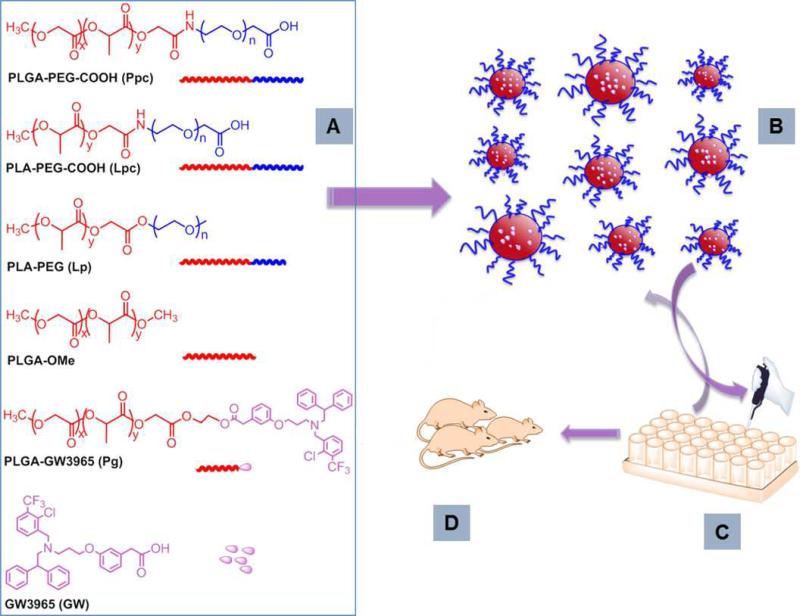
Figure 1.
Development of LXR-NP library, along with in vitro, and in vivo, evaluations. A) Chemical structure of polymers and polymer drug conjugates used in the generation of particle library. B) Synthesis and characterization of LXR-NPs prepared via self-assembly of polymers using nanoprecipitaion method. C) In vitro evaluation of LXR-NPs for anti-inflammatory action and cytotoxicity. D) In vivo assessment of NPs containing LXR agonist.
2. RESULTS AND DISCUSSION
2.1. Synthesis and Characterization of LXR-NP Library
To develop the library of LXR-NPs, we synthesized a series of biodegradable polymers: poly (D,L-lactide-co-glycolide)-b-poly(ethylene glycol), (PLGA43.5K-PEG3.4K-COOH, Ppc); poly (D,L-lactide)-b-poly(ethylene glycol), (PLA17K-PEG3.4K-COOH, Lpc); poly (D,L-lactide)-b-poly(ethylene glycol), (PLA9K–PEG2K, Lp); and poly (lactide-co-glycolide)- LXR agonist conjugate, (PLGA6.7K-GW3965, Pg). Diblock polymers with carboxyl terminal groups on PEG (Ppc, Lpc) were synthesized according to published procedures.[21] Briefly, the carboxy terminals of PLGA (MW 43.5K) or PLA (MW 17K) were activated by 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) activation methodology and reacted with the amino functionality of PEG (MW 3.4 K) to yield Ppc and Lpc, respectively. Diblock polymers in which PEG contains a terminal methyl group (Lp) were synthesized using a ring opening polymerization reaction.[22] Poly(ethylene glycol) methyl ether (CH3-PEG-OH, MW 2K), was used as an initiator of ring opening polymerization of lactide in the presence of Tin catalyst, yielding the desired Lp with ~11k MW. The PLGA-LXR agonist conjugate Pg was produced by first converting the terminal carboxyl groups of PLGA into hydroxyl groups (SI 1), followed by conjugation of LXR agonist GW3965 via EDC/NHS chemistry. All the products were purified as shown in the Materials and Methods section below and characterized by 1H NMR spectroscopy.
A combinatorial library of LXR-NPs with diverse physicochemical properties was synthesized through a single-step self-assembly process. Organic solutions (acetone or acetonitrile) containing GW3965 and polymeric components were added dropwise to an aqueous phase, resulting in encapsulation of GW3965 in solid particles. More than 70 formulations were synthesized by varying parameters of the polymeric units such as the molecular weight, concentration, and composition. All NPs in the library consisted of (i) a biodegradable PLGA- and/or PLA-based polymeric hydrophobic core that can encapsulate GW3965 and release it in a controlled manner; and (ii) a hydrophilic corona formed by PEG units to provide steric stabilization, stealth properties against protein absorption, and long blood circulation time. The LXR agonist GW3965 was either physically encapsulated and/or blended into the hydrophobic polymer core via drug-polymer conjugate Pg in all the LXR-NPs. Encapsulation efficiency and GW3965 loading were improved by blending Pg with either Ppc or Lp, and encapsulation of GW3965. By systematically blending different proportions of polymers and drugs, we obtained NP formulations with various sizes, surface charges, polymer/drug ratios, and lactide/glycolide ratios (Figure 2A). In order to get several different lactide/glycolide ratios, as well as MW variations, PLGA with a terminal methoxy group (PLGA-OMe) was blended with Ppc and/or Lp to produce the LXR-NPs. LXR-NPs with various hydrodynamic size and surface charges were obtained by mixing different proportions of Ppc/Lpc with Lp as it contains a neutral methoxy terminal group, while Ppc or Lpc contains carboxyl as a terminal end group. All the NPs were characterized with respect to particle size, surface charge, GW3965 loading, and encapsulation efficiency (Figure 2B).
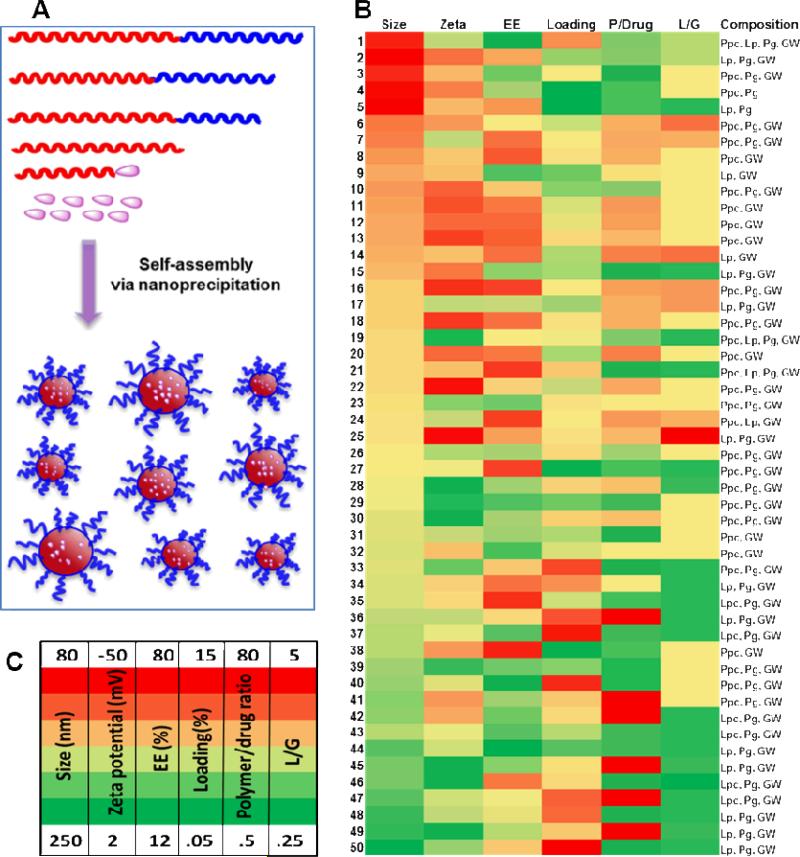
Figure 2.
A) Synthesis of NPs via nanoprecipitation method (Inset, red block: controlled release polymer, blue block: PEG). B) Assortment of NP library. Heat map illustrates formulation parameters (% of encapsulation efficiency, % of loading, and polymer/drug ratio) and physicochemical properties (hydrodynamic size, surface charge) of NPs. C) Heat map parameters.
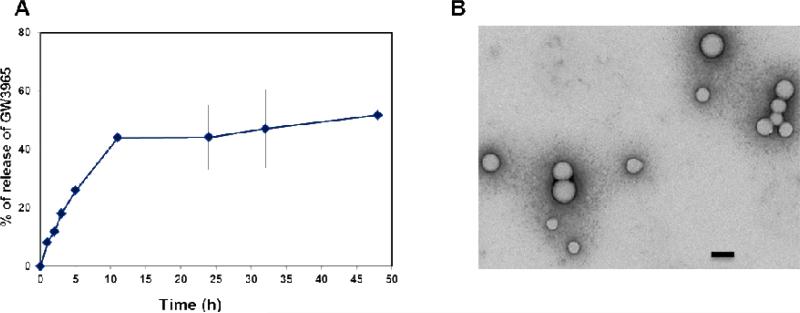
Hydrodynamic size of the NPs in this library varied from 80 to 250 nm, with surface charges ranging from 2 to -50 mV depending on the composition of polymers. Formulations containing Lp produced higher hydrodynamic size and lower surface charges compared with NPs containing Ppc. The NPs containing Lp as a major component had an average size of ~ 170 nm, while average size with Ppc as a major polymeric component was ~ 120 nm. The short PEG length (MW 2K) with a methyl terminal group of Lp may cause increase in NP size, as Ppc contains a longer PEG (MW 3.4K) group with terminal carboxyl functionality. The difference in length and terminal functionalities of PEG provided an extra tool to manipulate the surface charge and size of NPs. The percentage encapsulation efficiency (% EE) and loading efficiency were measured for all the formulations; EE as high as 80% (formulation #21) and 15% loading of GW3965 (formulation #37) were achieved (Figure 2B). Transmission electron microscopy (TEM) studies showed that these NPs were spherical with approximate size < 100 nm and uniform structure (Figure 3B). The release kinetic studies of GW3965 were performed by incubating NPs in PBS at 37 °C, and GW3965 concentrations in the NPs were measured at different time intervals using high-performance liquid chromatography (HPLC). The cumulative release curve (Figure 3A, and Figure S2) shows an initial fast release of GW3965 (27 %) from the NPs for the first 5 hours, followed by a slow and diffusion-controlled process, releasing 57% within 48 h.
Figure 3.
A) In vitro release profile of GW3965 from PLA-PEG/PLGA/GW NPs incubated at 37 °C is shown (mean ±SD, n = 3). The LXR agonist released from the NPs at different time points was quantified by HPLC. B) Transition electron microscopy (TEM) images of LXR agonist containing NPs formed by blending PLA-PEG-COOH and PLGA-GW polymers (Scale bar, 100 nm).
NPs synthesized by blending Pg with either Ppc or Lp with w/w ratios ranging from 1-20 resulted in hydrodynamic size 90-120 nm, which are suitable for in vitro and in vivo studies. Of these NPs, four formulations were chosen for further in vitro testing: PpcPgGW (# 6), LpPgGW (# 15), PpcGW (# 20), and LpGW (# 14) (GW 3965 is noted as GW in the formulations). Their hydrodynamic sizes were 100-116 nm, with a surface charge of -31 to -38 mV, ~ 80% EE, and 2.5-7% loading drug.
2.2. In-vitro Efficacy and Anti-inflammatory Effect of LXR-NPs on Peritoneal Macrophages
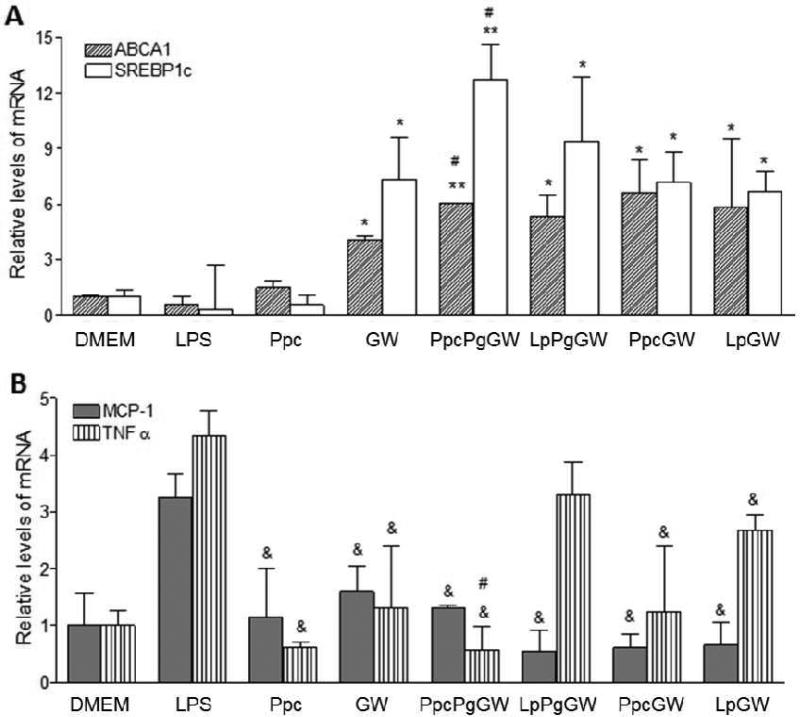
From the combinatorial library of LXR-NPs, formulations with ideal physicochemical properties were further tested in mouse peritoneal macrophages for in vitro efficacy in inducing LXR target genes and altering LPS-induced inflammatory responses through transrepression of NF-kB. Studies have shown that LXRs, in addition to suppressing the expression of macrophage inflammatory genes, can induce the expression of several genes involved in reverse cholesterol transport, as well as in lipid synthesis.[3] For our in vitro studies, we have focused on LXR target genes ABCA1 (ATP-binding cassette A1) and SREBP1c (sterol regulatory element binding protein) as efficacy markers for LXRs activation.[23] We also measured relative mRNA expression and protein levels of inflammatory cytokine mediators MCP-1 and TNFα, as NF-kB targets are suppressed upon LXR activation. Isolated murine peritoneal macrophages were pretreated for 18 h with 1 μM GW3965 (either in solution or encapsulated in NP formulation) followed by LPS (100ng/ml) treatment to induce a pro-inflammatory response. The effect of GW3965 (free or in NP form) on gene expression was determined using quantitative real-time PCR (qRT-PCR), and the mRNA expression levels of treated groups were compared to the untreated control. As shown in Figure 4A, ABCA1 and SREBP1c mRNA levels in macrophages treated with LXR-NPs were five- to six-fold and nine- to eleven-fold higher (p < 0.05) than empty NP- and LPS-treated macrophages, respectively. In addition, the PpcPgGW NPs produced better gene target expression than GW3965. These results suggest that NPs provide better macrophage uptake of the agonist, and combined with controlled release of GW3965, LXR-NPs were made to be more effective than free GW3965 alone.
Figure 4.
Peritoneal macrophages were treated with Ppc, GW 1μM, PpcPgGW 1μM, LpPgGW 1μM, PpcGW 1μM, or LpGW 1μM. Following 18 h, the cells were incubated with 100ng/ml LPS for 6 more hours. The gene expression was measured by real time quantitative PCR for ABCA1 and SREBP-1c (A) and TNFα and MCP-1 (B). Each bar represents the Mean ± SEM. *p < 0.05 vs DMEM, LPS and Ppc; #p < 0.05 vs GW; &p<0.05 vs LPS
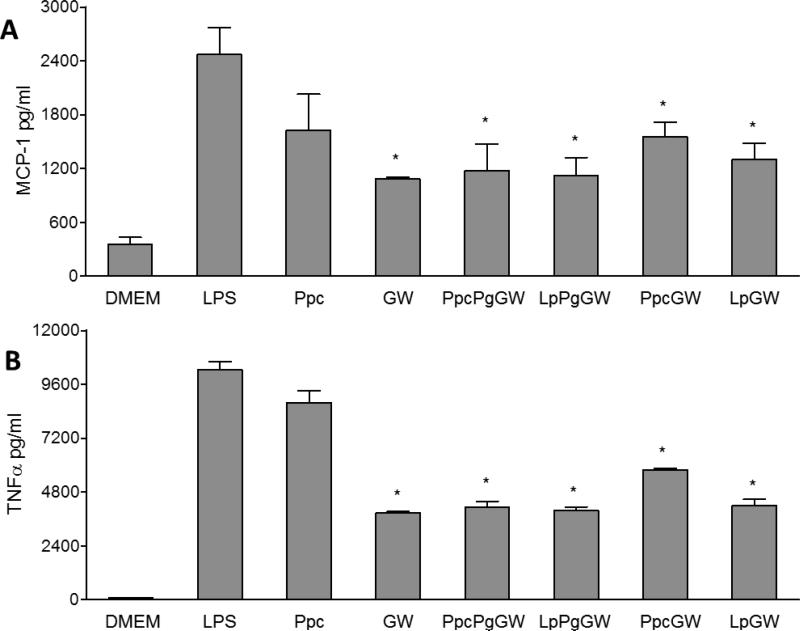
The expression and secretion of pro-inflammatory cytokines (MCP-1 and TNFα) were quantified by both qRT-PCR and ELISA. Macrophages treated with GW3965 or LXR-NPs significantly suppressed mRNA expression of MCP-1 and TNFα upon LPS stimulation (Figure 4B). When compared to LPS control, there was a 50 to 85% decrease in (p < 0.05) MCP-1 mRNA levels in cells treated with LXR-NPs, as compared to ~ 40% decrease in MCP-1 mRNA levels observed for (p < 0.05) for GW3965 alone. Moreover, macrophages treated with PpcPgGW NPs had significantly curbed TNFα mRNA levels compared to those treated with free GW3965 (88% to 60 % decrease, p < 0.05). Similarly, compared to LPS treatment, LXR-NP treatment resulted in lower MCP-1 (~1200 pg/ml, p < 0.05) and TNFα (~ 5000 pg/ml, p < 0.05) protein secretion as shown in Figure 5A and B. Based on the above results, PpcPgGW formulation has higher efficacy in reducing inflammatory gene and protein expression than the free LXR agonist and the other NPs tested. The anti-inflammatory effect of this formulation was further investigated in in vivo mouse models.
Figure 5.
Peritoneal macrophages were treated with Ppc, GW 1μM, PpcPgGW 1μM, LpPgGW 1μM, PpcGW 1μM, or LpGW 1μM. Following 18 h, the cells were incubated with 100ng/ml LPS for 6 more hours. The upper media was collected and tested for MCP-1 (A) and TNFα (B) secretion from cells by ELISA. Each bar represents the Mean ± SEM. *p<0.05 vs LPS.
2.3. In vivo Anti-inflammatory Effect of GW3965-Containing NPs
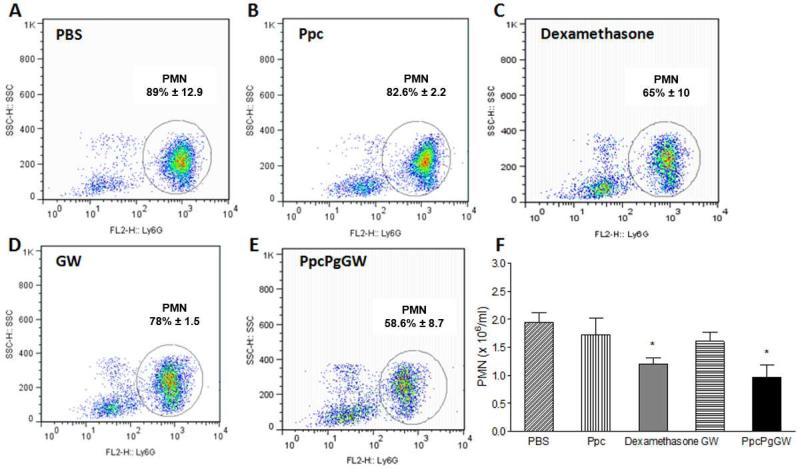
To determine whether LXR-NPs are anti-inflammatory and pro resolving in vivo, we used a standard zymosan-induced peritonitis model[24] to quantitatively assess the resolution of inflammation in wild-type C57BL/6 mice. The mice were intravenously (i.v.) treated with PBS, empty NPs (Ppc), GW3965 (8 mg/kg), dexamethasone (1mg/kg), and our selected LXR-NP formulation PpcPgGW (8 mg/kg GW3965). After one hour, 100 μg of zymosan was administrated intraperitoneally (i.p.) to each mouse, and leukocytic cell infiltration into the inflammation site was assessed 4 h later (Figure 6). We saw 50% reduction of zymosan-induced PMN cell recruitment into the inflammation site in the mice treated with PpcPgGW, while empty NPs Ppc had no protective effect. In addition, among mice treated with dexamethasone (a potent commercial steroid drug) or free GW3965, the PMN infiltration was blocked by only 38% or 17%, respectively. As empty NPs did not exert any protective effect, and free GW3965 blocked only 17% of PMN infiltration at the inflammation site, our findings demonstrate the enhanced anti-inflammatory activity of LXR agonist delivered via NP formulations. The improved pharmacokinetics and controlled release of GW3965 by LXR-NPs can be attributed to the greater bioactivity of GW3965.
Figure 6.
WT male mice (n=5) were administrated i.v with the respective regimens and after one hour mice were injected i.p. with Zymosan, (A) Zymosan + PBS, (B) Zymosan + Ppc, (C) Zymosan + Dexamethasone 1 mg/kg, (D) Zymosan + 8mg/kg GW, (E) Zymosan + PpcPgGW 8mg/kg. Four hours afterwards, the cells were harvested and analyzed by FACS (F). Each bar represents the Mean ± SEM. *p<0.05 vs PBS and Ppc
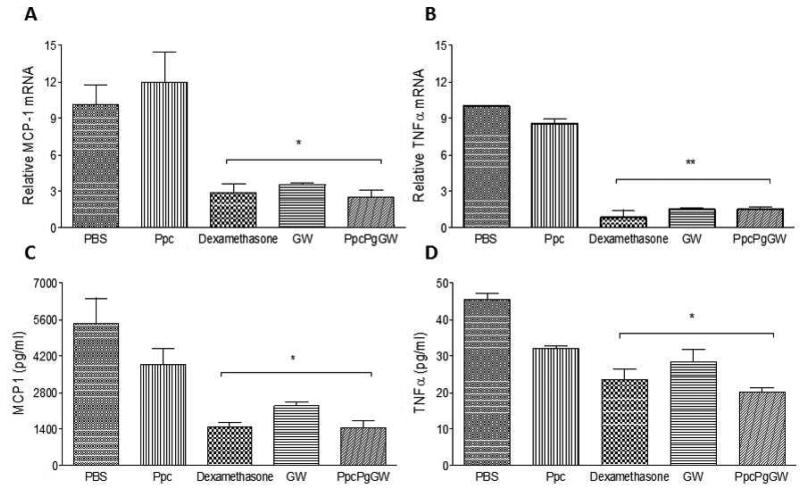
The in vivo pro-resolving action of LXR-NPs was further indicated by the suppression of MCP-1 and TNFα mRNA gene expression and protein secretion in the peritoneal exudates harvested from the mice. Peritoneal macrophages were harvested 4 h following zymosan injection, and the gene expression in these cells of MCP-1, TNFα and the protein content of the peritoneal exudates of these factors, was measured (Figure 7). Our results indicate that, comparable to dexamethasone treatment, treatment with GW3965 (either in solution or in NP form) significantly inhibited the secretion of both pro-inflammatory factors. We also saw 70% decrease in MCP-1 mRNA levels and ~ 80% decrease in TNFα mRNA levels for PpcPgGW treatment as compared to PBS treatment (control mice). The LXR-NP formulation PpcPgGW showed enhanced ability to resolve inflammation, as evidenced by major suppression in the secretion of both MCP-1(1400 pg/ml, p < 0.05) and TNFα (20 pg/ml, p < 0.05) when compared to PBS (MCP-1: 5600 pg/ml, TNFα: 45 pg/ml ) and Ppc (MCP-1:4200 pg/ml, TNFα:32 pg/ml). Also, compared to free GW3965 (MCP-1: 2200 pg/ml, TNFα:25 pg/ml) or dexamethasone (MCP-1 :1400 pg/ml, TNFα:25 pg/ml), secretion of MCP-1 and TNFα proteins was lower in the PpcPgGW group, highlighting its ability to suppress the inflammatory response in vivo. Our ELISA results are consistent with the zymosan-induced PMN recruitment experiments, further underscoring the in vivo anti-inflammatory effects of PpcPgGW, which exceed those of dexamethasone.
Figure 7.
WT male mice (n=5) were administrated i.v with the respective regimens PBS, Ppc, Dexamethasone 1 mg/kg, GW 8mg/kg, and PpcPgGW 8mg/kg, and after one hour, mice were injected i.p. with Zymosan. Four hours afterwards, the cells were harvested and the gene expression was measured by real time quantitative PCR for TNFα (A) and MCP-1 (B). The peritoneal exudates were tested for MCP-1 (C) and TNFα (D) release from cells by ELISA. Each bar represents the Mean ± SEM. *p<0.05, **p<0.01 vs PBS and Ppc.
The results presented here are in agreement with previously reported studies on the role of LXRs as anti-inflammatory mediators in acute and chronic inflammation. Specifically, our studies validate the capability of LXR-NP formulations to improve LXR agonist activity in vivo, and these proof-of-concept studies may pave the way for the development of LXR agonist-based anti-inflammatory nanotherapies.
3. Conclusions
We have synthesized a library of LXR agonist–containing NPs via a self-assembly process using polylactide- and glycolide-based biodegradable polymeric derivatives. Our combinatorial library consists of more than 70 formulations with variations in critical physicochemical parameters. Based on a number of criteria, the NPs selected for furher study demonstrated anti-inflammatory effects in vitro by inhibiting LPS-induced inflammatory responses in peritoneal macrophages, as evidenced by significant suppression of the secretion of pro-inflammatory mediators MCP-1 and TNFα. Furthermore, in the in vivo zymosan-induced peritonitis model, LXR-NPs considerably reduced PMN cell infiltration into the inflammation site (50%). LXR agonist activity was improved by administration through the LXR-NP formulations. These inflammatory proresolving LXR-NPs have the potential to be developed as therapeutics for the treatment of a variety of diseases where inflammation is underlying cause, including peritonitis.
4. Experimental section
Materials
Poly(D,L-lactide-co-glycolide) (50/50) with terminal carboxylate groups (PLGA, inherent viscosity 0.55-0.75 dL/g, and 0.15-0.25 dL/g) and DL lactide were obtained from Lactel (Pelham, AL, USA). N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), N-Hydroxysuccinimide (NHS), ethylene glycol, and poly(ethylene glycol) methyl ether with a molecular weight (MW) of 2000 ( (CH3-PEG-OH), tin(II) 2-ethylhexanoate, GW3965 hydrochloride, and lipopolysaccharide (LPS) were purchased from Sigma-Aldrich. A PEG polymer, MW 3,400, with a terminal amine and carboxylic group (NH2-PEG-COOH) was purchased from Laysan Bio, Inc. (Arab, AL, USA). PLGA-PEG-COOH was synthesized as previously described[25],[26]. 1H NMR spectra were recorded on a Bruker AVANCE-400 NMR spectrometer with a Spectro Spin superconducting magnet at Harvard Medical School. Drug quantification was carried out on Agilent HPLC using an RC18 column. The NP size was obtained with quasi-electric laser light scattering using a ZetaPALS dynamic light-scattering detector (15mW laser, incident beam ¼ 676nm; Brookhaven Instruments). For cell culture procedures, Dulbecco's modified Eagle's medium (DMEM) and fetal bovine serum (FBS) were purchased from Invitrogen, and thioglycolate medium was purchased from Fluka Analytical. For in vivo studies, Zymosan A and Dexamethasone were purchased from Sigma-Aldrich.
Synthesis of PLA-PEG
Synthesis of PLA-PEG was accomplished via ring-opening polymerization reaction as described previously.[27] Poly(ethylene glycol) methyl ether (MW 2000) (500 mg, 0.25 mmol), DL lactide (3g, 21mmol), and Tin(II) 2-ethylhexanoate (2 mol % from a stock solution in toluene) were added to a toluene solution (15 ml) under nitrogen and refluxed at 120°C for 10 hours. The reaction mixture was concentrated and dropped in ice-cold diethyleither:methanol (50:50) solution to form a white precipitate. The precipitate was further purified by repeated dissolution–reprecipitation using DCM-Ether/MeOH. Finally, the precipitate was dried under vacuum to obtain PLA-PEG polymer with ~70% yield. NMR (400 MHz, CDCl3): δ 5.0-5.22 (m, (-OCH(CH3)CO)), 3.58 (s, (-OCH2CH2O-), 1.4-1.6 (m, (-OCH(CH3)CO-) ppm.
Synthesis of PLGA-GW3965
PLGA-GW3965 was synthesized in a two-step procedure as shown in SI 1. In the first step, the terminal carboxylic groups of PLGA-COOH (I.V. 0.15-0.25 dL/g) were reacted with ethylene glycol to convert them to hydroxyl groups. 500mg of PLGA-COOH (~0.07 mmol) was dissolved in dichloromethane (DCM), and 57.5mg of EDC (0.3 mmol) and 35 mg of NHS (0.3 mmol) were added and stirred for one hour at room temperature. The reaction mixture was added dropwise to ice-cold methanol to form a white precipitate of PLGA-NHS. The precipitate was purified by dissolving in 500μL of DCM and then re-precipitating in ice-cold methanol. The precipitate was dried under vacuum and dissolved in DCM and DIEA (36 mg, 0.28 mmol). Subsequently, excess ethylene glycol was added, and the solution was stirred at room temperature overnight. The reaction mixture was concentrated under vacuum and added dropwise to ice-cold MeOH to form a precipitate. The precipitate was purified by repeated dissolution–reprecipitation using DCM-MeOH to obtain PLGA with hydroxyl end group and used for the second step without further purification. In the second step, 25mg of GW3965 (0.04 mmol), 20 mg of EDC (0.1 mmol), and 12 mg of NHS (0.1 mmol) were dissolved in DCM and stirred at room temperature for one hour. PLGA-OH in DCM and DIEA (11.5 mg, 0.09 mmol) was added to this reaction mixture and stirred overnight. The reaction mixture was concentrated under vacuum and added to ice-cold methanol to form a white precipitate of PLGA-GW3965. This precipitate was further purified by repeated dissolution–reprecipitation using DCM-MeOH to obtain pure PLGA-GW3965. 60% yield. 1H NMR (400 MHz, CDCl3): δ 5.1-5.3 (m, (-OCH(CH3)CO)), 4.6-4.9 (m, (-OCH2COO-)),4.29 (m, (-OCH2CH2 CO-GW3965), 1.49-1.6 (m, (-OCH(CH3)CO-) ppm. GW3965 peaks: 6.87-7.18, 6.30-6.73, 5.39, 3.88, 3.82, 3.49, 3.15, 2.83, and 2.02).
Synthesis and characterization of NPs
The library of LXR NPs was synthesized using a nanoprecipitation method. Briefly, all the polymers and GW3965 were dissolved in either acetonitrile (ACN) or acetone in a range of concentrations, such as, 10-20mg/mL for polymers and 1-2mg/mL for GW3965. These polymer and drug solutions were mixed together in desired ratios and added dropwise to nuclease-free water, while maintaining a 10:1 ratio of water to organic solvent. NPs were stirred overnight and filtered sequentially through sterile 0.45μm syringe filters (regenerated cellulose, 17mm, Cole Palmer Instruments). The NPs were concentrated by centrifugation using Amicon Ultra-15 centrifugal filter units (MWCO 100KDa, or 50KDa). The concentrated NPs were washed twice with de-ionized water and re-suspended in 1mL of either nuclease-free H2O or PBS.
Size and surface charge measurements of NPs were performed using dynamic light scattering; NPs were diluted 10-20 times in water for measurements. TEM experiments were carried out on a TecnaiTM G2 Spirit BioTWIN electron microscope equipped with an AMT 2k CCD camera and low-dose software (80kV, direct mag. 98000x). The TEM sample was prepared by depositing 10μL of NPs, freshly prepared in water, (1.0mg/mL) onto a carbon-coated copper grid. The excess solution was blotted, and the grids were immersed in a solution of 0.75% uranyl formate stain. The stain was blotted, and dried grids were used for imaging within one hour of the preparation of NPs.[14] Drug loading and encapsulation efficiency (EE) were determined by quantifying the amount of drug in the NPs. A calibration curve with known concentrations of LXR agonist was prepared by using HPLC, with 80:20 ACN: H2O mobile phase, while injected samples were prepared in in 1:1 ACN: H2O mixture. The amount of GW3965 in the NPs was measured using the calibration curve. A portion of the synthesized NPs was dissolved in 1:1 ACN: H2O with 10% of 0.01mM NaOH and vortexed for several hours to break down the NPs before injecting them into the HPLC. Drug loading is defined as the fraction of drug mass in the NPs, whereas EE is the fraction of initial drug that is encapsulated by the NPs.
GW3965 NP release profile study
To assess GW3965 NPs release kinetics, NPs were suspended in water and aliquoted (200μL) into several semi-permeable mini-dialysis tubes (molecular weight cutoff 10kDa; Pierce). The dialysis tubes were placed in 20L of PBS (pH 7.4) at 37°C. At defined time intervals, tubes were removed from dialysis and an aliquot of the NP suspension was collected. The remaining drug in NPs at different time points was quantified by using HPLC as described above.
Cell culture
Peritoneal macrophages were obtained from thyoglicolate-injected C57Bl/6J mice as described previously.[28] Cells (1 × 106) were plated on 6-well plates and cultured in DMEM supplemented with 10% FBS. For ligand treatments and anti-inflammatory effect experiments, cells were cultured for 18 h with the respective regimens at a 1μM concentration of the GW3965 LXR agonist (either in solution or in NP form), and then treated with 100ng/ml LPS for an additional 6 h.
Animals
All animals were obtained from Jackson Laboratories. The animals were allowed free access to sterile food pellets and water. All in vivo studies were performed in accordance with National Institutes of Health Animal Care guidelines.
Zymosan peritonitis
Male C57Bl/6J mice (6-8 weeks) were administrated i.v. with the respective regimens at a 8 mg/Kg concentration of the GW3965 (free GW or inside NPs) or 1mg/ml Dexamethasone. One hour later the mice were injected i.p. with Zymosan A (100μg/mouse) to induce peritonitis.[24] Peritoneal exudates were harvested 4 h post-zymosan administration.
qRT-PCR
Total RNA abundance was determined by qRT-PCR (TaqMan) using 100pg total RNA. The primer and probe sequences are described in Table 1. Peritoneal macrophage and in-vivo data were normalized to 28s and Cyclophilin (respectively) and were presented as the fold difference over controls.
Statistical analysis
Student's t-test or one-way ANOVA with post-hoc Tukey tests were used to determine significance. All error bars represent S.E.M.
Supplementary Material
Acknowledgments
This work was supported by a Program of Excellence in Nanotechnology (PEN) Award, Contract #HHSN268201000045C, from the National Heart, Lung, and Blood Institute, National Institutes of Health (NIH). This work was also supported by NIH grants CA151884 and the David Koch-Prostate Cancer Foundation Award in Nanotherapeutics. J.S. receives financial support from NCI R00CA160350. The authors thank Dr. David Greaves and Dr. Gabrielle Fredman for insightful comments and discussions.
Footnotes
Conflict of interest statement: In compliance with the Brigham and Women's Hospital and Harvard Medical School institutional guidelines, O.C.F. discloses his financial interest in BIND Therapeutics, Selecta Biosciences and Blend Therapeutics, three biotechnology companies developing nanoparticle technologies for medical applications. BIND, Selecta and Blend did not support the aforementioned research, and currently these companies have no rights to any technology or intellectual property developed as part of this research.
Contributor Information
Suresh Gadde, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02115, USA.
Orli Even-Or, Department of Cell Biology and the Leon H. Charney Division of Cardiology, Department of Medicine, New York University School of Medicine, New York, NY 10016..
Nazila Kamaly, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02115, USA.
Apoorva Hasija, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02115, USA.
Philippe G. Gagnon, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02115, USA
Krishna H. Adusumilli, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02115, USA
Andrea Erakovic, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02115, USA.
Anoop K. Pal, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02115, USA
Xue-Qing Zhang, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02115, USA.
Nagesh Kolishetti, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02115, USA.
Jinjun Shi, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02115, USA.
Edward A. Fisher, Department of Cell Biology and the Leon H. Charney Division of Cardiology, Department of Medicine, New York University School of Medicine, New York, NY 10016..
Omid C. Farokhzad, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02115, USA; King Abdulaziz University, Jeddah 21589, Saudi Arabia.
References
- 1.Calkin AC, Tontonoz P. Nat Rev Mol Cell Biol. 2012;13:213. doi: 10.1038/nrm3312. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zelcer N, Tontonoz P. J Clin Invest. 2006;116:607. doi: 10.1172/JCI27883. [DOI] [PMC free article] [PubMed] [Google Scholar]; Beaven SW, Matveyenko A, Wroblewski K, Chao L, Wilpitz D, Hsu TW, Lentz J, Drew B, Hevener AL, Tontonoz P. Cell Metab. 2013;18:106. doi: 10.1016/j.cmet.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hong C, Tontonoz P. Curr Opin Genet Dev. 2008;18:461. doi: 10.1016/j.gde.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joseph SB, Castrillo A, Laffitte BA, Mangelsdorf DJ, Tontonoz P. Nat Med. 2003;9:213. doi: 10.1038/nm820. [DOI] [PubMed] [Google Scholar]
- 4.Fowler AJ, Sheu MY, Schmuth M, Kao J, Fluhr JW, Rhein L, Collins JL, Willson TM, Mangelsdorf DJ, Elias PM, Feingold KR. J Invest Dermatol. 2003;120:246. doi: 10.1046/j.1523-1747.2003.12033.x. [DOI] [PubMed] [Google Scholar]
- 5.Hindinger C, Hinton DR, Kirwin SJ, Atkinson RD, Burnett ME, Bergmann CC, Stohlman SA. Journal of Neuroscience Research. 2006;84:1225. doi: 10.1002/jnr.21038. [DOI] [PubMed] [Google Scholar]
- 6.Zelcer N, Khanlou N, Clare R, Jiang Q, Reed-Geaghan EG, Landreth GE, Vinters HV, Tontonoz P. Proceedings of the National Academy of Sciences. 2007;104:10601. doi: 10.1073/pnas.0701096104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michael DR, Ashlin TG, Buckley ML, Ramji DP. Curr Atheroscler Rep. 2012;14:284. doi: 10.1007/s11883-012-0239-y. [DOI] [PubMed] [Google Scholar]
- 8.Bensinger SJ, Bradley MN, Joseph SB, Zelcer N, Janssen EM, Hausner MA, Shih R, Parks JS, Edwards PA, Jamieson BD, Tontonoz P. Cell. 2008;134:97. doi: 10.1016/j.cell.2008.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hanley TM, Blay Puryear W, Gummuluru S, Viglianti GA. PLoS Pathog. 2010;6:e1000981. doi: 10.1371/journal.ppat.1000981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, Yeh V, Molteni V. Expert Opin Ther Pat. 2010;20:535. doi: 10.1517/13543771003621269. [DOI] [PubMed] [Google Scholar]; Joseph SB, McKilligin E, Pei L, Watson MA, Collins AR, Laffitte BA, Chen M, Noh G, Goodman J, Hagger GN, Tran J, Tippin TK, Wang X, Lusis AJ, Hsueh WA, Law RE, Collins JL, Willson TM, Tontonoz P. Proc Natl Acad Sci U S A. 2002;99:7604. doi: 10.1073/pnas.112059299. [DOI] [PMC free article] [PubMed] [Google Scholar]; Levin N, Bischoff ED, Daige CL, Thomas D, Vu CT, Heyman RA, Tangirala RK, Schulman IG. Arterioscler Thromb Vasc Biol. 2005;25:135. doi: 10.1161/01.ATV.0000150044.84012.68. [DOI] [PubMed] [Google Scholar]
- 10.Kamaly N, Xiao Z, Valencia PM, Radovic-Moreno AF, Farokhzad OC. Chem Soc Rev. 2012;41:2971. doi: 10.1039/c2cs15344k. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shi J, Xiao Z, Kamaly N, Farokhzad OC. Acc Chem Res. 2011;44:1123. doi: 10.1021/ar200054n. [DOI] [PubMed] [Google Scholar]
- 11.Tang BC, Dawson M, Lai SK, Wang Y-Y, Suk JS, Yang M, Zeitlin P, Boyle MP, Fu J, Hanes J. Proceedings of the National Academy of Sciences. 2009;106:19268. doi: 10.1073/pnas.0905998106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radovic-Moreno AF, Lu TK, Puscasu VA, Yoon CJ, Langer R, Farokhzad OC. ACS Nano. 2012;6:4279. doi: 10.1021/nn3008383. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lobatto ME, Fuster V, Fayad ZA, Mulder WJ. Nat Rev Drug Discov. 2011;10:835. doi: 10.1038/nrd3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norling LV, Spite M, Yang R, Flower RJ, Perretti M, Serhan CN. J Immunol. 2011;186:5543. doi: 10.4049/jimmunol.1003865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamaly N, Fredman G, Subramanian M, Gadde S, Pesic A, Cheung L, Fayad ZA, Langer R, Tabas I, Farokhzad OC. Proc Natl Acad Sci U S A. 2013;110:6506. doi: 10.1073/pnas.1303377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh JK. Soft Matter. 2011;7:5096. [Google Scholar]; Gunn J, Zhang M. Trends Biotechnol. 2010;28:189. doi: 10.1016/j.tibtech.2009.12.006. [DOI] [PubMed] [Google Scholar]; Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Préat V. Journal of Controlled Release. 2012;161:505. doi: 10.1016/j.jconrel.2012.01.043. [DOI] [PubMed] [Google Scholar]
- 16.Avgoustakis K. Curr Drug Deliv. 2004;1:321. doi: 10.2174/1567201043334605. [DOI] [PubMed] [Google Scholar]
- 17.Makadia HK, Siegel SJ. Polymers. 2011;3:1377. doi: 10.3390/polym3031377. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kumari A, Yadav SK, Yadav SC. Colloids and Surfaces B: Biointerfaces. 2010;75:1. doi: 10.1016/j.colsurfb.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Siegwart DJ, Whitehead KA, Nuhn L, Sahay G, Cheng H, Jiang S, Ma M, Lytton-Jean A, Vegas A, Fenton P, Levins CG, Love KT, Lee H, Cortez C, Collins SP, Li YF, Jang J, Querbes W, Zurenko C, Novobrantseva T, Langer R, Anderson DG. Proc Natl Acad Sci U S A. 2011;108:12996. doi: 10.1073/pnas.1106379108. [DOI] [PMC free article] [PubMed] [Google Scholar]; Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, Qin J, Cantley W, Qin LL, Racie T, Frank- Kamenetsky M, Yip KN, Alvarez R, Sah DW, de Fougerolles A, Fitzgerald K, Koteliansky V, Akinc A, Langer R, Anderson DG. Proc Natl Acad Sci U S A. 2010;107:1864. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hrkach J, Von Hoff D, Mukkaram Ali M, Andrianova E, Auer J, Campbell T, De Witt D, Figa M, Figueiredo M, Horhota A, Low S, McDonnell K, Peeke E, Retnarajan B, Sabnis A, Schnipper E, Song JJ, Song YH, Summa J, Tompsett D, Troiano G, Van Geen Hoven T, Wright J, LoRusso P, Kantoff PW, Bander NH, Sweeney C, Farokhzad OC, Langer R, Zale S. Sci Transl Med. 2012;4:128ra39. doi: 10.1126/scitranslmed.3003651. [DOI] [PubMed] [Google Scholar]
- 20.Tsurufuji S, Sugio K, Takemasa F. Nature. 1979;280:408. doi: 10.1038/280408a0. [DOI] [PubMed] [Google Scholar]
- 21.Gu F, Langer R, Farokhzad OC. Methods Mol Biol. 2009;544:589. doi: 10.1007/978-1-59745-483-4_37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dutta S, Hung W-C, Huang B-H, Lin C-C. In: Synthetic Biodegradable Polymers. Rieger B, Künkel A, Coates GW, Reichardt R, Dinjus E, Zevaco TA, editors. Vol. 245. Springer; Berlin Heidelberg: 2012. p. 219. [Google Scholar]
- 23.Tontonoz P, Mangelsdorf DJ. Mol Endocrinol. 2003;17:985. doi: 10.1210/me.2003-0061. [DOI] [PubMed] [Google Scholar]
- 24.Cash JL, White GE, Greaves DR. Methods Enzymol. 2009;461:379. doi: 10.1016/S0076-6879(09)05417-2. [DOI] [PubMed] [Google Scholar]
- 25.Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW, Richie JP, Langer R. Proc Natl Acad Sci U S A. 2006;103:6315. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farokhzad OC, Jon S, Khademhosseini A, Tran TN, Lavan DA, Langer R. Cancer Res. 2004;64:7668. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- 27.Riley T, Govender T, Stolnik S, Xiong CD, Garnett MC, Illum L, Davis SS. Colloids and Surfaces B: Biointerfaces. 1999;16:147. [Google Scholar]
- 28.Piao ZH, Kim MS, Jeong M, Yun S, Lee SH, Sun HN, Song HY, Suh HW, Jung H, Yoon SR, Kim TD, Lee YH, Choi I. Cell Immunol. 2012;280:1. doi: 10.1016/j.cellimm.2012.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.