Abstract
Cre/loxP system-mediated site-specific recombination is utilized to study gene function in vivo. Successful conditional knockout of genes of interest is dependent on the availability of Cre-driver mice. We produced and characterized pancreatic β cell-specific Cre-driver mice for use in diabetes mellitus research. The gene encoding Cre was inserted into the second exon of mouse Ins1 in a bacterial artificial chromosome (BAC). Five founder mice were produced by microinjection of linearized BAC Ins1-cre. The transgene was integrated between Mafa and the telomere on chromosome 15 in one of the founders, BAC Ins1-cre25. To investigate Cre-loxP recombination, BAC Ins1-cre25 males were crossed with two different Cre-reporters, R26R and R26GRR females. On gross observation, reporter signal after Cre-loxP recombination was detected exclusively in the adult pancreatic islets in both F1 mice. Immunohistological analysis indicated that Cre-loxP recombination-mediated reporter signal was colocalized with insulin in pancreatic islet cells of both F1 mice, but not with glucagon. Moreover, Cre-loxP recombination signal was already observed in the pancreatic islets at E13.5 in both F1 fetuses. Finally, we investigated ectopic Cre-loxP recombination for Ins1, because the ortholog Ins2 is also expressed in the brain, in addition to the pancreas. However, there was no Cre-loxP recombination-mediated reporter signal in the brain of both F1 mice. Our data suggest that BAC Ins1-cre25 mice are a useful Cre-driver C57BL/6N for pancreatic β cell-specific Cre-loxP recombination, except for crossing with knock-in mice carrying floxed gene on chromosome 15.
Keywords: cre-driver mice, cre-loxP recombination, diabetes, insulin1, pancreatic β cells
Introduction
Diabetes mellitus is a functional disorder of glucose metabolism caused by insulin insufficiency. The results of the National Health and Nutrition Survey in Japan, 2007, estimated that the numbers of “individuals strongly suspected of having diabetes” and “individuals in whom diabetes cannot be ruled out” were approximately 8.9 and 22.1 million, respectively, based on data obtained by multiplying the survey results by the estimated population aged 20 years or older, broken down by gender and age classification (total population: approximately 0.14 billion) (http://www0.nih.go.jp/eiken/english/research/pdf/nhns2007.pdf). There are large numbers of diabetes mellitus patients in Japan as well as western countries. As insulin treatment is effective for type I and type II diabetes patients, determination of the functional molecular mechanism of pancreatic β cell development is essential.
Production and analysis of conditional knockout mice through breeding with transgenic mice expressing Cre protein in pancreatic β cells will be useful in determining the genetic functions of genes of interest in pancreatic β cells in vivo. The gene regulatory region of insulin, which is synthesized and secreted from pancreatic β cells, has been used as a pancreatic β cell-specific promoter for cre gene expression. RIP-cre mice are widely used for Cre-loxP recombination in pancreatic β cells. This is a transgenic mouse strain carrying a 668-bp rat insulin 2 promoter fused to cre with a nuclear transfer signal [4, 9, 14]. It is difficult to determine the tissues responsible for the anomalous phenotype in conditional knockout mice carrying RIP-cre transgenes, because RIP-cre mice also express Cre in the brain, including the hypothalamus, which is the region involved in the control of several endocrine functions [1, 5].
Although humans possess a single insulin gene, rodents have two insulin systems consisting of insulin 2 (Ins2), an ortholog to the insulin gene in other mammals, and insulin 1 (Ins1), a rodent-specific retrogene [16]. Both murine Ins1 and Ins2 are expressed in pancreatic β cells. Unlike Ins1, Ins2 is also expressed in the brain [2, 10]. Therefore, the gene regulatory region of Ins1 will provide a suitable promoter to permit pancreatic β cell-specific expression of exogenous genes. However, there have not been Ins1-cre driver mice available from laboratory mouse resource.
In the present study, we produced transgenic mice with pancreatic β cell-specific Cre expression using a bacterial artificial chromosome (BAC) containing the murine Ins1 gene to provide well-characterized Cre-driver mice for basic research into diabetes mellitus.
Materials and Methods
Generation of transgenic mice carrying BAC Ins1-cre
A nuclear location signal fused to the cre gene fragment with a polyadenylation signal, NLS-cre, was obtained from pCAG-NLS-cre. A BAC clone containing the entire mouse Ins1 gene, RP23-181I21, was purchased from Invitrogen (Carlsbad, CA). This BAC was composed of 150 kb of 5′-flanking region, 1.2 kb containing 2 exons and 1 intron, and 50 kb of 3′-flanking region. Using a Red/ET recombination system (Gene Bridges, Heidelberg, Germany), the NLS-cre gene was inserted by homologous recombination into the second exon with a translational initiation codon in the Ins1 BAC clone (Fig. 1A). To generate BAC transgenic mice, PI-SceI-linearized BAC Ins1-cre DNA was injected into the pronuclei of fertilized oocytes derived from C57BL/6N mice. The injected embryos were transferred into the uteri of pseudopregnant CD-1 females. C57BL/6N and CD-1 mice were purchased from Charles River Laboratories Japan (Atsugi, Japan). Genotypes were confirmed by PCR using the following primers: 5′-AGGCCATCTGGTCCCTTATTAAGAC-3′ and 5′-CTAATCGCCATCTTCCAGCAGG-3′ for Ins1-cre mice.
Fig. 1.
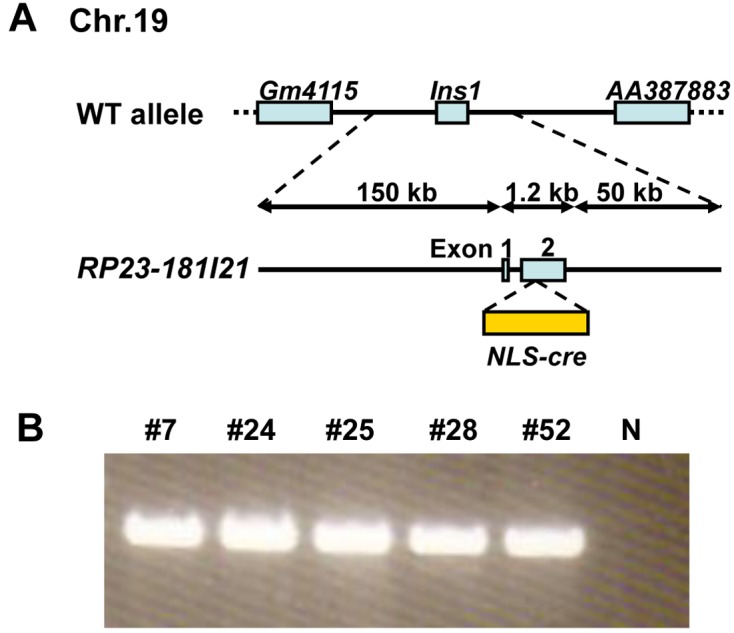
Construction of BAC Ins1-cre transgene. (A) Schematic representation of the transgene. (B) Transgene-specific amplification products of five founder mice determined by PCR analysis. Founder mice: #7, #24, #25, #28, and #52. N: negative control.
Preparation of Cre-reporter mice
To determine Cre recombination activity in the transgenic mice carrying BAC Ins1-cre gene, B6.129S4-Gt (ROSA) 26Sortm1Sor/J, R26R [17], and C57BL/6N-Gt (ROSA) 26Sor<tm1(CAG-EGFP,-tdsRed)Utr>/Rbrc, R26GRR [7], pairs were obtained from the Jackson Laboratory (Bar Harbor, ME) and RIKEN BioResource Center (Tsukuba, Japan) through the National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science, and Technology, Japan, respectively. Both Cre-reporter mouse strains were maintained as homozygous lines at the Laboratory Animal Resource Center, University of Tsukuba.
Animal care
Mice were kept in plastic cages under pathogen-free conditions in a room maintained at 23.5 ± 2.5°C and 52.5 ± 12.5% relative humidity under a 14-h light:10-h dark cycle. Mice had free access to commercial chow (MF diet; Oriental Yeast Co., Ltd.., Tokyo, Japan) and filtered water. All mouse experiments were performed under the approval of the University of Tsukuba Animal Experiment Committee.
Fluorescence in situ hybridization
R-banded chromosome preparations were made from the spleen lymphocytes of heterozygous BAC Ins1-cre25 mice as described previously [12]. The cultured cells were treated with BrdU during late S phase for differential replication banding. R-banded chromosomes were obtained by exposure of chromosome slides to UV light after staining with Hoechst 33258 (Sigma, St. Louis, MO). Two-color fluorescence in situ hybridization (FISH) analysis was performed according to the standard method using RP23-181I21 BAC DNA and RP23-94I16 BAC DNA as Ins1 and Mafa probe, respectively. BAC DNAs were purified by NucleoBond BAC 100 (Macherey-Nagel, Dueren, Germany). Ins1 and Mafa probes were labeled by nick translation (Roche, Penzberg, Germany) with biotin-dUTP (Roche) and Cy3-dUTP (GE Healthcare, Piscataway, NJ). Repeat sequences in BAC DNA probes were blocked with Cot1 DNA (Life Technologies, Gaithersburg, MD). Probes were denatured and hybridized in a standard hybridization mixture. Finally, chromosome samples were incubated with avidin-FITC (Roche).
Stereomicroscopic findings
For X-gal and fluorescence imaging during embryonic development, pregnant mice were euthanized by CO2 inhalation. Adult mice were anesthetized with pentobarbital and perfused with cold PBS. EGFP and tdsRed fluorescence were observed by fluorescence stereomicroscopy (M205FA; Leica, Wetzlar, Germany) provided with internal light sources and appropriate filter sets (excitation and emission: 470 ± 20 nm and 525 ± 25 nm and 545 ± 15 nm and 620 ± 30 nm band-pass filters for EGFP and tdsRed, respectively). Before X-gal staining, tissues were fixed with 0.2% glutaraldehyde in 0.1 M phosphate buffer (pH 7.3) containing 5 mM EGTA and 2 mM magnesium chloride for 30 min, and then washed with 0.1 M phosphate buffer (pH 7.3) containing 0.02% Nonidet-P40, 0.01% sodium deoxycholate, and 2 mM magnesium chloride. Staining was carried out overnight at 37°C in PBS containing 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 2 mM magnesium chloride, and 1 mg/ml X-gal. Embryos and whole-mount adult tissues were washed in PBS and examined under bright field illumination.
Immunohistological findings
For immunohistological demonstration of insulin and glucagon in islet tissue, mice were anesthetized with pentobarbital, perfused with cold PBS, and then perfused with 4% paraformaldehyde in cold PBS. Fixed samples were equilibrated in sucrose by placing the samples in 50-ml tubes with graded concentrations of sucrose (10%, 20%, and 30% in PBS). Samples were embedded in Tissue-Tek OCT (Fisher, Pittsburgh, PA) and frozen in liquid nitrogen. Frozen tissue blocks were brought to −20°C and sections 10 µm thick were cut and mounted on amino silane-coated slides. Slides were dried at room temperature (RT) overnight, and then probed with antibodies or stored at −80°C. For immunohistological demonstration of β-galactosidase expression, samples were fixed with 10% formalin and then embedded in paraffin. Five µm sections were cut and then deparaffinized with xylene. Tissue sections were incubated with rabbit anti-β-galactosidase antibody (Sigma), guinea pig anti-insulin antibody (Abcam, Cambridge, UK) and rabbit anti-glucagon antibody (Linco Research, St. Charles, MO) for 8 h at 4°C. The antigens were visualized using appropriate secondary antibodies conjugated with Alexa 488 or Alexa 647 with nuclear staining using diamidino-2-phenylinodole (DAPI) (Invitrogen). Fluorescence was examined by fluorescence microscopy (DMLB; Leica) with internal light sources and appropriate filter sets.
Results
Generation of BAC Ins1-cre25 mice
Sixty-three neonates were obtained from 340 transferred embryos injected with BAC Ins1-cre. After weaning, genotyping was carried out by PCR analysis of tail DNA. Five founder mice (#7, #24, #25, #28, and #52) carried the BAC Ins1-cre transgene (Fig. 1B). The pups derived from crossing founder #25 with wild-type C57BL/6N were characterized, because preliminary data indicated that R26R/#24 F1 and R26R/#25 F1 mice had stronger Cre activity (X gal-staining) in the adult pancreas than F1 mice from crosses between R26R mice and other founder lines (data not shown).
Chromosomal localization of BAC Ins1-cre
Next, we performed metaphase FISH analyses to determine the location of the transgene insertion site in BAC Ins1-cre25 mice. The BAC Ins1 DNAs conjugated with FITC were hybridized to hemizygous BAC Ins1-cre25 mouse chromosome spreads, and a green signal was detected at a single site on three chromosomes. Two of the three chromosomes were homologous chromosome 19, including the endogenous Ins1 locus. The other green signal was the BAC Ins1-cre transgene, which seemed to be located on chromosome 15 according to the R-banding pattern. To identify the chromosomal location of the BAC Ins1-cre transgene in detail, we performed two-color FISH using a FITC-labeled probe for the BAC Ins1-cre transgene, which was detected as a green signal, and a Cy3-labeled probe for Mafa on chromosome 15, which was detected as a red signal. The green signal for the transgene was clearly detected at a position distal to the red signal corresponding to Mafa on chromosome 15 (Fig. 2). Taken together, these observations indicated that the transgene was inserted into the middle region between the Mafa locus (75.7 Mb) and the telomere on chromosome 15.
Fig. 2.
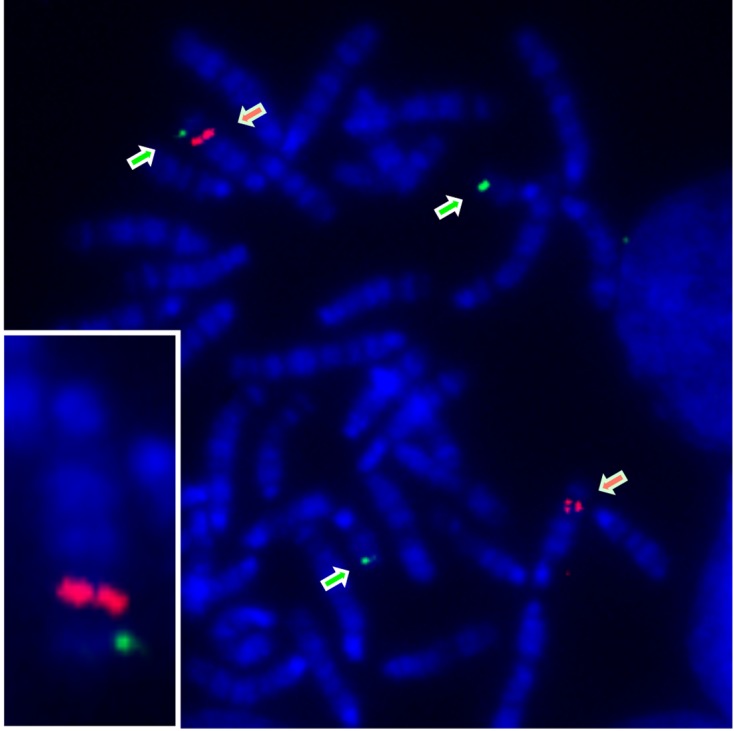
Metaphase chromosome spreads of hemizygous BAC Ins1-cre25 transgenic mice hybridized with Ins1 (RP23-181I21) and Mafa (RP23-94I16) conjugated with FITC and Cy3, respectively. Green arrows: Ins1; red arrows: Mafa. Bottom-left corner: chromosome 15 including BAC Ins1-cre transgene.
Cre activity in the adult pancreas
To investigate Cre activity in BAC Ins1-cre25 mice, we used two different Cre-reporter mouse strains, R26R (R26RKI/KI) and R26GRR (R26GRRKI/KI), in the present study. The R26R strain carries Gtrosa26tm1Sor, which includes a splice acceptor sequence, a neomycin-resistance (neo) expression cassette flanked by loxP sites, and the lacZ gene with a polyadenylation sequence in the ROSA26 locus [17]. When crossed with Cre-driver mice, lacZ is expressed in cells/tissues where cre is expressed. This strain is commonly used as a Cre-reporter mouse. However, it seems that nonspecific Cre activity was detected by endogenous galactoside and resident bacterial enzyme activity. Therefore, to evaluate Cre-loxP recombination in cells/tissues with different detection methods, we used a novel ROSA26 knock-in Cre-reporter mouse strain, R26GRR, exhibiting green fluorescence emission (EGFP) before and red fluorescence emission (tdsRed) after Cre-mediated recombination [7]. Constructions of two different Cre-reporter strains are shown in Supplement Fig. 1.
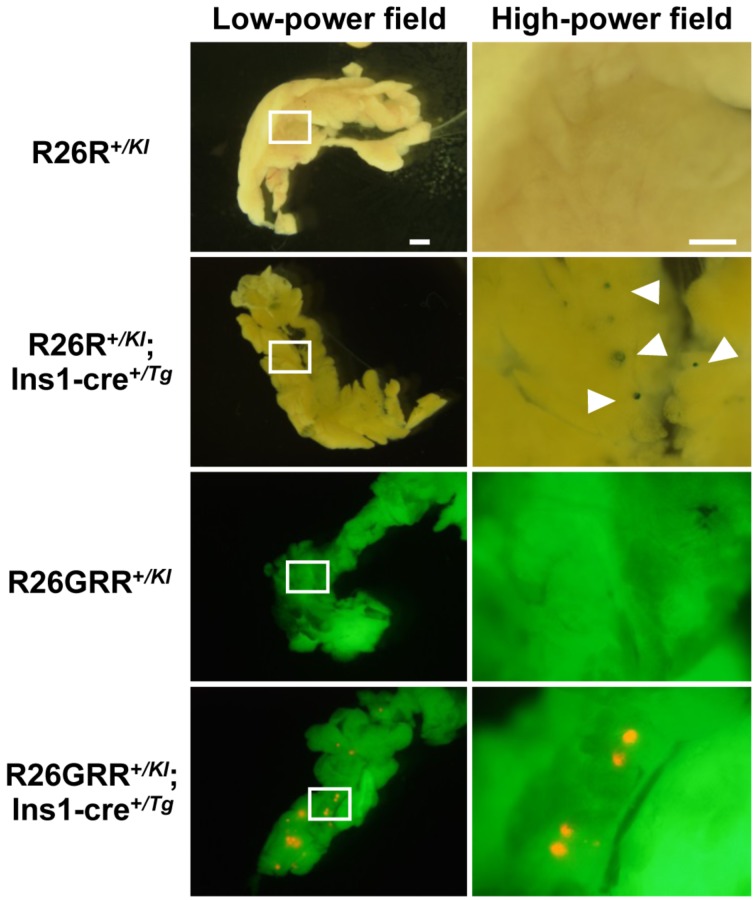
Hemizygous BAC Ins1-cre25 males were mated with homozygous R26R or R26GRR females and the adult pancreas was collected from their F1 progeny, R26R+/KI, R26R+/KI; Ins1-cre+/Tg, R26GRR+/KI, and R26GRR+/KI; Ins1-cre+/Tg mice. F1 mice without the cre gene were used as negative controls. First, we examined whole-mount preparations of the entire pancreas with X-gal staining and tdsRed fluorescence. No reporter signals for Cre-loxP recombination were observed in the pancreas from R26R+/KI mice or R26GRR+/KI mice. In contrast, X-gal staining in R26R+/KI; Ins1-cre+/Tg mice and fluorescence observation in R26GRR+/KI; Ins1-cre+/Tg mice indicated many blue and red dots scattered over the pancreas (Fig. 3). These results suggested that the BAC Ins1-cre transgene was capable of expressing Cre protein in pancreatic islets of BAC Ins1-cre25 mice.
Fig. 3.
Representative stereomicroscopic images of the pancreas in F1 mice, R26R+/KI, R26R+/KI; Ins1-cre+/Tg, R26GRR+/KI, or R26GRR+/KI; Ins1-cre +/Tg. X-gal staining: R26R+/KI mice and R26R+/KI; Ins1-cre+/Tg mice, EGFP & tdsRed fluorescence imaging: R26GRR+/KI mice and R26GRR+/KI; Ins1-cre+/Tg mice. Each experiment (n=6). Sale bar:1 mm (low-power field) and 500 µm (high-power field).
Insulin is synthesized, stored, and secreted by β cells of pancreatic islets. To determine whether Cre-mediated recombination occurred in β cells of pancreatic islets in more detail, we cut the pancreas into sections. Paraffin sections of the pancreas from adult R26R+/KI; Ins1-cre+/Tg mice were immunostained with anti-β-galactosidase antibody and anti-insulin antibody. β-Galactosidase was detected in all of the observed pancreatic islets, but not in other parts of the pancreas. The β-galactosidase-expressing cells were colocalized with insulin-expressing cells, and 90.0% ± 3.6% (n=3) of the insulin-expressing cells also expressed β-galactosidase. Similarly, the colocalization of insulin- and tdsRed-expressing cells in islets was confirmed in frozen sections from the adult R26GRR+/KI; Ins1-cre+/Tg mouse pancreas. Further, we found that the tdsRed-expressing cells were not colocalized with glucagon-expressing cells, which were immunostained with anti-glucagon antibody (Fig. 4). Taken together, the observations indicated that the transgene in BAC Ins1-cre25 mice has capacity for β cell-specific Cre-loxP recombination in the pancreas.
Fig. 4.
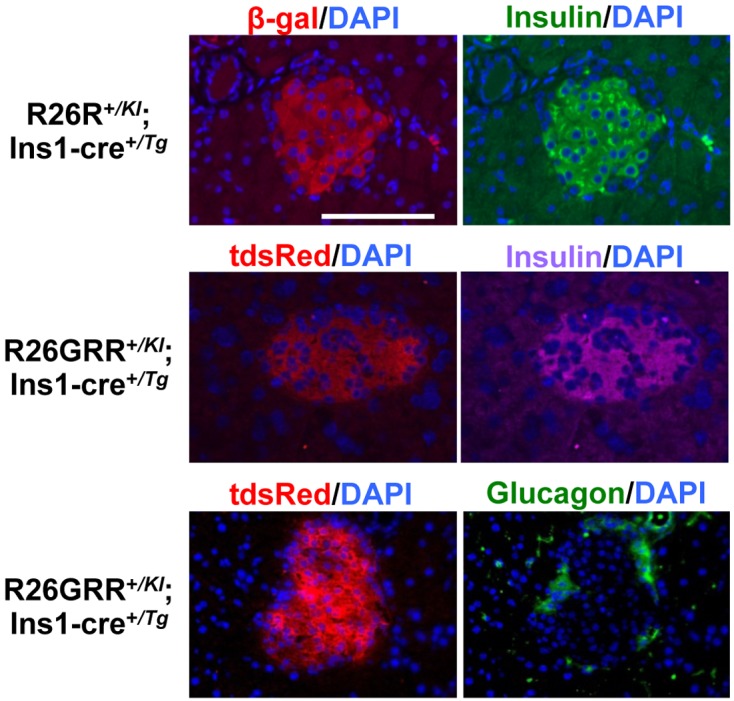
Representative pancreatic islet architecture of F1 mice, R26R+/KI; Ins1-cre+/Tg (paraffin sections) and R26GRR+/KI; Ins1-cre+/Tg (frozen sections), stained with anti-β-galactosidase antibody, anti-insulin antibody, anti-glucagon antibody, and diamidino-2-phenylindole (DAPI). Each experiment (n=3). Sale bar: 100 µm.
Cre activity during fetal pancreas development
Timed matings were set up between hemizygous BAC Ins1-cre25 males and homozygous R26R females or homozygous R26GRR females. Pregnant females were sacrificed to collect embryos on embryonic day (E) 11.5, E13.5, E16.5, and E18.5. There were no Cre-loxP recombination signals in the fetal pancreas in any of the embryos examined with R26R+/KI or R26GRR+/KI as negative controls. Indigo blue spots and tdsRed fluorescence were detected in the pancreas of E13.5 − E18.5 R26R+/KI; Ins1-cre+/Tg and R26GRR+/KI; Ins1-cre+/Tg embryos, respectively, suggesting the appearance of pancreatic islets. However, Cre-loxP recombination signals were not obtained in the pancreas from E11.5 embryos of both strains carrying cre and reporter genes (Fig. 5).
Fig. 5.
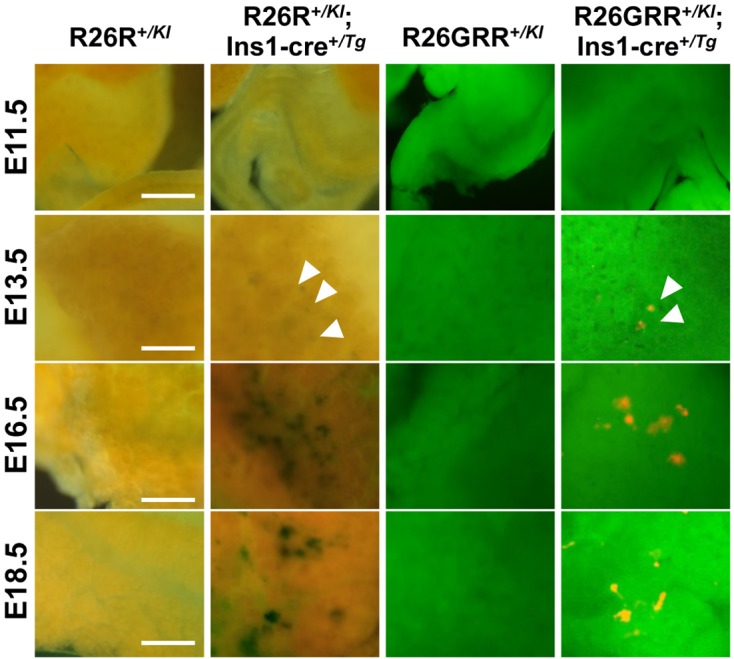
Representative stereomicroscopic imaging of the pancreas in F1 mice R26R+/KI, R26R+/KI; Ins1-cre+/Tg, R26GRR+/KI, and R26GRR+/KI; Ins1-cre+/Tg, during embryo development. X-gal staining: R26R+/KI mice and R26R+/KI; Ins1-cre+/Tg mice, EGFP & tdsRed florescence imaging: R26GRR+/KI mice and R26GRR+/KI; Ins1-cre+/Tg mice. Each experiment (average n=4). Sale bar: 500 µm (E11.5) and 100 µm (E13.5–18.5).
Cre activity in the brain
To investigate ectopic Cre-mediated recombination, we examined reporter signals for Cre-mediated recombination by stereomicroscopy in a variety of tissues from F1 adults obtained from crosses between hemizygous BAC Ins1-cre25 males and homozygous R26R females or homozygous R26GRR females. X-gal staining indicated reporter signals in the kidney, stomach, intestine, and testis of F1 mice carrying R26R+/KI; Ins1-cre+/Tg, similar to those of F1 mice carrying R26R+/KI. However, there were no signals in these tissues obtained from R26GRR+/KI; Ins1-cre+/Tg. These observations suggested that signals detected in mice carrying R26R+/KI; Ins1-cre+/Tg represented nonspecific Cre activity (Supplement Fig. 2).
Finally, we performed a more detailed stereomicroscopic analysis using sequential coronal sections of the entire brain from rostral to caudal in mice carrying R26R+/KI; Ins1-cre+/Tg and mice carrying R26GRR+/KI; Ins1-cre+/Tg, because Ins2, an ortholog of Ins1, is expressed not only in the pancreas but also in the brain. As expected, the reporter signal for Cre-loxP recombination was not detected in the brain of either set of F1 mice, similar to F1 mice carrying R26R+/KI or R26GRR+/KI (Fig. 6).
Fig. 6.
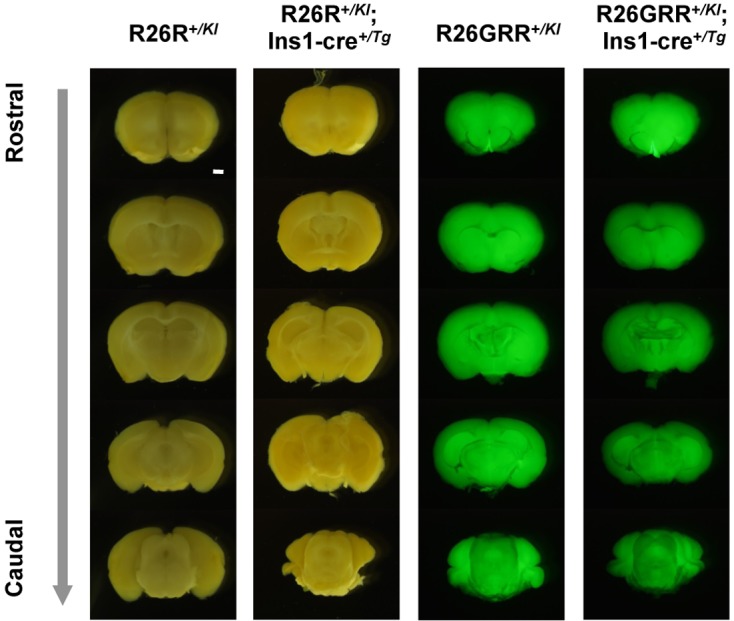
Representative stereomicroscopic sagittal imaging of the adult brain in F1 mice, R26R+/KI, R26R+/KI; Ins1-cre+/Tg, R26GRR+/KI, and R26GRR+/KI; Ins1-cre+/Tg. X-gal staining: R26R+/KI mice and R26R+/KI; Ins1-cre+/Tg mice, EGFP & tdsRed florescence imaging: R26GRR+/KI mice and R26GRR+/KI; Ins1-cre+/Tg mice. Each experiment (n=6). Sale bar: 1 mm.
Discussion
Pancreatic β cell-specific genome alteration using the Cre-loxP recombination system in mice is useful for understanding the biological functions of genes of interest for new diabetes treatments. We generated and characterized novel BAC Ins1-cre25 mice in which specifically expressed Cre under the control of Ins1 locus. Using two different Cre-reporter mice, we found that BAC Ins1-cre-mediated Cre-loxP recombination occurred in insulin-producing pancreatic islet β cells, but not in the brain and other organs. Further, the BAC Ins1-cre transgene already caused Cre-loxP recombination in pancreatic islets at E13.5. Moreover, we identified that the BAC Ins1-cre transgene was integrated into chromosome 15 in BAC Ins1-cre25 mice.
Two independent groups reported that STAT3lox/lox mice expressing the cre gene under the control of the Ins2 promoter exhibited glucose intolerance, impairment of early-phase insulin secretion, and mild obesity [1, 5]. Each group used RIP-cre mice for the production of β cell-specific STAT3 cKO mice. However, as RIP-cre mice have Cre activity in the brain, including hypothalamus, STAT3 deficiency occurs in neurons expressing leptin receptors in the hypothalamus in addition to pancreatic β cells. Therefore, it is unclear whether the metabolic disorders demonstrated in these strains were due to an independent function of STAT3 in pancreatic β cells. Recently, Wicksteed et al. [18] compared Cre activity in the brain in three different RIP-cre mouse strains, Tg (Ins2-cre)25Mgn, Tg (Ins2-cre)1Herr, and Tg (Ins2-cre/Esr)1Dam. Cre-loxP recombination signal was detected in the brain, including the hypothalamus, in each of these three commonly used RIP-cre mouse strains. Furthermore, although there are some other RIP-Cre mice [11], the Ins2 promoter appears to be unsuitable for exclusive pancreatic β cell-specific cre gene expression.
The regulatory region of Ins1 has been used for reporter gene expression to allow visualization of pancreatic β cells in mice. The 8.5-kb mouse Ins1 promoter (MIP) consisting of the region from −8.5 kb to +12 bp relative to the transcriptional initiation site has been used for the generation of transgenic mice in which pancreatic β cells are genetically tagged with GFP (MIP-GFP mice) [6]. Furthermore, transgenic mice expressing firefly luciferase under the control of the MIP (MIP-luc and MIP-luc VU) were generated to monitor β cell function in living animals with normal or altered metabolism [13, 20]. Similar to tissue-specific reporter gene expression in MIP-GFP mice, MIP-luc mice, and MIP-luc VU mice, Cre activity is observed in pancreatic β cells of transgenic mice in which tamoxifen-inducible Cre-loxP recombination is driven by MIP (Tg (Ins1-cre/ERT)1Lphl), but not in the brain [18]. The analysis of Cre-loxP recombination using BAC Ins1-cre25 mice confirmed that a certain regulatory region of Ins1 exclusively drives pancreatic β cell-specific Cre expression in mice lacking Cre expression in the brain. However, little information is available regarding the use Tg (Ins1-cre/ERT)1Lphl as Cre-driver mice.
Almost productions of Cre-driver mouse strains for pancreatic β cells has been based on the zygote microinjection using relatively short insulin gene promoter fragments [11]. The zygote microinjection results in both randomly integrated transgenes and variable transgene copy numbers, both of which can negatively influence the accuracy and duration of Cre expression. Further, short Cre-driver gene fragments may lack key cis-regulatory elements necessary for precise cell-or tissue-specific gene expression. To achieve high-faithful Cre-driver lines for specific Cre-loxP recombination in the pancreas, several preferred strategies were recently discussed by Magnuson and Ospipovich [11]. They recommended the use of BACs as an approach for obtaining high-fidelity Cre driver line expression [11]. The BAC Ins1-cre insert is large and therefore could carry the regulatory sequences necessary for spatially, temporally and quantitatively correct expression closely reflecting endogenous Ins1 expression independent of the integration site [3, 19]. In a recent study, we improved the intensity and background of luciferase activity in transgenic mice using an exogenous mouse 200-kb genomic fragment comprised of Ins1, the same fragment used in the present study, compared with MIP-luc VU mice in which luciferase expression is controlled by the 9.2-kb MIP [8]. Furthermore, the present study clearly indicated that BAC containing the Ins1locus allowed precise β cell-specific expression of cre gene in mice. Therefore, our data suggest that BAC-mediated cre gene expression is a useful approach for generating highly reliable Cre driver mouse strains.
In this study, we detected Cre-loxP recombination signals in the pancreases of embryos at E13.5 in both R26R+/KI; Ins1-cre+/Tg mice and R26GRR+/KI; Ins1-cre +/Tg mice. In the process of murine β cell development, a few insulin-expressing cells appear at E9.5 and then fully differentiated β cells first appear around E13 at the start of a massive wave of endocrine differentiation in the pancreas known as the “secondary transition” [15]. Our results suggested that Cre protein in BAC Ins1-cre25 mice excise the floxed gene segment in fetal pancreatic β cells at the secondary transition. However, it remains unclear why Cre activity is undetectable before the secondary transition in BAC Ins1-cre25 mice.
Due to chromosomal integration, the cre transgene can be passed from Cre-driver mice to floxed mice by cross-breeding to allow the production of conditional knockout mice. However, the transition of cre transgene is difficult when the cre transgene are physically close to the floxed gene on the same chromosome. Therefore, although the integration site of the cre transgene is important, the sites of cre transgene integration in all insulin-Cre drive mouse strains have not been determined. We showed that the cre transgene of BAC Ins1-cre25 mice is integrated into chromosome 15 at a position midway between Mafa and the telomere. Therefore, we recommend using BAC Ins1-cre25 mice as Cre-drivers for pancreatic β cell-specific Cre-loxP recombination, except for cross-breeding with floxed mice on chromosome 15. In future studies, we will characterize Cre-loxP recombination and perform chromosomal mapping of the transgene in BAC Ins1-cre24 mice (founder line of #24) to exclude this limitation on BAC Ins1-cre25 mice.
To our knowledge, BAC Ins1-cre25 is the first transgenic mouse line containing Cre driven by the Ins1 locus in a pure genetic background of C57BL/6N for exclusive pancreatic β cell-specific Cre/loxP recombination. The BAC Ins1-cre25 mouse strain, RBRC03934 C57BL/6N-Tg (Ins1-cre) 25Utr/Rbrc, is available from the RIKEN BioResource Center.
Supplementary
Acknowledgments
This work was supported by the RIKEN BRC program for R&D of Genetically Modified Mouse Strains (to K.Y.), and Grants-in-Aid for Scientific Research (S) (to S.T.) and Challenging Exploratory Research (to F.S.) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan. We thank the members of the Yagami Laboratory for helpful discussions and encouragement.
References
- 1.Cui Y., Huang L., Elefteriou F., Yang G., Shelton J.M., Giles J.E., Oz O.K., Pourbahrami T., Lu C.Y., Richardson J.A., Karsenty G., Li C.2004. Essential role of STAT3 in body weight and glucose homeostasis. Mol. Cell Biol. 24: 258–269. doi: 10.1128/MCB.24.1.258-269.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deltour L., Leduque P., Blume N., Madsen O., Dubois P., Jami J., Bucchini D.1993. Differential expression of the two nonallelic proinsulin genes in the developing mouse embryo. Proc. Natl. Acad. Sci. USA 90: 527–531. doi: 10.1073/pnas.90.2.527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gangalum R.K., Jing Z., Nagaoka Y., Jiang M., Bhat S.P., Bhat S.P.2011. Purification of BAC DNA for high-efficiency transgenesis. Biotechniques 51: 335–336, 338. doi: 10.2144/000113763 [DOI] [PubMed] [Google Scholar]
- 4.Gannon M., Shiota C., Postic C., Wright C.V., Magnuson M.2000. Analysis of the Cre-mediated recombination driven by rat insulin promoter in embryonic and adult mouse pancreas. Genesis 26: 139–142. doi: [DOI] [PubMed] [Google Scholar]
- 5.Gorogawa S., Fujitani Y., Kaneto H., Hazama Y., Watada H., Miyamoto Y., Takeda K., Akira S., Magnuson M.A., Yamasaki Y., Kajimoto Y., Hori M.2004. Insulin secretory defects and impaired islet architecture in pancreatic beta-cell-specific STAT3 knockout mice. Biochem. Biophys. Res. Commun. 319: 1159–1170. doi: 10.1016/j.bbrc.2004.05.095 [DOI] [PubMed] [Google Scholar]
- 6.Hara M., Dizon R.F., Glick B.S., Lee C.S., Kaestner K.H., Piston D.W., Bindokas V.P.2006. Imaging pancreatic beta-cells in the intact pancreas. Am. J. Physiol. Endocrinol. Metab. 290: E1041–E1047. doi: 10.1152/ajpendo.00365.2005 [DOI] [PubMed] [Google Scholar]
- 7.Hasegawa Y., Daitoku Y., Sekiguchi K., Tanimoto Y., Mizuno-Iijima S., Mizuno S., Kajiwara N., Ema M., Miwa Y., Mekada K., Yoshiki A., Takahashi S., Sugiyama F., Yagami K.2013. Novel ROSA26 Cre-reporter knock-in C57BL/6N mice exhibiting green emission before and red emission after Cre-mediated recombination. Exp. Anim. 62.: 295–304. doi: 10.1538/expanim.62.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katsumata T., Oishi H., Sekiguchi Y., Nagasaki H., Daassi D., Tai P.H., Ema M., Kudo T., Takahashi S.2013. Bioluminescence imaging of β cells and intrahepatic insulin gene activity under normal and pathological conditions. PLoS ONE 8: e60411. doi: 10.1371/journal.pone.0060411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulkarni R.N., Brüning J.C., Winnay J.N., Postic C., Magnuson M.A., Kahn C.R.1999. Tissue-specific knockout of the insulin receptor in pancreatic beta cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell 96: 329–339. doi: 10.1016/S0092-8674(00)80546-2 [DOI] [PubMed] [Google Scholar]
- 10.Madadi G., Dalvi P.S., Belsham D.D.2008. Regulation of brain insulin mRNA by glucose and glucagon-like peptide 1. Biochem. Biophys. Res. Commun. 376: 694–699. doi: 10.1016/j.bbrc.2008.09.054 [DOI] [PubMed] [Google Scholar]
- 11.Magnuson M.A., Osipovich A.B.2013. Pancreas-specific Cre driver lines and considerations for their prudent use. Cell Metab. 18: 9–20. doi: 10.1016/j.cmet.2013.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizuno S., Mizobuchi A., Iseki H., Iijima S., Matsuda Y., Kunita S., Sugiyama F., Yagami K.2010. A novel locus on proximal chromosome 18 associated with agenesis of the corpus callosum in mice. Mamm. Genome 21: 525–533. doi: 10.1007/s00335-010-9292-4 [DOI] [PubMed] [Google Scholar]
- 13.Park S.Y., Wang X., Chen Z., Powers A.C., Magnuson M.A., Head W.S., Piston D.W., Bell G.I.2005. Optical imaging of pancreatic beta cells in living mice expressing a mouse insulin I promoter-firefly luciferase transgene. Genesis 43: 80–86. doi: 10.1002/gene.20157 [DOI] [PubMed] [Google Scholar]
- 14.Postic C., Shiota M., Niswender K.D., Jetton T.L., Chen Y., Moates J.M., Shelton K.D., Lindner J., Cherrington A.D., Magnuson M.A.1999. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 274: 305–315. doi: 10.1074/jbc.274.1.305 [DOI] [PubMed] [Google Scholar]
- 15.Sander M., Sussel L., Conners J., Scheel D., Kalamaras J., Dela Cruz F., Schwitzgebel V., Hayes-Jordan A., German M.2000. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development. 127: 5533–5540. [DOI] [PubMed] [Google Scholar]
- 16.Soares M.B., Schon E., Henderson A., Karathanasis S.K., Cate R., Zeitlin S., Chirgwin J., Efstratiadis A.1985. RNA-mediated gene duplication: the rat preproinsulin I gene is a functional retroposon. Mol. Cell Biol. 5: 2090–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soriano P.1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21: 70–71. doi: 10.1038/5007 [DOI] [PubMed] [Google Scholar]
- 18.Wicksteed B., Brissova M., Yan W., Opland D.M., Plank J.L., Reinert R.B., Dickson L.M., Tamarina N.A., Philipson L.H., Shostak A., Bernal-Mizrachi E., Elghazi L., Roe M.W., Labosky P.A., Myers M.G., Jr, Gannon M., Powers A.C., Dempsey P.J.2010. Conditional gene targeting in mouse pancreatic ß-Cells: analysis of ectopic Cre transgene expression in the brain. Diabetes 59: 3090–3098. doi: 10.2337/db10-0624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Keuren M.L., Gavrilina G.B., Filipiak W.E., Zeidler M.G., Saunders T.L.2009. Generating transgenic mice from bacterial artificial chromosomes: transgenesis efficiency, integration and expression outcomes. Transgenic. Res. 18: 769–785. doi: 10.1007/s11248-009-9271-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Virostko J., Radhika A., Poffenberger G., Chen Z., Brissova M., Gilchrist J., Coleman B., Gannon M., Jansen E.D., Powers A.C.2010. Bioluminescence imaging in mouse models quantifies beta cell mass in the pancreas and after islet transplantation. Mol. Imaging Biol. 12: 42–53. doi: 10.1007/s11307-009-0240-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.