Abstract
The identification of cell types of origin for cancer has important implications for tumor stratification and personalized treatment. For prostate cancer, the cell of origin has been intensively studied, but it has remained unclear whether basal or luminal epithelial cells, or both, represent cells of origin under physiological conditions in vivo. Here, we use a novel lineage-tracing strategy to assess the cell of origin in a diverse range of mouse models, including Nkx3.1+/–; Pten+/–, Pten+/–, Hi-Myc, and TRAMP mice, as well as a hormonal carcinogenesis model. Our results show that luminal cells are consistently the observed cell of origin for each model in situ; however, explanted basal cells from these mice can generate tumors in grafts. Consequently, we propose that luminal cells are favored as cells of origin in many contexts, whereas basal cells only give rise to tumors after differentiation into luminal cells.
Keywords: prostate cancer, lineage-tracing, mouse models, hormonal carcinogenesis, cell of origin, cell of mutation
The identification of cell types of origin for cancer is significant since distinct cell populations within a tissue may give rise to different cancer subtypes distinguished by their histopathological phenotypes and patient outcomes (Blanpain, 2013; Visvader, 2009, 2011; Wang et al., 2013). Numerous studies have investigated the cell of origin by introducing an oncogenic insult within a defined cell type to determine whether these cells can give rise to cancer. However, such approaches are potentially limited as the cell type of origin may be dependent on the specific oncogenic insult and/or the model system. To date, no studies have systematically addressed which cell types can serve as cells of origin in multiple contexts of tumor initiation.
In human and mouse prostate epithelium, luminal and basal cells are the two major cell types, together with rare neuroendocrine cells (Shen and Abate-Shen, 2010). Lineage-tracing has shown that luminal and basal cells in the adult mouse prostate represent distinct populations that are mostly self-sustaining (Choi et al., 2012; Lu et al., 2013; Wang et al., 2013). Notably, lineage-marked basal cells rarely generate luminal cells during adult tissue homeostasis, but display plasticity under the influence of inductive embryonic urogenital mesenchyme in grafting assays, acquiring facultative progenitor properties and generating luminal cells (Choi et al., 2012; Lu et al., 2013; Wang et al., 2013).
For prostate cancer, previous studies have reached differing conclusions regarding the cell type(s) of origin (Goldstein and Witte, 2013; Wang and Shen, 2011; Xin, 2013). Although prostate adenocarcinoma has a luminal phenotype, both basal and luminal cells have been proposed to represent cells of origin. In particular, transformed human basal cells can give rise to prostate cancer in renal grafting models (Goldstein et al., 2010; Stoyanova et al., 2013; Taylor et al., 2012), whereas a luminal stem cell population identified in the regressed mouse prostate can act as a cell of origin in vivo (Wang et al., 2009). More recently, lineage-tracing in mice in which the Pten tumor suppressor was specifically deleted in either basal or luminal cells has shown that both cell types can act as cells of origin (Choi et al., 2012; Lu et al., 2013; Wang et al., 2013).
However, it remains unclear whether basal or luminal cells, or both, represent cell types of origin in the context of Pten deletion occurring throughout the prostate epithelium, or whether the cell of origin might vary depending upon specific oncogenic events. We have investigated this issue using a novel lineage-tracing strategy in a diverse range of mouse models that recapitulate important features of human prostate tumorigenesis. Our results indicate that luminal cells are consistently favored as cells of origin for prostate cancer.
Results
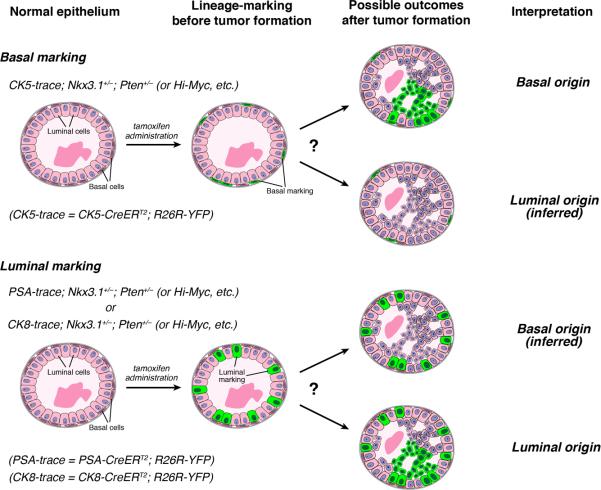
To determine the cell of origin for a mouse model of prostate cancer, we performed lineage-marking of either basal or luminal cells in apparently normal tissue to determine whether their progeny contribute to the tumors that subsequently arise (Figure 1). Since the lineage-tracing methodology uses inducible Cre recombinase, we analyzed mouse models in which the tumor phenotype is not driven by Cre. We used the CK5-CreERT2 driver (Rock et al., 2009) for lineage-tracing of basal cells, and the PSA-CreERT2 (Ratnacaram et al., 2008) or CK8-CreERT2 (Van Keymeulen et al., 2011) drivers for tracing of luminal cells, together with the R26R-YFP reporter (Srinivas et al., 2001). Tamoxifen induction for lineage-marking was performed in young adult male mice at seven weeks of age, when the basal and luminal lineages have been established as largely self-sustaining compartments (Choi et al., 2012; Ousset et al., 2012; Wang et al., 2013). Contribution of cells marked by the CK5-CreERT2 driver to tumors would imply that basal cells were the cell of origin, whereas tumor cells marked by the PSA-CreERT2 or CK8-CreERT2 drivers would indicate a luminal origin (Figure 1). Notably, our approach dissociates the time of lineage-marking from the onset of tumorigenesis, and allows multiple models to be analyzed using the same overall strategy.
Figure 1. Experimental design for analysis of cell of origin.
The inducible CK5-CreERT2 driver can lineage-mark basal cells by YFP expression in different prostate cancer models prior to overt cancer formation. Similarly, the inducible PSA-CreERT2 and CK8-CreERT2 drivers can mark luminal cells in phenotypically normal epithelium. The presence of YFP+ cell clusters in subsequent PIN/cancer lesions indicates that the marked cell type acts as the cell of origin in the mouse model analyzed.
In control experiments to examine the specificity of the inducible Cre drivers in a wild-type background, we found that CK5-CreERT2; R26R-YFP (which we denote CK5-trace) strictly marks basal cells with 23.6% efficiency, while PSA-CreERT2; R26R-YFP (PSA-trace) marks luminal cells with 11.5% efficiency, and CK8-CreERT2; R26R-YFP (CK8-trace) marks 4.1% of luminal cells (Table S1L, N, P), consistent with previous studies (Ousset et al., 2012; Ratnacaram et al., 2008; Wang et al., 2013). Importantly, the percentage of lineage-marked cells in the CK5-trace and PSA-trace mice does not change between two months of age, shortly after labeling, and six months of age, when our tumor analyses are mostly performed (Figure S1; Table S2A, B), indicating that the lineage-marked cell populations are stable in a nontumorigenic background.
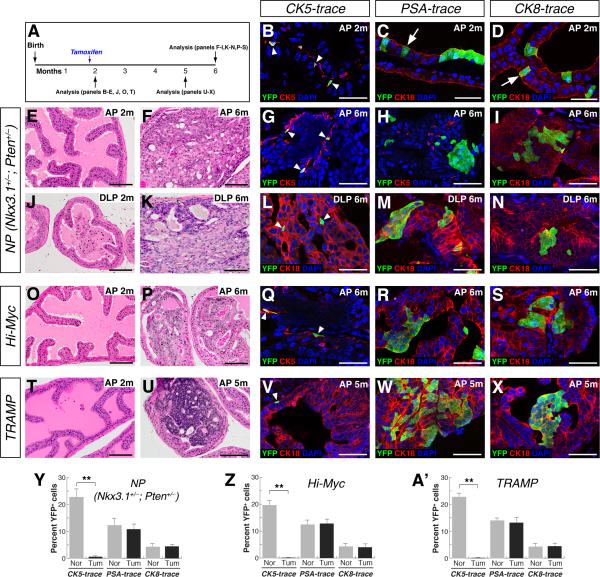
We first investigated the cell of origin for the high-grade prostatic intraepithelial neoplasia (PIN) lesions in the Nkx3.1+/–; Pten+/– (which we denote NP) model that is heterozygous for null alleles of the Nkx3.1 homeobox gene and of Pten (Kim et al., 2002). As reported previously, the anterior prostate (AP) and dorsolateral prostate (DLP) of NP mice appear normal at two months of age (Figure 2E, J), but frequently display high-grade PIN/carcinoma lesions at six months (Figure 2F, K). Quantitation of initial lineage-marking in CK5-trace; NP mice and PSA-trace; NP mice revealed similar efficiencies as mice with a wild-type background (Figure 2B, C, Y, Z; Table S1A, B). Notably, in tumor lesions of CK5-trace; NP mice at six months of age, we found that YFP+ cells in clusters (defined as containing at least three YFP+ cells) were rarely observed (0.5%, n=6 mice) (Figure 2G, L, Y; Figure S2A, D; Table S1A), while the percentage of YFP+ cells in untransformed regions was unaffected (Figure S3AC; Table S2C). In contrast, 10.8% of the cells in the tumor lesions of PSA-trace; NP mice (n=4) and 4.5% of the cells in tumor lesions of CK8-trace; NP mice (n=3) were YFP+ (Figure 2H, I, M, N, Y; Figure S2B, C, E, F; Table S1B, C, P). Furthermore, we found that YFP+ clusters were also rare in PIN lesions of six-month old CK5-trace; Pten+/– mice, whereas the frequency of YFP+ cells was unchanged in non-tumor regions (n=3) (Figure S3D, E; Figure S4A, B, D, E, G; Table S1D; Table S2D). However, the percentage of YFP+ cells in PIN lesions of PSA-trace; Pten+/– mice (n=3) was similar to the percentage initially marked by the PSA-CreERT2 inducible driver (Figure S4C, F, G; Table S1E).
Figure 2. Luminal cells are favored cells of origin in the Nkx3.1+/–; Pten+/– (NP), Hi-Myc, and TRAMP models.
(A) Experimental time course. (B) Lineage-marking of basal cells (arrowheads) in the AP of CK5-trace; NP mice at 2 months of age. (C, D) Marking of luminal cells (arrows) in the AP of PSA-trace; NP mice (C) or CK8-trace mice (D) at 2 months. (E, F, J, K) H&E staining of NP prostates shows normal histology at 2 months, and PIN/carcinoma lesions at 6 months. (G, L) Clusters of YFP+ cells are rarely detected in CK5-trace; NP tumor lesions at 6 months. (H, I, M, N) YFP+ cell clusters in tumor lesions of PSA-trace; NP (H, M) and CK8-trace; NP (I, N) mice at 6 months. (O, P) Normal AP histology in Hi-Myc mice at 2 months (O), and PIN/carcinoma lesions at 6 months (P). (Q) Absence of YFP+ cell clusters in CK5-trace; Hi-Myc tumor lesions in the AP at 6 months. (R, S) YFP+ cell clusters in tumor lesions of PSA-trace; Hi-Myc mice (R) and CK8-trace; Hi-Myc mice (S) at 6 months. (T, U) Normal histology of the AP in TRAMP mice at 2 months (T), and carcinoma at 5 months (U). (V) Absence of YFP+ cell clusters in CK5-trace; TRAMP AP tumor lesions at 5 months. (W, X) YFP+ clusters in AP tumor lesions of PSA-trace; TRAMP (W) and CK8-trace; TRAMP (X) mice at 5 months. (Y-A’) Percentage of YFP+ cells in NP (Y), Hi-Myc (Z), and TRAMP (A’) models; Nor = normal, Tum = tumor; ** p<0.001 by Student's t-test; error bars are one standard deviation. Arrowheads in G, L, Q, V indicate marked basal cells. Scale bars in B-D, G-I, L-N, Q-S, and V-X correspond to 50 microns and in E, F, J, K, O, P, T, U to 100 microns. See also Figures S1-S4.
Next, we examined the transgenic ARR2/probasin-Myc (Hi-Myc) model, in which expression of c-Myc is driven in both luminal and basal compartments, leading to invasive adenocarcinoma (Ellwood-Yen et al., 2003). Consistent with previous studies (Ellwood-Yen et al., 2003), the histology of the AP in Hi-Myc mice was mostly normal at two months of age (Figure 2O), although the DLP and ventral prostate (VP) were hyperplastic (Figure S4H, K). In the PIN/carcinoma lesions in the AP of CK5-trace; Hi-Myc mice at six months, YFP+ cell clusters were rare, whereas the percentage of YFP+ basal cells in untransformed regions was unaffected (n=5 mice) (Figure 2P, Q, Z; Figure S2G; Figure S3F, G; Table S1F; Table S2E). In contrast, 13.1% of the cells within the PIN/carcinoma lesions of six-month old PSA-trace; Hi-Myc mice (n=6) were YFP+, similar to the initial percentage (12.6%) of luminal cells marked at two months (Figure 2R, Z; Figure S2H; Table S1G). Similarly, YFP+ cells were present in PIN/carcinoma lesions of CK8-trace; Hi-Myc mice (n=4) in proportion to the initial luminal marking efficiency (Figure 2S, Z; Figure S2I; Table S1H, P). Similar results were found in the DLP and VP of CK5-trace; Hi-Myc and PSA-trace; Hi-Myc mice (Figure S4H-M).
We also investigated the TRAMP model, which expresses the SV40 large T antigen under the control of the probasin promoter, giving rise to aggressive tumors (Greenberg et al., 1995). We found that the AP in TRAMP mice appeared mostly normal at two months, but developed invasive, poorly differentiated adenocarcinoma by five months (Figure 2T, U). In tumor lesions of CK5-trace; TRAMP mice (n=4), YFP+ cell clusters were not observed, whereas the frequency of YFP+ cells in non-tumor regions was unaffected (Figure 2V, A’; Figure S2J; Figure S3H, I; Table S1I; Table S2F). However, YFP+ cell clusters were found in tumor lesions of PSA-trace; TRAMP mice (n=5) and CK8-trace; TRAMP mice (n=3) in percentages similar to the initial luminal marking efficiencies (Figure 2W, X, A’; Figure S2K, L; Table S1J, K, P). Similar results were observed in the DLP and VP of TRAMP mice, although these lobes were already hyperplastic at two months of age (Figure S4N-S). Taken together, these findings show that luminal cells are the favored cell of origin in each of the genetically-engineered mouse models examined.
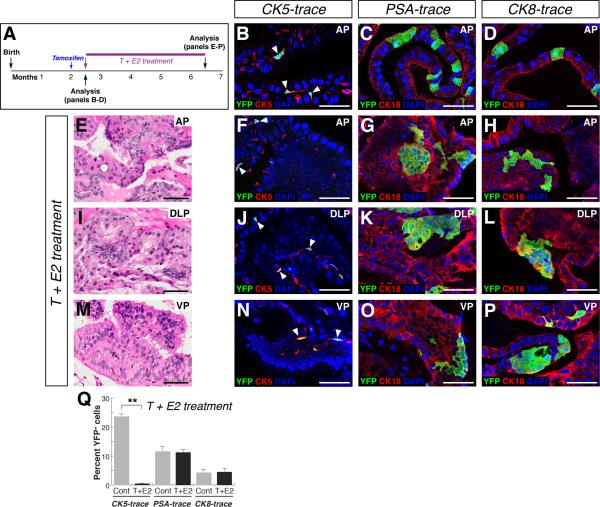
Given the potential caveat that cancer initiation might occur prior to adulthood in these genetically-engineered models, we investigated the cell of origin in a hormonal carcinogenesis paradigm (Ricke et al., 2008; Wang et al., 2000), in which lineage-marking unequivocally takes place prior to prostate tumor initiation. (Bosland et al., 1995; Noble, 1977; Ricke et al., 2008; Wang et al., 2000). After lineage-marking of basal cells in CK5-trace mice, or luminal cells in PSA-trace and CK8-trace mice (Figure 3A-D; Table S1L, N, P), we treated the mice with a combination of testosterone (T) and estradiol-17β (E2) for four months, resulting in formation of low-grade PIN lesions in all prostate lobes (Figure 3E, I, M). Using this protocol, we found that YFP+ clusters were rare in PIN lesions of CK5-trace mice (n=5), while the frequency of YFP+ cells was unaffected in untransformed regions (Figure 3F, J, N, Q; Figure S2M, P, S; Figure S3J, K; Table S1M; Table S2G). In contrast, YFP+ clusters were present in PIN lesions of PSA-trace (n=4) and CK8-trace mice (n=3) (Figure 3G,H, K, L, O, P; Figure S2N, O, Q, R, T, U), with the percentage of YFP+ cells similar to the initial efficiency of luminal cell marking (Figure 3Q; Table S1O, Q). These results indicate that carcinogenesis induced by T+E2 treatment leads to prostate cancer initiation from luminal cells.
Figure 3. Luminal cells are the favored cell of origin of tumors induced by T+E2 hormonal treatment.
(A) Experimental time course. (B-D) Lineage-marking of basal (arrowheads) and luminal cells in control CK5-trace (B), PSA-trace (C), and CK8-trace (D) mice. (E, I, M) PIN lesions in mice after T+E2 treatment. (F, J, N) YFP+ cell clusters are rarely detected in CK5-trace PIN lesions after T+E2 treatment; arrowheads indicate marked basal cells. (G, H, K, L, O, P) YFP+ clusters in PIN lesions of PSA-trace (G, K, O) and CK8-trace (H, L, P) mice after T+E2 treatment. (Q) Percentage of YFP+ cells; Cont = control untreated, T+E2 = treated; ** p<0.001 by Student's t-test; error bars are one standard deviation. Scale bars indicate 50 microns. See also Figures S2, S3.
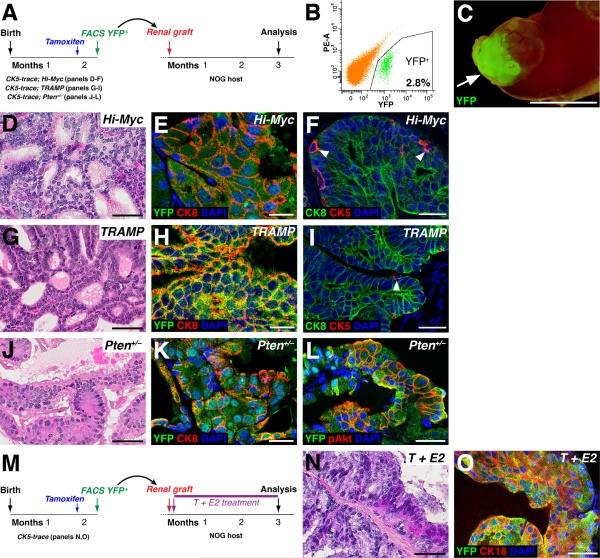
Previous studies have concluded that basal cells are cells of origin for human prostate cancer using renal grafting methods (Goldstein et al., 2010; Stoyanova et al., 2013; Taylor et al., 2012). To determine whether the potential discrepancy between these studies and our findings might be due to the different methodologies employed, we tested whether basal cells in our mouse models of prostate cancer could give rise to tumors after renal grafting. We performed tamoxifen induction of CK5-trace; Hi-Myc mice at seven weeks of age and isolated basal cells by flow-sorting for YFP (Figure 4A, B). The sorted basal cells were recombined with rat urogenital sinus mesenchyme and grafted under the renal capsule of immunodeficient NOG mice, followed by analysis after three months (Figure 4C). We observed extensive regions of YFP+ epithelium, which contained PIN lesions that were mostly comprised of luminal cells, indicating basal to luminal differentiation had taken place (Figure 4D-F). We obtained similar results for basal cells isolated from CK5-trace; TRAMP mice (Figure 4G-I) , as well as from CK5-trace; Pten+/– mice, in which the graft PIN lesions were also positive for phospho-Akt (Figure 4J-L). Finally, we performed renal grafting of YFP+ basal cells isolated from tamoxifen-induced CK5-trace mice, followed by treatment of the NOG graft recipients with T+E2 for three months (Figure 4M). In the resulting grafts, marked basal cells could give rise to PIN lesions that mostly contained luminal cells (Figure 4N, O). Taken together, our results show that prostate basal cells are not favored as the cell of origin in their native microenvironment for any of the mouse models analyzed, but nonetheless can give rise to tumors in renal grafts.
Figure 4. Basal cells can give rise to prostate cancer in renal grafts.
(A) Experimental design. (B) Representative flow-sort of YFP+ basal cells (2.8% of total prostate cells) from CK5-trace; Hi-Myc mice. (C) Kidney from recipient NOG mouse containing graft with YFP fluorescence (arrow). (D-I) Grafted basal cells from CK5-trace; Hi-Myc (D-F) or CK5-trace; TRAMP (G-I) mice generate PIN lesions (D, G), which contain mostly luminal cells (E, H) and some basal cells (arrowheads, F, I). (J-L) Basal cells from CK5-trace; Pten+/– mice generate PIN lesions (J) that contain mostly luminal cells (K) and express phospho-Akt (L). (M) Experimental design for T+E2 treatment of grafts. (N,O) Grafted basal cells give rise to PIN lesions (N) that contain mostly luminal cells (O) after T+E2 treatment. Scale bars in E, F, H, I, K correspond to 25 microns, in C to 5 mm, and in D, G, J, I, N, O to 50 microns.
Discussion
In principle, the cell of origin for cancer might be context-specific, depending upon the oncogenic pathways being activated. In our studies, we have employed a novel lineage-tracing methodology for systematic assessment of the cell of origin for prostate cancer in a diverse range of mouse models. Using this “agnostic” lineage-tracing approach, we have unexpectedly found that luminal epithelial cells are consistently observed as the cell of origin.
Overall, we have analyzed a representative sample of widely used mouse models of human prostate cancer (Irshad and Abate-Shen, 2013; Ittmann et al., 2013; Shappell et al., 2004). However, there may be specific caveats associated with each model; for example, tumor initiation might conceivably occur in basal cells prior to seven weeks of age in the transgenic models, resulting in early basal to luminal differentiation that would escape lineage-marking. This possibility seems unlikely since all tumor initiation would have to occur prior to seven weeks of age, to avoid detection of subsequent tumor formation from basal cells by lineage-tracing. Nonetheless, our analysis has yielded a remarkably consistent result that luminal cells are favored as the cell of origin, and consequently we believe that this finding is likely to reflect the biology of prostate cancer, rather than a coincidence of intrinsic biases in each model. However, we note that basal cells could nonetheless act as cells of origin for prostate adenocarcinoma in other experimental contexts. In addition, the ability of inflammation to enhance basal-to-luminal differentiation in vivo (Kwon et al., 2014) suggests that alterations of the tissue microenvironment could influence the cell of origin (Goldstein and Witte, 2013).
To date, the cell of origin has usually been assayed by conditional gene targeting to generate oncogenic insults within a specific cell type. However, if the targeted cell type is a stem/progenitor cell, it can be difficult to discern whether tumor initiation takes place within the stem/progenitor itself, or instead within its differentiated progeny. In this situation, it can be useful to distinguish between a “cell of origin” and a “cell of mutation” as distinct entities (Liu et al., 2011; Liu and Zong, 2012). In particular, a progenitor that initially acquires a mutation may not directly transform, and hence be a “cell of mutation”, while its lineage-restricted progeny may inherit the mutation and subsequently undergo oncogenic transformation, and thus would represent a “cell of origin”. For example, lineage-tracing of gliomas in a p53; Nf1 mouse model has shown that neural stem cells act as a cell of mutation, whereas their descendant oligodendrocyte progenitors correspond to the cell of origin (Liu et al., 2011).
In this regard, prostate basal cells removed from their normal tissue microenvironment can acquire facultative bipotential progenitor properties after combination with embryonic urogenital mesenchyme, resulting in the differentiation of luminal cells (Goldstein et al., 2008; Lawson et al., 2007; Lawson et al., 2010; Wang et al., 2013), while transformed basal cells give rise to luminal tumors in renal grafts (Goldstein et al., 2010; Stoyanova et al., 2013; Taylor et al., 2012). Our findings are consistent since lineage-marked basal cells in each of our mouse models can give rise to prostate cancer in the context of renal grafts. Consequently, we propose that mutated basal cells do not usually act as a cell of origin in prostate tissue in situ, but can function as a cell of mutation in renal grafts by acquiring facultative progenitor properties, and thereby generating luminal progeny that are authentic cells of origin.
Notably, previous studies have shown that targeted deletion of Pten in basal cells results in formation of tumors in situ, albeit with a temporal delay that appears to be associated with basal to luminal differentiation, and which are less aggressive than tumors arising from targeting of luminal cells (Choi et al., 2012; Wang et al., 2013). Interestingly, PIN lesions arose from targeted basal cells by three months of age, in contrast with the absence of contribution from lineage-marked basal cells in NP and Pten+/– mice. These findings are potentially consistent with a “competition” model, which is not mutually exclusive with the cell of mutation model. Thus, if Pten loss occurs in both luminal and basal cells, transformed luminal cells might emerge before basal cells can be transformed, and might suppress subsequent basal cell transformation in a non-cell autonomous manner.
Finally, our finding that luminal cells are the favored cell of origin in multiple mouse models raises the possibility that most human prostate adenocarcinomas arise from luminal cells. In particular, cytological examination of human PIN lesions suggests that early initiating events occur in luminal cells, including c-Myc up-regulation and telomere elongation (Gurel et al., 2008; Meeker et al., 2002). Moreover, human prostate luminal cells may be prone to cancer initiation due to a decreased DNA damage response (Jaamaa et al., 2010). Our results also imply that cell of origin analyses for human cancer may be inherently difficult using grafting assays, due to the plasticity of basal cells. Instead, approaches such as retrospective lineage-tracing using mitochondrial mutations may provide insight into human prostate cancer origins (Blackwood et al., 2011; Gaisa et al., 2011). Since the cell of origin may be a critical factor in conferring aggressiveness in prostate cancer (Wang et al., 2013), these and other approaches to identify cell types of origin are likely to be important for biomarker identification and disease prognosis.
Experimental Procedures
Mouse procedures
Mouse lines were maintained on an inbred C57BL/6N or mixed C57BL/6N-129S6/SvEvTac background. Primer sequences for genotyping are listed in Table S2. For tamoxifen induction, mice were administered 9 mg/40 g tamoxifen (Sigma) suspended in corn oil by oral gavage once daily for 4 consecutive days.
For T+E2 treatment, a 1.0 cm Silastic capsule (No. 602–305 Silastic tubing; 1.54 mm inside diameter, 3.18 mm outside diameter; Dow-Corning #2415569) filled with testosterone (Sigma) and a 0.4 cm Silastic capsule filled with estradiol-17β (Sigma) were implanted subcutaneously. Mice were treated with hormones for 4 months.
Tissue collection and flow cytometry
Prostate tissue dissection, fixation, and dissociation were performed as described (Wang et al., 2013). Cell sorting was performed based on YFP fluorescence on a BD FACS Aria II instrument in the Flow Cytometry Shared Resource of the Herbert Irving Comprehensive Cancer Center. We used SSC/FSC gating to exclude debris and doublets, followed by PE/YFP FITC-A gating to exclude auto-fluorescent double-positive cells and to collect the single-positive YFP-expressing cell population.
Renal grafting assay
For tissue recombinants, 1.0 × 104 dissociated YFP+ cells were mixed with 2.5 × 105 dissociated urogenital sinus mesenchyme cells from embryonic day 18.0 rat embryos. Tissue recombinants were cultured in DMEM/10% FBS/10-7 M DHT overnight, followed by transplantation under the kidney capsules of immunodeficient NOD.Cg-Prkdcscid Il2rgtmlSug/JicTac (NOG) mice (Taconic) and growth for 12 weeks.
Histology and immunostaining
H&E staining and immunofluorescence staining were performed (Wang et al., 2013) using the following primary antibodies: rabbit CK5 (Covance #PRB-160P, 1:1000), rabbit CK8 (Abcam #ab53280, 1:250), mouse CK18 (Abcam #ab668, 1:100), chick GFP (Abcam #ab13970, 1:2000), rabbit phospho-Akt (Cell Signaling #3787, 1:50). Samples were incubated with secondary antibodies (diluted 1:500 in PBST) labeled with Alexa Fluor 488, 555, or 647 (Invitrogen/Molecular Probes), and mounted with VECTASHIELD medium with DAPI (Vector Labs). Immunofluorescence was imaged using a Leica TCS SP5 spectral confocal microscope.
Data quantitation
Cell numbers were counted using confocal ×40 and ×63 photomicrographs. For histologically normal tissues at two months, the percentage of YFP+ cells (labeled “Nor” in Figures 2Y-A’, S1G, and “Cont” in Figure 3Q) represents the ratio of YFP+ cells to total basal or luminal cells. For tumor tissues at later ages, the percentage of YFP+ cells (labeled “Tum” in Figures 2Y-A’, S1G, and “T+E2” in Figure 3Q) represents the ratio of clustered YFP+ cells in tumor lesions to total epithelial cells within these lesions. Statistical analyses were performed using a two-sample t-test. At least three animals for each experiment or genotype were analyzed.
Supplementary Material
Acknowledgements
We thank Cory Abate-Shen for comments on the manuscript. This work was supported by post-doctoral fellowships from the DOD Prostate Cancer Research Program (PC101819 to Z.A.W.; PC131821 to R.T.), and by grants from the National Institutes of Health (P01CA154293 to M.M.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Z.A.W. and M.M.S. designed the study. Z.A.W. performed the experiments, with contributions from R.T. for H&E and immunostaining, and S.K.B. for renal grafting. P.C. provided PSA-CreERT2 mice. Z.A.W., R.T., and M.M.S. analyzed data and prepared the manuscript.
References
- Blackwood JK, Williamson SC, Greaves LC, Wilson L, Rigas AC, Sandher R, Pickard RS, Robson CN, Turnbull DM, Taylor RW, et al. In situ lineage tracking of human prostatic epithelial stem cell fate reveals a common clonal origin for basal and luminal cells. J Pathol. 2011;225:181–188. doi: 10.1002/path.2965. [DOI] [PubMed] [Google Scholar]
- Blanpain C. Tracing the cellular origin of cancer. Nat Cell Biol. 2013;15:126–134. doi: 10.1038/ncb2657. [DOI] [PubMed] [Google Scholar]
- Bosland MC, Ford H, Horton L. Induction at high incidence of ductal prostate adenocarcinomas in NBL/Cr and Sprague-Dawley Hsd:SD rats treated with a combination of testosterone and estradiol-17 beta or diethylstilbestrol. Carcinogenesis. 1995;16:1311–1317. doi: 10.1093/carcin/16.6.1311. [DOI] [PubMed] [Google Scholar]
- Choi N, Zhang B, Zhang L, Ittmann M, Xin L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell. 2012;21:253–265. doi: 10.1016/j.ccr.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, Thomas GV, Sawyers CL. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–238. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- Gaisa NT, Graham TA, McDonald SA, Poulsom R, Heidenreich A, Jakse G, Knuechel R, Wright NA. Clonal architecture of human prostatic epithelium in benign and malignant conditions. J Pathol. 2011;225:172–180. doi: 10.1002/path.2959. [DOI] [PubMed] [Google Scholar]
- Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON. Identification of a cell of origin for human prostate cancer. Science. 2010;329:568–571. doi: 10.1126/science.1189992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AS, Lawson DA, Cheng D, Sun W, Garraway IP, Witte ON. Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proc Natl Acad Sci USA. 2008;105:20882–20887. doi: 10.1073/pnas.0811411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AS, Witte ON. Does the microenvironment influence the cell types of origin for prostate cancer? Genes Dev. 2013;27:1539–1544. doi: 10.1101/gad.222380.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg NM, DeMayo F, Finegold MJ, Medina D, Tilley WD, Aspinall JO, Cunha GR, Donjacour AA, Matusik RJ, Rosen JM. Prostate cancer in a transgenic mouse. Proc Natl Acad Sci USA. 1995;92:3439–3443. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurel B, Iwata T, Koh CM, Jenkins RB, Lan F, Van Dang C, Hicks JL, Morgan J, Cornish TC, Sutcliffe S, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21:1156–1167. doi: 10.1038/modpathol.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irshad S, Abate-Shen C. Modeling prostate cancer in mice: something old, something new, something premalignant, something metastatic. Cancer Metastasis Rev. 2013;32:109–122. doi: 10.1007/s10555-012-9409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittmann M, Huang J, Radaelli E, Martin P, Signoretti S, Sullivan R, Simons BW, Ward JM, Robinson BD, Chu GC, et al. Animal models of human prostate cancer: the consensus report of the New York meeting of the Mouse Models of Human Cancers Consortium Prostate Pathology Committee. Cancer Res. 2013;73:2718–2736. doi: 10.1158/0008-5472.CAN-12-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaamaa S, Af Hallstrom TM, Sankila A, Rantanen V, Koistinen H, Stenman UH, Zhang Z, Yang Z, De Marzo AM, Taari K, et al. DNA damage recognition via activated ATM and p53 pathway in nonproliferating human prostate tissue. Cancer Res. 2010;70:8630–8641. doi: 10.1158/0008-5472.CAN-10-0937. [DOI] [PubMed] [Google Scholar]
- Kim MJ, Cardiff RD, Desai N, Banach-Petrosky WA, Parsons R, Shen MM, Abate-Shen C. Cooperativity of Nkx3.1 and Pten loss of function in a mouse model of prostate carcinogenesis. Proc Natl Acad Sci USA. 2002;99:2884–2889. doi: 10.1073/pnas.042688999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon OJ, Zhang L, Ittmann MM, Xin L. Prostatic inflammation enhances basal-to-luminal differentiation and accelerates initiation of prostate cancer with a basal cell origin. Proc Natl Acad Sci USA. 2014;111:E592–600. doi: 10.1073/pnas.1318157111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DA, Xin L, Lukacs RU, Cheng D, Witte ON. Isolation and functional characterization of murine prostate stem cells. Proc Natl Acad Sci USA. 2007;104:181–186. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DA, Zong Y, Memarzadeh S, Xin L, Huang J, Witte ON. Basal epithelial stem cells are efficient targets for prostate cancer initiation. Proc Natl Acad Sci USA. 2010;107:2610–2615. doi: 10.1073/pnas.0913873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, Foreman O, Bronson RT, Nishiyama A, Luo L, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Zong H. Developmental origins of brain tumors. Current opinion in neurobiology. 2012;22:844–849. doi: 10.1016/j.conb.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TL, Huang YF, You LR, Chao NC, Su FY, Chang JL, Chen CM. Conditionally ablated Pten in prostate basal cells promotes basal-to-luminal differentiation and causes invasive prostate cancer in mice. Am J Pathol. 2013;182:975–991. doi: 10.1016/j.ajpath.2012.11.025. [DOI] [PubMed] [Google Scholar]
- Meeker AK, Hicks JL, Platz EA, March GE, Bennett CJ, Delannoy MJ, De Marzo AM. Telomere shortening is an early somatic DNA alteration in human prostate tumorigenesis. Cancer Res. 2002;62:6405–6409. [PubMed] [Google Scholar]
- Noble RL. The development of prostatic adenocarcinoma in Nb rats following prolonged sex hormone administration. Cancer Res. 1977;37:1929–1933. [PubMed] [Google Scholar]
- Ousset M, Van Keymeulen A, Bouvencourt G, Sharma N, Achouri Y, Simons BD, Blanpain C. Multipotent and unipotent progenitors contribute to prostate postnatal development. Nat Cell Biol. 2012;14:1131–1138. doi: 10.1038/ncb2600. [DOI] [PubMed] [Google Scholar]
- Ratnacaram CK, Teletin M, Jiang M, Meng X, Chambon P, Metzger D. Temporally controlled ablation of PTEN in adult mouse prostate epithelium generates a model of invasive prostatic adenocarcinoma. Proc Natl Acad Sci USA. 2008;105:2521–2526. doi: 10.1073/pnas.0712021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke WA, McPherson SJ, Bianco JJ, Cunha GR, Wang Y, Risbridger GP. Prostatic hormonal carcinogenesis is mediated by in situ estrogen production and estrogen receptor alpha signaling. FASEB J. 2008;22:1512–1520. doi: 10.1096/fj.07-9526com. [DOI] [PubMed] [Google Scholar]
- Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BL. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shappell SB, Thomas GV, Roberts RL, Herbert R, Ittmann MM, Rubin MA, Humphrey PA, Sundberg JP, Rozengurt N, Barrios R, et al. Prostate pathology of genetically engineered mice: definitions and classification. The consensus report from the Bar Harbor meeting of the Mouse Models of Human Cancer Consortium Prostate Pathology Committee. Cancer Res. 2004;64:2270–2305. doi: 10.1158/0008-5472.can-03-0946. [DOI] [PubMed] [Google Scholar]
- Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoyanova T, Cooper AR, Drake JM, Liu X, Armstrong AJ, Pienta KJ, Zhang H, Kohn DB, Huang J, Witte ON, et al. Prostate cancer originating in basal cells progresses to adenocarcinoma propagated by luminal-like cells. Proc Natl Acad Sci USA. 2013;110:20111–20116. doi: 10.1073/pnas.1320565110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RA, Toivanen R, Frydenberg M, Pedersen J, Harewood L, Australian Prostate Cancer, B. Collins AT, Maitland NJ, Risbridger GP. Human epithelial basal cells are cells of origin of prostate cancer, independent of CD133 status. Stem Cells. 2012;30:1087–1096. doi: 10.1002/stem.1094. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, Sharma N, Dekoninck S, Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23:2563–2577. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- Wang X, Kruithof-de Julio M, Economides KD, Walker D, Yu H, Halili MV, Hu Y-P, Price SM, Abate-Shen C, Shen MM. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hayward SW, Donjacour AA, Young P, Jacks T, Sage J, Dahiya R, Cardiff RD, Day ML, Cunha GR. Sex hormone-induced carcinogenesis in Rb-deficient prostate tissue. Cancer Res. 2000;60:6008–6017. [PubMed] [Google Scholar]
- Wang ZA, Mitrofanova A, Bergren SK, Abate-Shen C, Cardiff RD, Califano A, Shen MM. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nature Cell Biology. 2013;15:274–283. doi: 10.1038/ncb2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZA, Shen MM. Revisiting the concept of cancer stem cells in prostate cancer. Oncogene. 2011;30:1261–1271. doi: 10.1038/onc.2010.530. [DOI] [PubMed] [Google Scholar]
- Xin L. Cells of origin for cancer: an updated view from prostate cancer. Oncogene. 2013;32:3655–3663. doi: 10.1038/onc.2012.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.