Abstract
Many studies have reported the EGFR mutations in Chinese non small cell lung cancer (NSCLC) patients and their relationship with clinicopathological characteristics. But the frequency and type of EGFR mutations are varied. The relationship between EGFR mutations and clinicopathological characteristics remains unclear. We systematically reviewed studies of EGFR mutations in mainland Chinese NSCLC patients. Standard statistical methods for meta-analysis were applied. A total of 5,442 patients from 21 studies were included in the meta-analysis. The overall EGFR mutation rate was 37.5% (2,039/5,442). Among those 1,935 patients with detailed data about EGFR mutation types, the most prevalent mutation type was L858R, which accounted for 38.3% of all EGFR mutations. EGFR-TKIs sensitive mutations occupied 88.5% of all EGFR mutations. While the acquired EGFR-TKIs resistant mutations (T790 M) could occur naturally at the frequency of 1.5% without exposure of EGFR-TKIs. Male patients had a lower mutation rate than female patients (32.6% versus 53.0%, OR=0.40, 95% CI: 0.34-0.48, P < 0.001). The EGFR mutation rate for smokers was 19.2%, lower than that for non-smokers (47.9%) (OR = 0.40, 95% CI: 0.34-0.48, P < 0.001). Patients with adenocarcinoma had a higher mutation rate than those with non-adenocarcinoma (50.2% versus 17.0%, OR = 4.84, 95% CI: 4.07-5.75, P < 0.001). EGFR mutation rate did not differ significantly between stage I and stage II/III/IV patients (35.8% versus 32.2%, OR = 1.08, 95% CI: 0.84-1.43, P = 0.580). Our results show that EGFR mutation rate is high in mainland Chinese NSCLC patients. EGFR-TKIs sensitive mutations are main types of mutations. EGFR mutations are significantly associated with gender, smoking history, histology type, but not stage.
Keywords: Non-small cell lung cancer, EGFR, mainland China, targeted therapy, clinicopathological features, mutation analysis
Introduction
Lung cancer is a leading cause of mortality in both the developing and developed worlds, with ~1.18 million deaths reported every year [1]. Non-small cell lung cancer (NSCLC) accounts for ~85% of all lung cancers and about 70% of patients with NSCLC are primarily diagnosed at late stage [1], which results in poor prognosis.
It was discovered that epidermal growth factor receptor (EGFR) mutations are related to the pathogenesis of NSCLC. EGFR, a member of receptor tyrosine kinases superfamily, can bind to extracellular ligands, dimerize with each other and auto-phosphorylate [2]. Then, EGFR acts as a key regulatory molecule in cellular signaling pathways, promoting tumor cell proliferation, invasion and metastasis [3]. In NSCLC, EGFR is over-expressed by 40 to 80% and the expression levels are correlated with the EGFR tyrosine kinase domain mutations [4]. Recently, EGFR tyrosine kinase inhibitors (TKIs) have been developed for the treatment of advanced NSCLC [5]. Unfortunately, only one part of lung cancer patients is sensible to EGFR-TKIs. In 2004, Lynch et al. reported that EGFR mutations are related to the sensitivity of NSCLC to gefitinib [6], which brings NSCLC into the realm of personalized medicine. Subsequently, many publications have revealed that somatic mutations of the EGFR gene constitutively enhanced EGFR tyrosine kinase activity [7,8].
Currently, EGFR-TKIs are recommended to be the second- or third-line agents for advanced NSCLC. Even EGFR-TKIs are recommended as the first-line agents for patients with EGFR mutations [9]. However, phase III trials with EGFR-TKIs in patients with NSCLC had demonstrated that gefitinib did not show any survival gain over placebo. Further analysis found that patients who had never smoked and were of Asian origin seemed to have a significant survival benefit from gefitinib [10]. One possible reason is that this study was conducted in unselected patients with NSCLC. Thus, detection of the EGFR mutation status in patients with NSCLC is necessary for an appropriate use of EGFR-TKIs.
About 10% of European patients with NSCLC harbor EGFR mutations and 31% in Asian [11-13]. However, several studies reported that the frequency of EGFR mutations in mainland China is varied ranged from 19.2% to 55.8% [14-16]. The mutations of EGFR in NSCLC had been founded to be associated with many demographic and clinicopathological factors, including race, gender, smoking status and tumor histology [14]. Shigematsu et al. reported that patients associated with female gender, never smokers, Asian ethnicity and adenocarcinoma histology are sensible to EGFR-TKIs [17]. But results of some publications from Mainland China are inconsistent with those findings [18-20]. So, the relationship between EGFR mutations and clinicopathological features is unclear in patients with NSCLC from mainland China. Given the current knowledge in the field, the EGFR mutation status of NSCLC patients in mainland China have to be considered in the use of EGFR-TKIs. Furthermore, most previous studies on EGFR mutations primarily focused on adenocarcinoma (ADC), few studies have evaluated the EGFR mutations in non-adenocarcinoma NSCLC (NADC) in large scale. Comprehensive review of EGFR mutations is essential for individualized molecular targeted therapy for NSCLC. We thus conducted this meta-analysis of updated data from mainland China to clarify EGFR mutation status and correlations between EGFR mutations and clinicopathologic characteristics in NSCLC patients.
Materials and methods
Search strategy and selection criteria
We searched the PubMed, EMBASE, Cochrane Library as well as Chinese biomedical literature databases to identify all the articles with the combination of the following keywords: “epidermal growth factor receptor mutation” or “EGFR mutation”, “lung” or “pulmonary”, “non-small cell lung cancer” or “NSCLC” or “adenocarcinoma” or “squamous cell carcinoma” and “China” or “Chinese”. Meeting abstracts from the American society of Clinical Oncology (ASCO) and World Congress of Lung Cancer (WCLC) were also hand searched. Reference lists of original articles and review articles were also examined. All articles were identified by use of the related-articles function in PubMed. The published languages were English or Chinese and the literature search was conducted up to January 5, 2014.
Results from the initial search that match the criteria below were eligible. (a) The studies must determine EGFR mutation status of primary NSCLC tissues at least including exons 18-21; (b) Individuals with NSCLC must be histologically confirmed; (c) Any patients did not receive chemotherapy or radiotherapy or treatment with EGFR-TKIs before detection of EGFR mutations; (c) The demographic characteristics of pathology, gender, age, smoking history and stage were measured; (d) Studies have no restrictions on the methods of obtaining the tissue specimens and EGFR mutation detection. The studies were excluded if (a) patients that were not from mainland China; (b) patients with small cell lung cancer or other malignancies or benign lung tumors; (c) number of patients with NSCLC less than 40. If the same patient population was used in more than one study, only the complete study would be included.
Data extraction
Two independent authors extracted all data from the identified studies using standardized data compilation forms and disagreements were resolved by discussion to validate the accuracy of extraction. The following information was extracted from all included studies: the first author, year of publication, number of included patients, age, EGFR mutation status, gender, smoking history, histology types, tumor-node- metastasis (TNM) stage, tissue sample and detection method of each study. The numbers of cases with EGFR mutations from each study were collected to evaluate mutation frequency among NSCLC, meanwhile, the mutation types were also collected from some studies who displayed the data.
Patients who had smoked < 100 cigarettes in their lifetime were identified as ‘non-smokers’. Patients who had smoked > 100 cigarettes in their lifetime were identified as ‘smokers’. Smoking status was based on records of original studies. The diagnosis of pulmonary adenocarcinoma (ADC) was based on the histopathological findings, and pulmonary non-adenocarcinoma NSCLC (NADC) included squamous carcinoma, adenosquamous carcinoma, large cell carcinoma, sarcomatoid carcinoma, and mucoepidermoid carcinoma. TNM staging was determined by the records of original studies.
Statistical analysis
The relationship between EGFR mutations and demographic and clinicopathological characteristics was analyzed by Meta-analysis. The X2-based Q test and I2 statistic were performed to quantify heterogeneity. I2 statistic with values > 50% and X2 test with P < 0.05 indicate strong heterogeneity between the studies [21]. The Mantel-Haenszel method and the DerSimonian and Laird method were used to determine the choice of fixed-effects model or the random-effects model. When P value was ≥ 0.05, the fixed-effects model was more appropriate. Otherwise, the random-effects model was used to provide wider confidence interval (CI). The pooled odds ratio (OR) and 95% CI were calculated. The meta-analysis results were displayed as forest plots. Sensitivity analysis was performed by excluding each study at a time individually and recalculating the ORs and CIs. Potential publication bias was analyzed by Begg’s funnel plots and Egger’s linear regression test [22]. P < 0.05 by t test was considered statistically significant publication bias. This meta-analysis was performed by using the software Stata 11.0 (Stata Corporation, College Station, TX). All the P values were two-sided. Differences were considered statistically significant at P < 0.05.
Results
Literature search and study characteristics
Our search in electronic database and meeting abstracts retrieved 463 references. The selection steps were summarized in the flow diagram shown in Figure 1. Twenty one studies were finally enrolled, including 8 studies of full publications in English [14,16,23-28], 9 studies of full publications in Chinese [29-37] and 4 studies of meeting abstracts [15,38-40]. Totally, 5,442 NSCLC patients with individual patient data were meta-analyzed for EGFR mutations in our study. Those patients came from twenty medical centers in Guangzhou, Beijing, Shanghai, Changchun, Tianjin, Nanjing, Urumqi, Xi’an, Xiamen and Chengdu.
Figure 1.
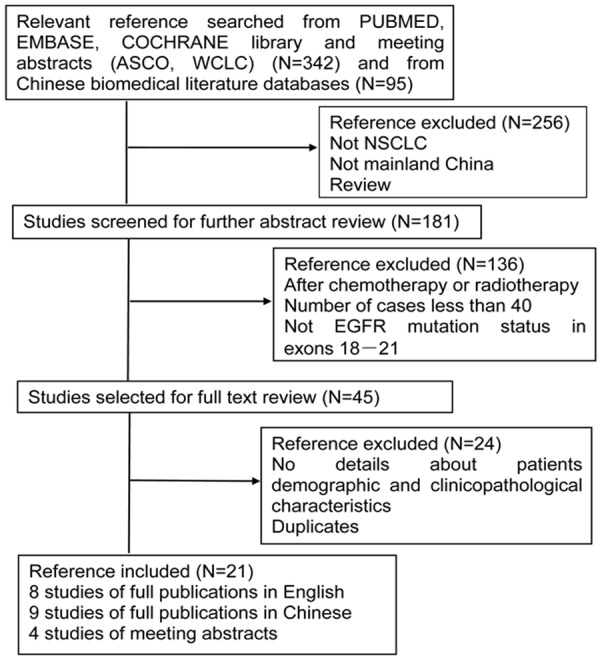
Study flow chart showing process for selecting eligible publications.
Generally, 5,442 enrolled patients included more males than females: the male: female ratio was 1.6:1. Median age was reported in 17 studies, which reported a median age of 55.3-65 years. There were more smokers than non smokers: the smoker: non smoker ratio was 1.1:1. There were also more ADC patients than NADC patients: ADC patients: NADC patient’s ratio was 2.0:1. The ratio of patients in stage I to patients in stage II/III/IV was 0.5:1. Specimens were obtained from formalin-fixed, paraffin-embedded tissues (FFPE) in 19 studies. Only 1 study used snap-frozen tissues in liquid nitrogen (SFILN) analysis and 1 study of fresh tumor tissues detection. As illustrated in Table 1, the most common detection method was sequencing: 14 studies carried out sequencing alone or with other methods. Seven studies used other detection methods: 5 studies of amplification mutation refractory system (ARMS) analysis, 1 study of high resolution melting (HRM) analysis and 1 study of bi-loop probe and specific primer quantitative PCR (BPSP-qPCR). Articles reporting data on EGFR mutations were examined to assess their scope. Screening of EGFR mutations was variable, however, all reports included exons 18-21, where more than 96% of mutations have been reported based on EGFR Mutations Database (http://somaticmutations-egfr.org/NSCLC.html).
Table 1.
Characteristics of studies included in the meta-analysis
| Author, Year | Cases | Age1 | EGFR | Gender | Smoking | Histology | TNM stage | Detection | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||
| M1 | W | M2 | F | Y | N | ADC | NADC | I | II/III/IV | Sample | Method | |||
| Qin B, 2005 [23] | 41 | NA | 10 | 31 | 30 | 11 | 21 | 20 | 17 | 24 | NA | NA | FFPE | SE |
| Xu J, 2011 [24] | 861 | 58 | 353 | 508 | 493 | 368 | 411 | 439 | 667 | 194 | NA | NA | FFPE | SE |
| An S, 2012 [25] | 524 | 59.3 | 147 | 377 | 361 | 163 | 232 | 292 | 354 | 170 | 143 | 381 | SFILN | SE |
| Li Y, 2013 [26] | 208 | NA | 51 | 157 | 147 | 61 | 130 | 78 | 95 | 113 | 49 | 159 | FFPE | SE |
| Jing C, 2013 [27] | 120 | 62 | 48 | 72 | 69 | 51 | NA | NA | 70 | 50 | NA | NA | FFPE | HRM |
| Liu Y, 2013 [16] | 251 | NA | 140 | 111 | 138 | 113 | NA | NA | 193 | 58 | NA | NA | FFPE | SE |
| Zhang Y, 2013 [28] | 99 | NA | 33 | 66 | 68 | 31 | 43 | 56 | 54 | 45 | 16 | 83 | FFPE | ARMS |
| Lai Y, 2013 [14] | 697 | 55.3 | 235 | 462 | 476 | 221 | 366 | 331 | 293 | 404 | 215 | 482 | Fresh tissues | ARMS |
| Zhang J, 2008 [29] | 82 | 61 | 42 | 40 | 48 | 34 | 40 | 42 | 59 | 23 | 44 | 38 | FFPE | ARMS |
| Liang Z, 2008 [30] | 290 | 58 | 121 | 169 | 145 | 145 | NA | NA | 244 | 46 | NA | NA | FFPE | ARMS |
| Yin X, 2010 [31] | 107 | 61 | 30 | 77 | 82 | 25 | 60 | 47 | 52 | 55 | 44 | 63 | FFPE | SE |
| Feng Q, 2011 [32] | 309 | 59.7 | 105 | 204 | 184 | 125 | 132 | 127 | 201 | 99 | NA | NA | FFPE | SE |
| Sun M, 2011 [33] | 443 | 57 | 189 | 254 | 243 | 200 | NA | NA | 346 | 97 | NA | NA | FFPE | SE |
| Zhang J, 2011 [34] | 454 | 59 | 219 | 235 | 256 | 198 | 279 | 175 | 340 | 114 | NA | NA | FFPE | SE |
| Wang L, 2011 [35] | 184 | 58.5 | 69 | 115 | 106 | 78 | 79 | 105 | 135 | 49 | NA | NA | FFPE | BPSP |
| Gao J, 2012 [36] | 120 | 65 | 44 | 76 | 77 | 43 | 60 | 60 | 93 | 27 | NA | NA | FFPE | ARMS |
| Sun L, 2013 [37] | 301 | 59 | 99 | 202 | 174 | 127 | 173 | 128 | 188 | 113 | NA | NA | FFPE | SE |
| Zhang X, 2005 [38] | 76 | 62.5 | 31 | 45 | 41 | 35 | NA | NA | 51 | 25 | NA | NA | FFPE | SE |
| Wu Y, 2005 [15] | 142 | 60 | 42 | 100 | 98 | 44 | 84 | 58 | 90 | 52 | 60 | 82 | FFPE | SE |
| Zhang L, 2005 [39] | 52 | 58 | 10 | 42 | 39 | 13 | 33 | 19 | 28 | 24 | NA | NA | FFPE | SE |
| Zhou C, 2005 [40] | 81 | 62 | 21 | 60 | 52 | 29 | 41 | 40 | 38 | 43 | 34 | 47 | FFPE | SE |
| Total | 5442 | — | 2039 | 3403 | 3327 | 2115 | 2184 | 2017 | 3608 | 1825 | 605 | 1335 | — | — |
Abbreviations: Age1: median age; M1: mutant type EGFR; W: wild type EGFR; M2: male; F: female; Y: smoker; N: non-smoker; ADC: adenocarcinoma; NADC: non-adenocarcinoma NSCLC; NA: not available; FFPE: formalin-fixed, paraffin-embedded tissues; SFILN: snap-frozen tissues in liquid nitrogen; SE: sequencing; HRM: high resolution melting analysis; ARMS: amplification mutation refractory system; BPSP: bi-loop probe and specific primer quantitative PCR.
EGFR mutation patterns
Mutations in the EGFR gene exons 18-21 were detected in 2,039 of 5,442 NSCLC patients. The overall EGFR mutation rate was 37.5%. The data from meeting abstracts lacked information about mutation types. Types of EGFR mutation were available from 1,935 patients. As shown in Figure 2A, the distribution of EGFR mutations was that 66 were in exon 18 (3.4%), 906 were in exon 19 (46.8%), 71 were in exon 20 (3.7%) and 812 were in exon 21 (42.0%). There were 80 patients (2.9%) that had double or multiple mutations. The most common type of double or multiple mutations was mutations in exons 19 and 21 (25.0%, 20/80). Details of the EGFR mutations are comprehensively reviewed from eligible studies. There were 73 kinds of EGFR mutations in exons 18-21. Figure 2B showed the most common 27 kinds of mutations. In mainland Chinese patients with NSCLC, the most prevalent mutation types were L858R in exon 21 and del(746-750) in exon 19, which accounted for 38.3% (741/1935) and 37.0% (716/1935) of all EGFR mutations, respectively. In exon 18, G719C was dominant mutation type (43.9%, 29/66). Del(746-A750), del(747-749)ins(S), del(747-751), del(746-752), del(747-749)ins(P) and del(747-753) were the six major in-frame deletions in exon 19, which occupied 90.4% (819/906) of exon 19 mutations. T790M and insertion mutations accounted for 71.8% of all exon 20 mutations. The four major mutations (87.5%, 792/812) in exon 21 were L858R, L858M, L861R and L861Q. The rate of EGFR-TKI sensitive mutations was 88.5% (1713/1935), including exon 19 deletion, L858R, L861Q, G719X (G719C, G719S, G719A). The rate of EGFR-TKI resistant mutations was 2.6% (76/1935), including exon 20 insertion and T790M.
Figure 2.
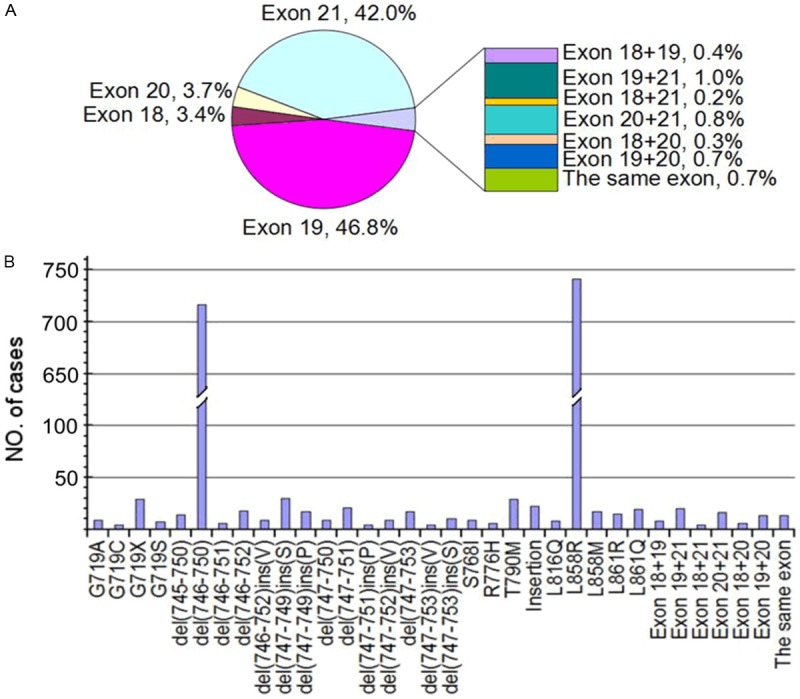
EGFR mutations and their frequency in NSCLC. A. Pie-chart of EGFR mutation frequency by exon in 2039 patients. B. EGFR mutation hotspots from exon 18 to 21 in 1935 patients.
EGFR mutations and patient characteristics
In meta-analysis, we chose 0.01 to 100 as the logarithmic scale (Figure 1). For the comparison between male patients and female patients, ten studies were excluded because patient’s data about gender is patchy. All the other eleven studies [15,16,23,27-30,33,34,36,37] reported a significant difference between male patients and female patients except Qin B, et al [23] and Wu Y, et al [15]. Heterogeneity analysis revealed that there was no significant between-study heterogeneity in those 11 studies (chi-squared = 8.51 (d.f. = 0), P = 0.579, I-squared = 0.0%). So, Meta-analysis was performed using fixed-effects model. Overall, male patients with NSCLC had a significantly lower EGFR mutation rate (32.6%, 439/1346) than female patients (53.0%, 548/1033). The pooled OR and 95% CI was 0.40 (0.34-0.48) (P < 0.001) (Figure 3A).
Figure 3.
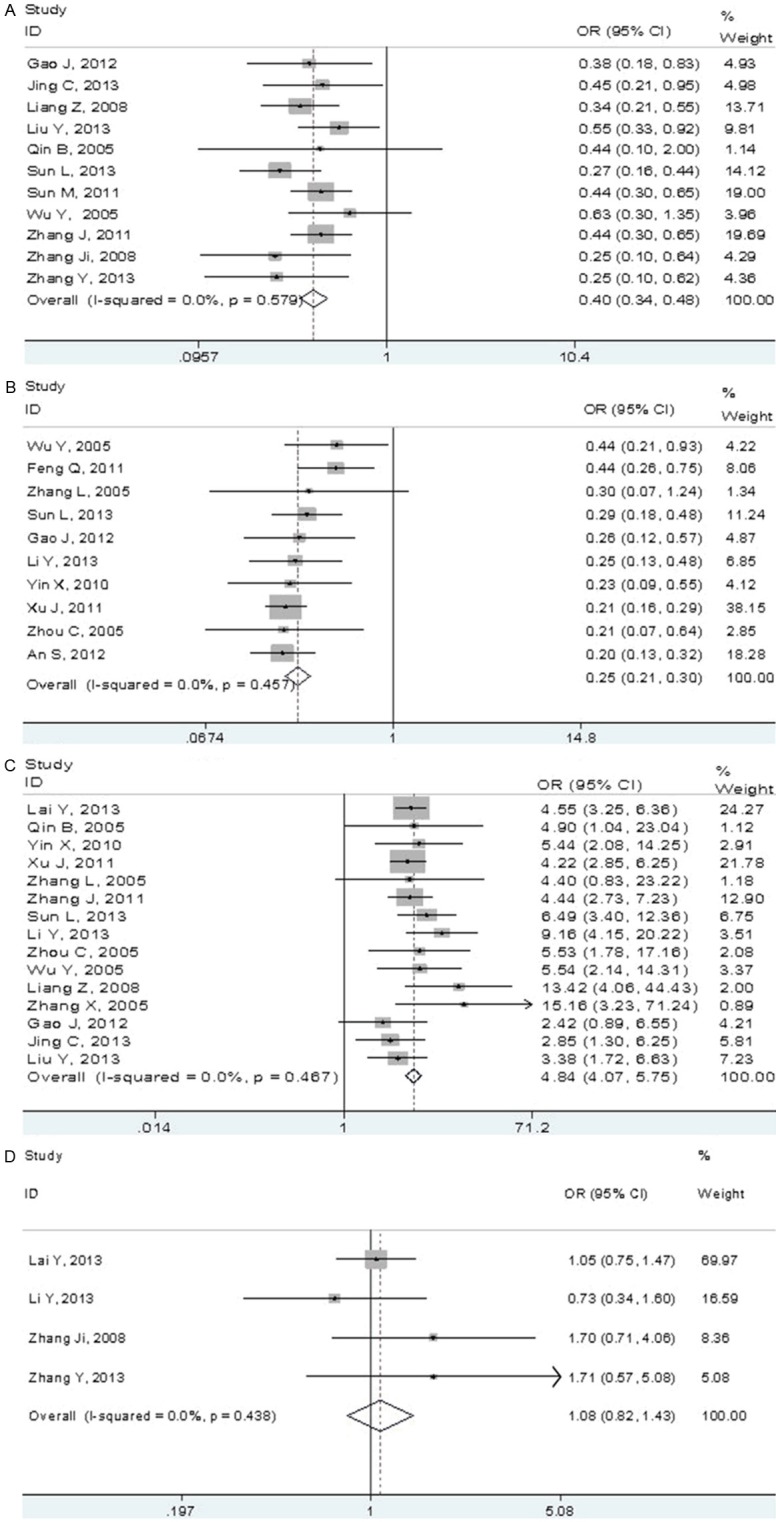
Forest plots of relationship between EGFR mutations and the NSCLC patient characteristics. A. Fixed-effects meta-analysis of EGFR mutations with gender (male versus female). B. Fixed-effects meta-analysis of EGFR mutations with smoking history (smoking versus non-smoking). C. Fixed-effects meta-analysis of EGFR mutations with histology type (adenocarcinoma versus non-adenocarcinoma). D. Fixed-effects meta-analysis of EGFR mutations with tumor stage (stage I versus stages II/III/IV).
Associations of EGFR mutations with smoking history were suggested in 10 studies, but 11 studies did not provide the required data. Ten studies were analyzed to examine the relationship between EGFR mutations and smoking history [15,24-26,31,32,36,37,39,40]. For heterogeneity test, chi-squared was 8.79 (d.f. = 9, P = 0.457) and I-squared was 0.0%. Smokers had a lower EGFR mutation rate (19.2%, 259/1350) than non-smokers (47.9%, 617/1287), with the OR and 95% CI being 0.25 (0.21-0.35) (P < 0.001) (Figure 3B).
Fifteen studies were eligible to estimate the relationship between EGFR mutations and histology type [14-16,23,24,26,27,30,31,34,36-40]. Six studies were excluded because of incomplete information. Between-study heterogeneity could be ignored (chi-squared = 13.77 (d.f. = 14), P = 0.467, I-squared =0.0%). Patients with ADC had a higher EGFR mutation rate (50.2%, 1238/2468) than NADC (17.0%, 226/1333). The pooled OR and 95% CI was 4.84 (4.07-5.75) (P < 0.001) (Figure 3C).
To further determine the relationship between EGFR mutations and clinic feature of patients, we then compared the frequencies of EGFR mutations in stage I patients and stage II/III/IV. All patients were broadly grouped into stage I or stage II/III/IV groups based on Zhang Y, et al. [28]. Four studies provided information in detail about stage of patients, of which a crude OR was calculated [14,26,28,29]. For heterogeneity test, chi-squared was 2.71 (d.f. = 3, P = 0.438) and I-squared was 0.0%. There were EGFR mutations in 116/324 (35.8%) of stage I patients, in 245/762 (32.2%) of stage II/III/IV patients. The pooled OR and 95% CI was 1.08 (0.82-1.43) (P = 0.580) (Figure 3D).
Sensitivity analyses
We performed sensitivity analysis to assess the stability of the results of this meta-analysis by sequentially omitting each study. The leave-one-out sensitivity analysis indicated that no individual study changed the pooled ORs qualitatively, suggesting that our results were stable and reliable (Figure 4).
Figure 4.
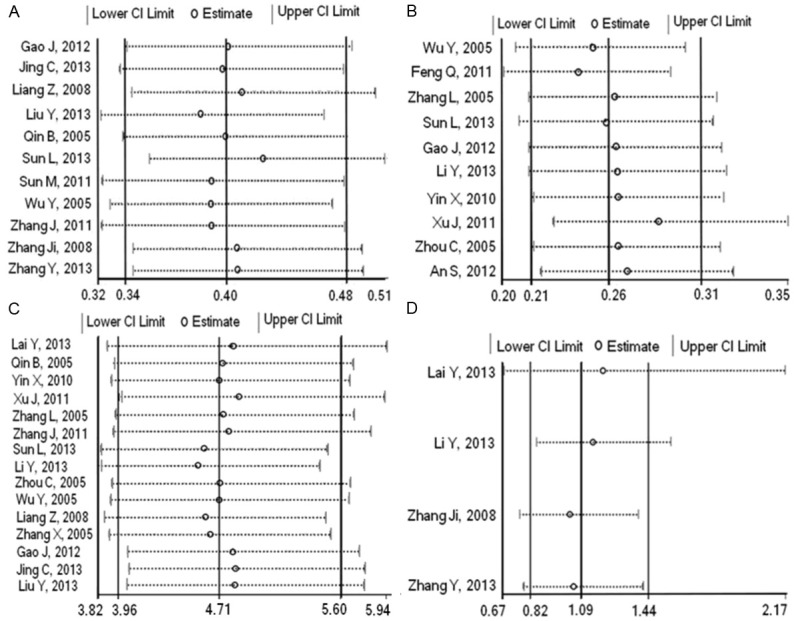
Results of sensitivity analysis. Sensitivity analysis of studies about association between EGFR mutations and gender (A), smoking history (B), histology type (C) and tumor stage (D).
Publication bias
The Begg’s funnel plot and Egger’s test were conducted to assess the publication bias of studies. The shapes of the funnel plots seemed symmetrical in all Meta-analyses, suggesting the absence of publication bias (Figure 5). Then, the Egger’s test was performed to provide statistical evidence of funnel plot symmetry (Table 2). The results indicated no significant evidence for publication bias of the current Meta-analyses. So, bias from publications might not have a significant influence on the results of our meta-analyses.
Figure 5.
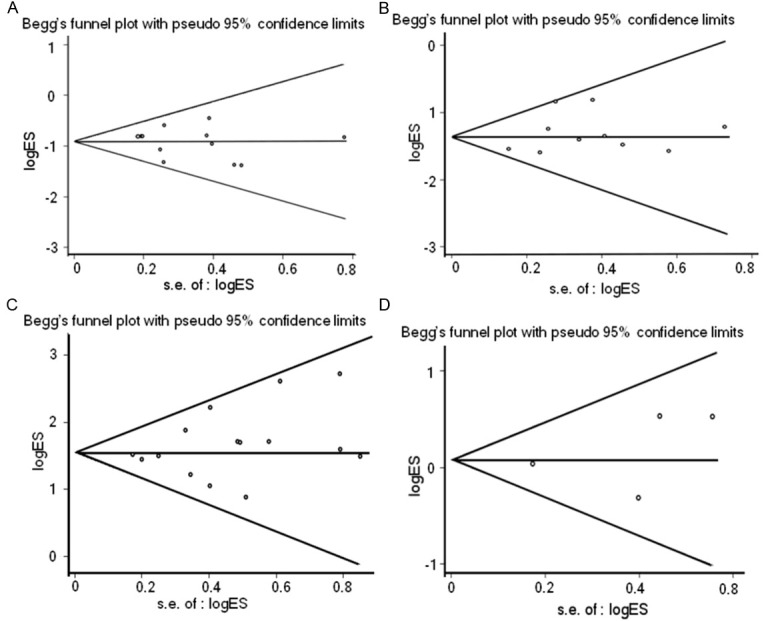
Results of Begg’s funnel plots. Begg’s funnel plots with pseudo 95% CIs of relationship between EGFR mutations and gender (A), smoking history (B), histology type (C) and tumor stage (D).
Table 2.
Summary of publication bias test
| Begg’s test | Egger’s test | |||
|---|---|---|---|---|
|
|
||||
| Z | P | t (bias) | P | |
| EGFR Mutations and gender | 0.78 | 0.436 | -0.60 | 0.564 |
| EGFR Mutations and smoking history | 0.36 | 0.721 | 1.02 | 0.339 |
| EGFR Mutations and histology type | 0.59 | 0.553 | 1.25 | 0.234 |
| EGFR Mutations and stage | 0.34 | 0.734 | 0.64 | 0.590 |
Discussion
The present report is a meta-analysis with updated data improving our understanding about the EGFR mutation status in NSCLC in mainland China, which may be helpful in targeting patients for better responses to EGFR-TKIs therapy. In this meta-analysis of 5,442 NSCLC cases, 2,039 patients were positive in EGFR mutations. The overall EGFR mutation rate was 37.5%. Thus far, many studies have reported mutational analyses for EGFR gene all over the world. Generally, patients of Asian origin have a higher prevalence compared with Europeans [11-13]. But overall EGFR mutation rate differed widely among Asian regions: 31% in Japan [13], 18.9% in South Korea [41], 27.5% in Taiwan [42], 34.3% in Hong Kong [43]. We found the EGFR mutation rate in unselected cases of NSCLC was 37.5% in mainland China, which was higher than the results from previous Asian population-based studies.
Among those 73 kinds of EGFR mutations in exons 18-21, L858R point mutation was the most major type of EGFR mutations in mainland Chinese patients with NSCLC. The frequency was consistent with report of Chan SK et al. [44]. But it is not consistent with previous Asian population-based studies [13], which demonstrated that del(746-750) mutation was more frequent. It could be due to ethnic variations between Chinese and other populations or sampling errors. Large-scale studies have demonstrated that patients with EGFR-mutant NSCLC had better therapeutic response to TKIs, because NSCLC patients harbor EGFR-TKI sensitive mutations such as exon 19 deletion, L858R, L861Q, G719X (such as G719C, G719S, G719A) [7]. In mainland Chinese NSCLC patients, EGFR-TKIs sensitive mutations occupied 88.5% (1713/1935) of all EGFR mutations. That is why patients of Chinese origin seemed to have a significant survival benefit from EGFR-TKIs [10,17].
It must be noted that two major EGFR-TKIs resistant mutations (exon 20 insertion and T790M) were found in mainland Chinese patients with NSCLC prior to any treatment, although they were much less common with 2.6% (76/1935) of EGFR mutations. Exon 20 insertion is a kind of primary TKIs resistance mutation. It is associated with activation of EGFR kinase domain, affinity of EGFR to TKIs, and carcinogenesis of NSCLC. Patients with NSCLC harboring exon 20 insertion mutations should receive irreversible inhibitors rather than gefitinib or erlotinib. T790M is a kind of acquired resistance mutation against TKIs, which could enhance the affinity of TKIs to ATP [45]. In our study, T790M mutation was found in mainland Chinese patients with NSCLC at the frequency of 1.5% (29/1935) before treatment with TKIs. It indicates that T790M might also lead to primary resistance against TKIs. Under the selective pressure imposed by TKIs, T790M mutation may expand and lead to TKIs resistance. Patients harboring T790M mutation had worse prognosis than those without T790M mutation [7]. Thus, the identification of resistant EGFR mutations is as important as the identification of TKIs sensitive ones. There were also many types of rare EGFR mutations, such as double or multiple mutations and specific mutations reported only in mainland China. However, it needs further research to confirm their function in the treatment of NSCLC.
We also found the EGFR mutations were associated with gender, smoking history, and histology type, but not stage of NSCLC patients (Figure 3). Although the included studies involved different detection methods and tissue samples and the number of cases varied widely, between-study heterogeneity did not have a substantial influence on the results of our meta-analysis (I-squared = 0.0%). Sensitivity analyses also suggested the stability of this meta-analysis (Figure 4). In concordance with previous reports [10,14,17], EGFR mutations are identified at high frequencies in females, never-smokers and adenocarcinoma patients. In mainland China, the EGFR mutations rate was lower in men (OR = 0.40, 95% CI, 0.34-0.48; P < 0.001), smokers (OR = 0.25, 95% CI, 0.21-0.30; P < 0.001), and higher in patients with ADC (OR = 4.84, 95% CI, 4.07-5.75; P < 0.001). This may own to the relationship between NSCLC and smoking, as well as the lifestyle pattern of Chinese women. About 94% mainland Chinese women were non-smokers. Eighty-three percent of Chinese women with lung cancer do not have smoking history [46]. Therefore, both gender and smoking history are confounding factors of affecting EGFR mutations. Moreover, the frequency of EGFR mutations decreased as pack-years increased. EGFR mutations were less common in patients who smoked for more than 15 pack-years or stopped smoking less than 25 years [47]. Thus, TKIs therapy should not be confined to non-smokers with NSCLC, but expanded to former smokers who are less than 15 pack-years or more than 25 years smoke-free. Patients with ADC harbored much higher mutation rate than those with NADC (50.2% versus 17.0%, P < 0.001). The pooled OR and 95% CI was 4.84 (4.07-5.75). So ADC may be used as the prior clinical predictor for EGFR mutations.
In addition, our study failed to find a significant association between the EGFR mutations and tumor stage (OR = 1.08, 95% CI, 0.82-1.43; P = 0.580), which was different from the study of Li Z et al. [19]. Li Z et al. reported that EGFR gene mutations were more common in Chinese NSCLC patients advanced stage [19]. Mutation detection methods, specimen types or sampling errors are likely to be the underlying causes of the discordance between our study and Li Z’s. However, our study was consistent with most previous reports [14,26,28].
Although Begg’s funnel plot and Egger’s test both indicated the publication bias was not significant (Figure 5, Table 2), it is possible that some relevant publications or unpublished researches or publications in other languages were missed, which might place a bias on our results. There is some evidence that age and tumor differentiation could alter the EGFR mutation rate [32,36]. However, due to the lack of original data in the publications, we didn’t analyze these factors. Accurate OR should be calculated by age, tumor differentiation and other exposure factors. Due to heterogeneses of EGFR detection methods and technical limitations, it is difficult to unify the depth of sequencing, and the quality of samples. Therefore, the findings of relationship between EGFR mutations and clinicopathological features in this meta-analysis still require further replication with more precise analysis and larger studies to avoid the drawbacks.
Given the heightened interest in targeted therapy against NSCLC, analysis of the EGFR mutation profile from mainland China should be significant. China has the largest pool of NSCLC patients harboring EGFR mutations in the world, representing a fascinating opportunity for the development of TKIs against NSCLC. Therapeutically, mainland Chinese patients carrying the sensitive mutations should respond well to TKIs. Furthermore, most mutations are naturally occurring without exposure to TKIs. Detection of sensitive or resistant EGFR mutations should help to elucidate how different mutations might affect function, dimerization and TKIs sensitivity. Identification of associations between EGFR mutations and clinicopathological features is also conducive to predict the EGFR mutations and identify patients who would respond favorably to EGFR inhibitors.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37:S9–S15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 3.Scaltriti M, Baselga J. The epidermal growth factor receptor pathway: a model for targeted therapy. Clin Cancer Res. 2006;12:5268–5272. doi: 10.1158/1078-0432.CCR-05-1554. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki M, Shigematsu H, Hiroshima K, Iizasa T, Nakatani Y, Minna JD, Gazdar AF, Fujisawa T. Epidermal growth factor receptor expression status in lung cancer correlates with its mutation. Hum Pathol. 2005;36:1127–1134. doi: 10.1016/j.humpath.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Kris MG, Natale RB, Herbst RS, Lynch TJ Jr, Prager D, Belani CP, Schiller JH, Kelly K, Spiridonidis H, Sandler A, Albain KS, Cella D, Wolf MK, Averbuch SD, Ochs JJ, Kay AC. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA. 2003;290:2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- 6.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non small cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 7.Mitsudomi T, Yatabe Y. Mutations of the epidermal growth factor receptor gene and related genes as determinants of epidermal growth factor receptor tyrosine kinase inhibitors sensitivity in lung cancer. Cancer Sci. 2007;98:1817–1824. doi: 10.1111/j.1349-7006.2007.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 9.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, Fujita Y, Okinaga S, Hirano H, Yoshimori K, Harada T, Ogura T, Ando M, Miyazawa H, Tanaka T, Saijo Y, Hagiwara K, Morita S, Nukiwa T North-East Japan Study Group. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 10.Thatcher N, Chang A, Parikh P, Rodrigues Pereira J, Ciuleanu T, von Pawel J, Thongprasert S, Tan EH, Pemberton K, Archer V, Carroll K. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer) Lancet. 2005;366:1527–1537. doi: 10.1016/S0140-6736(05)67625-8. [DOI] [PubMed] [Google Scholar]
- 11.Tsao AS, Tang XM, Sabloff B, Xiao L, Shigematsu H, Roth J, Spitz M, Hong WK, Gazdar A, Wistuba I. Clinicopathologic characteristics of the EGFR gene mutation in non-small cell lung cancer. J Thorac Oncol. 2006;1:231–239. doi: 10.1016/s1556-0864(15)31573-2. [DOI] [PubMed] [Google Scholar]
- 12.Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, Majem M, Lopez-Vivanco G, Isla D, Provencio M, Insa A, Massuti B, Gonzalez-Larriba JL, Paz-Ares L, Bover I, Garcia-Campelo R, Moreno MA, Catot S, Rolfo C, Reguart N, Palmero R, Sánchez JM, Bastus R, Mayo C, Bertran-Alamillo J, Molina MA, Sanchez JJ, Taron M Spanish Lung Cancer Group. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka T, Matsuoka M, Sutani A, Porta R, Cardenal F, Camps C, Majem M, Lopez-Vivanco G, Isla D, Provencio M, Insa A, Massuti B, Gonzalez-Larriba JL, Paz-Ares L, Bover I, Garcia-Campelo R, Moreno MA, Catot S, Rolfo C, Reguart N, Palmero R, Sánchez JM, Bastus R, Mayo C, Bertran-Alamillo J, Molina MA, Sanchez JJ, Taron M Spanish Lung Cancer Group. Frequency of and variables associated with the EGFR mutation and its subtypes. Int J Cancer. 2010;126:651–655. doi: 10.1002/ijc.24746. [DOI] [PubMed] [Google Scholar]
- 14.Lai Y, Zhang Z, Li J, Sun D, Zhou Y, Jiang T, Han Y, Huang L, Zhu Y, Li X, Yan X. EGFR Mutations in Surgically Resected Fresh Specimens from 697 Consecutive Chinese Patients with Non-Small Cell Lung Cancer and Their Relationships with Clinical Features. Int J Mol Sci. 2013;14:24549–24559. doi: 10.3390/ijms141224549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Y, Lin J, Wang K. EGFR mutations in lung cancers and sensitivity to gefitinib in Chinese (Abstract) J. Clin. Oncol. 2005;23:7089. [Google Scholar]
- 16.Liu Y, Wu BQ, Zhong HH, Hui P, Fang WG. Screening for EGFR and KRAS mutations in non-small cell lung carcinomas using DNA extraction by hydrothermal pressure coupled with PCR-based direct sequencing. Int J Clin Exp Pathol. 2013;6:1880–1889. [PMC free article] [PubMed] [Google Scholar]
- 17.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, Fong KM, Lee H, Toyooka S, Shimizu N, Fujisawa T, Feng Z, Roth JA, Herz J, Minna JD, Gazdar AF. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 18.Guo J, Zhou C, Zhang J, Tang L. Relationship between mutations of epidermal growth factor receptor in the plasma and pleural effusion and responses to gefitinib in advanced pretreated non-small cell lung cancer. Zhongguo Fei Ai Za Zhi. 2007;10:504–507. doi: 10.3779/j.issn.1009-3419.2007.06.12. [DOI] [PubMed] [Google Scholar]
- 19.Li Z, Zhang LJ, Wang WP, Guo K, Shao JY, Rong TH. Correlation between EGFR gene mutation and high copy number and their association with the clinicopathological features in Chinese patients with non-small cell lung cancer. Zhonghua Zhong Liu Za Zhi. 2011;33:666–670. [PubMed] [Google Scholar]
- 20.Han Y, Xu JM, Duan HQ, Song ST, Liu XQ, Zhang Y, Zhang JS. EGFR mutation predicts response and prognosis in iressa-treated advanced-stage non-small cell lung cancer. Zhonghua Zhong Liu Za Zhi. 2007;29:278–283. [PubMed] [Google Scholar]
- 21.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 22.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin BM, Chen X, Zhu JD, Pei DQ. Identification of EGFR kinase domain mutations among lung cancer patients in China: implication for targeted cancer therapy. Cell Res. 2005;15:212–217. doi: 10.1038/sj.cr.7290289. [DOI] [PubMed] [Google Scholar]
- 24.Xu J, He J, Yang H, Luo X, Liang Z, Chen J, Cai Z, Xu J, Ren-Heidenreich L. Somatic mutation analysis of EGFR, KRAS, BRAF and PIK3CA in 861 patients with non-small cell lung cancer. Cancer Biomark. 2011-2012;10:63–69. doi: 10.3233/CBM-2012-0233. [DOI] [PubMed] [Google Scholar]
- 25.An SJ, Chen ZH, Su J, Zhang XC, Zhong WZ, Yang JJ, Zhou Q, Yang XN, Huang L, Guan JL, Nie Q, Yan HH, Mok TS, Wu YL. Identification of enriched driver gene alterations in subgroups of non-small cell lung cancer patients based on histology and smoking status. PLoS One. 2012;7:e40109. doi: 10.1371/journal.pone.0040109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Li Y, Wang T, Wei S, Wang J, Wong M, Wang Y, Zhou Q, Liu H, Chen J. Clinical significance of EML4-ALK fusion gene and association with EGFR and KRAS gene mutations in 208 Chinese patients with non-small cell lung cancer. PLoS One. 2013;8:e52093. doi: 10.1371/journal.pone.0052093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jing CW, Wang Z, Cao HX, Ma R, Wu JZ. High resolution melting analysis for epidermal growth factor receptor mutations in formalin-fixed paraffin-embedded tissue and plasma free DNA from non-small cell lung cancer patients. Asian Pac J Cancer Prev. 2013;14:6619–6623. doi: 10.7314/apjcp.2013.14.11.6619. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Wang Q, Han ZG, Shan L. Differences in epidermal growth factor receptor gene mutations and relationship with clinicopathological features in NSCLC between Uygur and Han ethnic groups. Asian Pac J Cancer Prev. 2013;14:2879–2883. doi: 10.7314/apjcp.2013.14.5.2879. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J, Liang ZY, Zeng X, Wu SF, Gao J, Liu TH. Detection of epidermal growth factor receptor gene mutations in non-small cell lung cancers by real-time polymerase chain reaction using scorpion amplification refractory mutation system. Zhonghua Bing Li Xue Za Zhi. 2008;37:294–299. [PubMed] [Google Scholar]
- 30.Liang ZY, Zeng X, Zhang J, Wu SF, Gao J, Liu TH. Status of gene mutation and copy number of EGFR in 290 cases of non-small cell lung carcinoma. Zhonghua Bing Li Xue Za Zhi. 2008;37:654–659. [PubMed] [Google Scholar]
- 31.Yin XW, Jiang XT, Yuan YT, Du YP. Influence of mutations in epidermal growth factor receptor gene on growth, metastasis and survival rate of non-small cell lung carcinoma. Zhonghua Yi Xue Za Zhi. 2010;90:1808–1812. [PubMed] [Google Scholar]
- 32.Feng Q, Li XH, Chen Z, He JS, Wang CX, Zhou LX, Xue WC. Epidermal growth factor receptor gene mutations and clinicopathologic correlation in 309 patients with non-small cell lung cancer. Zhonghua Bing Li Xue Za Zhi. 2011;40:660–663. [PubMed] [Google Scholar]
- 33.Sun MH, Yang F, Shen L, Zhang L, Chen Y, Cai X, Zhu XL, Zhou XY. Detection of epidermal growth factor receptor mutations in non-small-cell lung carcinoma by direct sequencing and correlations with clinicopathological characteristics and sample types. Zhonghua Bing Li Xue Za Zhi. 2011;40:655–659. [PubMed] [Google Scholar]
- 34.Zhang J, Wu J, Gao H, Zhu L, Shao JC, Shi MP, Han BH. Epidermal growth factor receptor gene mutations, amplification and clinicopathologic correlation in patients with lung cancer. Zhonghua Bing Li Xue Za Zhi. 2011;40:675–678. [PubMed] [Google Scholar]
- 35.Wang LS, Zhang Y, Lu XJ, Lu HJ, Zhou L, Wang YS, Deng L, Huang MJ, Peng F, Wang J, Ren L, Hou M, Li L, Xu Y, Ying BW, Lu Y. Detection of epidermal growth factor receptor gene mutations in non-small cell lung cancer using bi-loop probe specific primer quantitative PCR. Zhonghua Bing Li Xue Za Zhi. 2011;40:667–670. [PubMed] [Google Scholar]
- 36.Gao J, Chen JQ, Zhang L, Liang ZY. Relationship between EGFR and KRAS mutations and prognosis in Chinese patients with non-small cell lung cancer: a mutation analysis with real-time polymerase chain reaction using scorpion amplification refractory mutation system. Zhonghua Bing Li Xue Za Zhi. 2012;41:652–656. doi: 10.3760/cma.j.issn.0529-5807.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Sun LN, Luan HL, Zang FL, Wang M, Dong N, Guo Y, Sun BC, Zhan ZL. Relationship between EGFR and K-ras mutations and clinicopathological characteristics and response to erlotinib treatment in 301 Chinese patients with non-small cell lung cancer. Zhonghua Zhong Liu Za Zhi. 2010;32:667–670. [PubMed] [Google Scholar]
- 38.Zhang XT, Li LY, Chang XY. Efficacy and correlation with predictive markers of gefitinib in pretreated Chinese patients with advanced NSCLC (Abstract) J. Clin. Oncol. 2005;23:7240. [Google Scholar]
- 39.Zhang L, Zhang X, Wang X. The status of epidermal growth factor receptor (EGFR) mutations at exon 19 and 21 in Chinese patients with NSCLC (Abstract) J. Clin. Oncol. 2005;23:7097. [Google Scholar]
- 40.Zhou C. Epidermal growth factor receptor mutations in Chinese patients with non-small cell lung cancer (Abstract) J. Clin. Oncol. 2005;23:7101. [Google Scholar]
- 41.Han SW, Kim TY, Hwang PG, Jeong S, Kim J, Choi IS, Oh DY, Kim JH, Kim DW, Chung DH, Im SA, Kim YT, Lee JS, Heo DS, Bang YJ, Kim NK. Predictive and prognostic impact of epidermal growth factor receptor mutation in nonsmall-cell lung cancer patients treated with gefitinib. J. Clin. Oncol. 2005;23:2493–2501. doi: 10.1200/JCO.2005.01.388. [DOI] [PubMed] [Google Scholar]
- 42.Wu JY, Yu CJ, Yang CH, Wu SG, Chiu YH, Gow CH, Chang YC, Hsu YC, Wei PF, Shih JY, Yang PC. First- or second-line therapy with gefitinib produces equal survival in non-small cell lung cancer. Am J Respir Crit Care Med. 2008;178:847–853. doi: 10.1164/rccm.200803-389OC. [DOI] [PubMed] [Google Scholar]
- 43.Yung TK, Chan KC, Mok TS, Tong J, To KF, Lo YM. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res. 2009;15:2076–2084. doi: 10.1158/1078-0432.CCR-08-2622. [DOI] [PubMed] [Google Scholar]
- 44.Chan SK, Gullick WJ, Hill ME. Mutations of the epidermal growth factor receptor in non-small cell lung cancer -- search and destroy. Eur J Cancer. 2006;42:17–23. doi: 10.1016/j.ejca.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 45.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang SY, Hu YL, Wu YL, Li X, Chi GB, Chen Y, Dai WS. A comparative study of the risk factors for lung cancer in Guangdong, China. Lung Cancer. 1996;14:S99–S105. doi: 10.1016/s0169-5002(96)90215-9. [DOI] [PubMed] [Google Scholar]
- 47.Pham D, Kris MG, Riely GJ, Sarkaria IS, McDonough T, Chuai S, Venkatraman ES, Miller VA, Ladanyi M, Pao W, Wilson RK, Singh B, Rusch VW. Use of cigarette-smoking history to estimate the likelihood of mutations in epidermal growth factor receptor gene exons 19 and 21 in lung adenocarcinomas. J. Clin. Oncol. 2006;24:1700–1704. doi: 10.1200/JCO.2005.04.3224. [DOI] [PubMed] [Google Scholar]


