SUMMARY
Posttranscriptional maturation is a critical step in miRNA biogenesis that determines mature miRNA levels. In addition to core components (Drosha and DGCR8) in the microprocessor, regulatory RNA-binding proteins may confer the specificity for recruiting and processing individual pri-miRNAs. Here, we identify DDX1 a regulatory protein that promotes the expression of a subset of miRNAs, including five members in the miR-200 family and four miRNAs in an 8-miRNA signature of a mesenchymal ovarian cancer subtype. A majority of DDX1-dependent miRNAs are induced after DNA damage. This induction is facilitated by the ATM-mediated phosphorylation of DDX1. Inhibiting DDX1 promotes ovarian tumor growth and metastasis in a syngeneic mouse model. Analysis of The Cancer Genome Atlas (TCGA) reveals that low DDX1 levels are associated with poor clinical outcome in patients with serous ovarian cancer. These findings suggest that DDX1 is a key modulator in miRNA maturation and ovarian tumor suppression.
INTRODUCTION
MicroRNAs (miRNAs) are small endogenous noncoding RNAs that regulate gene expression by repressing translation and/or promoting degradation of their target mRNAs. Universally expressed in nearly all metazoans, plants, and even DNA viruses, they are involved in many cellular processes such as proliferation, differentiation, stress responses, apoptosis, and development (Bartel, 2009; Siomi and Siomi, 2010). The biogenesis of miRNA is a tightly regulated multi-step process. In the nucleus, primary miRNAs (pri-miRNAs) are first processed by the microprocessor containing the RNase III enzyme Drosha and its cofactor DGCR8 (DiGeorge syndrome critical region gene 8) (Gregory et al., 2006). The processed products are precursor miRNAs (pre-miRNAs) that have a hairpin structure of ~70-nt. They are exported by Exportin-5 to the cytoplasm (Murchison and Hannon, 2004), where the stem-loops of pre-miRNAs are cleaved off by another RNase III, Dicer, resulting in the production of mature miRNAs (Lund and Dahlberg, 2006). The RNA-induced silencing complex (RISC) loaded with mature miRNA is subsequently guided by miRNA to pair with target mRNA transcripts at their 3’-untranslated region (3’-UTR) and induce mRNA degradation or inhibition of translation (Chong et al., 2010; Friedman et al., 2009).
Aberrant miRNA expression has been reported in a variety of human cancers (Iorio and Croce, 2012; Esteller, 2011). The first direct evidence was derived from studies for identification of tumor suppressor genes at chromosome 13q14 in chronic lymphocytic leukemia (CLL) (Calin et al., 2002; Calin et al., 2005). Deletion of miR-15a and miR-16-1 in the 13q14 region was found to be involved in the pathogenesis of human CLL. Since then, accumulating evidence has shown that many miRNA genes residing in the regions of chromosomal instability or nearby chromosomal breakpoints are prone to genomic alterations (Calin et al., 2004; Zhang et al., 2006). In addition to genomic abnormalities in the miRNA genes, defects in miRNA biogenesis machinery often result in miRNA dysregulation in human cancer, which include DNA/histone modifications, transcriptional activation/suppression, and pri-miRNA maturation (Wan et al., 2013a). In particular, post-transcriptional maturation, rather than transcriptional regulation, determines the levels of mature miRNAs in many cases. Suppression of global miRNA production was found in many types of human cancer due to down-regulated expression of Drosha or Dicer (Davalos and Esteller, 2010; Merritt et al., 2008). However, human cancer miRNome studies have demonstrated aberrant expression of many individual miRNAs (oncomiRs or tumor suppressive miRs) (Lee and Dutta, 2009), which obviously cannot be ascribed to a global shutdown of the miRNA processing machinery. It appears that regulatory components in the Drosha and Dicer complexes may confer miRNA specificity by controlling their processing activity in sequence- or structure- specific manners (Zhang and Lu, 2011). Several RNA-binding proteins, such as KSRP, TDP-43, p68 and p72 have been identified as such regulatory proteins that interact with miRNA processing complexes and modulate maturation of specific miRNAs (Fuller-Pace and Moore, 2011; Kawahara and Mieda-Sato, 2012; Trabucchi et al., 2009; Mori et al., 2014).
Among all cancer-associated miRNAs, the miR-200 family (miR-200a/b/c, miR-141 and miR-429) is believed to play an essential role in tumor suppression by inhibiting epithelial-mesenchymal transition (EMT), an initiating step of metastasis (Gregory et al., 2008; Korpal et al., 2008; Park et al., 2008). The miR-200s target the E-cadherin transcriptional repressors ZEB1 and ZEB2 (Korpal et al., 2008; Korpal and Kang, 2008). Knockdown of miR-141 and miR-200b was shown to reduce E-cadherin expression and thus increase cell motility and induce EMT (Pecot et al., 2013). Forced miR-200 expression inhibited the formation of distant metastasis in lung adenocarcinoma (Yang et al., 2011). However, miR-200 overexpression promoted metastatic colonization in mouse models probably through direct targeting of Sec23a (Korpal et al., 2011). The seemingly conflicting reports suggest the role of miR-200s in suppressing or promoting metastasis in cancer-dependent contexts. Our recent study demonstrated that in ovarian, lung, renal and basal-like breast adenocarcinomas, elevated miR-200 expression portends improved clinical outcome, in part, through secretion of metastasis-suppressive proteins (Pecot et al., 2013). We recently identified a miRNA-regulatory network that defines a mesenchymal subtype associated with poor overall survival in patients with serous ovarian cancer (Yang et al., 2013). Two miR-200 family members, miR-200a and miR-141, are included in this miRNA signature, supporting the role of miR-200s in EMT and cancer metastasis.
Equally as important as transcriptional regulation, the processing of premature miRNAs (pri-miRNAs and pre-miRNAs) is a critical rate-limiting step that controls mature miRNA levels. The first step in miRNA maturation is executed by the Drosha microprocessor in which Drosha and DGCR8 are the core components (Gregory et al., 2004; Lee et al., 2003). However, neither Drosha nor DGCR8 have binding specificity for individual pri-miRNAs. Differential expression levels of mature miRNAs, even those derived from the same primary transcript, suggesting that regulatory components in the complex may confer the specificity for recruiting and processing pri-miRNAs. Among a growing list of RNA-binding proteins identified from the microprocessor complex are the members of the DEAD-box helicase family, including p68, p72 and DDX1 (Gregory et al., 2004; Mori et al., 2014; Suzuki et al., 2009). In the present study, we show that DDX1 interacts with the Drosha complex and promotes the expression of a subset of miRNAs.
RESULTS
DDX1 interacts with Drosha microprocessor
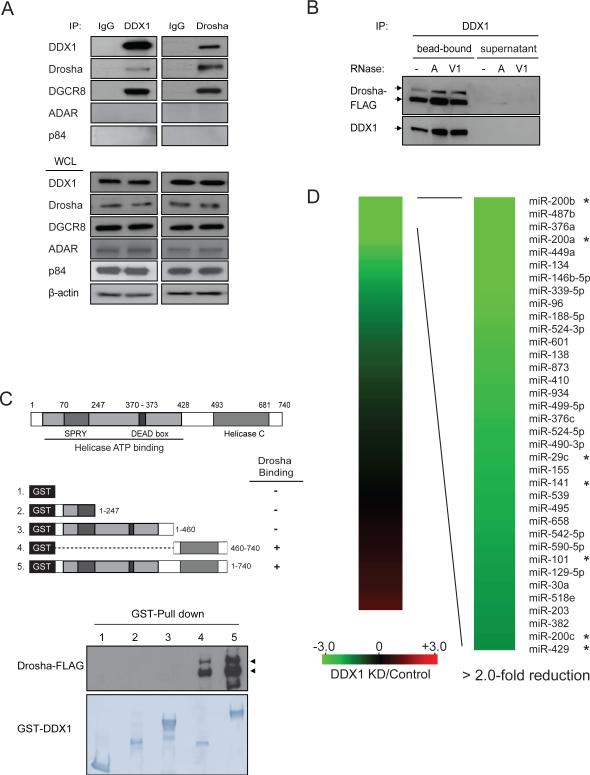
DDX1 was first identified in a high-molecular-mass complex containing a number of Drosha-associated polypeptides (Gregory et al., 2004). However, the functional role of DDX1 has yet to know. A majority of DDX1 is present in the cell nucleus and significant co-localization between DDX1 and Drosha was observed (Figure S1A). We determined the physical interaction of DDX1 with the Drosha microprocessor (Figure 1A). Endogenous DDX1 was detected in the Drosha immunoprecipitate and conversely, Drosha and DGCR8 were also identified in the DDX1 immunoprecipitate. Two negative control RNA-binding proteins ADAR and p84 were not detected in the Drosha complex (Figure 1A) (Kawahara and Mieda-Sato, 2012). To determine whether RNA molecules are involved in these interactions, the DDX1 immunoprecipitates were treated with RNase A (degrading single-stranded RNA) (Raines, 1998) or RNase V1 (degrading double-stranded RNA) (Kawai and Amano, 2012) to remove RNAs (Figure S1B). Regardless of RNase treatments, Drosha firmly interacted with endogenous DDX1 on the DDX1 antibody-conjugated beads and no Drosha was dissociated and released to the supernatant (Figure 1B). Therefore, the DDX1-Drosha interaction does not involve any RNA molecules. To determine the domain(s) of DDX1 responsible for the Drosha binding, full-length DDX1 or its deletion mutants were tested in the GST pull-down assay (Figure 1C). The carboxyl terminal sequence (amino acid residues 460-740) including a helicase domain (residues 493-681) was essential for DDX1 to interact with Drosha, whereas most part of DDX1 (residues 1-460) at the N-terminus was dispensable.
Figure 1. Interaction of DDX1 with Drosha microprocessor.
(A) Interaction between endogenous DDX1 and Drosha/DGCR8. Immunoprecipitation (IP) and Western blotting analyses were performed using indicated antibodies. Normal IgG was used as a negative control for IP. RNA-binding proteins ADAR and p84 were used as negative controls for the Drosha-binding proteins. WCL: whole cell lysate. (B) RNA-independent interaction between DDX1 and Drosha. DDX1 immunoprecipitates were treated with RNase A (10U) or RNase V1 (10U) and separated to supernatant and bead-bound fractions for Western blotting analyses. (C) Schematic representation of DDX1 domains (upper panel) and the interaction between Drosha and truncated forms of DDX1 (bottom panel). GST-fused DDX1 proteins were immobilized on glutathione Sepharose beads and mixed with the lysate of HEK293T cells expressing Drosha-FLAG. (D) Depletion of DDX1 inhibits the expression of a subset of miRNAs. Total RNAs from control and DDX1-knockdown (KD) were subject to global miRNA profiling analyses using q-PCR miRNA microarray. Green or red color on the heat map indicates a decrease or increase of miRNA level, and color intensities correspond to relative signal levels. Thirty-six miRNAs with over 2-fold reduction in the DDX1-knockdown cells were identified as DDX1-dependent miRNAs. The DDX1-dependent miRNAs in the miR-200 family and the 8-miRNA signature for mesenchymal subtype of ovarian cancer were marked with *. See also Figure S1.
DDX1 promotes the expression of a subset of miRNAs
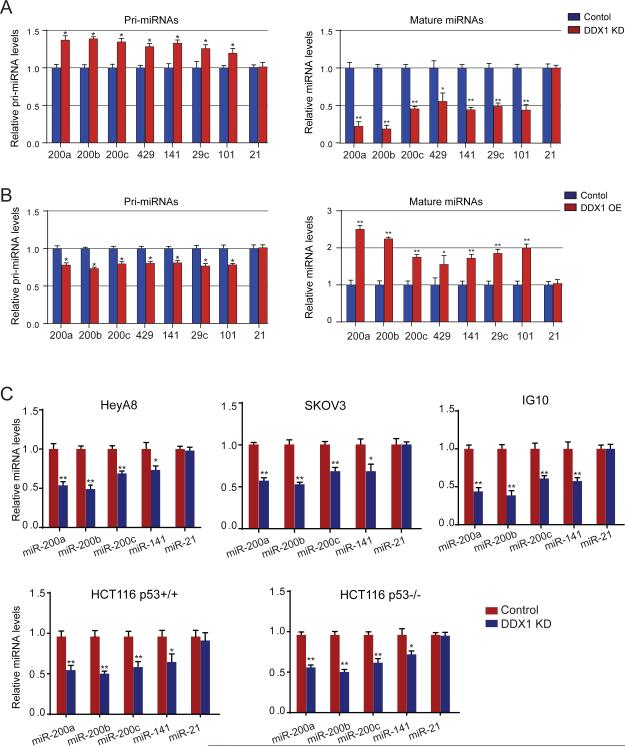
Observation of direct interaction between DDX1 and the microprocessor led us to investigate a potential role of DDX1 in miRNA biogenesis. Using a reverse transcription quantitative-PCR (qRT-PCR) human miRNA array, miRNA expression profiles in control and DDX1-knockdown U2OS cells were analyzed to determine the effect of DDX1 on global miRNA expression (Figures 1D). Depletion of DDX1 significantly decreased the expression levels of a subset of 36 miRNAs (cut-off > 2-fold) (GEO accession number GSE54990), including all five members in the miR-200 family (miR-200a, miR-200b, miR-200c, miR-141 and miR-429). Our recent study of ovarian cancer genomics revealed a 8-miRNA signature that defines a mesenchymal subtype of serous ovarian cancer(Yang et al., 2013). Among the eight miRNAs, four miRNAs (miR-200a, miR-29c, miR-141 and miR-101) are significantly dependent on DDX1, suggesting that DDX1 may play a role in ovarian tumor progression. To determine whether DDX1 regulates the miRNA expression at transcriptional or post-transcriptional levels, we performed nuclear run-on assays to measure the effect of DDX1 on pri-miRNA transcription. No notable differences were seen in the transcription of pri-miR-200s from the two miR-200 gene clusters (miR-200a/200b/429 and miR-200c/141) in the control and DDX1-silenced cells (Figure S2A). However, levels of mature DDX1-dependent miRNAs, but not control miR-21, were significantly decreased in the DDX1-knockdown U2OS cells (Figures 2A and S2B). Due to the potential inhibition of miRNA processing activity, primary transcripts of the DDX1-dependent miRNAs were accumulated. Conversely, these DDX1-dependent miRNAs were up-regulated in the DDX1-overexpressing cells (Figure 2B). These results suggest that DDX1 promotes the expression of specific miRNAs at the post-transcriptional level.
Figure 2. Posttranscriptional regulation of miRNA expression by DDX1.
(A) Levels of primary or mature forms of the DDX1-dependent miRNAs in control and DDX1-knockdown U2OS cells. (B) Levels of primary or mature forms of the DDX1-dependent miRNAs in control and DDX1-overexpressing U2OS cells. In both (A) and (B), miR-21 was used as a DDX1-independent control. Error bars represents the mean ± SD, * p < 0.05, ** p < 0.001. (C) Levels of DDX1-dependent miRNAs in control or DDX1-knockdown cell lines as indicated. HeyA8, SKOV3 are human ovarian cancer cell lines, and IG10 is a mouse ovarian cancer cell line. HCT116p53+/+ and HCT116p53−/− are a pair of isogenic colorectal cancer cell lines. U6 RNA was used for normalization in qPCR analyses and miR-21 was used as a DDX1-independent miRNA control. Error bars represents the mean ± SD, * p < 0.05, ** p < 0.001. See also Figure S2.
We next examined the effect of DDX1 knockdown on miRNA expression in three ovarian cancer cell lines (HeyA8, SKOV3 and IG10) and a pair of isogenic HCT116 cell lines (HCT116p53+/+ and HCT116p53−/−) (Figure 2C). DDX1 was efficiently knocked down in all the tested cell lines (Figure S2B). Levels of the DDX1-dependent miRNAs were measured by qPCR using specific primers for mature miRNAs. Despite some variations across cell lines, the DDX1-dependent miRNAs were consistently suppressed in the DDX1-silenced cells (Figures 2C, S2C and S2D). As a control, miR-21 levels were not influenced by the DDX1 knockdown. The level of mature miR-429 could not be determined due to low expression levels (high CT values in qPCR) in most of the tested cell lines (Figure S2C). While p53 was previously reported to transcriptionally activate miR-200 genes (Kim et al., 2011; Chang et al., 2011), our results from HCT116p53+/+ and HCT116p53−/− cells suggest that DDX1 regulates miRNA expression in a p53-independent manner (Figure 2C).
DDX1 directly binds specific pri-miRNAs and promotes their processing
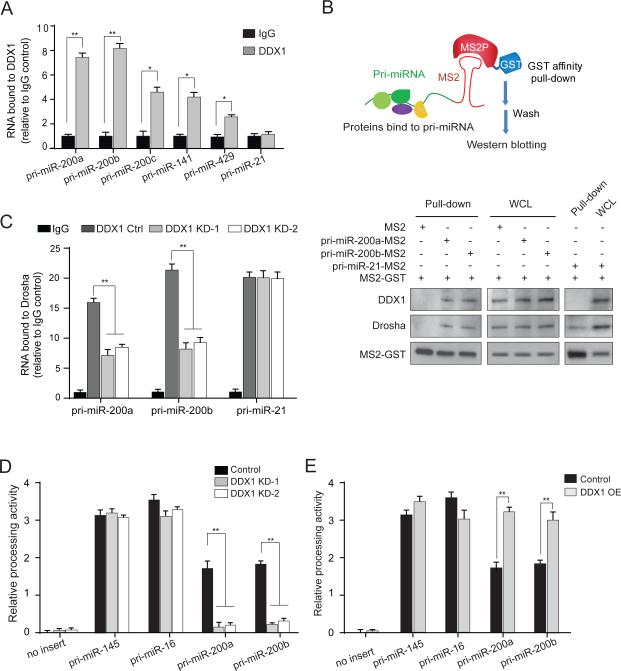
As a double-stranded RNA-binding protein, DDX1 may specifically bind target pri-miRNAs and recruit them to the microprocessor. RNA immunoprecipitation (RIP) assays showed specific interactions between endogenous DDX1 and five of the DDX1-dependent pri-miRNAs (Figures 3A, S3A and S3B). As a DDX1-independent negative control, pri-miR-21 had no interaction with DDX1. To further confirm the pri-miRNA-DDX1 interaction, we employed a MS2-TRAP (MS2-tagged RNA affinity purification) assay (Yoon et al., 2012). In this assay, MS2 is a 19-nucleotide bacteriophage RNA sequence that folds into a hairpin loop structure, which is recognized with high specificity and affinity by the bacteriophage capsid protein MS2P (Figure 3B). MS2-tagged pri-miR-200a/b was co-expressed with GST-MS2P in the cells. The pri-mRNA-protein complex was pulled down by anti-GST beads (Figure S3C). Both DDX1 and Drosha were detected in the pri-miR-200a/b immunoprecipitates. However, only Drosha, but not DDX1, was detected in the control pri-miR-21 immunoprecipitates (Figure 3B). Because DDX1 directly interacts with pri-miRNAs and its association with the Drosha complex is independent of RNA (Figure 1B), we postulated that DDX1 may enhance the recruitment of specific pri-miRNAs to the Drosha microprocessor. In the Drosha RIP assay, depletion of DDX1 specifically decreased the amounts of pri-miR-200a/b, but not pri-miR-21 (negative control), in the Drosha complex (Figure 3C). To determine the effect of DDX1 on the pri-miRNA processing, we conducted a luciferase-based in-vivo pri-miRNA processing assay (Kawai and Amano, 2012). Human U2OS cells were transfected with luciferase vectors carrying a pri-miRNA sequence between the open-reading frame of luciferase and polyadenylation signal. The Drosha-mediated cleavage of pri-miRNA results in loss of the polyadenylation tail, leading to instability of the luciferase transcript and decreased luciferase signals (Figure S3D). In the functionality test for the assay, the Drosha processing activity on each pri-miRNA was inversely correlated with the luciferase activity (Figure S3E). We assessed the processing activity of each pri-miRNA in the control, DDX1-overexpressing and -knockdown cells (Figures 3D and 3E). Overexpression of DDX1 significantly increased the Drosha-mediated processing activities on pri-miR-200a/b. By contrast, knockdown of DDX1 inhibited the processing of pri-miR-200a/b. As negative controls, the processing of pri-miR-16 and pri-miR-145 was not affected by altered DDX1 expression. These results suggest that DDX1 facilitates the Drosha-mediated pri-miRNA processing.
Figure 3. DDX1 recruits target pri-miRNAs to the Drosha microprocessor and promotes their processing.
(A) RNA immunoprecipitation (RIP) analysis reveals that primary forms of DDX1-dependent miRNAs are physically associated with DDX1 in U2OS cells. The DDX1-bound pri-miRNAs were analyzed by qRT-PCR. Control IgG was used as a negative control (* p < 0.05, ** p < 0.01). (B) Interaction between pri-miRNAs and DDX1 determined by the MS2-TRAP assay. Schematic illustration of MS2-TRAP assay is shown at the top. MS2-tagged pri-miRNAs and their associated protein complexes were pulled down by anti-GST beads and proteins were detected by Western blotting. (C) RIP analysis of the Drosha-interacting pri-miRNAs in control and DDX1-knockdown cells (** p < 0.01). Pri-miR-21 was used as a DDX1-independent control. (D-E) Pri-miRNA processing activity in DDX1- knockdown (D) and overexpressing (E) cells. The firefly luciferase signals are normalized by internal control Renilla luciferase readings and relative processing activity was shown as fold changes (** p < 0.01). Error bars represents the mean ± SD in this figure. See also Figures S3 & S4.
To understand sequential and structural requirements of pri-miRNAs for their interaction with DDX1, we analyzed the predicted secondary structures of the DDX1-dependent pri-miRNAs. We found two conserved residues, AA, near or in the loops of these pre-miRNAs (Table S1 and Figure S4A). Mutating the AA dilet to CC abolished the interaction between pri-miR-200a/b and DDX1 (Figure S4B), suggesting that the AA dilet is probably essential for the DDX1 interaction.
DDX1 promotes pri-miRNA processing in DNA damage response
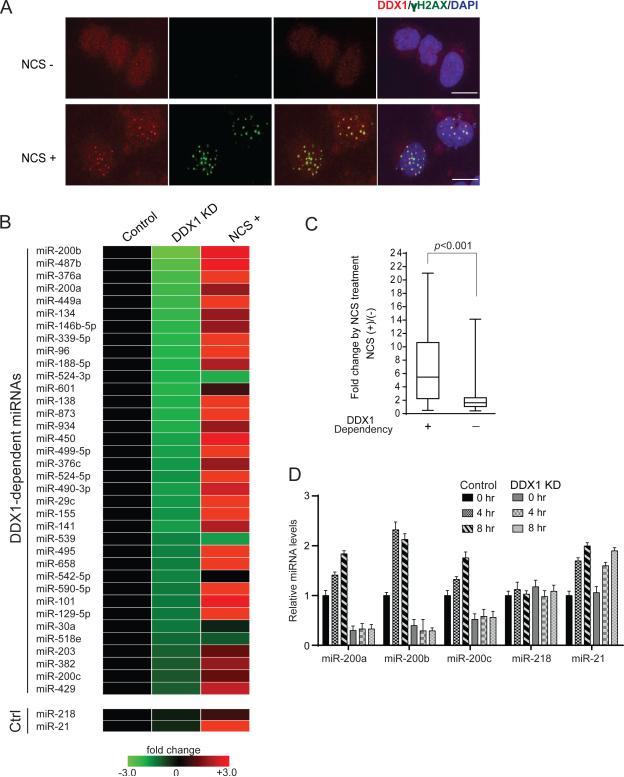
Recent studies have shown that DNA damage stress leads to a global change of miRNA expression profiles (Pothof et al., 2009; Wan et al., 2013b; Zhang et al., 2011; Ishii and Saito, 2006). Interestingly, a previous report showed that DDX1 was co-localized with ionizing radiation-induced DNA damage foci (Li et al., 2008). We attempted to determine whether DDX1 serves as a mediator that translates DNA damage signaling to miRNA biogenesis. DDX1 was primarily in the nucleus and translocalized into the DNA damage foci (indicated by γ-H2AX staining) after treatment of neocarzinostatin (NCS), a radiomimetic drug (Figure 4A). We analyzed global miRNA expression profiles in control and stable DDX1-knockdown U2OS cells treated with NCS, and identified DDX1-dependent and DNA damage-induced miRNA signatures (Figure 4B). Interestingly, a majority of the DDX1-dependent miRNAs (31 out 35) were significantly induced after DNA damage (Figures 4B and 4C), indicating the DNA damage-induced DDX1 activity in miRNA biogenesis. Consistent with the global miRNA expression data, levels of mature miR-200a/b/c were gradually increased after DNA damage (Figures 4D and S5A). However, knockdown of DDX1 severely suppressed their basal expression levels and abolished their induction after DNA damage. Our previous study showed that miR-21 is induced by phosphorylated KSRP after DNA damage (Zhang et al., 2011). Indeed, miR-21 was expressed and induced in a DDX1-independent manner (Figure 4D). The negative control miR-218 was not affected by DDX1 and DNA damage stress. Induction of miR-200s after DNA damage was independent of p53 status in the isogenic p53+/+ and p53−/− HCT116 cell lines (Figure S5B), but depletion of DDX1 or inhibiting ATM activity completely abolished the DNA damage-mediated induction of miR-200s (Figures S5C and S5D). While p53 was previously shown to transactivate miR-200 genes (Chang et al., 2011; Kim et al., 2011), our results suggest that DDX1-dependent post-transcriptional regulation is essential to determine miR-200 expression levels.
Figure 4. DNA damage-induced expression of DDX1-dependent miRNAs.
(A) DDX1 is translocalized to the DNA damage foci. U2OS cells were treated with NCS (500 ng/ml) for 30 min and analyzed by immunostaining using anti-γ-H2AX and anti-DDX1 antibodies. DAPI was used for nucleus staining. Scale bar: 10μm. (B) Expression of DDX1-dependent miRNAs is induced after DNA damage. Total RNAs were purified from the control and DDX1-knockdown cells treated with or without NCS (500 ng/ml, 6 h) and subject to global miRNA expression profiling analyses using human miRNA qPCR microarray. miR-218 and miR-21 were used as DDX1-independent controls. miR-21 is induced after DNA damage. (C) DNA damage induction of DDX1-dependent (DDX1-KD/Control < 0.50) and -independent (DDX1-KD/control > 0.50) miRNAs. (D) Levels of miR-200a/b/c were induced after NCS (500 ng/ml) treatment in control U2OS cells, but not in the DDX1-knockdown U2OS cells. miR-218 and miR-21 were used as DDX1-independent controls. Error bars represents the mean ± SD. See also Figure S5.
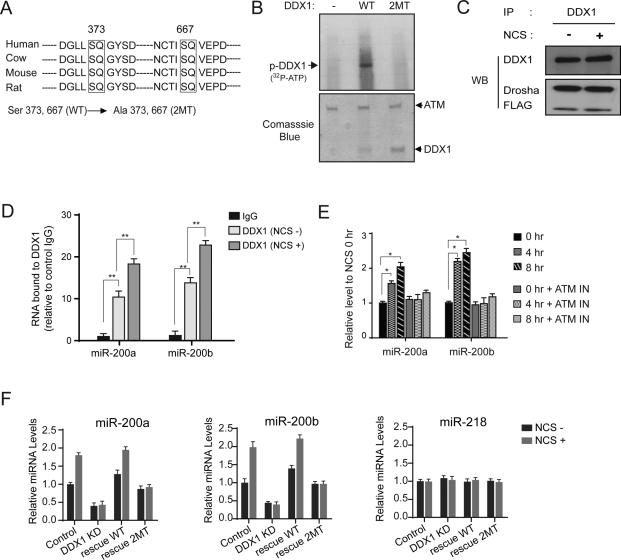
ATM phosphorylation facilitates DDX1 in the pri-miRNA processing
DDX1 is recruited to the DNA damage foci possibly via its interaction with RAD50 and ATM (Li et al., 2008). However, the levels of DDX1 and its interactions with those proteins in the DNA damage foci were not notably changed after DNA damage (Figures S6A and S6B), suggesting that DDX1 may not be essential for the initiation of DNA damage response. Actually, only minimal changes in the activity of homologous recombination DNA repair were observed in the DDX1-overexpressing or -knockdown cells (Figure S6C). Consistent with the previous report (Li et al., 2008), we confirmed that DDX1 is phosphorylated in an ATM-dependent manner (Figure S6D, upper panel). In silico analysis identified two consensus ATM phosphorylation sites on DDX1 in mammals (Figure 5A). We generated a phosphorylation-deficient mutant (S373A/S667A, shown as 2MT) of DDX1. In-vitro and in-vivo ATM kinase assays showed that this mutant DDX1 could not be phosphorylated by ATM (Figure 5B and the bottom panel of Figure S6D). We postulated that ATM-mediated phosphorylation of DDX1 may facilitate DDX1 in the pri-miRNA processing. We first investigated the physical interaction between DDX1 and Drosha in the presence or absence of DNA damage. No differences were observed on the protein levels of DDX1 and Drosha and their nuclear/cytoplasmic distributions after DNA damage (Figure S6E-G), and their interaction was not significantly changed as well (Figure 5C). These results suggest that phosphorylation of DDX1 does not enhance its interaction with the Drosha microprocessor. Next, we performed RIP assays to determine whether the pri-miRNA-binding activity of DDX1 was enhanced after DNA damage. Markedly increased levels of pri-miR-200a/b were bound by DDX1 after DNA damage (Figure 5D). As a result, the levels of mature miR-200a/b were increased after DNA damage, and this induction was abolished by inhibition of ATM (Figure 5E), suggesting that the ATM phosphorylation promotes DDX1 to more efficiently recruit pri-miRNAs to the Drosha complex. Next, we examined whether expression of wildtype (WT) or phosphorylation-deficient (2MT) DDX1 restored the DNA damage-induced miR-200a/b biogenesis in the DDX1-depleted cells. We generated shRNA-resistant DDX1 expression constructs (wildtype and mutant) by mutating shRNA-targeting nucleotides to synonymous ones. Overexpression of wildtype DDX1 restored the expression of mature miR-200a/b, and DNA damage further induced the levels of these miRNAs (Figure 5F). By contrast, the phospho-mutant DDX1 failed to induce miR-200a/b levels after NCS treatment (Figure 5F). However, this phospho-mutant form was capable of restoring basal levels of miR-200a/b in the DDX1-depleted cells. This result is consistent with the observation showing that DDX1's interaction with the Drosha microprocessor is independent of DNA damage stress (Figure 5C). In unstressed cells, the phospho-mutant DDX1 interacts with the Drosha complex and maintain its basic function in pri-miRNA processing. However, the ATM phosphorylation enhances DDX1's interaction with target pri-miRNAs and thus induces their processing after DNA damage.
Figure 5. ATM phosphorylation facilitates DDX1 in pri-miRNA processing.
(A) Two putative ATM phosphorylation sites are conserved in mammalian DDX1 genes. Phosphorylation-deficient mutant (2MT) was generated by mutating serine to alanine on both sites. (B) Wildtype (WT) DDX1, but not the 2MT, is phosphorylated by the ATM kinase. Purified wildtype or mutant DDX1 proteins were incubated with immunopurified ATM in a kinase buffer containing 32P-ATP. (C) Interaction between DDX1 and Drosha was unchanged after DNA damage. U2OS cells were treated with 500 ng/ml NCS for 4 h and cell lysates were harvested for immunoprecipitation and Western blotting analyses. (D) DNA damage treatment enhances the interaction of DDX1 with pri-miR-200a/b. RIP assays were performed using anti-DDX1 antibody and lysates of U2OS cells treated with or without NCS (** p < 0.01). (E) DNA damage-induced expression of miR-200a/b is dependent on ATM. ATM activity is inhibited by the inhibitor CGK733 (10 μM) (* p < 0.05). (F) Overexpression of wildtype DDX1, but not the phosphorylation-deficient DDX1, restored the induction of miR-200a/b expression after DNA damage. miR-218 was used as a negative control. Error bars represents the mean ± SD in this figure. See also Figure S6.
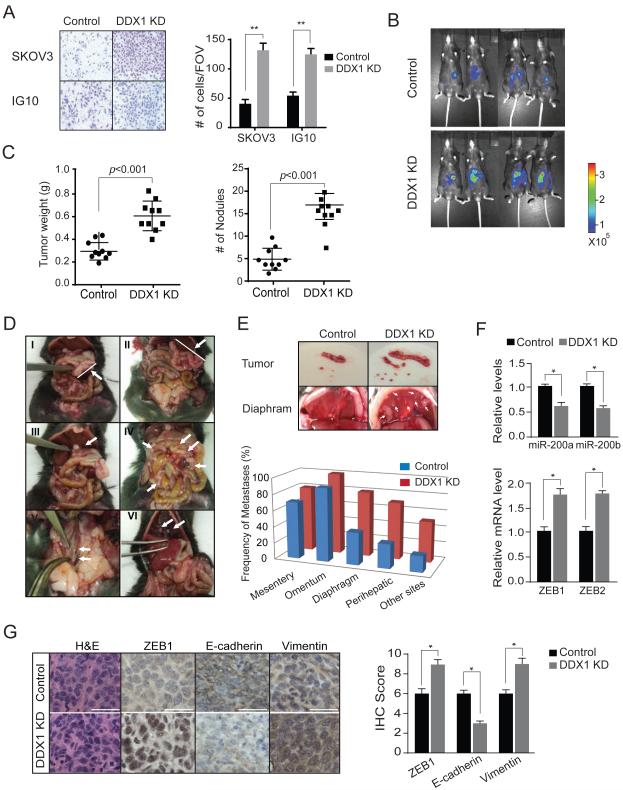
DDX1 inhibits ovarian tumor invasion and metastasis
We recently showed that elevated miR-200 expression is associated with good clinical outcomes in ovarian, renal and lung cancers (Pecot et al., 2013). Moreover, our study of ovarian cancer genomics revealed an 8-miRNA signature that defines a mesenchymal subtype of serous ovarian cancer (Yang et al., 2013). The DDX1-dependent miRNAs include not only all of the five members in the miR-200 family, but also four miRNAs (miR-200a, miR-29c, miR-141 and miR-101) in the 8-miRNA signature, suggesting that DDX1 may be a key player in ovarian tumor progression. We first examined the effect of DDX1 on ovarian cancer cell invasion. Results of in-vitro invasion assays demonstrated that ovarian cancer cells SKOV3 (human) and IG10 (mouse) had much higher activity of cell invasion when DDX1 was knocked down (Figure 6A), although their proliferation rates were not notably affected (data not shown). Next, we employed a syngeneic ovarian tumor model to examine the role of DDX1 on ovarian tumor progression in vivo. DDX1 was stably knocked down in the murine IG10 cells, which are transformed ovarian surface epithelial cells derived from C57BL/6 mice (Roby et al., 2000). The IG10 cells stably expressing luciferase were generated for monitoring tumor growth in vivo. Control and DDX1-silenced IG10 cells (1×106) were injected into peritoneal cavity of C57BL/6N mice (n = 10 per group). Tumor growth within the peritoneal cavity and build-up ascites were monitored. We observed dramatic increases of luminescence in the mice bearing tumors derived from the DDX1-silenced cells (Figure 6B). After 8 weeks, the animals were sacrificed and inspected for tumor weights and tumor nodules. Depletion of DDX1 resulted in profound increases in tumor weight (161.6% increase, p < 0.001) and number of nodules (188.2% increase, p < 0.001) (Figure 6C). Moreover, higher metastatic activity of the DDX1-silenced ovarian tumors was observed. These tumors showed highly frequent metastases to common metastatic sites (mesentery, omentum, diaphragm, perihepatic sites, pelvic and paraaortic lymph nodes, and kidney) of high-grade serous ovarian carcinoma (Figures 6D and 6E). In the DDX1-silenced tumors, the levels of miR-200a/b were significantly decreased while their targets ZEB1 and ZEB2 were induced (Figure 6F). To assess whether knockdown of DDX1 promotes EMT in vivo, we measured the levels of ZEB1, E-cadherin and Vimentin in control and DDX1-silenced tumors. In comparison with the control, the DDX1-silenced tumors exhibited elevated levels of ZEB1 (145% increase, p = 0.017) and Vimentin (150% increase, p = 0.011), and decreased levels of E-cadherin (61% reduction, p = 0.009) (Figure 6G). Taken together, these results suggest that suppression of DDX1 promotes ovarian tumor progression.
Figure 6. Silencing DDX1 promotes ovarian tumor cell invasion in vitro and ovarian tumor progression in vivo.
(A) Knockdown of DDX1 promotes ovarian tumor cell invasion. Matrigel invasion assays were performed on control or DDX1-knockdown SKOV3 and IG10 cells. Average numbers of invasive cells per field of view (FOV) were presented (**p < 0.001) (B) Knockdown of DDX1 promotes IG10 syngeneic tumor growth. Control and DDX1-knockdown IG10 cells expressing firefly luciferase were injected intraperitoneally into female C57BL/6N mice. Shown are the representative luciferase images of ovarian tumors. (C) Quantification of tumor weights and tumor nodules in mice (n = 10 for each group). Error bar represents the mean ± SEM. (D) Representative images of tumor nodules and metastases in the mice carrying syngeneic ovarian tumors derived from control and DDX1-knockdown IG10 cells. Shown here are tumor mass in omentum of a control tumor (I) and tumor mass in omentum (II), perihepatic area (III), mesentery (IV), lymph nodes (V), and diaphragm (VI) of DDX1-knockdown tumors. (E) Frequency of metastases to distant sites (mesentery, omentum, diaphragm, perihepatic and other sites). Other sites include paraaortic lymph nodes, kidney, and ovary. (F) Levels of miR-200a/b and their targets ZEB1 and ZEB2 in control and DDX-silenced tumors. Error bars represents the mean ± SD. (G) Immunohistochemical analysis of ZEB1, E-Cadherin and Vimentin in control and DDX1-knockdown ovarian tumors. Scale bar: 30 μm. Relative levels of ZEB1, E-Cadherin and Vimentin are shown in the graph (See Materials and Methods for IHC score, * p < 0.05). Error bars represents the mean ± SD.
The potential of DDX1 as a predictive marker of clinical outcome
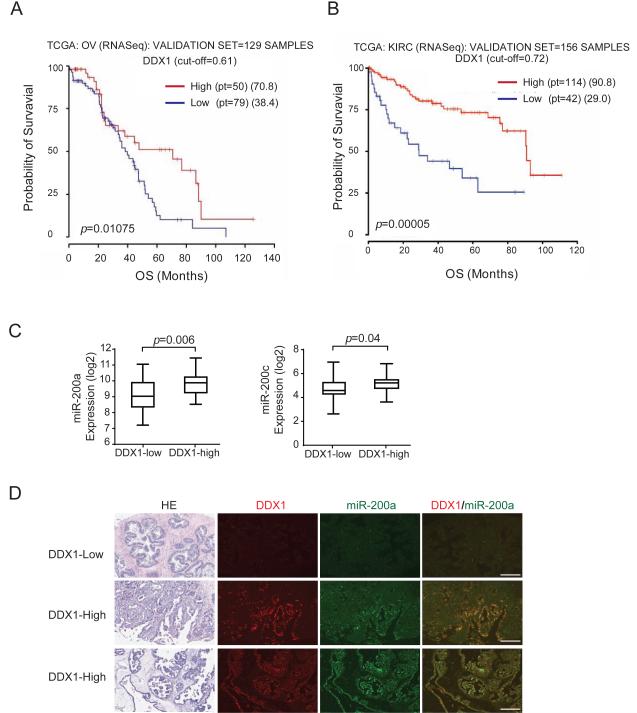
To determine the correlation between DDX1 levels and clinical outcome of cancer patients, we analyzed the expression of DDX1 and miRNAs and the correlation between DDX1 and overall patient survival in The Cancer Genome Atlas (TCGA). We randomly split the entire population of patients with certain type of cancer into training/validation cohorts. Analysis of both cohorts across cancer types revealed that low levels of DDX1 are significantly associated with poor overall survival in patients with serous ovarian adenocarcinoma and in patients with kidney renal clear cell carcinoma (Figure 7A-B and Figure S7A-B). In the validation cohort of ovarian adenocarcinomas, the median survival of patients with high-DDX1 ovarian cancer is 70.8 months, which is remarkably better than 38.4 months for patients with low-DDX1 ovarian cancer (p = 0.01075) (Figure 7A). Levels of miR-200a and miR-200c in DDX1-high ovarian tumors are 2.1-fold (p = 0.006) and 1.8-fold (p = 0.04) higher than those in DDX1-low tumors, respectively, in the validation data set (Figure 7C). The positive correlation between DDX1 and miR-200 levels was also observed in the training set (Figure S7C). Consistent with the TCGA data analysis, we observed that DDX1 levels were positively correlated with miR-200a levels from in-situ hybridization analyses of ovarian tumor tissue microarray (Figures 7D and S7D). These results suggest that DDX1 is potentially a predictive marker for clinical outcome of ovarian cancer patients.
Figure 7. DDX1 levels are positively correlated with clinical outcome in cancer patients.
(A) Low levels of DDX1 are associated with poor overall survival in patients with ovarian tumors. Kaplan-Meyer plots for overall survival (OS) of patients with ovarian serous adenocarcinoma (OV). Entire population (n = 388) was randomly split in training/validation cohorts (2/3, 1/3) and a correlation with the survival was determined as described in Supplemental Experimental Procedures. Analysis of validation cohort (n = 129) is shown here. (B) Low levels of DDX1 are associated with poor overall survival in patients with kidney renal clear cell carcinoma (KIRC). Analysis of validation cohort (n = 156) is shown here. (C) Relative expression levels of miR-200a (p = 0.006) and miR-200c (p = 0.04) in the DDX1-low and -high groups from the validation cohort of ovarian serous adenocarcinomas. Error bars represents the mean ± SD. (D) Immunohistochemical analysis and in situ hybridization of DDX1 and miR-200a in human ovarian tumor tissue microarray. Scale bar: 100μm. See also Figure S7.
DISCUSSION
The current study identifies DDX1 as a novel cofactor in the Drosha microprocessor, which governs post-transcriptional maturation of a subset of miRNAs. Drosha is an RNase III enzyme that binds and cleaves double-stranded RNA with no sequence specificity. A recent study showed that one nucleotide change in stem-loop of pri-miRNA inhibits the Drosha-mediated cleavage, suggesting that the stem-loop structure of pri-miRNA may be critical for the Drosha activity (Slezak-Prochazka et al., 2010). While all the pri-miRNAs share this common feature with one or more stem-loop structures, each pri-miRNA also has unique complexity in sequence and structure that may determine the specificity of the Drosha-mediated processing. Those RNA-binding proteins, such as DDX1, become perfect candidates to provide this type of specificity through their selective interaction with target pri-miRNAs. Moreover, in contrast to another RNase III endonuclease Dicer for pre-miRNA processing, Drosha does not have helicase activity (Welker et al., 2011). Interaction of DDX1 with the Drosha microprocessor may not only confer pri-miRNA-binding specificity, but also help resolve the complex structure of pri-miRNA to generate desirable stem-loop structure for efficient cleavage.
The key player in the DNA damage response, p53, is also involved in the regulation of miRNA expression. Whereas it is a defined transcriptional factor that transactivates miR-34, miR-192, and miR-215 (He et al., 2007; Braun et al., 2008), p53 also up-regulates the post-transcriptional processing of miR-16, miR-143 and miR-145 through association with p68 in the Drosha complex (Suzuki et al., 2009). In addition, p53 suppresses EMT by repressing the expression of ZEB1 and ZEB2 through direct transactivation of miR-200 genes in primary hepatocellular carcinomas and human mammary epithelial cells (Kim et al., 2011; Chang et al., 2011). In the current study, we found that most of the DDX1-dependent miRNAs are induced after DNA damage but they are not associated with p53 (Figure 1D), suggesting that DDX1 regulates miRNA biogenesis in a p53-independent manner. Regardless of p53 status, DDX1-dependent induction of miR-200s was observed in the isogenic p53+/+ and p53−/− HCT116 cell lines. However, the p53-induced miR-200 expression is completely dependent on DDX1 (Figure S5C). Thus, the ATM kinase not only induces the level of p53, but also phosphorylates DDX1 and facilitates the processing of pri-miRNAs in the DNA damage response. Our analysis of ovarian serous adenocarcinoma database in TCGA revealed that a majority (90.7% or 573 out of 632) of ovarian tumors possess mutant p53, indicating that DDX1-associated, but not p53-associated, mechanism may determine the miR-200 expression in ovarian cancer cells. We propose that the ATM-initiated DNA damage signaling promotes the expression of a subset of miRNAs and inhibits tumor cell invasion in a DDX1- and p53- dependent manner. However, tumor progression is often accompanied by inactivation of DNA damage response and p53 (Bartkova et al., 2006; Halazonetis et al., 2008). The protein level of DDX1 will determine the expression of miR-200s in those malignant tumor cells during invasion and metastasis.
We recently identified an 8-miRNA signature for the mesenchymal subtype of serous ovarian cancer. Interestingly, four (miR-200a, -101, -141 and -29c) out of the eight miRNAs in this signature are positively regulated by DDX1, suggesting that low levels of DDX1 may predict for high metastasis potential of human ovarian cancer. Indeed, analysis of TCGA data base revealed that low levels of DDX1 are significantly associated with poor clinical outcomes in patients with serous ovarian cancer. Depletion of DDX1 significantly promotes ovarian tumor metastasis in mouse syngeneic ovarian tumor model. While lower levels of DDX1 are correlated with poor clinical outcome in patients with ovarian tumors, it is unknown whether the function of DDX1 in tumor progression is completely mediated by miRNAs. As a RNA-binding protein with RNA helicase activity, DDX1 may regulate the stability and expression of many types of RNA molecules such as mRNA and noncoding RNAs. Identification of the DDX1-assoicated RNAs and analysis of these RNA structures will allow us to better understand biological functions of DDX1. In addition, it is also of great importance to define how DDX1 expression is regulated in normal and cancer cells. Transcriptional and post-transcriptional mechanisms need to be identified to interpret the suppressed expression of DDX1 in cancer cells. It will also be of great interest to determine whether other cellular stresses in tumors, such as hypoxia or reactive oxygen species, contribute to the activity of DDX1. Further studies of DDX1-dependent miRNAs will identify molecular interactions between DDX1, miRNAs and cancer signaling pathways.
EXPERIMENTAL PROCEDURES
Cell culture and treatments
U2OS and HEK293T cell lines were purchased from the American Type Culture Collection, and HCT116p53+/+ and HCT116p53−/− were obtained from B. Vogelstein at Johns Hopkins University. These cell lines were maintained in Dulbecco's Modified Eagle Medium (DMEM) with 10% FBS at 37°C in 5% CO2. HeyA8, SKOV3, IG10 cell lines were grown in RPMI medium with 15% FBS at 37°C in 5% CO2. To induce double-stranded DNA breaks, cells were treated with neocarzinostatin (NCS, #N9162, Sigma-Aldrich) at indicated concentrations and harvested at indicated time points after treatment for RNA and protein analyses. To inhibit ATM kinase activity, cells were treated with 10 μM ATM kinase inhibitor CGK733 (#118501, Calbiochem) or DMSO (mock treatment) 1 h prior to DNA damaging treatment.
Syngeneic ovarian tumour mouse model
Female C57BL/6N mice (6-8 weeks old) were purchased from Jackson Laboratories. All studies were approved and supervised by the Institutional Animal Care and Use Committee at the MD Anderson Cancer Center. After mice were anesthetized, control and DDX1-silenced murine IG10-luciferase cells (1×106) in 100 μl HBSS Hank's solution were intraperitoneally injected. Tumors was monitored by the IVIS imaging system after luciferin injection for 10 min. Mice were euthanized before they met the institutional euthanasia criteria for tumor burden and overall health condition.
Nuclear run-on assay
Nuclear run-on assay was performed as described previously (Wan et al., 2013b). Briefly, an aliquot (1μg) of single-stranded DNA fragment (complementary to each specific miRNA transcript) was amplified by PCR. DNA was plot-blotted onto 0.45 μm nitrocellulose membrane (Bio-rad). After isolation of the nucleus and run-on transcription reaction, RNA was isolated with Trizol reagent. Membranes were probed with 32P-labelled run-on RNA, exposed to film and analyzed for intensity with a scintillation counter.
MS2-TRAP assay
The MS2-TRAP assay was performed as described previously (Yoon et al., 2012). In brief, vectors expressing MS2P-GST and MS2-tagged pri-miRNA were transfected into HEK293T cells. Cells were lysed and the RNA-protein complexes were affinity-purified using GST agarose beads. Pri-miRNAs in the complexes was isolated and detected by qRT-PCR and the RNA binding proteins (DDX1 and Drosha) were detected by Western blotting analysis.
In vivo pri-miRNA processing assay
In vivo pri-miRNA processing was performed as previously described (Kawai and Amano, 2012). Briefly, pmirGLO-miR-200a, miR-200b, miR-16 or miR-145 expression vectors were transfected into HEK29T cells. After 48 hours, cell extracts were prepared and the ratios of firefly and Renilla luciferase were obtained using a Dual-Luciferase Reporter Assay kit (Promega). As a control, empty pmirGLO vector was used. Relative pri-miRNA processing activity = (Lucno-insert – Lucpri-insert)/Lucpri-insert. Lucno-insert and Lucpri-insert numbers were normalized with Renilla luciferase numbers.
Statistical analysis
Differences between groups were analysed using conventional Student's t test or ANOVA. Reported p values were two sided and considered significant at <0.05. Statistical calculations were executed using GraphPad Prism6.
Supplementary Material
HIGHLIGHTS.
DDX1 is a regulatory component of the Drosha microprocessor.
DDX1 promotes the expression of a subset of miRNAs.
Knockdown of DDX1 promotes ovarian tumor invasion and metastasis.
Levels of DDX1 are correlated with clinical outcome in ovarian cancer patients.
ACKNOWLEDGMENTS
The authors would like to thank Drs. V. Narry Kim (Seoul National University), Shinji Kawai and Atsuo Amano (Osaka University Graduate School of Dentistry), and Roseline Godbout (University of Alberta) for providing necessary reagents in this work. This work was supported by grants to X.L. from the National Institutes of Health (CA136549 and P50 CA083639) and the University of Texas Stars Plus Award, and grants to A.K.S. from the National Institutes of Health (UH2TR000943 and U54CA151668). C.H. was supported in part by the Odyssey Program and The Cockrell Foundation Award for Scientific Achievement at The University of Texas MD Anderson Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
X.L., X.Z. and A.K.S. conceived and supervised the project. C. H. designed and performed most of the experiments. Y.L. generated a number of vectors and performed DNA repair assays. G.W. performed MS2-TRAP assays. H.J.C. and L. Z. assisted in the in-vivo tumor model experiments. X.Z. performed in-situ hybridization assays. C.I. conducted bioinformatic and biostatistical analyses. X.H. provided intellectual input to the whole project and analyzed the results. C.H. and X.L. wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Accession numbers
The GEO accession number for global miRNA PCR array data is GSE54990.
REFERENCES
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, Takaoka M, Nakagawa H, Tort F, Fugger K, Johansson F, Sehested M, Andersen CL, Dyrskjot L, Orntoft T, Lukas J, Kittas C, Helleday T, Halazonetis TD, Bartek J, Gorgoulis VG. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, Orntoft TF, Andersen CL, Dobbelstein M. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68:10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di LG, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW, Yu WH, Rehman SK, Hsu JL, Lee HH, Liu M, Chen CT, Yu D, Hung MC. p53 regulates epithelial mesenchymal transition and stem cell properties through modulating miRNAs. Nat. Cell Biol. 2011;13:317–323. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MM, Zhang G, Cheloufi S, Neubert TA, Hannon GJ, Littman DR. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010;24:1951–1960. doi: 10.1101/gad.1953310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos V, Esteller M. MicroRNAs and cancer epigenetics: a macrorevolution. Curr. Opin. Oncol. 2010;22:35–45. doi: 10.1097/CCO.0b013e328333dcbb. [DOI] [PubMed] [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller-Pace FV, Moore HC. RNA helicases p68 and p72: multifunctional proteins with important implications for cancer development. Future. Oncol. 2011;7:239–251. doi: 10.2217/fon.11.1. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Shiekhattar R. MicroRNA biogenesis: isolation and characterization of the microprocessor complex. Methods Mol. Biol. 2006;342:33–47. doi: 10.1385/1-59745-123-1:33. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de SE, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii H, Saito T. Radiation-induced response of micro RNA expression in murine embryonic stem cells. Med. Chem. 2006;2:555–563. doi: 10.2174/1573406410602060555. [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc. Natl. Acad. Sci. U. S. A. 2012;109:3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai S, Amano A. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. J. Cell Biol. 2012;197:201–208. doi: 10.1083/jcb.201110008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Veronese A, Pichiorri F, Lee TJ, Jeon YJ, Volinia S, Pineau P, Marchio A, Palatini J, Suh SS, Alder H, Liu CG, Dejean A, Croce CM. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J. Exp. Med. 2011;208:875–883. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celia-Terrassa T, Mercatali L, Khan Z, Goodarzi H, Hua Y, Wei Y, Hu G, Garcia BA, Ragoussis J, Amadori D, Harris AL, Kang Y. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat. Med. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA. Biol. 2008;5:115–119. doi: 10.4161/rna.5.3.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee YS, Dutta A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Monckton EA, Godbout R. A role for DEAD box 1 at DNA double-strand breaks. Mol. Cell Biol. 2008;28:6413–6425. doi: 10.1128/MCB.01053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE. Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:59–66. doi: 10.1101/sqb.2006.71.050. [DOI] [PubMed] [Google Scholar]
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, Deavers MT, Mourad-Zeidan A, Wang H, Mueller P, Lenburg ME, Gray JW, Mok S, Birrer MJ, Lopez-Berestein G, Coleman RL, Bar-Eli M, Sood AK. Dicer, Drosha, and outcomes in patients with ovarian cancer. N. Engl. J. Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Triboulet R, Mohseni M, Schlegelmilch K, Shrestha K, Camargo FD, Gregory RI. Hippo Signaling Regulates Microprocessor and Links Cell-Density-Dependent miRNA Biogenesis to Cancer. Cell. 2014;156:893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Hannon GJ. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell Biol. 2004;16:223–229. doi: 10.1016/j.ceb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecot CV, Rupaimoole R, Yang D, Akbani R, Ivan C, Lu C, Wu S, Han HD, Shah MY, Rodriguez-Aguayo C, Bottsford-Miller J, Liu Y, Kim SB, Unruh A, Gonzalez-Villasana V, Huang L, Zand B, Moreno-Smith M, Mangala LS, Taylor M, Dalton HJ, Sehgal V, Wen Y, Kang Y, Baggerly KA, Lee JS, Ram PT, Ravoori MK, Kundra V, Zhang X, Ali-Fehmi R, Gonzalez-Angulo AM, Massion PP, Calin GA, Lopez-Berestein G, Zhang W, Sood AK. Tumour angiogenesis regulation by the miR-200 family. Nat. Commun. 2013;4:2427. doi: 10.1038/ncomms3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothof J, Verkaik NS, van IW, Wiemer EA, Ta VT, van der Horst GT, Jaspers NG, van Gent DC, Hoeijmakers JH, Persengiev SP. MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. EMBO J. 2009;28:2090–2099. doi: 10.1038/emboj.2009.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raines RT. Ribonuclease A. Chem. Rev. 1998;98:1045–1066. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- Roby KF, Taylor CC, Sweetwood JP, Cheng Y, Pace JL, Tawfik O, Persons DL, Smith PG, Terranova PF. Development of a syngeneic mouse model for events related to ovarian cancer. Carcinogenesis. 2000;21:585–591. doi: 10.1093/carcin/21.4.585. [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol. Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Slezak-Prochazka I, Durmus S, Kroesen BJ, van den Berg A. MicroRNAs, macrocontrol: regulation of miRNA processing. RNA. 2010;16:1087–1095. doi: 10.1261/rna.1804410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan G, Liu Y, Han C, Zhang X, Lu X. Noncoding RNAs in DNA Repair and Genome Integrity. Antioxid. Redox. Signal. 2013a doi: 10.1089/ars.2013.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan G, Zhang X, Langley RR, Liu Y, Hu X, Han C, Peng G, Ellis LM, Jones SN, Lu X. DNA-Damage-Induced Nuclear Export of Precursor MicroRNAs Is Regulated by the ATM-AKT Pathway. Cell Rep. 2013b doi: 10.1016/j.celrep.2013.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker NC, Maity TS, Ye X, Aruscavage PJ, Krauchuk AA, Liu Q, Bass BL. Dicer's helicase domain discriminates dsRNA termini to promote an altered reaction mode. Mol. Cell. 2011;41:589–599. doi: 10.1016/j.molcel.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Sun Y, Hu L, Zheng H, Ji P, Pecot CV, Zhao Y, Reynolds S, Cheng H, Rupaimoole R, Cogdell D, Nykter M, Broaddus R, Rodriguez-Aguayo C, Lopez-Berestein G, Liu J, Shmulevich I, Sood AK, Chen K, Zhang W. Integrated Analyses Identify a Master MicroRNA Regulatory Network for the Mesenchymal Subtype in Serous Ovarian Cancer. Cancer Cell. 2013;23:186–199. doi: 10.1016/j.ccr.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ahn YH, Gibbons DL, Zang Y, Lin W, Thilaganathan N, Alvarez CA, Moreira DC, Creighton CJ, Gregory PA, Goodall GJ, Kurie JM. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. J. Clin. Invest. 2011;121:1373–1385. doi: 10.1172/JCI42579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Srikantan S, Gorospe M. MS2-TRAP (MS2-tagged RNA affinity purification): tagging RNA to identify associated miRNAs. Methods. 2012;58:81–87. doi: 10.1016/j.ymeth.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, Yao G, Medina A, O'brien-Jenkins A, Katsaros D, Hatzigeorgiou A, Gimotty PA, Weber BL, Coukos G. microRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lu X. Posttranscriptional regulation of miRNAs in the DNA damage response. RNA. Biol. 2011;8:960–963. doi: 10.4161/rna.8.6.17337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wan G, Berger FG, He X, Lu X. The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol. Cell. 2011;41:371–383. doi: 10.1016/j.molcel.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.