Abstract
The transient inactivation of protein phosphatases contributes to the efficiency and temporal control of kinase-dependent signal transduction. In particular, members of the protein tyrosine phosphatase family are known to undergo reversible oxidation of their active site cysteine. The thiol oxidation step requires activation of co-localized NADPH oxidases and is mediated by locally produced ROS, in particular H2O2. How oxidized phosphatases are returned to the reduced active state is less well studied. Both major thiol reductive systems, the thioredoxin and the glutathione systems, have been implicated in the reactivation of phosphatases. Here, we show that the protein tyrosine phosphatase PTP1B and the dual-specificity phosphatase PTEN are preferentially reactivated by the thioredoxin system. We show that inducible depletion of TRX1 slows down PTEN re-activation in intact living cells. Finally, using a mechanism-based trapping approach we demonstrate direct thiol disulfide exchange between the active sites of thioredoxin and either phosphatase. The application of thioredoxin trapping mutants represents a complementary approach to direct assays of PTP oxidation in elucidating the significance of redox regulation of PTP function in the control of cell signaling.
Keywords: insulin signaling, phosphatase, reactive oxygen species, redox regulation, substrate trapping, thioredoxin 1
INTRODUCTION
Historically, reactive oxygen species (ROS) have been perceived predominantly as toxic and unwanted by-products of aerobic metabolism. It did not seem likely that ROS could have positive or even essential functions in cellular physiology. However, this view changed when ROS production was recognized as an immune defense mechanism [1, 2]. The phagocyte NADPH oxidase transfers electrons from NADPH to molecular oxygen, generating superoxide, which is then converted to H2O2. Subsequently, homologs of the phagocytic enzyme were found in all cell types, constituting the NOX and DUOX families of NADPH oxidases [1, 2]. Now it is well accepted that NOX-derived ROS also play crucial roles in signal transduction [3, 4]. Many growth factors and hormones, including epidermal growth factor (EGF) [5] platelet-derived growth factor [6] and insulin [7], induce the generation of H2O2, which acts as a second messenger.
H2O2 influences protein function by modifying thiol groups, the oxidation of which initially yields sulphenic acid (R-SOH). Further oxidation, which leads to sulphinic (R-SO2H) and sulphonic acid (R-SO3H), results in modifications that are generally irreversible. However, to prevent irreversible oxidation, the cysteinyl sulphenic acid can condense with a nearby thiol group to form intra- or intermolecular protein disulfide bonds (RSSR) or to become S-glutathionylated (RSSG). Alternatively, the cysteinyl sulphenic acid may also form a cyclic sulphenyl amide. In each case, these modifications are reversible by reduction, making them ideally suited for the transient control of protein function.
Although many signaling proteins, including kinases, are now known to be redox regulated, a particular focus of research has been on the regulation of signal transduction by transient oxidation and inactivation of protein phosphatases. All members of the protein tyrosine phosphatase (PTP) family share the same catalytic mechanism, which depends on a cysteine residue that is essential for catalysis and is located at the base of the active site cleft [8, 9]. Due to the architecture of the active site, this cysteine is deprotonated at physiological pH, which favors its function as the nucleophile in catalysis [8, 9]. Deprotonation is also a prerequisite for its susceptibility to oxidation by H2O2 [10, 11]. In the classical PTPs, oxidation of the active site thiolate promotes the formation of sulphenic acid, which is rapidly converted to a cyclic sulphenamide that induces a conformational change to expose the sulfur atom on the surface of the phosphatase [12]. Therefore, following this conformational change the oxidized sulphur atom becomes accessible to cellular reductants. Unlike most of the classical PTPs, the dual specificity phosphatases harbor a second cysteine in the active site, which condenses to form an intramolecular disulphide bond with the sulphenic acid. In either case, the catalytic cysteine is protected against irreversible oxidation and can be reconverted to its active form by reduction.
Oxidative inactivation of PTPs promotes tyrosine phosphorylation and thus enhances signaling responses. A range of PTPs, including representatives of both the classical and dual-specificity phosphatases, are oxidized transiently in response to a variety of extracellular stimuli, including growth factors, hormones, antigens and ECM components [8, 13]. For example, PTP1B, the prototypic member of the PTP family, was found to be oxidized in response to EGF [14] and insulin [7, 15, 16]. The dual specificity phosphatase PTEN is oxidized in response to LPS [17], PDGF, EGF [18, 19] and insulin [20]. The reduction and reactivation of such PTPs ensures the transient and reversible nature of the oxidative thiol modification, allowing PTPs to recover their activity as soon as H2O2 concentrations decrease and preventing uncontrolled stimulation of signaling pathways. Nevertheless, the exact reductive mechanisms by which PTPs become reactivated have been less well studied than the oxidative mechanisms by which they are inactivated, although both major thiol-reductive systems, the thioredoxin and the glutathione system, have been implicated in the reactivation of PTP1B and PTEN [14, 21–24]. In this study, we use a combination of biochemical analysis, mechanism-based trapping and RNAi-induced suppression to demonstrate the importance of thioredoxin for the reduction and reactivation of PTP1B and PTEN.
RESULTS
TRX1 reactivated oxidized PTP1B and PTEN in vitro
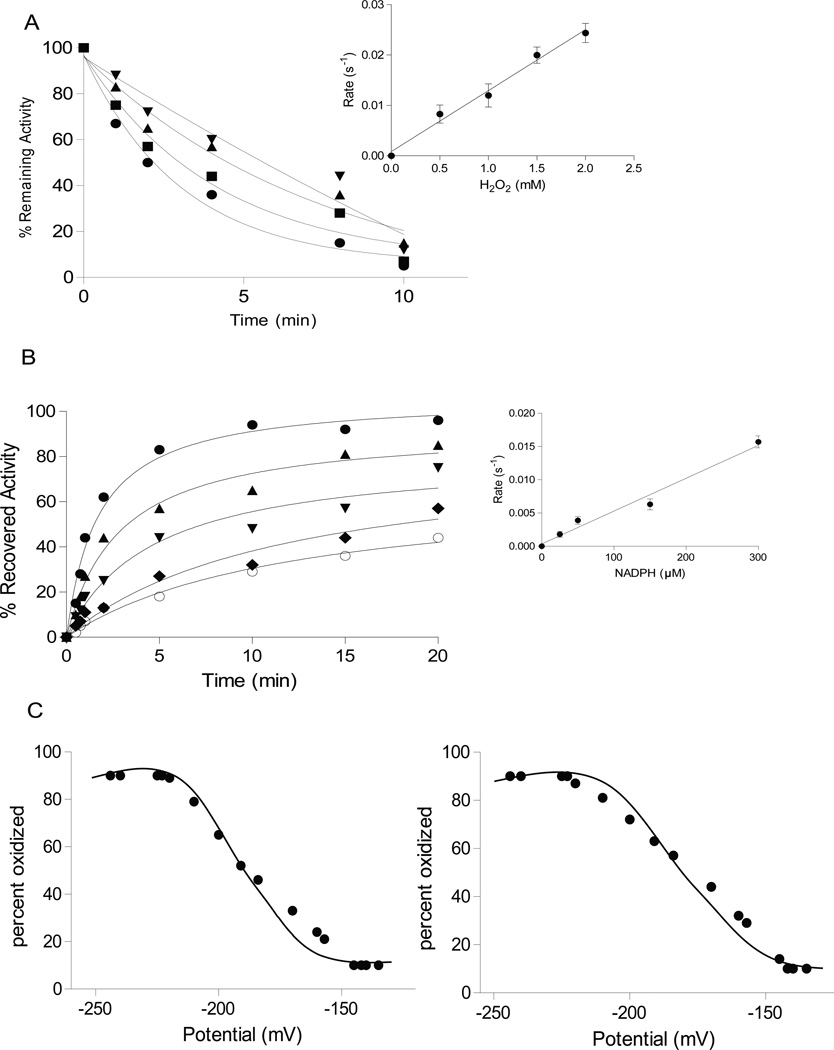
As a first step, we measured inhibition of phosphatase activity by H2O2 in vitro. For PTEN we obtained an apparent second order rate constant of 14 ± 0.5 M−1s−1 (Figure 1A), and a similar rate for PTP1B (Table 1). We then compared different reductants for their ability to reverse the oxidative inactivation: dithiothreitol (DTT), reduced glutathione (GSH), thioredoxin 1 plus thioredoxin reductase (TRX1/TR), and GSH plus glutaredoxin (GRX/GSH). In each case, the rate of reactivation depended on the concentration of the reductant (shown in Figure 1B for the TRX system) and corresponded to a second-order rate constant (Table 1). With all three reductants, more than 90 % of phosphatase activity could be recovered. However, the reactivation of either phosphatase by the TRX1/TR system was 20-fold faster than by GSH and 5-fold faster than by Grx/GSH (Table 1). Thus, the reactivation of H2O2-treated PTEN or PTP1B by the thioredoxin system was kinetically favored over reduction by the glutathione system.
Figure 1. TRX1 reactivated oxidized PTP1B and PTEN in vitro.
(A) Time dependent inactivation of PTEN by H2O2, 0.5 mM (▼), 1 mM (▲), 1.5 mM (■), 2 mM (●). Inset: concentration dependence of the rate of inactivation.
(B) Time dependent reactivation of PTEN by TRX1/TR. NADPH was used at 10 µM (○), 25 µM (◆), 50 µM (▼), 150 µM (▲) or 300 µM (●). Inset: concentration dependence of the rate of reactivation. Data are representative of three independent experiments.
(C) Analysis of the redox potential of PTEN and PTP1B. Recombinant PTEN (left) and PTP1B (right) were incubated in redox buffers with varying ratios of GSH/GSSG and the protein thiol status was determined from the emission spectrum of 5-flourescein maleimide.
Table 1.
Inactivation and reactivation rates of PTEN and PTP1B upon oxidation with H2O2 and reduction with either DTT, GSH, TRX1/TR or GRX/GSH
| Enzyme | H2O2 (M−1s−1) |
DTT (M−1s−1) |
GSH (M−1s−1) |
TRX1/TR (M−1s−1) |
GRX/GSH (M−1s−1) |
|---|---|---|---|---|---|
| PTEN | 14± 0.5 | 0.83± 0.12 | 0.036± 0.01 | 0.95± 0.15 | 0.14± 0.04 |
| PTP1B | 10± 0.8 | 0.55± 0.1 | 0.042± 0.01 | 0.75± 0.2 | 0.12± 0.03 |
We determined the midpoint redox potential of the active site cysteine of the two phosphatases. To this end, we equilibrated PTEN or PTP1B with redox buffers of varying redox potential and quantified free thiols with a fluorescent probe. The mid-point potential of PTEN was determined as −188 mV and that of PTP1B as −179 mV (Figure 1C). Since the midpoint potential of the dithiol-disulfide pair in the active site of human TRX1 has been reported as −230 mV [25], reduction of either PTEN or PTP1B by TRX1 is thermodynamically favorable.
TRX1 formed disulfide-exchange intermediates with PTP1B and PTEN
Having observed that the thioredoxin system is more efficient than the glutathione system in reactivating oxidized PTEN or PTP1B, we aimed to confirm a direct interaction between TRX1 and the phosphatases that would lead to disulfide exchange. To this end, we made use of a mechanism-based trapping approach. In wild type TRX1, the first cysteine in the CXXC motif forms a mixed disulfide intermediate with the target protein, which is subsequently resolved by the second cysteine [26]. Mutagenesis of the second cysteine to serine thus stabilizes the mixed disulfide intermediate, which then becomes amenable to purification and analysis. TRX1 harbours three additional cysteine residues, which are dispensable for catalytic activity, but can cause inactivation by intra- or intermolecular disulphide bond formation [27, 28]. Therefore, we used constructs in which these additional cysteines had been substituted by alanine, which had no effect on the trapping profile [27].
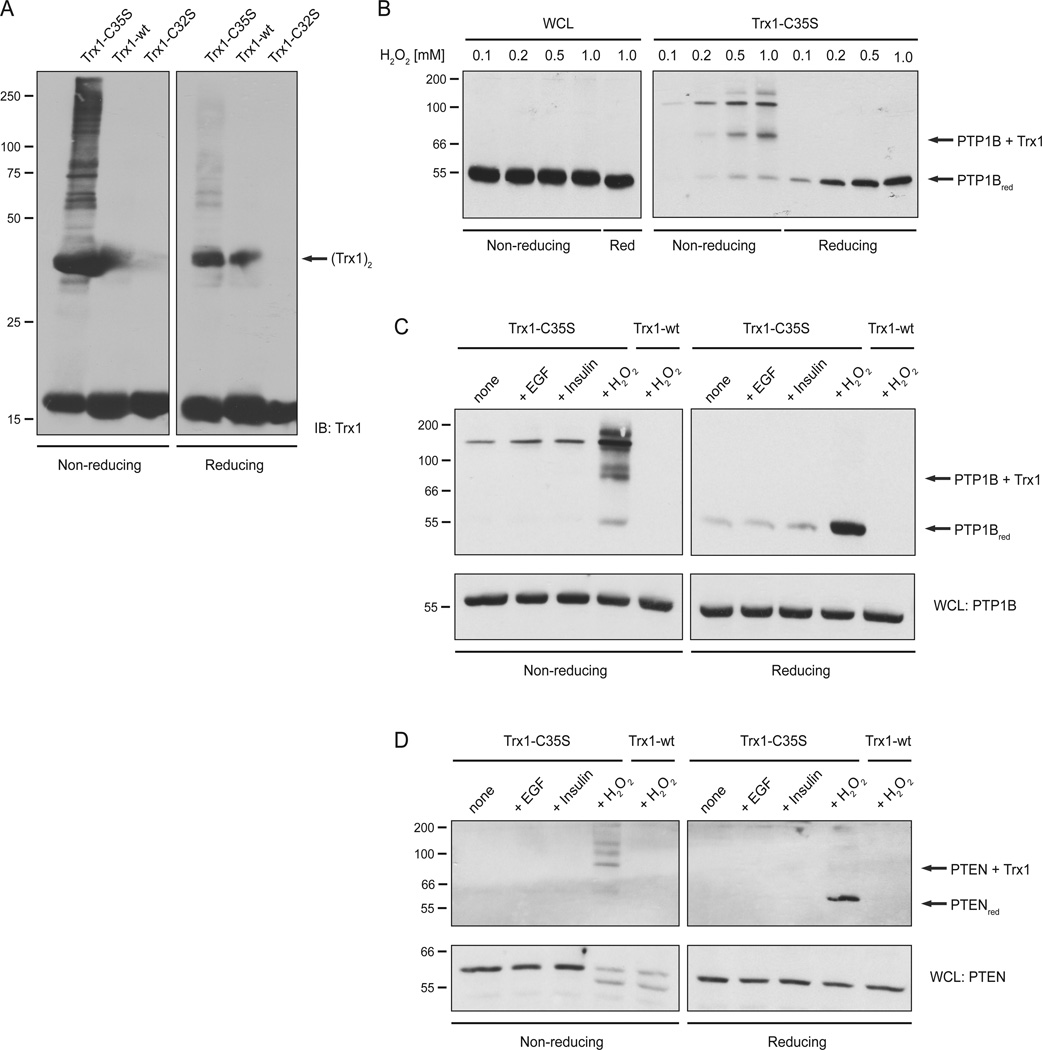
In a first step, we treated HeLa cells with a bolus of exogenous H2O2 to induce oxidation of endogenous proteins. Cells were lysed in the presence of the alkylating reagent N-ethyl maleimide (NEM) to prevent artificial thiol oxidation during or after lysis. Following removal of unreacted NEM, lysates were incubated with recombinant TRX1-C35S, TRX1-WT or TRX1-C32S. The SBP-tagged thioredoxins were collected with strepavidin beads, washed extensively and analyzed by SDS-PAGE and immunoblotting with TRX1 antibodies (Figure 2A). As expected, only the TRX1 trapping mutant (C35S) formed disulfide conjugates with several other proteins (left panel), which were disrupted upon reduction with DTT (right panel).
Figure 2. TRX1 formed disulfide-exchange intermediates with PTP1B and PTEN.
(A) HeLa cells were stimulated with 1 mM H2O2 and lysate samples were incubated with different recombinant TRX1-constructs. TRX1-substrate complexes were resolved by non-reducing and reducing SDS-PAGE and analyzed by anti-TRX1 immunoblotting. Dimerization of exogenously added TRX1 is indicated. The molecular marker is indicated in kDa.
(B) HeLa cells were stimulated with different concentrations of H2O2 and lysate samples were incubated with the TRX1 trapping mutant (TRX1-C35S). Samples of WCL (left panel) and TRX1-substrate complexes (right panel) were resolved by non-reducing and reducing SDS-PAGE and analyzed by anti-PTP1B immunoblotting. The disulfide-linked TRX1-PTP1B complex and reduced PTP1B are indicated. The molecular marker is indicated in kDa.
(C) HeLa cells were stimulated with 50 ng/ml EGF, 50 nM insulin or 1 mM H2O2 and lysate samples were incubated with the TRX1 trapping mutant (TRX1-C35S) or TRX1-wt as control. TRX1-substrate complexes and samples of WCL were analyzed as described in (A). The disulfide-linked TRX1-PTP1B complex and reduced PTP1B are indicated. The molecular marker is indicated in kDa.
(D) Samples from (B) were analyzed by PTEN-specific immunoblotting. The disulfide-linked TRX1-PTEN complex and reduced PTEN are indicated. The markers indicate kDa.
In the next step, we looked specifically for a disulfide exchange intermediate between TRX1 and PTP1B. HeLa cells were treated with increasing concentrations of H2O2 (0.1 – 1 mM) and analyzed by PTP1B-specific immunoblotting. A disulfide-linked PTP1B-TRX1 complex was detected at H2O2 concentrations as low as 0.2 mM (Figure 2B; right panel). In addition to the 1:1 complex, we also detected species of higher molecular weight suggesting that additional stoichiometries are possible (Figure 2C). Similarly, treatment of cells with H2O2 led to oxidation of PTEN, as visualized by electrophoretic gel mobility shifts; in this case we also detected conjugates of PTEN and the TRX1 (Figure 2D).
TRX1 contributed to PTEN reduction in living cells
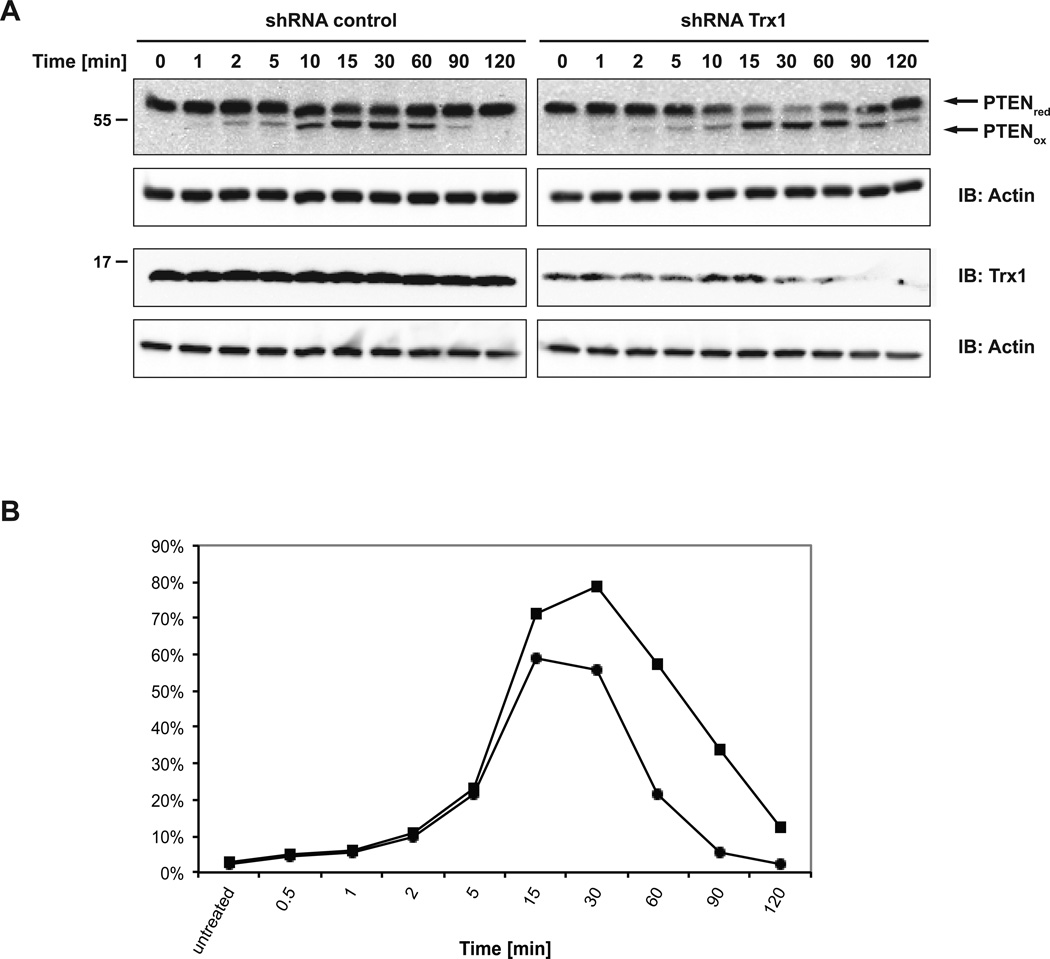
As endogenous TRX1 is an abundant protein, with a cytosolic concentration between 1 and 10 µM [29], it has the potential to be a formidable barrier to oxidation. Therefore, we tested the effects of expressing a tetracycline-inducible shRNA against TRX1. Depletion of TRX1 was induced by the addition of doxycycline for at least five days prior to the experiment. We treated HeLa cells that expressed either TRX1 shRNA or control shRNA with H2O2 and prepared lysate samples at different time points after treatment. Initially, we focused on an analysis of PTEN because the oxidized and reduced forms of this phosphatase can be easily differentiated by mobility on SDS PAGE gels. In cells expressing the control shRNA, most PTEN was converted back to its reduced form after 90 min; however, reduction was clearly delayed in cells expressing TRX1 shRNA, with significant amounts of oxidized PTEN still detectable after 120 min (Figure 3). This result suggested that TRX1 substantially contributes to PTEN reduction in intact living cells.
Figure 3. Effect of suppressing TRX1 on oxidation-reduction of PTEN in cells.
(A) HeLa T-REx-281 (shRNA control) or HeLa T-REx-252 (TRX1 shRNA) cells were treated with 1 µg/ml doxycycline for 7 days. Cells were treated with 500 µM H2O2 and lysate samples were prepared at the time points indicated. Samples were resolved by non-reducing (upper panel) or reducing SDS-PAGE and analyzed by anti-PTEN, anti TRX1 and anti-actin immunoblotting.
(B) Quantitation of a time course of H2O2-induced oxidation of PTEN in the presence (closed circles) and absence (closed squares) of RNAi-mediated suppression of TRX1. The y-axis represents the % of PTEN migrating as the oxidized form on SDS-PAGE compared to the total population of PTEN.
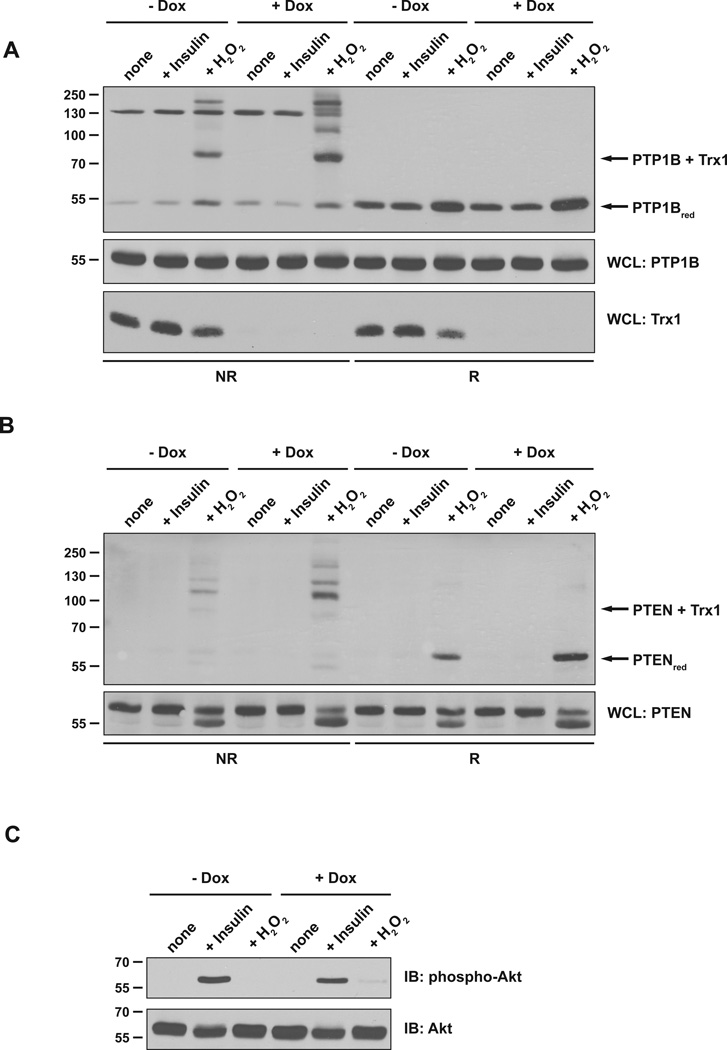
In addition, we considered the possibility that rapid reduction of oxidized phosphatases prior to cell lysis may limit the amount of disulfide-linked TRX1 conjugates detectable by the trapping approach. Therefore, we performed trapping experiments from lysates of HeLa cells in which TRX1 was suppressed by expressing the doxycycline-inducible shRNA. Cells in which TRX1 was efficiently depleted were then stimulated with exogenous H2O2. We observed trapping of both PTP1B (Figure 4A) and PTEN (Figure 4B), with the extent of complex formation significantly increased in lysates from cells with diminished levels of endogenous TRX1 relative to controls. Again, these data are consistent with an important role for endogenous TRX1 in reduction of PTEN and PTP1B in intact cells. Nevertheless, complex formation with the trapping mutant form of TRX1 was not detected in lysates of cells stimulated with insulin (Figure 4A & B), or EGF, despite confirmation of insulin-induced phosphorylation of AKT, consistent with activation of cell signaling (Figure 4C). This suggests that a threshold level of oxidation may be required before the TRX trapping mutants can form complexes with the target PTP in cell lysates.
Figure 4. TRX1 contributed to reduction of PTEN and PTP1B in cells.
(A) HeLa T-REx-252 cells were either left untreated or treated with 1 µg/ml doxycycline (Dox) for 5 days. Cells were stimulated with 100 nM insulin or 1 mM H2O2 and lysate samples were incubated with the TRX1 trapping mutant (TRX1-C35S). TRX1-substrate complexes (upper panel) and samples of WCL (middle panel) were resolved by non-reducing and reducing SDS-PAGE and analyzed by anti-PTP1B immunoblotting. The disulfide-linked TRX1-PTP1B complex and reduced PTP1B are indicated. WCL samples were analyzed by TRX1-specific immunoblotting to verify TRX1 depletion (lower panel).
(B) Samples as in (A) were analyzed by PTEN-specific immunoblotting. The disulfide-linked TRX1-PTEN complex and reduced PTEN are indicated. The markers indicate kDa.
(C) Samples of WCL were analyzed with anti-phospho-Akt (Ser473) antibody to confirm responsiveness of the cells to insulin.
An ectopically expressed TRX1 trapping mutant co-precipitated with PTP1B after insulin stimulation
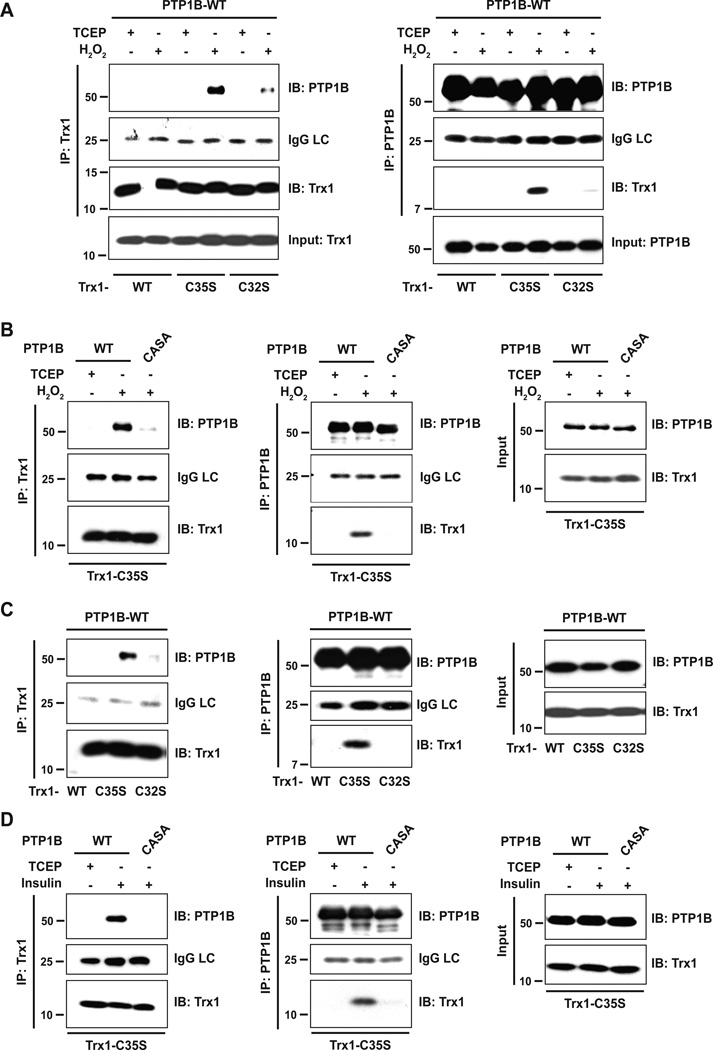
Since we were not able to detect trapping of PTP1B from lysates of EGF- or insulin-stimulated cells, we tested for interaction by co-expressing the TRX1 trapping mutant and PTP1B. To this end, 293T cells were co-transfected with FLAG-tagged wild type PTP1B and His6-tagged TRX1 (TRX1-C35S, TRX1-WT, TRX1-C32S). Cells were either left untreated or treated with H2O2. Lysates were subjected to immunoprecipitation with either His6 (TRX1)- or FLAG (PTP1B)-specific antibodies, and interactions were analyzed by TRX1- and PTP1B-specific immunoblotting. As expected, exogenously applied H2O2 induced disulfide bond formation between PTP1B and the TRX1 trapping mutant (TRX1-C35S) (Figure 5A). To confirm that the catalytic cysteine of PTP1B is essential for disulfide bond formation with TRX1, we used a PTP1B mutant that lacks the catalytic cysteine, but adopts the conformation of the oxidized protein in a stable manner (PTP1B-CASA) [16]. Indeed, PTB1B-CASA failed to form a conjugate with TRX1-C35S (Figure 5B). Using this approach, we also detected TRX1(C35S)-PTP1B complex formation following stimulation of the cells with a physiological trigger, in this case insulin (Figure 5C). Again, conjugation depended on the catalytic cysteine of PTP1B (Figure 5D).
Figure 5. An ectopically expressed TRX1 trapping mutant co-precipitated with PTP1B after insulin stimulation.
(A) Different His6-tagged TRX1 constructs were co-expressed with Flag-tagged PTP1B in 293T cells. Cells were treated with 1 mM H2O2 to oxidize intracellular proteins. Untreated cells were harvested in the presence of TCEP. TRX1 (left panel) or PTP1B (right panel) was immunoprecipitated from whole cell lysates and analyzed by immunoblotting. TRX1 and PTP1B expression was confirmed in input lysates. LC: IgG light chain
(B) TRX1-C35S and PTP1B-WT or the CASA mutant were co-expressed in 293T cells. TRX1 (left panel) or PTP1B (middle panel) was immunoprecipitated from lysates of reduced (TCEP) or H2O2-treated cells and immunoprecipitates were analyzed as described in (A). TRX1 and PTP1B expression was confirmed in input lysates (right panel).
(C) 293T cells co-expressing different His6-tagged TRX1 constructs and Flag-tagged PTP1B were treated with 100 nM insulin for 10 minutes, the time point at which insulin receptor β subunit (IRβ) and IRS-1 displayed maximum tyrosine phosphorylation [16]. TRX1 (left panel) or PTP1B (middle panel) was immunoprecipitated from whole cell lysates and immunoprecipitates were analyzed as described in (A). TRX1 and PTP1B expression was confirmed in input lysates (right panel).
(D) 293T cells co-expressing TRX1-C35S and PTP1B-WT or -CASA were treated with 100 nM insulin. PTP1B-WT or -CASA were immunoprecipitated and analyzed as described in (A). TRX1 and PTP1B expression was confirmed in input lysates (right panel).
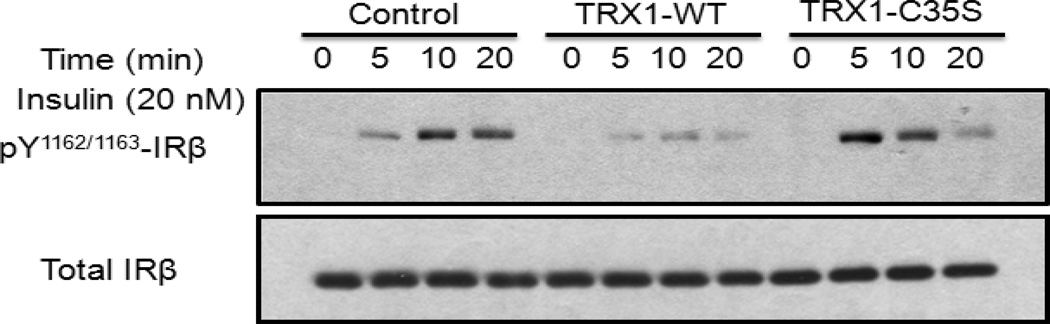
On the basis of these observations, one would predict that alteration of TRX1 expression would result in a change in tyrosine phosphorylation of PTP1B substrates. To test this, we examined a time course of insulin-induced phosphorylation of the insulin receptor β-subunit (IR-β) in the presence of TRX1 wild type or trapping mutant. The data illustrate that overexpression of wild type TRX1, which would be expected to lead to more reduced, active PTP1B, led to decreased phosphorylation of IR-β. In contrast, the TRX1-C35S trapping mutant led to enhanced phosphorylation of IR-β early in the time course (Figure 6). Together, these results further support the conclusion that TRX1 catalytically reactivates reversibly oxidized PTP1B in intact cells, also in response to physiological stimuli.
Figure 6. Effects of TRX1 on insulin signaling.
293T cells expressing vector only, His6-tagged TRX1-WT or His6-tagged TRX1-C35S constructs were treated with 20 nM insulin for 0, 5, 10, 20 minutes. Insulin-stimulated phosphorylation of the insulin receptor β-subunit was monitored by immunoblotting with anti-insulin receptor [pYpY1162/1163] phospho-specific antibody. Sample loading on the gel was standardized by immunoblotting with antibody to insulin receptor β-subunit.
DISCUSSION
Like the PTKs, the PTPs display remarkable specificity in their control of cell signaling and disruption of PTP function has been linked to major human diseases [8, 9]. As a barrier to PTK function, and as critical regulators of signaling in their own right, the activity of the PTP family of enzymes must be tightly regulated in vivo to permit a normal response to extracellular stimuli. Although this is achieved through multiple mechanisms, the recently described redox regulation of PTP function represents a powerful new tier of control over pTyr-dependent signaling. A unique feature of the PTP family is the presence of an essential reactive cysteine at the active site, which is exquisitely sensitive to oxidation with concomitant abrogation of its function as a nucleophile in catalysis. The reduction of molecular oxygen to produce superoxide is triggered using electrons generated by NOX enzymes or the mitochondrial transport chain in response to physiological stimuli, such as growth factors, hormones and cytokines [1–4]. After spontaneous or enzymatic conversion of superoxide to H2O2, which is the major ROS associated with this regulation of signaling, the inactivation of PTPs at defined subcellular locations permits fine-tuning of the signal-induced changes in tyrosine phosphorylation. A critical part of this fine-tuning is the reversible nature of PTP oxidation, in which reduction of the oxidized PTP permits restoration of activity and a return to the ground state. Therefore, in addition to the mechanisms underlying the control of ROS production, it is important to characterize the control of both the oxidation and reduction of the molecular targets of ROS, so as to understand the full scope of the effects of this covalent modification.
In the study of reversible protein phosphorylation, the initial emphasis of research was placed on the protein kinases and the addition of phosphate to target substrates, with the importance of protein phosphatases and the removal of phosphate not being fully appreciated until later. Interestingly, the study of redox regulation has followed a similar path. Considerable progress has been made in defining the mechanism of ROS production, the controls over the levels of ROS in cells and the scope of potential targets of this modification [10, 11]. Nevertheless, for oxidation and inactivation of the PTPs to represent an effective regulatory mechanism, it is essential that this is a reversible modification. Conversion of the active site Cys residue to higher oxidized, sulphinic or sulphonic acid forms would be essentially irreversible; however, there are protective mechanisms in place to favour production of the singly oxidized, reversible sulphenic acid. As demonstrated for PTP1B [11] and PTPα [30], and potentially applying broadly across the classical pTyr-specific PTPs, a condensation reaction converts the sulphenic acid to a cyclic sulphenamide, which induces a conformational change that exposes the oxidized cysteine on the surface of the protein where it may encounter cellular reducing agents. Several dual specificity phosphatases, including PTEN [23], cdc25 [31, 32] and PRL2 [33], and the classical PTP SHP2 [34], display an alternative protective mechanism in which a vicinal cysteinyl residue promotes formation of an intramolecular S-S bond with the sulphenic acid form of the active site cysteine. In addition, intermolecular S-S bond formation, such as glutathionylation, has been demonstrated for several PTPs [21, 22, 35]. The next step is the reduction and reactivation of the oxidized PTPs, the mechanism and importance of which remains relatively underexplored.
A wide variety of low Mr thiols, particularly those with low pKa, have been shown to reduce and reactivate member of the PTP family [36]. The biologically important thiol glutathione has been shown to act on several oxidized PTPs, with reports of glutathionylated forms of PTP1B [21, 22] and the Low Mr PTP [35]. Indeed, experiments with butathionine sulphoximine, to deplete GSH in cell and animal models, attest to its potential importance in vivo [6, 37]. In addition, thiol-containing enzymes, such as thioredoxin (TRX) have also been implicated in reduction of those oxidized PTPs that employ the S-S bond protective mechanism, including cdc25 [32], PRL2 [33] and PTEN [23]. Nevertheless, although PTEN and TRX have been reported to interact in a redox-dependent manner, at least one mode of this interaction involves S-S bond formation between the active site Cys of TRX1 and Cys-212 of the C2 domain of PTEN, which leads to inhibitory effects on phosphatase function that are indirect [38].
Although TRX is best known as a disulphide reductase [26], there have also been reports that it functions as a reducing agent for proteins that lack the second resolving Cys residue, and so do not form an S-S bond, such as methionine sulphoxide reductase [39]. Interaction of TRX with the classical PTPs SHP1 and CD45 has been implicated in the mechanism by which SOCS-1 suppresses ROS-mediated apoptosis [40]. In addition, there have been reports that TRX may also reduce PTP1B [14, 24], although a persulphide modification of the PTP active site Cys generated following sulphydration by H2S was shown to promote reduction by TRX relative to the oxidized form of the phosphatase [41]. These observations suggest that the variety of reducing systems not only represent a barrier to oxidation, but also the reduction and reactivation step may serve as an additional point at which control may be exerted over the redox regulation of PTP function.
In this study, we used a mechanism-based trapping approach, which previously led to identification of TRX target proteins in plants [42], in the secretory pathway of lymphocytes [43, 44], in mitochondria [45] and on the cell surface [27]. This strategy is similar to the approach pioneered by this lab involving substrate trapping mutants to investigate the specificity of PTPs in vivo [46]. We found that the trapping mutant of TRX1 formed mixed disulphide intermediates with both PTP1B and PTEN after H2O2 treatment of cells. The amount of phosphatase covalently captured by the trapping mutant depended on the concentration of exogenously applied H2O2.
In addition to the expected PTP1B-Trx1(CS) conjugate, we observed another anti-PTP1B-reactive species (~120kD), approximately twice the size of the PTP1B monomer (Figure 2B, right panel). We consider it likely that this band represents a covalent PTP1B dimer which was formed as a secondary product of the trapping reaction. PTP1B has been reported to form non-covalent dimers [47] and several of its 10 cysteines appear to be accessible on its surface [48]. Thus, PTP1B-TRX1(CS) conjugates are likely to coprecipitate non-covalently associate PTP1B molecules. These may engage in thiol-disulfide exchange with the mixed disulphide of the PTP1B-Trx1(CS) conjugates and, thereby, create covalent PTP1B dimers. This notion is supported by the observation that no 120kD band can be detected in whole cell lysates or when wild type TRX1 is used in the control trapping reaction. Moreover, as expected, in two of our experiments the intensity of the 120kD band was increased by H2O2 treatment (Fig. 2B, right panel; Fig. 2C, left panel). Although the 120kD band was not further enhanced by H2O2 treatment in another experiment (Figure 4A), we consider it unlikely that PTP1B is constitutively oxidized in these cells, because basal ROS levels in HeLa cells are low [49]. Instead, the observed variability regarding the intensity of 120kD band in the absence of H2O2 treatment is likely due to incomplete thiol blocking by NEM. Depending on cell type and culture conditions, very high NEM concentrations (100 mM or higher) may be needed for complete blocking [50]. Incomplete NEM blocking can be expected to lead to (variable) oxidation of PTP1B during cell lysis, which would entail increased trapping and thus formation of covalent PTP1B dimers. Thus, in our interpretation the observation of the 120kD band is consequential to and supportive of the PTP1B-TRX1 redox interaction.
Although an interaction was induced by H2O2, we were unable to detect trapping of PTP1B or PTEN in lysates from cells stimulated with growth factors. This is consistent with the documented low stoichiometry of oxidation of PTPs produced following acute stimulation with growth factors and hormones, the extent of which may be below the threshold for detection by application of TRX trapping mutants to whole cell lysates [16, 51]. In fact, even in macrophages induced to generate an oxidative burst, only a small fraction of PTEN (16%) became oxidized [17, 18, 20]. Furthermore, due to the fact that it is an abundant protein, endogenous TRX1 may exert a major influence on reduction of oxidized proteins during cell lysis and, therefore, may limit the amount of oxidized PTPs that can be captured by the trapping mutant. Therefore, to increase the endogenous steady state concentration of oxidized TRX1 substrates, we used cells in which TRX1 was depleted by RNA interference. Although the amount of PTP1B and PTEN trapped from lysates of H2O2-stimulated cells was increased after depletion of endogenous TRX1, we still did not detect trapping of PTPs in lysates from growth factor-stimulated cells. Nevertheless, depletion of endogenous TRX1 delayed the recovery of the reduced form of PTEN after H2O2 treatment, highlighting the importance of TRX1 as a mediator of PTEN reactivation for regulation of PIP3-dependent signals in intact cells.
In contrast, TRX trapping mutants were effective when expressed in cells so that complex formation occurred in situ. Their application in this context allowed us to define the basis for the TRX-PTP1B interaction in more detail (Figure 7). Following co-expression of the TRX1 trapping mutant with PTP1B, we observed that stimulation of the cells with insulin induced co-precipitation of PTP1B with the TRX1 trapping mutant, but not wild type or C32S mutant, in which the active site cysteine of TRX1 was changed to serine. In turn, a mutant form of PTP1B that adopts the structural features of the oxidized form of the enzyme, but lacks its catalytic cysteine, PTP1B-CASA [16], was not targeted by TRX1 after insulin stimulation. Thus, the interaction was based upon the catalytic cysteines of both proteins. Overall, in addition to highlighting the important role of thioredoxin in reduction and reactivation of PTPs, the TRX trapping mutants illustrate another approach to defining the importance of reversible oxidation in the regulation of PTP function in general and tyrosine phosphorylation-dependent signaling in a broad array of signaling contexts.
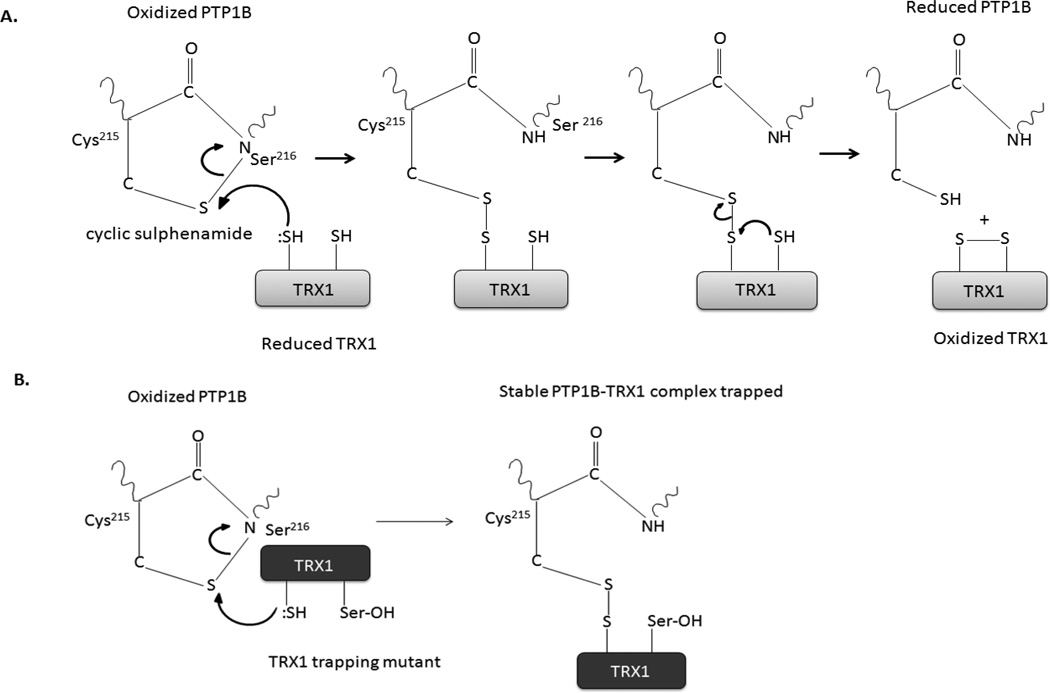
Figure 7. TRX1-mediated reduction of PTP1B.
(A) The proposed mechanism for reduction of the cyclic sulphenamide form of oxidized PTP1B by TRX1.
(B) The absence of the resolving cysteine (Cys-35) generates a trapping mutant form of TRX1 that forms a stable complex with oxidized PTP1B (lower panel).
EXPERIMENTAL PROCEDURES
Antibodies
The following antibodies were used for immunoblotting and immunoprecipitation: anti-His (Genescript), anti-actin and anti-Flag M2 (Sigma), anti-phospho-Akt (Ser473), anti-Akt, anti-PTEN and anti-TRX1 (Cell Signaling), anti-PTP1B (FG6; N.K. Tonks), rabbit polyclonal anti-insulin receptor [pYpY1162/1163] phospho-specific antibody (Invitrogen) and light chain-specific Anti-Mouse IgG (Jackson Immuno Research Laboratories).
Protein expression, purification and quantitation
Bacterial expression and purification of TRX1 (WT and mutant) and PTP1B were described previously [41, 52]. For mammalian expression TRX1 was subcloned into PTT3 vector using Xba1 (5´) and BamH1 (3´) restriction sites [53]. C32S-TRX1 and C35S-TRX1 mutants were generated using a Mutagenesis Kit (Stratagene) and the sequence was verified. The enzymes Thioredoxin Reductase and Glutaredoxin were obtained from Sigma-Aldrich. Throughout the study protein concentration was determined using Bradford reagent.
Inducible depletion of TRX1 by shRNA
To develop a doxycycline-dependent inducible TRX1 knockdown system in HeLa cells, the H1 promoter of the retroviral vector pSUPER-Retro-NeoGFP (OligoEngine) was replaced with the TetR-repressable modified H1 promoter of pTER (kindly provided by Hans Clevers, Utrecht, Netherlands). The following shRNAs were cloned into the newly generated pSUPER-Retro-TRE-NeoGFP vector using BglII and XhoI:
GATCCCGGATGTTGCTTCAGAGTGTTTCAAGAGAACACTCTGAAGCAACATCCTTTTTGGAAC GATCCCGCATGCCAACATTCCAGTTTTCAAGAGAAACTGGAATGTTGGCATGCTTTTTGGAAC.
The resulting constructs were used for retroviral transduction of HeLa T-Rex cells (Invitrogen) (HeLa T-REx-252 & HeLa T-REx-281). Positive clones were selected using G418 (1 mg/ml) and further sorted for GFP expression by flow cytometry. Analysis of TRX1 knockdown efficiency after 7 days of doxycycline (1 µg/ml) exposure revealed a 95% reduction of TRX1 protein levels in HeLa T-REx-252 cells. No effect on TRX1 levels was observed in HeLa T-REx-281 cells, which were therefore used as a negative control.
Cell culture, transient transfection and immunoprecipitation
Flag-tagged PTP1B and His6-tagged TRX-1 constructs were cotransfected into 293T cells using FuGENE 6 transfection reagent (Roche). After 48 hrs, transfected cells were serum starved in DMEM for 16 hours. Cells were treated with 100 nM insulin or 1 mM H2O2 for 10 min and 5 min, respectively, at 37°C. Untreated and treated cells were washed with PBS and lysed in de-gassed lysis buffer (25 mM HEPES, pH 7.4, 150 mM NaCl, 0.25 % (w/v) Deoxycholate, 1 % (v/v) Triton X-100, 25 mM NaF, 10 mM MgCl2, 1 mM EDTA, 10 % (v/v) glycerol, 0.5 mM PMSF, 10 mM benzamidine and EDTA-free Complete Mini Protease Inhibitor tablets (Roche)) for 1 h at 4°C. Soluble proteins were harvested by centrifugation at 13,000 × g for 10 min at 4°C and quantitated. TRX1 and PTP1B were immunoprecipitated with anti-His and anti-PTP1B or anti-Flag M2, respectively, using 400 µg total protein for 2 hrs at 4°C. Protein complexes were isolated with Protein A/G-Sepharose for 1 h at 4°C, washed once with lysis buffer and twice with IP wash buffer (PBS, pH 7.4, 0.05 % (w/v) BSA, 0.05 % (v/v) Tween-20 and protease inhibitors) and boiled in reducing SDS sample buffer.
To test the effects of TRX1 on insulin signaling, His6-tagged TRX-1 or vector only constructs were transfected into 293T cells using FuGENE 6 transfection reagent (Roche). After 48 hrs, transfected cells were serum starved in DMEM for 16 hours. Cells were treated with 20 nM insulin for various times (0, 5, 10, 20 minutes) at 37°C. Cells were washed with PBS and lysed in de-gassed lysis buffer as mentioned above. Soluble proteins were harvested by centrifugation at 13,000 × g for 10 min at 4°C and quantitated. Total proteins in the cell lysates were separated by SDS-PAGE and specific tyrosyl phosphorylation of the IR-β subunit was monitored using rabbit polyclonal anti-insulin receptor [pYpY1162/1163] phospho-specific antibody.
TRX1 substrate trapping
HeLa or HeLa T-REx-252 cells at 80–90 % confluency were serum-starved overnight in DMEM Low Glucose, w/o phenol red (Sigma). Cells were stimulated with human EGF (50 ng/ml; PeproTech), insulin (50 or 100 nM; Calbiochem) or H2O2 (1 mM; Sigma) for 5 min at room temperature. Cells were lysed using PBS with 1 % (v/v) Triton X-100, 100 mM N-ethyl maleimide (NEM) and Complete Mini Protease Inhibitors EDTA-free (Roche) for 20 min on ice. Soluble proteins were harvested by centrifugation at 13,000 × g for 10 min at 4°C and quantitated. Total protein (~25 µg) was used as lysate control. Total protein (0.5–1mg) was applied to Zeba Spin Desalting Columns (Thermo Scientific) and samples were collected by centrifugation at 1,000 × g for 5 min at 4°C. Streptavidin sepharose beads (GE Healthcare) were loaded with 100 µg of recombinant TRX1/sample in PBS with 20 mM DTT for 2 hrs at 4°C and subsequently washed to remove the reductant. The loaded beads were added to the lysate samples and incubated for 1 h at 4°C. The reaction was stopped with 20 mM NEM and beads were washed with buffer 1 (PBS with 1 % (v/v) Triton X-100, 10 mM NEM and 500 mM NaCl), buffer 2 (PBS with 1 % (v/v) Triton X-100, 1 mM NEM and 0.1 % (w/v) SDS) and PBS. Protein complexes were eluted in PBS with 2 mM biotin for 30 min at 4°C and equal parts were boiled in non-reducing or reducing (20 mM DTT) Laemmli sample buffer and subsequently treated with 40 mM NEM.
Time course of transient PTEN oxidation in HeLa cells
HeLa T-REx-281 (shRNA control) or −252 (shRNA TRX1) cells at 70–90 % confluency were stimulated with 500 µM H2O2. At the indicated time points, the supernatant was removed and cells were treated with 10 mM NEM in PBS for 5 min at 4°C. Cells were harvested in lysis buffer (TBS with 1 % (v/v) Triton X-100, 10 mM N-ethyl maleimide (NEM) and Complete Mini Protease Inhibitors EDTA-free (Roche)) and samples were incubated for 30 min at 4°C. Lysates were cleared by centrifugation at 13,000 × g for 15 min at 4°C. Equal parts of the lysates were boiled in non-reducing or reducing (20 mM DTT) Laemmli sample buffer.
Rate of inactivation and reactivation of PTP1B/ PTEN
PTEN or PTP1B [10 µM] was incubated with varying concentrations of H2O2 (0–5 mM) in 50 mM Tris pH 6.5, 100 mM NaCl and 0.1 % BSA at 25°C and the rate of inactivation was measured using 6,8-Difluoro-4-Methylumbelliferyl Phosphate (DiFMUP) as substrate. To measure the rate of reactivation, PTP1B or PTEN [10 µM] was oxidized and aliquots were diluted 2-fold into 50 mM Tris pH 6.5, 100 mM NaCl, 0.1 % BSA, and 1 mM EDTA containing varying amounts of DTT [10–200 mM], GSH [50–200 mM], TRX1/TR (0.2–5 equivalents of TRX1 to PTP1B or PTEN) or GSH and GR/GRX (10 mM GSH, together with glutathione reductase (GR) and glutaredoxin, both at 5 µM GRX) for varying time periods (1–60 min) and the phosphatase activity was monitored. For assays using TRX1/TR and GSH/Grx as reductants, NADPH (25–300 µM) was included at a ratio of 200:1.
Measurement of redox potentials
To determine the redox potential of the active site cysteine of PTP1B or PTEN, recombinant enzymes (20 µg) were incubated in 100 mM HEPES, pH 7.0 and 1 mM EDTA in the presence of varying concentrations of GSH/GSSG for 2 hrs at 25°C. Protein samples were precipitated using TCA (20 %) for 30 min at 4°C and the precipitants were centrifuged at 10,000 × g for 10 min and washed with acetone. Pellets were resuspended in 100 mM Hepes, pH 7.0, 1 mM EDTA and 1 % (w/v) SDS and alkylated using 0.1 mM 5-FAM (fluorescein-5-maleimide, Molecular Probes) for 2 hrs at 25°C. The unreacted 5-FAM was removed using PD-10 size exclusion columns (GE Healthcare) and the degree of labeling was calculated using the following equation (molar extinction coefficient (ε) of 5-FAM = 68000 M−1 cm−1):
moles fluor/moles protein = Emissionmax / ε X protein concentration
Fluorescence spectra were recorded and the fluorescence emission at 520 nm was used to quantitate the reduced and oxidized cysteine. The redox potential was calculated using the Nernst equation Eh = E0−RT/nf ln 2[GSH]/GSSG where E0 is the standard reduction potential (−240 mV).
ACKNOWLEDGEMENTS
We thank Darryl Pappin (Cold Spring Harbor Laboratory) for his advice on the proposed mechanism for reduction of PTP1B by TRX1. This research was supported by NIH grant GM55989 to N.K.T. and the CSHL Cancer Centre Support Grant CA45508. U.S. was the recipient of a postdoctoral fellowship from the Cold Spring Harbor Laboratory (CSHL). R.G was supported by a scholarship from the Studienstiftung des deutschen Volkes. N.K.T. is also grateful for support from the following foundations; The Gladowsky Breast Cancer Foundation, The Don Monti Memorial Research Foundation, Hansen Memorial Foundation, West Islip Breast Cancer Coalition for Long Island, Glen Cove CARES, Find a Cure Today (FACT), Constance Silveri, Robertson Research Fund and the Masthead Cove Yacht Club Carol Marcincuk Fund.
Abbreviations used
- DTT
dithiothreitol
- EGF
epidermal growth factor
- Grx
glutaredoxin
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- H2O2
hydrogen peroxide
- PBS
phosphate-buffered saline
- PTEN
phosphatase and tensin homolog
- PTP
protein tyrosine phosphatase
- ROS
reactive oxygen species
- TBS
Tris-buffered saline
- TR
thioredoxin reductase
- Trx1
thioredoxin 1
- WCL
whole cell lysate
Footnotes
Authors’ contributions
U.S., A.H., N.K., R.G. and L.W. performed experiments; U.S., A.H., N.K., R.G., L.W., T.D. and N.K.T. designed experiments and analyzed data. U.S and N.K.T. wrote the paper, with input from all the authors. N.K.T. directed the study.
REFERENCES
- 1.Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- 2.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 3.Droge W. Free radicals in the physiological control of cell function. Physiol Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 4.Finkel T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bae YS, Kang SW, Seo MS, Baines IC, Tekle E, Chock PB, Rhee SG. Epidermal growth factor (EGF)-induced generation of hydrogen peroxide. Role in EGF receptor-mediated tyrosine phosphorylation. J Biol Chem. 1997;272:217–221. [PubMed] [Google Scholar]
- 6.Meng TC, Fukada T, Tonks NK. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol Cell. 2002;9:387–399. doi: 10.1016/s1097-2765(02)00445-8. [DOI] [PubMed] [Google Scholar]
- 7.Meng TC, Buckley DA, Galic S, Tiganis T, Tonks NK. Regulation of insulin signaling through reversible oxidation of the protein-tyrosine phosphatases TC45 and PTP1B. J Biol Chem. 2004;279:37716–37725. doi: 10.1074/jbc.M404606200. [DOI] [PubMed] [Google Scholar]
- 8.Tonks NK. Protein tyrosine phosphatases--from housekeeping enzymes to master regulators of signal transduction. Febs J. 2013;280:346–378. doi: 10.1111/febs.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 10.Tonks NK. Redox redux: revisiting PTPs and the control of cell signaling. Cell. 2005;121:667–670. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 11.Ostman A, Frijhoff J, Sandin A, Bohmer FD. Regulation of protein tyrosine phosphatases by reversible oxidation. J Biochem. 2011;150:345–356. doi: 10.1093/jb/mvr104. [DOI] [PubMed] [Google Scholar]
- 12.Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, Tonks NK, Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 13.Frijhoff J, Dagnell M, Godfrey R, Ostman A. Regulation of Protein Tyrosine Phosphatase Oxidation in Cell Adhesion and Migration. Antioxid Redox Signal. 2013 doi: 10.1089/ars.2013.5643. [DOI] [PubMed] [Google Scholar]
- 14.Lee SR, Kwon KS, Kim SR, Rhee SG. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J Biol Chem. 1998;273:15366–15372. doi: 10.1074/jbc.273.25.15366. [DOI] [PubMed] [Google Scholar]
- 15.Mahadev K, Zilbering A, Zhu L, Goldstein BJ. Insulin-stimulated hydrogen peroxide reversibly inhibits protein-tyrosine phosphatase 1b in vivo and enhances the early insulin action cascade. J Biol Chem. 2001;276:21938–21942. doi: 10.1074/jbc.C100109200. [DOI] [PubMed] [Google Scholar]
- 16.Haque A, Andersen JN, Salmeen A, Barford D, Tonks NK. Conformation-sensing antibodies stabilize the oxidized form of PTP1B and inhibit its phosphatase activity. Cell. 2011;147:185–198. doi: 10.1016/j.cell.2011.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leslie NR, Bennett D, Lindsay YE, Stewart H, Gray A, Downes CP. Redox regulation of PI 3-kinase signalling via inactivation of PTEN. Embo J. 2003;22:5501–5510. doi: 10.1093/emboj/cdg513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon J, Lee SR, Yang KS, Ahn Y, Kim YJ, Stadtman ER, Rhee SG. Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. Proc Natl Acad Sci U S A. 2004;101:16419–16424. doi: 10.1073/pnas.0407396101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boivin B, Zhang S, Arbiser JL, Zhang ZY, Tonks NK. A modified cysteinyl-labeling assay reveals reversible oxidation of protein tyrosine phosphatases in angiomyolipoma cells. Proc Natl Acad Sci U S A. 2008;105:9959–9964. doi: 10.1073/pnas.0804336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seo JH, Ahn Y, Lee SR, Yeol Yeo C, Chung Hur K. The major target of the endogenously generated reactive oxygen species in response to insulin stimulation is phosphatase and tensin homolog and not phosphoinositide-3 kinase (PI-3 kinase) in the PI-3 kinase/Akt pathway. Mol Biol Cell. 2005;16:348–357. doi: 10.1091/mbc.E04-05-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett WC, DeGnore JP, Konig S, Fales HM, Keng YF, Zhang ZY, Yim MB, Chock PB. Regulation of PTP1B via glutathionylation of the active site cysteine 215. Biochemistry. 1999;38:6699–6705. doi: 10.1021/bi990240v. [DOI] [PubMed] [Google Scholar]
- 22.Barrett WC, DeGnore JP, Keng YF, Zhang ZY, Yim MB, Chock PB. Roles of superoxide radical anion in signal transduction mediated by reversible regulation of protein-tyrosine phosphatase 1B. J Biol Chem. 1999;274:34543–34546. doi: 10.1074/jbc.274.49.34543. [DOI] [PubMed] [Google Scholar]
- 23.Lee SR, Yang KS, Kwon J, Lee C, Jeong W, Rhee SG. Reversible inactivation of the tumor suppressor PTEN by H2O2. J Biol Chem. 2002;277:20336–20342. doi: 10.1074/jbc.M111899200. [DOI] [PubMed] [Google Scholar]
- 24.Dagnell M, Frijhoff J, Pader I, Augsten M, Boivin B, Xu J, Mandal PK, Tonks NK, Hellberg C, Conrad M, et al. Selective activation of oxidized PTP1B by the thioredoxin system modulates PDGF-beta receptor tyrosine kinase signaling. Proc Natl Acad Sci U S A. 2013;110:13398–13403. doi: 10.1073/pnas.1302891110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watson WH, Pohl J, Montfort WR, Stuchlik O, Reed MS, Powis G, Jones DP. Redox potential of human thioredoxin 1 and identification of a second dithiol/disulfide motif. J Biol Chem. 2003;278:33408–33415. doi: 10.1074/jbc.M211107200. [DOI] [PubMed] [Google Scholar]
- 26.Collet JF, Messens J. Structure, function, and mechanism of thioredoxin proteins. Antioxid Redox Signal. 2010;13:1205–1216. doi: 10.1089/ars.2010.3114. [DOI] [PubMed] [Google Scholar]
- 27.Schwertassek U, Balmer Y, Gutscher M, Weingarten L, Preuss M, Engelhard J, Winkler M, Dick TP. Selective redox regulation of cytokine receptor signaling by extracellular thioredoxin-1. Embo J. 2007;26:3086–3097. doi: 10.1038/sj.emboj.7601746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casagrande S, Bonetto V, Fratelli M, Gianazza E, Eberini I, Massignan T, Salmona M, Chang G, Holmgren A, Ghezzi P. Glutathionylation of human thioredoxin: a possible crosstalk between the glutathione and thioredoxin systems. Proc Natl Acad Sci U S A. 2002;99:9745–9749. doi: 10.1073/pnas.152168599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holmgren A, Luthman M. Tissue distrubution and subcellular localization of bovine thioredoxin determined by radioimmunoassay. Biochemistry. 1978;17:4071–4077. doi: 10.1021/bi00612a031. [DOI] [PubMed] [Google Scholar]
- 30.Yang J, Groen A, Lemeer S, Jans A, Slijper M, Roe SM, den Hertog J, Barford D. Reversible oxidation of the membrane distal domain of receptor PTPalpha is mediated by a cyclic sulfenamide. Biochemistry. 2007;46:709–719. doi: 10.1021/bi061546m. [DOI] [PubMed] [Google Scholar]
- 31.Savitsky PA, Finkel T. Redox regulation of Cdc25C. J Biol Chem. 2002;277:20535–20540. doi: 10.1074/jbc.M201589200. [DOI] [PubMed] [Google Scholar]
- 32.Sohn J, Rudolph J. Catalytic and chemical competence of regulation of cdc25 phosphatase by oxidation/reduction. Biochemistry. 2003;42:10060–10070. doi: 10.1021/bi0345081. [DOI] [PubMed] [Google Scholar]
- 33.Funato Y, Miki H. Reversible oxidation of PRL family protein-tyrosine phosphatases. Methods. 2014;65:184–189. doi: 10.1016/j.ymeth.2013.06.032. [DOI] [PubMed] [Google Scholar]
- 34.Chen CY, Willard D, Rudolph J. Redox regulation of SH2-domain-containing protein tyrosine phosphatases by two backdoor cysteines. Biochemistry. 2009;48:1399–1409. doi: 10.1021/bi801973z. [DOI] [PubMed] [Google Scholar]
- 35.Abdelsaid MA, El-Remessy AB. S-glutathionylation of LMW-PTP regulates VEGF-mediated FAK activation and endothelial cell migration. J Cell Sci. 2012;125:4751–4760. doi: 10.1242/jcs.103481. [DOI] [PubMed] [Google Scholar]
- 36.Parsons ZD, Gates KS. Thiol-dependent recovery of catalytic activity from oxidized protein tyrosine phosphatases. Biochemistry. 2013;52:6412–6423. doi: 10.1021/bi400451m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Limon-Pacheco JH, Hernandez NA, Fanjul-Moles ML, Gonsebatt ME. Glutathione depletion activates mitogen-activated protein kinase (MAPK) pathways that display organ-specific responses and brain protection in mice. Free Radic Biol Med. 2007;43:1335–1347. doi: 10.1016/j.freeradbiomed.2007.06.028. [DOI] [PubMed] [Google Scholar]
- 38.Meuillet EJ, Mahadevan D, Berggren M, Coon A, Powis G. Thioredoxin-1 binds to the C2 domain of PTEN inhibiting PTEN's lipid phosphatase activity and membrane binding: a mechanism for the functional loss of PTEN's tumor suppressor activity. Arch Biochem Biophys. 2004;429:123–133. doi: 10.1016/j.abb.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 39.Kim HY, Kim JR. Thioredoxin as a reducing agent for mammalian methionine sulfoxide reductases B lacking resolving cysteine. Biochem Biophys Res Commun. 2008;371:490–494. doi: 10.1016/j.bbrc.2008.04.101. [DOI] [PubMed] [Google Scholar]
- 40.Oh J, Hur MW, Lee CE. SOCS1 protects protein tyrosine phosphatases by thioredoxin upregulation and attenuates Jaks to suppress ROS-mediated apoptosis. Oncogene. 2009;28:3145–3156. doi: 10.1038/onc.2009.169. [DOI] [PubMed] [Google Scholar]
- 41.Krishnan N, Fu C, Pappin DJ, Tonks NK. H2S-Induced sulfhydration of the phosphatase PTP1B and its role in the endoplasmic reticulum stress response. Sci Signal. 2011;4:ra86. doi: 10.1126/scisignal.2002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Motohashi K, Kondoh A, Stumpp MT, Hisabori T. Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc Natl Acad Sci U S A. 2001;98:11224–11229. doi: 10.1073/pnas.191282098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dick TP, Bangia N, Peaper DR, Cresswell P. Disulfide bond isomerization and the assembly of MHC class I-peptide complexes. Immunity. 2002;16:87–98. doi: 10.1016/s1074-7613(02)00263-7. [DOI] [PubMed] [Google Scholar]
- 44.Dick TP, Cresswell P. Thiol oxidation and reduction in major histocompatibility complex class I-restricted antigen processing and presentation. Methods Enzymol. 2002;348:49–54. doi: 10.1016/s0076-6879(02)48625-9. [DOI] [PubMed] [Google Scholar]
- 45.Engelhard J, Christian BE, Weingarten L, Kuntz G, Spremulli LL, Dick TP. In situ kinetic trapping reveals a fingerprint of reversible protein thiol oxidation in the mitochondrial matrix. Free Radic Biol Med. 2011;50:1234–1241. doi: 10.1016/j.freeradbiomed.2011.01.035. [DOI] [PubMed] [Google Scholar]
- 46.Flint AJ, Tiganis T, Barford D, Tonks NK. Development of "substrate-trapping" mutants to identify physiological substrates of protein tyrosine phosphatases. Proc Natl Acad Sci U S A. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderie I, Schulz I, Schmid A. Characterization of the C-terminal ER membrane anchor of PTP1B. Experimental Cell Research. 2007;313:3189–3197. doi: 10.1016/j.yexcr.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 48.Hansen SK, Cancilla MT, Shiau TP, Kung J, Chen T, Erlanson DA. Allosteric inhibition of PTP1B activity by selective modification of a non-active site cysteine residue. Biochemistry. 2005;31:7704–7712. doi: 10.1021/bi047417s. [DOI] [PubMed] [Google Scholar]
- 49.Lou YW, Chen YY, Hsu SF, Chen RK, Lee CL, Khoo KH, Tonks NK, Meng TC. Redox regulation of the protein tyrosine phosphatase PTP1B in cancer cells. Febs J. 2008;275:69–88. doi: 10.1111/j.1742-4658.2007.06173.x. [DOI] [PubMed] [Google Scholar]
- 50.Sobotta MC, Barata AG, Schmidt U, Mueller S, Millonig G, Dick TP. Exposing cells to H2O2: a quantitative comparison between continuous low-dose and one-time high-dose treatments. Free Radic Biol Med. 2013;60:325–335. doi: 10.1016/j.freeradbiomed.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 51.Karisch R, Neel BG. Methods to monitor classical protein-tyrosine phosphatase oxidation. Febs J. 2012 doi: 10.1111/j.1742-4658.2012.08626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwertassek U, Weingarten L, Dick TP. Identification of redox-active cell-surface proteins by mechanism-based kinetic trapping. Sci STKE. 2007;2007:pl8. doi: 10.1126/stke.4172007pl8. [DOI] [PubMed] [Google Scholar]
- 53.Durocher Y, Perret S, Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30:E9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]