Abstract
In vivo studies of implanted acellular biological scaffolds in experimental animals have shown constructive remodeling mediated by anti-inflammatory macrophages. Little is known about the human macrophage response to such biomaterials, or the nature of the signaling mechanisms that govern the macrophage phenotype in this environment. The cellular events at the interface of a tissue and implanted decellularized biomaterial were examined by establishing a novel ex vivo tissue culture model in which surgically excised human urinary tract tissue was combined with porcine acellular bladder matrix (PABM). Evaluation of the tissue–biomaterial interface showed a time-dependent infiltration of the biomaterial by CD68+ CD80− macrophages. The migration of CD68+ cells from the tissue to the interface was accompanied by maturation to a CD163hi phenotype, suggesting that factor(s) associated with the biomaterial or the wound edge was/were responsible for the active recruitment and polarization of local macrophages. Glucocorticoid receptor (GR) and peroxisome proliferator activated receptor gamma (PPARγ) signaling was investigated as candidate pathways for integrating inflammatory responses; both showed intense nuclear labeling in interface macrophages. GR and PPARγ activation polarized peripheral blood-derived macrophages from a default M1 (CD80+) toward an M2 (CD163+) phenotype, but PPARγ signaling predominated, as its antagonism blocked any GR-mediated effect. Seeding on PABM was effective at polarizing peripheral blood-derived macrophages from a default CD80+ phenotype on glass to a CD80− phenotype, with intense nuclear localization of PPARγ. These results endorse in vivo observations that the infiltration of decellularized biological scaffolds, exemplified here by PABM, is pioneered by macrophages. Thus, it appears that natural factors present in PABM are involved in the active recruitment and polarization of macrophages to a CD163+ phenotype, with activation of PPARγ identified as the candidate pathway. The harnessing of these natural matrix-associated factors may be useful in enhancing the integration of synthetic and other natural biomaterials by polarizing macrophage activation toward an M2 regulatory phenotype.
Introduction
The attributes of tissue-derived acellular biological scaffolds have been reviewed extensively over the last decade.1 However, there remains a lack of understanding of the molecular and cellular mechanisms that facilitate the integration of biological scaffolds following implantation. It is axiomatic that surgical implantation of any biomaterial will initiate an innate immunological response: the intensity, duration, and outcome of which will be dependent upon the nature of the biomaterial. The innate response to biomaterials is coordinated by macrophages. Widely recognized for their role in the innate immune response to microorganisms, macrophages have also been identified as the key cells infiltrating sites of tissue injury and have been shown to play an orchestrating role in wound healing processes.2 These cells promote healing by responding to tissue damage, stimulating angiogenesis and inducing collagen synthesis.3
The functional plasticity of the macrophage response to a diverse range of protective, pathological, and tissue remodeling processes has resulted in the recognition of different activation states.4–6 As first proposed, macrophages were considered to polarize between a “classically activated” pro-inflammatory phenotype and an “alternatively activated” anti-inflammatory phenotype,7 later referred to as M1 and M2, respectively.5 Studies in which tissue-derived acellular biological scaffolds have been implanted in vivo have observed that macrophages displaying an anti-inflammatory or “M2” phenotype are associated with remodeling and integration, while pro-inflammatory or “M1” macrophages are associated with encapsulation.8–10 Transcriptional profiling of this M1/M2 dichotomy has identified a range of molecules that can be employed to characterize macrophage polarization.11
It is now recognized that by integrating responses to different environmental cues, macrophages display a plasticity of phenotypes across a spectrum of activation states encompassing host defense, wound healing, tissue pathology, and immune regulation, which are dynamic and overlapping.6 There is a paucity of research aimed at understanding the signaling mechanisms that direct the macrophage phenotype in the context of the biological scaffold. Defining the factors that govern the response of the macrophage to the unique environment of the implanted acellular biological scaffold may help direct the future development of functional natural and synthetic biomaterial scaffolds. Current models for the assessment of macrophage responses to biomaterials are limited either to in vivo implantation in rodents, or simple cell culture systems, often using immortalized “macrophage” cell lines that may not exhibit the plasticity of their primary mononuclear phagocyte counterparts. Coupled with this is a need to develop relevant preclinical models that are able to investigate and predict biomaterial performance. Although a few individual studies have incorporated biomaterials into human organ culture systems,12–14 these reports have not considered the response of tissue-derived immunological cells to the biomaterials.
The aim of the present study was to develop an ex vivo biomaterial–human tissue interface at which to examine the early cell integration events and from this identify potential signaling mechanisms that govern the human macrophage phenotype, as informed by in vitro studies of peripheral-blood-monocyte-derived macrophages.
Materials and Methods
Harvest and preparation of porcine acellular bladder matrix
Porcine bladders were obtained from the local abattoir and transported in transport medium15 consisting of HBSS (with Ca2+ and Mg2+) containing 10 mM HEPES (pH 7.6), 20 KIU·mL−1 Trasylol® (Bayer Healthcare Pharmaceuticals), 20 U·mL−1 penicillin, and 20 μg·mL−1 streptomycin. Preparation of porcine acellular bladder matrix (PABM) was performed by decellularizing the bladder by distension and immersion in a series of buffers containing EDTA, SDS, and DNAse/RNase, and with terminal disinfection in 0.1% (v/v) peracetic acid (pH 7.2) for 3 h at 37°C, followed by repeated washing in phosphate-buffered saline (PBS), as previously detailed.16 All solutions were sterilized prior to use and processing was performed in a class II biological safety cabinet following standard aseptic technique. Complete decellularization of the PABM was examined by staining with Hoechst 33258 (H33258) to confirm absence of nuclear material.16
For gamma irradiation, PABM was cut aseptically into ∼1-cm2 pieces before being sandwiched within surgical mesh and packaged into a foil pouch. The pouch was then inserted into a Tyvek pouch and heat-sealed. The packaged biomaterial was transferred to Isotron Limited (Swindon) for gamma irradiation at 30 kGy.
Organ culture
An organ culture system17 was adapted whereby a slit was introduced into a 1-cm2 piece of PABM biomaterial to accommodate a 0.5-cm2 piece of fresh urinary tract tissue. To establish the system, split-thickness fresh porcine bladder tissue (consisting of lamina propria and epithelium) was derived by incising horizontally between the lamina propria and the detrusor smooth muscle of the urinary bladder. Following optimization, a human tissue–biomaterial organ culture model was developed using surgically resected pieces of full-thickness human urinary tract tissue; this tissue was obtained as surplus tissue from surgery and had informed patient consent and research ethics committee approval for use in research.
The tissue–biomaterial constructs were cultured on the membrane of a cell culture well insert with 3-μm pores (catalogue number 734-0034; BD Falcon) within a six-well plate (Corning® Costar®; Fig. 1A). Constructs were maintained at an air–liquid interface in Waymouth's medium (Life Technologies) supplemented with 10% (v/v) fetal bovine serum (FBS; Sera Laboratories International), 300 μg·mL−1 L-ascorbic acid (Sigma-Aldrich), 2 μg·mL−1 hydrocortisone hemisuccinate (Sigma-Aldrich), and 450 ng·mL−1 ferrous sulfate (Fisons), as described for maintaining organ cultures.17 Omission of hydrocortisone hemisuccinate or ferrous sulfate from the medium was carried out in control experiments to preclude their influence on the results. Medium was exchanged every 48 h.
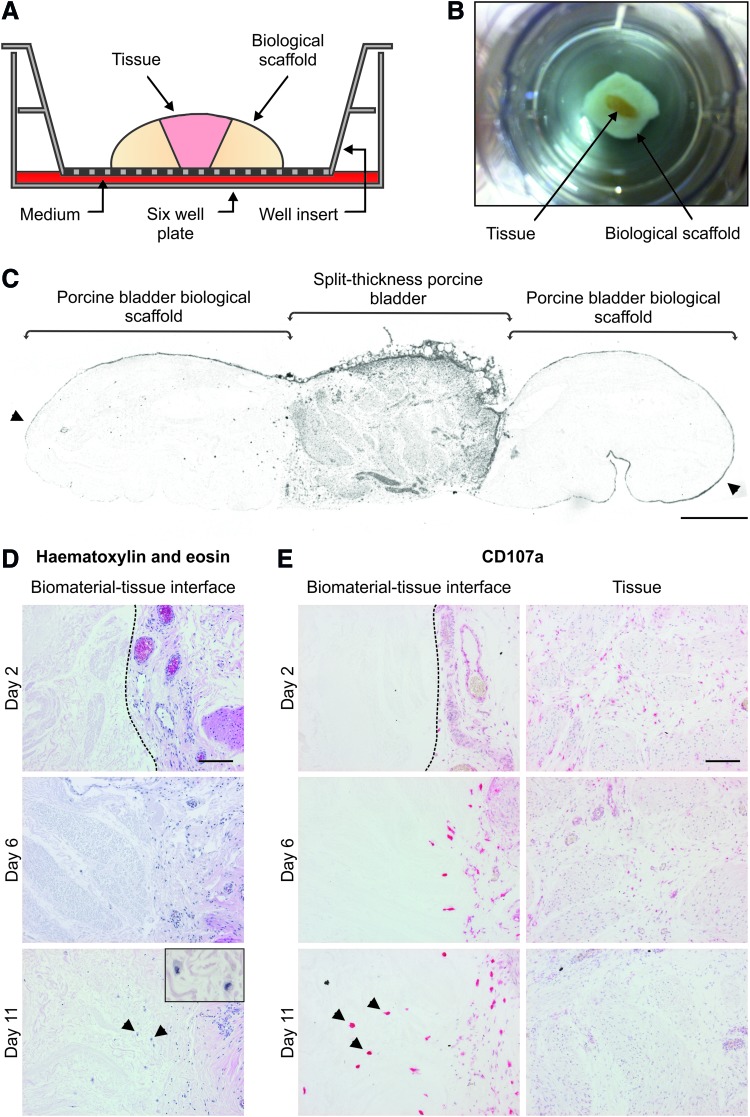
FIG. 1.
(A) Schematic cross-section of the biomaterial–organ culture system. Urinary tract tissue was inserted within the porcine bladder biological scaffold and cultured for up to 11 days. (B) Photograph of the apical aspect of the biomaterial–organ culture construct within the culture well insert. Porcine split-thickness tissue appears yellow in color and is surrounded by the white PABM. For scale, the base of the well insert has an effective growth area of 4.2 cm2. (C) Grayscale image of a whole-section slide scan of a biological scaffold–porcine bladder tissue section. The tissue can be seen within the central region of the section and is flanked either side by porcine bladder biological scaffold. Arrows indicate bladder epithelial cells covering the surface of the PABM. Scale bar represents 700 μm. (D) Hematoxylin-and-eosin-stained sections of the allogeneic porcine biomaterial–organ culture model. Histological staining of sections of the allogeneic biological scaffold–organ culture constructs at days 2, 6, and 11 showed a gradual increase of cells within the biological scaffold. Inset image on day 11 highlights that cells present in the PABM have a mononuclear phagocyte-like morphology (arrows). Scale bars represent 100 μm. (E) Alkaline phosphatase immunolabeling for porcine CD107a expression in tissue sections from the allogeneic biomaterial–organ culture system. Images were captured at the PABM–tissue interface and within the central regions of the tissue. Cells that were present in the biological scaffold were intensely labeled for porcine CD107a (arrows). Dotted lines represent biomaterial–tissue interface. Scale bars represent 100 μm. PABM, porcine acellular bladder matrix. Color images available online at www.liebertpub.com/tea
Isolation and culture of human peripheral blood mononuclear cells
Peripheral blood was obtained with informed consent from healthy volunteers at the York Clinical Research Facility based at York Teaching Hospital. Blood was collected into lithium heparin Monovette tubes (Sarstedt) and peripheral blood mononuclear cells (PBMCs) were isolated over Lymphoprep (Axis-Shields) before phagocytic mononuclear cells were identified and enumerated using latex bead ingestion. Monocytes were isolated by selective adherence to glass and cultured in RPMI1640 supplemented with 10% (v/v) heat-inactivated FBS and 2 mM L-glutamine in the presence of an agonist, antagonist, or vehicle control. The agonists used were as follows: 250 nM dexamethasone (1126; Tocris), 100 nM phorbol 12-myristate 13-acetate (PMA; Sigma), and 1 μM troglitazone (T2573; Sigma-Aldrich). The peroxisome proliferator activated receptor gamma (PPARγ) antagonist was 5 μM T0070907 (2301; Tocris). Appropriate vehicle controls were included in all experiments. Monocytes were seeded on the PABM within a seeding ring and isolated by selective adherence before culturing the constructs in RPMI1640 supplemented with 10% (v/v) heat-inactivated FBS and 2 mM L-glutamine.
Culture of THP-1 cells
The human acute monocytic leukemia–derived THP-1 cell line (ATCC® number TIB-202™) was cultured in suspension in RPMI1640 supplemented with 10% (v/v) heat-inactivated FBS, 2 mM L-glutamine, and 0.05 mM β-mercaptoethanol (Sigma-Aldrich). THP-1 cells were differentiated to adherent macrophage-like cells by culture in 100 nM PMA for 48 h.
Primary antibodies
The phenotype of mononuclear phagocytes was assessed using mouse monoclonal antibodies against CD11b (Clone 238446, MAB16991; R&D Systems), CD11b-APC (Clone 238446, FAB16991A; R&D Systems), CD68 (Clone PG-M1, M0876; Dako), CD80 (Clone 37711, MAB140; R&D Systems), CD107a (Clone 4E9/11, MCA2315; Serotec), CD163 (Clone EDhu-1, MCA1853; Serotec), CD163-FITC (Clone EDhu-1, MCA1853F; Serotec), glucocorticoid receptor (GR, Clone 4H2, NCL-GCR; Novacastra), and PPARγ (Clone E-8, SC-7273; Santa Cruz). Antibody concentrations were determined by titration.
Histology and immunohistochemistry
Organ culture constructs and PABM were fixed in either zinc salts or 10% (v/v) neutral-buffered formalin and processed into paraffin wax. Tissue sections (5 μm) were cut onto Superfrost™ microscope slides and stained with hematoxylin and eosin. For immunolabeling, endogenous peroxidase was blocked with 3% (v/v) H2O2, before heat retrieval of antigens by boiling for 10 min in 10 mM citric acid buffer (pH 6.0), or 1 mM EDTA buffer (pH 8.0), or enzymatic retrieval by incubating in 0.1% (w/v) porcine trypsin in 0.1% (w/v) CaCl2 at 37°C for 1 min followed by heat retrieval in 10 mM citric acid buffer (pH 6.0). Endogenous avidin and biotin were blocked (Vector Laboratories), followed by blocking of Fc receptors with 10% (v/v) rabbit serum (Dako). Primary antibodies were applied overnight at 4°C. After washing, rabbit anti-mouse IgG biotinylated secondary antibody (E0354; Dako) was applied to each slide for 30 min at ambient temperature. Secondary antibody was detected using the Vector Stain ABC streptavidin-horseradish peroxidase amplification kit (Vector Laboratories), followed by incubation with diaminobenzidine substrate (Sigma-Aldrich), or the Vector Stain streptavidin-alkaline phosphatase amplification kit followed by incubation with alkaline phosphatase substrate (Vector Laboratories). Immunolabeled tissue sections were counterstained in Mayer's hematoxylin and mounted in DPX. Immunoperoxidase labeling of antigens expressed at low levels was performed using the catalyzed signal amplification kit (Dako), according to manufacturer's instructions. Irrelevant primary antibody controls and positive control tissues were included in all experiments. Images were either captured on an Olympus BX60 microscope using Image Pro Plus software (version 4.5.1.29; Media Cybernetics) or whole tissue sections were captured on a slide scanner (MetaSystems). Where antigen expression was quantified, analysis regions were set using Image J Software (National Institute of Health).
Immunoblotting
Cells were lysed with 50 μL of 2% (w/v) SDS lysis buffer containing 13 mM dithiothreitol (Sigma-Aldrich) and protease inhibitor cocktail (Sigma-Aldrich). Protein (20 μg) from each sample was separated on reduced 3–8% (w/v) Tris-acetate NuPAGE® gels (Life Technologies) and electrotransferred onto a PVDF membrane (Millipore). The membrane was blocked in 50% (v/v) Odyssey blocking buffer (Li-Cor Biosciences) in 10 mM Tris-buffered saline (pH 7.4; TBS) for 1 h and probed sequentially with primary antibody overnight at 4°C and then Alexa Fluor® 680 conjugated goat anti-mouse IgG secondary antibody (A21057; Molecular Probes), with washing between, before imaging on an Odyssey Infrared scanner (Li-Cor Biosciences).
Flow cytometry
Adherent monocyte-derived macrophages were harvested from the culture substrate by incubation in 1% (w/v) EDTA for 10 min and scraping into ice-cold PBS. PBMCs and adherent monocyte-derived macrophages in suspension were adjusted to 1×106 cells·mL−1 in ice-cold PBS and incubated with LIVE/DEAD® Fixable Dead Cell Stain (Molecular Probes) for 30 min at ambient temperature in order to identify and exclude dead cells from the analysis. The cell suspensions were labeled with 11 μL of fluorescent-conjugated primary antibody or isotype control for 30 min on ice before washing and fixing in 1% (v/v) neutral-buffered formalin overnight. Immunolabeled-fixed cells were analyzed on a three-laser CyAn flow cytometer (Dako) and the flow cytometry data were interpreted using the Summit software package (version 4.3; Dako).
Immunofluorescence microscopy
Goat serum (10% v/v) was applied to methanol–acetone-fixed cultures for 1 h before addition of primary antibody and incubation at 4°C overnight in a humidified environment. Alexa Fluor® 594-conjugated goat anti-IgG secondary antibody (A11005; Molecular Probes) was added for 1 h at ambient temperature, before washing, nuclear counterstaining with 0.1 μg·mL−1 bis-benzimide (33258; Hoechst), and mounting in 0.1% (w/v) p-phenylendiaminodihydrochloride (Sigma-Aldrich) in 90% (w/v) glycerol in PBS (pH 8). Immunolabeling was assessed by epifluorescence on an Olympus BX60 microscope. Gray-scale images (32-bit) were merged to produce a color composite image using Image J software.
Statistical analysis
All statistical analyses were performed using InStat® statistical software version 3.05 (GraphPad). Mann–Whitney U tests were performed to test significance (p<0.05) between data sets with a small or unequal number of replicates.
Results
Examination of the allogeneic ex vivo biomaterial–organ culture model
Incorporation of the PABM into replicate organ cultures provided a system for investigating the biomaterial–tissue interface over multiple days. Porcine split-thickness urinary tract tissue was initially used as the live component in order to establish the model. The apical aspect of the constructs showed a “fried egg”-like appearance within the culture well insert (Fig. 1B). Visualization of the histological section through the whole construct showed that the live urinary tract tissue component was flanked on either side by the PABM to produce two ex vivo biomaterial–tissue interfaces (Fig. 1C). Histological analysis of constructs harvested from culture at days 2, 6, and 11 revealed that the biomaterial–tissue interface was maintained throughout the culture period (Fig. 1D). There was an increase in the number of cells within the PABM over time, from which it was inferred that cells had migrated from the live tissue into the PABM. The infiltrating cells had a large cytoplasmic volume and a kidney-shaped nucleus, indicative of mononuclear phagocytes (Fig. 1D). Immunolabeling of the tissue sections for the porcine macrophage-associated marker CD107a showed a time-associated increase in the number of intense CD107a+ cells at the biomaterial–tissue interface and infiltrating the PABM (Fig. 1E). CD107ahi cells with an equivalent morphology were not observed within the central regions of the live urinary tract tissue component of the constructs, although smaller CD107alo cells were apparent in the central regions at early time points, suggesting cell migration and maturation over time (Fig. 1E). Use of gamma-irradiated PABM in the biomaterial–organ culture model showed inhibited migration of CD107ahi cells into the matrix, with CD107ahi cells lining up along the tissue–biomaterial interface (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/tea).
Histological analysis of biomaterial–human tissue interface
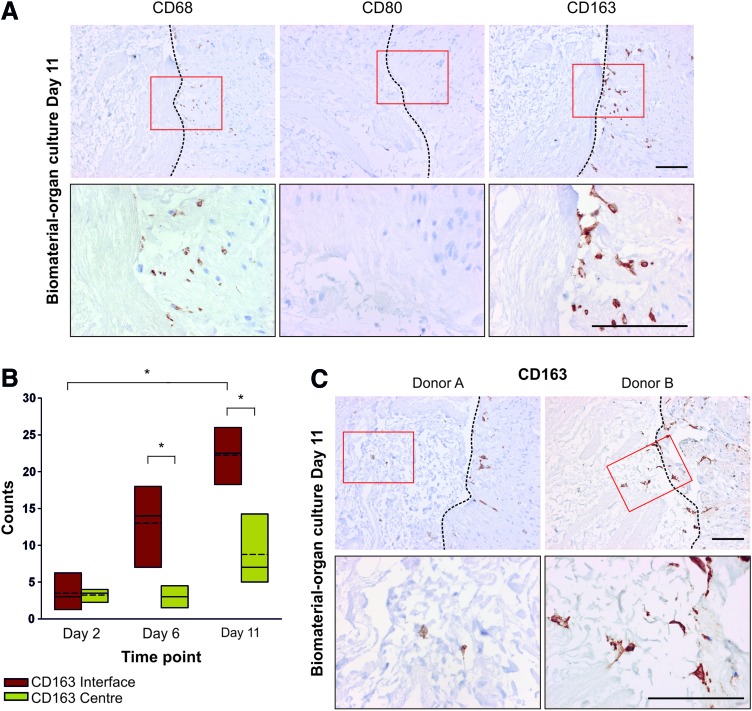
Culture of surgically-resected human urinary tract tissue in the biomaterial–organ culture model produced an ex vivo xenogeneic biomaterial–human tissue interface. Replicate constructs were harvested at days 2, 6, and 11 for histological analysis. Immunolabeling of histological sections of the biomaterial–human organ culture constructs showed CD68+ cells at the biomaterial–tissue interface and within the PABM at day 11, indicating interaction and infiltration by human mononuclear phagocytes (Fig. 2A). Investigation of the expression of macrophage functional markers showed an absence of the proinflammatory macrophage marker CD80, both at the biomaterial–human tissue interface (Fig. 2A) and within the central region of the live human tissue, although the antibody was shown to be immunoreactive on control human colon tissue (data not shown). Conversely, immunolabeling of the biomaterial–human organ culture constructs with an antibody to the anti-inflammatory macrophage marker CD163 showed striking expression of the antigen at the PABM–human tissue interface at day 11 of culture (Fig. 2A). Quantification of CD163 expression in multiple histological sections taken at different levels through the tissue constructs showed that the number of CD163+ cells at the biomaterial–tissue interface increased between days 2 and 11, which reached significance at days 6 and 11 (Fig. 2B). The presence of CD163+ cells at the biomaterial–tissue interface and infiltration of macrophages into the PABM by day 11 was demonstrated in constructs developed using tissue from all three independent donors (Fig. 2A, C).
FIG. 2.
(A) Analysis of human macrophage-associated marker expression at the biomaterial–human tissue interface. Biomaterial–organ culture constructs created using human ureteric tissue were harvested at day 11 and processed for immunohistochemistry. Images show the PABM on the left and the tissue on the right of each image. Boxes indicated the areas of the higher magnification images. All scale bars represent 100 μm. (B) Quantification of CD163 immunolabeling throughout the xenogeneic biomaterial–organ culture constructs. Three sections separated by >100 μm throughout the biomaterial–organ culture constructs harvested at days 2, 6, and 11 were immunolabeled for CD163 expression. The symbol * represents a significant difference (p<0.05) between median values as determined by the nonparametric Mann–Whitney U test. (C) Infiltration of the PABM by CD163+ cells was reproducible in multiple donors. Evaluation of the biomaterial–human tissue interface showed that CD163+ cells migrated from the tissue into the PABM. Boxes indicated the areas of the higher magnification images. Dotted lines represent biomaterial–tissue interface. All scale bars represent 100 μm. Color images available online at www.liebertpub.com/tea
CD163 expression by human macrophages
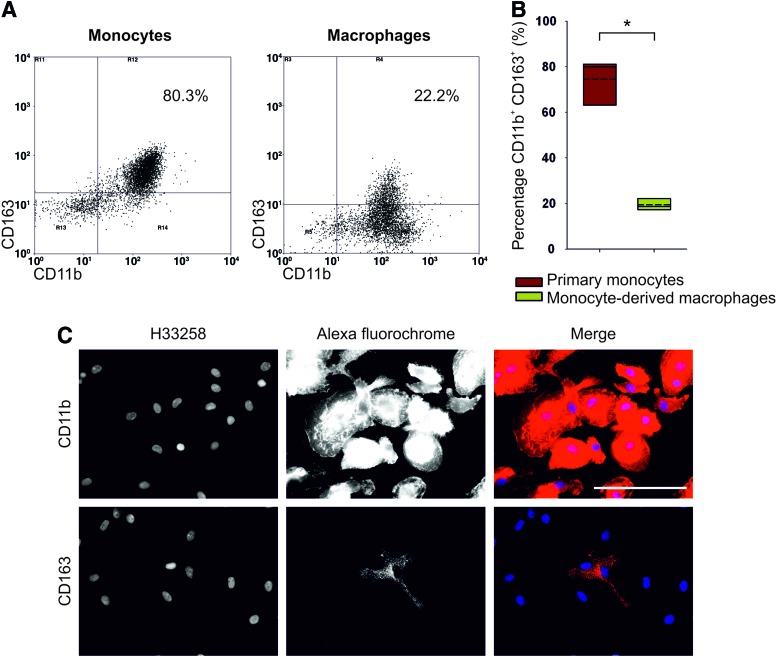
To gain understanding of CD163 regulation in human macrophages, peripheral blood monocytes and monocyte-derived macrophages were labeled for CD163. Human peripheral blood monocytes were identified by CD11b expression, of which ∼80% (n=6) expressed CD163 (Fig. 3A, B); this was in agreement with recent analysis.18 Immunofluorescence imaging of mature human monocyte-derived macrophages cultured on glass for 11 days showed that all mononuclear phagocytes maintained expression of CD11b as they matured from monocytes to macrophages. However, expression of CD163 was restricted to a subpopulation of monocyte-derived macrophages (Fig. 3C). Flow cytometric analysis of CD163 expression by CD11b+ monocyte-derived macrophages cultured on a glass substrate for 11 days showed a gradient of CD163 expression, with some 18% (n=3) of CD11b+ monocyte-derived macrophages defined as CD163hi (Fig. 3B). This indicated that in the absence of exogenous stimulation, CD163 was expressed constitutively by a subpopulation of peripheral-blood-derived macrophages.
FIG. 3.
(A) Flow cytometric analysis of CD163 expression by CD11b+ peripheral blood monocytes (left flow cytometry dot plot) and day 11 monocyte-derived CD11b+ macrophages (right flow cytometry dot plot). Regions (R3, R4, R5, R6, R11, R12, R13, R14) of analysis were determined based on isotype control labeling. (B) Change in expression of CD163 protein upon differentiation of CD11b+ peripheral blood monocytes (n=6 donors) to CD11b+ monocyte-derived macrophages (n=3 donors). Regions of analysis were assigned based on isotype control labeling. The symbol * represents a significant difference (p<0.05) in the percentage of CD163+ cells between peripheral blood monocytes and monocyte-derived macrophages as determined by the nonparametric Mann–Whitney U test. (C) Representative immunofluorescent imaging of human peripheral blood monocyte–derived macrophages immunolabeled with anti-CD11b or anti-CD163. Scale bar represents 100 μm. Color images available online at www.liebertpub.com/tea
Nuclear receptor modulation of CD163 expression on macrophages
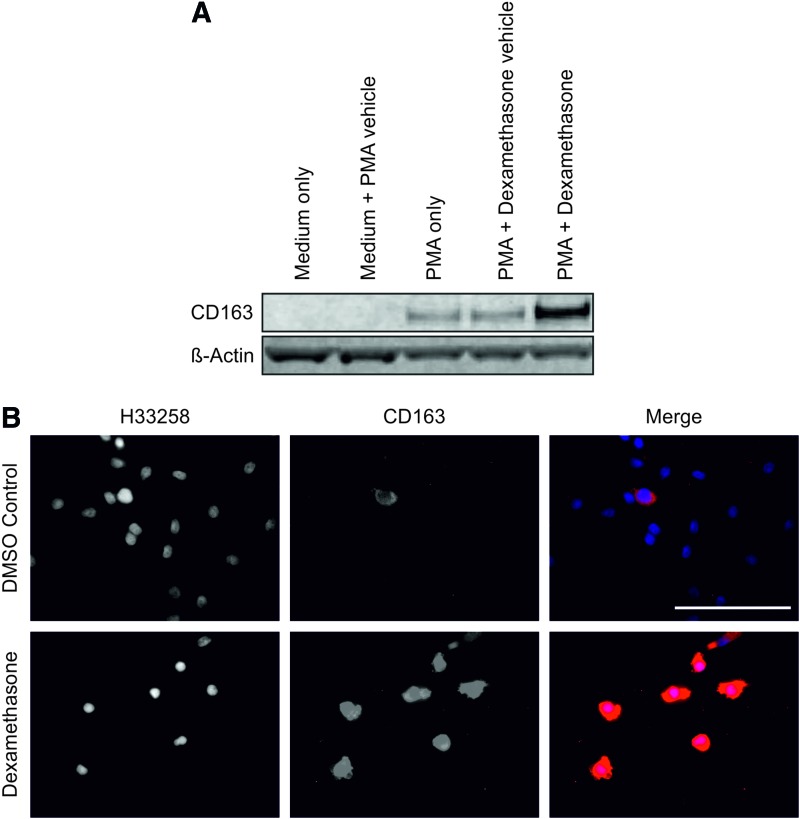
CD163 expression has been reported to be regulated by anti-inflammatory glucocorticoids.19 We used the synthetic glucocorticoid dexamethasone to activate the GR in the monocyte/macrophage THP-1 cell line and in human-peripheral-blood-derived monocyte–derived macrophages (Fig. 4). Monocytic THP-1 cells did not express CD163, but differentiation to adherent macrophage-like cells with PMA resulted in de novo expression of the CD163 antigen, which was further enhanced by stimulation with dexamethasone (Fig. 4A). Similarly, culture of human-monocyte-derived macrophages with dexamethasone for 6 days resulted in CD163 expression by all cells (Fig. 4B). The nuclear receptor PPARγ has been implicated in mediating an anti-inflammatory phenotype in macrophages and has also been shown to play a role in orchestrating wound healing processes following tissue injury20 (reviewed in Michalik and Wahli21). The hypothesis that PPARγ activation would modulate expression of CD163 by monocyte-derived macrophages was therefore tested.
FIG. 4.
(A) De novo expression of CD163 by THP-1 cells when differentiated into adherent macrophage-like cells by PMA and further enhancement in response to the GR agonist dexamethasone (250 nM) treatment for 48 h. β-Actin was used as a loading control. (B) Human peripheral blood monocyte–derived macrophages at day 6 in either the presence of 250 nM dexamethasone or the DMSO vehicle control. Scale bar represents 100 μm. GR, glucocorticoid receptor; PMA, phorbol 12-myristate 13-acetate. Color images available online at www.liebertpub.com/tea
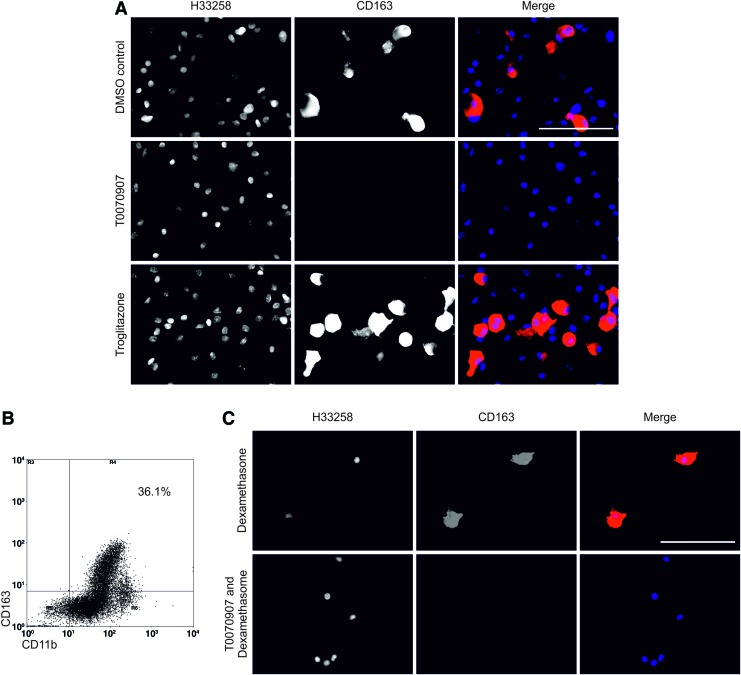
Activation of PPARγ with troglitazone resulted in a qualitative increase in the number of CD163+ macrophages at 11 days (Fig. 5A). This was confirmed by flow cytometry, which showed that activation of PPARγ increased the CD163hi population from 18% to 36% at day 11 (Fig. 5B). By contrast, inhibition of PPARγ activation with T0070907 resulted in a total loss of expression of CD163 by all monocyte-derived macrophages (Fig. 5A). A population of CD11b+ monocyte-derived macrophages that did not express CD163 in response to the PPARγ agonist was observed by immunofluorescence and flow cytometry, indicating that not all macrophages responded to the activation of PPARγ in the same way (Fig. 5B). The PPARγ antagonist T0070907 inhibited the glucocorticoid-mediated upregulation of CD163 by monocyte-derived macrophages, indicating an interdependency of the two signaling pathways (Fig. 5C).
FIG. 5.
(A) The regulation of CD163 expression by PPARγ. As observed previously, a subpopulation of monocyte-derived macrophages expressed CD163 in the presence of the DMSO vehicle control. Inhibition of PPARγ by culture of monocyte-derived macrophages in the presence of 5 μM T0070907 for 11 days showed an absence of CD163+ subpopulation. Activation of PPARγ by culture of the monocyte-derived macrophages in the presence of the PPARγ agonist troglitazone at 1 μM for 48 h, followed by culture in the DMSO vehicle control medium for 9 days, showed an increase in the CD163+ subpopulation. Scale bar represents 100 μm. (B) Flow cytometric analysis of monocyte-derived macrophages activated with troglitazone for 48 h followed by culture in control medium for a further 9 days. Cells were harvested and immunolabeled with anti-CD163-FITC and anti-CD11b-APC. Regions (R3, R4, R5, R6) of analysis were assigned based on isotype control labeling. Percentage of CD163+ CD11b+ monocyte-derived macrophages is shown. (C) Monocyte-derived macrophages were cultured in the presence of 250 nM dexamethasone or 250 nM dexamethasone and 5 μM T0070907. Inhibition of PPARγ by T0070907 completely ablated dexamethasone-induced CD163 expression. Scale bar represents 100 μm. PPARγ, peroxisome proliferator activated receptor gamma. Color images available online at www.liebertpub.com/tea
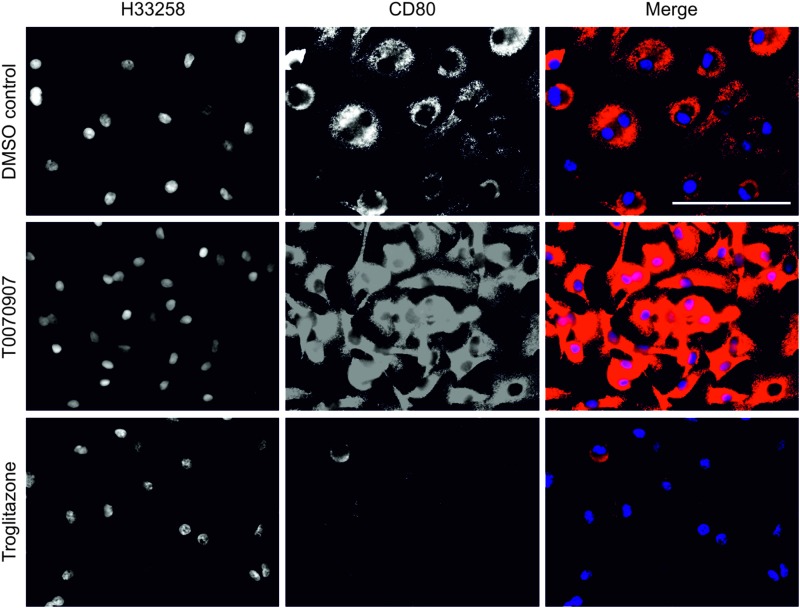
To confirm the role of PPARγ in inducing an anti-inflammatory macrophage phenotype, the effects of PPARγ on the proinflammatory macrophage marker CD80 were also investigated. All monocyte-derived macrophages showed a basal level of CD80 expression when cultured on glass for 11 days in control culture conditions (Fig. 6). Antagonism of PPARγ with T0070907 resulted in a major increase in the intensity of CD80 immunolabeling by peripheral-blood-monocyte-derived macrophages, while activation of PPARγ totally ablated CD80 expression (Fig. 6).
FIG. 6.
The PPARγ-dependent regulation of the macrophage costimulatory receptor CD80. Human peripheral blood monocyte–derived macrophages expressed a basal level of CD80 when in the presence of the DMSO vehicle control for 11 days. Inhibition of PPARγ by culture of monocyte-derived macrophages in the presence of 5 μM T0070907 for 11 days on a glass substrate showed a qualitative increase in CD80 expression. Activation of PPARγ by culture of the monocyte-derived macrophages in the presence of the PPARγ agonist troglitazone at 1 μM for 48 h, followed by culture in the DMSO vehicle control medium for 9 days, showed a loss in CD80 expression by monocyte-derived macrophages. Scale bar represents 100 μm. Color images available online at www.liebertpub.com/tea
Nuclear receptor expression and localization in the xenogeneic biomaterial–organ culture model
Immunohistochemical evaluation of the PABM–human tissue organ culture constructs demonstrated intense nuclear localization of both PPARγ and GR in the cells present at the biomaterial–tissue interface and in cells that had infiltrated the PABM, indicating potential activation. PPARγ+ cells present within the bladder matrix material exhibited kidney-shaped nuclei indicative of mononuclear phagocytes (Fig. 7).
FIG. 7.
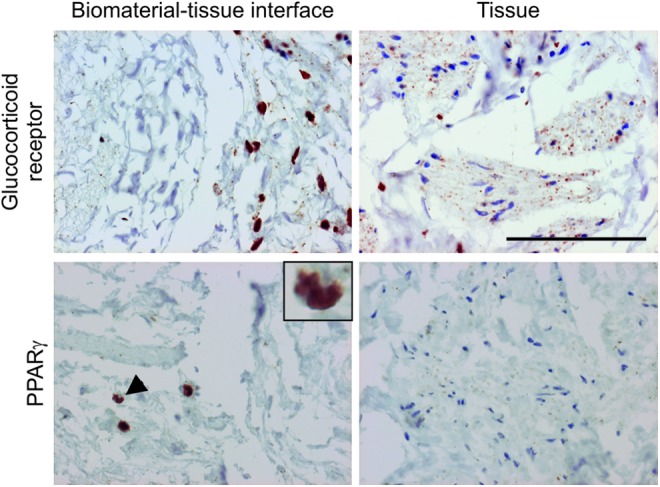
The GR and PPARγ show intense nuclear localization at the biomaterial–tissue interface but not within the central regions of the tissue. Day-11 biomaterial–organ culture constructs were immunolabeled for GR and PPARγ expression in the central regions of the tissue compared with the biomaterial–tissue interface. PPARγ+ cells within the biological scaffold showed a large cytoplasm-to-nuclei ratio and a kidney-shaped nuclei indicative of mononuclear phagocytes (arrow and inset image). Scale bar represents 100 μm. Color images available online at www.liebertpub.com/tea
CD80 expression and PPARγ localization in macrophages cultured on different substrates
Macrophages seeded onto glass showed a basal level of CD80 expression as indicated earlier; however, when the macrophages were seeded directly onto the PABM, they showed an absence of CD80 expression and intense nuclear localization of PPARγ (Fig. 8). Conversely, macrophages cultured on a glass substrate showed a combination of cytoplasmic and nuclear PPARγ localization (Fig. 8).
FIG. 8.
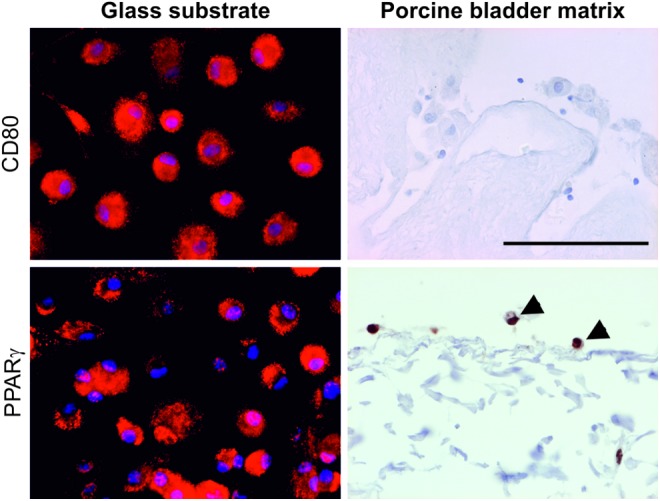
Expression of CD80 and localization of PPARγ in monocyte-derived macrophages seeded onto either a glass substrate or the PABM. Arrows denote macrophages on PABM showing nuclear PPARγ localization. Scale bar represents 100 μm. Color images available online at www.liebertpub.com/tea
Discussion
We have developed a novel ex vivo culture model to enable examination of the cellular events that occur at the interface between a tissue and implanted decellularized biomaterial. The biomaterial–organ culture system facilitates the study of biomaterial–tissue interfaces in an organ-specific context using allogeneic or xenogeneic biomaterial combinations. By examining the interface between urinary tract tissue and PABM, we have demonstrated that infiltration of the biomaterial was mediated by a pioneering population of cells identified as macrophages on the basis of kidney-shaped nuclear morphology and expression of CD107a+ (in porcine) and CD68+ (in human) tissue systems. Gamma irradiation of the PABM inhibited the migration of CD107a+ cells from the tissue into the matrix, suggesting that the native matrix structure facilitated macrophage infiltration. In the human system, the phenotype of the infiltrating macrophages was further defined as CD163hi in conjunction with intense nuclear expression of PPARγ and GR.
Most commonly, biomaterials are tested by subcutaneous implantation in rodents and it has been proposed that the resultant macrophage phenotype may be used to predict the outcome of biomaterial implantation in man.9 However, it is well known in the toxicity field that the extrapolation from rodents to humans can be inaccurate.22 Porcine models are suggested to be superior to rodent systems for studies of immunological mechanisms, due in part to similarities between porcine and human macrophages in terms of nitric oxide induction in response to lipopolysaccharide and the regulation of arginine metabolism.23 In addition, pigs are useful as preclinical models for experimental surgery (e.g., for reconstructive urology24) due to their similar size and organ arrangement to humans. A disadvantage of porcine systems, however, is a dearth of well-characterized macrophage-associated markers for porcine cells and thus the potential to develop human ex vivo models is of immediate advantage and relevance.
It is well established that damage caused by a wounding event will recruit leukocytes to the site of injury.2 The identification of CD68+ cells with distinct kidney-shaped nuclei at the tissue–PABM interface revealed that macrophages were the first cells to interact with the decellularized bladder matrix. Further characterization of the interface CD68+ macrophages showed that these cells were negative for the proinflammatory marker CD80, whereas CD163 was expressed. CD163 has been identified as a macrophage marker associated with biomaterial integration; for example, CD80− CD163+ M2 macrophages have been demonstrated at the site of constructive tissue remodeling following implantation of a porcine-derived biological scaffold into Sprague–Dawley rats in vivo.25 Interestingly, in our own studies of synthetic polycaprolactone scaffolds implanted in a rat model, the CD68+ macrophages that colonized and coated the biomaterial were CD163 negative despite a noninflammatory remodeling immune response.26 In the context of the interface between a tissue wound and decellularized natural matrix, the undetermined question was whether the expression of CD163 by macrophages reflected a passive, default phenotype (e.g., in the absence of proinflammatory stimuli), or was the result of a more active process.
From the immunohistochemical analysis it was possible to infer that CD68+ cells migrated to the interface region over time and accompanying this, there was maturation to a CD163hi phenotype. This suggested that factor(s) associated with either the biomaterial or the wound edge may be responsible for the local recruitment and maturation of macrophages. GR and PPARγ are members of the nuclear receptor superfamily and both are candidates for mediating anti-inflammatory responses by acting as ligand-activated transcription factors for specific gene expression programs. Emerging evidence of cross-talk between PPARγ and GR has suggested that the nuclear receptors probably function in combination to integrate local and systemic responses to inflammation.27 By immunohistochemistry, intense nuclear labeling of GR and PPARγ by macrophages at the interface initiated a more detailed examination of the potential role of these pathways in PBMCs and derived mononuclear phagocyte cell cultures. The results using PBMCs showed that it was possible to derive cells of an equivalent phenotype to those seen at the biomaterial interface and, employing CD80 and CD163 as markers, we confirmed that PBMC-derived macrophages could be polarized between classical M1 (CD80+) and M2 (CD163+) phenotypes by modulating the activation of these anti-inflammatory regulators. These two markers represent well-characterized polar responses on a spectrum of macrophage activation states and have previously been used in combination to define an integrative response.25 The study presented here demonstrates a novel regulatory mechanism behind the CD80/CD163 switch that could potentially be harnessed to polarize macrophage phenotype toward an anti-inflammatory or regulatory response during biomaterial integration.
Despite the biomaterial–organ culture model providing new insight into the human response to decellularized biological scaffolds, we recognize the limitation of extrapolating the ex vivo approach to an in vivo implantation model with regards to recruitment of inflammatory cells and fully modeling an integrative response. One unresolved question from the tissue culture model is whether the observed CD68+ cells represented a tissue-resident macrophage population or were derived from circulating monocytes captured in the peripheral vasculature of the tissue at the time of excision? Nevertheless, we revealed that the majority of circulating CD11b+ monocytes showed constitutive expression of CD163. We further observed heterogeneity in the monocyte-derived macrophage population by identifying a minority of PBMC-derived macrophages that retained CD163 expression even after culture in noninducing conditions. We are unaware of any other study that documents this in vitro CD163+ subpopulation, although Philippidis et al. documented increased CD163 expression by human-monocyte-derived macrophages in culture28 and Eligini et al. recently described the spontaneous differentiation of distinct macrophage phenotypes in vitro where CD163 expression was greater in monocyte-derived macrophages with a rounded, rather than spindle, morphology.29 Porta et al. described a population of macrophages that are “tolerant” in response to endotoxin and display an inherent skewing toward the M2 phenotype with impaired M1 responses.30 It is intriguing to postulate that the CD163+ population maintained in culture represents monocyte-derived macrophages with a greater propensity toward anti-inflammatory/M2 macrophage function.
Here, stimulation of GR with dexamethasone induced upregulation of CD163 by all cells in culture, whereas PPARγ activation enhanced only the size of the constitutive CD163+ subpopulation. By contrast, antagonism of PPARγ ablated all CD163 expression and de-repressed CD80 expression. This latter is in agreement with other studies that have shown PPARγ activation of human-monocyte-derived dendritic cells to repress the lipopolysaccharide-induced expression of CD80.31,32 Finally, we established a dependency for GR signaling on PPARγ activity, as inhibition of PPARγ function inhibited the glucocorticoid-dependent upregulation of CD163.
In regard to the context of the tissue–biomaterial interface, the question is whether the activation of PPARγ and GR was effected through factors in the wound environment or in the decellularized biomaterial? Here, the decellularized biomaterial was found to be effective in polarizing the macrophage from a basal CD80+ phenotype (the default phenotype on a glass substrate) to a CD80− phenotype with intense nuclear localization of PPARγ (when seeded on PABM). These results indicated that, as well as the factors in the wound environment, there may be natural agonists present in the PABM that provide an inherent advantage to natural biomaterials, as supported by studies of T-cell polarization.33 Future work will examine this hypothesis and further explore the consequence of crosslinking and terminal sterilization on the effectiveness of PABM and other natural biomaterials to act through PPARγ and GR to mediate an integrative CD80− CD163+ macrophage response.
Conclusions
The observation of anti-inflammatory human macrophages with intense nuclear localization of nuclear receptors at biomaterial–tissue interface provides new insight into the factors determining the host response to biomaterials by indicating a potential role for nuclear receptor signaling at a biomaterial–human tissue interface. Natural factors incorporated into biomaterials may help polarize the macrophage phenotype and be used to promote and enhance the efficacy and integration of biomaterials into the host.
Supplementary Material
Acknowledgments
This work was supported by a BBSRC CASE-funded PhD studentship to S.J.B. in partnership with Tissue Regenix Group (grant number BB\D527026\1). J.S. is funded by York Against Cancer. E.I. receives funding through WELMEC, a Centre of Excellence in Medical Engineering funded by the Wellcome Trust and EPSRC, under grant number WT 088908/Z/09/Z and from the NIHR-funded Leeds Musculoskeletal Biomedical Research Centre (LMBRU). The authors acknowledge the York Clinical Research Facility for recruiting volunteers, obtaining consent, and taking blood. The authors thank Tissue Regenix Group for their support of the project and in particular Dr. Helen Berry for helpful discussions in her role as industrial supervisor.
Disclosure Statement
E. Ingham is a share holder in and advisor to Tissue Regenix group PLC. No other competing financial interests exist.
References
- 1.Badylak S.F., Freytes D.O., and Gilbert T.W.Extracellular matrix as a biological scaffold material: structure and function. Acta Biomater 5,1, 2009 [DOI] [PubMed] [Google Scholar]
- 2.Martin P., and Leibovich S.J.Inflammatory cells during wound repair: the good, the bad and the ugly. Trends Cell Biol 15,599, 2005 [DOI] [PubMed] [Google Scholar]
- 3.Hunt T.K., Knighton D.R., Thakral K.K., Goodson W.H., and Andrews W.S.Studies on inflammation and wound healing: angiogenesis and collagen synthesis stimulated in vivo by resident and activated wound macrophages. Surgery 96,48, 1984 [PubMed] [Google Scholar]
- 4.Gordon S., and Martinez F.O.Alternative activation of macrophages: mechanism and functions. Immunity 32,593, 2010 [DOI] [PubMed] [Google Scholar]
- 5.Mantovani A., Sica A., Sozzani S., Allavena P., Vecchi A., and Locati M.The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol 25,677, 2004 [DOI] [PubMed] [Google Scholar]
- 6.Mosser D.M., and Edwards J.P.Exploring the full spectrum of macrophage activation. Nat Rev Immunol 8,958, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stein M., Keshav S., Harris N., and Gordon S.Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med 176,287, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown B.N., Valentin J.E., Stewart-Akers A.M., Mccabe G.P., and Badylak S.F.Macrophage phenotype and remodeling outcomes in response to biologic scaffolds with and without a cellular component. Biomaterials 30,1482, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown B.N., Londono R., Tottey S., Zhang L., Kukla K.A., Wolf M.T., et al. Macrophage phenotype as a predictor of constructive remodeling following the implantation of biologically derived surgical mesh materials. Acta Biomater 8,978, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown B.N., Ratner B.D., Goodman S.B., Amar S., and Badylak S.F.Macrophage polarization: an opportunity for improved outcomes in biomaterials and regenerative medicine. Biomaterials 33,3792, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez F.O., Gordon S., Locati M., and Mantovani A.Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol 177,7303, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Fukano Y., Knowles N.G., Usui M.L., Underwood R.A., Hauch K.D., Marshall A.J., et al. Characterization of an in vitro model for evaluating the interface between skin and percutaneous biomaterials. Wound Repair Regen 14,484, 2006 [DOI] [PubMed] [Google Scholar]
- 13.Beckstead B.L., Tung J.C., Liang K.J., Tavakkol Z., Usui M.L., Olerud J.E., et al. Methods to promote Notch signaling at the biomaterial interface and evaluation in a rafted organ culture model. J Biomed Mater Res 91,436, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peramo A., Marcelo C.L., Goldstein S.A., and Martin D.C.Novel organotypic cultures of human skin explants with an implant-tissue biomaterial interface. Ann Biomed Eng 37,401, 2009 [DOI] [PubMed] [Google Scholar]
- 15.Southgate J., Hutton K.A., Thomas D.F., and Trejdosiewicz L.K.Normal human urothelial cells in vitro: proliferation and induction of stratification. Lab Invest 71,583, 1994 [PubMed] [Google Scholar]
- 16.Bolland F., Korossis S., Wilshaw S-P., Ingham E., Fisher J., Kearney J.N., et al. Development and characterisation of a full-thickness acellular porcine bladder matrix for tissue engineering. Biomaterials 28,1061, 2007 [DOI] [PubMed] [Google Scholar]
- 17.Scriven S.D., Booth C., Thomas D.F., Trejdosiewicz L.K., and Southgate J.Reconstitution of human urothelium from monolayer cultures. J Urol 158(3 Pt 2),1147, 1997 [DOI] [PubMed] [Google Scholar]
- 18.Maniecki M.B., Etzerodt A., Moestrup S.K., Møller H.J., and Graversen J.H.Comparative assessment of the recognition of domain-specific CD163 monoclonal antibodies in human monocytes explains wide discrepancy in reported levels of cellular surface CD163 expression. Immunobiology 216,882, 2011 [DOI] [PubMed] [Google Scholar]
- 19.Högger P., Dreier J., Droste A., Buck F., and Sorg C.Identification of the integral membrane protein RM3/1 on human monocytes as a glucocorticoid-inducible member of the scavenger receptor cysteine-rich family (CD163). J Immunol 161,1883, 1998 [PubMed] [Google Scholar]
- 20.Kapoor M., Kojima F., Yang L., and Crofford L.J.Sequential induction of pro- and anti-inflammatory prostaglandins and peroxisome proliferators-activated receptor-gamma during normal wound healing: a time course study. Prostaglandins Leukot Essent Fatty Acids 76,103, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michalik L., and Wahli W.Involvement of PPAR nuclear receptors in tissue injury and wound repair. J Clin Invest 116,598, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perel P., Roberts I., Sena E., Wheble P., Briscoe C., Sandercock P., et al. Comparison of treatment effects between animal experiments and clinical trials: systematic review. BMJ 334, 197, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapetanovic R., Fairbairn L., Beraldi D., Sester D.P., Archibald A.L., Tuggle C.K., et al. Pig bone marrow-derived macrophages resemble human macrophages in their response to bacterial lipopolysaccharide. J Immunol 188,3382, 2012 [DOI] [PubMed] [Google Scholar]
- 24.Turner A., Subramanian R., Thomas D.F.M., Hinley J., Abbas S.K., Stahlschmidt J., et al. Transplantation of autologous differentiated urothelium in an experimental model of composite cystoplasty. Eur Urol 59,447, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Badylak S.F., Valentin J.E., Ravindra A.K., Mccabe G.P., and Stewart-Akers A.M.Macrophage phenotype as a determinant of biologic scaffold remodeling. Tissue Eng Part A 14,1835, 2008 [DOI] [PubMed] [Google Scholar]
- 26.Baker S.C., Rohman G., Hinley J., Stahlschmidt J., Cameron N.R., and Southgate J.Cellular integration and vascularisation promoted by a resorbable, particulate-leached, cross-linked poly(ɛ-caprolactone) scaffold. Macromol Biosci 11,618, 2011 [DOI] [PubMed] [Google Scholar]
- 27.Glass C.K., and Ogawa S.Combinatorial roles of nuclear receptors in inflammation and immunity. Nat Rev Immunol 6,44, 2006 [DOI] [PubMed] [Google Scholar]
- 28.Philippidis P., Mason J.C., Evans B.J., Nadra I., Taylor K.M., Haskard D.O., et al. Hemoglobin scavenger receptor CD163 mediates interleukin-10 release and heme oxygenase-1 synthesis: antiinflammatory monocyte-macrophage responses in vitro, in resolving skin blisters in vivo, and after cardiopulmonary bypass surgery. Circ Res 94,119, 2004 [DOI] [PubMed] [Google Scholar]
- 29.Eligini S., Crisci M., Bono E., Songia P., Tremoli E., Colombo G.I., et al. Human monocyte-derived macrophages spontaneously differentiated in vitro show distinct phenotypes. J Cell Physiol 228,1464, 2013 [DOI] [PubMed] [Google Scholar]
- 30.Porta C., Rimoldi M., Raes G., Brys L., Ghezzi P., Di Liberto D., et al. Tolerance and M2 (alternative) macrophage polarization are related processes orchestrated by p50 nuclear factor kappaB. Proc Natl Acad Sci U S A 106,14978, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gosset P., Charbonnier A.S., Delerive P., Fontaine J., Staels B, Pestel J., et al. Peroxisome proliferator-activated receptor gamma activators affect the maturation of human monocyte-derived dendritic cells. Eur J Immunol 31,2857, 2001 [DOI] [PubMed] [Google Scholar]
- 32.Nencioni A., Grünebach F., Zobywlaski A., Denzlinger C., Brugger W., and Brossart P.Dendritic cell immunogenicity is regulated by peroxisome proliferator-activated receptor γ. J Immunol Am Assoc Immnol 169,1228, 2002 [DOI] [PubMed] [Google Scholar]
- 33.Fishman J.M., Lowdell M.W., Urbani L., Ansari T., Burns A.J., Turmaine M., et al. Immunomodulatory effect of a decellularized skeletal muscle scaffold in a discordant xenotransplantation model. Proc Natl Acad Sci U S A 110,14360, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.