Abstract
This study describes the development and testing of a magnetic microfluidic chip (MMC) for trapping and isolating cells tagged with superparamagnetic beads (SPBs) in a microfluidic environment for selective treatment and analysis. The trapping and isolation are done in two separate steps; first, the trapping of the tagged cells in a main channel is achieved by soft ferromagnetic disks and second, the transportation of the cells into side chambers for isolation is executed by tapered conductive paths made of Gold (Au). Numerical simulations were performed to analyze the magnetic flux and force distributions of the disks and conducting paths, for trapping and transporting SPBs. The MMC was fabricated using standard microfabrication processes. Experiments were performed with E. coli (K12 strand) tagged with 2.8 μm SPBs. The results showed that E. coli can be separated from a sample solution by trapping them at the disk sites, and then isolated into chambers by transporting them along the tapered conducting paths. Once the E. coli was trapped inside the side chambers, two selective treatments were performed. In one chamber, a solution with minimal nutrition content was added and, in another chamber, a solution with essential nutrition was added. The results showed that the growth of bacteria cultured in the second chamber containing nutrient was significantly higher, demonstrating that the E. coli was not affected by the magnetically driven transportation and the feasibility of performing different treatments on selectively isolated cells on a single microfluidic platform.
I. INTRODUCTION
In recent years, a large amount of research has been conducted on micro devices, which aim to integrate single or multiple lab processes into micro-sized chips.1–5 The feasibility of utilizing micro-chips for cell analysis stems from the fact that the sizes of those targets are in the micrometer range or below. The miniaturization of analytical devices results in lower sample and reagent consumption, reduced risk of contamination and faster reaction times.1–4
Numerous microdevices have been developed in the last decade for cell analysis.5,6 In this paper, magnetic principles are utilized for trapping, transporting, and isolating cells tagged with superparamagnetic beads (SPBs). SPBs consist of nano-sized iron oxide crystals encapsulated in an organic (polymeric) or inorganic matrix.7 They are magnetized only in the presence of a magnetic field. In the absence of a field, the magnetization direction of SPBs flips randomly, due to thermal fluctuations, resulting in an average net magnetic moment of zero and no residual magnetic force between SPBs. This means they can be manipulated by a magnetic field, yet their dispersion is ensured. The organic/inorganic coating may be modified with functional groups, depending on the applications, based on specific interactions; therefore, a large variety of cells can be attached to SPBs.8
Different micro-chips have recently been studied, which have magnetic elements integrated with microfluidic systems, to trap, transport and isolate cells.8,9 There are also commercially available products such as MACS® from Miltenyi, Cell Search® from Veridex, LLC, BeadRetriever™ from Applied Biosystems®, and magnetic tweezers, such as from PicoTwist that are able to separate cells tagged with SPBs in heterogeneous solutions. Recently, novel methods for purification of cells have been explored, which deviate from the standard macro-scale magnetic separation approach employed by commercial products. A device consisting of a column wrapped around with Ni wire placed between magnets sorted live cardiomyocytes from a heterogeneous solution in a label-free microfluidic approach with 93% purity.10 Osman et al. separated Jurkat cells labeled with magnetic nanoparticles from a solution spiked with human embryonic kidney under a continuous flow utilizing an integrated flat micro-patterned hard magnetic film.11 They obtained purification levels greater than MACS and Dynal separators. Darabi et al. employed an array of thin nickel stripes on a glass substrate excited by an array of external magnets to separate CD4 + T cells from peripheral blood with 90% purity.12
The magnetic elements can be either passive or active; passive elements are usually soft-ferromagnetic structures,13–18 while active elements are microelectromagnets.19–25 Soft-ferromagnetic structures can be magnetized by applying an external magnetic field and demagnetized by removing the field. They usually provide stronger magnetic fields than micro-electromagnets and are employed to spatially concentrate magnetic fields. The benefit of using micro-electromagnets is that the magnetic field can be simply controlled by varying the amplitude of the electric current, leading to precise local control of magnetic forces. The magnetic microfluidic chip (MMC) presented in this research combines the advantages of soft-ferromagnetic structures and micro-electromagnets. We utilize Permalloy (Ni80Fe20) disks to trap SPBs and tapered conducting paths made of gold (Au) to transport the SPBs. This method ensures that a single SPB can be trapped at a single disk in a microfluidic channel, and then transported to a side chamber by the tapered conducting path for isolation. Inside the chamber, the SPB is again immobilized at a disk, enabling the application of selective treatments on the cells and monitoring their effects.
The design of the MMC technically allows for cells tagged with SPBs to be separated in solutions with unlabeled cells; cells tagged with SPBs can be trapped at the disks, unlabeled cells can be removed from the channel, and then the labeled cells can be moved by tapered conducting paths to chambers for further analysis. However, the separation of tagged cells in solutions with unlabeled cells has been widely explored in the papers mentioned above, and can be achieved with high purity by the commercially available products. Therefore, in this research, the emphasis is on the capacity of the MMC to isolate and perform selective treatments on cells tagged with SPBs on a simple and integrated platform. The approach we have taken in this paper differs from our previous work26–34 and from other work in terms of simplicity of fabrication, operation of the device as well as using living cells and also by the integration of all components into a microfluidic chip. Moreover, compared to the commercially available products, the MMC is an integrated chip of small size (3.1 × 2.3 cm2 chip area) that enables both cell separation and experiments on-chip. Such a device has the potential to become a powerful tool for drug testing, for genetic engineering, and for studying different biological processes.
II. ISOLATION MECHANISM USING THE MAGNETIC MICROFLUIDIC CHIP
A. Theory
Magnetic manipulation of SPBs requires a magnetic force strong enough to overcome the dominating viscous effects.35 An SPB, in a liquid medium, travelling through a region with a magnetic flux gradient, experiences a magnetic force Fm. Fm can be expressed as
| (1) |
where V is the volume of the spherical SPB (m3), Δχ is the difference in volume susceptibility between the SPB and the surrounding liquid medium, μ0 = 4π × 10−7 Vs/Am is the permeability of free space, and B is magnetic flux density (T). Δχ in a water medium is equal to the χ of the SPB, since χ of water is ≪ χ of an SPB.
The velocity, vm, of an SPB in a liquid medium and a magnetic flux gradient is
| (2) |
where fd = 6πηr is the drag coefficient, η is the viscosity of the liquid medium, and r is the hydrodynamic radius of the SPB. The gravitational and buoyancy forces are neglected in this study, since the Reynolds number is ≪ 1 and hence, viscous terms dominate inertial terms.
B. Device architecture
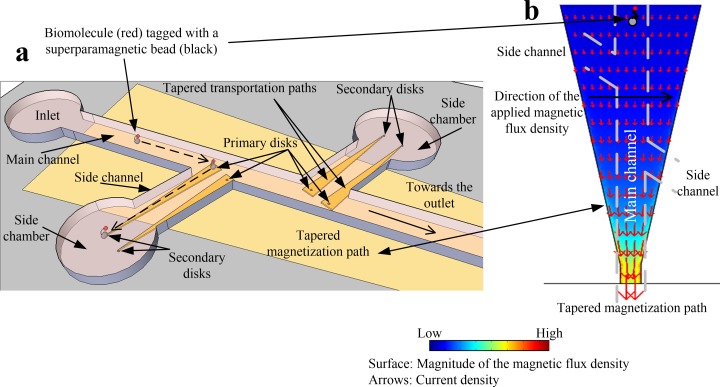
The MMC architecture is shown in Fig. 1(a) and has one main channel, two side channels, and 2 side chambers. The microfluidic system is made from Polydimethylsiloxane (PDMS). 2-D numerical analyses were performed with commercial finite-element software (COMSOL®) to aid in the design of the MMC. Biomolecules tagged with SPBs are injected at the inlet and a current through the tapered magnetization path, made from Au, underneath the channel magnetizes the disks and aids the flow of SPBs towards the primary disks (magnetization path in Fig. 1(a)).
FIG. 1.
Schematic of the magnetic microfluidic chip. (a) Chip design displaying the trapping of superparamagnetic beads by primary disks, and transportation and isolation of beads by tapered transportation paths. (b) Numerical results of the integrated electromagnet (tapered magnetization path) that is utilized to magnetize the disks as well as aid in the flow of beads towards the disks.
The magnetization path provides an increasing value of the magnitude of the magnetic flux density B towards the narrow section (Fig. 1(b)), causing SPBs to move away from the inlet along the main channel. The inhomogeneous B produced by the primary disks gives rise to a magnetic flux density gradient that exerts an attractive force on the SPBs. Once the SPBs are trapped on top of the primary disks, the current to the magnetization path is turned off and applied to the tapered transportation paths that extend from the main channel through the side channels to the side chambers (Fig. 1(a)). This causes the SPBs to move to the side chambers, where they are trapped at secondary disks. Those are magnetized by the magnetic flux of the tapered transportation paths. In the current design, the MMC consists of 2 primary and 2 secondary disks per side chamber, two side chambers, and a main channel.
C. Numerical analysis
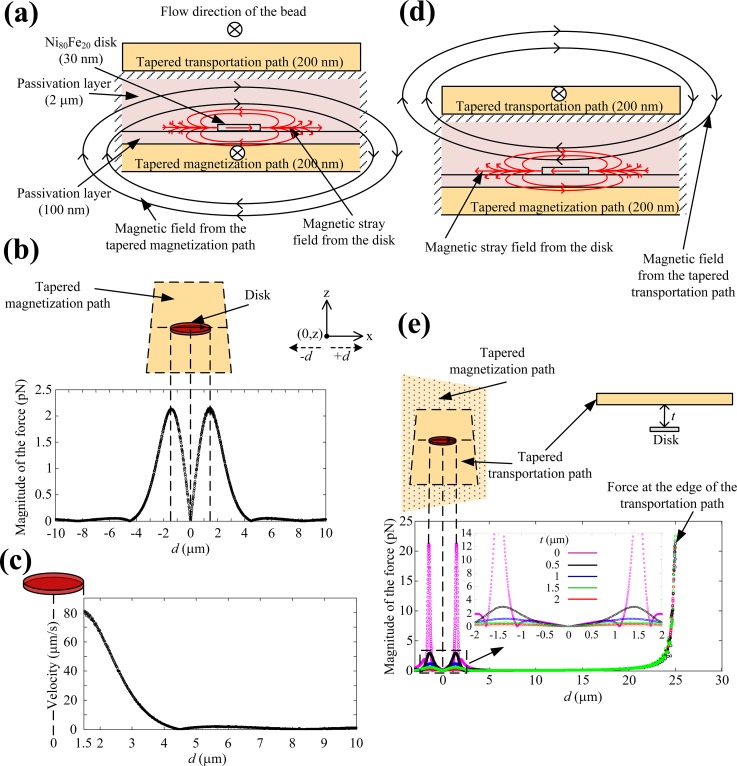
Fig. 2(a) shows the cross-section of the MMC at a primary disk. The disk is made of Ni80Fe20 and the magnetization path is separated from the disk by a 100 nm SiN3 passivation layer. The parameters for Ni80Fe20 were taken from previous studies,36–42 the diameter of the disk is 3 μm, and the thickness of the disk is 30 nm. The disk's diameter influences several parameters like the magnetic properties or the number of SPBs that can be trapped at a disk. In general, the smaller the disk, the fewer SPBs will be trapped at it; hence, the disk diameter is a parameter to be optimized for specific applications. By choosing the disk diameter to be approximately the same as the diameter of the SPBs, only one SPB is usually trapped at a disk,43,44 and only one strand of E. coli is bound to an SPB due to steric hindrance,45 as observed during many experiments. This provided better control over the experiments and enhanced the visualization of the SPB-cell complex, making it easier to quantify the number of SPBs and cells. The current through the magnetization path magnetizes the disk, which generates a magnetic stray field as shown in Fig. 2(a). The magnetization direction is transverse to the direction of the SPB motion. Due to the stray fields of the disk, B is reduced above the disk and changes sharply at its borders. This change attracts the SPBs to the disks (Eq. (1)), which acts like a trap, at a velocity defined by Eq. (2). Fig. 2(b) shows the magnitude of the magnetic force Fm, along the center of the disk at a height of 2.2 μm above the disk, when a current of 500 mA was applied to the magnetization path. Fm, calculated for an SPB with a radius of 1.4 μm and a susceptibility of 1.246 in a solution with a viscosity of 1 × 10−3 Pa·s, has a maximum value of 2.2 pN at the edges of the disk, which is sufficient to trap SPBs.8,9,13,15,20–22,26–35,47
FIG. 2.
(a) Cross-section of the MMC showing the magnetic fields of the magnetization path (black) and the stray fields produced by the disk (red) due to magnetization. (b) The magnitude of the magnetic force produced by the disk on a 2.8 μm SPB in the magnetization direction, along the center, and 2.2 μm above the disk. (c) The magnitude of the velocity of a 2.8 μm SPB attracted by the magnetized disk along the center and 2.2 μm above the disk. (d) Cross-section of the disk and tapered transportation path showing the magnetic fields of the transportation path (black) and the stray fields produced by the disk (red) due to magnetization. (e) Force distribution at the disk and the transportation path at different distances above the disk.
Fig. 2(c) shows the velocity of an SPB during the trapping process. The beads are attracted from a distance of about 4 μm with the velocity increasing exponentially and reaching a value of 80 μm/s at the edge of the disk, where the bead eventually gets trapped. This is in agreement with our observations during experiments, in which SPBs were attracted to a disk as long as they were in close proximity to a disk. It is worth to mention that a fluid flow could be applied to aid the movement of the SPBs along the main channel. In this case, the critical flow velocities at 0.5, 1, 1.5, 2, and 2.5 μm from a disk are 68, 43, 24, 11, and 3.5 μm/s, respectively. When the current is switched from the magnetization path to the transportation path, the magnetic flux (Fig. 2(c)) exerts a force on the SPB pointing from the primary disk to the secondary disk in the side chamber. However, this magnetic flux also magnetizes the disk and the stray field of the disk produces a force on the SPB that opposes its motion along the transportation path as shown in Fig. 2(c). Fig. 2(d) shows the forces, at the surface of the transportation path, trapping the SPB at the primary disk, at different thicknesses t, between the top of the disk and the bottom of the transportation path, when a current of 100 mA is applied to the transportation path with a cross-section of 50 μm × 200 nm. It can be seen that the influence of the stray field diminishes with increasing t above the disk, and becomes insignificant at a distance of 2 μm. Therefore, the disk was separated from the transportation path with a 2 μm SiN3 passivation layer. Once the SPB is removed from the disk, it moves towards the edge of the transportation path, where the force is highest, as shown in Fig. 2(d), and towards the secondary disk in the side chamber.
III. EXPERIMENT
A. Fabrication of the magnetic microfluidic chip
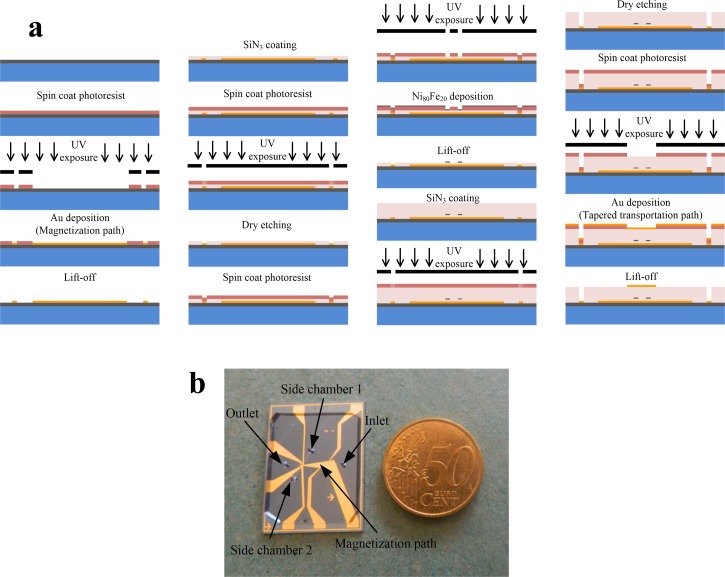
The full fabrication process is described in Fig. 3(a). A 4″ silicon wafer was first treated with Hexamethyldisilazane for better adhesion of the photoresist. Then, positive photoresist AZ 1512HS (micro resist technology GmbH) was spin coated (3000 rpm, 30 s), baked (100 °C, 60 s) and exposed to UV light (EVG 6200 UV contact aligner, exposure dose: 30 mJ/cm2) through a chrome mask followed by photoresist development (AZ 726, 25 s). The thickness of the patterned photoresist was approximately 1.2 μm. Those parameters were used for all lithography steps. On top of the patterned photoresist, 200 nm of Au were sputter deposited as the first metal layer, and the magnetization path was patterned by a lift-off process. To achieve the lift-off process, the wafer was left in acetone followed by sonication and thorough rinsing. The widest and narrowest sections of the magnetization path are 1850 and 80 μm, respectively. Then, SiN3 was deposited (100 nm thick, using a combination of Silane SiH4 and Ammonia NH3) to form an insulation layer, and vias to the Au layer were opened, through a lithographically defined mask, by dry etching (a combination of Fluoroform CHF3 and Sulfur hexafluoride SF6).
FIG. 3.
(a) Fabrication of the magnetic microfluidic chip. (b) Image of the MMC with PDMS microchannels.
Next, the Ni80Fe20 disks were defined by UV lithography followed by dry etching the SiN3 and depositing a 30 nm thick layer of Ni80Fe20 using e-beam evaporation. Before the deposition of the second Au layer, a SiN3 insulation layer (2 μm thick) was deposited and etched as described before. The second metal layer (200 nm thick), the transportation path, was then deposited following the same procedure as for the first layer. The widest and the narrowest sections of the transportation path are 115 and 4 μm, respectively.
Finally, the microfluidic channels were fabricated from PDMS (Polydimethylsiloxane) employing a soft lithography technique.48–50 A mold was fabricated using a 7.5 μm thick layer of AZ 9260 that was spin coated on a silicon wafer, exposed to UV light and developed. Liquid PDMS was spread over the entire surface of the mold, cured at 75 °C for 2 h and then gently peeled off. Finally, the PDMS layer was bonded to the MMC chip using plasma oxygen bonding (YES R3 Plasma cleaner). The main channel is 1 cm long, 100 μm wide and 7.5 μm high, the side channels are 0.3 cm long. Fig. 3(b) shows a photo of a fabricated MMC chip and its size compared to a coin.
B. Labeling of E. coli with SPBs
Streptavidin coated SPBs (Dynabeads® M-270 from Life Technologies™) with 2.8 μm in diameter and Rabbit polyclonal to E. coli (Biotin) (ab20640, from abcam®) were placed in ice for 30 min. 100 μl of SPBs in a tube were placed on a magnet for 2 min to separate the beads from the supernatant. 100 μl of 1 × phosphate buffered solution (PBS) and 0.05% Tween-20 were added along the insides of the tube, where the beads were collected for washing and to remove preservatives. This step was repeated three times. Next, 20 μl of the antibody (Biotin) were added and incubated for 30 min at room temperature with gentle rotation. After that, the beads and antibodies were re-suspended in 100 μl of PBS and Tween. Following this, 2 ml of Terrific Broth (TB) modified were incubated with 5 μl of E. coli K12 (SHuffle® T7 lysY Competent) at 37 °C for 16 h and then washed three times using PBS and Tween. Subsequently, 10 μl of SPBs were mixed with 100 μl of E. coli K12 and incubated at room temperature for 60 min with gentle rotation. Finally, SPBs and E. coli were washed three times to remove extra E. coli and re-suspended in 200 μl of PBS and Tween.
C. Experimental setup for trapping and isolation of E. coli
Experiments were performed with the aid of a probe station equipped with a microscope. Seven probes were connected to a DC power supply to provide a current to the magnetization path, and the transportation paths extending from the main channel to the side chambers. Syringe pumps were used to load samples into the main channel or nutrients into the side channels with a slow flow rate (less than 3 μl/min).
D. Staining of E. coli
Vybrant® CFDA/SE Cell Tracer Kit (carboxyfluorescein diacetate, succinimidyl ester, Invitrogen), a lipophilic dye that is taken up by cell membranes, was used to stain the E. coli. The bacterial suspension was washed twice with PBS followed by centrifugation at 4900 G for 3 min at 4 °C. Then, E. coli were incubated at 37 °C with CFDA/SE for 30 min in the dark. CFDA/SE stained cells were harvested by centrifugation, washed and incubated for another 30 min at 37 °C. The fluorescent imaging was performed using a ZEISS LSM710 confocal microscope.
IV. EXPERIMENTAL RESULTS AND DISCUSSION
A. Characterization of soft ferromagnetic disks
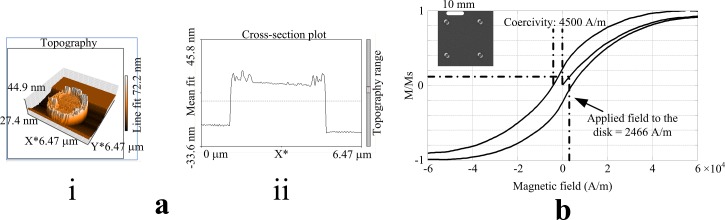
Atomic Force Microscopy (AFM) was used to obtain surface information of the fabricated disks. Fig. 4(a) shows a disk that is 3 μm in diameter and 30 nm in thickness. As remainder of the lift-off fabrication process, the edges show some roughness with a height of about 5 nm (Figs. 4(a) and 4(a). If SPBs would be trapped directly on top of such a disk, they might get stuck. In case of the developed design, this is not an issue, since the disks are located below the transportation paths, and the two layers (passivation and transportation path) between the chip's surface and the disks smoothen the surface and prevent the direct contact of SPBs with the disks.
FIG. 4.
Characterization of the disks. (a) Atomic force microscopy results for a disk with 3 μm in diameter and 30 nm in thickness. (i) 3-D profile. (ii) Cross-section profile. (b) Magnetization curve of a 125 × 125 disk array.
An Alternating Gradient Magnetometer (AGM) was utilized to obtain the magnetic characteristics of the disks. Fig. 4(b) shows the magnetization curve in the in-plane direction for an array of 125 × 125 Permalloy disks. The inset in Fig. 4(b) shows four disks from this array, which are separated by a distance of 20 μm. It can be seen from Fig. 4(b) that the applied magnetic field used in the numerical analysis magnetizes the disk to 15% of its saturation value. This is ideal for our experiments, since the demagnetization procedure could be challenging, when disks are fully magnetized.36–42 For our experiments, it is important that the disks are magnetized and demagnetized by simply turning the current on and off.
The magnetic field required for saturation is 7 × 104 A/m and the coercivity is 4500 A/m. These values agree well with previous studies on permalloy disks.36,37,42 The saturation field can also be estimated by taking into account the shape anisotropy given by the aspect ratio s = d/t, where d and t are the diameter and thickness of the disk, respectively. The magnetic field inside the disk Hdisk is a function of the applied field Happ and the demagnetizing field Hd
| (3) |
where Hd = −NipMs. Ms is 8 × 105 A/m and Nip is the in-plane demagnetizing factor of the disk.
When the disks are approaching saturation, Eq. (3) can be re-written as
| (4) |
where Hs = Happ at saturation and Nip for a disk with s ≫ 1 is expressed as51
| (5) |
Substituting s = 100 into Eq. (5), a value of 7.8 × 104 A/m is obtained for Hs from Eq. (4), which agrees well with the one found by measurement.
B. Optimization of chip surface and channel walls
Friction and adhesion between the SPBs and the walls/surface can have a considerable effect on the movement of SPBs and significantly influence the transportation of SPBs along the magnetization and transportation paths. In order to prevent non-specific binding and cell adhesion to the walls of the PDMS structures and to the Au surface, the channels were first treated with 10 mg/ml BSA (Bovine serum albumin) and 0.1% (v/v) SDS (Sodium dodecyl sulfate) solution for 15 min. BSA is known to inhibit cell adhesion and efficiently prevent adsorption on the walls of PDMS channels,52–54 whereas the ionic surfactant (SDS) prevents binding of SPBs or undesired waste to silicon and photoresist surfaces55–57 like the MMC surface. This treatment also removes all dead cells or other small particles left from previous experiments. However, since the SDS is a strong surfactant,58 a final washing step was performed to prevent any side effects, especially on cell vitality. For this purpose, the channels were washed for 15 min using a moderate concentration of 0.2% (v/v) of Tween 20, which is a non-ionic, gentle surfactant that protects the native state of proteins and constitutions of cells and prevents specific adsorption. It also has long term stability. However, using of Tween 20 will also decrease the fluorescent background (important for E. coli staining—see Sec. III D).59–61 The procedure mentioned above significantly enhanced the transportation of SPBs compared to the case where the procedure was not performed prior to experiments.
C. Trapping and selective isolation of E. coli
E. coli bacteria were chosen for the experimental verification of the MMC concept because it is of high relevance with some strands being pathogenic. It is also a simple organism that is easy to culture in the laboratory. In this research, we aim to selectively isolate E. coli with the MMC to enable observation of their response to different cell culture media.
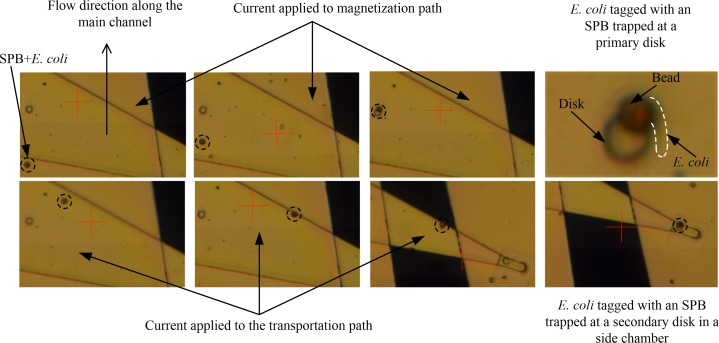
First, the channels were washed by applying a mixture of BSA (Bovine serum albumin), SDS (Sodium dodecyl sulfate), and Tween 20 for 30 min at the inlet of the main channel with all outlets open. Second, 10 μl of the solution containing E. coli (see Sec. III B) tagged with SPBs was injected into the inlet of the main channel, while only the outlet of this channel was open. Next, a current of 500 mA was applied to the magnetization path to move the SPBs along the main channel and trap them at the disks. This process is shown in Fig. 5, where SPBs moves along the main channel and gets trapped at the edge of the disk, confirming the result of the numerical analysis, which showed the trapping site at the surface of the disk, is at the edge of the disk. Depending on distance between the SPB and the disk after injecting the sample solution, it would take up to 15 min. Once an SPB with E. coli was trapped at a disk, the current to the magnetization path was turned off, and a current of 100 mA was applied to the transportation path. Fig. 5 shows that this step caused the SPB to be dislodged from the primary disk and move towards the edge of the transportation path and through the side channel. A significant increase in the velocity of the SPB was observed during this movement, due to the increasing magnetic field gradient. Finally, the SPB was trapped at the secondary disk located at the end of the transportation path. Again, the SPB was trapped at the edge of the disk. The time required for an SPB to travel along the transportation path was generally 30–40 s. Using the same procedure as above, another SPB attached to E. coli was isolated into the second side chamber.
FIG. 5.
Experimental results demonstrating the trapping, transporting and selective isolation of E. coli tagged with a superparamagnetic bead by the magnetic microfluidic chip. Note, for better visibility, the E. coli is encircled with a dashed line.
Generally, when a bead was moving directly across a disk, as shown in Fig. 5, it always got trapped at the disk and it stayed trapped until current to the magnetization path was turned off. When a bead passed a disk at some distance, it could usually be attracted from approximately 5 μm as shown in Fig. 6 for Bead 1. Beads 2 and 3 were too far away to be trapped. The time taken for Bead 1 to get trapped at the disk was approximately 4 s. An increase in velocity of Bead 1 as it approached the disk was clearly observed during the experiments. In the case, a bare SPB was trapped at a disk, the current to the magnetizing path was turned off to allow the SPB to dislodge and move away from the disk.
FIG. 6.
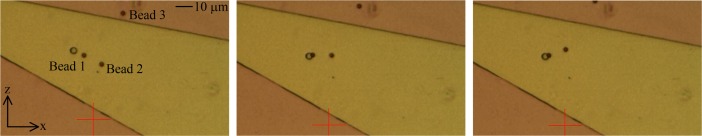
Trapping of an SPB from a distance of about 5 μm.
D. Culturing E. coli in side chambers
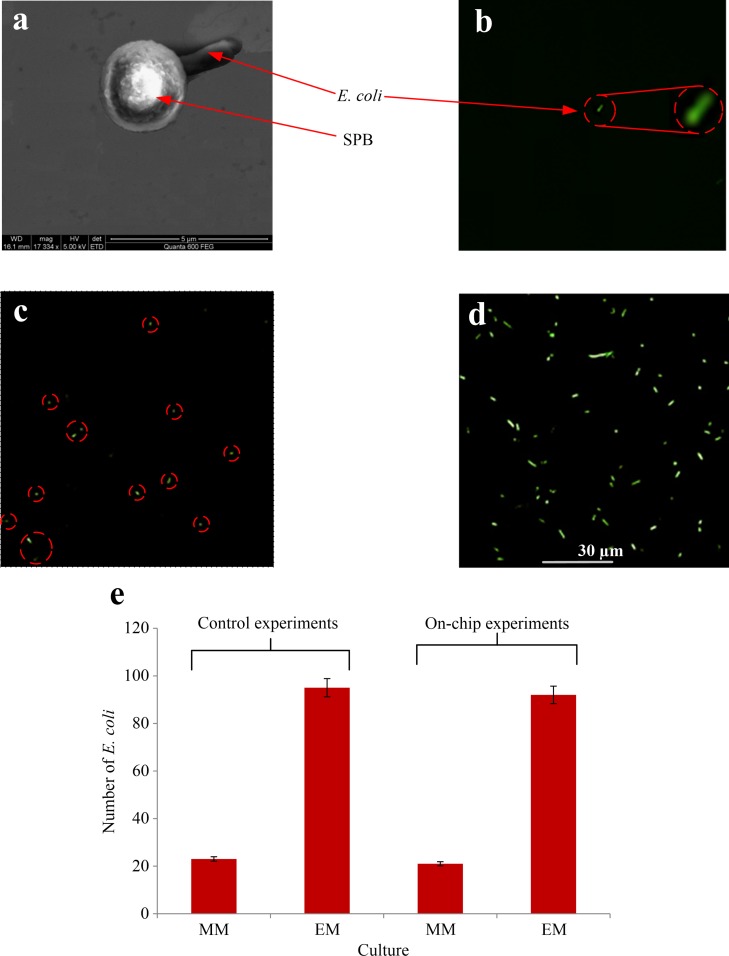
At the beginning of the culturing experiment, the chip was washed by applying BSA (Bovine serum albumin), SDS (Sodium dodecyl sulfate), and Tween 20 (as described in Sec. IV B) for 30 min from the inlet of the main channel with all outlets open. Then, the chambers were flushed for 5 min with a solution of 0.1% glycerol. The first washing step served to remove any unwanted particles in the side chambers and any binding effect of the SPBs or any undesired waste to the surface or walls of the MMC. The second step was to prepare a suitable culture environment for E. coli and to remove any undesirable effects of the previously added additives. In order to ease the observation of E. coli, they were stained using CFDA/SE dye (see Sec. III D). It can be observed from Figs. 7(a) to 7(b) that only a single E. coli attached to an SPB was present in each chamber. During the culture experiments, which lasted for 6 h, two different treatments were performed on the E. coli isolated in the side chambers.
FIG. 7.
(a) SEM image of single E. coli attached to one SPB. (b) Fluorescent image of a single E. coli at the beginning of the culturing experiment. (c) and (d) Fluorescent images of side chambers 1 (minimal media) and 2 (enriched media), respectively, taken after 6 h of culturing. (e) The number of E. coli after 6 h culturing, using minimal media (MM) and enriched media (EM). The results in columns 1 and 2 are obtained from control experiments. Columns 3 and 4 show the results obtained from E. coli tagged with an SPB and isolated with the microfluidic chip into side chamber 1 (minimal media) and 2 (enriched media).
A media containing minimal content of nutrition, which is a solution of 0.1% (v/v) glycerol, and 0.1% glucose, was added to both chambers through the side chamber inlets, and an additional 1% of Terrific Broth, used as enriching media, was added only to chamber 2. The entire chambers were scanned using optical and fluorescent microscopy to ensure that no contaminations were found at this point, which demonstrates that the subsequently obtained culture results were initiated from a single E. coli in each chamber. All experiments were performed inside an incubator at 37 °C, to maintain the same pH, oxygen, and temperature conditions. The objective of these culture experiments was not to find or use the optimal culture conditions of E. coli, but to test whether the E. coli can survive the procedures applied so far (exposure to magnetic fields, transportation along the channels during trapping and isolation steps and washing). Figs. 7(c) and 7(d) show fluorescent images of the side chambers after 6 h (further details in the supplementary figure Fig. S162). In both chambers, the single E. coli was able to divide, and in chamber 2, where the enriched media was used, the number of E. coli was more than 4 times higher than their number in chamber 1, where minimal media were used. The average numbers of cells obtained from a single E. coli during similar experiments using 3 different MMCs are shown in columns 3 and 4 of the table in Fig. 7(e). Comparing these results with the control experiments (columns 1 and 2 in the same table) reveal that the isolated E. coli is not affected by the manipulations they undergo in the MMC, and that they can adapt to the modified culturing conditions.
V. CONCLUSION
A magnetic microfluidic chip was presented in this paper, which enabled the trapping and isolation of E. coli by tagging them with SPBs. Trapping of individual SPBs inside a main channel at specific locations was realized by Permalloy (Ni80Fe20) disks, which were magnetized by the help of an integrated conducting path. This tapered path also provided the magnetic field gradient to move the SPBs from the inlet of the main channel to the trapping sites. Controlled motion of the individual SPBs from the trapping sites into side chambers was enabled by tapered transportation paths. The specific arrangement of the paths and Permalloy disks allowed trapping of SPBs with current applied to the magnetization path yet released the SPBs and moved them to the side chambers when the current was switched to the transportation path. AGM analyses of the disks provided their magnetic characteristics, namely, saturation magnetization (8 × 105 A/m), saturation field (7 × 104 A/m), and coercivity (4500 A/m), which agreed well with theoretical results based on shape anisotropy calculations as well as previous works on permalloy disks. The MMC was fabricated using standard microfabrication techniques. E. coli (K12 strand) tagged with 2.8 μm SPBs were trapped and separated individually into side chambers, and fluorescent images showed that E. coli were successfully cultured starting form a single E. coli. The experiments imply that the procedures employed to trap and manipulate E. coli tagged with SPBs did not affect the isolated E. coli. The growth rate of E. coli in a solution containing Terrific Broth in one chamber was significantly higher (more than 4 times) than the one in another chamber, which did not contain Terrific Broth.
In this work, a specific design of the MMC was implemented to trap four SPBs (with E. coli) and move them individually into two side chambers, where they were exposed to different environments. The motion of the SPBs was solely controlled by magnetic forces, i.e., electric currents, which could potentially lead to devices with simple on-chip operation. The presented concepts can readily be employed and modified for different designs to meet various demands.
ACKNOWLEDGMENTS
The authors acknowledge Dr. Zhihong Wang, Basil Chew, Ahad Syed, and Xiang Yu for their help with microfabrication and material characterization. The authors would also like to thank Professor Nicole Pamme, Professor Quentin Pankhurst, Professor Martin Gijs, Professor Horst Hoffmann, and Professor Menno Prins, and Dr. Mark Tarn, Dr. Chengxun Liu, and Dr. Shu-I Tu for fruitful discussions on the research.
Research reported in this publication was supported by the King Abdullah University of Science and Technology (KAUST).
REFERENCES
- 1.Ng A. H. C., Uddayasankar U., and Wheeler A. R., Anal. Bioanal. Chem. 397(3), 991 (2010). 10.1007/s00216-010-3678-8 [DOI] [PubMed] [Google Scholar]
- 2.El-Ali J., Sorger P. K., and Jensen K. F., Nature 442(7101), 403 (2006). 10.1038/nature05063 [DOI] [PubMed] [Google Scholar]
- 3.Whitesides G. M., Nature 442(7101), 368 (2006). 10.1038/nature05058 [DOI] [PubMed] [Google Scholar]
- 4.Yeo L. Y., Chang H.-C., Chan P. P. Y., and Friend J. R., Small 7(1), 12 (2011). 10.1002/smll.201000946 [DOI] [PubMed] [Google Scholar]
- 5.Dharmasiri U., Witek M. A., Adams A. A., and Soper S. A., in Annual Review of Analytical Chemistry, edited by Yeung E. S. and Zare R. N. (Annual Reviews, Palo Alto, CA, 2010), Vol. 3, p. 409 [DOI] [PubMed] [Google Scholar]
- 6.Yun H., Kim K., and Lee W. G., Biofabrication 5(2), 022001 (2013). 10.1088/1758-5082/5/2/022001 [DOI] [PubMed] [Google Scholar]
- 7.Pankhurst Q. A., Connolly J., Jones S. K., and Dobson J., J. Phys. D: Appl. Phys. 36(13), R167 (2003). 10.1088/0022-3727/36/13/201 [DOI] [Google Scholar]
- 8.Gijs M. A. M., Lacharme F., and Lehmann U., Chem. Rev. 110(3), 1518 (2010). 10.1021/cr9001929 [DOI] [PubMed] [Google Scholar]
- 9.Pamme N., Lab Chip 6(1), 24 (2006). 10.1039/b513005k [DOI] [PubMed] [Google Scholar]
- 10.Sofla A., Cirkovic B., Hsieh A., Miklas J. W., Filipovic N., and Radisic M., Biomicrofluidics 7(1), 014110 (2013). 10.1063/1.4791649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osman O., Toru S., Dumas-Bouchiat F., Dempsey N. M., Haddour N., Zanini L. F., Buret F., Reyne G., and Frenea-Robin M., Biomicrofluidics 7(5), 054115 (2013). 10.1063/1.4825395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darabi J. and Guo C., Biomicrofluidics 7(5), 054106 (2013). 10.1063/1.4821628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inglis D. W., Riehn R., Austin R. H., and Sturm J. C., Appl. Phys. Lett. 85(21), 5093 (2004). 10.1063/1.1823015 [DOI] [Google Scholar]
- 14.Gooneratne C. P. and Kosel J., in Proceedings of the 2012 Sixth International Conference on Sensing Technology (IEEE, 2012), p. 97 [Google Scholar]
- 15.Anandakumar S., Rani V. S., Oh S., Sinha B. L., Takahashi M., and Kim C., Biosens. Bioelectron. 26(4), 1755 (2010). 10.1016/j.bios.2010.08.033 [DOI] [PubMed] [Google Scholar]
- 16.Morimoto Y., Abe M., Hatakayama M., Handa H., and Sandhu A., IEEE Trans. Magn. 45(6), 2871 (2009). 10.1109/TMAG.2009.2018861 [DOI] [Google Scholar]
- 17.Rapoport E., Montana D., and Beach G. S. D., Lab Chip 12(21), 4433 (2012). 10.1039/c2lc40715a [DOI] [PubMed] [Google Scholar]
- 18.Donolato M., Vavassori P., Gobbi M., Deryabina M., Hansen M. F., Metlushko V., Ilic B., Cantoni M., Petti D., Brivio S., and Bertacco R., Adv. Mater. 22(24), 2706 (2010). 10.1002/adma.201000146 [DOI] [PubMed] [Google Scholar]
- 19.Graham D. L., Ferreira H. A., Feliciano N., Freitas P. P., Clarke L. A., and Amaral M. D., Sens. Actuators, B 107(2), 936 (2005). 10.1016/j.snb.2004.12.071 [DOI] [Google Scholar]
- 20.Derec C., Wilhelm C., Servais J., and Bacri J.-C., Microfluid Nanofluid 8(1), 123 (2010). 10.1007/s10404-009-0486-6 [DOI] [Google Scholar]
- 21.Lee H., Purdon A. M., and Westervelt R. M., IEEE Trans. Magn. 40(4), 2991 (2004). 10.1109/TMAG.2004.829179 [DOI] [Google Scholar]
- 22.Wirix-Speetjens R., Fyen W., Xu K. D., Boeck J. D., and Borghs G., IEEE Trans. Magn. 41(10), 4128 (2005). 10.1109/TMAG.2005.855345 [DOI] [Google Scholar]
- 23.Beveridge J. S., Stephens J. R., Latham A. H., and Williams M. E., Anal. Chem. 81(23), 9618 (2009). 10.1021/ac9016456 [DOI] [PubMed] [Google Scholar]
- 24.Chaves R. C., Bensimon D., and Freitas P. P., J. Appl. Phys. 109(6), 064702 (2011). 10.1063/1.3560853 [DOI] [Google Scholar]
- 25.Kokkinis G., Keplinger F., and Giouroudi I., Biomicrofluidics 7(5), 054117 (2013). 10.1063/1.4826546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gooneratne C. P., Liang C., Giouroudi I., and Kosel J., J. Appl. Phys. 111(7), 07B327 (2012). 10.1063/1.3678303 [DOI] [Google Scholar]
- 27.Gooneratne C. P., Liang C., and Kosel J., Microelectron. Eng. 88(8), 1757 (2011). 10.1016/j.mee.2010.12.068 [DOI] [Google Scholar]
- 28.Gooneratne C. P., Giouroudi I., Liang C., and Kosel J., J. Appl. Phys. 109(7), 07E517 (2011). 10.1063/1.3536822 [DOI] [Google Scholar]
- 29.Gooneratne C. P., Giouroudi I., and Kosel J., Sens. Lett. 10(3–4), 770 (2012). 10.1166/sl.2012.2583 [DOI] [Google Scholar]
- 30.Li F., Gooneratne C., and Kosel J., Magnetic Biosensor System to Detect Biological Targets (IEEE, Piscataway, NJ, 2012), p. 1238 [Google Scholar]
- 31.Li F., Giouroudi I., and Kosel J., J. Appl. Phys. 111(7), 07B328 (2012). 10.1063/1.3678304 [DOI] [Google Scholar]
- 32.Giouroudi I., van den Driesche S., Kosel J., Groessinger R., and Vellekoop M. J., J. Appl. Phys. 109(7), 07B304 (2011). 10.1063/1.3556952 [DOI] [Google Scholar]
- 33.Li F. and Kosel J., IEEE Trans. Magn. 48(11), 2854 (2012). 10.1109/TMAG.2012.2202644 [DOI] [Google Scholar]
- 34.Gooneratne C. P., Giouroudi I., and Kosel J., in Advancement in Sensing Technology, edited by Mukhopadhyay S. C., Jayasundera K. P., and Fuchs A. (Springer, Berlin, Heidelberg, 2013), Vol. 1, p. 121 [Google Scholar]
- 35.Liu C., Stakenborg T., Peeters S., and Lagae L., J. Appl. Phys. 105(10), 102014 (2009). 10.1063/1.3116091 [DOI] [Google Scholar]
- 36.Schneider M. and Hoffmann H., J. Appl. Phys. 86(8), 4539 (1999). 10.1063/1.371399 [DOI] [Google Scholar]
- 37.Smyth J. F., Schultz S., Kern D., Schmid H., and Yee D., J. Appl. Phys. 63(8), 4237 (1988). 10.1063/1.340217 [DOI] [Google Scholar]
- 38.Gubbiotti G., Carlotti G., Nizzoli F., Zivieri R., Okuno T., and Shinjo T., IEEE Trans. Magn. 38(5), 2532 (2002). 10.1109/TMAG.2002.801920 [DOI] [Google Scholar]
- 39.Grebenshchikov Y. B. and Usov N. A., J. Appl. Phys. 93(8), 4810 (2003). 10.1063/1.1559635 [DOI] [Google Scholar]
- 40.Kikuchi N., Okamoto S., Kitakami O., Shimada Y., Kim S. G., Otani Y., and Fukamichi K., IEEE Trans. Magn. 37(4), 2082 (2001). 10.1109/20.951060 [DOI] [Google Scholar]
- 41.Dimitrov D., Halianov I., Kassabov J., and Marinov S., J. Phys.: Condens. Matter 5(9), 1257 (1993). 10.1088/0953-8984/5/9/010 [DOI] [Google Scholar]
- 42.Svalov A. V., Aseguinolaza I. R., Garcia-Arribas A., Orue I., Barandiaran J. M., Alonso J., Fernandez-Gubieda M. L., and Kurlyandskaya G. V., IEEE Trans. Magn. 46(2), 333 (2010). 10.1109/TMAG.2009.2032519 [DOI] [Google Scholar]
- 43.Calderon F. L., Stora T., Monval O. M., Poulin P., and Bibette J., Phys. Rev. Lett. 72(18), 2959 (1994). 10.1103/PhysRevLett.72.2959 [DOI] [PubMed] [Google Scholar]
- 44.Wang M. S., He L., and Yin Y. D., Mater. Today 16(4), 110 (2013). 10.1016/j.mattod.2013.04.008 [DOI] [Google Scholar]
- 45.Tu S. I., Uknalis J., Patterson D., and Gehring A. G., J. Rapid Methods Autom. Microbiol. 6(4), 259 (1998). 10.1111/j.1745-4581.1998.tb00207.x [DOI] [Google Scholar]
- 46.van Reenen A., Gao Y., Bos A. H., de Jong A. M., Hulsen M. A., den Toonder J. M. J., and Prins M. W. J., Appl. Phys. Lett. 103(4), 043704 (2013). 10.1063/1.4816429 [DOI] [Google Scholar]
- 47.van Ommering K., Lamers C. C. H., Nieuwenhuis J. H., van Ijzendoorn L. J., and Prins M. W. J., J. Appl. Phys. 105(10), 104905 (2009). 10.1063/1.3118500 [DOI] [Google Scholar]
- 48.Yassine O., Morin P., Dispagne O., Renaud L., Denoroy L., Kleimann P., Faure K., Rocca J. L., Ouaini N., and Ferrigno R., Anal. Chim. Acta 609(2), 215 (2008). 10.1016/j.aca.2007.12.045 [DOI] [PubMed] [Google Scholar]
- 49.Renaud L., Yassine O., Kleimann P., Deman A. L., Chateaux J. F., Morin P., Ouaini N., and Ferrigno R., Exp. Heat Transfer 23(1), 63 (2009). 10.1080/08916150903402740 [DOI] [Google Scholar]
- 50.Faure K., Bias M., Yassine O., Delaunay N., Cretier G., Albert M., and Rocca J. L., Electrophoresis 28(11), 1668 (2007). 10.1002/elps.200600566 [DOI] [PubMed] [Google Scholar]
- 51.Osborn J. A., Phys. Rev. 67(11–1), 351 (1945). 10.1103/PhysRev.67.351 [DOI] [Google Scholar]
- 52.Long Z., Nugent E., Javer A., Cicuta P., Sclavi B., Lagomarsino M. C., and Dorfman K. D., Lab Chip 13(5), 947 (2013). 10.1039/c2lc41196b [DOI] [PubMed] [Google Scholar]
- 53.Lu H., Koo L. Y., Wang W. C. M., Lauffenburger D. A., Griffith L. G., and Jensen K. F., Anal. Chem. 76(18), 5257 (2004). 10.1021/ac049837t [DOI] [PubMed] [Google Scholar]
- 54.Wang S., Inci F., Chaunzwa T. L., Ramanujam A., Vasudevan A., Subramanian S., Ip A. C., Sridharan B., Gurkan U. A., and Demirci U., Int. J. Nanomed. 7, 2591 (2012). 10.2147/IJN.S29629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sikanen T., Wiedmer S. K., Heikkila L., Franssila S., Kostiainen R., and Kotiaho T., Electrophoresis 31(15), 2566 (2010). 10.1002/elps.201000130 [DOI] [PubMed] [Google Scholar]
- 56.Zare R. N. and Kim S., Ann. Rev. Biomed. Eng. 12, 187 (2010). 10.1146/annurev-bioeng-070909-105238 [DOI] [PubMed] [Google Scholar]
- 57.Roman G. T., McDaniel K., and Culbertson C. T., Analyst 131(2), 194 (2006). 10.1039/b510765b [DOI] [PubMed] [Google Scholar]
- 58.Heo J., Thomas K. J., Seong G. H., and Crooks R. M., Anal. Chem. 75(1), 22 (2003). 10.1021/ac0259717 [DOI] [PubMed] [Google Scholar]
- 59.Lutz S., Weber P., Focke M., Faltin B., Hoffmann J., Muller C., Mark D., Roth G., Munday P., Armes N., Piepenburg O., Zengerle R., and von Stetten F., Lab Chip 10(7), 887 (2010). 10.1039/b921140c [DOI] [PubMed] [Google Scholar]
- 60.Wong I. and Ho C. M., Microfluid Nanofluid 7(3), 291 (2009). 10.1007/s10404-009-0443-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cox W. G. and Singer V. L., J. Histochem. Cytochem. 47(11), 1443 (1999). 10.1177/002215549904701110 [DOI] [PubMed] [Google Scholar]
- 62. See supplementary material at http://dx.doi.org/10.1063/1.4883855E-BIOMGB-8-022403 for fluorescent images showing the growth of E. coli
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4883855E-BIOMGB-8-022403 for fluorescent images showing the growth of E. coli