Abstract
The main cells of the adipose tissue of animals, adipocytes, are characterized by the presence of large cytosolic lipid droplets (LDs) that store triglyceride (TG) and cholesterol. However, most cells have LDs and the ability to store lipids. LDs have a well-known central role in storage and provision of fatty acids and cholesterol. However, the complexity of the regulation of lipid metabolism on the surface of the LDs is still a matter of intense study. Beyond this role, a number of recent studies have suggested that LDs have major functions in other cellular processes, such as protein storage and degradation, infection, and immunity. Thus, our perception of LDs has been radically transformed from simple globules of fat to highly dynamic organelles of unexpected complexity. Here, we compiled some recent evidence supporting the emerging view that LDs act as platforms connecting a number of relevant metabolic and cellular functions.
Keywords: lipid droplet, lipid metabolism, perilipin, fat body, triglycerides, lipoprotein
Introduction
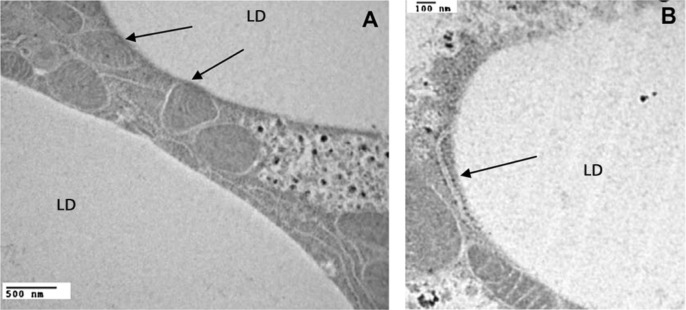
The major metabolic fates of fatty acids (FA) entering the cell are esterification into triglycerides (TG), synthesis of phospholipids for membrane biosynthesis, and beta-oxidation for ATP production in mitochondria. Adipocytes, however, capture and store the excess of FAs as TG in cytosolic lipid droplets (LDs), which are large quasi-spherical structures readily visualized by light microscopy.1–3 LDs are in physical contact with other LDs and also with other cellular structures such as endoplasmic reticulum (ER), mitochondria, peroxisomes, and endosomes. These interactions are frequently observed by electron microscopy4 (Fig. 1). Interestingly, live imaging revealed that LDs also undergo intracellular movements in the confines of the cytoplasm.5,6 LDs can be isolated from cell homogenates by density gradient ultracentrifugation7–10 and purified LDs retain their shape and functions, such as the ability to store TG11 or serve as substrate for lipases12 when tested in vitro.
Figure 1.
Interaction among LDs, mitochondria, and ER. The transmission electron micrographs of M. sexta fat body show the physical contacts (indicated with arrows) observed between LD and mitochondria (A), or between LD and ER (B). This figure was adapted from a previous report10 and is reproduced with permission.
The structure of LDs is commonly described as formed by a core of neutral lipids, predominantly TG and sterol esters, surrounded by a monolayer of phospholipids and a variety of proteins.13–15 TGs are the main lipids found in LDs. Other lipids are diglycerides (DG), retinyl- and cholesteryl esters (CEs), and ether lipids.16–20 A high content of CEs is found in LDs from macrophages and adrenal cells.21–24 The LD surface consists of phospholipids, lysophospholipids, and cholesterol.16 The phospholipid monolayer is the boundary that separates the hydrophobic core of LDs from the aqueous environment.25,26 It could also be involved in the differential recruitment of lipid droplet proteins.25 The protein coat is composed of numerous proteins; it can comprise up to hundreds of proteins.27 A distinctive group of proteins, the perilipins or PAT proteins, are always associated with LDs as shown in all LD proteomes.7–10,28–38 This is a small group of proteins composed by perilipin (PLIN1), ADRP (PLIN2), TIP47 (PLIN3), S3–12 (PLIN4), and OXPAT (PLIN5).39,40 The PAT family of proteins is evolutionary conserved and relevant studies on the biology of LDs and the function of PLIN proteins are being conducted not only in vertebrates but also in other eukaryotic systems such as insects6,10,12,41–49 and yeast.50,51 Early studies on TG metabolism in adipocytes have shown significant conservation between mammals and insects.52 PLIN1 and PLIN2 occur in insects and were originally named LSD1 and LSD2.53 PLIN proteins have in common a conserved region of ~100 amino acids toward the N-terminal called the PAT domain.54 The function of this region remains unknown but it is not required for binding to LD.52 PLIN proteins play important roles in the processes of lipid storage and mobilization, however the underlying mechanisms of function are not completely known in any system, yet. The occurrence of PLIN proteins depends on a variety of conditions such as type of tissue and the developmental and metabolic stage of the cell. PLIN proteins may also differ in the subcellular localization; while PLIN1 is always found associated with LDs for which they are classified as constitutive LD proteins, PLIN3–5, which are also found in the cytosol, are considered exchangeable LD-proteins.52 PLIN1 plays major roles in the storage and degradation of TG in adipocytes as indicated by studies in vertebrates55,56 and in insects.52,57 PLIN1 blocks lipolysis under basal conditions, but enables the action of lipases under hormone stimulated lipolysis.39 So far, only Drosophila’s PLIN1 has been purified and reconstituted in lipid droplet-like particles allowing some unique structural and functional studies.49,58
Since the hallmark discovery that PLIN1 regulates the storage of lipids,59 the study of LDs has seen a steady increase in attention by the scientific community. In the past decade, LDs have emerged as heterogeneous organelles with great dynamics in size, abundance, and composition in regard to both proteins and lipids.60 Recent studies have provided more details on LD structure, biogenesis, function, and molecular regulation.3,13,15,61,62 Given the conservation among the basic mechanisms of control of lipid metabolism,63 studies in insects41 and other systems have been instrumental to the advancement of this field. LDs are currently associated with multiple cellular roles beyond the storage of lipids.27 Here we compiled some of the most recent studies with the goal of presenting the LDs as a major platform for the regulation and execution of multiple cellular functions.
Heterogeneity, A Feature of LDs
With the exception of mature white adipocytes from mammalian cells in which a single supersized LD (>100 μm)64 is present, LDs of a broad range of sizes (1–100 μm) are observed in both vertebrate and invertebrate adipocytes.15 Almost all types of cells are able to store TG in LDs, and in general, droplets in non-adipocyte cells are smaller.65 LDs are subjected to fast and substantial variations in size and number in response to hormonal changes and the availability of nutrients. For instance, a thirty-fold increase in LD-volume is observed within hours when Drosophila S2 cells are incubated in the presence of oleic acid.66 In mammalian cells, LD size can increase by the transfer of lipids between droplets, which is possible through contact sites and the cooperative action of FSP27 and PLIN1.67 This process leads to the formation of the single LD in mature adipocytes.68 Conversely, TG hydrolysis induces LD fragmentation and shrinking69,70 with a rapid formation of small and micro LDs (<1 μm) in adipocytes.71–73 Although, both small and micro LD originate during lipolysis, small LDs would be generated from the fission of large LDs, whereas micro LDs would be formed with TG produced by the re-esterification of FAs.72 Micro LDs and small LDs are considered physiologically relevant in the response to energy demands.72,73
Enzymes of lipid synthesis are mostly integral membrane proteins of ER.74 Therefore, the site of the biogenesis of LDs is assumed to be the ER membrane, which harbors the diacylglycerol acyltransferases (DGAT) that are needed to catalyze the last reaction of TG synthesis. The hydrophobicity and low solubility of TG in phospholipids indicate that they would start accumulating in the bilayer core promoting the formation of intramembrane lipid globules that would eventually bud off from the ER to the cytoplasm.62,75 Once the LD is formed, it could accept additional lipids. Live-cell microscopy studies suggested that at least part of TG synthesis takes place in the immediate vicinity of LDs,11 and that intracellular lipids continuously exit and re-enter LDs.76
Some studies suggest that the newly formed LDs that appear in adipocytes incubated with FA localize on the cytosolic side of the plasma membrane. These LDs are smaller and coated with different types of perilipins unlike the larger and older cytosolic droplets that are coated with perilipin 1.77–79 Likewise, studies in Drosophila have shown that the fat body presents small LDs (1 μm) containing LSD2 and LSD1 and larger LDs that only express LSD1.42 The occurrence of subpopulations of LD is in line with previous studies showing the presence of different pools of neutral lipids in adipocytes from mammalian80 and insect81 adipose tissue. The incorporation of FA into these tissues leads to the accumulation of diacylglycerides (DG). In both systems, 20–50% of the FA are accumulated as DG, and the newly synthesized DG pool has a relatively long half-life.80,82–84 This pool of DG remained inaccessible to the TG biosynthetic pathway for several hours, and was preferentially mobilized under hormonally-stimulated lipolysis.81 It is possible that these lipids are stored in the newly formed LDs at the periphery of the cell.
LDs are Organelles
The quasi-spherical geometry and the presence of a large lipid core that characterizes both lipoproteins and LDs suggest that they could be compared. In fact, until not long ago, LDs were seen as simple intracellular lipoproteins. It may then be appropriate to point out some of the differences between these two structures. Lipoproteins are assembled in the ER lumen and secreted by the secretory pathway into the circulatory system.85 LDs also originate from the ER, and although the process of LD assembly is not understood yet, it is clear that nascent LD segregates from the cytoplasmic face of the ER. The synthesis of lipoproteins is restricted to certain types of cells (eg hepatocytes, enterocytes).86 However, LDs are common organelles present in most types of cells. Some tissues, such as the liver in vertebrates and the fat body of insects store lipids as cytoplasmic LDs and also synthesize and secrete lipoproteins to circulation.87 Lipoproteins transport lipids among tissues, whereas LDs hold lipids in the cytoplasm. The function of lipoproteins requires a structure that limits their interactions with many cellular and extracellular components, and thus, allows the particle to reach the target tissues. Conversely, LDs serve as a scaffold that recruits proteins involved in lipid metabolism and other metabolic processes. LDs are in contact with elements of cytoskeleton as shown in all LD proteomes.7–10,28–38 The intermediate filament vimentin was one of the first LD-associated proteins identified.88 Early studies showed a tight association between LDs and vimentin filaments enduring extensive washes with 1% Triton X-100.89 It has been shown that vimentin is required for proper lipolysis90,91 and for the provision of LD-cholesterol to the mitochondria for steroidogenesis.92 Cholesterol stored in the LDs of adrenal cells must be delivered to mitochondria, where the first step of steroid synthesis takes place. Genetic ablation of vimentin in mice resulted in a marked defect in steroidogenesis.92 Vimentin binds to LDs and proteins that are involved in LD metabolism, such as the hormone sensitive lipase, HSL,91 and the steroidogenic acute regulatory protein that targets mitochondria.93 Vimentin is considered an important component of the network that facilitates lipolysis and the movement of LDs toward mitochondria.93
In Drosophila embryos, a bidirectional movement of LDs along cytoskeletal tracks6 has been demonstrated.94 These movements are driven by the cytoplasmic motors dynein and kinesin-1 and controlled by Klar, a highly abundant LD protein in fly embryos. Klar binds to LDs through a specific domain localized in the C-terminal region of the protein.95 Deletion of this Klar domain impairs binding of Klar to LDs and the ability to control droplet motion.96 The interaction of Klar with LD is controlled by the phosphorylation of LSD2, a LD protein from the PLIN family.94 As LSD2 controls lipid storage,43 it was proposed that phosphorylation of LSD2 controls both lipid metabolism and droplet motion. LD mobility could be associated with the delivery of lipids to other cellular organelles.6
The number of proteins associated with the surface of LDs is remarkably large, whereas circulating lipoproteins only contain a discrete number of specific proteins (structural and exchangeable apolipoproteins). Given the variety of proteins associated with LDs, different types of interactions and mechanisms of association of proteins with the LD surface are expected. PLIN1, the best characterized LD-associated protein so far, is synthesized in free ribosomes and it is supposed to localize to LD surface from the cytosol.97–99 More recent information showed that PLIN1 also can relocate to the ER from where the protein moves back and forth between ER and LD.100 These observations are in accordance with the tight association between LD and ER observed by electron microscopy,51,101,102 and the functional association between LDs and ER that has been demonstrated in yeast Saccharomyces cerevisiae.103 Insect PLIN1 or LSD1 is not soluble in the hydrophilic environment and can only be maintained in solution when reconstituted in lipoprotein particles.49 It is then expected that its intracellular movement and localization to LD be mediated by small lipoprotein complexes. The involvement of coatomer-dependent mechanisms for proper localization of proteins at the LD surface has also been demonstrated.104,105
Underlining the dynamic nature of these organelles is the fact that the protein coat of LD undergoes major changes in response to the metabolic state of the cell.8,10 As an illustration, Figure 2 shows protein profiles of LDs isolated from Manduca sexta fat body adipocytes under opposite physiological conditions: high lipogenesis and high lipolysis, which are characteristic of the larval and adult stages, respectively. The differences in protein composition that can be readily observed in Figure 2 were confirmed by mass spectrometry.10 The differences affected proteins such as perilipins, but also tens of other proteins that are not necessarily related to lipid metabolism. Lipoproteins also undergo changes in protein composition that have major impacts in metabolism. However, overall, the composition of lipoproteins is kept simple and includes a discrete set of proteins, mostly the apolipoproteins. The protein profile of purified insect lipoprotein, lipophorin is shown in Figure 2. Lipophorin is also synthesized in the fat body and, as LDs, was also purified by ultracentrifugation in a density gradient.
Figure 2.
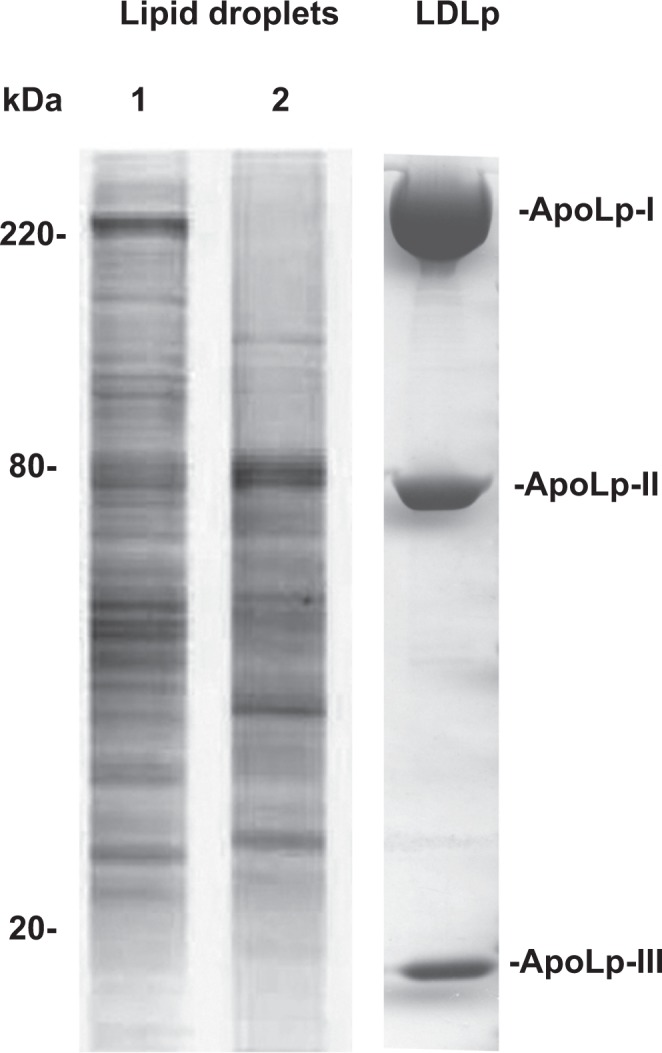
Changes in the protein composition of LDs from the same tissue. Protein profiles of purified LDs from the fat bodies of Manduca sexta larva and adult insects obtained by SDS-PAGE are shown in lanes 1 and 2, respectively. Additional information on the protein compositions of the LDs was previously reported.10 For comparison, the protein profile of Manduca sexta low density lipoprotein purified by ultracentrifugation in a density gradient180 is shown in the lane labeled LDLp. ApoLp-I, -II and -III refer to the protein components of the insect low density lipoprotein. Parts of the figure are reproduced from previous reports10,180 with permission.
LD Proteomes: First Look at the LD Network
Proteomic studies of purified LD have shown that, in addition to the PLIN proteins, the protein coat of LDs includes a large number of significantly abundant proteins. A complex mixture of proteins from multiple subcellular compartments including cytosol, mitochondria, lysosomes, and ER are found associated with purified LDs. Proteins of the PLIN family, lipid metabolism, nucleus, ribosomes, chaperones, cytoskeleton, ER, membrane trafficking, and cell signaling are commonly identified in LD proteomes.7–10,28–38 The possibility of contamination of the LD preparations is often raised.3,13,52 However, the consistency on these findings in preparations of LDs from different systems clearly argues against that possibility. Electron micrographs from many systems have demonstrated tight physical contact of LDs with other subcellular compartments.10,101,106,107 Most recently, reports of functional interaction between LDs and ER,103 and LDs and mitochondria,108 and LDs and peroxisomes61 add support that these proteomes for the most part are a reflection of the complexity of LDs. Interestingly, LD proteomes show proteins that are known to possess integral membrane regions. This common observation suggests that the LD envelope may have bilayer domains enriched in ER or mitochondrial proteins or, alternatively, may contain domains that are fragments of ER or mitochondrial membranes. It seems obvious now that LDs cannot be simply described as a fat globule surrounded by a monolayer of phospholipid and a discrete number of different proteins anchored to its surface. Electron microscopy has provided evidence consistent with the presence of monolayer structure.2,109 However, that evidence may not apply to some LD population. The complexity of the protein composition of LDs that has been provided by the proteomic studies suggests that LDs have an ample spectrum of interactions with other organelles and a number of cytosolic proteins and protein complexes. This picture of LDs is in line with the increasing evidence supporting a role of LDs in a number of cellular functions. Recent studies showing that structural and functional interactions between LDs and other organelles are of physiological relevance support this notion.50
LDs as Signaling Platforms Involved in Multiple Cellular Functions
The array of functions related to LDs can be separated into classical and non-classical functions. The former are directly or indirectly linked to lipid storage and mobilization, whereas the second group of functions involving “refugee” proteins are related to vesicular trafficking, protein folding, protein storage, autophagy, immunity, and virus replication.44 “Refugee” or non-resident proteins are proteins characteristic of other cellular compartment that nonetheless associate with LDs. Numerous refugee proteins have been identified in independent biochemical and proteomic studies.7–10,28–38 An increasing number of mostly recent studies are shedding light into why those proteins are present in LDs and what metabolic and/or cellular conditions cause their association with the organelle.
LDs and whole-body energy homeostasis
The adipose tissue in vertebrates and the fat body in insects play a major role in maintaining the whole body energy homeostasis. Under an energy demand, FA are mobilized from these tissues to supply fuel for other organs. Mammalian adipocytes secrete free FA, whereas insects mobilize FA as DG, which are transported in circulation by lipophorin.110 On the surface of the lipid droplet is PLIN1, which serves as a gatekeeper controlling the access of lipases to the lipids inside the droplet. Catecholamines in vertebrates111 and adipokinetic hormones in insects87,112 control the mobilization of FA through a cAMP/PKA signaling cascade. PKA-mediated phosphorylation of PLIN1 in vertebrates113 and LSD1 in insects12 activates TG hydrolysis. In vertebrates, lipolysis is catalyzed by the concerted action of at least PLIN1 proteins, three lipases (ATGL, HSL, MGL), a co-lipase (CGI58), and two separate pools of PKA.24,114 PKA proximal to plasma membrane phosphorylates HSL triggering the translocation to the surface of LD.22,115 The second PKA seems to be on the surface of LD116,117 apparently recruited by the protein OPA1.118 A new study showed that this protein, which is known to be a mitochondrial protein, is a putative A-kinase anchoring protein (AKAP) on LDs.119 Under stimulated conditions, OPA1 seems to operate on the surface of LDs recruiting PKA in connection to PLIN1 phosphorylation and activation of lipolysis.119
Other kinases such as ERK, p38 MAP, and AMP kinases are also activated by catecholamines120–122 and adipokinetic hormones.123 For example, HSL can be phosphorylated by ERK which increases its activity,124,125 whereas phosphorylation by AMP kinase is inhibitory.126,127 PLIN1 and HSL have multiple phosphorylation sites.56 The modulation of the lipolytic response may rely on differential phosphorylation levels of these proteins.128
The lipolytic activity of a cell must respond to short and long term requirements of its own metabolism, such as FA oxidation and membrane synthesis, but also of the whole body metabolism, for instance for the synthesis of steroid hormones, for the synthesis of lipoproteins, and for the provision of FA to other tissues. Since FA and cholesterol are mobilized from the surface of the LDs, it is not surprising that the LDs harbor a number of lipases, kinases, and regulatory proteins. This complexity is needed to allow the coordination of multiple signals originated from the metabolic requirements of both the cell and the whole organism.
LDs as transient platforms for protein storage
LDs are highly abundant in the oocytes of insects and play an essential role in the reproduction of insects by providing fuel and FA for the development of the embryo. The accumulation of histones in LDs has been detected in proteomic study in early Drosophila embryos.9 More importantly, these histones were transferred to the nuclei during development.9 Histone proteins are needed to assemble chromatin. As early embryogenesis involves several rounds of DNA-replication without translation, pre-made histones are required for chromatin packaging. In Drosophila, maternal histone protein is synthesized and temporarily stored in the nurse cells and then transferred to the oocyte.129,130 Maternal histone H2A, H2B, and H2Av proteins as well as those proteins that are generated by maternal histone mRNA accumulates in the LDs. Now we know that histones are recruited by the LDs through a LD-associated protein called Jabba, which forms protein complexes with histones.131 The sequestration of histones to LDs by Jabba helps their stabilization. Since, in the absence of Jabba, there is no storage of maternal histone H2A, H2B, and H2Av in the oocytes or early embryos, this process could be important to preserve histones from degradation. Like histones, Jabba is maternally provided.
It has been proposed that other proteins target LDs for temporary shelter. The common presence of ATP-synthase subunits in LDs is intriguing.7–10,132,133 A new study showed that three ATP-synthase subunits interact with invadolysin,134 which also is a LD-associated protein.135 Invadolysin is a conserved metalloproteinase required for proper mitochondrial function.134 It is possible that following their synthesis in the cytoplasm, the inactive ATP-synthase subunits are recruited by the LDs. The interaction with invadolysin on the LD surface could be needed as part of a mechanism of folding and activation of ATP-synthase subunits.
LDs as transient platforms in the path of protein degradation
Misfolded proteins in the ER lumen are removed from the ER via dislocation to the cytosol, where they are degraded by the ubiquitin/proteasome system. This process, which is called ER-associated degradation (ERAD), covers a range of different mechanisms that include protein adaptors whose function is to recruit misfolded proteins to the ubiquitination machinery.30,33 Proteasomes are distributed in the vicinity of LDs,136 and LDs are involved in the degradation of at least two substrates: lipidated ApoB100137 and 3-hydroxy-3-methylglutaryl CoA (HMG CoA) reductase,138 the rate-limiting enzyme of cholesterol synthesis. A role for LDs in ERAD was originally suggested when proteasome inhibition caused the accumulation of ApoB100 at the surface of LDs.137 Furthermore, LDs might play a broader role in the degradation of misfolded ER proteins since pharmacological inhibition of LD formation impaired the dislocation of other proteins.145 To eliminate lipidated ApoB100 from the ER lumen, the lipidated particle is transported to the LD surface in a process that involves the proteins Derlin-1 and Ubxd8.139 It has been suggested that Derlin-1 in the ER membrane and Ubxd8 on the LD surface interact in an ER–LD juncture allowing the passage of ApoB100 to the LD surface for ubiquitination.139 Ubxd8, a LD-associated protein140–142 recruits VCP/p97, the ATP driven chaperone required to process ubiquitin-labeled proteins.143 The ubiquitination machinery also involves E3 ubiquitin ligases that use adaptor proteins for substrate recognition. Ancient ubiquitous protein 1 (AUP1), a highly conserved protein that localizes to ER and LDs34,38 and is found in LD proteoms,8 acts as an adaptor recruiting different ubiquitin ligases such as E2 conjugase Ube2g2,144 an E3 ligase AMFR/gp78.138,145,146
AUP1 also mediates the sterol-induced ubiquitination of HMG-CoA reductase.146 AUP1 recruits the ubiquitin-conjugating enzyme Ubc7 to LDs and facilitates its binding to both gp78 and Trc8. The LD mediated degradation of the rate-limiting HMG CoA reductase is important for the control of cholesterol synthesis in mammalian cells.138
Apart from degradation of ERAD proteins, the abundance of LD-associated proteins PLIN1 and 2 are also controlled by the ubiquitin/proteasome system.147 In this regard, it has been shown that the multifunctional protein spartin acts as an adaptor needed for the degradation of ADRP that takes place during adipocyte maturation.148 Housekeeping chaperones such as Hsp70, which are found in all LD proteomes,8–10,31,149,150 may also serve as adaptors to recruit misfolded proteins for proteasomal degradation.
Although the number of examples is limited, the reported studies suggest that degradation of ER proteins on the surface of LDs could be a mechanism controlling the fate of a large number of proteins. The study reported by Klemm and coworkers145 certainly suggest that this could be the case. As observed for HMG-CoA reductase, the abundance of other membrane enzymes, such as other enzymes involved lipid synthesis could also be controlled by ubiquitination and proteasome activity on the LD surface.
LDs recruit autophagic machinery leading to lipophagy
A number of factors, such as cellular starvation and cellular infection lead to the activation of autophagy. In this process, cytoplasmic components are first engulfed in a vesicular structure and then fused with lysosomes for degradation.70,151 LDs are targets of autophagy and one of the required structural components of the autophagosomes is the protein LC3 covalently conjugated to phosphatidylethanolamine (PE). LC3 is recruited to the surface of LDs, where it is conjugated with PE.152 Other required proteins of the autophagic machinery, Atg2, Atg5, and Atg7, are also recruited at the LD surface to help the formation of autophagosomes.153,154 In the starved cell, small LDs or portions of larger LDs are directed to lysosomes via autophagosome vesicles in a process called lipophagy.155,156 This process may be important to eliminate misfolded proteins stored in LDs or fragments of membrane proteins that remain in the LD after the proteasomal activity. It may also be important to provide FA through the action of lysosomal acid lipases.152,157 Some studies have shown that alteration of the autophagy process through lysosomal inhibition or knockdown of the expression of the Atg5 gene decreases TG breakdown and increases LD number and size.153 Although much remains to be known, current information suggests that LDs play an active role in the degradation of cellular components. To this end, LDs seem to provide a platform for the association of both the degradation targets and the degradation machineries of the proteasomal complex and autophagy.
On the role of LDs in cellular infection and immunity
A range of pathogens such as hepatitis C virus (HCV),158 dengue virus,159 bacteria Mycobacterium leprae,160 and the protozoan Trypanosoma cruzi161 target LDs and manipulate host cell lipid metabolism. Pathogens may target host-derived LDs either for gaining nutrients or for escaping host immune systems.162 They also exploit host-derived LDs as the sites for their protein assembly.163 HCV assembles virions at lipid droplet surface.164 HCV core protein localizes to LDs and initiates production of viral particles at LD-associated membranes of the ER. During infection, some nonstructural proteins of HCV, such as NS2, NS3, and NS5A and viral replication RNA complexes are recruited on the LD membrane surface.163 A critical role of DGAT1, the enzyme that synthesizes TGs in the ER, in recruiting the virus to LDs has been recently reported.165 DGAT1 would serve as a cellular hub for HCV core and NS5A proteins, guiding both onto the surface of the same subset of LDs. Binding of HCV core protein to LD causes a redistribution of the LDs around the nucleus and displacement of ADRP from the LD surface.166 The interaction between HCV core protein and LDs also leads to the accumulation of TG and the formation of larger LDs. This interaction could be associated with the development of steatosis.167 HCV core protein can also slowly affect LD localization by controlling the directionality of LD movement on microtubules.168
LDs play an important role in the synthesis of eicosanoids,169 important mediators of the inflammatory response. LDs of macrophages and mast cells store arachidonic acid,170,171 which is the precursor for the synthesis of eicosanoids. LDs of these cells are considered central organelles of the inflammatory response because they are the place for storage and synthesis of prostaglandins and leukotrienes that are secreted during infective processes.162 Interestingly, LDs of macrophages seem to be structurally different from other LDs.172 In addition to the protein coat surrounding the droplet, these LDs appear to have internal structures consisting of membranous structures and proteins in the core of the particle.173–175 To what extent this is a unique structural feature of the LDs of macrophages, or a more general case remains to be elucidated. Several pathogens induce the accumulation of LDs in immune cells.161,172,176–179 T. cruzi infection promotes the accumulation of LDs concomitantly with a higher production of prostaglandin PGE2.161 Furthermore, during the in vivo infection with T. cruzi, LDs associate with phagosomes, the vacuole in which the pathogen is internalized. This complicated picture suggests that some pathogens may be able to manipulate host-derived LDs to extend their survival, even when the cells are producing greater amounts of the inflammatory mediators.176 It is not clear whether the phenomena observed are more important for pathogen survival or for the defense of the host. However, increasing evidence suggests that LDs are playing a role in infection/immunity processes.
Conclusion
From simple globules of fat to highly dynamic organelles of unexpected complexity, our perception of LDs has been radically transformed in the last years. Now we recognize LDs as organelles involved in a number of cellular processes beyond their role as site for lipid storage. The broad range of LD functions implicates specific interactions with other cellular compartments and in some conditions the concerted movement of LDs along microtubule tracks. Although the details of the mechanisms for any of the cellular functions in which LDs are involved are limited, protein–protein interactions involving LD-associated proteins emerge as vital components of LD biology. Knowing the full spectrum of LD interactions and signaling mechanisms is essential to understand LD biology and lipid metabolism.
Footnotes
Author Contributions
Wrote the first draft of the manuscript: FZS, ELA. Contributed to the writing of the manuscript: JLS. Made critical revisions and approved final version: ELA, JLS. All authors reviewed and approved of the final manuscript.
DISCLOSURES AND ETHICS
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests. Provenance: the authors were invited to submit this paper.
ACADEMIC EDITOR: Tim Levine, Editor in Chief
FUNDING: This project was supported by Oklahoma Agricultural Experiment Station, Oklahoma State University and by Grant R01GM064677 from the National Institute of General Medical Sciences of the National Institute of Health. FZS is supported in part by a J.W. Fulbright foreign scholarship.
DISCLAIMER: The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
REFERENCES
- 1.Digel M, Ehehalt R, Fuellekrug J. Lipid droplets lighting up: insights from live microscopy. FEBS Lett. 2010;584(11):2168–2175. doi: 10.1016/j.febslet.2010.03.035. [DOI] [PubMed] [Google Scholar]
- 2.Fujimoto T, Ohsaki Y, Suzuki M, Cheng JL. Imaging lipid droplets by electron microscopy. Methods Cell Biol. 2013;116:227–251. doi: 10.1016/B978-0-12-408051-5.00012-7. [DOI] [PubMed] [Google Scholar]
- 3.Ohsaki Y, Suzuki M, Fujimoto T. Open questions in lipid droplet biology. Chem Biol. 2014;21(1):86–96. doi: 10.1016/j.chembiol.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Murphy S, Martin S, Parton RG. Lipid droplet-organelle interactions; sharing the fats. Biochim Biophys Acta. 2009;1791(6):441–447. doi: 10.1016/j.bbalip.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Targett-Adams P, Chambers D, Gledhill S, et al. Live cell analysis and targeting of the lipid droplet-binding adipocyte differentiation-related protein. J Biol Chem. 2003;278(18):15998–16007. doi: 10.1074/jbc.M211289200. [DOI] [PubMed] [Google Scholar]
- 6.Welte MA. Fat on the move: intracellular motion of lipid droplets. Biochem Soc Trans. 2009;37:991–996. doi: 10.1042/BST0370991. [DOI] [PubMed] [Google Scholar]
- 7.Beller M, Riedel D, Jänsch L, et al. Characterization of the Drosophila lipid droplet subproteome. Mol Cell Proteomics. 2006;5(6):1082–1094. doi: 10.1074/mcp.M600011-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J Biol Chem. 2004;279(45):46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- 9.Cermelli S, Guo Y, Gross SP, Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr Biol. 2006;16(18):1783–1795. doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- 10.Soulages JL, Firdaus SJ, Hartson S, Chen X, Howard AD, Arrese EL. Developmental changes in the protein composition of Manduca sexta lipid droplets. Insect Biochem Mol Biol. 2012;42(5):305–320. doi: 10.1016/j.ibmb.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuerschner L, Moessinger C, Thiele C. Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic. 2008;9(3):338–352. doi: 10.1111/j.1600-0854.2007.00689.x. [DOI] [PubMed] [Google Scholar]
- 12.Patel RT, Soulages JL, Hariharasundaram B, Arrese EL. Activation of the lipid droplet controls the rate of lipolysis of triglycerides in the insect fat body. J Biol Chem. 2005;280(24):22624–22631. doi: 10.1074/jbc.M413128200. [DOI] [PubMed] [Google Scholar]
- 13.Fujimoto T, Parton RG. Not just fat: the structure and function of the lipid droplet. Cold Spring Harbor Perspectives in Biology. 2011;3(3) doi: 10.1101/cshperspect.a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Y, Cordes KR, Farese RV, Walther TC. Lipid droplets at a glance. J Cell Sci. 2009;122(6):749–752. doi: 10.1242/jcs.037630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walther TC, Farese RV., Jr Lipid droplets and cellular lipid metabolism. Annu Rev Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartz R, Li WH, Venables B, et al. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res. 2007;48(4):837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Fujimoto T, Ohsaki Y, Cheng J, Suzuki M, Shinohara Y. Lipid droplets: a classic organelle with new outfits. Histochem Cell Biol. 2008;130(2):263–279. doi: 10.1007/s00418-008-0449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy DJ. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res. 2001;40(5):325–438. doi: 10.1016/s0163-7827(01)00013-3. [DOI] [PubMed] [Google Scholar]
- 19.Prattes S, Hörl G, Hammer A, et al. Intracellular distribution and mobilization of unesterified cholesterol in adipocytes: triglyceride droplets are surrounded by cholesterol-rich ER-like surface layer structures. J Cell Sci. 2000;113(17):2977–2989. doi: 10.1242/jcs.113.17.2977. [DOI] [PubMed] [Google Scholar]
- 20.Zweytick D, Athenstaedt K, Daum G. Intracellular lipid particles of eukaryotic cells. Biochim Biophys Acta. 2000;1469(2):101–120. doi: 10.1016/s0005-2736(00)00294-7. [DOI] [PubMed] [Google Scholar]
- 21.Fong TH, Yang CC, Greenberg AS, Wang SM. Immunocytochemical studies on lipid droplet-surface proteins in adrenal cells. J Cell Biochem. 2002;86(3):432–439. doi: 10.1002/jcb.10222. [DOI] [PubMed] [Google Scholar]
- 22.Krintel C, Morgelin M, Logan DT, Holm C. Phosphorylation of hormone-sensitive lipase by protein kinase A in vitro promotes an increase in its hydrophobic surface area. FEBS J. 2009;276(17):4752–4762. doi: 10.1111/j.1742-4658.2009.07172.x. [DOI] [PubMed] [Google Scholar]
- 23.Servetnick DA, Brasaemle DL, Gruiagray J, Kimmel AR, Wolff J, Londos C. Perilipins are associated with cholesteryl ester droplets in steroidogenic adrenalcortical and Leydig-cells. J Biol Chem. 1995;270(28):16970–16973. doi: 10.1074/jbc.270.28.16970. [DOI] [PubMed] [Google Scholar]
- 24.Zechner R, Zimmermann R, Eichmann TO, et al. FAT SIGNALS—lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012;15(3):279–291. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penno A, Hackenbroich G, Thiele C. Phospholipids and lipid droplets. Biochim Biophys Acta. 2013;1831(3):589–594. doi: 10.1016/j.bbalip.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Storey SM, McIntosh AL, Senthivinayagam S, Moon KC, Atshaves BP. The phospholipid monolayer associated with perilipin-enriched lipid droplets is a highly organized rigid membrane structure. Am J Physiol Endocrinol Metab. 2011;301(5):E991–E1003. doi: 10.1152/ajpendo.00109.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hodges BDM, Wu CC. Proteomic insights into an expanded cellular role for cytoplasmic lipid droplets. J Lipid Res. 2010;51(2):262–273. doi: 10.1194/jlr.R003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartz R, Zehmer JK, Zhu M, et al. Dynamic activity of lipid droplets: protein phosphorylation and GTP-Mediated protein translocation. J Proteome Res. 2007;6(8):3256–3265. doi: 10.1021/pr070158j. [DOI] [PubMed] [Google Scholar]
- 29.Beilstein F, Bouchoux J, Rousset M, Demignot S. Proteomic analysis of lipid droplets from Caco-2/TC7 enterocytes identifies novel modulators of lipid secretion. PLoS One. 2013;8(1):e53017–e53017. doi: 10.1371/journal.pone.0053017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brodsky JL, Wojcikiewicz RJH. Substrate-specific mediators of ER associated degradation (ERAD) Curr Opin Cell Biol. 2009;21(4):516–521. doi: 10.1016/j.ceb.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding Y, Wu Y, Zeng R, Liao K. Proteomic profiling of lipid droplet-associated proteins in primary adipocytes of normal and obese mouse. Chin J Biochem Biophys. 2012;44(5):394–406. doi: 10.1093/abbs/gms008. [DOI] [PubMed] [Google Scholar]
- 32.Fujimoto Y, Itabe H, Sakai J, et al. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim Biophys Acta. 2004;1644(1):47–59. doi: 10.1016/j.bbamcr.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 33.Hampton RY. ER-associated degradation in protein quality control and cellular regulation. Curr Opin Cell Biol. 2002;14(4):476–482. doi: 10.1016/s0955-0674(02)00358-7. [DOI] [PubMed] [Google Scholar]
- 34.Larsson S, Resjo S, Gomez MF, James P, Holm C. Characterization of the lipid droplet proteome of a clonal insulin-producing beta-cell line (INS-1 832/13) J Proteome Res. 2012;11(2):1264–1273. doi: 10.1021/pr200957p. [DOI] [PubMed] [Google Scholar]
- 35.Leonard DA, Chen HW. ATP-dependent degradation of 3-hydroxy-3-methylglutaryl coenzyme-A reductase in permeabilized cells. J Biol Chem. 1987;262(16):7914–7919. [PubMed] [Google Scholar]
- 36.Sato S, Fukasawa M, Yamakawa Y, et al. Proteomic profiling of lipid droplet proteins in hepatoma cell lines expressing hepatitis C virus core protein. J Biochem. 2006;139(5):921–930. doi: 10.1093/jb/mvj104. [DOI] [PubMed] [Google Scholar]
- 37.Turró S, Ingelmo-Torres M, Estanyol JM, et al. Identification and characterization of associated with lipid droplet protein 1: a novel membrane-associated protein that resides on hepatic lipid droplets. Traffic. 2006;7(9):1254–1269. doi: 10.1111/j.1600-0854.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- 38.Wan H-C, Melo RCN, Jin Z, Dvorak AM, Weller PF. Roles and origins of leukocyte lipid bodies: proteomic and ultrastructural studies. FASEB J. 2007;21(1):167–178. doi: 10.1096/fj.06-6711com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brasaemle DL. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res. 2007;48(12):2547–2559. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- 40.Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J Lipid Res. 2010;51(3):468–471. doi: 10.1194/jlr.R000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuhnlein RP. The contribution of the Drosophila model to lipid droplet research. Prog Lipid Res. 2011;50(4):348–356. doi: 10.1016/j.plipres.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 42.Bi J, Xiang Y, Chen H, et al. Opposite and redundant roles of the two Drosophila perilipins in lipid mobilization. J Cell Sci. 2012;125(pt 15):3568–3577. doi: 10.1242/jcs.101329. [DOI] [PubMed] [Google Scholar]
- 43.Teixeira L, Rabouille C, Rorth P, Ephrussi A, Vanzo NF. Drosophila Perilipin/ADRP homologue Lsd2 regulates lipid metabolism. Mech Dev. 2003;120(9):1071–1081. doi: 10.1016/s0925-4773(03)00158-8. [DOI] [PubMed] [Google Scholar]
- 44.Welte MA. Proteins under new management: lipid droplets deliver. Trends Cell Biol. 2007;17(8):363–369. doi: 10.1016/j.tcb.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 45.Gronke S, Beller M, Fellert S, Ramakrishnan H, Jackle H, Kuhnlein RP. Control of fat storage by a Drosophila PAT domain protein. Curr Biol. 2003;13(7):603–606. doi: 10.1016/s0960-9822(03)00175-1. [DOI] [PubMed] [Google Scholar]
- 46.Ohnishi A, Hull JJ, Kaji M, et al. Hormone signaling linked to silkmoth sex pheromone biosynthesis involves Ca2+/calmodulin-dependent protein kinase II-mediated phosphorylation of the insect PAT family protein Bombyx mori lipid storage droplet protein-1 (BmLsd1) J Biol Chem. 2011;286(27):24101–24112. doi: 10.1074/jbc.M111.250555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arrese EL, Mirza S, Rivera L, Howard AD, Chetty PS, Soulages JL. Expression of lipid storage droplet protein-1 may define the role of AKH as a lipid mobilizing hormone in. Manduca sexta Insect Biochem Mol Biol. 2008;38(11):993–1000. doi: 10.1016/j.ibmb.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arrese EL, Rivera L, Hamada M, Soulages JL. Purification and characterization of recombinant lipid storage protein-2 from. Drosophila melanogaster Protein Pept Lett. 2008;15(9):1027–1032. doi: 10.2174/092986608785849191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arrese EL, Rivera L, Hamada M, et al. Function and structure of lipid storage droplet protein 1 studied in lipoprotein complexes. Arch Biochem Biophys. 2008;473(1):42–47. doi: 10.1016/j.abb.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goodman JM. Demonstrated and inferred metabolism associated with cytosolic lipid droplets. J Lipid Res. 2009;50(11):2148–2156. doi: 10.1194/jlr.R001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szymanski KM, Binns D, Bartz R, et al. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci U S A. 2007;104(52):20890–20895. doi: 10.1073/pnas.0704154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta. 2009;1791(6):419–440. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miura S, Gan JW, Brzostowski J, et al. Functional conservation for lipid storage droplet association among perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J Biol Chem. 2002;277(35):32253–32257. doi: 10.1074/jbc.M204410200. [DOI] [PubMed] [Google Scholar]
- 54.Lu X, Gruia-Gray J, Copeland NG, et al. The murine perilipin gene: the lipid droplet-associated perilipins derive from tissue-specific, mRNA splice variants and define a gene family of ancient origin. Mamm Genome. 2001;12(9):741–749. doi: 10.1007/s00335-01-2055-5. [DOI] [PubMed] [Google Scholar]
- 55.Ducharme NA, Bickel PE. Minireview: lipid droplets in lipogenesis and lipolysis. Endocrinology. 2008;149(3):942–949. doi: 10.1210/en.2007-1713. [DOI] [PubMed] [Google Scholar]
- 56.Brasaemle DL. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res. 2007;48(12):2547–2559. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- 57.Beller M, Bulankina AV, Hsiao HH, Urlaub H, Jackle H, Kuhnlein RP. PERILIPIN-dependent control of lipid droplet structure and fat storage in. Drosophila Cell Metab. 2010;12(5):521–532. doi: 10.1016/j.cmet.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 58.Lin P, Chen X, Moktan H, et al. Membrane attachment and structure models of lipid storage droplet protein 1. Biochim Biophys Acta. 2014;1838(3):874–881. doi: 10.1016/j.bbamem.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Greenberg AS, Egan JJ, Wek SA, Garty NB, Blanchettemackie EJ, Londos C. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J Biol Chem. 1991;266(17):11341–11346. [PubMed] [Google Scholar]
- 60.Farese RV, Jr, Walther TC. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 2009;139(5):855–860. doi: 10.1016/j.cell.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brasaemle DL, Wolins NE. Packaging of fat: an evolving model of lipid droplet assembly and expansion. J Biol Chem. 2012;287(4):2273–2279. doi: 10.1074/jbc.R111.309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thiam AR, Farese RV, Jr, Walther TC. The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol. 2013;14(12):775–786. doi: 10.1038/nrm3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuhnlein RP. Thematic review series: lipid droplet synthesis and metabolism: from yeast to man. Lipid droplet-based storage fat metabolism in. Drosophila J Lipid Res. 2012;53(8):1430–1436. doi: 10.1194/jlr.R024299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang H, Galea A, Sytnyk V, Crossley M. Controlling the size of lipid droplets: lipid and protein factors. Curr Opin Cell Biol. 2012;24(4):509–516. doi: 10.1016/j.ceb.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 65.Suzuki M, Shinohara Y, Ohsaki Y, Fujimoto T. Lipid droplets: size matters. J Electron Microsc. 2011;60:S101–S116. doi: 10.1093/jmicro/dfr016. [DOI] [PubMed] [Google Scholar]
- 66.Guo Y, Walther TC, Rao M, et al. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453(7195):657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grahn TH, Zhang Y, Lee MJ, et al. FSP27 and PLIN1 interaction promotes the formation of large lipid droplets in human adipocytes. Biochem Biophys Res Commun. 2013;432(2):296–301. doi: 10.1016/j.bbrc.2013.01.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun Z, Gong J, Wu H, et al. Perilipin1 promotes unilocular lipid droplet formation through the activation of Fsp27 in adipocytes. Nat Commun. 2013;4:1594–1594. doi: 10.1038/ncomms2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marcinkiewicz A, Gauthier D, Garcia A, Brasaemle DL. The phosphorylation of serine 492 of perilipin A directs lipid droplet fragmentation and dispersion. J Biol Chem. 2006;281(17):11901–11909. doi: 10.1074/jbc.M600171200. [DOI] [PubMed] [Google Scholar]
- 70.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21(22):2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 71.Ariotti N, Murphy S, Hamilton NA, et al. Postlipolytic insulin-dependent remodeling of micro lipid droplets in adipocytes. Mol Biol Cell. 2012;23(10):1826–1837. doi: 10.1091/mbc.E11-10-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hashimoto T, Segawa H, Okuno M, et al. Active involvement of micro-lipid droplets and lipid-droplet-associated proteins in hormone-stimulated lipolysis in adipocytes. J Cell Sci. 2012;125(24):6127–6136. doi: 10.1242/jcs.113084. [DOI] [PubMed] [Google Scholar]
- 73.Paar M, Jüngst C, Steiner NA, et al. Remodeling of lipid droplets during lipolysis and growth in adipocytes. J Biol Chem. 2012;287(14):11164–11173. doi: 10.1074/jbc.M111.316794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buhman KK, Chen HC, Farese RV. The enzymes of neutral lipid synthesis. J Biol Chem. 2001;276(44):40369–40372. doi: 10.1074/jbc.R100050200. [DOI] [PubMed] [Google Scholar]
- 75.Walther TC, Farese RV., Jr The life of lipid droplets. Biochim Biophys Acta. 2009;1791(6):459–466. doi: 10.1016/j.bbalip.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Somwar R, Roberts CT, Jr, Varlamov O. Live-cell imaging demonstrates rapid cargo exchange between lipid droplets in adipocytes. FEBS Lett. 2011;585(12):1946–1950. doi: 10.1016/j.febslet.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 77.Wolins NE, Quaynor BK, Skinner JR, Schoenfish MJ, Tzekov A, Bickel P. S3–12, Adipophilin, and TIP47 package lipid in adipocytes. J Biol Chem. 2005;280(19):19146–19155. doi: 10.1074/jbc.M500978200. [DOI] [PubMed] [Google Scholar]
- 78.Wolins NE, Skinner JR, Schoenfish MJ, Tzekov A, Bensch KG, Bickel PE. Adipocyte protein S3–12 coats nascent lipid droplets. J Biol Chem. 2003;278(39):37713–37721. doi: 10.1074/jbc.M304025200. [DOI] [PubMed] [Google Scholar]
- 79.Hsieh K, Lee YK, Londos C, Raaka BM, Dalen KT, Kimmel AR. Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J Cell Sci. 2012;125(pt 17):4067–4076. doi: 10.1242/jcs.104943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Angel A. Studies on the compartmentation of lipid in adipose cells. I: subcellular distribution, composition, and transport of newly synthesized lipid: liposomes. J Lipid Res. 1970;11(5):420–432. [PubMed] [Google Scholar]
- 81.Arrese EL, Gazard JL, Flowers MT, Soulages JL, Wells MA. Diacylglycerol transport in the insect fat body: evidence of involvement of lipid droplets and the cytosolic fraction. J Lipid Res. 2001;42(2):225–234. [PubMed] [Google Scholar]
- 82.Zinder O, Eisenberg E, Shapiro B. Compartmentation of glycerides in adipose tissue cells. I. The mechanism of free fatty acid release. J Biol Chem. 1973;248(22):7673–7676. [PubMed] [Google Scholar]
- 83.Winand J, Furnelle J, Wodon C, Christophe J. Spectrum of fatty acids synthesized in situ and metabolic heterogeneity of free fatty acids and glycerides within isolated rat adipocytes. Biochim Biophys Acta. 1971;239(2):142–153. doi: 10.1016/0005-2760(71)90160-3. [DOI] [PubMed] [Google Scholar]
- 84.Edens NK, Leibel RL, Hirsch J. Lipolytic effects on diacylglycerol accumulation in human adipose tissue in vitro. J Lipid Res. 1990;31(8):1351–1359. [PubMed] [Google Scholar]
- 85.Tiwari S, Siddiqi SA. Intracellular trafficking and secretion of VLDL. Arterioscler Thromb Vasc Biol. 2012;32(5):1079–1086. doi: 10.1161/ATVBAHA.111.241471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vance JE. Assembly and secretion of lipoproteins. In: Vance DEV, Vance JE, editors. Biochemistry of Lipids, Lipoproteins and Membranes. 4th ed. Elsevier Science BV; 2002. pp. 505–526. [Google Scholar]
- 87.Arrese EL, Soulages JL. Insect fat body: energy, metabolism, and regulation. Annu Rev Entomol. 2010;55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Franke WW, Hergt M, Grund C. Rearrangement of the vimentin cytoskeleton during adipose conversion—formation of an intermediate filament cage around lipid globules. Cell. 1987;49(1):131–141. doi: 10.1016/0092-8674(87)90763-x. [DOI] [PubMed] [Google Scholar]
- 89.Almahbobi G, Hall PF. The role of intermediate filaments in adrenal steroidogenesis. J Cell Sci. 1990;97(pt 4):679–687. doi: 10.1242/jcs.97.4.679. [DOI] [PubMed] [Google Scholar]
- 90.Kumar N, Robidoux J, Daniel KW, Guzman G, Floering LM, Collins S. Requirement of vimentin filament assembly for beta(3)-adrenergic receptor activation of ERK MAP kinase and lipolysis. J Biol Chem. 2007;282(12):9244–9250. doi: 10.1074/jbc.M605571200. [DOI] [PubMed] [Google Scholar]
- 91.Shen WJ, Patel S, Eriksson JE, Kraemer FB. Vimentin is a functional partner of hormone sensitive lipase and facilitates lipolysis. J Proteome Res. 2010;9(4):1786–1794. doi: 10.1021/pr900909t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shen WJ, Zaidi SK, Patel S, et al. Ablation of vimentin results in defective steroidogenesis. Endocrinology. 2012;153(7):3249–3257. doi: 10.1210/en.2012-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kraemer FB, Khor VK, Shen W-J, Azhar S. Cholesterol ester droplets and steroidogenesis. Mol Cell Endocrinol. 2013;371:1–2. 15–19. doi: 10.1016/j.mce.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Welte MA, Cermelli S, Griner J, et al. Regulation of lipid-droplet transport by the perilipin homolog LSD2. Curr Biol. 2005;15(14):1266–1275. doi: 10.1016/j.cub.2005.06.062. [DOI] [PubMed] [Google Scholar]
- 95.Guo Y, Jangi S, Welte MA. Organelle-specific control of intracellular transport: distinctly targeted isoforms of the regulator Klar. Mol Biol Cell. 2005;16(3):1406–1416. doi: 10.1091/mbc.E04-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yu YV, Li Z, Rizzo NP, Einstein J, Welte MA. Targeting the motor regulator Klar to lipid droplets. BMC Cell Biol. 2011;12 doi: 10.1186/1471-2121-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Londos C, Brasaemle DL, Schultz CJ, Segrest JP, Kimmel AR, Perilipins ADRP. other proteins that associate with intracellular neutral lipid droplets in animal cells. Semin Cell Dev Biol. 1999;10(1):51–58. doi: 10.1006/scdb.1998.0275. [DOI] [PubMed] [Google Scholar]
- 98.Wolins NE, Brasaemle DL, Bickel PE. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 2006;580(23):5484–5491. doi: 10.1016/j.febslet.2006.08.040. [DOI] [PubMed] [Google Scholar]
- 99.Brasaemle DL, Barber T, Kimmel AR, Londos C. Post-translational regulation of perilipin expression. Stabilization by stored intracellular neutral lipids. J Biol Chem. 1997;272(14):9378–9387. doi: 10.1074/jbc.272.14.9378. [DOI] [PubMed] [Google Scholar]
- 100.Skinner JR, Harris LA, Shew TM, Abumrad NA, Wolins NE. Perilipin 1 moves between the fat droplet and the endoplasmic reticulum. Adipocyte. 2013;2(2):80–86. doi: 10.4161/adip.22864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Blanchette-Mackie EJ, Dwyer NK, Barber T, et al. Perilipin is located on the surface layer of intracellular lipid droplets in adipocytes. J Lipid Res. 1995;36(6):1211–1226. [PubMed] [Google Scholar]
- 102.Skinner JR, Shew TM, Schwartz DM, et al. Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J Biol Chem. 2009;284(45):30941–30948. doi: 10.1074/jbc.M109.013995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R. Lipid droplets are functionally connected to the endoplasmic reticulum in. Saccharomyces cerevisiae J Cell Sci. 2011;124(14):2424–2437. doi: 10.1242/jcs.076836. [DOI] [PubMed] [Google Scholar]
- 104.Beller M, Sztalryd C, Southall N, et al. COPI complex is a regulator of lipid homeostasis. PLoS Biol. 2008;6(11):2530–2549. doi: 10.1371/journal.pbio.0060292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Soni KG, Mardones GA, Sougrat R, Smirnova E, Jackson CL, Bonifacino JS. Coatomer-dependent protein delivery to lipid droplets. J Cell Sci. 2009;122(11):1834–1841. doi: 10.1242/jcs.045849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Novikoff AB, Novikoff PM, Rosen OM, Rubin CS. Organelle relationships in cultured 3T3-L1 preadipocytes. J Cell Biol. 1980;87(1):180–196. doi: 10.1083/jcb.87.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Blanchette-Mackie EJ, Scow RO. Movement of lipolytic products to mitochondria in brown adipose tissue of young rats: an electron microscope study. J Lipid Res. 1983;24(3):229–244. [PubMed] [Google Scholar]
- 108.Wang H, Sreenivasan U, Hu H, et al. Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J Lipid Res. 2011;52(12):2159–2168. doi: 10.1194/jlr.M017939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J Biol Chem. 2002;277(46):44507–44512. doi: 10.1074/jbc.M207712200. [DOI] [PubMed] [Google Scholar]
- 110.Soulages JL, Wells MA. Lipophorin: the structure of an insect lipoprotein and its role in lipid transport in insects. Adv Protein Chem. 1994;45:371–415. doi: 10.1016/s0065-3233(08)60644-0. [DOI] [PubMed] [Google Scholar]
- 111.Collins S. beta-Adrenoceptor signaling networks in adipocytes for recruiting stored fat and energy expenditure. Front Endocrinol. 2011;2:102. doi: 10.3389/fendo.2011.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Arrese EL, Flowers MT, Gazard JL, Wells MA. Calcium and cAMP are second messengers in the adipokinetic hormone-induced lipolysis of triacylglycerols in Manduca sexta fat body. J Lipid Res. 1999;40(3):556–564. [PubMed] [Google Scholar]
- 113.Souza SC, Muliro KV, Liscum L, et al. Modulation of hormone-sensitive lipase and protein kinase A-mediated lipolysis by perilipin A in an adenoviral reconstituted system. J Biol Chem. 2002;277(10):8267–8272. doi: 10.1074/jbc.M108329200. [DOI] [PubMed] [Google Scholar]
- 114.Bridges D, MacDonald JA, Wadzinski B, Moorhead GBG. Identification and characterization of D-AKAP1 as a major adipocyte PKA and PP1 binding protein. Biochem Biophys Res Commun. 2006;346(1):351–357. doi: 10.1016/j.bbrc.2006.05.138. [DOI] [PubMed] [Google Scholar]
- 115.Su CL, Sztalryd C, Contreras JA, Holm C, Kimmel AR, Londos C. Mutational analysis of the hormone-sensitive lipase translocation reaction in adipocytes. J Biol Chem. 2003;278(44):43615–43619. doi: 10.1074/jbc.M301809200. [DOI] [PubMed] [Google Scholar]
- 116.Nomura S, Kawanami H, Ueda H, Kizaki T, Ohno H, Izawa T. Possible mechanisms by which adipocyte lipolysis is enhanced in exercise-trained rats. Biochem Biophys Res Commun. 2002;295(2):236–242. doi: 10.1016/s0006-291x(02)00664-2. [DOI] [PubMed] [Google Scholar]
- 117.Zhang J, Hupfeld CJ, Taylor SS, Olefsky JM, Tsien RY. Insulin disrupts beta-adrenergic signalling to protein kinase A in adipocytes. Nature. 2005;437(7058):569–573. doi: 10.1038/nature04140. [DOI] [PubMed] [Google Scholar]
- 118.Greenberg AS, Kraemer FB, Soni KG, et al. Lipid droplet meets a mitochondrial protein to regulate adipocyte lipolysis. EMBO J. 2011;30(21):4337–4339. doi: 10.1038/emboj.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pidoux G, Witczak O, Jarnæss E, et al. Optic atrophy 1 is an A-kinase anchoring protein on lipid droplets that mediates adrenergic control of lipolysis. EMBO J. 2011;30(21):4371–4386. doi: 10.1038/emboj.2011.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Collins S, Cao WH, Robidoux J. Learning new tricks from old dogs: beta-adrenergic receptors teach new lessons on firing up adipose tissue metabolism. Mol Endocrinol. 2004;18(9):2123–2131. doi: 10.1210/me.2004-0193. [DOI] [PubMed] [Google Scholar]
- 121.Gauthier MS, Miyoshi H, Souza SC, et al. AMP-activated protein kinase is activated as a consequence of lipolysis in the adipocyte—potential mechanism and physiological relevance. J Biol Chem. 2008;283(24):16514–16524. doi: 10.1074/jbc.M708177200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Omar B, Zmuda-Trzebiatowska E, Manganiello V, Goransson O, Degerman E. Regulation of AMP-activated protein kinase by cAMP in adipocytes: roles for phosphodiesterases, protein kinase B, protein kinase A, Epac and lipolysis. Cell Signal. 2009;21(5):760–766. doi: 10.1016/j.cellsig.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gade G, Auerswald L. Mode of action of neuropeptides from the adipokinetic hormone family. Gen Comp Endocrinol. 2003;132(1):10–20. doi: 10.1016/s0016-6480(03)00159-x. [DOI] [PubMed] [Google Scholar]
- 124.Greenberg AS, Shen WJ, Muliro K, et al. Stimulation of lipolysis and hormone-sensitive lipase via the extracellular signal-regulated kinase pathway. J Biol Chem. 2001;276(48):45456–45461. doi: 10.1074/jbc.M104436200. [DOI] [PubMed] [Google Scholar]
- 125.Langfort J, Donsmark M, Ploug T, Holm C, Galbo H. Hormone-sensitive lipase in skeletal muscle: regulatory mechanisms. Acta Physiol Scand. 2003;178(4):397–403. doi: 10.1046/j.1365-201X.2003.01155.x. [DOI] [PubMed] [Google Scholar]
- 126.Holm C. Molecular mechanisms regulating hormone-sensitive lipase and lipolysis. Biochem Soc Trans. 2003;31(pt 6):1120–1124. doi: 10.1042/bst0311120. [DOI] [PubMed] [Google Scholar]
- 127.Garton AJ, Campbell DG, Carling D, Hardie DG, Colbran RJ, Yeaman SJ. Phosphorylation of bovine hormone-sensitive lipase by the AMP-activated protein kinase. A possible antilipolytic mechanism. Eur J Biochem. 1989;179(1):249–254. doi: 10.1111/j.1432-1033.1989.tb14548.x. [DOI] [PubMed] [Google Scholar]
- 128.McDonough PM, Maciejewski-Lenoir D, Hartig SM, et al. Differential phosphorylation of perilipin 1A at the initiation of lipolysis revealed by novel monoclonal antibodies and high content analysis. PLoS One. 2013;8(2):e55511. doi: 10.1371/journal.pone.0055511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hammond MP, Laird CD. Chromosome structure and DNA-replication in nurse and follicle cells of. Drosophila melanogaster Chromosoma. 1985;91(3–4):267–278. doi: 10.1007/BF00328222. [DOI] [PubMed] [Google Scholar]
- 130.Ruddell A, Jacobslorena M. Biphasic pattern of histone gene-expression during Drosophila oogenesis. Proc Natl Acad Sci U S A. 1985;82(10):3316–3319. doi: 10.1073/pnas.82.10.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li Z, Thiel K, Thul PJ, Beller M, Kuehnlein RP, Welte MA. Lipid droplets control the maternal histone supply of Drosophila embryos. Curr Biol. 2012;22(22):2104–2113. doi: 10.1016/j.cub.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.DeLany JP, Floyd ZE, Zvonic S, et al. Proteomic analysis of primary cultures of human adipose-derived stem cells—modulation by adipogenesis. Mol Cell Proteomics. 2005;4(6):731–740. doi: 10.1074/mcp.M400198-MCP200. [DOI] [PubMed] [Google Scholar]
- 133.Dugail I, Hajduch E. A new look at adipocyte lipid droplets: towards a role in the sensing of triacylglycerol stores? Cell Mol Life Sci. 2007;64:19–20. 2452–2458. doi: 10.1007/s00018-007-7277-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Di Cara F, Duca E, Dunbar DR, Cagney G, Heck MMS. Invadolysin, a conserved lipid-droplet-associated metalloproteinase, is required for mitochondrial function in. Drosophila J Cell Sci. 2013;126(20):4769–4781. doi: 10.1242/jcs.133306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cobbe N, Marshall KM, Gururaja Rao S, et al. The conserved metalloprotease invadolysin localizes to the surface of lipid droplets. J Cell Sci. 2009;122(18):3414–3423. doi: 10.1242/jcs.044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ohsaki Y, Cheng J, Fujita A, Tokumoto T, Fujimoto T. Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B. Mol Biol Cell. 2006;17(6):2674–2683. doi: 10.1091/mbc.E05-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ohsaki Y, Cheng J, Suzuki M, Fujita A, Fujimoto T. Lipid droplets are arrested in the ER membrane by tight binding of lipidated apolipoprotein B-100. J Cell Sci. 2008;121(14):2415–2422. doi: 10.1242/jcs.025452. [DOI] [PubMed] [Google Scholar]
- 138.Hartman IZ, Liu P, Zehmer JK, et al. Sterol-induced dislocation of 3-hydroxy-3-methylglutaryl coenzyme A reductase from endoplasmic reticulum membranes into the cytosol through a subcellular compartment resembling lipid droplets. J Biol Chem. 2010;285(25):19288–19298. doi: 10.1074/jbc.M110.134213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Suzuki M, Otsuka T, Ohsaki Y, et al. Derlin-1 and UBXD8 are engaged in dislocation and degradation of lipidated ApoB-100 at lipid droplets. Mol Biol Cell. 2012;23(5):800–810. doi: 10.1091/mbc.E11-11-0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu PS, Ying YS, Zhao YM, Mundy DI, Zhu MF, Anderson RGW. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J Biol Chem. 2004;279(5):3787–3792. doi: 10.1074/jbc.M311945200. [DOI] [PubMed] [Google Scholar]
- 141.Schuberth C, Buchberger A. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci. 2008;65(15):2360–2371. doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bartz R, Seemann J, Zehmer JK, et al. Evidence that mono-ADP-ribosylation of CtBP1/BARS regulates lipid storage. Mol Biol Cell. 2007;18(8):3015–3025. doi: 10.1091/mbc.E06-09-0869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol. 2012;14(2):117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 144.Spandl J, Lohmann D, Kuerschner L, Moessinger C, Thiele C. Ancient ubiquitous protein 1 (AUP1) localizes to lipid droplets and binds the E2 ubiquitin conjugase G2 (Ube2g2) via its G2 binding region. J Biol Chem. 2011;286(7):5599–5606. doi: 10.1074/jbc.M110.190785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Klemm EJ, Spooner E, Ploegh HL. Dual role of ancient ubiquitous protein 1 (AUP1) in lipid droplet accumulation and endoplasmic reticulum (ER) protein quality control. J Biol Chem. 2011;286(43):37602–37614. doi: 10.1074/jbc.M111.284794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jo Y, Hartman IZ, DeBose-Boyd RA. Ancient ubiquitous protein-1 mediates sterol-induced ubiquitination of 3-hydroxy-3-methylglutaryl CoA reductase in lipid droplet-associated endoplasmic reticulum membranes. Mol Biol Cell. 2013;24(3):169–183. doi: 10.1091/mbc.E12-07-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xu GH, Sztalryd C, Londos C. Degradation of perilipin is mediated through ubiquitination-proteasome pathway. Biochim Biophys Acta. 2006;1761(1):83–90. doi: 10.1016/j.bbalip.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 148.Hooper C, Puttamadappa SS, Loring Z, Shekhtman A, Bakowska JC. Spartin activates atrophin-1-interacting protein 4 (AIP4) E3 ubiquitin ligase and promotes ubiquitination of adipophilin on lipid droplets. BMC Biol. 2010;8 doi: 10.1186/1741-7007-8-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jiang H, He J, Pu S, Tang C, Xu G. Heat shock protein 70 is translocated to lipid droplets in rat adipocytes upon heat stimulation. Biochim Biophys Acta. 2007;1771(1):66–74. doi: 10.1016/j.bbalip.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 150.Wu CC, Howell KE, Neville MC, Yates JR, McManaman JL. Proteomics reveal a link between the endoplasmic reticulum and lipid secretory mechanisms in mammary epithelial cells. Electrophoresis. 2000;21(16):3470–3482. doi: 10.1002/1522-2683(20001001)21:16<3470::AID-ELPS3470>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 151.Dugail I. Lysosome/lipid droplet interplay in metabolic diseases. Biochimie. 2014;96:102–105. doi: 10.1016/j.biochi.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 152.Singh R, Cuervo AM. Lipophagy: connecting autophagy and lipid metabolism. Int J Cell Biol. 2012;2012:282041–282041. doi: 10.1155/2012/282041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Singh R, Kaushik S, Wang YJ, et al. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–U1164. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Velikkakath AKG, Nishimura T, Oita E, Ishihara N, Mizushima N. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol Biol Cell. 2012;23(5):896–909. doi: 10.1091/mbc.E11-09-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dong H, Czaja MJ. Regulation of lipid droplets by autophagy. Trends Endocrinol Metab. 2011;22(6):234–240. doi: 10.1016/j.tem.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu K, Czaja MJ. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013;20(1):3–11. doi: 10.1038/cdd.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Finn PF, Dice JF. Proteolytic and lipolytic responses to starvation. Nutrition. 2006;22:7–8. 830–844. doi: 10.1016/j.nut.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 158.Barba G, Harper F, Harada T, et al. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc Natl Acad Sci U S A. 1997;94(4):1200–1205. doi: 10.1073/pnas.94.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Samsa MM, Mondotte JA, Iglesias NG, et al. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 2009;5(10):e1000632. doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mattos KA, D’Avila H, Rodrigues LS, et al. Lipid droplet formation in leprosy: Toll-like receptor-regulated organelles involved in eicosanoid formation and Mycobacterium leprae pathogenesis. J Leukoc Biol. 2010;87(3):371–384. doi: 10.1189/jlb.0609433. [DOI] [PubMed] [Google Scholar]
- 161.Melo RCN, D’Avila H, Fabrino DL, Almeida PE, Bozza PT. Macrophage lipid body induction by Chagas disease in vivo: putative intracellular domains for eicosanoid formation during infection. Tissue Cell. 2003;35(1):59–67. doi: 10.1016/s0040-8166(02)00105-2. [DOI] [PubMed] [Google Scholar]
- 162.Saka HA, Valdivia R. Emerging roles for lipid droplets in immunity and host-pathogen interactions. Annu Rev Cell Dev Biol. 2012;28:411–437. doi: 10.1146/annurev-cellbio-092910-153958. [DOI] [PubMed] [Google Scholar]
- 163.Herker E, Ott M. Emerging role of lipid droplets in host/pathogen interactions. J Biol Chem. 2012;287(4):2280–2287. doi: 10.1074/jbc.R111.300202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Miyanari Y, Atsuzawa K, Usuda N, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9(9):1089–U1074. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 165.Herker E, Harris C, Hernandez C, et al. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat Med. 2010;16(11):1295–1298. doi: 10.1038/nm.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Boulant S, Douglas MW, Moody L, Budkowska A, Targett-Adams P, McLauchlan J. Hepatitis C virus core protein induces lipid droplet redistribution in a microtubule- and dynein-dependent manner. Traffic. 2008;9(8):1268–1282. doi: 10.1111/j.1600-0854.2008.00767.x. [DOI] [PubMed] [Google Scholar]
- 167.Syed GH, Amako Y, Siddiqui A. Hepatitis C virus hijacks host lipid metabolism. Trends Endocrinol Metab. 2010;21(1):33–40. doi: 10.1016/j.tem.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lyn RK, Hope G, Sherratt AR, McLauchlan J, Pezacki JP. Bidirectional lipid droplet velocities are controlled by differential binding strengths of HCV core DII protein. PLoS One. 2013;8(11):e78065. doi: 10.1371/journal.pone.0078065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bozza PT, Bakker-Abreu I, Navarro-Xavier RA, Bandeira-Melo C. Lipid body function in eicosanoid synthesis: an update. Prostaglandins Leukot Essent Fatty Acids. 2011;85(5):205–213. doi: 10.1016/j.plefa.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 170.Dichlberger A, Kovanen PT, Schneider WJ. Mast cells: from lipid droplets to lipid mediators. Clin Sci (Lond) 2013;125(3):121–130. doi: 10.1042/CS20120602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Dvorak AM, Dvorak HF, Peters SP, et al. Lipid bodies—cytoplasmic organelles important to arachidonate metabolism in macrophages and mast-cells. J Immunol. 1983;131(6):2965–2976. [PubMed] [Google Scholar]
- 172.Melo RCN, D’Avila H, Wan H-C, Bozza PT, Dvorak AM, Weller PF. Lipid bodies in inflammatory cells: structure, function, and current imaging techniques. J Histochem Cytochem. 2011;59(5):540–556. doi: 10.1369/0022155411404073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Robenek H, Robenek MJ, Troyer D. PAT family proteins pervade lipid droplet cores. J Lipid Res. 2005;46(6):1331–1338. doi: 10.1194/jlr.M400323-JLR200. [DOI] [PubMed] [Google Scholar]
- 174.Robenek H, Severs NJ. Recent advances in freeze-fracture electron microscopy: the replica immunolabeling technique. Biol Proced Online. 2008;10:9–19. doi: 10.1251/bpo138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Robenek H, Buers I, Hofnagel O, Robenek MJ, Troyer D, Severs NJ. Compartmentalization of proteins in lipid droplet biogenesis. Biochim Biophys Acta. 2009;1791(6):408–418. doi: 10.1016/j.bbalip.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 176.Melo RCN, Dvorak AM. Lipid body-phagosome interaction in macrophages during infectious diseases: host defense or pathogen survival strategy? PLoS Pathog. 2012;8(7):e1002729. doi: 10.1371/journal.ppat.1002729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Melo RCN, Fabrino DL, Dias FF, Parreira GG. Lipid bodies: structural markers of inflammatory macrophages in innate immunity. Inflamm Res. 2006;55(8):342–348. doi: 10.1007/s00011-006-5205-0. [DOI] [PubMed] [Google Scholar]
- 178.D’Avila H, Freire-de-Lima CG, Roque NR, et al. Host cell lipid bodies triggered by Trypanosoma cruzi infection and enhanced by the uptake of apoptotic cells are associated with prostaglandin E-2 generation and increased parasite growth. J Infect Dis. 2011;204(6):951–961. doi: 10.1093/infdis/jir432. [DOI] [PubMed] [Google Scholar]
- 179.D’Avila H, Toledo DAM, Melo RCN. Lipid bodies: inflammatory organelles implicated in host-Trypanosoma cruzi interplay during innate immune responses. Mediators Inflamm. 2012;2012 doi: 10.1155/2012/478601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wu Z, Soulages JL, Joshi BD, Daniel SM, Hager ZJ, Arrese EL. TGL-mediated lipolysis in Manduca sexta fat body: possible roles for lipoamide-dehydrogenase (LipDH) and high-density lipophorin (HDLp) Insect Biochem Mol Biol. 2014;45:58–68. doi: 10.1016/j.ibmb.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]