ABSTRACT
Food-borne pathogens are ongoing problems, and new pathogens are emerging. The impact of fungi, however, is largely underestimated. Recently, commercial yogurts contaminated with Mucor circinelloides were sold, and >200 consumers became ill with nausea, vomiting, and diarrhea. Mucoralean fungi cause the fatal fungal infection mucormycosis, whose incidence has been continuously increasing. In this study, we isolated an M. circinelloides strain from a yogurt container, and multilocus sequence typing identified the strain as Mucor circinelloides f. circinelloides. M. circinelloides f. circinelloides is the most virulent M. circinelloides subspecies and is commonly associated with human infections, whereas M. circinelloides f. lusitanicus and M. circinelloides f. griseocyanus are less common causes of infection. Whole-genome analysis of the yogurt isolate confirmed it as being close to the M. circinelloides f. circinelloides subgroup, with a higher percentage of divergence with the M. circinelloides f. lusitanicus subgroup. In mating assays, the yogurt isolate formed sexual zygospores with the (−) M. circinelloides f. circinelloides tester strain, which is congruent with its sex locus encoding SexP, the (+) mating type sex determinant. The yogurt isolate was virulent in murine and wax moth larva host systems. In a murine gastromucormycosis model, Mucor was recovered from fecal samples of infected mice for up to 10 days, indicating that Mucor can survive transit through the GI tract. In interactions with human immune cells, M. circinelloides f. lusitanicus induced proinflammatory cytokines but M. circinelloides f. circinelloides did not, which may explain the different levels of virulence in mammalian hosts. This study demonstrates that M. circinelloides can spoil food products and cause gastrointestinal illness in consumers and may pose a particular risk to immunocompromised patients.
IMPORTANCE
The U.S. FDA reported that yogurt products were contaminated with M. circinelloides, a mucoralean fungal pathogen, and >200 consumers complained of symptoms, including vomiting, nausea, and diarrhea. The manufacturer voluntarily withdrew the affected yogurt products from the market. Compared to other food-borne pathogens, including bacteria, viruses, and parasites, less focus has been placed on the risk of fungal pathogens. This study evaluates the potential risk from the food-borne fungal pathogen M. circinelloides that was isolated from the contaminated commercial yogurt. We successfully cultured an M. circinelloides isolate and found that the isolate belongs to the species M. circinelloides f. circinelloides, which is often associated with human infections. In murine and insect host models, the isolate was virulent. While information disseminated in the popular press would suggest this fungal contaminant poses little or no risk to consumers, our results show instead that it is capable of causing significant infections in animals.
INTRODUCTION
Daily essentials, such as food, are exposed to contamination/infestation by pathogenic microbes and eventually provide a source of infections. Food-borne intestinal infectious disease is a continuous problem in the health care system and causes considerable social and economic burdens. Among others, bacteria are normally considered prominent pathogens, and three well-known food-borne bacteria include Salmonella spp., Campylobacter spp., and Escherichia coli (reviewed in reference 1). Listeria monocytogenes is an emerging bacterial pathogen associated with food (2). Viral pathogens also cause food-borne illness (3, 4). Noroviruses are one of the most common agents for gastroenteritis and cause especially severe symptoms in immunocompromised patients (4–6). Other examples of food-borne viral pathogens include hepatitis A virus, hepatitis E virus, and rotavirus (1). In addition, parasites can cause food-borne infectious diseases, and currently, ~300 parasitic worms and ~70 protozoan species are known to infect humans and animals (reviewed in reference 1).
However, studies evaluating fungi as food-borne pathogens and assessing associated risks are limited. In September 2013, there was a food-borne illness outbreak after consumers ingested yogurts contaminated with mold. More than 200 individuals suffered from vomiting, nausea, and diarrhea after consumption of the yogurt (7, 8). The U.S. Food and Drug Administration (U.S. FDA) immediately analyzed the responsible mold and identified it as Mucor circinelloides.
M. circinelloides belongs to the order Mucorales, among the lineages of early-diverging fungi known as zygomycetes, and is one of the causal agents of the fungal infection mucormycosis. The increase of immunocompromised cohorts in recent years caused by, for example, transplantation, malignancies, HIV/AIDS, steroids, diabetes, and neutropenia, has been mirrored by an increase in the incidence of mucormycosis. Significantly, ~15% of patients with severe neutropenia develop mucormycosis (9, 10). The mortality from this fungal infection is high, ranging from 68 to 100% (9, 11). The main infection sites are the lungs, the sinuses, soft tissues, skin, and the bloodstream (12, 13). Gastrointestinal (GI) mucormycosis causes symptoms that include nonspecific abdominal tenderness and distention with nausea and vomiting, and case reports (14) have been published (15–18). Transplant recipients are especially susceptible to gastrointestinal mucormycosis following ingestion of causative fungal species (17). One such case was reported in a bone marrow transplant recipient who ingested naturopathic medicine contaminated with Mucor, resulting in the development of gastrointestinal mucormycosis (16). The mortality rate of gastrointestinal mucormycosis is as high as 85%, and the infection is often disseminated, resulting in higher rates of mortality (15).
Mucor species are the second most common mucoralean fungus causing mucormycosis, surpassed only by Rhizopus species (19, 20). According to the U.S. FDA report, M. circinelloides was associated with the outbreak of food-borne illnesses after consumption of the contaminated yogurt. The M. circinelloides species complex consists of several distinct species or subspecies: M. circinelloides f. lusitanicus, M. circinelloides f. circinelloides, and M. circinelloides f. griseocyanus (21, 22). M. circinelloides f. circinelloides isolates are more often associated with patients and display higher virulence in the murine mucormycosis model (21).
In this study, we obtained a plain Chobani yogurt that was within the manufacturer’s voluntary date recall range and also in the production lot subject to recall. This sample was provided by a couple in Texas who both consumed the contaminated product. Both individuals developed nausea and diarrhea, and one also developed vomiting. From the container, we isolated an M. circinelloides strain, designated Mucho, and subsequently identified the isolate as belonging to the M. circinelloides f. circinelloides subgroup. Whole-genome analysis further supports that the yogurt isolate belongs to M. circinelloides f. circinelloides and is distinct from the M. circinelloides f. lusitanicus and M. circinelloides f. griseocyanus subgroups. In virulence tests with two mucormycosis virulence models, we found that the isolate infects animals and causes mortality. Our results demonstrate that M. circinelloides is a food-borne pathogen that can cause lethal mucormycosis and suggest that caution should be exercised with respect to fungal pathogens in food, particularly for individuals who are immunocompromised.
RESULTS
Isolation of a mold from a yogurt within the company recall range.
The U.S. Food and Drug Administration (FDA) released a report that there was a recall of yogurt made by Chobani from a factory located in Twin Falls, ID. The recalled yogurts were labeled as those with a best by date between 11 September and 7 October 2013. The IMS code assigned was 16-012 (7). It was reported to the FDA that more than 300 individuals who consumed yogurts produced by the company experienced gastrointestinal discomfort, including nausea, cramps, vomiting, and diarrhea. An immediate investigation found that the yogurts were contaminated with M. circinelloides (23).
A couple in Corpus Christi, TX, experienced moderate to moderately severe illness following consumption of part of a plain yogurt from Chobani. One of them experienced repeated vomiting and diarrhea for two entire days with two days of missed work; the other was severely nauseated with diarrhea for a few days without vomiting (see Text S1 for the note from the couple). According to the couple, an apparent mold grew in the yogurt placed in the refrigerator. We obtained the yogurt container, and it was dated for expiration on 30 September 2013, which was within the recall date period, and had the IMS code 16-012 (Fig. S1). We isolated a mold from the yogurt container, and the mold displayed the typical growth features that are observed in Mucor species, i.e., formation of aseptate hyphae, aerial hyphae decorated with a ball-like sporangium, and a sporangium containing thousands of asexual spores (data not shown).
Identification and phylogenetic analyses of the yogurt isolate.
For identification, a multilocus sequence typing (MLST) analysis was conducted with three genes used in the Fungal Tree of Life Project (24). Initially, seven isolates were cultured from seven different locations within the suspect container. An intragenic spacer region (ITS), a large subunit rRNA gene (LSU rRNA), and an RNA polymerase subunit gene (RPB1) were amplified and sequenced from the genomes of each isolate. All sequences for each of the three genes from the seven isolates were identical (data not shown), indicating that, at least at these loci, the seven isolates are indistinguishable. One of the seven isolates was designated Mucho and selected for further analysis.
We also tested 16 other yogurt products from Chobani with different “best by” dates and IMS codes (see Table S1 in the supplemental material). None of them were found to be contaminated with M. circinelloides. However, we isolated a Yarrowia lipolytica isolate from an opened yogurt product, which is not known as a pathogen and is associated with lipid production as an industrial microorganism (reviewed in references 25 and 26).
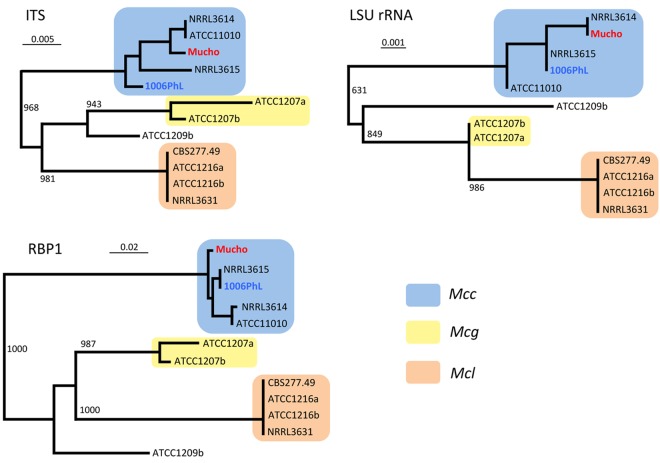
The M. circinelloides species complex includes at least three species or subspecies: M. circinelloides f. circinelloides, M. circinelloides f. lusitanicus, and M. circinelloides f. griseocyanus (21, 22). Maximum likelihood trees were constructed with the three genes obtained and known sequences for the same three genes from representative M. circinelloides f. circinelloides (NRRL3614, NRRL3615, and ATCC11010), M. circinelloides f. lusitanicus (CBS277.49, ATCC1216a, ATCC1216b, and NRRL3631), and M. circinelloides f. griseocyanus (ATCC1207a and ATCC1207b) isolates (see reference 21 and references therein ) (Fig. 1). The three individual gene trees displayed similar patterns, indicating that there are three distinct groups represented by M. circinelloides f. circinelloides, M. circinelloides f. lusitanicus, and M. circinelloides f. griseocyanus. No examples of phylogenetic incongruence were observed, and it is therefore evident that Mucho belongs to the M. circinelloides f. circinelloides subgroup. We also included an M. circinelloides f. circinelloides isolate, 1006PhL, recently sequenced at the Broad Institute (www.broadinstitute.org/annotation /genome/rhizopus_oryzae/MultiHome.html) (27), in this analysis and conclude that this isolate also belongs to the M. circinelloides f. circinelloides subgroup. The M. circinelloides f. circinelloides 1006PhL isolate was obtained from the skin of a normal human volunteer during a skin mycobiome study (27).
FIG 1 .
Phylogenetic analyses of the yogurt isolate Mucho. Three genes (ITS, LSU rRNA, and RPB1) were used to construct the phylogenetic trees. In all cases, there are three distinct groups, represented by M. circinelloides f. circinelloides (Mcc), M. circinelloides f. lusitanicus (Mcl), and M. circinelloides f. griseocyanus (Mcg). No phylogenetic incongruence was found. The three trees show that Mucho and 1006PhL belong to M. circinelloides f. circinelloides. The bootstrap support is 1,000, and the scales indicate base substitutions per position.
Whole-genome comparisons: Mucho versus M. circinelloides f. lusitanicus and Mucho versus 1006PhL (M. circinelloides f. circinelloides).
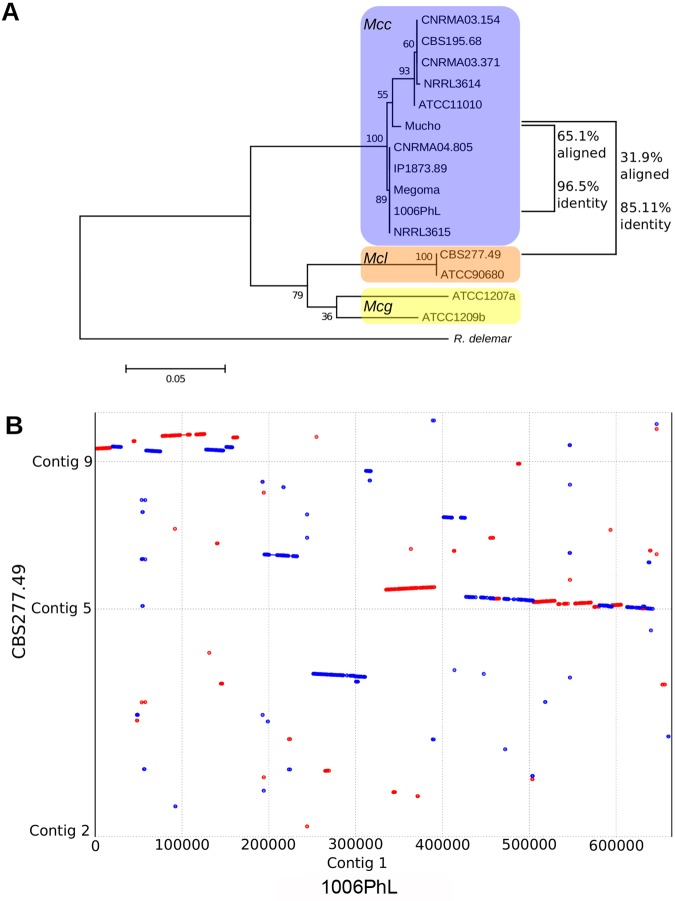
Whole-genome sequencing reveals that the genome of Mucho is much more similar to the M. circinelloides f. circinelloides 1006PhL isolate than to the M. circinelloides f. lusitanicus CBS277.49 isolate (http://jgi.doe.gov/Mucci2/Mucci2.home.html). However, there are still substantial polymorphisms differentiating Mucho from 1006PhL (Fig. 2A). The genome of 1006PhL shares only 85.03% identity with CBS277.49 over the 14 megabases successfully aligned using Nucmer (part of the MUMmer package), while the genome of Mucho shares 85.11% identity with CBS277.49 over 11.2 megabases. The Mucho and 1006PhL isolates share substantially more of their sequences, with 65.1% of the genomes aligning with 96.5% identity using Nucmer. Using a reference-based assembly instead, the Mucho genome is still differentiated from the 1006PhL genome by approximately 630,000 single-nucleotide polymorphisms (SNPs) over the 72.2% of the genome that was callable (97.6% identity).
FIG 2 .
Comparison of the Mucho genome to two other M. circinelloides genomes. (A) Maximum likelihood tree of the phylogeny of the Mucor species complex derived from RPB1 with 500 bootstrap replicates. Percentages of coverage and identity between the entire genome of Mucho and each available sequenced genome were determined with Nucmer and are indicated to the right of the tree. Scale bar indicates 0.05 substitutions per nucleotide position (see reference 21 and references therein for the information about the strains used). (B) A dot plot was constructed using Promer and visualized using MUMmerplot. This plot compares the structures of contig 1 of the 1006PhL genome from the M. circinelloides f. circinelloides group and contigs of the more distant M. circinelloides f. lusitanicus isolate CBS277.49. A similar result was obtained when Mucho contig 1, mapped based on contig 1 of the 1006PhL genome, was compared to the CBS277.49 genome. Red indicates alignment in the forward direction, while blue indicates alignment in the reverse direction.
Interestingly, the M. circinelloides f. lusitanicus genome was not suitable as a reference for genome assembly of Mucho using the short-read component of the Burrows-Wheeler aligner (BWA) (12.2% coverage over 5×); M. circinelloides f. circinelloides was much more compatible (87.5% coverage over 5×). This appears to be the result of substantial rearrangement of the genomes between M. circinelloides f. circinelloides and M. circinelloides f. lusitanicus, as well as, likely, a result of low sequence identity. An alignment of the first contig from 1006PhL with the entire genome of CBS277.49 maps to three different M. circinelloides f. lusitanicus scaffolds and includes at least six large inversions and a number of regions that were not aligned between the two genomes (Fig. 2B).
Taken together, these findings suggest that M. circinelloides f. circinelloides, M. circinelloides f. lusitanicus, and M. circinelloides f. griseocyanus are different enough to be three distinct species rather than simply subspecies. Moreover, even with the M. circinelloides f. circinelloides subgroup, the finding that isolates differ by as much as 3.5% at the whole-genome level suggests substantial diversity compared to that of other well defined fungal species.
Mucho is sexually fertile.
The mating ability of Mucho was tested with the M. circinelloides f. lusitanicus [(−) CBS277.49 and (+) NRRL3631], M. circinelloides f. griseocyanus [(+) ATCC1207a and (−) ATCC1207b], and M. circinelloides f. circinelloides [(−) NRRL3614 and (+) NRRL3615] isolates. The formation of zygospores was monitored after 4 weeks of coculture in the dark (Fig. 3). Mucho mated with the (−) M. circinelloides f. circinelloides strain NRRL3614 but not with the (+) NRRL3615 isolate, indicating that Mucho is a (+) mating type strain. No intersubspecies mating with either M. circinelloides f. lusitanicus or M. circinelloides f. griseocyanus isolates was observed (data not shown). M. circinelloides has a sex locus encoding a high mobility group (HMG) domain protein as the sex determinant, and each mating type carries an allelic HMG transcription factor gene, sexP for (+) and sexM for (−) mating type (28). The Mucho genome was found to contain the sexP gene from the sex locus, which is in full accord with its (+) mating specificity.
FIG 3 .
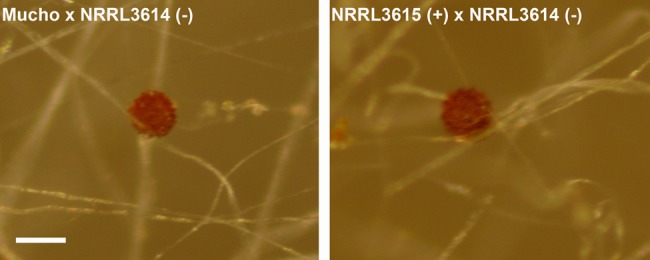
Mucho is sexually compatible with an M. circinelloides f. circinelloides isolate. Zygospore formation between Mucho and (−) NRRL3614 was observed, whereas Mucho did not mate with (+) NRRL3615 (data not shown). The results indicate that Mucho is the (+) mating type. Its mating ability with the other M. circinelloides f. circinelloides isolate also supports that Mucho is M. circinelloides f. circinelloides. Scale is 100 µm.
Mucho is virulent in systemic infections in murine and Galleria hosts.
Among the three M. circinelloides species or subspecies, the M. circinelloides f. circinelloides subgroup is the most commonly associated with clinical infections (21). Our previous findings indicate that M. circinelloides f. circinelloides is the most virulent in the murine host model system, whereas the M. circinelloides f. lusitanicus and M. circinelloides f. griseocyanus isolates are less virulent/avirulent. However, different levels of virulence are also observed among M. circinelloides f. circinelloides isolates (21). To assess the virulence of Mucho and 1006PhL, we employed two host systems: larvae of the wax moth Galleria mellonella and mice (Fig. 4).
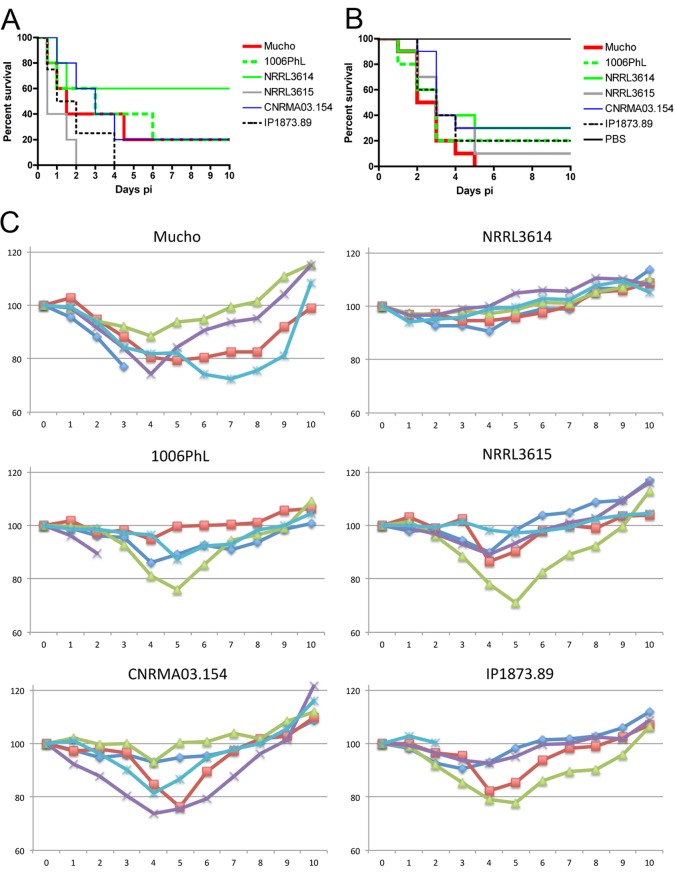
FIG 4 .
Virulence of Mucho in two host models. Fungal spores were injected by tail vein infection into murine hosts and via pseudopods for wax moth larvae. (A) When male BALB/c mice (~20 g) were used as hosts, Mucho displayed moderate virulence in comparison to the results for the most virulent isolate, NRRL3615, and the least virulent isolate, NRRL3614. The difference in virulence between NRRL3615 and NRRL3614 is significant (P = 0.0404); however, the differences in virulence of Mucho and NRRL3615 or NRRL3614 are not statistically significant (P = 0.2064 and P = 0.2404, respectively). Similar results were observed for the virulence of the 1006PhL strain (see text for details). (B) All of the M. circinelloides f. circinelloides isolates tested were significantly virulent in the wax moth larva host model compared to the results for the PBS control (P < 0.0001); however, the difference in virulence between the isolates was not significant (P = 0.7071). (C) When male CD1 mice (~30 g) were used as hosts, mortality from the M. circinelloides f. circinelloides strain infection was lower; for example, under the given conditions, one mouse each died from infection with Mucho, 1006PhL, or IP1873.89. However, substantial weight loss was observed in all of the mice infected with the M. circinelloides f. circinelloides strains, with the exception of mice infected with NRRL3614 (x axis, days postinfection; y axis, percentage compared to initial weight). Between days 2 and 6 p.i., the infected mice underwent weight loss of up to ~30% of the initial weight at the time of infection. The infected mice that survived eventually regained their preinfection body weight. The NRRL3614 isolate that exhibited the least virulence in the BALB/c mouse host model also displayed lower virulence than the other M. circinelloides f. circinelloides isolates tested, and all of the infected mice exhibited only subtle weight loss between days 2 and 4 p.i.
Four M. circinelloides f. circinelloides isolates (NRRL3614, NRRL3615, IP1873.89, and CNRMA03.154) were tested and compared with Mucho and 1006PhL. Male BALB/c mice (~20 g) were infected through tail vein injection with 106 spores resuspended in 200 µl of sterile phosphate-buffered saline (PBS). In accord with our previous results (21), NRRL3614 was significantly less virulent than NRRL3615 (P = 0.0404). The trend of the killing curves may indicate that Mucho is more virulent than the least virulent NRRL3614 strain and less virulent than the most virulent NRRL3615 strain (Fig. 4A). A statistical analysis, however, did not support that the survival curve of mice infected with Mucho was significantly different from that of either isolate (P = 0.2062 for Mucho versus NRRL3615 and P = 0.2404 for Mucho versus NRRL3614). However, it is clearly apparent that Mucho is virulent in the murine tail vein injection model. Similar results were obtained with the human skin isolate 1006PhL: no significant difference in virulence was observed for 1006PhL versus NRRL3615 (P = 0.0788) or 1006PhL versus NRRL3614 (P = 0.2813). In the Galleria larva host model, all M. circinelloides f. circinelloides isolates (including Mucho and 1006PhL) displayed significant virulence compared to the results for the PBS control (for example, P < 0.0001 for Mucho versus PBS) (Fig. 4B). No significant difference in the virulence of the M. circinelloides f. circinelloides strains was observed (P = 0.7071).
We also tested the virulence of these strains with male CD1 (~30 g) mice as hosts (Fig. 4C). Spores (106) were inoculated via tail vein injection, and all conditions and methods of monitoring were the same as described above for BALB/c mice (see also Materials and Methods). The mortality of CD1 mice after infection with the M. circinelloides f. circinelloides strains was lower than that of BALB/c mice. For example, only one mouse died after infection with Mucho, 1006PhL, or IP1873.89. We found in monitoring body weight postinfection (p.i.) that the cohort of mice infected with Mucho underwent significant weight loss by 2 days p.i.; for example, three mice lost ~30% of their body weight by 3, 4, or 6 days p.i., one of which progressed to imminent mortality and was sacrificed on day 3 p.i. Infections with the 1006PhL, NRRL3615, CNRMA03.154, and IP1873.89 isolates that were virulent in BALB/c mice (Fig. 4A) all resulted in obvious weight loss between 2 and 6 days p.i. On the other hand, the least virulent strain, NRRL3614, caused less apparent weight loss, which is in accord with the results of the virulence tests with BALB/c mice. In all cases, the infected mice eventually recovered their initial body weight, indicating that the weight difference between the two mouse strains tested (~20 g for BALB/c and ~30 g for CD1) might contribute to different levels of virulence or that the CD1 mouse background may be less susceptible to M. circinelloides infection than the BALB/c background.
Gastrointestinal colonization of Mucho in the murine host.
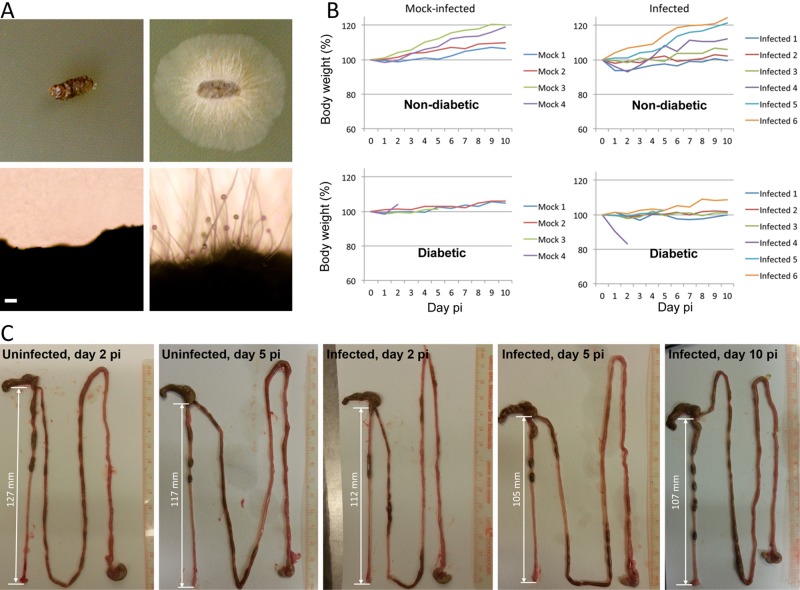
We developed a murine gastrointestinal host model to assess the risk from ingestion of Mucor. Diabetic and nondiabetic murine host models were tested by using the BALB/c mouse strain. Groups of diabetic and nondiabetic mice were infected with Mucho via oral gavage. From each mouse, we collected feces daily (three mice tested from each diabetic or nondiabetic mouse host) and placed them on yeast extract-peptone-dextrose (YPD) medium. Interestingly, fecal samples up to day 10 p.i. displayed Mucor growth after 24 h of incubation at 30°C, which indicates that Mucor can survive passage through the mouse GI tract (Fig. 5A; see Table S2 in the supplemental material). Only one diabetic mouse was found to lose significant weight (~20% of body weight at day 2 p.i.); however, overall, we did not observe apparent weight loss from oral infection with Mucho (Fig. 5B). Interestingly, the colons from infected mice sacrificed at days 2, 5, and 10 p.i. for GI dissection and histopathology tended to be shorter than those from noninfected mice (Fig. 5C); however, this trend is not statistically supported due to the small sample size from each day. In histopathological analysis, we did not observe apparent inflammatory symptom development or fungal invasion in the colons or ceca (data not shown).
FIG 5 .
Gastrointestinal infection of Mucho in the murine host. (A) Groups of BALB/c mice were infected with the Mucho isolate (106) via oral gavage. The fecal samples from three infected diabetic mice displayed Mucor mycelia after 24 h of incubation at 30°C, indicating that Mucor can survive the passage through the GI tract. The figure depicts feces from noninfected (left) and infected (right) diabetic mice on YPD medium at day 1 p.i. (incubated for 24 h at 30°C). Scale = 100 µm. (B) Body weights of mice after fungal spore challenge did not exhibit apparent differences from those of uninfected controls. Top, nondiabetic mice; bottom, mice rendered diabetic with streptozocin treatment. (C) The colons from infected mice (2, 5, and 10 days postinfection) tended to be shorter than those from uninfected mice (2 and 5 days p.i.). However, due to the low numbers of samples, this result is not supported statistically.
Mucho induces cytokine production from immune cells.
The fact that M. circinelloides f. circinelloides isolates display greater virulence than M. circinelloides f. lusitanicus isolates suggests that the two species or subspecies may interact differentially with immune cells. To test this hypothesis, we examined the cytokine responses of THP-1 human monocytes following exposure to M. circinelloides f. circinelloides (Mucho) and M. circinelloides f. lusitanicus (R7B) strains. Of 41 cytokines examined (see Table S3 in the supplemental material), one cytokine, alpha interferon 2 (IFN-α2), appeared to increase moderately in response to all three strains (Fig. S2), indicating activation of the interferon response by this fungal species. Surprisingly, the proinflammatory chemokines interleukin-8 (IL-8), monocyte chemoattractant protein 1 (MCP-1), and macrophage inflammatory protein 1α (MIP-1α), key factors in the recruitment of neutrophils and other immune cells, were only induced by the M. circinelloides f. lusitanicus isolate (Fig. 6 and Fig. S2). Interestingly, another related Mucorales fungal pathogen, Rhizopus delemar (RA99-880), also did not induce the proinflammatory cytokines tested (Fig. 6 and Fig. S2). This suggests that the M. circinelloides f. circinelloides and R. delemar strains fail to trigger or inhibit proinflammatory chemokine production, which may contribute to the greater virulence of the M. circinelloides f. circinelloides subspecies. Two other pathogenic fungi, Aspergillus fumigatus and Candida albicans, were also tested; in these cases, A. fumigatus induced both IL-8 and MIP-1α, whereas C. albicans only induced MIP-1α.
FIG 6 .
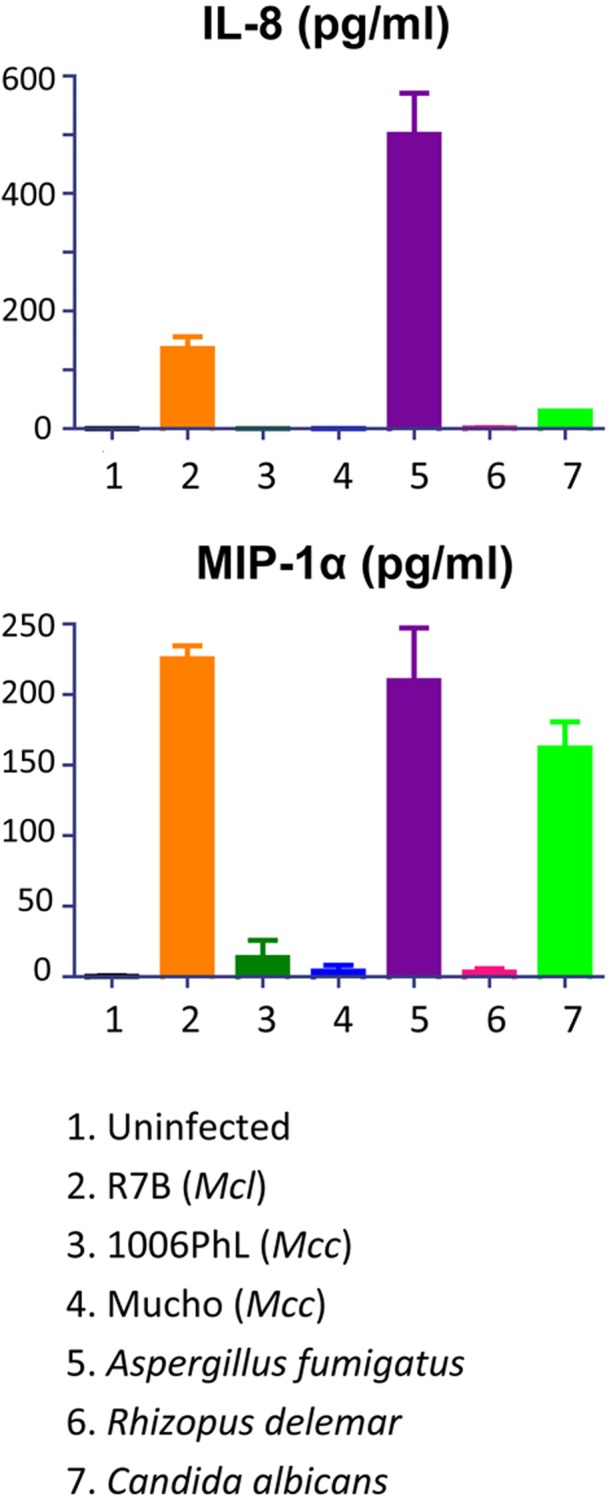
Proinflammatory cytokine production by Mucor and other pathogenic fungi. The R7B M. circinelloides f. lusitanicus strain induced IL-8 and MIP-1α from THP-1 cells; however, two M. circinelloides f. circinelloides strains (Mucho and 1006PhL) did not, indicating that different host responses to M. circinelloides f. lusitanicus and M. circinelloides f. circinelloides might contribute to the different virulence outcomes in mammalian hosts. The R. delemar isolate RA99-880, which is also a mucoralean fungus and is virulent in mammalian hosts, had results similar to those for the two M. circinelloides f. circinelloides strains. A. fumigatus induced both IL-8 and MIP-1α, but C. albicans only induced MIP-1α, indicating that these pathogens result in host-pathogen interactions that are distinguishable from those of the mucoralean fungi.
DISCUSSION
Fungal infections have increased as the cohorts immunocompromised by infections and/or medical conditions mount. Fungal pathogens as food-borne pathogens, however, have been neglected compared to other well known food-borne pathogens, including bacteria, viruses, and parasites. Food-borne infectious diseases are a serious ongoing problem for the health care system and cause tremendous economic and social burdens (1). New pathogens are emerging unexpectedly. It is therefore reasonable to raise precautionary concerns with regard to fungi as food-borne infectious disease agents.
There was a recent outbreak of illnesses related to M. circinelloides contamination in commercial yogurt products. Consumers complained of discomfort after eating the yogurt products, according to a U.S. FDA report. As a result, the manufacturer voluntarily withdrew their products that had “best by” dates within a certain time range (best by dates of 11 September to 7 October 2013 with the IMS code 16-012). These events prompted us to investigate the mold responsible for this recall and potential health concerns related to it by investigating the causative agent in animal host models.
The mold isolated from the contaminated yogurt, designated Mucho, was identified as M. circinelloides f. circinelloides by MLST, whole-genome comparisons, and mating analyses (Fig. 1, 2, and 3). There are at least three species or subspecies in the M. circinelloides species complex, M. circinelloides f. circinelloides, M. circinelloides f. lusitanicus, and M. circinelloides f. griseocyanus. Comparison of the M. circinelloides f. lusitanicus and M. circinelloides f. circinelloides genomes reveals dramatic rearrangements, as well as significant differences in sequence identity. It is highly likely that, as a result, there is a substantial reproductive barrier separating these two lineages. The Mucho isolate mated only with a (+) M. circinelloides f. circinelloides strain and not with (+) M. circinelloides f. lusitanicus or (+) M. circinelloides f. griseocyanus isolates. Even if an M. circinelloides f. circinelloides strain may be able to mate with an M. circinelloides f. lusitanicus strain, the progeny are likely to be either less fit or inviable. This suggests that there may be a species barrier between the M. circinelloides f. circinelloides and M. circinelloides f. lusitanicus subtypes, consistent with their divergent and rearranged genome sequences.
Among the species or subspecies, M. circinelloides f. circinelloides is known to be more virulent in the murine host model and, in particular, is more frequently associated with human infections (21). Different outcomes during interactions with cultured immune cells may explain the differences in virulence (Fig. 6), where an M. circinelloides f. lusitanicus isolate induced proinflammatory cytokines but two M. circinelloides f. circinelloides isolates did not. Our subsequent virulence tests found that the Mucho strain is virulent in animal hosts (Fig. 4), which may be explained by differential induction of proinflammatory cytokines by M. circinelloides f. lusitanicus and M. circinelloides f. circinelloides. However, in the oral gavage model experiment (Fig. 5), the Mucho isolate did not cause apparent disease symptoms based on weight monitoring postinfection (except in one mouse). Nevertheless, the Mucho strain survived passage through the mouse GI tract based on recovery of the fungus from fecal samples, indicating that M. circinelloides can survive passage through the GI tract. These results implicate Mucor species as potential causal agents for serious food-borne illness, especially for immunocompromised patients. A series of case reports of gastrointestinal mucormycosis further supports that Mucor can cause fatal fungal infections through ingestion of contaminated foods or medicines (15–18, 29).
It is as yet unclear whether Mucor infection is directly responsible for the illnesses reported in the M. circinelloides outbreak related to the yogurts. An alternative explanation is that intoxication with secondary metabolites, such as mycotoxins, could be responsible for this incident. Possibly the contaminated yogurts could contain harmful toxins produced by Mucor. Ergot fungi provide an example. Ergot toxin is an alkaloid fungal toxin produced by Claviceps species, and the consumption of rye and other cereals contaminated with ergot toxin causes ergotism (30, 31). The symptoms caused by the toxin include gangrene of the skin and are associated with the tragic Salem witch trials of 1692 that were held in Salem Village, MA. This food-borne ergotism was misinterpreted as a symptom of witchcraft, resulting in the persecution and execution of 20 Puritans (32, 33). Another example of a well known mycotoxin is aflatoxin, produced by Aspergillus species (30). The responsible fungal species colonize grains and peanuts and contaminate them with aflatoxin; the toxin was first known as the source of turkey X disease (34). Aflatoxin is a carcinogen posing a serious medical problem for the health care system (35). In addition, T-2 toxin produced by Fusarium spp. causes alimentary toxic aleukia, and consumption of bread contaminated with the toxin resulted in the deaths of ~100,000 people in the Soviet Union from 1942 to 1948 (36).
The genomes of M. circinelloides f. circinelloides Mucho and 1006PhL and M. circinelloides f. lusitanicus CBS277.49 contain genes predicted to be involved in the production of secondary metabolites, which indicates that Mucor might produce harmful toxins (Table 1; see Fig. S3 in the supplemental material). Each of the genes identified in these pathways from 1006PhL is present in the Mucho genome, and some of the proteins encoded are clearly more conserved than others. Genes predicted to encode proteins involved in the synthesis of terpenes and bacteriocins were particularly well conserved. Although the related species Rhizopus microsporus produces rhizoxin via the endosymbiotic bacterium Burkholderia rhizoxinica, it is not known whether any Mucor species harbor endosymbionts or produce mycotoxins (37). However, our whole-genome sequencing did not reveal sequences of endosymbiotic bacteria.
TABLE 1 .
Secondary metabolite genes in the genomes of Mucho, 1006PhL, and CBS277.49 strains
| Category | Gene accession numbera |
Putative function | ||
|---|---|---|---|---|
| 1006PhL | Mucho | CBS277.49 | ||
| Terpene | HMPREF 1544_00936 | KJ999696 | Mucci1.e_gw1.6.146.1 | 3-Methylcrotonyl-coenzyme A (CoA) carboxylase biotin-containing subunit |
| HMPREF 1544_00937 | KJ999697 | estExt_Genewise1Plus.C_070865 | Farnesyltranstransferase | |
| HMPREF 1544_01407 | KJ999698 | e_gw1.02.1791.1 | Farnesyl-diphosphate farnesyltransferase | |
| HMPREF 1544_07394 | KJ999705 | e_gw1.05.994.1 | Lanosterol synthase | |
| HMPREF 1544_08078 | KJ999706 | Genemark1.6702_g | Lycopene cyclase | |
| HMPREF 1544_08719 | KJ999709 | e_gw1.01.1613.1 | Tripeptidyl-peptidase 2 | |
| HMPREF 1544_08721 | KJ999710 | fgenesh1_pm.01_#_453 | d-Arabinitol 2-dehydrogenase | |
| HMPREF 1544_08722 | KJ999711 | fgenesh1_kg.01_#_372_#_535_1_CCIA_CCIB_EXTA | Geranylgeranyl pyrophosphate synthase | |
| HMPREF 1544_08724 | KJ999712 | Mucci1.e_gw1.1.1753.1 | α-1,2-Mannosyltransferase | |
| HMPREF 1544_08726 | KJ999713 | fgenesh1_kg.01_#_373_#_1623_1_CCIA_CCIB_EXTA | α-Keto reductase | |
| HMPREF 1544_10222 | KJ999717 | Mucci1.e_gw1.1.244.1 | Phytoene dehydrogenase | |
| HMPREF 1544_10223 | KJ999718 | fgenesh1_kg.01_#_90_#_233_1_CCIA_CCIB_EXTA | Bifunctional lycopene cyclase/phytoene synthase | |
| HMPREF 1544_10229 | KJ999719 | Mucci1.fgeneshMC_pm.1_#_622 | Taurine dioxygenase | |
| HMPREF 1544_10230 | KJ999720 | fgenesh1_pg.01_#_386 | Taurine dioxygenase | |
| HMPREF 1544_11141 | KJ999722 | fgenesh1_pm.03_#_326 | NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 | |
| NRPS | HMPREF 1544_02563 | KJ999699 | estExt_Genewise1Plus.C_080395 | Homocitrate synthase |
| HMPREF 1544_02567 | KJ999700 | fgenesh1_pg.08_#_263 | l-Aminoadipate-semialdehyde dehydrogenase | |
| HMPREF 1544_02572 | KJ999701 | fgenesh1_kg.08_#_57_#_1752_1_CCIA_CCIB_EXTA | Methylmalonate-semialdehyde dehydrogenase (acylating) | |
| Siderophore | HMPREF 1544_03746 | KJ999702 | fgenesh1_kg.01_#_25_#_201_1_CCIA_CCIB_EXTA | Ferric iron reductase FhuF-like transporter |
| Bacteriocin | HMPREF 1544_10032 | KJ999713 | fgenesh1_pm.01_#_98 | Adenosinetriphosphatase |
| HMPREF 1544_10037 | KJ999715 | Mucci1.fgeneshMC_pg.1_#_1479 | Cytochrome P450 | |
| HMPREF 1544_10038 | KJ999716 | estExt_fgenesh1_pm.C_010092 | Amidohydrolase | |
| Other | HMPREF 1544_00134 | KJ999695 | Mucci1.fgeneshMC_pg.5_#_71 | Acyl-CoA synthetases (AMP-forming)/AMP-acid ligases II (lipid metabolism) |
| HMPREF 1544_07114 | KJ999703 | fgenesh1_pg.03_#_1066 | Glycosyltransferase | |
| HMPREF 1544_07116 | KJ999704 | Mucci1.fgeneshMC_pg.3_#_1109 | Acyl-CoA synthetases (AMP-forming)/AMP-acid ligases II (lipid metabolism) | |
| HMPREF 1544_08550 | KJ999707 | Mucci1.e_gw1.13.44.1 | Acyl-CoA oxidase | |
| HMPREF 1544_08551 | KJ999708 | Mucci1.fgeneshMC_pg.13_#_93 | Acyl-CoA synthetases (AMP-forming)/AMP-acid ligases II (lipid metabolism) | |
| HMPREF 1544_11114 | KJ999721 | Genemark1.7267_g | Acyl-CoA synthetases (AMP-forming)/AMP-acid ligases II (lipid metabolism) | |
There are other reports that fungal contamination associated with other daily essentials or medications can cause serious medical threats. A fungal meningitis outbreak occurred after epidural injection of methylprednisolone contaminated with the ascomycete fungus Exserohilum rostratum (38–40). A survey study of 328 patients who had been administered the steroid contaminated with this fungus and who then developed infections revealed that 81% developed central nervous system (CNS) infections and the remaining 19% suffered from non-CNS infections (29). Multiple fungal keratitis outbreaks were reported between 2004 and 2006 among contact lens wearers in Hong Kong, Singapore, France, and the United States (41–44). The fungus Fusarium that contaminated contact lens solutions was the responsible pathogen, which resulted in a withdrawal of ReNu with MoistureLoc (Bausch & Lomb) from the world market on 15 May 2006 (42). Fungal keratitis is an inflammation of the cornea and can cause permanent loss of eyesight.
It is obvious that fungal pathogens pose a severe problem for the health care system, and the ensuing outcomes of infection often cause a serious loss of quality of life or even blindness or death. However, compared to other pathogens, less attention has been afforded to fungal pathogens as contaminants of our daily essentials, such as foods, medical devices, medications, and the facilities that manufacture them. Prevention of fungal contamination and careful examination should be practiced. Our results in this study provide evidence that understudied Mucor species can cause a potentially serious illness following ingestion with foods.
MATERIALS AND METHODS
Strains and culture conditions.
A plain Chobani yogurt dated best by 30 September 2013 and with the IMS code 16-012 was obtained from the batch that was subject to recall (see Fig. S1 in the supplemental material). Yogurt samples from seven different spots within the suspect container were transferred with sterile applicators onto potato dextrose agar (PDA) medium containing 50 µg/ml ampicillin and kanamycin to control residual live bacteria. After 3 days of incubation at 30°C, mold mycelia grew out on the plates. After two cycles of streak purification, spores from each plate were collected. For spore production from all M. circinelloides f. circinelloides isolates, each strain was inoculated with a sterile toothpick into the center of a PDA agar plate. After 4 days of incubation in the light at room temperature, sterile distilled water was added to the plates and spores were collected. For further virulence tests, the spore suspensions were washed with sterile PBS.
For mating, V8 medium (pH 7) was inoculated with two strains approximately 1 in. apart from each other. After 4 weeks of coculture in the dark, the mating plates were observed by using a Nikon Eclipse E400 microscope equipped with a Nikon DXM1200F camera.
Phylogenetic analysis.
ITS, LSU rRNA, and RPB1 used for the Fungal Tree of Life Project (45) were amplified from genomic DNA of the Mucho isolate. The primers used for ITS were ITS1, TCCGTAGGTGAACCTGCGG, and ITS4, TCCTCCGCTTATTGATATGC; for LSU rRNA, the primers were D1/D2 LRDNA, GCATATCAATAAGCGGAGGAAAAG, and LR3, GGTCCGTGTTTCAAGACGG; and for RPB1, they were RPB1-Ac, GARTGYCCDGGDCAYTTYGG, and RPB1-Cr, CCNGCDATNTCRTTRTCCATRTA. PCR products were obtained for each gene, sequenced, and aligned with CLUSTALW. The aligned DNA sequences were used to construct phylogenetic trees by using the PhyML 3.0 software (46). The three genes of the Yarrowia lipolytica isolate were also sequenced, and the sequences obtained were analyzed with Blast to identify the species. The GenBank nucleotide accession numbers for the Mucho ITS, LSU rRNA, and RPB1 sequences are KJ588204, KJ588205, and KJ588205, respectively.
Whole-genome analysis
A reference genome was generated for the 1006PhL isolate of M. circinelloides f. circinelloides, isolated from the skin of a healthy volunteer (27). Genomic DNA was used to construct two libraries, 180-base fragments and 2- to 3-kilobase (kb) jumps, and each was sequenced on the Illumina HiSeq 2000 platform. The 101-base-pair Illumina reads were assembled using ALLPATHS-LG (47) (build R43527) with the default parameters, using roughly a 50-fold depth of fragment reads and 50-fold depth of jumping reads. The resulting 36.4-Mb assembly consisted of 470 scaffolds and 1,459 contigs. The sequences were submitted to NCBI under accession number AOCY00000000.
A single-end TruSeq library was constructed with Mucho genomic DNA, and the genome was sequenced using 50 base reads on the HiSeq 2000 platform. The reads were mapped to the 1006PhL reference genome using the short-read component of BWA (4). SNPs were called using the Genome Analysis Toolkit (GATK version 2.4-9) pipeline and the Unified Genotyper with the haploid setting (48). Comparison with the JGI-sequenced Mucor isolate CBS277.49 was carried out using whole-genome alignments generated using Nucmer, part of the MUMmer package. An artificial genome was constructed based on the SNPs identified from the reference-based assembly using GATK’s FastaAlternateReferenceMaker. This enabled a more accurate comparison of both 1006PhL and Mucho with CBS277.49 for some of the analysis, as it removed the assembly quality as a variable. Phylogenies constructed using whole-genome data were aligned and constructed using MEGA5 (49). The reads were submitted to NCBI under the project accession number PRJNA244237.
Virulence tests in animal models.
Spores for animal infections were resuspended in sterile PBS, and the numbers of spores were counted using a hemocytometer. PBS containing 50,000 spores or PBS alone was injected into a cohort of wax moth larvae for each strain (10 larvae per strain), as described previously (21, 50, 51). For the murine systemic infection model, groups of male BALB/c and CD1 mice (5 per each strain) were infected with 106 spores in 200 µl of sterile PBS through tail vein injection. Survival of the host was examined twice a day, and body weight was monitored daily. Animals that appeared moribund or in pain were sacrificed appropriately. Mortality data were evaluated with Kaplan-Meier survival curves by using PRISM (GraphPad Software, Inc.).
Gastrointestinal colonization was accomplished by oral gavage either with spores (106) of the Mucho strain resuspended in sterile water or with sterile water only. Diabetic and nondiabetic mouse host models were used; for the diabetic mouse host model, the mice were rendered diabetic by injecting streptozocin (190 mg per kg of body weight) as previously described (52, 53). Three male BALB/c mice (4 to 6 weeks old) were infected with the Mucho strain, and two mice were mock infected with sterile water as a control. Body weight was measured daily. Fecal samples were collected daily from each mouse separately and placed onto YPD medium to examine the growth of Mucor. The fungus from the feces from day 1 was confirmed by sequencing the ITS, and later recovery of Mucho was confirmed by morphological analysis, including observation of aseptated hyphae and sporangium formation, which are hallmarks of the Mucorales fungi. An absence of fungal burden was detected from uninfected mouse controls. The experiments were repeated in duplicate, and all mice were sacrificed appropriately at day 10 p.i. or at day 2, 5, or 10 p.i. for GI dissection and histopathology.
Interactions with immune cells.
THP-1 cells were purchased from the American Type Culture Collection (ATCC). Cells were maintained at 37°C in a 5% CO2 atmosphere. THP-1 cells were grown in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum. For Mucor infection experiments, cells were plated at 1 × 105 cells in 100 µl of medium and infected with spores at a multiplicity of infection of 1. Supernatants were collected for cytokine analysis at 24 h. Forty-one cytokines were measured using the Milliplex MAP (multianalyte panel) human cytokine/chemokine magnetic bead panel (Millipore) by the Duke Immune Reconstitution & Biomarker Analysis Shared Resource. IL-8 and MIP-1α enzyme-linked immunosorbent assays (R&D Systems) to verify changes seen with Luminex were conducted on supernatants according to the manufacturer’s instructions. Three other pathogenic fungus strains, RA99-880 (R. delemar), AF293 (A. fumigatus), and SC5314 (C. albicans), were included in addition to the R7B (M. circinelloides f. lusitanicus), Mucho (M. circinelloides f. circinelloides), and 1006PhL (M. circinelloides f. circinelloides) strains.
The murine animal studies were conducted at the Duke University Medical Center in full compliance with all of the guidelines of the Duke University Medical Center Institutional Animal Care and Use Committee (IACUC) and in full compliance with the United States Animal Welfare Act (Public Law 98–198). The Duke University Medical Center IACUC approved all of the vertebrate animal studies under protocol number A061-12-03. The studies were conducted in the Division of Laboratory Animal Resources (DLAR) facilities that are accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC).
SUPPLEMENTAL MATERIAL
Note from a couple in Corpus Christi, TX, who consumed the mold-contaminated yogurt. Download
A plain Chobani yogurt contaminated with M. circinelloides. The yogurt was obtained from Corpus Christi, TX. The small rectangular insert shows the yogurt “best by” date to be 30 September 2013 with IMS code 16-012. This was within the time range for yogurt subject to recall by the manufacturer. Download
Inflammatory cytokine production from the THP-1 cell line induced by Mucor. Luminex analysis revealed that the M. circinelloides f. lusitanicus isolate R7B induced the production of IFN-α2, IL-8, MCP-1, and MIP-1α. On the other hand, the 2 M. circinelloides f. circinelloides isolates Mucho and 1006PhL only induced IFN-α2 and not IL-8, MCP-1, or MIP-1α, indicating that different host responses might be involved in the different virulence outcomes in mammalian hosts. Download
Putative genes for secondary metabolites conserved in 1006PhL and Mucho. Genes encoding putative secondary metabolites are found in the 1006PhL and Mucho genomes. Several genes, predominantly encoding those involved in terpene biosynthesis (HMPREF1544_00936, HMPREF1544_00937, HMPREF1544_01407, HMPREF1544_02563, HMPREF1544_08724, HMPREF1544_08726, and HMPREF1544_10038) are highly conserved between the two genomes, with only synonymous SNPs or one nonsynonymous SNP. However, other putative proteins encoded in the Mucho genome vary, with substantial nonsynonymous SNPs compared to those of 1006PhL. SNPs in introns are not depicted, and several genes had changes in a stop or start codon location by 1 or 2 amino acids that did not alter the reading frame (HMPREF1544_08551, HMPREF1544_08721, HMPREF1544_08722, HMPREF 1544_08551, and HMPREF_1544). Clustered genes are arranged together in the figure. Nrps, nonribosomal polyketide synthetase. Download
Yogurt samples and fungal isolates
Recovery of Mucho from feces of infected mice.
Cytokines tested in this study.
ACKNOWLEDGMENTS
We thank the three reviewers and appreciate their constructive comments on this study. We thank Anna Averette for technical support. We thank Keisha Findley, Clayton Deming, and Julie Segre for providing the M. circinelloides f. circinelloides isolate 1006PhL and Bill Steinbach and Praveen Juvvadi for providing the A. fumigatus AF293 strain. We also thank the Joint Genome Institute and Santiago Torres-Martinez for making the CBS277.49 genome sequence available and John Perfect for discussions and key insights.
This work was funded by NIH/NIAID grant R21 AI085331 to S.C.L. and J.H. The 1006PhL genome sequencing project was supported in part by the Human Microbiome Project, grant U54HG004969.
Footnotes
Citation Lee SC, Billmyre RB, Li A, Carson S, Sykes SM, Huh EY, Mieczkowski P, Ko DC, Cuomo CA, Heitman J. 2014. Analysis of a food-borne fungal pathogen outbreak: virulence and genome of a Mucor circinelloides isolate from yogurt. mBio 5(4):e01390-14. doi:10.1128/mBio.01390-14.
REFERENCES
- 1. Newell DG, Koopmans M, Verhoef L, Duizer E, Aidara-Kane A, Sprong H, Opsteegh M, Langelaar M, Threfall J, Scheutz F, van der Giessen J, Kruse H. 2010. Food-borne diseases—the challenges of 20 years ago still persist while new ones continue to emerge. Int. J. Food Microbiol. 139(Suppl 1):S3–S15. 10.1016/j.ijfoodmicro.2010.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Denny J, McLauchlin J. 2008. Human Listeria monocytogenes infections in Europe—an opportunity for improved European surveillance. Euro Surveill. 13(13):8082 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=8082 [PubMed] [Google Scholar]
- 3. Koopmans M, Vennema H, Heersma H, van Strien E, van Duynhoven Y, Brown D, Reacher M, Lopman B, European Consortium. on Foodborne Viruses 2003. Early identification of common-source foodborne virus outbreaks in Europe. Emerg. Infect. Dis. 9:1136–1142. 10.3201/eid0909.020766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lopman B, Vennema H, Kohli E, Pothier P, Sanchez A, Negredo A, Buesa J, Schreier E, Reacher M, Brown D, Gray J, Iturriza M, Gallimore C, Bottiger B, Hedlund KO, Torvén M, von Bonsdorff CH, Maunula L, Poljsak-Prijatelj M, Zimsek J, Reuter G, Szücs G, Melegh B, Svennson L, van Duijnhoven Y, Koopmans M. 2004. Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet 363:682–688. 10.1016/S0140-6736(04)15641-9 [DOI] [PubMed] [Google Scholar]
- 5. Kroneman A, Verhoef L, Harris J, Vennema H, Duizer E, van Duynhoven Y, Gray J, Iturriza M, Böttiger B, Falkenhorst G, Johnsen C, von Bonsdorff C-H, Maunula L, Kuusi M, Pothier P, Gallay A, Schreier E, Höhne M, Koch J, Szücs G, Reuter G, Krisztalovics K, Lynch M, McKeown P, Foley B, Coughlan S, Ruggeri FM, Bartolo I, Vainio K, Isakbaeva E, Poljsak-Prijatelj M, Grom AH, Mijovski JZ, Bosch A, Buesa J, Fauquier AS, Hernandéz-Pezzi G, Hedlund K-O, Koopmans M. 2008. Analysis of integrated virological and epidemiological reports of norovirus outbreaks collected within the Foodborne Viruses in Europe Network from 1 July 2001 to 30 June 2006. J. Clin. Microbiol. 46:2959–2965. 10.1128/JCM.00499-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Siebenga J, Kroneman A, Vennema H, Duizer E, Koopmans M, Food-Borne Viruses in Europe Network 2008. Food-Borne Viruses in Europe Network report: the norovirus GII.4 2006b (for US named Minerva-like, for Japan Kobe034-like, for UK V6) variant now dominant in early seasonal surveillance. Euro Surveill. 10(2):8009 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=8009 [PubMed] [Google Scholar]
- 7. FDA 5 September 2013. Chobani, Inc. Voluntarily Recalls Greek Yogurt Because of Product Concerns. Press release. U.S. FDA, Washington, DC: http://www.fda.gov/safety/recalls/ucm367298.htm [Google Scholar]
- 8. Today USA. 10 September 2013. FDA receives dozens of reports of illness from yogurt USA Today, McLean, VA http://www.usatoday.com/story/news/nation/2013/09/10/fda-illness-yogurt/2792291/.
- 9. Chayakulkeeree M, Ghannoum MA, Perfect JR. 2006. Zygomycosis: the re-emerging fungal infection. Eur. J. Clin. Microbiol. Infect. Dis. 25:215–229. 10.1007/s10096-006-0107-1 [DOI] [PubMed] [Google Scholar]
- 10. Ibrahim AS, Spellberg B, Walsh TJ, Kontoyiannis DP. 2012. Pathogenesis of mucormycosis. Clin. Infect. Dis. 54:S16–S22. 10.1093/cid/cir865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roden MM, Zaoutis TE, Buchanan WL, Knudsen TA, Sarkisova TA, Schaufele RL, Sein M, Sein T, Chiou CC, Chu JH, Kontoyiannis DP, Walsh TJ. 2005. Epidemiology and outcome of Zygomycosis: a review of 929 reported cases. Clin. Infect. Dis. 41:634–653. 10.1086/432579 [DOI] [PubMed] [Google Scholar]
- 12. Dromer F, McGinnis MR. 2003. Zygomycosis, p. 297–308. In Annaissie EJ, McGinnis MR, Pfaller MA. (ed), Clinical mycology. Churchill Livingstone, New York, NY. [Google Scholar]
- 13. Ibrahim AS, Spellberg B. 2006. Zygomycetes as agents of infectious disease in humans, p 429–440 In Heitman J, Filler SG, Edwards JE, Jr, Mitchell AP. (ed), Molecular principles of fungal pathogenesis. ASM Press, Washington, DC [Google Scholar]
- 14. Spellberg B. 2012. Gastrointestinal mucormycosis: an evolving disease. Gastroenterol. Hepatol. 8:140–142 [PMC free article] [PubMed] [Google Scholar]
- 15. Aboltins CA, Pratt WA, Solano TR. 2006. Fungemia secondary to gastrointestinal Mucor indicus infection. Clin. Infect. Dis. 42:154–155. 10.1086/498751 [DOI] [PubMed] [Google Scholar]
- 16. Oliver MR, Van Voorhis WC, Boeckh M, Mattson D, Bowden RA. 1996. Hepatic mucormycosis in a bone marrow transplant recipient who ingested naturopathic medicine. Clin. Infect. Dis. 22:521–524. 10.1093/clinids/22.3.521 [DOI] [PubMed] [Google Scholar]
- 17. Singh N, Gayowski T, Singh J, Yu VL. 1995. Invasive gastrointestinal zygomycosis in a liver transplant recipient: case report and review of zygomycosis in solid-organ transplant recipients. Clin. Infect. Dis. 20:617–620. 10.1093/clinids/20.3.617 [DOI] [PubMed] [Google Scholar]
- 18. Thomson SR, Bade PG, Taams M, Chrystal V. 1991. Gastrointestinal mucormycosis. Br. J. Surg. 78:952–954. 10.1002/bjs.1800780819 [DOI] [PubMed] [Google Scholar]
- 19. Petrikkos G, Skiada A, Lortholary O, Roilides E, Walsh TJ, Kontoyiannis DP. 2012. Epidemiology and clinical manifestations of mucormycosis. Clin. Infect. Dis. 54:S23–S34. 10.1093/cid/cir866 [DOI] [PubMed] [Google Scholar]
- 20. Singh N, Aguado JM, Bonatti H, Forrest G, Gupta KL, Safdar N, John GT, Pursell KJ, Muñoz P, Patel R, Fortun J, Martin-Davila P, Philippe B, Philit F, Tabah A, Terzi N, Chatelet VR, Kusne S, Clark N, Blumberg E, Julia MB, Humar A, Houston S, Lass-Flörl C, Johnson L, Dubberke ER, Barron MA, Lortholary O. 2009. Zygomycosis in solid organ transplant recipients: a prospective, matched case-control study to assess risks for disease and outcome. J. Infect. Dis. 200:1002–1011. 10.1086/605445 [DOI] [PubMed] [Google Scholar]
- 21. Li CH, Cervantes M, Springer DJ, Boekhout T, Ruiz-Vazquez RM, Torres-Martinez SR, Heitman J, Lee SC. 2011. Sporangiospore size dimorphism is linked to virulence of Mucor circinelloides. PLoS Pathog. 7:e1002086. 10.1371/journal.ppat.1002086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schipper MAA. 1976. On Mucor circinelloides, Mucor racemosus, and related species. Stud. Mycol. 12:1–40 [Google Scholar]
- 23. FDA 18 September 2013. FDA establishment inspection report, Chobani Idaho. FEI 3009726115. U.S. FDA, Washington, DC: http://www.fda.gov/downloads/AboutFDA/CentersOffices/OfficeofGlobalRegulatoryOperationsandPolicy/ORA/ORAElectronicReadingRoom/UCM376634.pdf [Google Scholar]
- 24. James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung GH, Johnson D, O’Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schüssler A, Longcore JE, O’Donnell K, Mozley-Standridge S, Porter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, et al. 2006. Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature 443:818–822. 10.1038/nature05110 [DOI] [PubMed] [Google Scholar]
- 25. Beopoulos A, Cescut J, Haddouche R, Uribelarrea JL, Molina-Jouve C, Nicaud JM. 2009. Yarrowia lipolytica as a model for bio-oil production. Prog. Lipid Res. 48:375–387. 10.1016/j.plipres.2009.08.005 [DOI] [PubMed] [Google Scholar]
- 26. Papanikolaou S, Aggelis G. 2010. Yarrowia lipolytica: a model microorganism used for the production of tailor-made lipids. Eur. J. Lipid Sci. Technol. 112:639–654. 10.1002/ejlt.200900197 [DOI] [Google Scholar]
- 27. Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, Kong HH, Segre JA, Segre JA. 2013. Topographic diversity of fungal and bacterial communities in human skin. Nature 498:367–370. 10.1038/nature12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee SC, Corradi N, Byrnes EJ, Torres-Martinez S, Dietrich FS, Keeling PJ, Heitman J. 2008. Microsporidia evolved from ancestral sexual fungi. Curr. Biol. 18:1675–1679. 10.1016/j.cub.2008.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chiller TM, Roy M, Nguyen D, Guh A, Malani AN, Latham R, Peglow S, Kerkering T, Kaufman D, McFadden J, Collins J, Kainer M, Duwve J, Trump D, Blackmore C, Tan C, Cleveland AA, MacCannell T, Muehlenbachs A, Zaki SR, Brandt ME, Jernigan JA, Multistate Fungal Infection Clinical Investigation Team 2013. Clinical findings for fungal infections caused by methylprednisolone injections. N. Engl. J. Med. 369:1610–1619. 10.1056/NEJMoa1304879 [DOI] [PubMed] [Google Scholar]
- 30. Keller NP, Turner G, Bennett JW. 2005. Fungal secondary metabolism—from biochemistry to genomics. Nat. Rev. Microbiol. 3:937–947. 10.1038/nrmicro1286 [DOI] [PubMed] [Google Scholar]
- 31. Panaccione DG. 2005. Origins and significance of ergot alkaloid diversity in fungi. FEMS Microbiol. Lett. 251:9–17. 10.1016/j.femsle.2005.07.039 [DOI] [PubMed] [Google Scholar]
- 32. Caporael LR. 1976. Ergotism: the Satan loosed in Salem? Science 192:21–26. 10.1126/science.769159 [DOI] [PubMed] [Google Scholar]
- 33. Flotte TJ, Bell DA. 1989. Role of skin lesions in the Salem witchcraft trials. Am. J. Dermatopathol. 11:582–587. 10.1097/00000372-198912000-00014 [DOI] [PubMed] [Google Scholar]
- 34. Wannop CC. 1961. The histopathology of turkey “X” disease in Great Britain. Avian Dis. 5:371–381. 10.2307/1587768 [DOI] [Google Scholar]
- 35. Amaike S, Keller NP. 2011. Aspergillus flavus. Annu. Rev. Phytopathol. 49:107–133. 10.1146/annurev-phyto-072910-095221 [DOI] [PubMed] [Google Scholar]
- 36. Joff AZ. 1978. Fusarium poae and F. sporotrichioides as principal causal agents of alimentary toxic aleukia, p 21–86 In Wyllie TD, Morehouse LG. (ed), Mycotoxic fungi, mycotoxins, mycotoxicoses: an encyclopedic handbook, vol 3 Marcel Dekker, New York, NY. [Google Scholar]
- 37. Partida-Martinez LP, Hertweck C. 2005. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437:884–888. 10.1038/nature03997 [DOI] [PubMed] [Google Scholar]
- 38. Kleinfeld K, Jones P, Riebau D, Beck A, Paueksakon P, Abel T, Claassen DO. 2013. Vascular complications of fungal meningitis attributed to injections of contaminated methylprednisolone acetate. JAMA Neurol. 70:1173–1176. 10.1001/jamaneurol.2013.3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kuehn BM. 2013. CDC probes new outbreak associated with compounded steroids. JAMA 309:2541. 10.1001/jama.2013.7329 [DOI] [PubMed] [Google Scholar]
- 40. Smith RM, Tipple M, Chaudry MN, Schaefer MK, Park BJ. 2013. Relapse of fungal meningitis associated with contaminated methylprednisolone. N. Engl. J. Med. 368:2535–2536. 10.1056/NEJMc1306560 [DOI] [PubMed] [Google Scholar]
- 41. Bullock JD. 2008. An outbreak of Fusarium keratitis associated with contact lens use in the northeastern United States. Cornea 27:973–974. 10.1097/ICO.0b013e318177011a [DOI] [PubMed] [Google Scholar]
- 42. Bullock JD, Elder BL, Warwar RE, Snyder SA, Sizemore IE. 2014. Mechanism of drug failure in Fusarium keratitis, 2004-2006. N. Engl. J. Med. 370:88–89. 10.1056/NEJMc1304053 [DOI] [PubMed] [Google Scholar]
- 43. Chang DC, Grant GB, O’Donnell K, Wannemuehler KA, Noble-Wang J, Rao CY, Jacobson LM, Crowell CS, Sneed RS, Lewis FM, Schaffzin JK, Kainer MA, Genese CA, Alfonso EC, Jones DB, Srinivasan A, Fridkin SK, Park BJ, Fusarium Keratitis Investigation Team 2006. Multistate outbreak of Fusarium keratitis associated with use of a contact lens solution. JAMA 296:953–963. 10.1001/jama.296.8.953 [DOI] [PubMed] [Google Scholar]
- 44. Khor WB, Aung T, Saw SM, Wong TY, Tambyah PA, Tan AL, Beuerman R, Lim L, Chan WK, Heng WJ, Lim J, Loh RS, Lee SB, Tan DT. 2006. An outbreak of Fusarium keratitis associated with contact lens wear in Singapore. JAMA 295:2867–2873. 10.1001/jama.295.24.2867 [DOI] [PubMed] [Google Scholar]
- 45. Lutzoni F, Kauff F, Cox CJ, McLaughlin D, Celio G, Dentinger B, Padamsee M, Hibbett D, James TY, Baloch E, Grube M, Reeb V, Hofstetter V, Schoch C, Arnold AE, Miadlikowska J, Spatafora J, Johnson D, Hambleton S, Crockett M, Shoemaker R, Sung G-H, Lücking R, Lumbsch T, O’Donnell K, Binder M, Diederich P, Ertz D, Gueidan C, Hansen K, Harris RC, Hosaka K, Lim YW, Matheny B, Nishida H, Pfister D, Rogers J, Rossman A, Schmitt I, Sipman H, Stone J, Sugiyama J, Yahr R, Vilgalys R. 2004. Assembling the fungal tree of life: progress, classification, and evolution of subcellular traits. Am. J. Bot. 91:1446–1480. 10.3732/ajb.91.10.1446 [DOI] [PubMed] [Google Scholar]
- 46. Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- 47. Gnerre S, MacCallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, Sharpe T, Hall G, Shea TP, Sykes S, Berlin AM, Aird D, Costello M, Daza R, Williams L, Nicol R, Gnirke A, Nusbaum C, Lander ES, Jaffe DB. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. U. S. A. 108:1513–1518. 10.1073/pnas.1017351108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43:491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee SC, Li A, Calo S, Heitman J. 2013. Calcineurin plays key roles in the dimorphic transition and virulence of the human pathogenic zygomycete Mucor circinelloides. PLoS Pathog. 9:e1003625. 10.1371/journal.ppat.1003625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mylonakis E, Moreno R, El Khoury JB, Idnurm A, Heitman J, Calderwood SB, Ausubel FM, Diener A. 2005. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect. Immun. 73:3842–3850. 10.1128/IAI.73.7.3842-3850.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ibrahim AS, Avanessian V, Spellberg B, Edwards JE., Jr. 2003. Liposomal amphotericin B, and not amphotericin B deoxycholate, improves survival of diabetic mice infected with Rhizopus oryzae. Antimicrob. Agents Chemother. 47:3343–3344. 10.1128/AAC.47.10.3343-3344.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Waldorf AR, Ruderman N, Diamond RD. 1984. Specific susceptibility to mucormycosis in murine diabetes and bronchoalveolar macrophage defense against Rhizopus. J. Clin. Invest. 74:150–160. 10.1172/JCI111395 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note from a couple in Corpus Christi, TX, who consumed the mold-contaminated yogurt. Download
A plain Chobani yogurt contaminated with M. circinelloides. The yogurt was obtained from Corpus Christi, TX. The small rectangular insert shows the yogurt “best by” date to be 30 September 2013 with IMS code 16-012. This was within the time range for yogurt subject to recall by the manufacturer. Download
Inflammatory cytokine production from the THP-1 cell line induced by Mucor. Luminex analysis revealed that the M. circinelloides f. lusitanicus isolate R7B induced the production of IFN-α2, IL-8, MCP-1, and MIP-1α. On the other hand, the 2 M. circinelloides f. circinelloides isolates Mucho and 1006PhL only induced IFN-α2 and not IL-8, MCP-1, or MIP-1α, indicating that different host responses might be involved in the different virulence outcomes in mammalian hosts. Download
Putative genes for secondary metabolites conserved in 1006PhL and Mucho. Genes encoding putative secondary metabolites are found in the 1006PhL and Mucho genomes. Several genes, predominantly encoding those involved in terpene biosynthesis (HMPREF1544_00936, HMPREF1544_00937, HMPREF1544_01407, HMPREF1544_02563, HMPREF1544_08724, HMPREF1544_08726, and HMPREF1544_10038) are highly conserved between the two genomes, with only synonymous SNPs or one nonsynonymous SNP. However, other putative proteins encoded in the Mucho genome vary, with substantial nonsynonymous SNPs compared to those of 1006PhL. SNPs in introns are not depicted, and several genes had changes in a stop or start codon location by 1 or 2 amino acids that did not alter the reading frame (HMPREF1544_08551, HMPREF1544_08721, HMPREF1544_08722, HMPREF 1544_08551, and HMPREF_1544). Clustered genes are arranged together in the figure. Nrps, nonribosomal polyketide synthetase. Download
Yogurt samples and fungal isolates
Recovery of Mucho from feces of infected mice.
Cytokines tested in this study.