Abstract
Background: MicroRNAs (miRNAs) regulate genes in animals and plants and can be synthesized endogenously. In milk, miRNAs are encapsulated in exosomes, thereby conferring protection against degradation and facilitating uptake by endocytosis. The majority of bovine miRNAs have nucleotide sequences complementary to human gene transcripts, suggesting that miRNAs in milk might regulate human genes.
Objectives: We tested the hypotheses that humans absorb biologically meaningful amounts of miRNAs from nutritionally relevant doses of milk, milk-borne miRNAs regulate human gene expression, and mammals cannot compensate for dietary miRNA depletion by endogenous miRNA synthesis.
Methods: Healthy adults (3 men, 2 women; aged 26–49 y) consumed 0.25, 0.5, and 1.0 L of milk in a randomized crossover design. Gene expression studies and milk miRNA depletion studies were conducted in human cell cultures and mice, respectively. For comparison, feeding studies with plant miRNAs from broccoli were conducted in humans.
Results: Postprandial concentration time curves suggest that meaningful amounts of miRNA (miR)-29b and miR-200c were absorbed; plasma concentrations of miR-1 did not change (negative control). The expression of runt-related transcription factor 2 (RUNX2), a known target of miR-29b, increased by 31% in blood mononuclear cells after milk consumption compared with baseline. When milk exosomes were added to cell culture media, mimicking postprandial concentrations of miR-29b and miR-200c, reporter gene activities significantly decreased by 44% and 17%, respectively, compared with vehicle controls in human embryonic kidney 293 cells. When C57BL/6J mice were fed a milk miRNA-depleted diet for 4 wk, plasma miR-29b concentrations were significantly decreased by 61% compared with miRNA-sufficient controls, i.e., endogenous synthesis did not compensate for dietary depletion. Broccoli sprout feeding studies were conducted as a control and elicited no detectable increase in Brassica-specific miRNAs.
Conclusion: We conclude that miRNAs in milk are bioactive food compounds that regulate human genes.
Introduction
MicroRNAs (miRNAs)6 are small, noncoding RNAs (∼22 nucleotides in mature miRNAs) that may silence genes via destabilizing complementary mRNA sequences or preventing translation of mRNAs (1, 2). The nucleotide sequence in the seed region in miRNAs (nucleotides 2–7) is of particular importance for binding to target mRNA (3); imperfect pairing of sequences in the seed region in miRNA to mRNA impairs gene downregulation at the protein or RNA level (4). miRNAs are encoded by their own genes, introns, or exons of long nonprotein-coding transcripts (5). Traditionally, miRNAs are considered endogenous regulators of genes, i.e., miRNAs synthesized by human cells regulate the expression of genes in that host. We challenged this paradigm and tested the hypothesis that humans absorb miRNAs from milk in meaningful quantities and milk-borne miRNAs elicit biologically meaningful changes in human gene expression.
There is precedent for the bioavailability of dietary miRNAs in humans. A recent report suggested that miRNA (miR)-168a from rice (Oryza sativa; osa-miR-168a) is detectable in human and animal sera, and osa-miR-168a decreases the expression of LDL receptor adapter protein 1 mRNA, thereby inhibiting LDL receptor expression in mouse liver (6). Note that the serum concentration of osa-miR-168a is only ∼3 fmol/L in humans and that the bioavailability of plant-borne miRNAs in humans is controversial (6–10).
This study focused on milk as a source of dietary miRNAs on the basis of the rationale that a large fraction of miRNAs in milk is contained in exosomes, conferring protection against degradation (11, 12), Americans consume 88 L of milk annually (13), and cow milk contains large quantities of 245 miRNAs (14, 15). This study of milk-borne miRNAs was modeled primarily on miR-29b on the basis of the rationale that the nucleotide sequence of bovine miR-29b is identical to that of human miR-29b (16), and miR-29b increases bone mineralization in humans through promoting a gain in osteoblast differentiation (17) and a loss in osteoclast differentiation and function (18). In select experiments, miR-200c was included because its concentration is 8 times that of miR-29b in cow milk (15). The nucleotide sequence of bovine miR-200c is identical to that of human miR-200c (16), and miR-200c decreases cancer risk by targeting the transcription factor zinc finger E-box binding homeobox 1 (ZEB1); miR-200c–dependent loss of ZEB1 induces E-cadherin expression, thereby limiting epithelial-to-mesenchymal transition, a key event in metastasis (19, 20).
This study was guided by the following aims. First, we determined whether humans absorb quantitatively meaningful amounts of miRNAs from nutritionally relevant doses of milk and characterized the bioavailability of milk miRNAs by using pharmacokinetics protocols. Second, we assessed the effects of physiologically relevant miRNA concentrations on the expression of endogenous genes in human peripheral blood mononuclear cells (PBMCs) and reporter genes in human cell cultures. Third, we determined whether mammals compensate for dietary miRNA deficiency by an increased synthesis of endogenous miRNAs in a mouse miRNA depletion study. Fourth, we conducted a broccoli feeding study to determine whether plant-borne miRNAs are bioavailable in humans.
Materials and Methods
Human feeding study.
Five apparently healthy adults (3 men, 2 women) participated in a milk feeding study that used 3 doses of milk in a randomized crossover design with a washout period of at least 1 wk between doses. Exclusion criteria included pregnancy, smoking, milk allergies, and self-reported health problems. Preliminary studies suggested that plasma miRNA concentrations remain higher than baseline concentrations until 9 h after milk consumption; participants were instructed not to consume milk and dairy products for 12 h before the milk meal and during the period in which blood samples were collected. Doses of milk were normalized by total body water of participants, which is the suspected volume of distribution for miRNAs. The estimation of total body water was calculated as previously described (21). Normalization of doses by body water resulted in doses (means ± SEMs) of 0.218 ± 0.018, 0.436 ± 0.037, and 0.872 ± 0.073 L, representing the equivalent of 0.25, 0.5, and 1.0 L in a 26-y-old reference male (75 kg weight, 1.83 m height). Twenty milliliters of blood was collected before milk consumption (baseline; time = 0 h) and at timed intervals (1–3, 6, 9, and 24 h) after the milk meal. PBMCs and plasma were collected by using gradient centrifugation as described previously (22) and frozen at −80°C until analysis. This protocol was approved by the University of Nebraska–Lincoln Institutional Review Board, and all participants provided signed informed consent forms before participation.
Real-time qPCR.
The sequences of mature bovine miR-29b (bta-miR-29), miR-200c (bta-miR-200c), and miR-1 (bta-miR-1; control) are identical to their human and murine orthologs, and therefore real-time qPCR amplification targeted the mature human miR-29b (hsa-miR-29b-3p), miR-200c (hsa-miR-200c-3p), and miR-1 (hsa-miR-1-3p) that originate in the 3′ p arm of pre-miRNA (23). RNA was isolated from human and murine (see below) plasma by using the NucleoSpin miRNA plasma kit (Machery-Nagel). Reverse transcription was performed by using the miScript II RT kit (Qiagen). Real-time qPCR was performed by using miScript SYBR Green (Qiagen) and the universal reverse primer included in the kit plus primers specific for individual miRNA (Supplemental Table 1). miR-1 is not detectable in milk (data not shown) and served as a negative control. Five attomoles of synthetic miRNA, miSPIKE (IDT DNA), was added to each sample after denaturation of plasma with lysis buffer and served as an external standard and for calibration of real-time qPCR.
Pharmacokinetics analysis.
AUCs for plasma miRNAs were calculated by using the linear trapezoidal rule to assess the apparent bioavailability of miRNAs (24). Plasma concentrations of miR-29b and miR-200c returned to baseline values 9 h after consumption of the 2 lowest doses of milk. For the calculation of AUC, baseline values were subtracted from postprandial concentrations and the plasma concentrations from the first 9 h after milk meals were used. The maximal plasma concentration (Cmax) and the time of peak concentration (tmax) were obtained by visual inspection of the plasma time curves measured for miR-29b and miR-200c. Cmax of an orally administered compound is a marker of bioavailability and rate of absorption, whereas tmax is a marker of the approximate absorption site (25).
Gene expression.
The effects of milk-borne miRNAs on the expression of human genes were assessed in cell cultures and PBMCs from the human milk feeding studies. For studies in cell cultures, human embryonic kidney (HEK)-293 cells (American Type Culture Collection) were cultured in MEM containing 10% exosome-depleted FBS, 0.1% sodium pyruvate, 100,000 U/L penicillin, and 100 mg/L streptomycin for 10 d. Bovine serum was depleted of exosomes by ultracentrifugation at 130,000 × g for 4 h, leading to the removal of 97% and 81% of miR-29b and miR-200c, respectively. Cells were transfected with miRNA reporter genes as described previously (26). Forty-eight hours after transfection, exosome-depleted media were replaced with exosome-sufficient media at final concentrations of 600 fmol/L miR-29b or 1000 fmol/L miR-200c. Exosome-sufficient media were prepared by using exosomes collected from cow milk as previously described (27). The concentrations of miR-29b and miR-200c in exosomes were quantified by using real-time qPCR, and exosomes were added back to culture media to produce the desired concentrations of miRNAs. Reporter genes for miR-29b and miR-200c were created by inserting the 3′-untranslated regions (3′-UTRs) from genes collagen, type I, α1 (COL1A1; 3 miR-29b binding sites), and ZEB1 (2 miR-200c binding sites), respectively, downstream of the luciferase (LUC) reading frame, driven by a cytomegalovirus promoter, thereby creating plasmids LUC-mir-29b and LUC-mir-200c (Supplemental Fig. 1). LUC-mir-200c was obtained from Dr. Thomas Brabletz (University of Freiburg, Germany) and is denoted as ZEB1 3′UTR-Luc in the original publication (20). LUC-mir-29b was created by digesting LUC-mir-200c with MluI and HindIII to remove the ZEB1 sequence. The 3′-UTRs of COL1A1 were amplified by PCR (Supplemental Table 1) by using IMR-90 human lung fibroblast DNA as a template and ligated into the reporter plasmid by using MluI and HindIII. Cells were cotransfected with plasmid Rous sarcoma virus-β-galactosidase and luciferase plasmids to assess transfection efficiency (28). Reporter gene activities were normalized by transfection efficiency. Our rationale for choosing HEK-293 cells was that these cells can be transfected with near 100% efficiency and express COL1A1 in meaningful quantities (29, 30).
For studies in human PBMCs, a preliminary time-response screen was conducted to determine the time point when the effects of milk on the expression of runt-related transcription factor 2 (RUNX2) and ZEB1 mRNAs were maximal. RUNX2 is a downstream target of miR-29b–dependent signaling pathways; note that miR-29b is a positive regulator of the expression of RUNX2 (17). Our preliminary assessment suggested that changes in mRNA abundance were maximal 6 h after milk consumption (Supplemental Fig. 2; positive and negative changes for RUNX2 and ZEB1 mRNAs, respectively). Therefore, subsequent assays of RUNX2 and ZEB1 mRNAs were conducted by using samples from the time point 6 h after the milk meal by using real-time qPCR (Supplemental Table 1) and the ΔΔCt method (31); GAPDH was used to normalize for amplification efficiency (32). The abundance of miRNAs was measured as described for plasma samples, except that normalization was performed by using U6 rather than miSPIKE.
Mouse feeding study.
An miRNA depletion study was conducted in mice to determine whether endogenous miRNA synthesis compensates for dietary deficiency. Ten female C57BL/6J mice (Jackson Laboratory; stock 000664), aged 3 wk, were randomly divided between a milk miRNA-depleted treatment group (denoted ExoMinus) and a milk miRNA-sufficient control group (ExoPlus). Diets were based on the AIN-93G formula with the following modifications and were fed for 4 wk (33). In the ExoMinus diet, exosome-depleted fat-free cow milk was substituted for casein and cornstarch in the AIN-93G diet to provide 10% of total calories (Supplemental Table 2); the remainder of casein in the AIN-93G diet was replaced with soy protein (Bob’s Red Mill) to eliminate any milk-borne miRNAs. Milk was depleted of exosomes by ultrasonication for 60 min (VWR Aquasonic 250T) and incubation at 37°C for 60 min before lyophilization. Ten percent of calories represent a consumption of ∼0.5 L of fat-free milk by an adult on the basis of 352 kcal/L milk and an energy intake of 1760 kcal (34). The ExoPlus control diet was prepared by using exosome-containing milk powder but was otherwise identical to the ExoMinus diet. Components were blended and dried at 40°C overnight. No miR-29b was detectable in the ExoMinus diet, whereas the ExoPlus diet contained ∼83 fmol/g miR-29b, assessed by real-time qPCR. All mouse protocols used in this study were approved by the University of Nebraska–Lincoln Institutional Animal Care and Use Committee. After 4 wk of feeding miRNA-defined diets, mice were killed by carbon dioxide, and blood was collected in EDTA-coated collection tubes. Livers were excised, rinsed with cold saline, and flash-frozen in liquid nitrogen. Ten milligrams of liver was homogenized by using lysis buffer in the NucleoSpin miRNA kit (Machery-Nagel), and mRNA was purified following the manufacturer’s instructions. Reverse transcription was performed with the High Capacity RNA-to-cDNA kit (Life Technologies) for subsequent analysis of Kruppel-like factor 8 (KFL8) expression using real-time qPCR (Supplemental Table 1).
Broccoli-borne miRNAs in humans.
The bioavailability of plant-borne miRNAs is controversial (6–10). We used plasma samples archived from a previous broccoli sprout feeding study in humans to determine whether broccoli-borne miRNAs are detectable in prandial plasma samples (35). In this previous study, 8 healthy adults were provided 34, 68, and 102 g of broccoli sprouts, and timed samples were collected at t = 0, 2, 4, 8, and 24 h. Here, we analyzed samples collected at t = 0 and 4 h from the highest dose of broccoli sprouts for miR-824 and miR-167a in 4 randomly selected participants. Total RNA was isolated from 75 mg of broccoli sprouts by using Trizol. Samples were analyzed for miR-824 and miR-167a by using real-time qPCR (Supplemental Table 1). As of today, there is no evidence that humans synthesize miR-824 and miR-167a (36), i.e., the Brassica-specific miR-824 and plant-specific miR-167a are good markers of broccoli-borne miRNAs in human blood.
Statistical analyses.
Homogeneity of variances was tested by using Bartlett’s test. Variances were heterogeneous for miRNAs in mouse plasma, i.e., murine plasma data were log-transformed before subsequent statistical analysis. AUCs were calculated by using GraphPad Prism 6 (GraphPad Software). Pharmacokinetics data were analyzed by using repeated-measures ANOVA and Fisher’s protected least significant difference test for post hoc comparisons (37). PBMC gene expression data were analyzed by using the paired t test, whereas plasma miRNA concentrations and liver gene expression in mice were analyzed by using the Wilcoxon signed-rank test. StatView 5.0.1 (SAS Institute) was used for statistical analyses. Differences were considered significant if P < 0.05.
Results
Bioavailability of milk-borne miRNAs in humans.
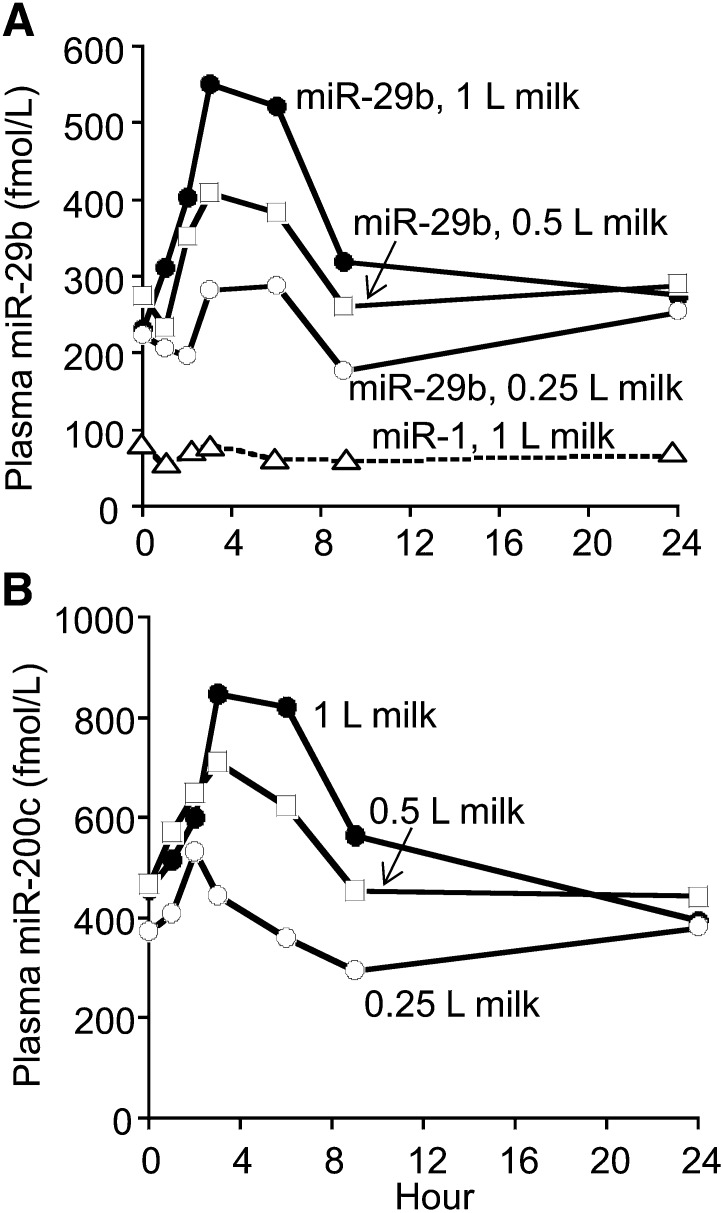
Cow milk (1% fat) contained 148 ± 42 and 680 ± 151 pmol/L of miR-29b and miR-200c, respectively. Humans absorbed considerable amounts of miR-29b from the cow milk (Fig. 1A). In contrast, the plasma concentration of miR-1 did not change after the consumption of 1 L of milk (negative control). Plasma miR-29b concentrations returned to baseline concentrations 9 h after the milk meals for the 0.25- and 0.5-L doses and 24 h after the 1-L dose. The AUC for miR-29b exhibited a linear dose-response relation regarding the amount of milk consumed (Table 1). Likewise, Cmax increased linearly with the amount of milk consumed, if Cmax was corrected for baseline concentrations of miR-29b. The tmax occurred 3.4 to 4.2 h after the milk meal. The miRNA miR-200c is another miRNA present in cow milk. Humans also absorbed considerable amounts of miR-200c from cow milk (Fig. 1B). Although the postprandial AUC for miR-200c was significantly higher for the 0.5- and 1.0-L doses of milk than for the 0.25-L dose, the AUC did not exhibit a linear dose-response relation; the AUCs were not significantly different between the 0.5- and the 1.0-L doses (Table 1).
FIGURE 1.
Plasma time curves of miR-29b (A), miR-1 (negative control; A), and miR-200c (B) after a milk meal in healthy adults. Values are means, n = 5. SEMs were omitted for clarity (compare with Table 1). miR, microRNA.
TABLE 1.
Pharmacokinetics analysis of plasma time curves of microRNAs after milk meals in healthy adults1
| Milk dose (L) |
|||||||
| miR-29b |
miR-200c |
miR-1 | |||||
| Variable | 0.25 | 0.5 | 1.0 | 0.25 | 0.5 | 1.0 | 1.0 |
| Baseline,2 fmol/L | 224 ± 43 | 274 ± 37 | 232 ± 51 | 375 ± 81 | 468 ± 103 | 458 ± 37 | 80 ± 21 |
| Cmax, fmol/L | 372 ± 40a | 484 ± 82a,b | 624 ± 83b | 632 ± 104a | 819 ± 123a,b | 924 ± 121b | 91 ± 18 |
| tmax, h | 4.2 ± 1.5 | 3.4 ± 0.7 | 3.6 ± 0.6 | 2.6 ± 0.9 | 3.8 ± 1.0 | 3.6 ± 0.6 | — |
| AUC,3 fmol/L × h | 327 ± 249a | 672 ± 231a | 1900 ± 275b | 646 ± 187a | 1800 ± 566b | 2260 ± 555b | — |
Values are means ± SEMs, n = 5. Within a variable for the same microRNA, means without a common letter differ (P < 0.05, n = 5). Cmax, maximal plasma concentration; miR, microRNA; tmax, time of peak concentration.
Plasma concentration at time 0 h.
For hours 0–9.
Effects of milk miRNAs on the concentrations of miRNAs in human PBMCs.
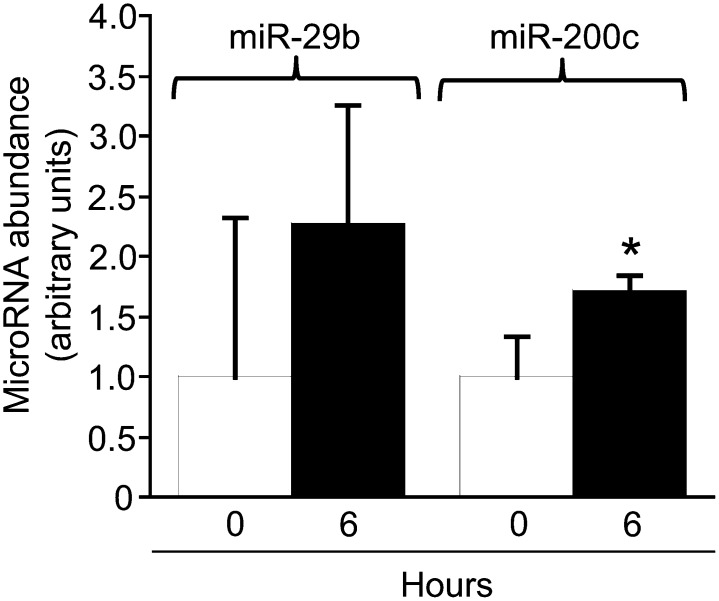
Milk feeding elicited an increase in the concentrations of miRNAs in human PBMCs that was significant only for miR-200c and trended toward significance for miR-29b (P = 0.09) (Fig. 2).
FIGURE 2.
Effects of milk microRNAs on the abundance of microRNAs in human peripheral blood mononuclear cells. Values are means ± SEMs, n = 5. *Different from hour 0, P < 0.05. miR, microRNA.
Effects of milk miRNAs on human gene expression.
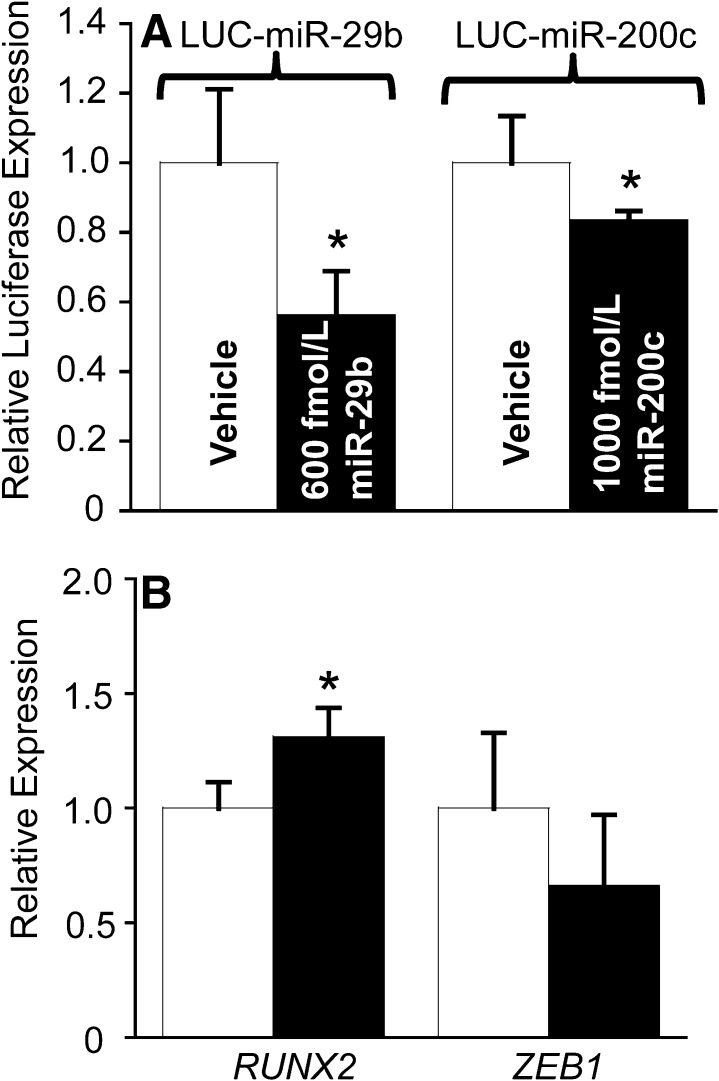
miRNA-containing milk exosomes affected the activity of reporter gene plasmids. When HEK-293 cells were cultured in media supplemented with milk exosomes providing 600 fmol/L miR-29b for 4 h, the activity of the miR-29b–dependent reporter gene LUC-mir-29b decreased by 44 ± 13% (P = 0.04) compared with cells cultured in exosome-depleted culture media (Fig. 3A). Likewise, when HEK-293 cells were cultured in media supplemented with milk exosomes providing 1000 fmol/L miR-200c for 4 h, the activity of the miR-200c–dependent reporter gene LUC-mir-200c decreased by 17 ± 3% (P = 0.02) compared with cells cultured in exosome-depleted culture media.
FIGURE 3.
Effects of milk microRNAs on gene expression in humans. Activities of microRNA reporter genes in HEK-293 cells (A). Expression of endogenous RUNX2 and ZEB1 genes in PBMCs from healthy adults (B). Values are means ± SEMs; n = 4 (LUC-miR-29b), n = 6 (LUC-miR-200c), and n = 5 (PBMCs). *Different from vehicle, P < 0.05. HEK, human embryonic kidney; LUC, luciferase; miR, microRNA; PBMC, peripheral blood mononuclear cell; RUNX2, runt-related transcription factor 2; ZEB1, zinc finger E-box binding homeobox 1.
Milk feeding caused changes in miRNA target gene expression in human PBMCs. The expression of RUNX2 was 31 ± 13% higher 6 h after the milk meal than at time 0 h (Fig. 3B; P = 0.04), consistent with the role of miR-29b as a positive regulator of RUNX2 (17). The difference in the expression of ZEB1 was not significantly lower 6 h after the milk meal than at time 0 h (Fig. 3B).
miRNA depletion studies in mice.
Endogenous miRNA transcription does not compensate for dietary deficiency in C57BL/6 mice. When mice were fed an miRNA-defined diet for 4 wk, the miR-29b plasma concentration decreased by 61% from 152 ± 79 fmol/L in the miRNA-sufficient ExoPlus group to 60 ± 27 fmol/L in the miRNA-depleted ExoMinus group (P = 0.04). The expression of Kruppel-like factor 8 (KLF8) mRNA was 64 ± 32% lower in miRNA-depleted mice than in miRNA-sufficient mice (P = 0.04).
Bioavailability of broccoli miRNAs in humans.
Broccoli sprouts contained 53 ± 16 and 42 ± 10 pmol/kg of miR-167a and miR-824, respectively. No change was observed in miR-167a plasma concentration in healthy adults after consumption of a broccoli sprout meal (13 ± 4 fmol/L at t = 0 h vs. 15 ± 7 fmol/L at t = 4 h; n = 4; P = 0.85). The concentration of miR-824 was below the detection limit (<1 fmol/L) at baseline or 4 h after broccoli sprout feeding in all of the 4 participants who were randomly chosen from our archived plasma samples (35).
Discussion
Some noncoding RNAs, including miRNAs, play essential roles in gene regulation in plant and animal kingdoms. To the best of our knowledge, this is the first study to report that humans absorb quantitatively meaningful amounts of miRNAs from nutritionally relevant amounts of cow milk. Our study provides unambiguous evidence that the amounts of miRNAs absorbed from milk are sufficient to alter human gene expression, i.e., miRNAs from 1 mammalian species can affect gene networks in another species. Our observation that endogenous miRNA synthesis cannot compensate for dietary deficiency is of particular interest, because it implies that a regular dietary miRNA intake may be important to prevent aberrant gene regulation.
Our observations are important from the perspective of maintaining human health on the basis of the following rationale. First, 245 miRNAs have been identified in cow milk (14). A preliminary analysis by sequence alignment suggests that the majority of the nucleotide sequences in bovine milk match the sequences of their human orthologs. We are in the process of developing algorithms for predicting human gene targets for bovine miRNAs, and initial assessments suggest that the number of target genes will exceed 11,000. Second, there is unambiguous evidence linking miRNAs with human health. For example, miR-29b promotes bone health through altering osteoblast and osteoclast differentiation (17, 18), miR-200c decreases cancer risk by targeting the transcription factor ZEB1 (19, 20), and miR-15b, miR-21, miR-27b, miR-34a, miR-106b, miR-130a, miR-155, miR-200c, and miR-223 have been implicated in immune function and Crohn disease (15, 38). Third, supplementation and depletion of milk altered miRNA concentrations and gene expression in human PBMCs and in mouse liver. For example, the concentration of miR-200c was greater 6 h after a milk meal than at time zero in human PBMCs. Likewise, the expression of KLF8 mRNA was greater in livers from milk miRNA-sufficient mice than in those from milk miRNA-depleted mice. Fourth, milk is an important staple in many Western diets. For example, Americans consume large quantities of milk, despite a steady decline from ∼237 pounds (108 kg) in 1987 to 195 pounds (88 kg) in 2012 (13, 39). Note that a large fraction of miRNAs in milk is contained in exosomes, providing protection against degradation (11, 12). Fifth, mice can be depleted of miRNAs by feeding an miRNA-depleted diet.
A recent report suggests that miR-168a from rice is detectable in human and animal sera, and osa-miR-168a decreases the expression of LDL receptor adapter protein 1 in mouse liver (6). The bioavailability of plant-borne miRNAs in humans is controversial (6–10). Note that this research focuses on milk-borne miRNAs for which postprandial plasma concentrations are 200–300 times higher than those reported for osa-miR-168a in humans (6), depending on the species of milk miRNA. We are skeptical of the bioavailability and biologic activity of plant-borne miRNAs and conducted a preliminary screen of miRNAs after a meal providing large amounts of broccoli sprouts. In our broccoli sprouts feeding study, we did not observe a postprandial increase in the Brassica-specific miR-824 or miR-167a. We speculate that the absence of effect in broccoli feeding studies might be due to 1 or some of the following factors. First, exosomes carry numerous surface proteins that might be important for cellular uptake (40, 41). The identity and amino acid sequence of surface proteins implicated in the cellular uptake of exosomes differ between plants and mammals, which might adversely affect the uptake of plant exosomes by human intestinal cells. Second, it is possible that the methylation of the 3′-terminal ribose in plant miRNAs by the methyl transferase HEN1 (42) impairs the intestinal transport of miRNAs. Third, the concentrations of miRNAs in broccoli sprouts are moderately below the concentrations present in milk. It is also unclear whether plant-based miRNAs are encapsulated in vesicles that provide protection against enzymatic and nonenzymatic degradation during food processing. Fourth, it is possible that the number of human genes with sequences complementary to plant miRNAs might be less than those in mammalian miRNAs. We acknowledge that our broccoli feeding study is associated with some uncertainties, e.g., random selection of 2 miRNAs and 1 postprandial time point. Notwithstanding these uncertainties, the analysis of samples from the broccoli feeding study provides a valuable negative control for our studies in milk.
There are some uncertainties associated with our studies of milk miRNAs. First, this study assessed the apparent bioavailability of miRNAs in milk, without taking into account metabolism and degradation in intestinal cells and liver. Ongoing studies of miRNA transport mechanisms suggest that a substantial fraction of some miRNA is degraded in intestinal cells. Therefore, our bioavailability data probably underestimate the true extent of miRNA absorption in humans, because only a fraction of the absorbed miRNAs will appear in the peripheral circulation. Second, our pharmacokinetics analysis suggests that postprandial plasma miRNA concentrations peak at ∼3–4 h after milk consumption. The tmax values for miRNAs are slightly later than the tmax observed for the vitamin riboflavin (tmax = ∼2 h) (24), which is absorbed in the duodenum (43). Considering that milk meals delay gastric emptying (44), we speculate that milk-borne miRNAs are absorbed primarily in the upper intestine. Third, this study focused on miR-29b, miR-200c, and miR-1 (negative control) and did not formally study other miRNAs in milk. We speculate that milk exosomes enter the human intestine by a mechanism that is shared by all exosomal miRNAs, and that any discrimination among miRNAs would only occur after intestinal uptake.
Ongoing and planned activities in our laboratory include the characterization of intestinal miRNA transport mechanisms, including metabolism and basolateral secretion, and the characterization of milk miRNA-dependent gene networks in humans with the use of computational biology approaches.
Supplementary Material
Acknowledgments
S.R.B. and J.Z. designed the research and wrote the manuscript; S.R.B. conducted the research, analyzed the data, and conducted the statistical and pharmacokinetics analyses; C.N., F.X., and J.R.W. designed and conducted the research; and J.Z. had primary responsibility for the final content. Each author contributed to the development of this work, and all authors read and approved the final manuscript.
Footnotes
Abbreviations used: Cmax, maximal plasma concentration; COL1A1, collagen, type I, α1; HEK, human embryonic kidney; KLF8, Kruppel-like factor 8; miRNA (miR), microRNA; LUC, luciferase; PBMC, peripheral blood mononuclear cell; RUNX2, runt-related transcription factor 2; tmax, time of peak concentration; UTR, untranslated region; ZEB1, zinc finger E-box binding homeobox 1.
References
- 1.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 2005;120:623–34. [DOI] [PubMed] [Google Scholar]
- 2.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 2012;336:237–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15–20. [DOI] [PubMed] [Google Scholar]
- 4.Wang X. Composition of seed sequence is a major determinant of microRNA targeting patterns. Bioinformatics 2014;30:1377–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res 2004;14: 10A:1902–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, Li J, Bian Z, Liang X, Cai X, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res 2012;22:107–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snow JW, Hale AE, Isaacs SK, Baggish AL, Chan SY. Ineffective delivery of diet-derived microRNAs to recipient animal organisms. RNA Biol 2013;10:1107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickinson B, Zhang Y, Petrick JS, Heck G, Ivashuta S, Marshall WS. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol 2013;31:965–7. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Zen K, Zhang CY. Reply to Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol 2013;31:967–9. [DOI] [PubMed] [Google Scholar]
- 10.Wang K, Li H, Yuan Y, Etheridge A, Zhou Y, Huang D, Wilmes P, Galas D. The complex exogenous RNA spectra in human plasma: an interface with human gut biota? PLoS ONE 2012;7:e51009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Q, Li M, Wang X, Li Q, Wang T, Zhu Q, Zhou X, Wang X, Gao X, Li X. Immune-related microRNAs are abundant in breast milk exosomes. Int J Biol Sci 2012;8:118–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kosaka N, Izumi H, Sekine K, Ochiya T. microRNA as a new immune-regulatory agent in breast milk. Silence 2010;1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.USDA. Dairy data. 1975 [cited 2013 Oct 21]. Available from: http://www.ers.usda.gov/data-products/dairy-data.aspx#.UmVuAmwo5aQ.
- 14.Chen X, Gao C, Li H, Huang L, Sun Q, Dong Y, Tian C, Gao S, Dong H, Guan D, et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res 2010;20:1128–37. [DOI] [PubMed] [Google Scholar]
- 15.Izumi H, Kosaka N, Shimizu T, Sekine K, Ochiya T, Takase M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J Dairy Sci 2012;95:4831–41. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths-Jones S. miRBase: microRNA sequences and annotation. Curr Protoc Bioinformatics 2010;29(12):1–10. [DOI] [PubMed]
- 17.Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem 2009;284:15676–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossi M, Pitari MR, Amodio N, Di Martino MT, Conforti F, Leone E, Botta C, Paolino FM, Del Giudice T, Iuliano E, et al. miR-29b negatively regulates human osteoclastic cell differentiation and function: Implications for the treatment of multiple myeloma-related bone disease. J Cell Physiol 2013;228:1506–15. [DOI] [PubMed] [Google Scholar]
- 19.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 2008;283:14910–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep 2008;9:582–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watson PE, Watson ID, Batt RD. Total body water volumes for adult males and females estimated from simple anthropometric measurements. Am J Clin Nutr 1980;33:27–39. [DOI] [PubMed] [Google Scholar]
- 22.Zempleni J, Mock DM. Uptake and metabolism of biotin by human peripheral blood mononuclear cells. Am J Physiol 1998;275:C382–8. [DOI] [PubMed] [Google Scholar]
- 23.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. A uniform system for microRNA annotation. RNA 2003;9:277–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zempleni J, Galloway JR, McCormick DB. Pharmacokinetics and utilization of orally and intravenously administered riboflavin in healthy humans. Am J Clin Nutr 1996;63:54–66. [DOI] [PubMed] [Google Scholar]
- 25.Bois FY, Tozer TN, Hauck WW, Chen ML, Patnaik R, Williams RL. Bioequivalence: performance of several measures of extent of absorption. Pharm Res 1994;11:715–22. [DOI] [PubMed] [Google Scholar]
- 26.Xia M, Malkaram SA, Zempleni J. Three promoters regulate the transcriptional activity of the human holocarboxylase synthetase gene. J Nutr Biochem 2013;24:1963–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hata T, Murakami K, Nakatani H, Yamamoto Y, Matsuda T, Aoki N. Isolation of bovine milk-derived microvesicles carrying mRNAs and microRNAs. Biochem Biophys Res Commun 2010;396:528–33. [DOI] [PubMed] [Google Scholar]
- 28.Griffin JB, Rodriguez-Melendez R, Zempleni J. The nuclear abundance of transcription factors Sp1 and Sp3 depends on biotin in Jurkat cells. J Nutr 2003;133:3409–15. [DOI] [PubMed] [Google Scholar]
- 29.Maurisse R, De Semir D, Emamekhoo H, Bedayat B, Abdolmohammadi A, Parsi H, Gruenert DC. Comparative transfection of DNA into primary and transformed mammalian cells from different lineages. BMC Biotechnol 2010;10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weizmann Institute of Science. GeneCards. 2013 [cited 2013 Dec 17]. Available from: http://www.genecards.org/cgi-bin/carddisp.pl?gene=COL1A1.
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔ CT Method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 32.Pestinger V, Wijeratne SSK, Rodriguez-Melendez R, Zempleni J. Novel histone biotinylation marks are enriched in repeat regions and participate in repression of transcriptionally competent genes. J Nutr Biochem 2011;22:328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reeves PG, Nielsen FH, Fahey GC., Jr AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition Ad Hoc Writing Committee on the reformulation of the AIN-76A rodent diet. J Nutr 1993;123:1939–51. [DOI] [PubMed] [Google Scholar]
- 34.USDA. National Nutrient Database for Standard Reference. Nutrient Data Laboratory, Beltsville Human Nutrition Research Center. 2011 [cited 2014 May 1]. Available from: http://ndb.nal.usda.gov/.
- 35.Baier SR, Zbasnik R, Schlegel V, Zempleni J. Off-target effects of sulforaphane include the derepression of long terminal repeats through histone acetylation events. J Nutr Biochem 2014;25:665–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kutter C, Schob H, Stadler M, Meins F, Jr, Si-Ammour A. MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell 2007;19:2417–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abacus Concepts. StatView. Berkeley (CA): Abacus Concepts; 1996.
- 38.Arnold CN, Pirie E, Dosenovic P, McInerney GM, Xia Y, Wang N, Li X, Siggs OM, Karlsson Hedestam GB, Beutler B. A forward genetic screen reveals roles for Nfkbid, Zeb1, and Ruvbl2 in humoral immunity. Proc Natl Acad Sci USA 2012;109:12286–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wisconsin Milk Marketing Board. Dairy statistics. 2013 [cited 2013 Oct 20]. Available from: http://media.eatwisconsincheese.com/dairyimpact/statistics/dairyStatistics.aspx.
- 40.Svensson KJ, Christianson HC, Wittrup A, Bourseau-Guilmain E, Lindqvist E, Svensson LM, Morgelin M, Belting M. Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J Biol Chem 2013;288:17713–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hazawa M, Tomiyama K, Saotome-Nakamura A, Obara C, Yasuda T, Gotoh T, Tanaka I, Yakumaru H, Ishihara H, Tajima K. Radiation increases the cellular uptake of exosomes through CD29/CD81 complex formation. Biochem Biophys Res Commun 2014;446:1165–71. [DOI] [PubMed] [Google Scholar]
- 42.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science 2005;307:932–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Said HM. Recent advances in carrier-mediated intestinal absorption of water-soluble vitamins. Annu Rev Physiol 2004;66:419–46. [DOI] [PubMed] [Google Scholar]
- 44.Calbet JA, MacLean DA. Role of caloric content on gastric emptying in humans. J Physiol 1997;498:553–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.