Abstract
β-D-glucosyl-5-hydroxymethyluracil (base J) is a hyper-modified nucleobase found in the nuclear DNA of kinetoplastid parasites. With replacement of a fraction of thymine in DNA, J is localized primarily in telomeric regions of all organisms carrying this modified base. The biosynthesis of J occurs in two putative steps: First, a specific thymine in DNA is recognized and converted into 5-hydroxymethyluracil (5-HmU) by J-binding proteins (JBP1 and JBP2); a glucosyl transferase (GT) subsequently glucosylates the 5-HmU to yield J. Although several recent studies revealed the roles of internal J in regulating transcription in kinetoplastids, functions of telomeric J and proteins involved in J synthesis remain elusive. Assessing the functions of base J and understanding fully its biosynthesis necessitate the measurement of its level in cells and organisms. In this study, we reported a reversed-phase HPLC coupled with tandem mass spectrometry (LC-MS/MS) method, together with the use of a surrogate internal standard (β-D-glucosyl-5-hydroxymethyl-2′-deoxycytidine, 5-gHmdC), for the accurate detection of β-D-glucosyl-5-hydroxymethyl-2′-deoxyuridine (dJ) in Trypanosoma brucei DNA. For comparison, we also measured the level of the precursor for dJ synthesis, i.e. 5-hydroxymethyl-2′-deoxyuridine (5-HmdU). We found that base J was not detectable in the JBP-null cells while it replaced approximately 0.5% thymine in wild-type cells, which was accompanied with a markedly decreased level of 5-HmdU in JBP1/JBP2-null strain relative to the wild-type strain. These results provided direct evidence supporting that JBP proteins play an important role in oxidizing thymidine to form 5-HmdU, which facilitated the generation of dJ. This is the first report about the application of LC-MS/MS for the quantification of base J. The analytical method built a solid foundation for dissecting the molecular mechanisms of J biosynthesis and assessing the biological functions of base J in the future.
Introduction
β-D-glucosyl-5-hydroxymethyluracil (base J) is the first hyper-modified base discovered in eukaryotic DNA [1]. Over the past two decades, this unique modified base has been found within members of unicellular kinetoplastids family, such as Trypanosoma and Leishmania species [2], and in the related unicellular flagellate protist Euglena gracilis [3], where it replaces a fraction of thymine in the genome. In contrast to its discovery in unicellular protozoa, base J was not detectable in animals, plants, or fungi tested, nor in a range of other simple eukaryotes [2]. In all kinetoplastid flagellates analyzed, base J is localized primarily in telomeric repeat regions (telomeric J) [4-6] and with a small portion present in other repetitive DNA sequences [7] and in sequences between transcription units (internal J) [8,9]. In the parasite Trypanosoma brucei, it also appears in the sub-telomeric variant surface glycoprotein (VSG) gene expression sites that are involved in producing a VSG coat on cell surface to evade host immune response [10]. The restricted presence of J in the genome of kinetoplastids but not other higher eukaryotes renders base J, and perhaps J biosynthesis, a potential target for parasite-specific chemotherapy [11].
Indirect evidence indicates that biosynthesis of J involves two steps, which starts with de novo hydroxylation of a specific thymidine residue to 5-hydroxymethyl-2′-deoxyuridine (5-HmdU) [11]. The intermediate 5-HmdU is then converted to β-D-glucosyl-5-hydroxymethyl-2′-deoxyuridine (dJ) by a yet unidentified glucosyl transferase (GT) [11]. Two enzymes involved in catalyzing the oxidation of thymidine were identified in trypanosomes, namely, J-binding proteins 1 and 2 (JBP1 and JBP2) [9]. Both of them contain a N-terminal thymine hydroxylase (TH) domain [12], but only JBP1 can bind to J in DNA via a particular J-binding domain in its C-terminal half [13]. On the other hand, JBP2 contains a SWI2/SNF2 domain homologous to ATPase/DNA helicases, which has been thought to be important for its activity [14]. Recently, the JBP proteins have been grouped together with mammalian TET proteins into the new TET/JBP subfamily of Fe(II)- and 2-oxoglutarate (2OG)-dependent dioxygenases [15]. Being able to oxidize 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-HmC) in mammalian DNA [16], TET enzymes are identified as the only homologue of JBP1/2 TH domain in eukaryotes and may play a very important role in active cytosine demethylation in mammals [17].
While much is known about J biosynthesis, the function of base J has long been elusive. However, ice was broken when recent studies showed that J is an epigenetic factor regulating transcription in kinetoplastids. Unusual for eukaryotes, the protein-coding genes in kinetoplastids are arranged in polycistronic gene clusters transcribed by RNA polymerase II (RNAP II) [18]. Genome-wide analysis in trypanosomes revealed an enrichment of internal J at chromosomal regions flanking polycistronic transcription [8]. In JBP1/2-knockout Trypanosoma cruzi, the loss of J leads to a major increase in transcription initiation and a loose chromatin structure at divergent transcription start regions [19]. Differently, loss of internal J, following the deletion of JBP proteins in Leishmania, resulted in massive readthrough at RNAP II transcriptional termination sites [20]. Although J may assume different functions in different species, the underlying mechanisms could be similar as more repressive chromatin structures are created with the synthesis of J. Furthermore, given the predominant presence of J at telomeric repeats among kinetoplastids analyzed, questions regarding whether J has a telomeric function were addressed, but remained unresolved thus far [11].
To help further dissect the functions of J and verify its presence in other organisms, highly sensitive and accurate detection methods are required. So far several analytical techniques have been employed for detecting base J, including 32P-postlabeling [21], two-dimensional thin-layer chromatography [3], immunoprecipitation experiments [10], immunoblots and Southern blots [6]. These methods, however, are limited by their relatively low sensitivity or accuracy and their failure to provide any structural information about J. van Leeuwen et al. [21] studied the efficiency of postlabeling, and found that it could only recover 50% of the total J owing to the poor digestion of this bulky DNA modification during analysis. The following production of rabbit polyclonal antisera against protein-coupled J-deoxyribosemonophosphate (dJMP) raised the sensitivity of J detection [2], but the yield was limited and it was difficult to generate more antisera [11]. Since the first application of mass spectrometry techniques in nucleic acid research, HPLC coupled with tandem mass spectrometry (LC-MS/MS) has become one of the standard methods for quantifying DNA modifications [22]. For instance, an LC-MS/MS coupled with the stable isotope-dilution method allowed for the examination of the roles of repair proteins in removing bulky DNA lesions from mammalian genome [23]. It has also been extensively used for the analysis of 5-mC and its oxidation products, which may be involved in epigenetic regulation of a broad range of biological processes and diseases [24]. LC-MS/MS, however, has not been employed for base J detection in any organisms. Herein, we sought to develop a bio-analytical method by using a surrogate internal standard (β-D-glucosyl-5-hydroxymethyl-2′-deoxycytidine, 5-gHmdC) and LC-MS/MS for the accurate measurement of dJ in wild-type T. brucei and the isogenic strain that is JBP-null. For comparison, we quantified the levels of 5-HmdU in these strains simultaneously.
Experimental Section
Materials
All chemicals and enzymes, unless otherwise noted, were purchased from Sigma-Aldrich (St. Louis, MO) and New England Biolabs (Ipswich, MA). Erythro-9-(2-hydroxy-3-nonyl)adenine (EHNA) hydrochloride was from Tocris Bioscience (Ellisville, MO). [1,3-15N2-2′-D]-5-HmdU were previously synthesized [25,26].
Preparation of Standard
Single-stranded oligodeoxyribonucleotide (ODN, 5′-ATGGCGXGCTAT-3′) housing a 5-hydroxymethyl-2′-deoxycytidine (5-HmdC) at the X position was incubated with uridine diphosphate (UDP)-glucose and T4 phage β-glucosyltransferase (β-GT) to generate the corresponding 5-gHmdC-containing ODN. The desired ODNs were purified on HPLC and its identity verified by MS and MS/MS. The J-containing ODN was prepared and characterized in a similar fashion.
DNA Extraction and Enzymatic Digestion
Blood stream form T. brucei cell line 221a of strain 427 (Wild-type and JBP-null) were cultured as previously described [27]. Total genomic DNA was isolated as described [28]. To the resulting DNA sample (1 μg) were added 100 fmol of the aforementioned 5-gHmdC-containing ODN, which was the surrogate internal standard for LC-MS/MS analysis, and a cocktail of enzymes including nuclease P1, phosphodiesterases 1 and 2, and alkaline phosphatase, as described previously [29]. To inhibit adenine deaminase during digestion, EHNA was also added to the digestion mixture until its final concentration reached 2.5 mM. To the resultant nucleoside mixture was then added 2 pmol [1,3-15N2-2′-D]-5-HmdU, and the enzymes in the digestion mixture were removed by chloroform extraction. The target nucleosides, dJ, 5-gHmdC, and 5-HmdU, were enriched via off-line HPLC.
HPLC Enrichment
The enrichment was performed on a Beckman HPLC system with pump module 125 and a UV detector (module 126). An Aeris WIDEPORE C18 column (4.6 × 250 mm, 3.6 μm beads, 200 Å pore size, Phenomenex, Torrance, CA) was used. An isocratic elution of 10 mM ammonium formate (pH 8.5) was employed at a flow rate of 0.8 mL/min. The HPLC fractions for dJ and 5-gHmdC were collected together, whereas the fraction for 5-HmdU was collected separately for subsequent LC-MS/MS analysis.
Mass Spectrometric Analysis
LC-MS/MS measurement of dJ was conducted on an LTQ XL linear ion trap mass spectrometer equipped with a nanoelectrospray ionization source coupled to an EASY-nLC II system (Thermo Fisher Scientific, San Jose, CA). Samples were automatically loaded onto a home-made trapping column (150 μm × 40 mm) packed with Magic AQ reversed-phase C18 material (5 μm beads, 120 Å pore size, Michrom BioResources, Auburn, CA) at a flow rate of 3 μL/min and eluted to an in-house packed Magic C18 AQ column (75 μm × 200 mm, 5 μm beads, 300 Å pore size). A solution of 0.1% (v/v) formic acid in water (solution A) and a solution of 0.1% (v/v) formic acid in acetonitrile (solution B) were used as mobile phases. The modified nucleosides were separated with a gradient of 40 min 0-10% B and at a flow rate of 300 nL/min. The temperature for the ion transport tube was maintained at 275°C, the activation Q was 0.25, and activation time was 30 ms. In the positive-ion mode, the spray, capillary and tube lens voltages were 2.0 kV, 12 V and 100 V, respectively.
The enriched fractions containing 5-HmdU were separated on an Agilent 1200 capillary HPLC using a 0.5 × 250 mm Zorbax SB-C18 column (5 μm beads, 80 Å pore size, Agilent Technologies, Santa Clara, CA). The eluent was directed to an LTQ linear ion-trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA) following the previously reported procedures [30].
Results and Discussion
The use of a 5-gHmdC-containing ODN as a surrogate internal standard for base J quantification
LC-MS/MS coupled with the isotope-dilution technique constitutes the most reliable analytical method for the quantification of modified nucleosides in cellular and tissue DNA [31]. Although synthetic methods were previously developed for the synthesis of dJ and for its incorporation into ODNs [32-34], the methods involve a multi-step synthesis and are not readily amenable for the preparation of stable isotope-labeled dJ. Thus, we decided to take an alternative approach for the LC-MS/MS quantification of dJ in cellular DNA, where we employed a 5-gHmdC-bearing single-stranded ODN as a surrogate internal standard. In this context, it is important to note that 5-gHmdC is absent in genomic DNA of T. brucei, as revealed by LC-MS/MS analysis. We synthesized the 5-gHmdC-containing ODN using T4 phage β-GT following previously published procedures [35]. In this vein, the T4 β-GT-catalyzed glucosylation of 5-HmdC has been utilized for the measurement of global 5-HmdC levels in vivo [35-37]. In our method, the 5-gHmdC-carrying standard ODN was digested together with DNA isolated from T. brucei prior to HPLC enrichment, which corrects for incomplete release of dJ from DNA during enzymatic hydrolysis and potential loss of the analyte during other stages of sample preparation process, thereby providing more accurate quantification of dJ.
The identity of the standard ODN was verified by ESI-MS and MS/MS analyses (Figure S1A and S1B). The MS/MS of the [M-3H]3- ion (m/z 1283.5) yielded typical [an-base] and wn series ions [38]. In particular, we observed a 481.5-Da difference between the w5 (m/z 1556.1) and w62- (m/z 1018.3) ions. This mass difference is consistent with the residue mass of a 5-gHmdC monophosphate, thereby supporting the presence of this modified nucleotide at the 7th position counting from the 5′ terminus of the ODN. The similar conclusion can be drawn from the mass difference between the [a7-X]2- (m/z 994.8) and [a8-G]2- (m/z 1235.8) ions.
Analytical strategy for simultaneous quantification of dJ and 5-HmdU
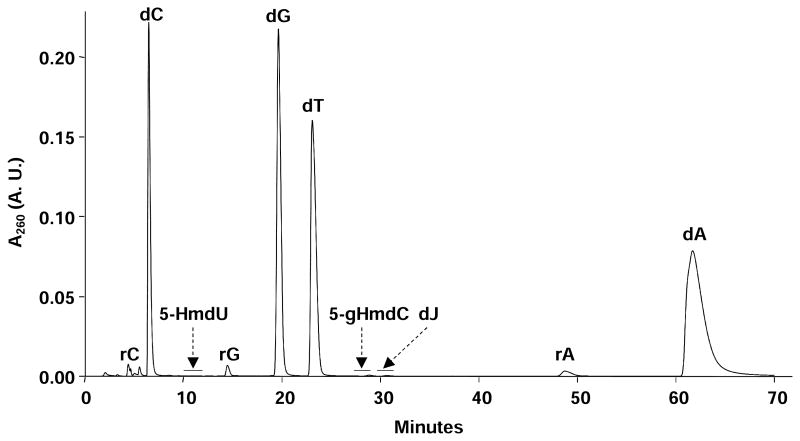
To improve the detection sensitivity of dJ and 5-HmdU in isolated cellular DNA, we employed off-line HPLC enrichment to remove the unmodified nucleosides and buffer salts employed in the enzymatic digestion. An example HPLC trace is shown in Figure 1, where the three modified nucleosides, 5-HmdU, 5-gHmdC and dJ, eluting at 11 min, 28 min, and 30 min, respectively, were well resolved from each other and from the canonical 2′-deoxyribonucleosides and residual ribonucleosides. In this vein, the amounts of RNA contamination in the DNA samples were corrected based on the peak areas of the 2′-deoxyribonucleosides and ribonucleosides found in the chromatograms for the HPLC enrichment, as described previously [30].
Figure 1.
A representative HPLC trace for the off-line enrichment of 5-HmdU, 5-gHmdC, and dJ from the enzymatic digestion mixture of genomic DNA isolated from wild-type T. brucei.
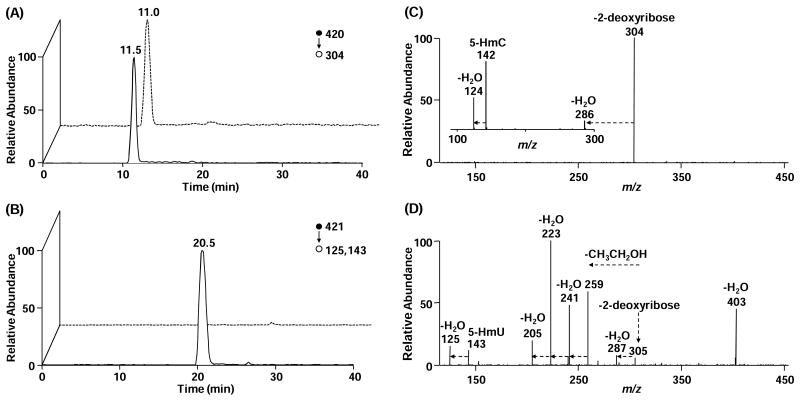
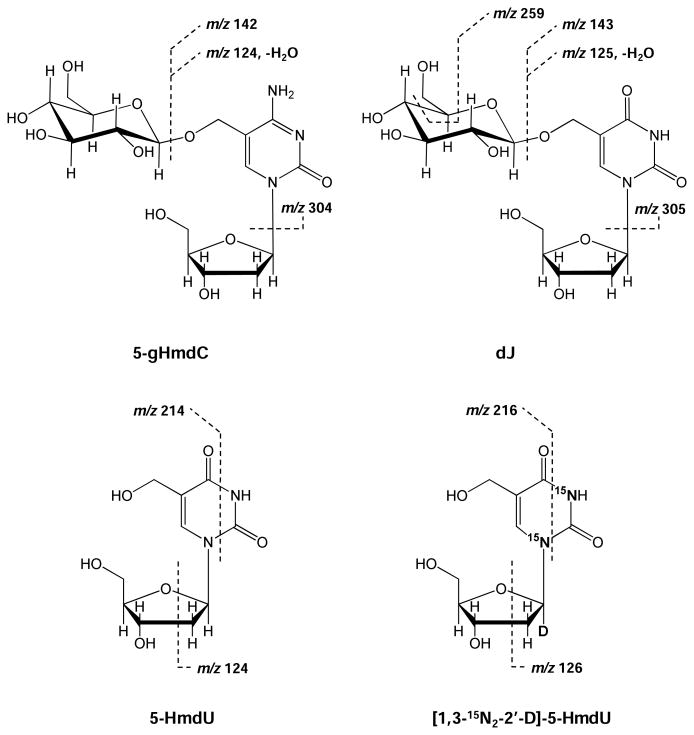
To achieve unambiguous identification and quantification of dJ, we subjected the dJ- and 5-gHmdC-containing fractions to nano-scale LC (nanoLC)-MS2 analysis on an LTQ linear ion-trap mass spectrometer. As displayed in Figure 2A and 2B, a peak was observed in the selected-ion chromatogram (SIC) for monitoring the m/z 420 → 304 transition for 5-gHmdC or m/z 421 → 125, 143 transition for dJ. Relative to offline-HPLC enrichment, a much slower gradient was employed to online-HPLC separation in order to avoid the interference between surrogate standard and analyte, the molecular weights of which differ by only 1-Da. The identities of the components eluting at 11 min and 20 min were further confirmed to be 5-gHmdC and dJ from MS/MS measurements. The chemical structures and the proposed major fragmentation pathways for the [M+H]+ ions of 5-gHmdC and dJ found in MS/MS are depicted in Scheme 1. Upon collisional activation, the N-glycosidic linkage in the [M+H]+ ion of 5-gHmdC (m/z 420) was cleaved readily to produce the most abundant product ion of m/z 304 in MS/MS (Figure 2C). Further collisional activation of the ion of m/z 304 led to the generation of three major fragment ions (m/z 124, 142, and 286) in MS3, as a consequence from neutral losses of glucose, part of glucose [C6H10O5], and H2O, respectively (see inset of Figure 2C). Likewise, collisional activation of the [M+H]+ ions of dJ (m/z 421) yielded fragment ions of m/z 305, emanating from the elimination of a 2-deoxyribose moiety, and m/z 125, 143, and 287, which are attributed to the subsequent losses of glucose, C6H10O5, and H2O from base J portion (Figure 2D). Apart from these fragment ions, the MS/MS of the [M+H]+ ion of dJ gave additional fragment ions that are ascribed to the partial losses of the glucosyl unit on J, i.e., the ions of m/z 205, 223, 241, and 259 (Figure 2D). Because the fragmentation behaviors differed between 5-gHmdC and dJ, we selected unique transitions for monitoring the surrogate internal standard (5-gHmdC, m/z 420 → 304) and the target analyte (dJ, m/z 421 → 125, 143), which allows for unequivocal identification and reliable quantification of base J in cellular DNA.
Figure 2.
Representative LC-MS/MS results for the quantification of dJ. Shown in left panels are the selected-ion chromatograms for monitoring the indicated transitions for the surrogate internal standard (5-gHmdC, A) and the analyte (dJ, B), and solid and dashed lines represent samples from wild-type and JBP-null T. brucei, respectively. Right panels give the MS/MS for 5-gHmdC (C) and dJ (D), and the inset (C) gives the MS/MS/MS for 5-gHmdC.
Scheme 1.
Chemical structures of the modified nucleosides measured in this study and the cleavages for the formation of major fragment ions observed in the MS2 of the [M+H]+ ion of 5-gHmdC and dJ as well as in the MS3 of the [M-H]- ion of 5-HmdU. The sites of 15N and deuterium (D) are indicated in the structure of the isotope-labeled 5-HmdU.
A calibration curve was constructed for the quantification of dJ under identical conditions as described for sample analysis (See Experimental Section and Figure S2). Along this line, known amounts of the dJ-containing ODNs and 100 fmol of 5-gHmdC-harboring ODN were digested together with calf thymus DNA at the same amount as the genomic DNA, and then enriched via offline-HPLC. The addition of calf thymus DNA to the calibration mixture serves to mimic the digestion conditions for genomic DNA. This, together with the analysis of 5-gHmdC- and dJ-containing HPLC fractions under the same LC-MS/MS conditions, facilitated the accurate quantification of dJ. It is worth noting that, from the calibration curve, over 100-fold difference was found for the efficiencies in the formation of the ion of m/z 304 for 5-gHmdC and those for the formation of the ions of m/z 125 and 143 for dJ. At least two factors may account for this difference. First, the ion of m/z 304 is the only product ion found in the MS/MS of the [M+H]+ ion of 5-gHmdC, whereas a large number of fragment ions are observed in the MS/MS of the [M+H]+ ion of dJ (Figure 2C and 2D). Second, a previous study by Liguori and coworkers [39] suggested that the proton affinity of 2′-deoxyuridine is significantly lower than that of 2′-deoxycytidine. Therefore, the glucosylated 5-HmdC may possess a higher proton affinity than glucosylated 5-HmdU, which may contribute to the more facile ionization of the former nucleoside. In spite of this difference, according to the data gained on three separate days, we obtained excellent linearity for the calibration curve (R2 = 0.998).
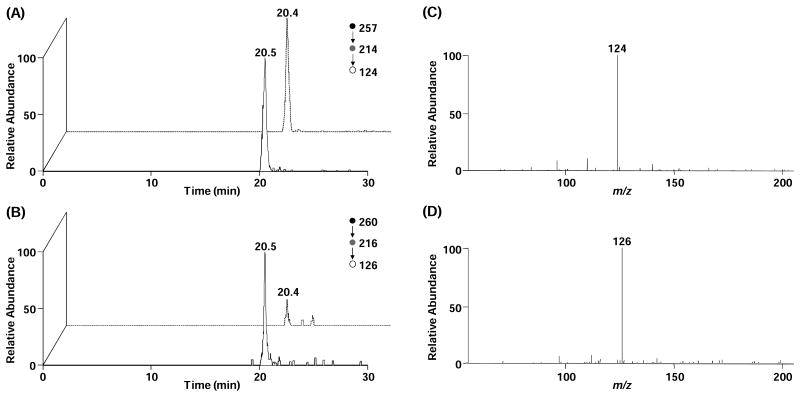
Because the sensitivity for the detection of 5-HmdU is better in the negative- than the positive-ion mode, we quantified 5-HmdU by using LC-MS3 in the negative-ion mode, as previously reported [30]. The co-elution of 5-HmdU analyte and standard (Figure 3A and 3B), together with their identical fragmentation patterns (Figure 3C and 3D), provides solid evidence for the identification of the component and leads to accurate quantification of 5-HmdU.
Figure 3.
Representative LC-MS/MS/MS results for the quantification of 5-HmdU. Shown in left panels are the selected-ion chromatograms for monitoring the indicated transitions for unlabeled 5-HmdU (A) and isotope-labeled 5-HmdU (B), and solid and dashed lines represent samples from wild-type and JBP-null T. brucei, respectively. Right panels give the MS/MS/MS for 5-HmdU (C) and labeled 5-HmdU (D).
Quantification of dJ and 5-HmdU in T. brucei
With the use of this method, we measured simultaneously the levels of dJ and 5-HmdU in T. brucei. To our knowledge, this is the first rigorous quantification of base J and its intermediate product 5-HmU in the genome of kinetoplastids. It is worth noting that the employment of nanoLC coupled to mass spectrometer decreases the amount of DNA required for J detection (∼200 ng for wild-type cells), while in the meantime the detection sensitivity is improved by at least 10 fold relative to the use of regular ESI and a capillary HPLC operating at a flow rate of 8 μL/min. The limit of quantitation (LOQ) and limit of detection (LOD), which referred to the amounts of dJ giving rise to signal-to-noise ratios of 10 and 3, were 46.8 fmol and 15.6 fmol, respectively. This level of sensitivity renders the method highly powerful for the detection of J in cellular DNA, which will be very useful for dissecting the roles of various protein factors in J biosynthesis and for interrogating the biological functions of base J.
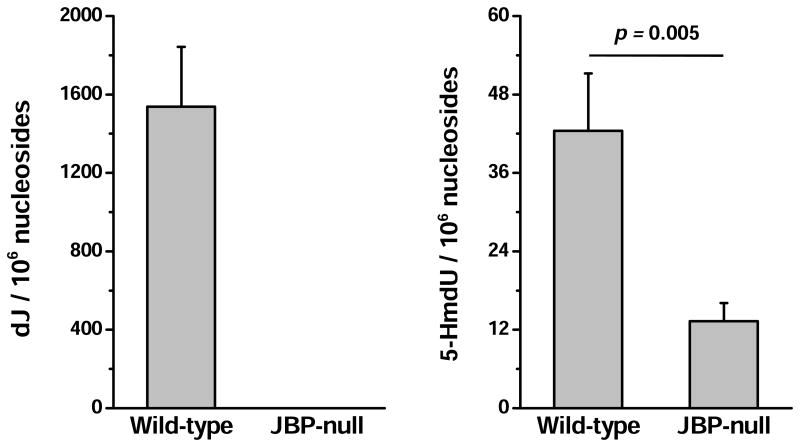
Our quantitative result revealed that the level of dJ in T. brucei genome was ∼1500 modifications per 106 nucleosides (Figures 2 and 4A). Based on the peak areas of the four natural 2′-deoxyribonucleosides found in the HPLC trace with consideration of their extinction coefficients at 260 nm, the percentage of thymidine was quantified to be approximately 29.1%. Thus, the level of dJ represented 0.53% of dT in the wild-type strain, which is consistent with the levels reported previously [3,4]. Conversely, J was not detectable in the JBP-null strain (Figure 4A). To confirm that this observation was based on the absence of J but not due to the limit of detection, we increased the amount of DNA used for the analysis by 40 fold. However, there was still no signal exhibited in SICs for the analyte (data not shown). A remarkable difference in the SICs for monitoring dJ in wild-type and mutant T. brucei was displayed in Figure 2B. Similarly, we found that the level of 5-HmdU in the wild-type strain was 42.5 modifications per 106 nucleosides, while it was present at a level that is 3.2 times lower in the JBP-null strain (13.3 modifications per 106 nucleosides, Figures 3 and 4B).
Figure 4.
Frequencies of dJ and 5-HmdU in T. brucei DNA (n=3). The data represent the mean and standard deviation of the measurement results. The p values were calculated using unpaired two-tailed t test.
The significant decreases in the levels of dJ and 5-HmdU in JBP-null T. brucei are in keeping with the important roles of JBP proteins in the biosynthesis of 5-HmU and J. In this study, we found that the level of 5-HmdU in JBP-null cells was approximately one third of that in wild-type cells. However, considering the LOD and the amount of DNA used for detection, base J, if exists, would be present in the mutant strain at a level that is at least 12,500 times lower than that in the wild-type strain. This is in line with the previous observation that deletion of both JBP1 and JBP2 results in lack of base J in cells [9]. The absence of base J in JBP-null background is primarily attributed to the lack of JBP-mediated biosynthesis of the precursor of base J (i.e., 5-HmU). In this vein, all available evidence now supports that base J synthesis is initiated by JBP1- and JBP2-mediated de novo oxidation of dT to 5-HmdU [40-43]. Additionally, it was hypothesized that JBP1 can bind to the initially synthesized J via its specific J-binding domain and then oxidize adjacent thymidines followed by GT-catalyzed glucosylation [11]. Therefore, the lack of local amplification of J synthesis may also contribute to the absence of J in the trypanosome.
In the presence of 5-HmU, the biosynthesis of J may potentially enter into the second stage where a glucose moiety is transferred onto 5-HmU by the putative GT [11]. Thus, one may expect that some of the 5-HmU may be converted to base J in the JBP-null background. Several factors may account for our observation of the absence of base J, but the presence of a significant level of 5-HmdU in the JBP-null T. brucei. First, JBPs and putative GT may function together as a protein complex, the loss of function of one (JBPs) may compromise the expression and/or the function of the other (GT). Second, there could be other enzymes involved in the oxidation of thymine to 5-HmU in T. brucei and the resulting 5-HmU may not be converted to base J. Moreover, some of the 5-HmdU detected in wild-type and JBP-null T. brucei may arise from oxidatively induced DNA damage. Further studies are necessary for a better understanding of this observation.
Taken together, the mass spectrometry-based analytical method developed in this study provides the first accurate and simultaneous measurements of dJ and its intermediate product 5-HmdU in kinetoplastid flagellates. Our results are consistent with the important roles of JBP proteins in J synthesis. It can be envisaged that the analytical method will be generally applicable for examining the roles of other proteins involved in J synthesis, such as the putative GT that has not yet been identified. Additionally, being the only hyper-modified base discovered in eukaryotic genome, J has been found in unicellular kinetoplastids but not in animals. Given that TET enzymes are the only homologue of JBP1/2 in mammals, it is still an open question whether hyper-modifications, such as J derivatives, are synthesized in higher organisms. In this context, the sensitive method reported here could be adapted readily for exploring the potential presence of other hyper-modifications and their biological functions in other eukaryotes.
Supplementary Material
Acknowledgments
The authors would like to thank the National Institutes of Health for supporting this research (R01 CA101864 to Y.W. and 2R56AI1063523-07A1 to R.S.)
Abbreviations
- J
β-D-glucosyl-5-hydroxymethyluracil
- dJ
β-D-glucosyl-5-hydroxymethyl-2′-deoxyuridine
- 5-gHmdC
β-D-glucosyl-5-hydroxymethyl-2′-deoxycytidine
- 5-HmU
5-hydroxymethyluracil
- 5-HmdU
5-hydroxymethyl-2′-deoxyuridine
- JBP
J-binding protein
- TH
thymine hydroxylase
- GT
glucosyl transferase
- ODN
oligodeoxyribonucleotide
- ESI-MS
electrospray ionization-mass spectrometry
- MS/MS
tandem MS
References
- 1.Gommers-Ampt JH, Van Leeuwen F, de Beer AL, Vliegenthart JF, Dizdaroglu M, Kowalak JA, Crain PF, Borst P. beta-D-glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell. 1993;75:1129–1136. doi: 10.1016/0092-8674(93)90322-h. [DOI] [PubMed] [Google Scholar]
- 2.van Leeuwen F, Taylor MC, Mondragon A, Moreau H, Gibson W, Kieft R, Borst P. beta-D-glucosyl-hydroxymethyluracil is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres. Proc Natl Acad Sci USA. 1998;95:2366–2371. doi: 10.1073/pnas.95.5.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dooijes D, Chaves I, Kieft R, Dirks-Mulder A, Martin W, Borst P. Base J originally found in kinetoplastida is also a minor constituent of nuclear DNA of Euglena gracilis. Nucleic Acids Res. 2000;28:3017–3021. doi: 10.1093/nar/28.16.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Leeuwen F, Wijsman ER, Kuyl-Yeheskiely E, van der Marel GA, van Boom JH, Borst P. The telomeric GGGTTA repeats of Trypanosoma brucei contain the hypermodified base J in both strands. Nucleic Acids Res. 1996;24:2476–2482. doi: 10.1093/nar/24.13.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Genest PA, Ter Riet B, Cijsouw T, van Luenen HG, Borst P. Telomeric localization of the modified DNA base J in the genome of the protozoan parasite Leishmania. Nucleic Acids Res. 2007;35:2116–2124. doi: 10.1093/nar/gkm050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ekanayake DK, Cipriano MJ, Sabatini R. Telomeric co-localization of the modified base J and contingency genes in the protozoan parasite Trypanosoma cruzi. Nucleic Acids Res. 2007;35:6367–6377. doi: 10.1093/nar/gkm693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Leeuwen F, Kieft R, Cross M, Borst P. Tandemly repeated DNA is a target for the partial replacement of thymine by beta-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei. Mol Biochem Parasitol. 2000;109:133–145. doi: 10.1016/s0166-6851(00)00247-4. [DOI] [PubMed] [Google Scholar]
- 8.Cliffe LJ, Siegel TN, Marshall M, Cross GA, Sabatini R. Two thymidine hydroxylases differentially regulate the formation of glucosylated DNA at regions flanking polymerase II polycistronic transcription units throughout the genome of Trypanosoma brucei. Nucleic Acids Res. 2010;38:3923–3935. doi: 10.1093/nar/gkq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cliffe LJ, Kieft R, Southern T, Birkeland SR, Marshall M, Sweeney K, Sabatini R. JBP1 and JBP2 are two distinct thymidine hydroxylases involved in J biosynthesis in genomic DNA of African trypanosomes. Nucleic Acids Res. 2009;37:1452–1462. doi: 10.1093/nar/gkn1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Leeuwen F, Wijsman ER, Kieft R, van der Marel GA, van Boom JH, Borst P. Localization of the modified base J in telomeric VSG gene expression sites of Trypanosoma brucei. Genes Dev. 1997;11:3232–3241. doi: 10.1101/gad.11.23.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borst P, Sabatini R. Base J: discovery, biosynthesis, and possible functions. Annu Rev Microbiol. 2008;62:235–251. doi: 10.1146/annurev.micro.62.081307.162750. [DOI] [PubMed] [Google Scholar]
- 12.Yu Z, Genest PA, ter Riet B, Sweeney K, DiPaolo C, Kieft R, Christodoulou E, Perrakis A, Simmons JM, Hausinger RP, van Luenen HG, Rigden DJ, Sabatini R, Borst P. The protein that binds to DNA base J in trypanosomatids has features of a thymidine hydroxylase. Nucleic Acids Res. 2007;35:2107–2115. doi: 10.1093/nar/gkm049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cross M, Kieft R, Sabatini R, Wilm M, de Kort M, van der Marel GA, van Boom JH, van Leeuwen F, Borst P. The modified base J is the target for a novel DNA-binding protein in kinetoplastid protozoans. EMBO J. 1999;18:6573–6581. doi: 10.1093/emboj/18.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieft R, Brand V, Ekanayake DK, Sweeney K, DiPaolo C, Reznikoff WS, Sabatini R. JBP2, a SWI2/SNF2-like protein, regulates de novo telomeric DNA glycosylation in bloodstream form Trypanosoma brucei. Mol Biochem Parasitol. 2007;156:24–31. doi: 10.1016/j.molbiopara.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ekanayake D, Sabatini R. Epigenetic regulation of polymerase II transcription initiation in Trypanosoma cruzi: modulation of nucleosome abundance, histone modification, and polymerase occupancy by O-linked thymine DNA glucosylation. Eukaryot Cell. 2011;10:1465–1472. doi: 10.1128/EC.05185-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ekanayake DK, Minning T, Weatherly B, Gunasekera K, Nilsson D, Tarleton R, Ochsenreiter T, Sabatini R. Epigenetic regulation of transcription and virulence in Trypanosoma cruzi by O-linked thymine glucosylation of DNA. Mol Cell Biol. 2011;31:1690–1700. doi: 10.1128/MCB.01277-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Luenen HG, Farris C, Jan S, Genest PA, Tripathi P, Velds A, Kerkhoven RM, Nieuwland M, Haydock A, Ramasamy G, Vainio S, Heidebrecht T, Perrakis A, Pagie L, van Steensel B, Myler PJ, Borst P. Glucosylated hydroxymethyluracil, DNA base j, prevents transcriptional readthrough in leishmania. Cell. 2012;150:909–921. doi: 10.1016/j.cell.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Leeuwen F, de Kort M, van der Marel GA, van Boom JH, Borst P. The modified DNA base beta-D-glucosylhydroxymethyluracil confers resistance to micrococcal nuclease and is incompletely recovered by 32P-postlabeling. Anal Biochem. 1998;258:223–229. doi: 10.1006/abio.1998.2587. [DOI] [PubMed] [Google Scholar]
- 22.Dudley E, Bond L. Mass spectrometry analysis of nucleosides and nucleotides. Mass Spectrom Rev. 2013 doi: 10.1002/mas.21388. [DOI] [PubMed] [Google Scholar]
- 23.Liu S, Wang Y. A quantitative mass spectrometry-based approach for assessing the repair of 8-methoxypsoralen-induced DNA interstrand cross-links and monoadducts in mammalian cells. Anal Chem. 2013;85:6732–6739. doi: 10.1021/ac4012232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan BF, Feng YQ. Recent advances in the analysis of 5-methylcytosine and its oxidation products. TrAC Trends Anal Chem. 2014;54:24–35. [Google Scholar]
- 25.Cao H, Wang Y. Collisionally activated dissociation of protonated 2′-deoxycytidine, 2′-deoxyuridine, and their oxidatively damaged derivatives. J Am Soc Mass Spectrom. 2006;17:1335–1341. doi: 10.1016/j.jasms.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 26.Hong H, Cao H, Wang Y. Identification and quantification of a guanine-thymine intrastrand cross-link lesion induced by Cu(II)/H2O2/ascorbate. Chem Res Toxicol. 2006;19:614–621. doi: 10.1021/tx060025x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiPaolo C, Kieft R, Cross M, Sabatini R. Regulation of trypanosome DNA glycosylation by a SWI2/SNF2-like protein. Mol Cell. 2005;17:441–451. doi: 10.1016/j.molcel.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 28.Bernards A, Van der Ploeg LH, Frasch AC, Borst P, Boothroyd JC, Coleman S, Cross GA. Activation of trypanosome surface glycoprotein genes involves a duplication-transposition leading to an altered 3′ end. Cell. 1981;27:497–505. doi: 10.1016/0092-8674(81)90391-3. [DOI] [PubMed] [Google Scholar]
- 29.Liu S, Dunwell TL, Pfeifer GP, Dunwell JM, Ullah I, Wang YS. Detection of Oxidation Products of 5-Methyl-2′-Deoxycytidine in Arabidopsis DNA. PLoS One. 2013;8 doi: 10.1371/journal.pone.0084620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S, Wang J, Su Y, Guerrero C, Zeng Y, Mitra D, Brooks PJ, Fisher DE, Song H, Wang Y. Quantitative assessment of Tet-induced oxidation products of 5-methylcytosine in cellular and tissue DNA. Nucleic Acids Res. 2013;41:6421–6429. doi: 10.1093/nar/gkt360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tretyakova N, Goggin M, Sangaraju D, Janis G. Quantitation of DNA adducts by stable isotope dilution mass spectrometry. Chem Res Toxicol. 2012;25:2007–2035. doi: 10.1021/tx3002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Kort M, Ebrahimi E, Wijsman ER, van der Marel GA, van Boom JH. Synthesis of Oligodeoxynucleotides Containing 5-(β-D-Glucopyranosyloxymethyl)-2′-deoxyuridine, a Modified Nucleoside in the DNA of Trypanosoma Brucei. Eur J Org Chem. 1999;1999:2337–2344. [Google Scholar]
- 33.Turner John J, Meeuwenoord Nico J, Rood A, Borst P, van der Marel Gijs A, van Boom Jacques H. Reinvestigation into the Synthesis of Oligonucleotides Containing 5-(β-D-Glucopyranosyloxymethyl)-2′-deoxyuridine. Eur J Org Chem. 2003;2003:3832–3839. [Google Scholar]
- 34.Grover RK, Pond SJ, Cui Q, Subramaniam P, Case DA, Millar DP, Wentworth P., Jr O-glycoside orientation is an essential aspect of base J recognition by the kinetoplastid DNA-binding protein JBP1. Angew Chem Int Ed Engl. 2007;46:2839–2843. doi: 10.1002/anie.200604635. [DOI] [PubMed] [Google Scholar]
- 35.Terragni J, Bitinaite J, Zheng Y, Pradhan S. Biochemical characterization of recombinant beta-glucosyltransferase and analysis of global 5-hydroxymethylcytosine in unique genomes. Biochemistry. 2012;51:1009–1019. doi: 10.1021/bi2014739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen ML, Shen F, Huang W, Qi JH, Wang Y, Feng YQ, Liu SM, Yuan BF. Quantification of 5-methylcytosine and 5-hydroxymethylcytosine in genomic DNA from hepatocellular carcinoma tissues by capillary hydrophilic-interaction liquid chromatography/quadrupole TOF mass spectrometry. Clin Chem. 2013;59:824–832. doi: 10.1373/clinchem.2012.193938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McLuckey SA, Van Berkel GJ, Glish GL. Tandem mass spectrometry of small, multiply charged oligonucleotides. J Am Soc Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 39.Liguori A, Napoli A, Sindona G. Determination of substituent effects on the proton affinities of natural nucleosides by the kinetic method. Rapid Commun Mass Spectrom. 1994;8:89–93. doi: 10.1002/rcm.1290080117. [DOI] [PubMed] [Google Scholar]
- 40.Heidebrecht T, Fish A, von Castelmur E, Johnson KA, Zaccai G, Borst P, Perrakis A. Binding of the J-binding protein to DNA containing glucosylated hmU (base J) or 5-hmC: evidence for a rapid conformational change upon DNA binding. J Am Chem Soc. 2012;134:13357–13365. doi: 10.1021/ja303423t. [DOI] [PubMed] [Google Scholar]
- 41.Heidebrecht T, Christodoulou E, Chalmers MJ, Jan S, Ter Riet B, Grover RK, Joosten RP, Littler D, van Luenen H, Griffin PR, Wentworth P, Jr, Borst P, Perrakis A. The structural basis for recognition of base J containing DNA by a novel DNA binding domain in JBP1. Nucleic Acids Res. 2011;39:5715–5728. doi: 10.1093/nar/gkr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vainio S, Genest PA, ter Riet B, van Luenen H, Borst P. Evidence that J-binding protein 2 is a thymidine hydroxylase catalyzing the first step in the biosynthesis of DNA base. J Mol Biochem Parasitol. 2009;164:157–161. doi: 10.1016/j.molbiopara.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Cliffe LJ, Hirsch G, Wang J, Ekanayake D, Bullard W, Hu M, Wang Y, Sabatini R. JBP1 and JBP2 Proteins Are Fe2+/2-Oxoglutarate-dependent Dioxygenases Regulating Hydroxylation of Thymidine Residues in Trypanosome DNA. J Biol Chem. 2012;287:19886–19895. doi: 10.1074/jbc.M112.341974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.