Abstract
Radix Astragali is a popular herb used in traditional Chinese medicine for its proimmune and antidiabetic properties. However, methods are needed to help distinguish Radix Astragali from its varied adulterants. DNA barcoding is a widely applicable molecular method used to identify medicinal plants. Yet, its use has been hampered by genetic distance, base variation, and limitations of the bio-NJ tree. Herein, we report the validation of an integrated analysis method for plant species identification using DNA barcoding that focuses on genetic distance, identification efficiency, inter- and intraspecific variation, and barcoding gap. We collected 478 sequences from six candidate DNA barcodes (ITS2, ITS, psbA-trnH, rbcL, matK, and COI) from 29 species of Radix Astragali and adulterants. The internal transcribed spacer (ITS) sequence was demonstrated as the optimal barcode for identifying Radix Astragali and its adulterants. This new analysis method is helpful in identifying Radix Astragali and expedites the utilization and data mining of DNA barcoding.
1. Introduction
Radix Astragali (Huang Qi), a commonly used Chinese medicinal material, is mainly sourced from the plants of Astragalus membranaceus and Astragalus mongholicus according to Chinese Pharmacopoeia (2010 edition). Radix Astragali is widely used for its antiperspirant, antidiuretic, and antidiabetic properties and as a tonic drug [1–3]. It possesses various beneficial compounds, including astragalosides, isoflavonoids, isoflavones, isoflavan, and pterocarpan glycosides [4–6].
Due to the high market demand for Radix Astragali, a diverse group of adulterants with similar-morphological characteristics from genuses, such as Astragalus, Hedysarum, and Malva are often used in its stead [7]. The traditional methods used to identify Radix Astragali for use as a medicinal material, such as morphological and microscopic identification [8], thin-layer chromatography and Ultraviolet spectroscopy [9], Fourier Transform infrared spectroscopy (FTIR) [10], and high performance liquid chromatography (HPLC) [11], all, require specialized equipment and training. Several PCR-based molecular methods have been developed, providing an alternative means of identification. Multiplex PCR methods of DNA fragment analysis, such as randomly amplified polymorphic DNA (RAPD) [12] or amplified fragment length polymorphism (AFLP) [13], are unstable for the results to identify. DNA barcoding is a widely used molecular marker technology, first proposed by Hebert et al. [14, 15]. It uses a standardized and conserved, but diverse, DNA sequence to identify species and uncover biological diversity [16, 17]. In previous studies, various coding sequences for identifying Radix Astragali and its adulterants have been used, such as the 5S-rRNA spacer domain [18], 3′ untranslated region (3′ UTR) [19], ITS (internal transcribed spacer region) and 18S rRNA [3, 20, 21], ITS2 [22], ITS1 [6], matK (maturase K) and rbcL (ribulose 1, 5-bisphosphate carboxylase) of chloroplast genome, and coxI (cytochrome c oxidase 1) of the mitochondrial genome [23]. However, sequence analysis was mainly focused on genetic distance, variable sites, amplified polymorphisms, and the use of a modified neighbor-joining (NJ) algorithm, Bio-NJ tree, which were basic analyses limited to particular species. A more effective method of molecular identification is necessary. The current study evaluates the identification reliability and efficiency of DNA barcoding for the identification of Radix Astragali using six indicators of genetic distance, identification efficiency, intra- and interspecific variation, gap rate, and barcoding gap. Six barcodes were selected for identification because they are commonly used in plant, especially in medicinal plant. We collected Radix Astragaliand several of its adulterants reported in previous research and downloaded the genetic sequences from the GenBank database. A total of 29 species (including 19 species of Astragalus) and 478 sequences from six barcodes were used to validate the new method for identifying Radix Astragali and adulterants and to accelerate the data utilization of DNA barcoding.
2. Materials and Methods
2.1. Materials Information
A total of 77 specimens were collected from two origins of Radix Astragali, along with seven adulterants. Radix Astragali specimens were collected from Inner Mongolia, Shaan xi, and Gan su provinces in the People's Republic of China, which are the main producing areas. The collection information is shown in Table 1. All corresponding voucher specimens were deposited in the Herbarium of the Institute of Medicinal Plant Development at the Chinese Academy of Medical Sciences in Beijing, China. The GenBank accession number of the ITS2 in this experiment was orderly KJ999296–KJ999344, the accession number of ITS sequences was orderly KJ999345–KJ999416, and the accession number of psbA-trnH was orderly KJ999256–KJ999295. The sequences added in the subsequent analysis, including ITS, ITS2, psbA-trnH, matK, and rbcL, were downloaded from the GenBank database.
Table 1.
Taxon sampling information of astragalus and its adulterants.
| Experiment number | species | Sampling spot | |
|---|---|---|---|
| S1-S5 | Astragalus membranaceus | Shaanxi | China |
| SD1-SD9 | Astragalus membranaceus | Shaanxi | China |
| GS1-GS6 | Astragalus mongholicus | Gansu | China |
| NM1-NM10 | Astragalus mongholicus | Neimeng | China |
| SX1-SX10 | Astragalus mongholicus | Shanxi | China |
| HHQ1-HHQ7 | Astragalus chinensis | Beijing | China |
| CY1-CY6 | Astragalus scaberrimus | Beijing | China |
| JK1-JK3 | Malva pusilla | Shaanxi | China |
| MX | Medicago sativa | Shaanxi | China |
| HH1-HH7 | Melilotus officinalis | Shaanxi | China |
| HQ1-HQ12 | Hedysarum polybotrys | Gansu | China |
| XJ | Astragalus adsurgens | Beijing | China |
2.2. DNA Extraction, PCR Amplification, and Sequencing
The material specimens were naturally dried and 30 mg of dried plant material was used for the DNA extraction. Samples were rubbed for two minutes at a frequency of 30 r/s in a FastPrep bead mill (Retsch MM400, Germany), and total genomic DNA was isolated from the crushed material according to the manufacturer's instructions (Plant Genomic DNA Kit, Tiangen Biotech Co., China). We made the following modifications to the protocol: chloroform was diluted with isoamyl alcohol (24 : 1 in the same volume) and buffer solution GP2 with isopropanol (same volume). The powder, 700 μL of 65°C GP1, and 1 μL β-mercaptoethanol were mixed for 10–20 s before being incubated for 60 minutes at 65°C. Then, 700 μL of the chloroform:isoamyl alcohol mixture was added and the solution was centrifuged for 5 minutes at 12000 rpm (~13400 ×g). Supernatant was removed and placed into a new tube before adding 700 μL isopropanol and blending for 15–20 minutes. The mixture was centrifuged in CB3 spin columns for 40 s at 12000 rpm. The filtrate was discarded and 500 μL GD (adding quantitative anhydrous ethanol before use) was added before centrifuging at 12000 rpm for 40 s. The filtrate was discarded and 700 μL PW (adding quantitative anhydrous ethanol before use) was used to wash the membrane before centrifuging for 40 s at 12000 rpm. This step was repeated with 500 μL PW, followed by a final centrifuge for 2 minutes at 12000 rpm to remove residual wash buffer. The spin column was dried at room temperature for 3–5 minutes and then centrifuged for 2 minutes at 12000 rpm to obtain the total DNA.
General PCR reaction conditions and universal DNA barcode primers were used for the ITS, ITS2, and psbA-trnH barcodes, as presented in Table 2 [24–26]. PCR amplification was performed on 25-μL reaction mixtures containing 2 μL DNA template (20–100 ng), 8.5 μL ddH2O, 12.5 μL 2× Taq PCR Master Mix (Beijing TransGen Biotech Co., China), and 1/1-μL forward/reverse (F/R) primers (2.5 μM). The reaction mixtures were amplified in a 9700 GeneAmp PCR system (Applied Biosystems, USA). Amplicons were visualized by electrophoresis on 1% agarose gels. Purified PCR products were sequenced in both directions using the ABI 3730XL sequencer (Applied Biosystems, USA).
Table 2.
Primers and PCR reaction conditions.
| Primer name | Primer sequences (5′-3′) | PCR reaction condition |
|---|---|---|
| ITS2 | ||
| 2F | ATGCGATACTTGGTGTGAAT | 94°C 5 min; |
| 3R | GACGCTTCTCCAGACTACAAT | 94°C 30 s, 56°C 30 s, 72°C 45 s, 40 cycles; 72°C 10 min; |
| ITS | ||
| 4R | TCCTCCGCTTATTGATATGC | 94°C 5 min; |
| 5F | GGAAGTAAAAGTCGTAACAAGG | 94°C 1 min, 50°C 1 min, 72°C 1.5 min + 3 s/cycle, 30 cycles; 72°C 7 min; |
| psbA | ||
| fwdPA | GTTATGCATGAACGTAATGCTC | 94°C 4 min; |
| trnH | ||
| rev TH | CGCGCATGGTGGATTCACAATCC | 94°C 30 s, 55°C 1 min, 72°C 1 min, 35 cycles; 72°C 10 min; |
2.3. Sequence Assembly, Alignment, and Analysis
Sequencing peak diagrams were obtained and proofread, and then contigs were assembled using a CodonCode Aligner 5.0.1 (CodonCode Co., USA). Complete ITS2 sequences were obtained using the HMMer annotation method, based on the Hidden Markov model (HMM) [27]. All of the sequences were aligned using ClustalW, in combination with 317 sequences from six commonly used barcodes (ITS2, ITS, psbA-trnH, matK, rbcL, and COI), which were downloaded from the GenBank database (Table 3). Sequence genetic distance and GC content were calculated using the maximum composite likelihood model. Maximum likelihood (ML) trees were constructed based on the Tamura-Nei model, and bootstrap tests were conducted using 1000 repeats to assess the confidence of the phylogenetic relationships by MEGA 6.0 software [28]. The barcoding gap, defined as the spacer region between intra- and interspecific genetic variations, and identification efficiency, based on BLAST1 and K2P nearest distance, were performed by the Perl language algorithm (Putty) [25, 29, 30].
Table 3.
Sequences from GenBank for identifying Astragalus and its adulterants.
| Region | Family | Species | Accession number |
|---|---|---|---|
| ITS2 | Fabaceae | Melilotus officinalis | U50765, Z97687 |
| Fabaceae | Astragalus adsurgens | L10757, GU217639, GU217640, GU217641 | |
| Fabaceae | Astragalus chinensis | GQ434365, GQ434366 | |
| Fabaceae | Hedysarum polybotrys | GQ434367 | |
| Fabaceae | Astragalus mongholicus | GQ434368, GU217643 | |
| Fabaceae | Astragalus mongholicus var. dahuricus | GU217635 | |
| Fabaceae | Astragalus membranaceus | GU217642, JF421475 | |
| Fabaceae | Caragana sinica | GU217654 | |
| Fabaceae | Medicago sativa | GU217662, Z99236, AF028417, JN617208 | |
| Fabaceae | Medicago sativa subsp. caerulea | AF028418 | |
| Fabaceae | Medicago sativa subsp. glomerata | AF028419 | |
| Fabaceae | Medicago falcata | AF028420 | |
| Malvaceae | Alcea rosea | AF303023 | |
|
| |||
| ITS | Fabaceae | Astragalus membranaceus | AF359749, EF685968, EU852042, FJ572044, GU289659 |
| GU289660, GU289661, GU289662, GU289663, GU289664 | |||
| HM142272, HM142273, HM142274, HM142275, HM142276 | |||
| HM142277, HM142278, HM142279, HM142280, HM142281 | |||
| HQ891827, JX017320, JX017321, JX017322, JX017323 | |||
| JX017324, JX017325, JX017326, JX017327, JX017328 | |||
| JX017329, JX017330, JX017331, JX017332, AF121675 | |||
| Fabaceae | Astragalus mongholicus | AF359750, EF685969, HM142282, HM142283, HM142284 | |
| HM142285, HM142286, HM142287, HM142288, HM142289 | |||
| HM142290, JF736665, JF736666, JF736667, JF736668 | |||
| JF736669, AB787166 | |||
| Fabaceae | Astragalus propinquus | AF359751 | |
| Fabaceae | Astragalus lepsensis | AF359752 | |
| Fabaceae | Astragalus aksuensis | AF359753, AB231091 | |
| Fabaceae | Astragalus hoantchy | AF359754, AF521952 | |
| Fabaceae | Astragalus hoantchy subsp. dshimensis | AF359755 | |
| Fabaceae | Astragalus lehmannianus | AF359756 | |
| Fabaceae | Astragalus sieversianus | AF359757 | |
| Fabaceae | Astragalus austrosibiricus | AF359758 | |
| Fabaceae | Astragalus uliginosus | EF685970 | |
| Fabaceae | Astragalus scaberrimus | AB051988 | |
| Fabaceae | Astragalus chinensis | FJ980292, HM142297, AF121681 | |
| Fabaceae | Astragalus borealimongolicus | HM142291, HM142292, HM142293, HM142294, HM142295 | |
| HM142296 | |||
| Fabaceae | Astragalus adsurgens | HM142298, HM142299, HQ199326 | |
| Fabaceae | Astragalus mongholicus var. dahuricus | HM142300, KC262199 | |
| Fabaceae | Astragalus zacharensis | HM142301 | |
| Fabaceae | Astragalus melilotoides | HM142302 | |
| Fabaceae | Astragalus scaberrimus | HM142303 | |
| Fabaceae | Astragalus sieversianus | AB741299 | |
| Fabaceae | Oxytropis anertii | EF685971 | |
| Fabaceae | Caragana sinica | DQ914785, FJ537284, GQ338283 | |
| Fabaceae | Glycyrrhiza pallidiflora | EU591998, GQ246130 | |
| Fabaceae | Melilotus officinalis | AB546796, JF461307, JF461308, JF461309, DQ311985 | |
| Fabaceae | Medicago sativa | GQ488541, AF053142, AY256392, JX017335, JX017336 | |
| JX017337, KF938697 | |||
| Fabaceae | Oxytropis caerulea | GU217599, HQ199316 | |
| Fabaceae | Hedysarum vicioides | HM142304, HM142305 | |
| Fabaceae | Hedysarum polybotrys | JX017333, JX017334, KF032294 | |
| Malvaceae | Malva neglecta | EF419478, EF419479 | |
| Malvaceae | Alcea rosea | AH010172, EF419544, EF679714, JX017319 | |
|
| |||
| psbA-trnH | Fabaceae | Astragalus membranaceus f. pallidipurpureus | GQ139474 |
| Fabaceae | Astragalus adsurgens | GU396749, GU396750, GU396751, KF011553 | |
| Fabaceae | Astragalus mongholicus | GU396754, AB787167 | |
| Fabaceae | Astragalus membranaceus | GQ139475, GQ139476, GQ139477, GQ139478, GQ139479 | |
| GQ139480, GQ139481, GQ139482, GQ139483, GU396752 | |||
| GU396753 | |||
| Fabaceae | Caragana sinica | GU396767, KJ025053 | |
| Fabaceae | Oxytropis caerulea | GU396771 | |
| Fabaceae | Medicago sativa | GU396781, HQ596768, HE966707 | |
| Fabaceae | Glycyrrhiza pallidiflora | GU396807 | |
| Fabaceae | Melilotus officinalis | HE966710 | |
| Malvaceae | Malva neglecta | EF419597, EF419598, HQ596765, HQ596765 | |
| Malvaceae | Alcea rosea | EF419662, EF679744 | |
|
| |||
| matK | Fabaceae | Astragalus membranaceus | EF685992, HM142232, HM142233, HM142234, HM142235 |
| HM142236, HM142237, HM142238, HM142239, HM142240 | |||
| HM142254 | |||
| Fabaceae | Astragalus mongholicus | EF685993, HM142241, HM142242, HM142243, HM142244 | |
| HM142245, HM142246, HM142247, HM142255, HM142256 | |||
| Fabaceae | Astragalus uliginosus | EF685994, HM142262 | |
| Fabaceae | Astragalus mongholicus var. dahuricus | HM049531, HM142260 | |
| Fabaceae | Astragalus chinensis | HM049533, HM142263 | |
| Fabaceae | Astragalus adsurgens | HM049537, HM142258, HM142259, AY920437 | |
| Fabaceae | Astragalus borealimongolicus | HM142248, HM142249, HM142250, HM142251, HM142252 | |
| HM142253 | |||
| Fabaceae | Astragalus zacharensis | HM142261 | |
| Fabaceae | Astragalus melilotoides | HM142264 | |
| Fabaceae | Astragalus scaberrimus | HM142265 | |
| Fabaceae | Astragalus sieversianus | AB741343 | |
| Fabaceae | Medicago sativa | AF522108, HQ593363, HM851138, AY386881, HE967439 | |
| AF169289 | |||
| Fabaceae | Oxytropis anertii | EF685995, HM142266 | |
| Fabaceae | Oxytropis caerulea | HM049544 | |
| Fabaceae | Glycyrrhiza pallidiflora | EF685997, HM142269, JQ619944 | |
| Fabaceae | Hedysarum vicioides | EF685996, HM142257, HM142267 | |
| Fabaceae | Caragana sinica | HM049541 | |
| Fabaceae | Melilotus officinalis | HE970723 | |
| Malvaceae | Malva neglecta | EU346788, HQ593360, JN894566, JN894571, JN895781 | |
| JQ412262, | |||
| Malvaceae | Alcea rosea | EU346805 | |
|
| |||
| rbcL | Fabaceae | Medicago sativa | Z70173 |
| Fabaceae | Astragalus membranaceus | EF685978, HM142199, HM142200, HM142201, HM142202 | |
| HM142203, HM142204, HM142205, HM142206, HM142207 | |||
| HM142221 | |||
| Fabaceae | Astragalus mongholicus | EF685979, HM142208, HM142209, HM142210, HM142211 | |
| HM142212, HM142213, HM142214, HM142222, HM142223 | |||
| Fabaceae | Astragalus uliginosus | EF685980, HM142225 | |
| Fabaceae | Hedysarum vicioides | EF685982, U74246, HM142224, HM142227, | |
| Fabaceae | Astragalus adsurgens | EF685984 | |
| Fabaceae | Astragalus borealimongolicus | HM142215, HM142216, HM142217, HM142218, HM142219 | |
| HM142220, | |||
| Fabaceae | Oxytropis anertii | EF685981, HM142226 | |
| Fabaceae | Glycyrrhiza pallidiflora | EF685983, AB012129, HM142228 | |
| Fabaceae | Caragana sinica | FJ537233 | |
| Fabaceae | Melilotus officinalis | JQ933405, JX848463 | |
3. Results
3.1. Sequence Information and Identification Efficiency
A total of 478 sequences for six barcodes were analyzed, from which 161 sequences were obtained from Astragalus Radix and its adulterants. Sequence information and identification success rates are listed in Table 4. The average GC content of six barcodes was discrepant, and ITS and ITS2 regions from nuclear ribosomal DNA performed higher than other barcodes (52.97% versus 50.80%). Among the six barcodes, ITS2 provided the largest average genetic distance (1.0792), and rbcL was the smallest (0.0349). All of the six barcodes obtained a zero value for the minimum genetic distance. In terms of identification efficiency, the nearest distance method was superior to the BLAST1 method for all of the six barcodes. Moreover, ITS and the psbA-trnH and matK regions provided a higher rate of success than the other three barcodes using the BLAST1 method. However, matK, ITS, and psbA-trnH performed better than the other three barcodes, based on the nearest distance method. ITS and psbA-trnH obtained higher genetic distances, so the matK, ITS, and psbA-trnH barcodes were the preferable methods for identifying Radix Astragali and its adulterants based on superior sequencing efficiency and identification efficiency.
Table 4.
The information of identification efficiency for six barcodes.
| Markers | COI | ITS2 | ITS | matK | rbcL | psbA-trnH |
|---|---|---|---|---|---|---|
| Number of sequences | 39 | 72 | 185 | 65 | 43 | 74 |
| Average GC content/% | 43.29 | 50.80 | 52.97 | 31.14 | 42.88 | 21.77 |
| Genetic distance | ||||||
| Min | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| Max | 0.0086 | 7.9494 | 5.3130 | 0.2801 | 0.0349 | 2.2701 |
| Average | 0.0019 | 1.0792 | 0.3508 | 0.0711 | 0.0116 | 0.5080 |
| Identification efficiency/% | ||||||
| BLAST 1/% | 10.26 | 12.50 | 30.81 | 29.23 | 23.26 | 29.73 |
| Nearest distance/% | 33.33 | 27.78 | 52.43 | 66.15 | 37.21 | 41.89 |
3.2. Intra- and Interspecific Variation Analysis Using Six Parameters
Six parameters to analyze intraspecific variation and interspecific divergence were employed to assess the utility of six DNA barcodes (Table 5). We expected the “minimum interspecific distance” would be higher than the “coalescent depth” (maximum intraspecific distance). Therefore, we first utilized the “gap rate” to indicate the distinctness, calculated by the formula: (minimum interspecific distance − maximum intraspecific distance)/minimum interspecific distance. Results show that the ITS2, COI, matK, and rbcL regions outperformed the ITS and psbA-trnH regions for gap rates. However, when we compared all of the average inter- and intraspecific distances, the ITS2, rbcL, matK, and psbA-trnH regions performed better than the ITS and COI regions. Therefore, in terms of intra- and interspecific variation, ITS2, matK, and rbcL are the preferable options for identifying Radix Astragali and its adulterants.
Table 5.
Analysis of interspecific divergence and intraspecific variation for six barcodes.
| Marker (Mean ± SD) | COI | ITS2 | ITS | matK | rbcL | psbA-trnH |
|---|---|---|---|---|---|---|
| Theta | 2.2260 ± 6.2961 | 0.0030 ± 0.0046 | 0.0271 ± 0.0404 | 0.0021 ± 0.0035 | 0.0011 ± 0.0020 | 0.2415 ± 0.4777 |
| Coalescent depth | 0.0001 ± 0.0004 | 0.0040 ± 0.0046 | 0.1423 ± 0.3958 | 0.0032 ± 0.0050 | 0.0016 ± 0.0030 | 0.4109 ± 0.5683 |
| All intraspecific distance | 9.3280 ± 0.0003 | 0.0021 ± 0.0024 | 0.1153 ± 0.3051 | 0.0014 ± 0.0022 | 0.0002 ± 0.0011 | 0.3093 ± 0.4300 |
| Theta prime | 0.0012 ± 0.0008 | 0.0617 ± 0.0302 | 0.0603 ± 0.0371 | 0.0091 ± 0.0061 | 0.0024 ± 0.0035 | 0.3083 ± 0.2887 |
| Minimum interspecific distance | 0.0008 ± 0.0010 | 0.0440 ± 0.0386 | 0.0168 ± 0.0196 | 0.0066 ± 0.0066 | 0.0023 ± 0.0035 | 0.0423 ± 0.0380 |
| All interspecific distance | 0.0007 ± 0.0010 | 0.0343 ± 0.0389 | 0.1066 ± 0.2833 | 0.0071 ± 0.0064 | 0.0015 ± 0.0029 | 0.3166 ± 0.4070 |
| Gap rate/% | 87.50 | 90.91 | / | 51.52 | 30.43 | / |
3.3. Barcoding Gap Analysis
Analysis of the DNA barcoding gap presents the divergence of inter- and intraspecies and indicates separate, nonoverlapping distribution between specimens in an ideal situation [25]. In our study (Figure 1), the rbcL, COI, ITS, and matK regions possessed less relative distribution of inter- and intraspecific variation than psbA-trnH and ITS2, although there were no nonoverlapping regions for the six barcodes. Hence, the rbcL, COI, ITS, and matK regions are more successful at identifying Radix Astragali and its adulterants, from the standpoint of barcoding gap analysis.
Figure 1.
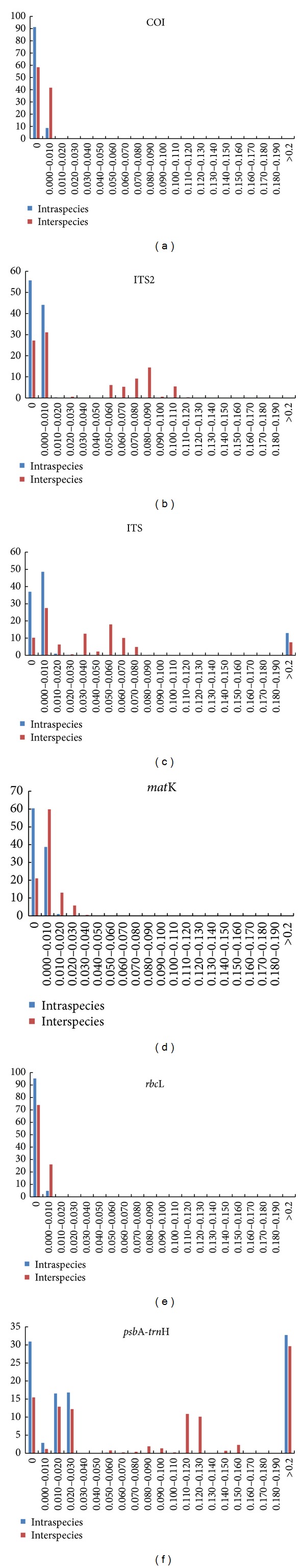
Barcoding gap for six barcodes.
3.4. ML Tree Analysis
Maximum likelihood (ML) is a general statistical criterion in widespread use for the inference of molecular phylogenies [31]. An ML tree visually revealed the relationship between species. As the results show (Figure 2), psbA-trnH successfully differentiated Radix Astragali and its adulterants. Furthermore, it produced areas of obvious separation for Radix Astragali. The remaining five barcodes also differentiated Radix Astragali and its adulterants. Each species clustered together, separate from other species. Considering the difficult amplification and sequencing and fast and accurate identification purpose of DNA barcoding, we did not add all the sequence data of ITS2 and psbA-trnH to build ML tree and subsequent analysis.
Figure 2.
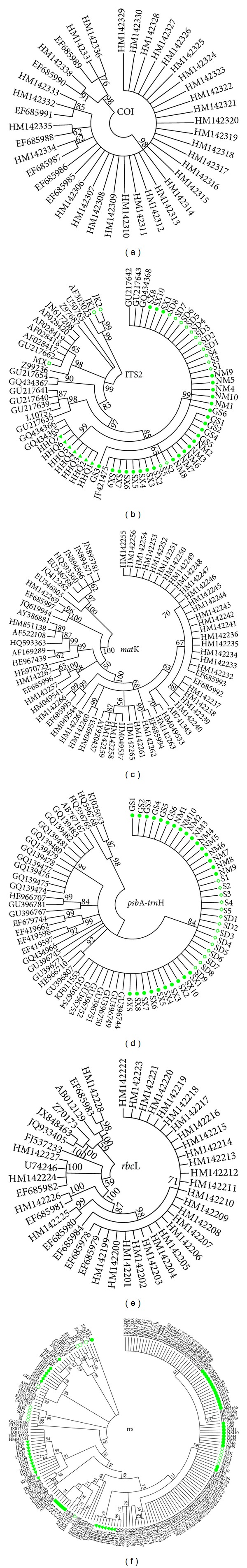
ML tree for six barcodes. ∗The different color and shape for different species in clusters presented the identification of different barcodes.
4. Discussion and Conclusions
Radix Astragali is reported to possess 47 bioactive compounds and has many bioactive properties [32–37]. Various Radix Astragali preparations are commercially available, not only in China as a TCM component, but also in the United States, as dietary supplements [38]. However, due to increasing demand, substitutes and adulterants have flooded the market. Traditional identification methods, such as morphological and microscopic methods, are limited by the lack of explicit criteria for character selection or coding and, thus, mainly depend on subjective assessments. Although chemical methods are able to distinguish between different species, it is difficult to differentiate sibling species that possess similar chemical compositions. In addition, chemical methods are unable to provide accurate species authentication. Several types of molecular markers for characterizing genotypes are useful in identifying plant species. For example, RAPD has been used to estimate genetic diversity in plant populations based on amplification of random DNA fragments and comparisons of common polymorphisms. DNA barcoding is advocated for species identification, due to its universal applicability, simplicity, and scientific accuracy. However, the analysis methods for DNA barcodes were limited. With the development of molecular biology and bioinformatics, a more improved analytic method for DNA barcoding can be established to identify Radix Astragali and closely related species.
In this study, we validated a new analytical method for identifying Radix Astragali using DNA barcoding. Seventy-seven specimens of Radix Astragali and its adulterants were collected, and the sequences of 29 species reported in the literature were downloaded from the GenBank database. Based on the 478 sequences for six barcodes (ITS2, ITS from nuclear genome; psbA-trnH, rbcL, and matK from chloroplast genome; COI from mitochondrial genome), genetic distance and ML Tree were calculated by MEGA 6.0 software, and identification efficiency, intra- and interspecific variation, and barcoding gap were calculated using the Perl language algorithm. Results of the six indicators assessed are shown in Table 6. ITS and psbA-trnH outperformed other barcodes in terms of identification efficiency. ITS2 performed better in terms of genetic distance, gap rate, and inter- and intraspecific variation. RbcL performed better in terms of barcoding gap and inter- and intraspecific variation. Although ITS2 was part of the ITS sequence, it performed poorly in identification efficiency. Therefore, we suggest that the ITS sequence is the optimal barcode, and that the psbA-trnH region is a complementary barcode for identifying Radix Astragali and its adulterants.
Table 6.
Six indicators assessed for DNA barcoding.
| DNA barcodes | Parameters | ||||||
|---|---|---|---|---|---|---|---|
| Average genetic distance | Identification efficiency | Gap rate | Inter- to intraspecific variation | Barcoding gap | Total score | ||
| BLAST1 | Nearest distances | ||||||
| ITS2 | 8 | 12 | 8 | 8 | 8 | 4 | 48 |
| ITS | 6 | 28 | 22 | 0 | 0 | 6 | 62 |
| psbA-trnH | 6 | 26 | 18 | 0 | 2 | 2 | 54 |
| rbcL | 4 | 12 | 14 | 4 | 6 | 8 | 48 |
| matK | 4 | 14 | 24 | 4 | 4 | 2 | 52 |
| COI | 2 | 6 | 10 | 6 | 0 | 6 | 30 |
∗The total score of six parameters was set by 10, 30, 30, 10, 10, and 10 in order. Identification efficiency based on two methods was set by 30 score because of its importance for identification.
In conclusion, we describe a new analytical method for the use of DNA barcoding in the identification of Radix Astragali. Six indicators, including average genetic distance, BLAST1 and the nearest distance method for identification efficiency, inter- and intraspecific variation, and gap rate were tested to evaluate six DNA barcodes using bioinformatics software and the Perl language algorithm. The ITS sequence was the optimal barcode for identifying Radix Astragali and its adulterants. This method provides a novel means for accurate identification of Radix Astragali and its adulterants and improves the utilization of DNA barcoding in identifying medicinal plant species.
Acknowledgments
Thanks are due to the National Natural Science Foundation of China (nos. 81274013, 8130069, and 81473315) and the National Science and Technology Major Projects for “Major New Drugs Innovation and Development” (no. 2011BAI07B01).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Kuo Y, Tsai W, Loke S, Wu T, Chiou W. Astragalus membranaceus flavonoids (AMF) ameliorate chronic fatigue syndrome induced by food intake restriction plus forced swimming. Journal of Ethnopharmacology. 2009;122(1):28–34. doi: 10.1016/j.jep.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 2.Cho WCS, Leung KN. In vitro and in vivo immunomodulating and immunorestorative effects of Astragalus membranaceus . Journal of Ethnopharmacology. 2007;113(1):132–141. doi: 10.1016/j.jep.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 3.Dong TTX, Ma XQ, Clarke C, et al. Phylogeny of Astragalus in China: molecular evidence f rom the DNA sequences of 5S rRNA spacer, ITS, and 18S rRNA. Journal of Agricultural and Food Chemistry. 2003;51(23):6709–6714. doi: 10.1021/jf034278x. [DOI] [PubMed] [Google Scholar]
- 4.Ma X, Tu P, Chen Y, Zhang T, Wei Y, Ito Y. Preparative isolation and purification of two isoflavones from Astragalus membranaceus Bge. var. mongholicus (Bge.) Hsiao by high-speed counter-current chromatography. Journal of Chromatography A. 2003;992(1-2):193–197. doi: 10.1016/s0021-9673(03)00315-7. [DOI] [PubMed] [Google Scholar]
- 5.Ma X, Tu P, Chen Y, Zhang T, Wei Y, Ito Y. Preparative isolation and purification of isoflavan and pterocarpan glycosides from Astragalus membranaceus Bge. var. mongholicus (Bge.) Hsiao by high-speed counter-current chromatography. Journal of Chromatography A. 2004;1023(2):311–315. doi: 10.1016/j.chroma.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 6.Yip PY, Kwan HS. Molecular identification of Astragalus membranaceus at the species and locality levels. Journal of Ethnopharmacology. 2006;106(2):222–229. doi: 10.1016/j.jep.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 7.Zhao YZ. Investigation the source and distribution of Radix Astragli. Chinese Traditional and Herbal Drugs. 2004;35(10):1189–1190. [Google Scholar]
- 8.Zhang YH, Zhang LM, Liu XB, et al. Study on morphological and microscopic identification for different producing areas of Radix Astragali. Journal of Chinese Medicinal Materials. 2013;36(10):1602–1604. [Google Scholar]
- 9.Wei L, Zeng FT. Using thin-layer chromatography and ultra-violet spectroscopy to identify Radix Astragali and its adulterants. Journal of Chinese Medicinal Materials. 1993;16(12):14–17. [Google Scholar]
- 10.Li G, Zhao H, Liu Y, et al. Study on Chinese herb astragalus membranceus by FTIR fingerprint. Spectroscopy and Spectral Analysis. 2010;30(6):1493–1497. [PubMed] [Google Scholar]
- 11.Ma XQ, Shi Q, Duan JA, Dong TTX, Tsim KWK. Chemical analysis of Radix Astragali (Huangqi) in China: a comparison with its adulterants and seasonal variations. Journal of Agricultural and Food Chemistry. 2002;50(17):4861–4866. doi: 10.1021/jf0202279. [DOI] [PubMed] [Google Scholar]
- 12.Na HJ, Um JY, Kim SC, et al. Molecular discrimination of medicinal Astragali radix by RAPD analysis. Immunopharmacology and Immunotoxicology. 2004;26(2):265–272. doi: 10.1081/iph-120037723. [DOI] [PubMed] [Google Scholar]
- 13.Duan LX, Chen TL, Li M, et al. Use of the metabolomics approach to characterize chinese medicinal material Huangqi. Molecular Plant. 2012;5(2):376–386. doi: 10.1093/mp/ssr093. [DOI] [PubMed] [Google Scholar]
- 14.Hebert PDN, Cywinska A, Ball SL, DeWaard JR. Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences. 2003;270(1512):313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(41):14812–14817. doi: 10.1073/pnas.0406166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng SH, Jiang X, Wu LB, Wang ZH, Huang LF. Chemical and genetic discrimination of cistanches herba based on UPLC-QTOF/MS and DNA barcoding. PLoS ONE. 2014;9(5) doi: 10.1371/journal.pone.0098061.e98061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang LF, Zheng SH, Wu LB, Jiang X, Chen SL. Ecotypes of Cistanche deserticolabased on chemical component and molecular traits. Scientia Sinica Vitae. 2014;44(3):318–328. [Google Scholar]
- 18.Ma XQ, Duan JA, Zhu DY, Dong TTX, Tsim KWK. Species identification of Radix Astragali (Huangqi) by DNA sequence of its 5S-rRNA spacer domain. Phytochemistry. 2000;54(4):363–368. doi: 10.1016/s0031-9422(00)00111-4. [DOI] [PubMed] [Google Scholar]
- 19.Chen G, Wang XL, Wong WS, Liu XD, Xia B, Li N. Application of 3′ Untranslated Region (UTR) sequence-based amplified polymorphism analysis in the rapid authentication of Radix astragali. Journal of Agricultural and Food Chemistry. 2005;53(22):8551–8556. doi: 10.1021/jf051334g. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Chen H-B, Gou B-L, Zhao Z-Z, Liang Z-T, Yi T. Study of the relationship between genetics and geography in determining the quality of Astragali Radix. Biological and Pharmaceutical Bulletin. 2011;34(9):1404–1412. doi: 10.1248/bpb.34.1404. [DOI] [PubMed] [Google Scholar]
- 21.Cui ZH, Li Y, Yuan QJ, Zhou L, Li M. Molecular identification of Astragali Radix and its adulterants by ITS sequences. China Journal of Chinese Materia Medica. 2012;37(24):3773–3776. [PubMed] [Google Scholar]
- 22.Gao T, Yao H, Ma XY, Zhu YJ, Song JY. Identification of Astragalus plants in China using the region ITS2. World Science and Technology/Modernization of Traditional Chinese Medicine and Materia Medica. 2010;12(2):222–227. [Google Scholar]
- 23.Guo H-Y, Wang W-W, Yang N, et al. DNA barcoding provides distinction between Radix Astragali and its adulterants. Science China Life Sciences. 2010;53(8):992–999. doi: 10.1007/s11427-010-4044-y. [DOI] [PubMed] [Google Scholar]
- 24.CBOL Plant Working Group. A DNA barcode for land plants. Proceedings of the National Academy of Sciences of United States of America. 2009;106(31):12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen SL, Yao H, Han JP, et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE. 2010;5(1) doi: 10.1371/journal.pone.0008613.e8613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(23):8369–8374. doi: 10.1073/pnas.0503123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller A, Schleicher T, Schultz J, Müller T, Dandekar T, Wolf M. 5.8S-28S rRNA interaction and HMM-based ITS2 annotation. Gene. 2009;430(1-2):50–57. doi: 10.1016/j.gene.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 28.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evoluationaay genetics analysis version 6.0. Molecular Biology and Evolution. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer CP, Paulay G. DNA barcoding: error rates based on comprehensive sampling. PLoS Biology. 2005;3(12, article e422) doi: 10.1371/journal.pbio.0030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross HA, Murugan S, Li WLS. Testing the reliability of genetic methods of species identification via simulation. Systematic Biology. 2008;57(2):216–230. doi: 10.1080/10635150802032990. [DOI] [PubMed] [Google Scholar]
- 31.Morrison DA. Increasing the efficiency of searches for the maximum likelihood tree in a phylogenetic analysis of up to 150 nucleotide sequences. Systematic Biology. 2007;56(6):988–1010. doi: 10.1080/10635150701779808. [DOI] [PubMed] [Google Scholar]
- 32.Zhang YP, Nie MK, Shi SY, et al. Integration of magnetic solid phase fishing and off-line two-dimensional high-performance liquid chromatographydiode array detectormass spectrometry for screening and identification of human serum albumin binders from Radix Astragali. Food Chemistry. 2014;146(1):56–64. doi: 10.1016/j.foodchem.2013.09.030. [DOI] [PubMed] [Google Scholar]
- 33.Liu XH, Zhao LG, Liang J, et al. Component analysis and structure identification of active substances for anti-gastric ulcer effects in Radix Astragali by liquid chromatography and tandem mass spectrometry. Journal of Chromatography B. 2014;960(1):43–51. doi: 10.1016/j.jchromb.2014.04.020. [DOI] [PubMed] [Google Scholar]
- 34.Chu C, Cai H-X, Ren M-T, et al. Characterization of novel astragaloside malonates from Radix Astragali by HPLC with ESI quadrupole TOF MS. Journal of Separation Science. 2010;33(4-5):570–581. doi: 10.1002/jssc.200900687. [DOI] [PubMed] [Google Scholar]
- 35.Fu J, Huang LF, Zhang HT, Yang SH, Chen SL. Structural features of a polysaccharide from Astragalus membranaceus (Fisch.) Bge. var. mongholicus (Bge.) Hsiao. Journal of Asian Natural Products Research. 2013;15(6):687–692. doi: 10.1080/10286020.2013.778832. [DOI] [PubMed] [Google Scholar]
- 36.Huang X, Liu Y, Song F, Liu Z, Liu S. Studies on principal components and antioxidant activity of different Radix Astragali samples using high-performance liquid chromatography/electrospray ionization multiple-stage tandem mass spectrometry. Talanta. 2009;78(3):1090–1101. doi: 10.1016/j.talanta.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 37.Nalbantsoy A, Nesil T, Yilmaz-Dilsiz Ö, Aksu G, Khan S, Bedir E. Evaluation of the immunomodulatory properties in mice and in vitro anti-inflammatory activity of cycloartane type saponins from Astragalus species. Journal of Ethnopharmacology. 2012;139(2):574–581. doi: 10.1016/j.jep.2011.11.053. [DOI] [PubMed] [Google Scholar]
- 38.Xiao WL, Motley TJ, Unachukwu UJ, et al. Chemical and genetic assessment of variability in commercial Radix Astragali (Astragalus spp.) by ion trap LC-MS and nuclear ribosomal DNA barcoding sequence analyses. Journal of Agricultural and Food Chemistry. 2011;59(5):1548–1556. doi: 10.1021/jf1028174. [DOI] [PMC free article] [PubMed] [Google Scholar]


