Abstract
Genetic information contains a record of the history of our species, and technological advances have transformed our ability to access this record. Many studies have used genome-wide data from populations today to learn about the peopling of the globe and subsequent adaptation to local conditions. Implicit in this research is the assumption that the geographic locations of people today are informative about the geographic locations of their ancestors in the distant past. However, it is now clear that long-range migration, admixture and population replacement subsequent to the initial out-of-Africa expansion have altered the genetic structure of most of the world’s human populations. In light of this, we argue that it is time to critically re-evaluate current models of the peopling of the globe, as well as the importance of natural selection in determining the geographic distribution of phenotypes. We specifically highlight the transformative potential of ancient DNA. By accessing the genetic make-up of populations living at archaeologically-known times and places, ancient DNA makes it possible to directly track migrations and responses to natural selection.
Introduction
Within the past 100,000 years, anatomically modern humans have expanded to occupy every habitable area of the globe. The history of this expansion has been explored with tools from a number of disciplines, including linguistics, archaeology, physical anthropology, and genetics. These disciplines all can be used to ask the same question: how did we get to where we are today?
Attempts to answer this question have often taken the form of a dialectic between two hypotheses. On the one hand are arguments in favor of demographic stasis, which propose that the inhabitants of a region are the descendants of the first people to arrive there. On the other side are the arguments in favor of rapid demographic change, which propose that the present-day inhabitants of a region descend from people who arrived during periods of technological or cultural change, replacing the previous inhabitants.
In archaeology, this debate has played out around the issue of whether sudden changes in material culture apparent in the archaeological record can be attributed to the spread of culture or to population movements: “pots versus people” [1]. In physical anthropology, the debate has played out around the issue of whether changes in morphological characters over time are due to in situ evolution or to the arrival of new populations (e.g., [2]).
The same debate has also played out in genetics. On the side of population replacements, there are the “wave of advance” and “demic diffusion” models, first proposed to describe the spread of agriculture through Europe. In these models, the Neolithic transition was accompanied by the spread of farmers from the Near East across Europe, who partially or completely replaced resident hunter-gatherers [3–6]. On the side of stasis, there are the “serial founder effect” models [7,8], which proposed that populations have remained in the locations they first colonized after the out-of-Africa expansion, exchanging migrants only at a low rate with their immediate neighbors until the long-range migrations of the last 500 years [9–12].
These genetic models – the wave-of-advance models on the one hand, and the serial founder effect models on the other – were proposed prior to the availability of large-scale genomic data. The great synthesis of genetic data with historical, archaeological and linguistic information, “The History and Geography of Human Genes” [13], was written based on data from around one hundred protein polymorphisms, and the papers that popularized the notion of a serial founder effect model were written based on data from around 1,000 microsatellites. However, it is now possible to genotype millions of polymorphisms in thousands of individuals using high-throughput sequencing. Because of these technological advances, the last few years have seen a dramatic increase in the quantity of data available for learning about human history. Equally important has been rapid innovation in methods for making inferences from these data. Here, we argue that the technological breakthroughs of the past few years motivate a systematic re-evaluation of human history using modern genomic tools—a new “History and Geography of Human Genes” that exploits many orders of magnitude more data than the original synthesis.
In the first section of the paper, we summarize what we see as major lessons from the recent literature. In particular, it is now clear that the data contradict any model in which the genetic structure of the world today is approximately the same as it was immediately following the out-of-Africa expansion. Instead, the last 50,000 years of human history have witnessed major upheavals, such that much of the geographic information about the first human migrations has been overwritten by subsequent population movements. However, the data also often contradict models of population replacement: when two distinct population groups come together during demographic expansions, the result is often genetic admixture rather than complete replacement. This suggests that new types of models—with admixture at their center—are necessary for describing key aspects of human history (for early examples of admixture models, see [14–16]).
In the second section of the paper, we sketch out a way forward for data-driven construction of these models. We specifically highlight the potential of ancient DNA studies of individuals from archaeologically-important cultures. Such studies in principle provide a source of information about history that bypasses some fundamental ambiguities in the interpretation of genetic, archaeological, or anthropological evidence alone. We discuss a number of potential applications of this technology to outstanding questions in human history.
There are a number of excellent articles that have reviewed the literature on genome-wide studies of human history [17–23]. Here our focus is not on providing a comprehensive review but instead on highlighting promising directions for future research.
Re-evaluation of the “serial founder effect” model
We begin with an illustration of some of the ambiguities in interpretation of genetic data in the context of the “serial founder effect” model. This model, initially proposed by Harpending, Eller, and Rogers [24,25], gained popularity with the publication of two papers [7,8] that observed that heterozygosity (Glossary) declines approximately linearly with geographic distance from Africa. This pattern, initially identified in genome-wide microsatellite genotypes from around 50 worldwide human populations, was subsequently confirmed based on patterns of haplotype diversity in large single nucleotide polymorphism (SNP) datasets [26].
The observation of a smooth decline in human diversity with increasing geographic distance from Africa was interpreted as a window into demographic events deep in our species’ past. Specifically, in the serial founder effect and related models (Figure 1A, [12]), the peopling of the globe proceeded by an iterative process in which small bands of individuals pushed into unoccupied territory, experienced population expansions, and subsequently gave rise to new small bands of individuals who then pushed further into unoccupied territory. This model has two important features: a large number of expansions into new territory by small groups of individuals (and concurrent bottlenecks), and little subsequent migration [7–12]. As Prugnolle et al. (2005) wrote: “what is clear…is that [this] pattern of constant loss of genetic diversity along colonisation routes could only have arisen through successive bottlenecks of small amplitude as the range of our species increased… The pattern we observe also suggests that subsequent migration was limited or at least very localised”. This qualitative conclusion was followed by more quantitative ones; in an explicit fit of the serial founder effect model to data, Deshpande et al. (2009) concluded that “incorporation into existing models of exchange between neighbouring populations is essential, but at a very low rate”.
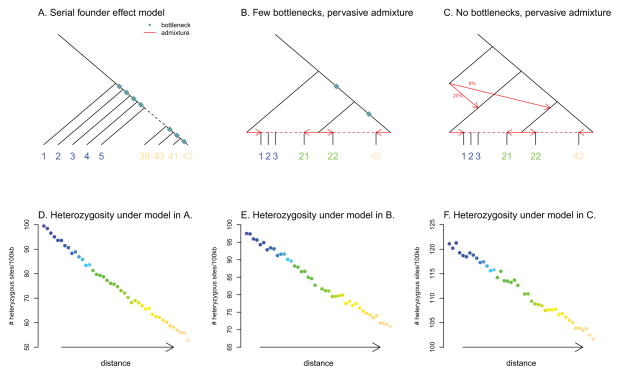
Figure 1. A negative correlation between heterozygosity and geographic distance from a source population can be generated by qualitatively different, historically plausible demographic models.
We simulated genetic data under different demographic models and calculated the average heterozygosity in each simulated population. A. Schematic of a serial founder effect model. B. Schematic of a demographic model with two bottlenecks and extensive admixture. C. Schematic of a demographic model with no bottlenecks and extensive admixture. D,E,F. Average heterozygosity in each population simulated under the demographic models in A,B, and C respectively. Each point represents a population, ordered along the x-axis according as in A.
This model has been influential in many fields. If it is correct, then the difficult problem of identifying the geographic origin of all modern humans is reduced to the simpler problem of finding the geographic region where people have the most genetic diversity [8]. This idea has been used extensively in discussions of human origins (e.g., [27,28]). The model also provides a null hypothesis of limited migration against which alternatives can be tested [29,30]. Outside of genetics, the serial founder effect model has been used as a framework to interpret data from linguistics [31], physical anthropology [32–35], material culture [36], and economics [37].
The serial founder effect model, however, is only one of many models that can produce qualitatively similar patterns of genetic diversity (e.g., [38,39]). Producing the empirical pattern of a smooth decline in diversity with distance from Africa simply requires that the average time to the most recent common ancestor between two chromosomes in a population depend on the distance of that population from Africa [12,39].
To illustrate this point, we constructed two models that produce smooth declines in diversity with distance from Africa just like the serial founder effect model. However, these models differ qualitatively from the serial founder model in that this pattern is driven by admixture rather than bottlenecks in the distant past. These are 1) a model with two severe bottlenecks and extensive subsequent population admixture (Figure 1B) and 2) a model without bottlenecks but with archaic admixture (from an anciently diverged population like Neanderthals) as well as extensive recent population admixture (Figure 1C). Details of the model specifications and simulation parameters are in the Supplementary Information. Using ms [40], we simulated 1,000 regions of 100kb under the serial founder effect model and each of these two models, and plotted the average heterozygosity in each population. In all three cases (Figure 1D,E,F) we recapitulated a smooth linear decline in heterozygosity with distance from a reference population (in the serial founder model, this reference population can be interpreted as the source of the expansion, but there is no analogous interpretation of this population in the other models). Qualitative patterns of linkage disequilibrium were also similar in all scenarios (Supplementary Figure 3).
These simulations show that the main observation that has been marshaled in support of the serial founder effect model is also consistent with very different histories (see also [29,38,39,41]). Specifically, in the absence of additional data, the smooth linear decline in heterozygosity away from Africa could represent a signal of many population bottlenecks during the initial out-of-Africa expansion tens of thousands of years ago, or it could represent a signal of extensive population mixture within the last few thousand years (or, of course, a combination of these or many other models that we have not considered). Because the data are compatible with both, arguing for one over the other involves a subjective determination of which class of model is more likely a priori. Perhaps the most important issue affecting this determination is how important migration has been over the last 50,000 years of human history. How representative are populations today of the populations that lived in the same locations after the out-of-Africa expansion?
Empirical data have shown that the current inhabitants of a region are often poor representatives of the populations that lived there in the distant past
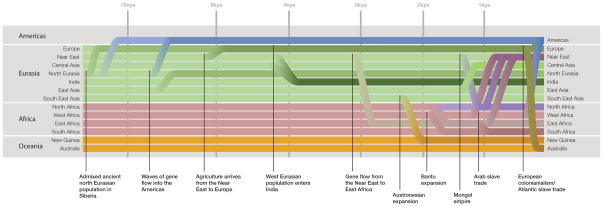
The answer to the question posed above has been the subject of considerable research over the past several years. In our opinion one finding is already clear: long-range migration and concomitant population replacement or admixture have occurred often enough in recent human history that the present-day inhabitants of many places in the world are rarely related in a simple manner to the more ancient peoples of the same region (Figure 2).
Figure 2.
A rough guide to genetically documented population movements in the history of anatomically modern humans.
The Americas over the last 500 years present one recent example. The Americas experienced massive demographic change after the arrival of Europeans and Africans, such that most of the ancestry of the Americas is not derived from the Native Americans who were the sole inhabitants of the region half a millennium ago [42–47]. Another recent example is Australia, where European migration over the last couple of hundred years is the main source of the genetic material in the region today [48].
An example from further back in time comes from the present-day hunter-gatherer and pastoralist populations of Siberia, which are often treated as surrogates for the populations that crossed the Bering land bridge to people the Americas beginning more than 15,000 years ago (e.g., [47,49–51]). DNA sequences from two individuals who lived in the Lake Baikal region of Siberia ~24,000 years before the present and ~17,000 years before the present, respectively, have indicated that this assumption is unfounded. These two ancient individuals (from time points prior to the time the Americas were thought to be peopled) are more closely related to present-day Native Americans than are present-day Siberians [52]. They appear to have been members of an “Ancient North Eurasian” population that no longer exists in unmixed form, but that admixed substantially with the ancestors of both present-day Europeans and Native Americans [52–54]. By contrast, most of the present-day indigenous populations of Siberia are more closely related to populations currently living in East Asia, indicating that the present-day indigenous population of Siberia is descended in large part from populations that arrived in the region after the end of the last ice age [52].
Ancient DNA studies have also made considerable progress in resolving the major debate about whether the arrival of agriculture in Europe involved the spread of people or technology [53,55–63]. A key observation comes from genome-wide data from hunter-gatherer and agriculturalist populations that lived around 5,000 years ago in present-day Sweden [62,63]. The farmer population appears most genetically similar to southern Europeans today, whereas the hunter-gatherers are more similar to northern Europeans (and, notably in light of the discussion of the serial founder effect model above, have levels of genetic diversity lower than modern European and East Asian populations [63]). Thus, at least in Scandinavia, the spread of agriculture was accompanied by the spread of people. The outcome of this spread of people was not population replacement, but rather admixture, such that European populations today trace some of their ancestry to both ancestral populations [53,62]. Mitochondrial DNA (mtDNA) studies suggest that this dynamic is characteristic of the arrival of agriculture throughout Europe [55–58].
The arrival of farmers was not the end of pre-historic migration in Europe (even putting aside discussion of migrations since the invention of writing; see [64–67]). In a single geographic region in present day Germany, mtDNA has been obtained from hundreds of human samples from archaeological cultures ranging from the early Neolithic to the Bronze Age [56]. There is an apparent genetic discontinuity between people of early and late Neolithic cultures. In particular, people of late Neolithic cultures bear more relatedness to the present-day populations of Eastern Europe and Russia than do people of early Neolithic cultures. Thus, demographic turnover has apparently occurred at least twice over the course of the last eight thousand years of European prehistory. This makes inferences about the inhabitants of Europe tens of thousands of years ago based on the locations of people today unreliable.
Evidence of major population mixture in the last several thousand years has also accumulated in parts of the world where no ancient DNA is (yet) available. A series of studies have detected and dated admixture in a range of human populations. These studies make it clear that there are multiple distinct sources of ancestry in most populations. We caution, however, that without ancient DNA, it is not possible to be confident which populations lived in a region before the admixture:
In India, nearly all people today are admixed between two distinct groups, one most closely related to present-day Europeans, Central Asians and Near Easterners, and one most closely related to isolated populations in the Andaman islands [68]. Much of this admixture occurred within the last 4,000 years [69].
In North Africa, nearly all people today descend from admixture between populations related to those present today in western Africa and in the Near East [70–72]. Some of this mixture can be dated to within the last few thousand years [65], indicating that much of the ancestry from these populations does not descend continuously from the Stone Age peoples of North Africa.
In sub-Saharan Africa, genetic studies have documented multiple examples of populations with ancestry from disparate sources. Many populations across sub-Saharan Africa trace some fraction of their ancestry to admixture in the last several thousand years with populations related to those in western Africa [65]. Further, populations in eastern and southern Africa have been influenced by gene flow from west Eurasian-related populations in the last 3,000 years [73,74]. And in Madagascar, all populations derive approximately half of their ancestry from populations related to those currently living in southeast Asia [75].
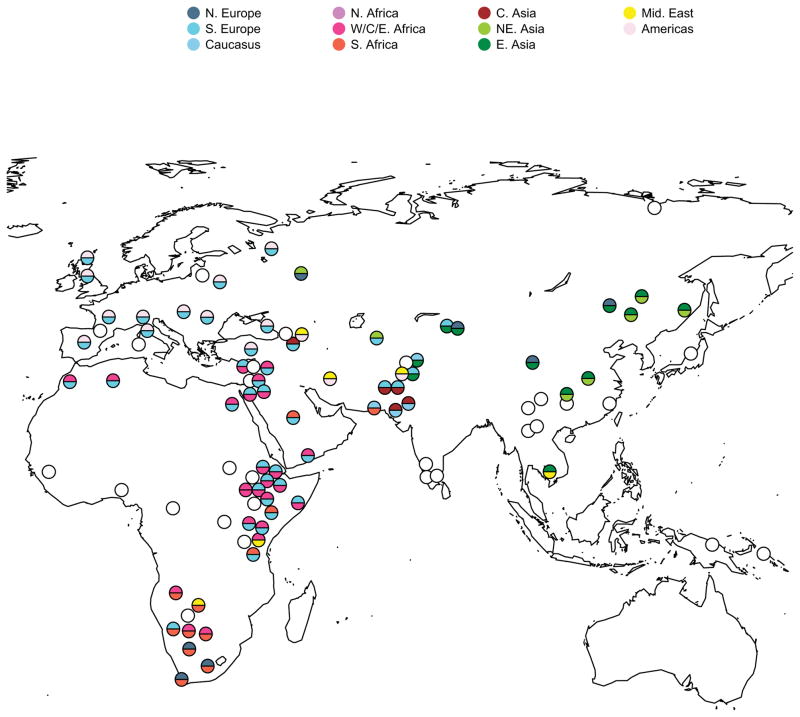
Beyond case studies, several groups have applied tests for admixture to diverse populations from around the globe and found that nearly all populations show evidence of admixture [54,65,76]. To illustrate the degree to which this admixture involved populations whose closest genetic relatives today are geographically distant from each other, in Figure 3 we present an analysis based on combining data for 103 worldwide populations from a number of sources [26–28,73,77,78] and running a simple three-population test for admixture [68] on all populations (see Supplementary Information for details). It is important to recognize that this test is not perfectly sensitive—for example, large amounts of genetic drift in a population’s history can mask the statistical signal of an admixture event—but when the test does detect a signal it provides incontrovertible evidence of gene flow into the ancestors of the population [54]. For all populations with statistically significant evidence of admixture, we identified the present-day populations that share the most genetic drift with the admixing ancestral populations.
Figure 3. A birds-eye view of admixture in human populations.
We performed a three-population test for admixture (Reich et al., 2009) on 103 worldwide populations. Circles show the approximate current geographic locations of all tested populations (except for populations in the Americas, which are not plotted for ease of display). Filled circles represent populations identified as admixed, and the colors represent the current geographic labels of the inferred admixing populations. Empty circles represent populations with no statistically-significant evidence for admixture in this test.
Figure 3 shows the geographic locations of all the populations, along with the locations of the best present-day proxies of their ancestral populations. Admixture between populations related to ones that are now geographically distant is evident in most populations of the world. For example, Native American-related ancestry is present throughout Europe [54], likely reflecting the genetic input from the Ancient Northern Eurasian population related to Upper Paleolithic Siberians [53,54] both into the Americas (most likely prior to 15,000 years ago) and into Europe [52,53]. Also, ancestry from a population related to those living in the Near East is found in Cambodia [30], likely due to mixture from an ancestral South Asian population that was itself an admixed population containing ancestry related to present-day Near Easterners [65]. The test we use as the basis for Figure 3 detects only one signal of admixture per population, and cannot detect complete population replacement. The true population history is thus likely to have been even more complex.
These examples show that the populations in a given region today are rarely descended in a simple manner from the inhabitants in the distant past. This provides further evidence that the serial founder effect model is no longer a reasonable null model for the relationship between present-day populations and their ancestors. Instead, clines in genetic diversity observed in data may often be better modeled as outcomes of admixture (as in Figures 1B and 1C) rather than a series of bottlenecks.
Ancient DNA: a transformative source of information about the past
The types of models that are most useful for making sense of human history are ones that specify geographic as well as temporal information; that is, those that make statements of the form “a population from location X moved to location Y during time period Z”. For example, one might wish to test whether the first pastoralists in southern Africa arrived as migrants from eastern Africa after around 2,500 years ago (the time when evidence of pastoralism in southern Africa appears in the archaeological record), against the alternative hypothesis that a pastoralist lifestyle was adopted by indigenous people who learned it through cultural transmission. Testing this hypothesis using DNA from individuals today involves assuming that populations in southern and eastern Africa today are representative of the populations in southern and eastern Africa at the times of interest [74,79,80]. This assumption is often difficult or impossible to test.
Studies of DNA from ancient human remains offer a way around this limitation. Rather than studying the past by the traces it has left in present-day people—which is problematic as human individuals and even whole populations are capable of migrating hundreds or thousands of kilometers in a lifetime—ancient DNA offers the ability to analyze the genetic patterns that existed at a particular time and geographical location. This allows direct inference about the relationships of historical populations to each other and to populations living today. Box 1 lists some surprising findings about human history based on ancient DNA, which would have been difficult or impossible to obtain without this source of information.
Box 1. Surprising findings about human history illuminated by ancient DNA.
There was archaic Neanderthal gene flow into anatomically modern humans outside Africa 37,000–85,000 years ago [81]. This gene flow contributed on the order of 2% of the genetic ancestry of non-Africans [82]. The first evidence of ancient admixture in non-Africans was suggested based on analysis of present-day humans, without access to any ancient DNA [83]. However, the consensus about whether admixture occurred only changed after ancient DNA evidence showed that the deeply diverged segments in present-day non-Africans are related to Neanderthals [82].
A previously unknown archaic population that was neither Neanderthal nor modern human was present in Siberia before 50,000 years ago [84,85]. The discovery of the “Denisovans” shows how ancient DNA can reveal “genomes in search of a fossil”; populations whose existence was not expected based on the archaeological or fossil record.
There was gene flow from a population related to the Denisovans into the ancestors of present-day aboriginal people from New Guinea, Australia, and the Philippines [85–89]. These populations all live in Oceania, far from where the Denisovan bone was found in Siberia, a finding that again would not have been expected in the absence of ancient DNA data.
Native Americans are admixed between an Ancient North Eurasian population and a population related to present-day East Asians, due to events prior to the diversification of Native American populations in the New World [52].
There were at least two important migration events in central Europe over the last 9,000 years [53,55–60,62].
Populations in the Americas have experienced multiple episodes of turnover, such that an individual that lived in Greenland around 4,000 years ago is more closely related to populations currently in Siberia than to present-day Greenland Inuit populations [90], and an individual that lived in North America around 12,000 years ago is more closely related to present-day Native South American populations than to present-day Native North American populations [91].
Ancient DNA results are so regularly surprising that almost any measurement is interesting: new historical discoveries have been made in virtually every ancient DNA study that has been carried out. The reason why ancient DNA studies are so informative is that the technology provides a tool to measure quantities that were previously unmeasurable. In this sense, the value of ancient DNA technology as a window into ancient migrations is analogous to the 17th century invention of the light microscope as a window into the world of microbes and cells.
Scientific opportunities for ancient DNA studies
Moving forward, ancient DNA studies afford major opportunities in two areas: studies of population history, and studies of natural selection.
The literature on ancient mtDNA contains a number of promising study designs. For example, one might sample multiple individuals from different archaeological cultures at a single time point [60]. Such a “horizontal time slice” allows a snapshot of population structure over a broad geographic region, which can then be compared to the relatively complete picture of population structure today. An alternative study design (like that used by [56]) is to take a single geographic location and sample individuals from multiple time points. This “vertical time slice” allows direct quantification of changes in population composition over time.
mtDNA studies have major drawbacks compared with analysis of the whole genome, however [92]. First, mtDNA is inherited maternally, and thus does not capture any information about the history of males (which may differ from that of females due to sex-biased demographic processes). More importantly, a study of a single locus (or two loci if the Y chromosome is included) has less statistical resolution for studies of history than do studies of the nuclear genome. The reason is that whole-genome studies of an individual contain information about thousands of that individual’s ancestors, not just information about those from a single lineage. It is thus important that the more advanced study designs based on mtDNA be combined with analysis of the more informative autosomal DNA.
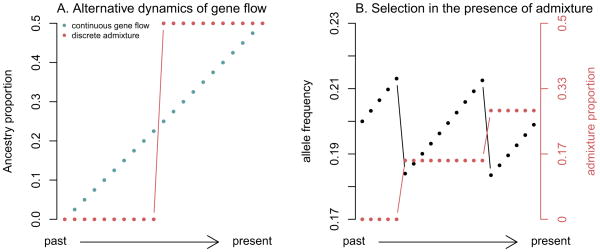
What outstanding questions in population history can be addressed with these designs? One question is whether changes in populations over time are typically gradual—due to consistent, low-level gene flow between neighboring populations—or punctate, with migration events rapidly altering the genetic composition of a region. One line of work on modeling human history explicitly assumes the latter [30,54,65,76,93]. If this assumption is unfounded, however, other models that accommodate continuous gene flow (e.g., [94,95]) may be more appropriate. In principle, distinguishing between these possibilities with a time series of ancient DNA is straightforward; in Figure 4A we show how the predictions differ.
Figure 4. Time series allow tests of different models of human history.
A. Different scenarios for the dynamics of gene flow. We show idealized time series of admixture proportions in a population under models of continuous gene flow and discrete admixture. B. Selection in the presence of admixture. We show idealized time series of the allele frequencies at a selected allele and admixture proportions in a population. Note that despite selection the allele does not show any net change in frequency.
Ancient DNA is also a promising way to address ongoing historical debates about the origins of different populations. Do linguistic isolates like the Basques in Europe have different mixtures of ancestries than their neighbors [96]? When did the west Eurasian-related population(s) that admixed with most Indian populations first appear in South Asia [68]? Did the first modern human inhabitants of East Asia descend from an earlier out-of-Africa migration than the populations living in East Asia today [89,97]? Answering these questions will require a time series of snapshots of human genetic structure, combining the two types of study design mentioned above. If experience is a guide, this type of information will uncover additional unexpected aspects of human history.
The admixture and population replacements identified by ancient DNA also have implications for studies of natural selection. It is often assumed that populations have been in their current geographic locations long enough to adapt to their local conditions. This is explicit in approaches that test for correlations between environmental variables and allele frequencies (e.g., [98–100]) and implicit in studies that interpret selected loci in terms of the current locations of populations (e.g., [101]).
To what extent are the geographic distributions of selected alleles today indicative of the geographic distributions of selective pressures? (The answer to this question may depend on the selective pressure in question). In individual cases, these two distributions are highly correlated; the classic example is the correlation between malaria incidence and disorders of hemoglobin [102]. For other cases, the correlation is imperfect. For example, alleles causing light skin pigmentation are at high frequency in northern Africa [103]. If the selective pressure causing light skin is indeed a relative lack of ultraviolet radiation [104], it seems reasonable to expect that this pressure has not affected people living around the Sahara desert. It is thus likely that the high frequency of alleles causing light skin pigmentation in north Africa was caused by the arrival of lightly-pigmented people [70].
More generally, it has been observed that the geographic distributions of alleles under natural selection in humans tend to match the distribution of neutral population structure rather than any obvious geographic variation in selection pressures [105,106]. One factor contributing to this observation may be that selection pressures on individual loci are relatively weak due to the quantitative nature of phenotypes [107–110]. Another contributing factor may be that population movements over the last several thousand years have to some extent decoupled the geographic distributions of selected alleles from the geographic distributions of selective pressures. From the point of view of an individual allele, the movement of populations acts as a form of fluctuating selection; as populations move from environment to environment, the selection coefficient on an allele may change in both sign and magnitude (assuming a fixed selection coefficient in a given environment). This means that the environment that dominated the allele frequency trajectory may not be the environment that the allele is found in today.
Ancient DNA is also a potentially transformative tool for understanding human adaptation more generally. Nearly all methods for detecting positive selection at the genetic level are based around the principle that a selected allele changes frequency more quickly than a neutral locus. Tracking the trajectories of individuals over time allows direct access to this information, and makes it possible to infer precisely when in time (and with more certainty where geographically) genetic changes began to arise [111]. In fact, in the presence of admixture it is simple to create scenarios where the frequency trajectory of an allele over time allows one to identify selection despite there being no net change in allele frequency (Figure 4B). To date, studies of selection over time have been limited either to small sample sizes [53,112] or small numbers of sites [111,113–115]. However, whole-genome technologies should make it possible to interrogate many thousands of phenotypically-relevant variants simultaneously.
Taming the wild west of ancient DNA
Realizing the potential of ancient DNA studies will require systematic approaches. However, ancient DNA studies today are often more spectacular than systematic. The paradigmatic approach to ancient DNA research in the era of high-throughput sequencing involves identifying a “golden” archaeological sample that yields usable DNA and obtaining a complete or partial genome sequence from it (e.g., [52,59,82,87,88,90,116]). To some extent, the discovery of analyzable samples has been the main driver of the scientific questions asked. However, the future of ancient DNA research requires a more systematic approach: hypothesis-driven sampling across time and space, and analysis of much larger sample sizes.
Ancient DNA research has three major experimental challenges and two computational challenges that need to be jointly addressed by researchers who wish to access this transformative technology.
The first experimental challenge is the danger of contamination from archaeologists or laboratory researchers who handle the sample. This is a recurring problem [117–124]. Although experimental guidelines (e.g., [122,125,126]) as well as laboratory methods for reducing the possibility of contamination [82] and empirically assessing the authenticity of ancient DNA sequences [127] have greatly improved the situation, in practice this concern will never disappear. The importance of controlling for the possibility of contamination is the single most important reason why, to date, convincing ancient DNA research has been dominated by a small number of specialist laboratories with the expertise and facilities to control contamination. All laboratories that carry out this type of work will need to maintain the same level of vigilance currently maintained at specialist laboratories. To reduce contamination at the source, it is also important that archaeologists excavating new samples become trained in handing samples in ways that minimize contamination. Valuable measures include wearing sterile gloves and a protective suit while excavating remains, not washing the remains, and immediately placing the remains in a sterile plastic bag and refrigerating them prior to shipment to an ancient DNA laboratory.
A second experimental challenge is the difficulty of identifying human remains that contain preserved DNA. This often requires screening dozens of individuals from multiple sites, with success rates in DNA extraction depending strongly on the conditions experienced by the sample: its age, temperature, humidity, acidity, the part of the body from which it derives, the speed with which it dried after death, whether or not it is from a sample that was rapidly defleshed, and other factors that are not currently understood. Modern methods have increased the efficiency of DNA extraction by capturing the short molecules that make up the great majority of any ancient sample [128,129]. In addition, improved library preparation methods have increased the fraction of molecules in an extract that are amenable to sequencing [87,116]. Nevertheless, it is still the case that there is great variability in success. The secret of successful ancient DNA research is not luck, but hard work: screening many carefully chosen and prepared samples until a subset are identified that perform well.
A third experimental challenge is the difficulty of finding a sample that has a sufficiently high proportion of DNA from the bone itself to be economically analyzed. Concretely, the challenge is that for many ancient DNA extracts, the proportion of endogenous DNA is very low, on the order of one percent or less. For example, a recent study on a ~40,000 year-old sample from the Tianyuan site of Northern China worked successfully with a sample that had an endogenous DNA proportion of 0.02% [116]. For such samples, important as they are, it is not economical to simply carry out brute-force sequencing of random genomic fragments and expect to obtain sufficient coverage to make meaningful inferences.
The challenges of ancient DNA research do not end once the sample preparation is complete, as the datasets that are generated pose formidable computational and analytical challenges. The first challenge is that once useable samples are obtained and sequenced, the data need to be processed. Ancient DNA laboratories, which traditionally have their strongest expertise in archaeology, physical anthropology, or biochemistry, often lack the bioinformatics expertise, data processing power and data storage solutions necessary to handle the millions or even billions of sequences that are generated by modern ancient DNA studies. Moreover, ancient DNA data also require tailored bioinformatics tools for handling the short sequences that are characteristic of old samples [130]; one cannot simply use existing tools such as SAMtools [131] or the Genome Analysis Toolkit [132] with the default settings. For example, the sequenced DNA fragments are usually short and degraded, and are expected to have C→T and G→A errors at the ends of the molecules due to cytosine deamination. The data additionally need to be computationally assessed for evidence of contamination, for example by checking the rate of molecules that do not map to the mtDNA consensus sequence obtained for the sample [116]. If a sample is determined to be contaminated, the characteristic ancient DNA errors can be leveraged to reduce the level of contamination (assuming that the contamination is not old) by restricting the analysis to sequences that contain differences from the human reference genome sequence that are hallmarks of errors due to ancient DNA degradation [133,134].
A second computational challenge for ancient DNA research is that once the data are processed and a sample is determined as likely to be authentic, the data also need to be analyzed using statistical methods that infer population history. Many methods that have been developed to make inferences about population relationships are not substantially biased by the fact that ancient DNA is old and error-prone [52,59,82,87,88,90,116]. However, these methods must still be implemented carefully to produce meaningful results. In particular, the methods need to handle complications of ancient DNA, such as the fact that there is rarely deep sequencing data, so that unambiguous determination of genotypes at each position in the genome is often unreliable.
At present, few laboratories have the experimental and computational expertise to address all these challenges simultaneously. As a result, ancient DNA research has been dominated by a few vertically integrated and well-funded laboratories that combine all these skills under the same roof. The high barriers to entry have meant that the full potential of ancient DNA has been largely untapped. In what follows, we sketch out a way forward that we expect will make ancient DNA analysis more accessible to the broad research community, including archaeologists.
Democratization of ancient DNA technology
The usual paradigm in ancient DNA analysis of the nuclear genome has been to identify a sample that has a high enough proportion of DNA to deeply sequence. Such golden samples are rare, and are typically identified only after laborious screening of many dozens of samples that have low proportions of human DNA. Once a sample is found that has an appreciable proportion of human DNA, it is typically sequenced to as high a coverage as possible (sometimes the limited number of starting molecules in many ancient DNA libraries is the main factor limiting sequencing depth). All of this is very expensive, and has been an important barrier to entry into ancient DNA work for less well-funded laboratories.
Whole-genome sequences, however, are not required for most historical inferences. In a paper that was important not only for what it showed about history but also for what it showed about the quantity of information that can be extracted from small amounts of data, Skoglund et al. (2012)[62] found that even low levels of genome coverage per sample – 1–5% of genomic bases covered in their case – were sufficient to support profound historical inferences.
A promising way to make ancient DNA analysis accessible to a much larger number of laboratories is to use targeted capture approaches that enrich a sample for human DNA. In one approach to target enrichment [135] it was shown that it is possible to take RNA baits obtained by transcribing a whole human genome and hybridize them in solution to an ancient DNA library, thus increasing the fraction of human DNA to be sequenced by more than an order of magnitude. Another group [116] showed that it is possible to synthesize oligonucleotide DNA baits targeted at a specified subset of the genome—the entire mtDNA sequence, or the coding sequences of all genes—and to hybridize to ancient DNA libraries in solution to enrich a library for molecules from the targeted subset of the genome. This strategy was used to obtain a high quality mitochondrial genome sequence from a ~400,000 year-old archaic human [133], as well as approximately 2-fold redundant coverage of chromosome 21 from the ~40,000 year old Tianyuan sample from northern China [116].
We ourselves are particularly enthusiastic about the possibility of adapting a technology like that described above to enrich human samples for panels of several hundred thousand SNPs that have already been genotyped on present-day samples. This is a sufficient number of SNPs that it would allow for high-resolution analysis of how an ancient sample relates to present-day as well as other ancient samples. The strategy has two potential advantages. First, through enrichment, it allows analysis of samples with much less than 10% human DNA, which are not economical for whole-genome sequencing studies. Second, assuming that it works, it requires about two orders of magnitude less sequencing per sample to saturate all its targets (Table 1). We caution that there are some questions—for example, estimation of population divergence times—that rely on identification of sample-specific mutations and may be better addressed with whole-genome sequencing data. Nevertheless, we believe that a capture experiment can answer the substantial majority of questions related to population history or natural selection that are addressable with genetic data, while allowing larger numbers of samples to be analyzed for the same cost.
Table 1.
Comparison of two strategies for ancient DNA studies of history
| Whole genome sequencing | Capture and sequencing of 300,000 SNPs | |
|---|---|---|
|
| ||
| Number of samples that need to be screened* | 1 | 1/10 |
| Amount of sequencing that needs to be performed* | 1 | 1/100 |
|
| ||
| High resolution inference of pop. relationships | Yes | Yes |
| Allows studies of selection | Yes | Yes |
| Variants specific to sample can be discovered | Yes | No |
| Works for samples with low % of human DNA | No | Yes |
These are rough estimates based on discussion with colleagues. The estimated amount of work required for capturing 300,000 SNPs is expressed as a fraction of that required for whole-genome sequencing.
An important goal for the coming years should be to make ancient DNA a tool that will become fully accessible not just to smaller laboratories but also to archaeologists. In this regard, it is important to encourage interaction and collaboration between the genetic and archaeologist communities to develop standards for the interpretation and use of genetic data in answering questions relevant to archaeology.
Members of the archaeology community are already sophisticated consumers of other scientific technologies for analyzing ancient biological remains such as carbon 14 analysis (for date estimation) and stable isotope analysis (for making inferences about diet). Currently, when archaeologists have a sample that they wish to analyze, they send it to specialist (sometimes commercial) laboratories, which then provide a carbon date and/or an isotope analysis, along with a report giving interpretation. We envision a future in which ancient DNA analysis could similarly become available as a service to archaeologists. Concretely, archaeologists would be able to send skeletal material to a specialist laboratory for analysis, where it could be tested for ancient DNA. If ancient DNA is detected, a whole mitochondrial sequence could be produced and a sex determined. For ancient samples with evidence of uncontaminated DNA, whole-genome data could be produced. The relationships of the mtDNA to others that have been previously generated could be summarized in the form of a tree, and the population’s affinities could be summarized in the form of a method like Principal Component Analysis.
To make such a future possible, it will be necessary to build up a database of present-day human and ancient DNA samples all genotyped at the same set of SNPs to which any new sample can be compared. In addition, it will be necessary to write software to automatically compare a new sample to samples in the database [54,136–138], which could be used as the basis for producing a report for archaeologists on how the sample relates to diverse present-day humans and to other ancient samples. Because of the subtleties of interpreting genetic data, it will be necessary for archaeologists to work collaboratively with population geneticists to provide in-depth interpretation of the results of such studies. Such a report would, however, be a useful tool for giving archaeologists a first impression of the biological ancestry of their samples, including information on the sex and plausibility of geographic origin.
We particularly wish to highlight the potential of ancient DNA as a direct tool for elucidating the population structure and patterns of relatedness within a particular archaeological site. We mention here two types of analysis of particular interest:
Correlating genetic findings—sex, relatedness and ancestry—to archaeological information derived from grave goods, status symbols, and other objects.
Identification of outlier individuals who are unusual in terms of their ancestry relative to others at nearby sites.
Concluding remarks
We have argued that it will likely to be fruitful to re-examine many aspects of human population history and natural selection from a perspective in which population movements and admixture play a central role. Moreover, we have shown that ancient DNA has emerged as transformative tool for addressing questions about human history – it is not just an interesting side show in terms of insights that it can bring to understanding of the human past, but a tremendous leap forward beyond what has been possible through analysis of DNA from present-day humans. What has already been discovered about human history from whole-genome analysis and ancient DNA is just the tip of the iceberg, and we expect that the coming decade will bring even more important discoveries. We conclude by listing a number of questions that are likely to be addressable (Outstanding Questions).
Supplementary Material
Outstanding Questions.
What was the genetic makeup of humans in eastern Africa before the arrival of Bantu speakers? The population genetic structure of sub-Saharan Africa has been transformed by the expansion of Bantu-speaking agriculturalists over the last 3,000 years. As a concrete example, the population structure of eastern Africa prior to this expansion remains controversial [139,140]. Ancient DNA from samples prior to the Bantu expansion could settle this debate. Although the climate in much, but not all, of sub-Saharan Africa is less favorable for preservation of DNA than that of northern Eurasia, it is not clear whether enough African samples have been tested to determine whether or not ancient DNA analysis works. Moreover, the time scale of the Bantu expansion is only several thousand years rather than tens of thousands of years. In light of continuing technological progress in ancient DNA extraction (including the successful extraction of DNA from a ~400,000 year old sample in Spain [133]), we are hopeful that ancient DNA studies of some African samples within the last few thousand years may become possible.
When were the geographic distributions of alleles under positive selection established? Loci under positive selection in humans have allele frequencies with characteristic geographic patterns, which have been interpreted as “west Eurasian”, “east Asian” and “non-African” selective sweeps [105]. Were these patterns established tens of thousands of years ago during the initial out-of-Africa by modern humans? Or were they established after later population movements? Resolving these questions will become possible once substantial numbers of ancient samples are genotyped. Work in this area is only just beginning [111].
Was the spread of Indo-European languages caused by large-scale migrations of people or did language shifts occur without extensive population replacement? For times prior to the invention of writing, it will never be possible to directly relate the language that people spoke to their remains. Nevertheless, genetic data may still be informative about the historical events that accompany linguistic expansions. The origins of the Indo-European languages—spoken 500 years ago across Europe and West, Central, and South Asia and even more widely distributed today—is a particularly important question [141,142]. These languages likely spread across Eurasia and diversified within the last 5–10 thousand years. Was this spread of languages caused by a large-scale movement of people? It now may be possible to obtain ancient DNA from cultures known to have spoken Indo-European languages—for example, Hittites and Tocharians—and to compare these populations with their neighbors to determine whether they harbor a genetic signature specific to the Indo-European speakers. It may also be possible to search for the spread of (currently hypothetical) Indo-European genetic signatures in Europe and India at the times when various hypotheses have suggested that they may have arrived.
Highlights.
Migration and population mixture have been pervasive in human history
Ancient DNA provides a new perspective on human history and adaptation
Acknowledgments
We thank Henry Harpending and Alan Rogers for a helpful discussion about the origins of the serial founder effect model. We are grateful for discussions and comments on the manuscript from David Anthony, Joachim Burger, Graham Coop, Qiaomei Fu, Razib Khan, Alexander Kim, Iosif Lazaridis, Swapan Mallick, Iain Mathieson, Mike McCormick, Nick Patterson, Ben Peter, Molly Przeworski, Jenny Raff, Nadin Rohland, Pontus Skoglund, Johannes Krause, Richard Meadow and Wolfgang Haak. DR was supported by NSF HOMINID grant BCS-1032255 and NIH grant GM100233 and is an Investigator of the Howard Hughes Medical Institute.
Glossary
- Admixture
A sudden increase in gene flow between two differentiated populations
- Bottleneck
A temporary decrease in population size in the history of a population
- Gene flow
The exchange of genes between two populations due to interbreeding
- Heterozygosity
the number of differences between two random copies of a genome in a population
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Renfrew C, Bahn PG. Archaeology: theories, methods, and practice. Thames and Hudson; 1996. [Google Scholar]
- 2.Powell JF, Neves WA. Craniofacial morphology of the first Americans: Pattern and process in the peopling of the New World. Am J Phys Anthropol Suppl. 1999;29:153–188. doi: 10.1002/(sici)1096-8644(1999)110:29+<153::aid-ajpa6>3.3.co;2-c. [DOI] [PubMed] [Google Scholar]
- 3.Ammerman AJ, Cavalli-Sforza LL. Measuring the rate of spread of early farming in Europe. Man 1971 [Google Scholar]
- 4.Ammerman AJ, Cavalli-Sforza LL. The neolithic transition and the genetics of populations in Europe. Princeton University Press; 1984. [Google Scholar]
- 5.Menozzi P, et al. Synthetic maps of human gene frequencies in Europeans. Science. 1978;201:786–92. doi: 10.1126/science.356262. [DOI] [PubMed] [Google Scholar]
- 6.Sokal RR, et al. Genetic evidence for the spread of agriculture in Europe by demic diffusion. 1991. [DOI] [PubMed] [Google Scholar]
- 7.Prugnolle F, et al. Geography predicts neutral genetic diversity of human populations. Curr Biol. 2005;15:R159–60. doi: 10.1016/j.cub.2005.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramachandran S, et al. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15942. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deshpande O, et al. A serial founder effect model for human settlement out of Africa. Proceedings of the Royal Society B: Biological Sciences. 2009;276:291–300. doi: 10.1098/rspb.2008.0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henn BM, et al. The great human expansion. Proc Natl Acad Sci U S A. 2012;109:17758–64. doi: 10.1073/pnas.1212380109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H, et al. A Geographically Explicit Genetic Model of Worldwide Human-Settlement History. The American Journal of Human Genetics. 2006;79:230–237. doi: 10.1086/505436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeGiorgio M, et al. Coalescence-time distributions in a serial founder model of human evolutionary history. Genetics. 2011;189:579–593. doi: 10.1534/genetics.111.129296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavalli-Sforza LLL, et al. The history and geography of human genes. Princeton university press; 1994. [Google Scholar]
- 14.Lathrop GM. Evolutionary trees and admixture: phylogenetic inference when some populations are hybridized. Ann Hum Genet. 1982;46:245–55. doi: 10.1111/j.1469-1809.1982.tb00716.x. [DOI] [PubMed] [Google Scholar]
- 15.Cavalli-Sforza LL, Piazza A. Analysis of evolution: Evolutionary rates, independence and treeness. Theoretical Population Biology. 1975;8:127–165. doi: 10.1016/0040-5809(75)90029-5. [DOI] [PubMed] [Google Scholar]
- 16.Bowcock AM, et al. Drift, admixture, and selection in human evolution: a study with DNA polymorphisms. Proc Natl Acad Sci USA. 1991;88:839–843. doi: 10.1073/pnas.88.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alves I, et al. Genomic data reveal a complex making of humans. PLoS genetics. 2012;8:e1002837. doi: 10.1371/journal.pgen.1002837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colonna V, et al. A world in a grain of sand: human history from genetic data. Genome Biol. 2011;12:234. doi: 10.1186/gb-2011-12-11-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novembre J, Ramachandran S. Perspectives on human population structure at the cusp of the sequencing era. Annu Rev Genomics Hum Genet. 2011;12:245–74. doi: 10.1146/annurev-genom-090810-183123. [DOI] [PubMed] [Google Scholar]
- 20.Pinhasi R, et al. The genetic history of Europeans. Trends Genet. 2012;28:496–505. doi: 10.1016/j.tig.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Stoneking M, Krause J. Learning about human population history from ancient and modern genomes. Nat Rev Genet. 2011;12:603–14. doi: 10.1038/nrg3029. [DOI] [PubMed] [Google Scholar]
- 22.Veeramah KR, Hammer MF. The impact of whole-genome sequencing on the reconstruction of human population history. Nature Reviews Genetics. 2014;15:149–162. doi: 10.1038/nrg3625. [DOI] [PubMed] [Google Scholar]
- 23.Wall JD, Slatkin M. Paleopopulation genetics. Annu Rev Genet. 2012;46:635–49. doi: 10.1146/annurev-genet-110711-155557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harpending HC, Eller E. The Biology of biodiversity. Springer; 2000. Human diversity and its history; pp. 301–314. [Google Scholar]
- 25.Harpending H, Rogers A. Genetic perspectives on human origins and differentiation. Annu Rev Genomics Hum Genet. 2000;1:361–85. doi: 10.1146/annurev.genom.1.1.361. [DOI] [PubMed] [Google Scholar]
- 26.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 27.Henn BM, et al. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc Natl Acad Sci U S A. 2011;108:5154–62. doi: 10.1073/pnas.1017511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlebusch CM, et al. Genomic variation in seven Khoe-San groups reveals adaptation and complex African history. Science. 2012;338:374–9. doi: 10.1126/science.1227721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hellenthal G, et al. Inferring human colonization history using a copying model. PLoS Genet. 2008;4:e1000078. doi: 10.1371/journal.pgen.1000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atkinson QD. Phonemic diversity supports a serial founder effect model of language expansion from Africa. Science. 2011;332:346–9. doi: 10.1126/science.1199295. [DOI] [PubMed] [Google Scholar]
- 32.Betti L, et al. Human pelvis and long bones reveal differential preservation of ancient population history and migration out of Africa. Hum Biol. 2012;84:139–152. doi: 10.3378/027.084.0203. [DOI] [PubMed] [Google Scholar]
- 33.Von Cramon-Taubadel N, Lycett SJ. Brief communication: human cranial variation fits iterative founder effect model with African origin. Am J Phys Anthropol. 2008;136:108–13. doi: 10.1002/ajpa.20775. [DOI] [PubMed] [Google Scholar]
- 34.Hanihara T. Morphological variation of major human populations based on nonmetric dental traits. Am J Phys Anthropol. 2008;136:169–182. doi: 10.1002/ajpa.20792. [DOI] [PubMed] [Google Scholar]
- 35.Manica A, et al. The effect of ancient population bottlenecks on human phenotypic variation. Nature. 2007;448:346–8. doi: 10.1038/nature05951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogers DS, et al. Inferring population histories using cultural data. Proc Biol Sci. 2009;276:3835–3843. doi: 10.1098/rspb.2009.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashraf Q, Galor O. The “Out of Africa” Hypothesis, Human Genetic Diversity, and Comparative Economic Development. The American Economic Review. 2013:103. doi: 10.1257/aer.103.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amos W, Hoffman JI. Evidence that two main bottleneck events shaped modern human genetic diversity. Proc Biol Sci. 2010;277:131–7. doi: 10.1098/rspb.2009.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeGiorgio M, et al. Out of Africa: modern human origins special feature: explaining worldwide patterns of human genetic variation using a coalescent-based serial founder model of migration outward from Africa. Proc Natl Acad Sci U S A. 2009;106:16057–62. doi: 10.1073/pnas.0903341106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–8. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 41.Hunley KL, et al. The global pattern of gene identity variation reveals a history of long-range migrations, bottlenecks, and local mate exchange: implications for biological race. Am J Phys Anthropol. 2009;139:35–46. doi: 10.1002/ajpa.20932. [DOI] [PubMed] [Google Scholar]
- 42.Bryc K, et al. Colloquium paper: genome-wide patterns of population structure and admixture among Hispanic/Latino populations. Proc Natl Acad Sci U S A. 2010;107(Suppl 2):8954–61. doi: 10.1073/pnas.0914618107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gravel S, et al. Reconstructing Native American Migrations from Whole-genome and Whole-exome Data. 2013 doi: 10.1371/journal.pgen.1004023. arXiv preprint arXiv:1306.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson NA, et al. Ancestral components of admixed genomes in a Mexican cohort. PLoS Genet. 2011;7:e1002410. doi: 10.1371/journal.pgen.1002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreno-Estrada A, et al. Reconstructing the population genetic history of the Caribbean. PLoS Genet. 2013;9:e1003925. doi: 10.1371/journal.pgen.1003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price AL, et al. A Genomewide Admixture Map for Latino Populations. The American Journal of Human Genetics. 2007;80:1024–1036. doi: 10.1086/518313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang S, et al. Genetic variation and population structure in native Americans. PLoS Genet. 2007;3:e185. doi: 10.1371/journal.pgen.0030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McEvoy BP, et al. Whole-genome genetic diversity in a sample of Australians with deep Aboriginal ancestry. Am J Hum Genet. 2010;87:297–305. doi: 10.1016/j.ajhg.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lell JT, et al. The dual origin and Siberian affinities of Native American Y chromosomes. Am J Hum Genet. 2002;70:192–206. doi: 10.1086/338457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santos FR, et al. The central Siberian origin for native American Y chromosomes. Am J Hum Genet. 1999;64:619–28. doi: 10.1086/302242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Starikovskaya EB, et al. Mitochondrial DNA diversity in indigenous populations of the southern extent of Siberia, and the origins of Native American haplogroups. Ann Hum Genet. 2005;69:67–89. doi: 10.1046/j.1529-8817.2003.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raghavan M, et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature. 2013 doi: 10.1038/nature12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lazaridis I, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. 2013 doi: 10.1038/nature13673. arXiv preprint arXiv:1312.6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patterson N, et al. Ancient admixture in human history. Genetics. 2012;192:1065–93. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bramanti B, et al. Genetic discontinuity between local hunter-gatherers and central Europe’s first farmers. Science. 2009;326:137–40. doi: 10.1126/science.1176869. [DOI] [PubMed] [Google Scholar]
- 56.Brandt G, et al. Ancient DNA reveals key stages in the formation of central European mitochondrial genetic diversity. Science. 2013;342:257–61. doi: 10.1126/science.1241844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haak W, et al. Ancient DNA from the first European farmers in 7500-year-old Neolithic sites. Science. 2005;310:1016–8. doi: 10.1126/science.1118725. [DOI] [PubMed] [Google Scholar]
- 58.Haak W, et al. Ancient DNA from European early neolithic farmers reveals their near eastern affinities. PLoS Biol. 2010;8:e1000536. doi: 10.1371/journal.pbio.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keller A, et al. New insights into the Tyrolean Iceman’s origin and phenotype as inferred by whole-genome sequencing. Nat Commun. 2012;3:698. doi: 10.1038/ncomms1701. [DOI] [PubMed] [Google Scholar]
- 60.Malmström H, et al. Ancient DNA reveals lack of continuity between neolithic hunter-gatherers and contemporary Scandinavians. Curr Biol. 2009;19:1758–62. doi: 10.1016/j.cub.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 61.Sikora M, et al. Population Genomic Analysis of Ancient and Modern Genomes Yields New Insights into the Genetic Ancestry of the Tyrolean Iceman and the Genetic Structure of Europe. PLoS Genetics. 2014;10:e1004353. doi: 10.1371/journal.pgen.1004353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skoglund P, et al. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science. 2012;336:466–9. doi: 10.1126/science.1216304. [DOI] [PubMed] [Google Scholar]
- 63.Skoglund P, et al. Genomic Diversity and Admixture Differs for Stone-Age Scandinavian Foragers and Farmers. Science. 2014;344:747–750. doi: 10.1126/science.1253448. [DOI] [PubMed] [Google Scholar]
- 64.Davies N. Europe: a history. HarperPerennial; 1998. [Google Scholar]
- 65.Hellenthal G, et al. A genetic atlas of human admixture history. Science. 2014;343:747–751. doi: 10.1126/science.1243518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moorjani P, et al. The history of African gene flow into Southern Europeans, Levantines, and Jews. PLoS Genet. 2011;7:e1001373. doi: 10.1371/journal.pgen.1001373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ralph P, Coop G. The geography of recent genetic ancestry across Europe. PLoS Biol. 2013;11:e1001555. doi: 10.1371/journal.pbio.1001555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reich D, et al. Reconstructing Indian population history. Nature. 2009;461:489–94. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moorjani P, et al. Genetic evidence for recent population mixture in India. Am J Hum Genet. 2013;93:422–38. doi: 10.1016/j.ajhg.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Henn BM, et al. Genomic Ancestry of North Africans Supports Back-to-Africa Migrations. PLoS Genet. 2012;8:e1002397. doi: 10.1371/journal.pgen.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rosenberg NA, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 72.Tishkoff SA, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pagani L, et al. Ethiopian genetic diversity reveals linguistic stratification and complex influences on the Ethiopian gene pool. Am J Hum Genet. 2012;91:83–96. doi: 10.1016/j.ajhg.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pickrell JK, et al. Ancient west Eurasian ancestry in southern and eastern Africa. Proc Natl Acad Sci USA. 2014;111:2632–2637. doi: 10.1073/pnas.1313787111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pierron D, et al. Genome-wide evidence of Austronesian-Bantu admixture and cultural reversion in a hunter-gatherer group of Madagascar. Proc Natl Acad Sci USA. 2014;111:936–941. doi: 10.1073/pnas.1321860111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Loh PR, et al. Inferring admixture histories of human populations using linkage disequilibrium. Genetics. 2013;193:1233–54. doi: 10.1534/genetics.112.147330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Altshuler DM, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Behar DM, et al. The genome-wide structure of the Jewish people. Nature. 2010;466:238–242. doi: 10.1038/nature09103. [DOI] [PubMed] [Google Scholar]
- 79.Breton G, et al. Lactase persistence alleles reveal partial East african ancestry of southern african Khoe pastoralists. Curr Biol. 2014;24:852–858. doi: 10.1016/j.cub.2014.02.041. [DOI] [PubMed] [Google Scholar]
- 80.Macholdt E, et al. Tracing pastoralist migrations to southern Africa with lactase persistence alleles. Curr Biol. 2014;24:875–879. doi: 10.1016/j.cub.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sankararaman S, et al. The date of interbreeding between Neandertals and modern humans. PLoS Genet. 2012;8:e1002947. doi: 10.1371/journal.pgen.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–22. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Plagnol V, Wall JD. Possible ancestral structure in human populations. PLoS Genet. 2006;2:e105. doi: 10.1371/journal.pgen.0020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Krause J, et al. The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature. 2010;464:894–897. doi: 10.1038/nature08976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reich D, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–60. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cooper A, Stringer CB. Paleontology. Did the Denisovans cross Wallace’s Line? Science. 2013;342:321–323. doi: 10.1126/science.1244869. [DOI] [PubMed] [Google Scholar]
- 87.Meyer M, et al. A High-Coverage Genome Sequence from an Archaic Denisovan Individual. Science. 2012 doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Prüfer K, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reich D, et al. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am J Hum Genet. 2011;89:516–28. doi: 10.1016/j.ajhg.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rasmussen M, et al. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010;463:757–62. doi: 10.1038/nature08835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rasmussen M, et al. The genome of a Late Pleistocene human from a Clovis burial site in western Montana. Nature. 2014;506:225–229. doi: 10.1038/nature13025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Mol Ecol. 2004;13:729–44. doi: 10.1046/j.1365-294x.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- 93.Lipson M, et al. Efficient moment-based inference of admixture parameters and sources of gene flow. Mol Biol Evol. 2013;30:1788–802. doi: 10.1093/molbev/mst099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Excoffier L, et al. Robust demographic inference from genomic and SNP data. PLoS Genet. 2013;9:e1003905. doi: 10.1371/journal.pgen.1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gutenkunst RN, et al. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Behar DM, et al. The Basque paradigm: genetic evidence of a maternal continuity in the Franco-Cantabrian region since pre-Neolithic times. Am J Hum Genet. 2012;90:486–93. doi: 10.1016/j.ajhg.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rasmussen M, et al. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011;334:94–8. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Coop G, et al. Using environmental correlations to identify loci underlying local adaptation. Genetics. 2010;185:1411–23. doi: 10.1534/genetics.110.114819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hancock AM, et al. Colloquium paper: human adaptations to diet, subsistence, and ecoregion are due to subtle shifts in allele frequency. Proc Natl Acad Sci U S A. 2010;107(Suppl 2):8924–30. doi: 10.1073/pnas.0914625107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huerta-Sánchez E, et al. Genetic signatures reveal high-altitude adaptation in a set of ethiopian populations. Mol Biol Evol. 2013;30:1877–88. doi: 10.1093/molbev/mst089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Simonson TS, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329:72–5. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- 102.Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005;77:171–192. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Norton HL, et al. Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol Biol Evol. 2007;24:710–722. doi: 10.1093/molbev/msl203. [DOI] [PubMed] [Google Scholar]
- 104.Jablonski NG, Chaplin G. The evolution of human skin coloration. J Hum Evol. 2000;39:57–106. doi: 10.1006/jhev.2000.0403. [DOI] [PubMed] [Google Scholar]
- 105.Coop G, et al. The role of geography in human adaptation. PLoS Genet. 2009;5:e1000500. doi: 10.1371/journal.pgen.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Granka JM, et al. Limited evidence for classic selective sweeps in African populations. Genetics. 2012;192:1049–64. doi: 10.1534/genetics.112.144071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hancock AM, et al. Adaptations to new environments in humans: the role of subtle allele frequency shifts. Philos Trans R Soc Lond B Biol Sci. 2010;365:2459–68. doi: 10.1098/rstb.2010.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hernandez RD, et al. Classic selective sweeps were rare in recent human evolution. Science. 2011;331:920–4. doi: 10.1126/science.1198878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pritchard JK, et al. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr Biol. 2010;20:R208–15. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Turchin MC, et al. Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nat Genet. 2012;44:1015–9. doi: 10.1038/ng.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wilde S, et al. Direct evidence for positive selection of skin, hair, and eye pigmentation in Europeans during the last 5,000 y. Proc Natl Acad Sci USA. 2014 doi: 10.1073/pnas.1316513111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Olalde I, et al. Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature. 2014 doi: 10.1038/nature12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Krüttli A, et al. Ancient DNA analysis reveals high frequency of European lactase persistence allele (T-13910) in medieval central europe. PLoS ONE. 2014;9:e86251. doi: 10.1371/journal.pone.0086251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Malmström H, et al. High frequency of lactose intolerance in a prehistoric hunter-gatherer population in northern Europe. BMC Evol Biol. 2010;10:89. doi: 10.1186/1471-2148-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Plantinga TS, et al. Low prevalence of lactase persistence in Neolithic South-West Europe. Eur J Hum Genet. 2012;20:778–782. doi: 10.1038/ejhg.2011.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fu Q, et al. DNA analysis of an early modern human from Tianyuan Cave, China. Proc Natl Acad Sci U S A. 2013;110:2223–7. doi: 10.1073/pnas.1221359110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cooper A, Poinar HN. Ancient DNA: do it right or not at all. Science. 2000;289:1139. doi: 10.1126/science.289.5482.1139b. [DOI] [PubMed] [Google Scholar]
- 118.Green RE, et al. Analysis of one million base pairs of Neanderthal DNA. Nature. 2006;444:330–336. doi: 10.1038/nature05336. [DOI] [PubMed] [Google Scholar]
- 119.Green RE, et al. The Neandertal genome and ancient DNA authenticity. The EMBO Journal. 2009;28:2494–2502. doi: 10.1038/emboj.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hedges SB, Schweitzer MH. Detecting dinosaur DNA. Science. 1995;268:1191–1192. doi: 10.1126/science.7761839. author reply 1194. [DOI] [PubMed] [Google Scholar]
- 121.Pääbo S. Molecular cloning of Ancient Egyptian mummy DNA. Nature. 1985;314:644–645. doi: 10.1038/314644a0. [DOI] [PubMed] [Google Scholar]
- 122.Pääbo S, et al. Genetic analyses from ancient DNA. Annu Rev Genet. 2004;38:645–679. doi: 10.1146/annurev.genet.37.110801.143214. [DOI] [PubMed] [Google Scholar]
- 123.Wall JD, Kim SK. Inconsistencies in Neanderthal genomic DNA sequences. PLoS Genet. 2007;3:1862–1866. doi: 10.1371/journal.pgen.0030175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Willerslev E, et al. Isolation of nucleic acids and cultures from fossil ice and permafrost. Trends Ecol Evol (Amst) 2004;19:141–147. doi: 10.1016/j.tree.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 125.Gilbert MTP, et al. Assessing ancient DNA studies. Trends Ecol Evol (Amst) 2005;20:541–544. doi: 10.1016/j.tree.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 126.Hofreiter M, et al. Ancient DNA. Nat Rev Genet. 2001;2:353–359. doi: 10.1038/35072071. [DOI] [PubMed] [Google Scholar]
- 127.Sawyer S, et al. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLoS ONE. 2012;7:e34131. doi: 10.1371/journal.pone.0034131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dabney J, et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc Natl Acad Sci USA. 2013;110:15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rohland N. DNA extraction of ancient animal hard tissue samples via adsorption to silica particles. Methods Mol Biol. 2012;840:21–28. doi: 10.1007/978-1-61779-516-9_3. [DOI] [PubMed] [Google Scholar]
- 130.Kircher M. Analysis of high-throughput ancient DNA sequencing data. Methods Mol Biol. 2012;840:197–228. doi: 10.1007/978-1-61779-516-9_23. [DOI] [PubMed] [Google Scholar]
- 131.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Meyer M, et al. A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature. 2013 doi: 10.1038/nature12788. [DOI] [PubMed] [Google Scholar]
- 134.Skoglund P, et al. Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. Proc Natl Acad Sci USA. 2014;111:2229–2234. doi: 10.1073/pnas.1318934111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Carpenter ML, et al. Pulling out the 1%: Whole-Genome Capture for the Targeted Enrichment of Ancient DNA Sequencing Libraries. The American Journal of Human Genetics. 2013;93:852–864. doi: 10.1016/j.ajhg.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Alexander DH, et al. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–64. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 138.Raj A, et al. Variational Inference of Population Structure in Large SNP Datasets. bioRxiv. 2013 doi: 10.1101/001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Morris AG. The myth of the East African’ Bushmen’. The South African Archaeological Bulletin 2003 [Google Scholar]
- 140.Schepartz LA. Who were the latter Pleistocene eastern Africans? African archaeological review. 1988;6:57–72. [Google Scholar]
- 141.Anthony DW. The horse, the wheel, and language: how bronze-age riders from the Eurasian steppes shaped the modern world. Princeton University Press; 2007. [Google Scholar]
- 142.Mallory J. The homelands of the Indo-Europeans. In: Blench R, Spriggs M, editors. Archaeology and language. Routledge; 1997. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.